Significance
Micro RNAs (miRNAs) are small RNA molecules that regulate gene expression posttranscriptionally in a process known as gene silencing. Fine-tuning the production of miRNAs is essential for correct silencing of their targets, which in turn is important for homeostasis and development. To fine-tune the production of miRNAs, plants deploy a combination of proteins that act as cofactors of the miRNA-processing machinery. Here, we describe REGULATOR OF CBF GENE EXPRESSION 3 (RCF3) as a tissue-specific regulator of miRNA biogenesis in plants. RCF3 interacts with the phosphatases C-TERMINAL DOMAIN PHOSPHATASE-LIKE1 and 2 (CPL1 and CPL2), ultimately affecting the phosphorylation of one of the main DICER-LIKE1 (DCL1) accessory proteins, HYPONASTIC LEAVES1 (HYL1), with a concomitant effect on miRNA production.
Keywords: micro RNA biogenesis, Arabidopsis thaliana, HYL1, phosphorylation, gene silencing
Abstract
The biogenesis of microRNAs (miRNAs), which regulate mRNA abundance through posttranscriptional silencing, comprises multiple well-orchestrated processing steps. We have identified the Arabidopsis thaliana K homology (KH) domain protein REGULATOR OF CBF GENE EXPRESSION 3 (RCF3) as a cofactor affecting miRNA biogenesis in specific plant tissues. MiRNA and miRNA-target levels were reduced in apex-enriched samples of rcf3 mutants, but not in other tissues. Mechanistically, RCF3 affects miRNA biogenesis through nuclear interactions with the phosphatases C-TERMINAL DOMAIN PHOSPHATASE-LIKE1 and 2 (CPL1 and CPL2). These interactions are essential to regulate the phosphorylation status, and thus the activity, of the double-stranded RNA binding protein and DICER-LIKE1 (DCL1) cofactor HYPONASTIC LEAVES1 (HYL1).
Micro RNAs (miRNAs) are short 21- to 24-nucleotide (nt)-long single-stranded RNAs that play an important role in posttranscriptional gene regulation in many multicellular organisms. In plants, after transcription of an MIRNA gene by RNA polymerase II, the primary miRNA (pri-miRNA) is incorporated into nuclear bodies known as D-bodies (1). There, it undergoes a two-step maturation process orchestrated by the ribonuclease DICER-LIKE1 (DCL1) (2). In a first step, DCL1, aided by cofactors including the C2H2-zinc finger protein SERRATE (SE), the nuclear cap-binding complex (CBC), the double-stranded RNA-binding protein HYPONASTIC LEAVES1 (HYL1), and the HYL1 phosphatase C-TERMINAL DOMAIN PHOSPHATASE-LIKE1 (CPL1), removes the two single-stranded RNA tails of the pri-miRNAs to form a largely double-stranded miRNA precursor (premiRNA) (2–5). For accurate excision of the final miRNA:miRNA* duplex and subsequent sorting of the active strand into specific effector complexes, the interaction between HYL1, SE, and DCL1 is crucial. In this context, it has been shown that dephosphorylation of HYL1 by CPL1 is important for correct processing and strand selection of the mature miRNAs (3). In the cytoplasm, the miRNAs associate with an ARGONAUTE (AGO) protein to form the active RNA-induced silencing complex (RISC). Guided by the sequence complementarity of the miRNA to its target, the RISC either induces cleavage or inhibits the translation of target mRNAs (6).
Mutations in genes encoding plant miRNA-related proteins cause a broad range of phenotypes, from embryo lethality of dcl1 and se null mutants to various developmental and physiological defects in hyl1, ago1, and hen1 mutants (4, 7–9). It is unclear how much of this variation in phenotypes reflects genetic redundancy, different processing requirements for different miRNA precursors, or tissue- and stage-specific activity of miRNA-related proteins. In animal systems, several protein complexes and cofactors that regulate different steps of miRNA biogenesis and function have been isolated and characterized (10). Only recently, an increasing number of miRNA biogenesis cofactors have been identified in plants. Most of the specific regulatory events controlled by these cofactors are not yet fully understood (3, 5, 11–16).
A number of genetic screens have been carried out to fill in the remaining blanks in the plant miRNA biogenesis pathway (3, 17–19). We have used an assay that exploits silencing of luciferase by an artificial miRNA as a reporter for miRNA activity to identify candidates for factors affecting miRNA biogenesis or function (3). Among the isolated mutants were two new alleles of REGULATOR OF CBF GENE EXPRESSION 3 (RCF3), also known as SHINY1 and HIGH OSMOTIC STRESS GENE EXPRESSION 5 (HOS5). RCF3 encodes one of the 26 K-homology (KH) domain proteins in Arabidopsis thaliana. KH domains are also found in heterogeneous nuclear ribonucleoprotein K (hnRNP K) and poly-r(C)-binding proteins (PCBPs) and are predicted to bind RNA (20–22). RCF3 contributes to tolerance against various stresses, including low temperature and osmotic stress and response to the stress hormone abscisic acid (21–23). RCF3 and CPL1 both interact with the splicing factors RS40 and RS41, and rcf3 mutants show aberrant intron retention (21). In association with CPL1, RCF3 also seems to inhibit transcription of a number of genes by preventing mRNA capping and thereby disabling the transition from transcription initiation to elongation (22).
Here, we identify RCF3 as a regulator of CPL1-mediated HYL1 phosphorylation. We report that RCF3 localizes to similar nuclear speckles as other miRNA factors, including CPL1, DCL1, and SE. RCF3 interacts with CPL1 and its close homolog CPL2. Inactivation of RCF3 causes a shift of HYL1 phosphoisoforms toward the less active, hyperphosphorylated version. Accordingly, rcf3 mutant defects can be corrected by overexpression of a hypophosphorylation mimic of HYL1. Unlike other known plant miRNA cofactors, RCF3 regulation of HYL1 phosphorylation and miRNA biogenesis takes place in specific niches in the regions spanning the vegetative and reproductive apices. Tissue-biased regulation of miRNA biogenesis is thus a phenomenon shared by plants and animals.
Results
Identification of Two RCF3 Mutant Alleles as miRNA-Deficient Mutants.
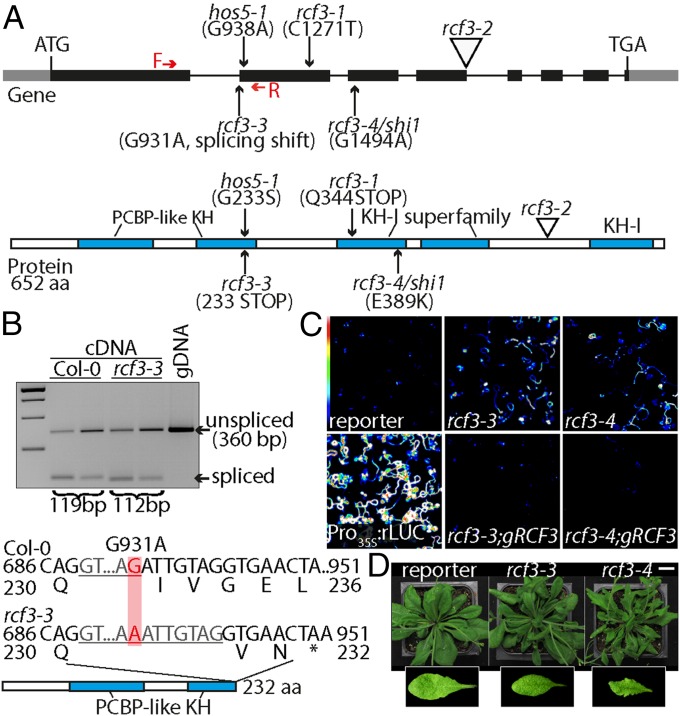
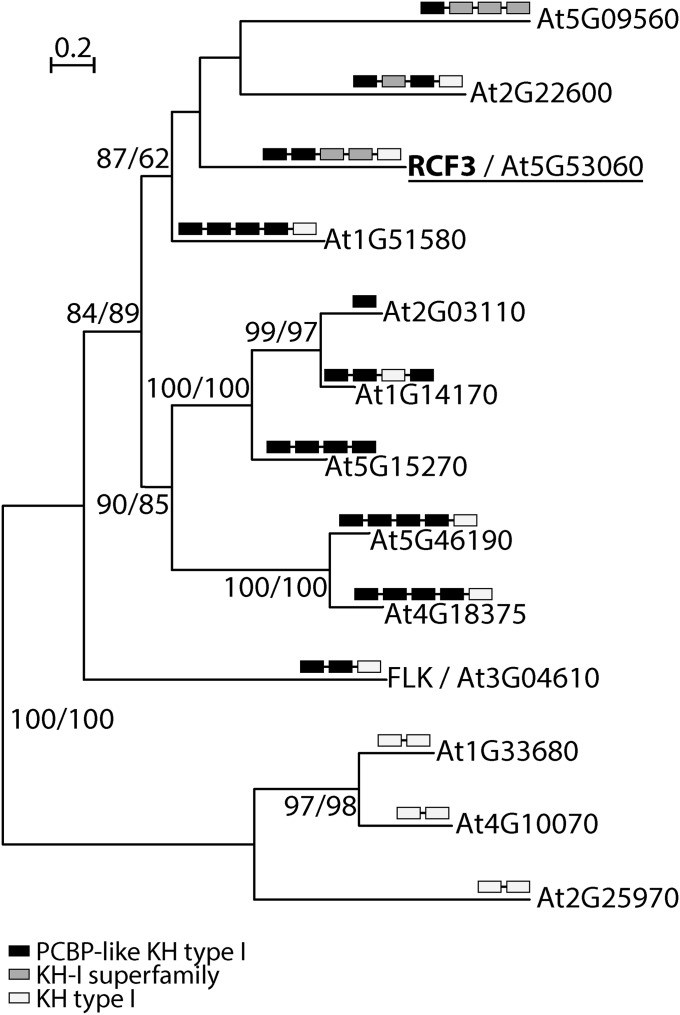
We identified several candidate genes required for miRNA biogenesis or function in a mutant screen using an artificial miRNA (amiRNA) that silences luciferase (3). Using whole genome sequencing and SHORE mapping (24), we mapped the locus responsible for apparent reduction of amiRNA activity, evident through reactivation of luciferase activity, in one of the isolated mutants to a region on chromosome 5 where we identified a mutation in the RCF3 (At5g53060) gene (Fig. 1A and Fig. S1A). This allele, rcf3-3, contains a polymorphism that disrupts the first splice acceptor site in the middle of the sequence encoding the second of five KH domains (Fig. 1A). The mutation causes the splicing machinery to use an alternative cryptic acceptor site 7 nt downstream, as revealed by cloning and sequencing of mutant RCF3 cDNA. This aberrant splicing leads to a frame shift and premature termination of translation (Fig. 1B). An additional allele isolated in our laboratory from an unrelated miRNA genetic screen, rcf3-4, contained a nonsynonymous substitution that affected the third KH domain (Fig. 1A and Fig. S1A). The exact same mutation has previously been identified in another screen (22). Transformation of the mutants with a genomic fragment of the WT RCF3 locus restored silencing of the luciferase reporter and reverted the morphological phenotype in both rcf3-3 and rcf3-4, confirming that the mutations in RCF3 were the cause of the observed phenotypes (Fig. 1C and Fig. S1B). Although rcf3-3 mutants have only subtle morphological defects, rcf3-4 mutants were visibly impaired in growth and development, with elongated, rolled leaves and overall bushy growth (Fig. 1D and Fig. S1 C and D). The fact that rcf3-4, a missense allele, presents stronger defects than rcf3-3, a potential null mutant, suggests that redundantly acting factors might substitute for RCF3 when it is completely absent, as observed in other cases of unusual genetic redundancy (3). A database search for potential RCF3 homologs revealed that 12 of the 26 KH domain proteins encoded in the A. thaliana genome are closely related to RCF3 and are candidates for such redundant action (20) (Fig. S2).
Fig. 1.
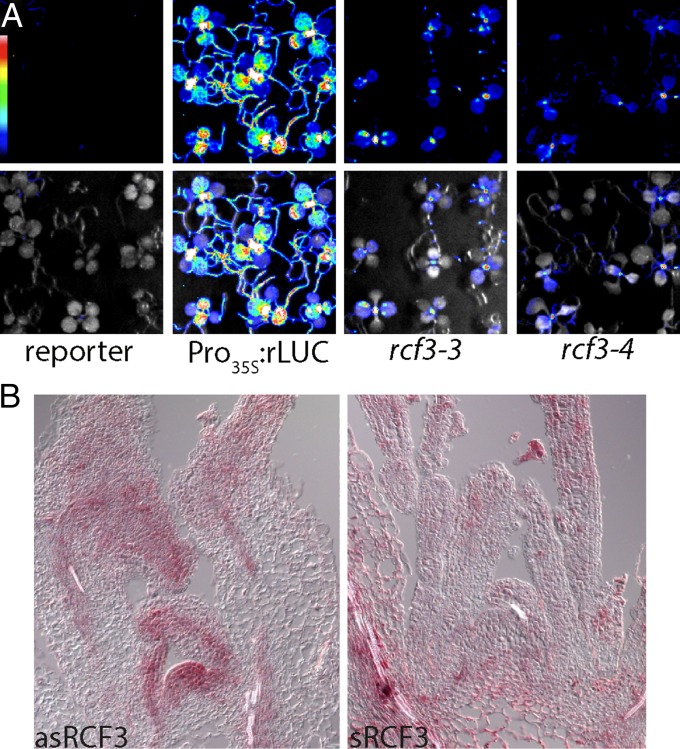
Characterization of rcf3 mutants. (A) Location and effects of mutations on the RCF3 protein. Red arrows note the forward and reverse primers used in B to detect the effect of rcf3-3 mutation on splicing. The five KH domains are marked in blue. (B) RT-PCR analysis of rcf3-3, with genomic DNA (gDNA) for comparison. Sequencing revealed use of a cryptic splice acceptor site in the mutant, leading to a frame shift containing an early stop codon. (C) Bioluminescence phenotype of rcf3 mutants, complemented mutants, amiRNA-activity reporter line (reporter), and Pro35S:rLUC controls. Colored scale indicates low (blue) to high (white) luminescence. (D) Morphological defects in rcf3 mutants. (Scale bar: 2 cm.)
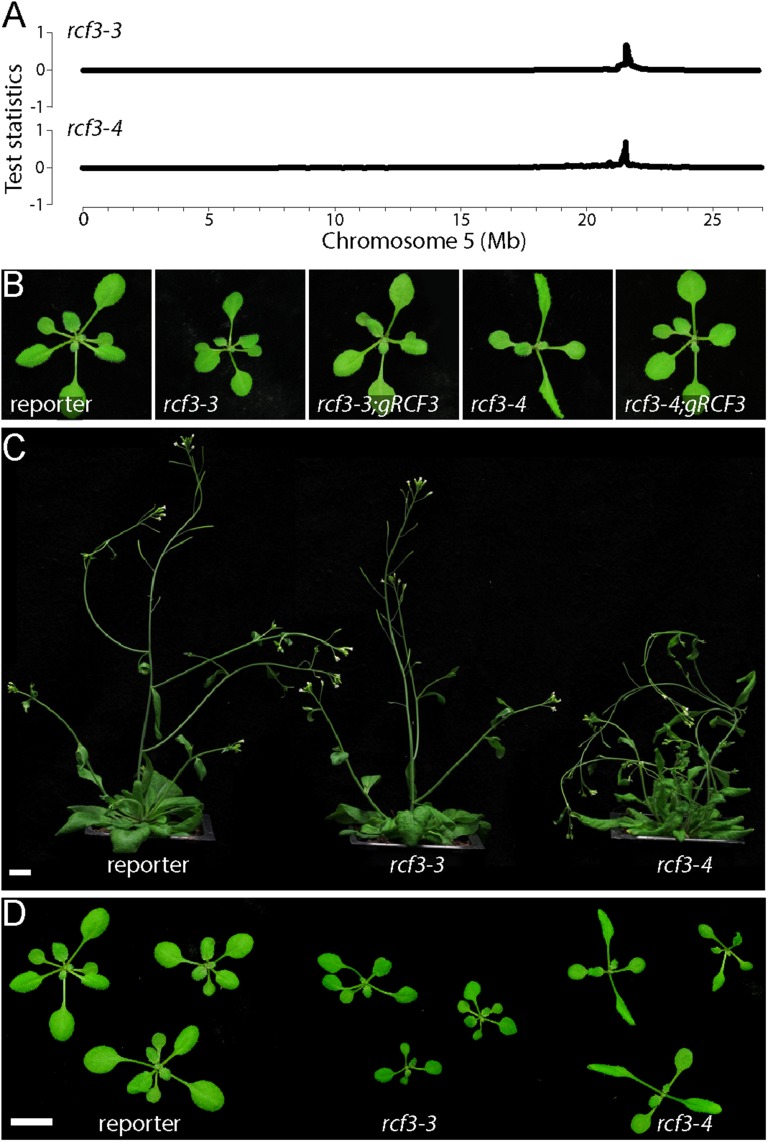
Fig. S1.
Phenotype of rcf3 mutant and rescued plants. (A) Chromosome 5 SHOREmap results for rcf3-3 and rcf3-4 mutants. (B) The 14-days-old rcf3 mutants with and without a genomic RCF3 rescue construct. (C and D) The 32- and 14-days-old rcf3 mutant plants. In both panels, plants were imaged individually and mounted in a single black background square to facilitate the comparison and observation. (Scale bars: 1 cm.)
Fig. S2.
Phylogenetic analysis of RCF3 homologs. RCF3-like KH domain proteins from A. thaliana. Approximate likelihood fraction (aLRT) and bootstrap values (BT) larger than 50% are shown at nodes. Black, gray, and white boxes indicate types of KH domains. The full-length RCF3 protein sequence was compared by BLASTP against the A. thaliana RefSeq protein database. All hits were analyzed for the identity of their annotated functional domains using both the National Center for Biotechnology Information (NCBI) conserved domain search engine (www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and Pfam version 27.0 (pfam.xfam.org/search/sequence). With MEGA version 5, complete protein sequences were aligned using Muscle, and a phylogenetic tree was estimated with the Maximum Likelihood method. FASTA-formatted alignments were loaded into SeaView Version 4.5.3 for computation of trees, using the best fitted model (WAG) including the approximate likelihood fraction (aLRT) and bootstrapping (100×).
RCF3 Regulates miRNA Biogenesis Predominantly in Specific Plant Tissues.
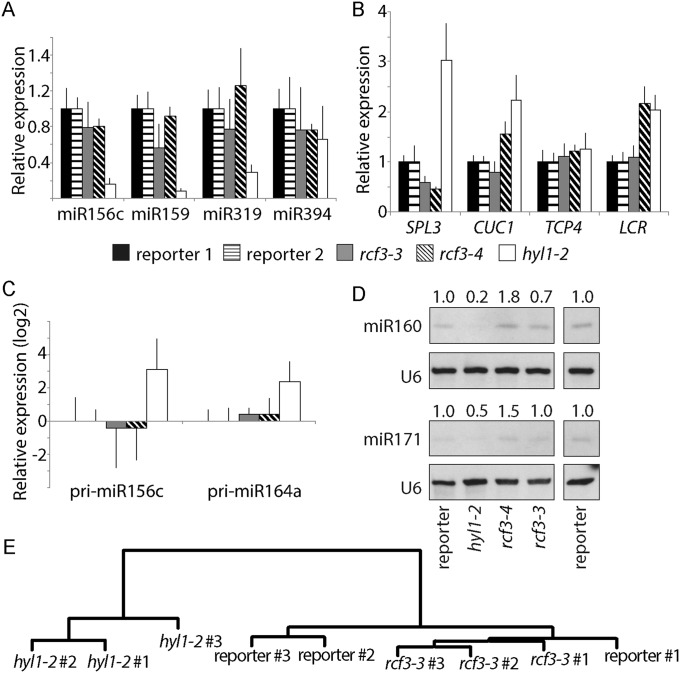
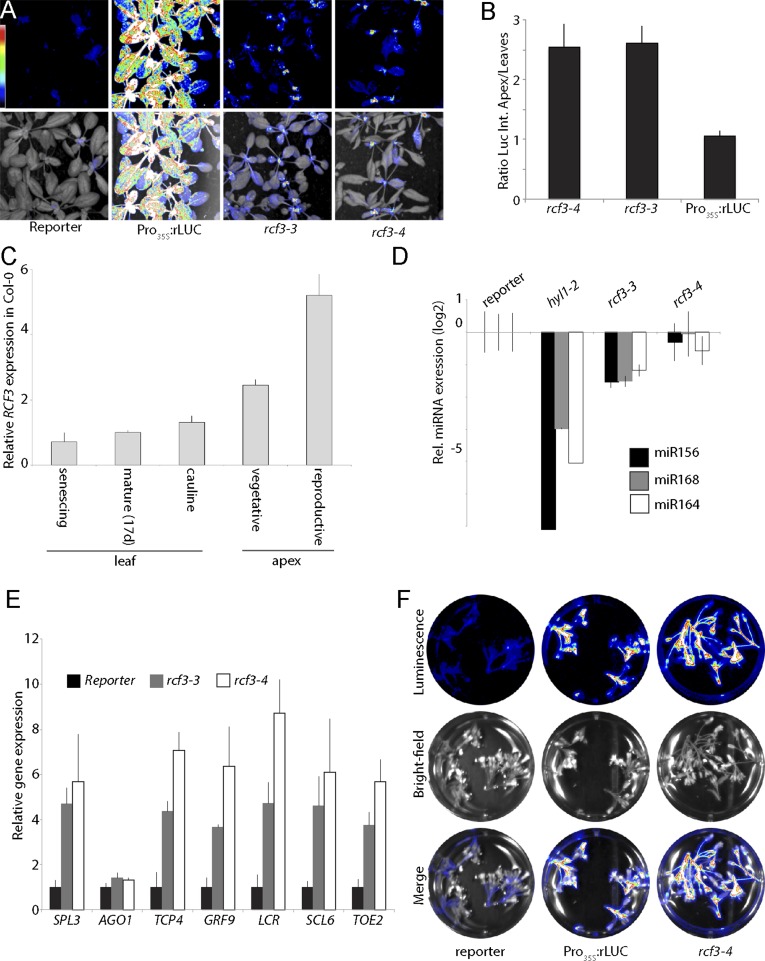
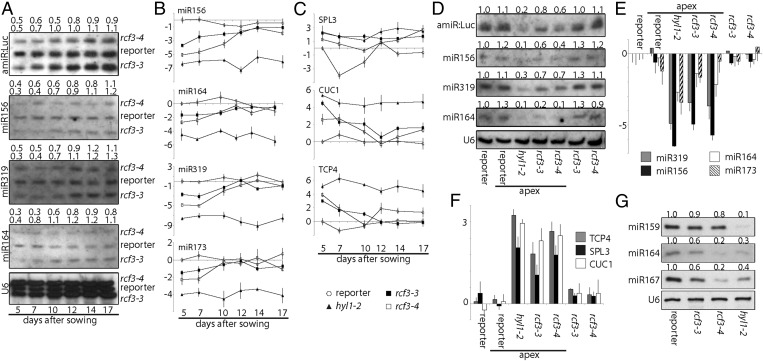
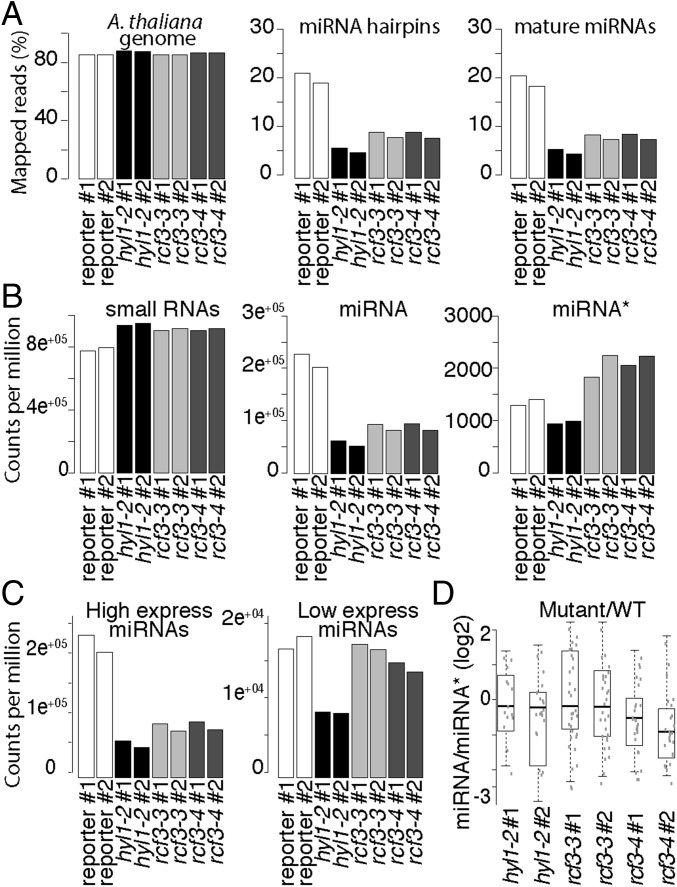
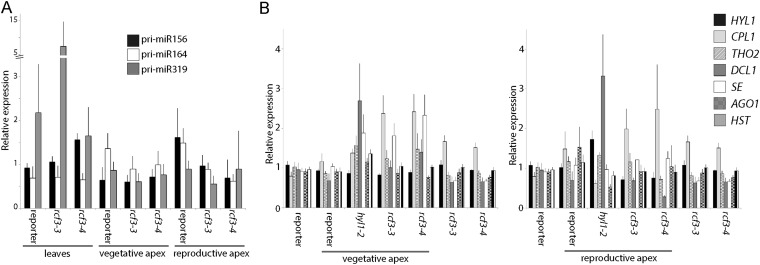
To determine whether RCF3 is required for miRNA biogenesis or for miRNA function, we evaluated the steady-state levels of mature miRNAs, miRNA precursors, and miRNA-targeted mRNAs in rcf3 mutants. MiRNAs and their precursors, as well as miRNA-targeted mRNAs, seemed largely unaffected in whole 14-d-old seedlings, as determined by quantitative RT-PCR (RT-qPCR) (Fig. S3 A–C). MiRNA levels were also only mildly affected in rosette leaves of 25-d-old plants, as determined by RNA blots (Fig. S3D). Similarly, deep sequencing analysis of small RNAs extracted from 25-d-old leaves did not show significant differences in the abundance of miRNAs or miRNA*s between mutants and WT plants. Hierarchical clustering of genome-wide small RNA coverage profiles within 20 bases of either side of mature miRNAs revealed only very subtle misprocessing of miRNA precursors in rcf3, albeit sufficient to allow mutant samples to cluster together (Fig. S3E). These observations were at first glance surprising, considering that the new rcf3 mutant alleles were isolated from miRNA activity-reporting screens. A closer inspection of the mutant plants with a CCD camera revealed that the activity of luciferase was largely confined to the vegetative apical region (Fig. 2A and Fig. S4 A and B). This observation suggested that RCF3 might act, at least in the miRNA pathway, predominantly in this region of the plant. In agreement with this hypothesis, in situ hybridization showed that RCF3 mRNA is abundant in the vegetative shoot apical meristem, young leaf primordia, and newly emerging leaves, a group of tissues hereinafter referred to as “vegetative apex” (Fig. 2B). Also in agreement with a spatially limited role of RCF3 in miRNA biogenesis, a clear reduction in the levels of several miRNAs was seen in very young (5- to 10-days-old) seedlings, which are enriched in vegetative apex tissue, whereas these differences largely disappeared in older plants (Fig. 3 A and B). The reduction in miRNA levels was paralleled by higher accumulation of several target mRNAs (Fig. 3C). The tissue-preferential effect of RCF3 on miRNA accumulation and activity was confirmed with RNA isolated from 14-days-old vegetative apices, in which reduction in mature miRNAs and corresponding overaccumulation of miRNA-targeted mRNAs was much more evident (Fig. 3 D–F) than in age-matched whole-plant samples (Fig. S3 A, B, and D). Supporting the role of RCF3 in the miRNA pathway, a genome-wide analysis of small RNA levels in 14-days-old vegetative apices showed a clear reduction in miRNA accumulation without a detectable change in the abundance of other small RNAs (Fig. 4 A and B). This effect was particularly pronounced for highly expressed miRNAs, such as miR158a, miR166, miR159/319, and miR165 (Fig. 4C and Dataset S1).
Fig. S3.
Effects of rcf3 mutation on miRNA accumulation and action in fully expanded leaves. (A–C) Expression of mature miRNAs, miRNA targets, and miRNA precursors as measured by RT-qPCR. Error bars indicate 2× SEM. (D) RNA blots for detection of mature miR160 and miR171, with U6 as loading control. Signal intensity was calculated with ImageJ and normalized to U6. Above each gel, the ratio of signal intensities of mutants to reporter control is noted. (E) Hierarchical clustering of normalized genome-wide small RNA coverage profiles within miRNA regions extending 20 bases either side of all mature miRNAs.
Fig. 2.
RCF3 expression and activity. (A) Bioluminescence activity in 10-d-old mutant plants, indicating preferential restoration of reporter activity in young tissue around the shoot apex. (Top) Luminescence. (Bottom) Luminescence merged with bright field image. Colored scale indicates low (blue) to high (white) luminescence. (B) Expression of RCF3 visualized by in situ hybridization. (Left) Antisense probe. (Right) Sense probe.
Fig. S4.
RCF3 expression and activity. (A) Bioluminescence activity in 17-d-old mutant plants, indicating preferential restoration of reporter activity in younger tissue around the shoot apex. (Top) Luminescence. (Bottom) Luminescence merged with bright field image. Colored scale indicates low (blue) to high (white) luminescence. (B) Ratio of luminescence intensity between shoot apex and leaves. Error bars indicate 2× SEM. (C) Expression of RCF3 mRNA as measured by RT-qPCR. Error bars indicate 2× SEM. (D and E) Expression of mature miRNAs and miRNA targets as measured by RT-qPCR in inflorescences. Error bars indicate 2× SEM. (F) Bioluminescence activity in inflorescences, indicating restoration of reporter activity in reproductive tissues. (Top) Luminescence. (Middle) Bright field image. (Bottom) Merge. Colored scale indicates low (blue) to high (white) luminescence.
Fig. 3.
miRNA levels and activity. (A) RNA blots for detection of mature endogenous miRNAs and amiRNA against luciferase (amiR:Luc) in a time course of 5- to 17-days-old whole seedlings. U6 was used as loading control. Samples were loaded on the same gel at 12-min intervals. Signal intensity was calculated with ImageJ and normalized to U6. Above each gel, the ratio of signal intensities of mutants to reporter control is noted. (B and C) Expression of mature miRNAs and miRNA targets as measured by RT-qPCR. Error bars indicate 2× SEM. The y axis shows the log2 of the relative expression. (D) RNA blots for detection of mature miRNAs and amiR:Luc in samples collected from 14-d-old whole plants or dissected vegetative apices. Expression relative to the reporter line is given on top. (E and F) Expression of mature miRNAs and miRNA targets as measured by RT-qPCR in the same samples as in D. Error bars indicate 2x SEM. The y axis shows the log2 values of relative expression. (G) RNA blots for detection of mature miRNAs in samples collected from inflorescences. Band intensity relative to the reporter line is given on top.
Fig. 4.
sRNA, miRNA, and miRNA* levels in rcf3 vegetative apices. (A) Fraction of reads mapping to the A. thaliana genome (TAIR 10), miRNA hairpins, and mature miRNAs. (B) Normalized counts per million mapped reads of all small RNAs, mature miRNAs, and miRNA*s. (C) Normalized counts per million mapped reads of highly and lowly expressed miRNAs. (D) Ratio of miRNA/miRNA* between mutants and control plants. For unique miRNAs (i.e., 156a-f), the ratio was calculated using the sum of all associated miRNA*s. The ratio was calculated only for those miRNAs that had greater than 2 counts per million in at least two samples. To compare the mutant to WT ratios, we first averaged the ratios of the WT replicates and then divided the mutant ratios by the mean WT ratio. The data were plotted as the log2 of the ratio mutant/WT showing misprocessed miRNAs as values below 0. Two biological replicates for each genotype: control miRNA-activity reporter line (reporter), hyl1-2, rcf3-3, rcf3-4.
We also observed, in the genome-wide small RNA analysis, an overaccumulation of miRNA* in rcf3 mutants compared with WT vegetative apices, indicating a shift in miRNA precursor processing activity or altered AGO1 strand retention in rcf3 mutants (Fig. 4B). A similar shift toward miRNA* accumulation has been described in cpl1 and hyl1 mutants, where misprocessing leads to altered miRNA/miRNA* ratios for a large portion of miRNAs. Our finding of a similar defect in rcf3 mutants (Fig. 4D) suggests that RCF3 might act through CPL1 and HYL1, which would be in line with the reported protein–protein interactions between RCF3 and CPL1 and between CPL1 and HYL1 (3, 21, 22).
Available expression data for RCF3 (25) report a peak of expression not only in the vegetative apex, but also in the reproductive apex, which we confirmed by RT-qPCR (Fig. S4C). We therefore monitored miRNA accumulation also in young rcf3 inflorescences (samples, from here on referred to as “reproductive apex,” included only young closed flower buds). Northern blot and RT-qPCR measurement of mature miRNAs revealed that rcf3 mutant reproductive tissues accumulate lower levels of miRNAs, similarly to what we observed in vegetative apices (Fig. 3G and Fig. S4D). We also detected an overaccumulation of miRNA-targeted mRNAs and a particularly high reactivation of the amiRNA-activity reporter in the inflorescences (Fig. S4 E and F). Together, these findings suggest that RCF3, due to its predominant expression in the reproductive and vegetative apical regions, affects miRNA accumulation and activity unevenly across plant tissues. RT-qPCR revealed no significant change of pri-miRNAs and known miRNA-related factors in rcf3 mutants, excluding that a potential transcription or splicing alteration was the cause of the observed phenotype (Fig. S5 A and B). However, we saw a slight overaccumulation of CPL1 mRNAs in the mutant plants (Fig. S5B). Because this protein is a functional partner of RCF3 (this work and ref. 26), we speculate that the increase in transcript levels is probably a feedback response compensating for the reduction in RCF3 activity.
Fig. S5.
Expression levels of pri-miRNAs (A) and miRNA processing factors (B) as measured by qRT-PCR in leaves and vegetative and reproductive apices. Error bars indicate 2× SEM.
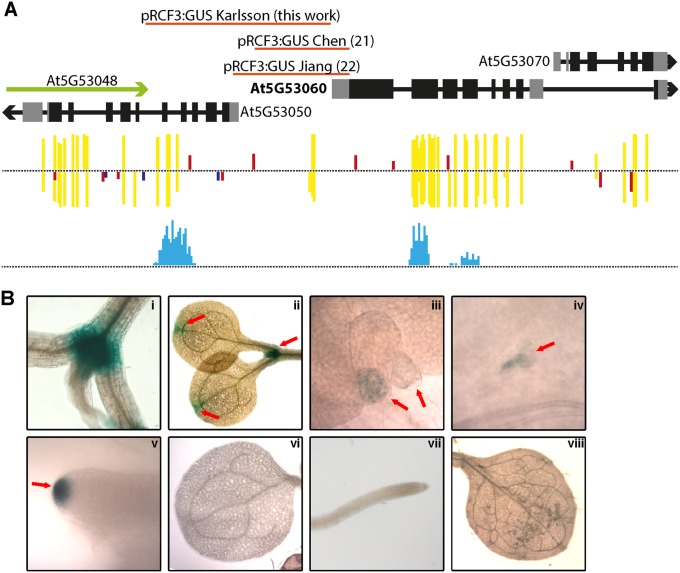
It has been previously shown that RCF3 is also expressed in other tissues besides the vegetative and reproductive apices (21, 22). An analysis of the RCF3 genomic location showed that the gene is located in a dense region of chromosome 5, with 11 genes in a 20-kb window. There are two natural antisense genes (At5G53050 and At5G53048) located 3,175 bp upstream of the RCF3 transcription start site (TSS) (Fig. S6A). Antisense genes can trigger transcriptional gene silencing (TGS) through small RNA-mediated DNA methylation. A bioinformatics search revealed the presence of DNA methylation marks and a peak of small RNAs mapping at ∼2,500 bp upstream of the RCF3 TSS (Fig. S6A). Notably, in previous reports where a promoter:GUS fusion was generated to determine the RCF3 expression pattern, the promoter fragments used did not extend into this particular region. To check whether the region influences the expression of RCF3, we cloned a larger fragment of the promoter. To our surprise, the new construct showed a far less promiscuous expression compared with the reported activity of the shorter constructs (Fig. S6B). Supporting the idea of RCF3 being subject to transcriptional gene silencing (TGS), T1 plants carrying the reporter construct presented stronger GUS signal compared with their T2 counterparts. These data suggested a new layer of regulation on RCF3 expression that will require furthers studies to dissect.
Fig. S6.
RCF3 genomic location. (A) Schematic representation of the RCF3 genomic neighborhood. Red lines represent the promoter fragments used in this work and in refs. 21 and 22. In yellow, red, and blue, CG, CHH, and CHG methylcytosine marks are noted as detected by ref. 38. Light blue indicates the region where small RNAs mapped. (B) Histochemical detection of the ProRCF3:GUS reporter activity. (i and ii) Specific GUS activity detected in T1 transgenic plants. (iii–v) Reduced and localized activity in transgenic T2 apices. (vi–viii) No detectable GUS activity in in T2 cotyledons, root tip, and leaves.
RCF3 Interacts with CPL1 and CPL2 to Regulate HYL1 Activity.
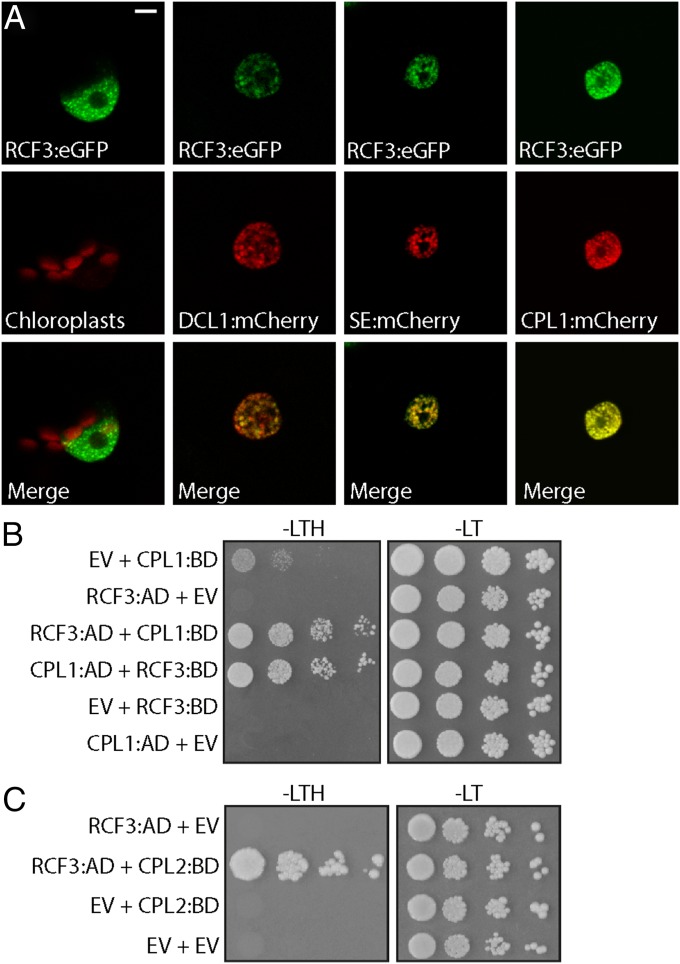
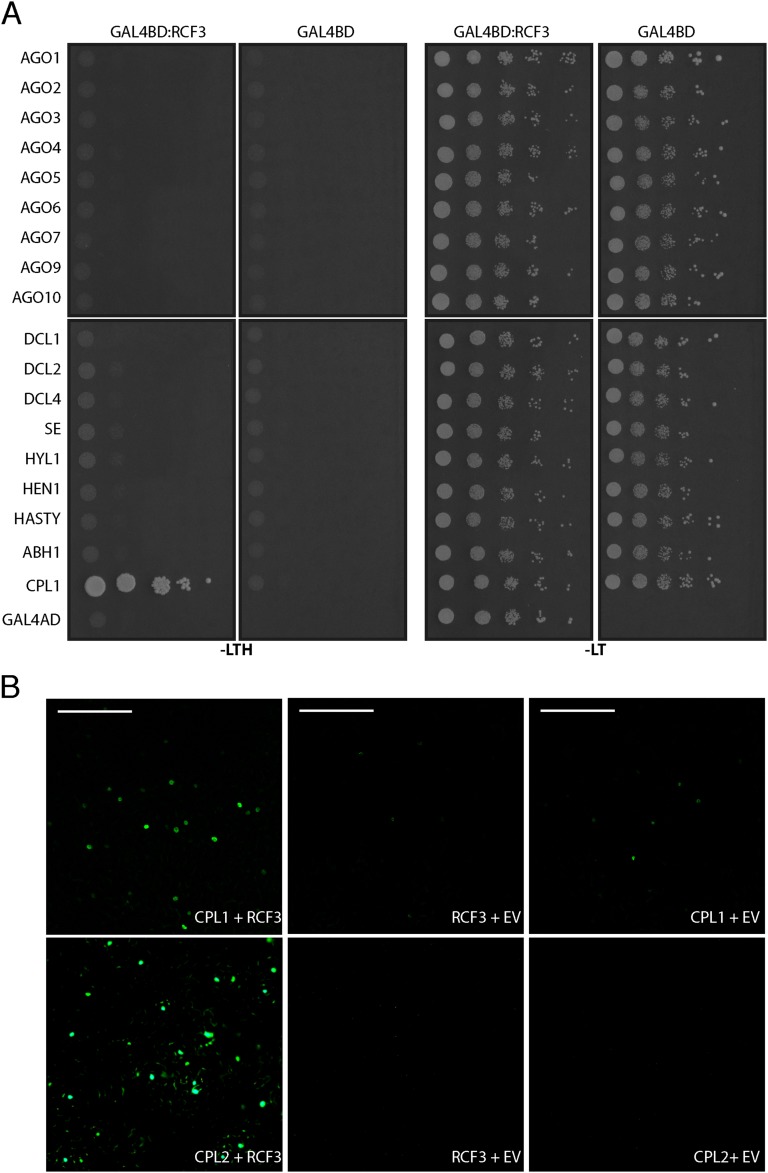
Using transient expression of fluorescent-tagged fusion proteins, we found that RCF3 accumulated in nuclear speckles, where it colocalized with DCL1, SE, and CPL1, supporting a role for RCF3 as a miRNA biogenesis cofactor (Fig. 5A). To identify direct partners of RCF3 in the miRNA pathway, we performed a yeast two-hybrid (Y2H) interaction screen with RCF3 fused to GAL4-BD against a collection of 18 small RNA-related proteins. Among these proteins, RCF3 interacted only with CPL1 (Fig. 5B and Fig. S7A); this interaction has been previously reported (21, 22, 27) and confirmed in planta by bimolecular fluorescence complementation (Fig. S7B). RCF3 also interacted with CPL2 in yeast and plants (Fig. 5C and Fig. S7B), consistent with a partly redundant function of CPL1 and CPL2 (3). This interaction was not seen in another study (27), which, different from our experiments, did not use the full-length CPL2 protein. The interaction of RCF3 with CPL1/CPL2, together with the overaccumulation of miRNA* molecules in rcf3 and cpl1 mutants, suggested that RCF3 acts in the miRNA pathway through CPL1/2.
Fig. 5.
Subcellular localization of RCF3. (A) Nuclear localization of RCF3:eGFP and colocalization with DCL1, SE, and CPL1 tagged with mCherry in transiently transformed N. benthamiana leaves. (Scale bar: 5 μm.) (B and C) Interaction of RCF3 with CPL1 and CPL2 in yeast two-hybrid assays. AD, GAL4 activation domain fusions; BD, GAL4 DNA binding domain fusions; EV, AD, or BD empty vectors; −LT, medium without leucine and tryptophan; −LTH, without leucine, tryptophan, and histidine. Shown are 1:10 serial dilutions.
Fig. S7.
RCF3 Y2H interaction screen with miRNA factors. (A) Yeast transformants were plated on selective medium without leucine, tryptophan (−LT), and histidine (−LTH). All miRNA-related factors were cloned fused to the GAL4 activation domain (AD) whereas RCF3 to the GAL4 binding domain (BD). Serial dilutions of the yeast were plated to ensure even initial yeast density. (B) BiFC assay in N. benthamiana cells. Empty vector (EV) cotransformation was used as negative control. (Scale bars: 0.2 mm.)
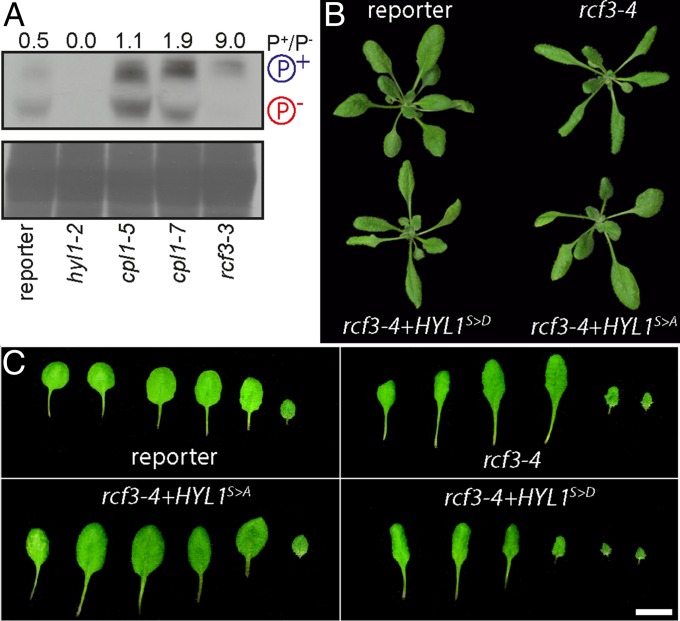
The miRNA biogenesis cofactor HYL1 is subject to phosphorylation, which regulates its activity (3, 28). HYL1 phosphorylation depends on CPL1, CPL2, and MITOGEN-ACTIVATED PROTEIN KINASE 3 (MPK3). In cpl1 mutant plants, HYL1 is hyperphosphorylated, and miRNA processing and strand selection are impaired (3). Because RCF3 interacted with CPL1 and CPL2, but not with HYL1, we tested whether lack of functional RCF3 had an indirect effect on HYL1 phosphorylation. The different phosphoisoforms of HYL1 were detected with the help of Phos-tag, a chemical compound that reduces the mobility of phosphorylated proteins in polyacrylamide gels. A strong shift toward hyperphosphorylated HYL1 was detected in 7-days-old rcf3 mutant seedlings, even more so than in cpl1 mutants (Fig. 6A). Identity of the HYL1 bands was verified with total protein samples extracted from hyl1-2 mutants (Fig. 6A, lane 2). In agreement with RCF3 being primarily active in very young tissues, we did not detect any change in HYL1 phosphorylation in old plants or fully expanded leaves.
Fig. 6.
Effect of RCF3 on HYL1 phosphorylation. (A) Phosphoprotein mobility shift assay, using PhosTag and anti-HYL1 antibodies. Hypo- and hyperphosphorylated HYL1 forms are indicated as P− and P+, respectively. Coomassie staining below as loading control. The intensity of the bands was measured with ImageJ and is expressed as the ratio of the hyper-/hypophosphorylated forms. (B and C) 14-days-old plants and leaf series, showing phenotypic rescue after transformation with HYL1 hyper- (S > D) and hypo- (S > A) phosphomimics. In B, plants were imaged individually and mounted in a single black background square to facilitate the comparison and observation.
If excessive HYL1 phosphorylation contributes to the morphological defects in rcf3 mutants, it should be possible to ameliorate these phenotypes by expressing a version of HYL1 that cannot be phosphorylated, mimicking the hypophosphorylated form of the protein. To test this hypothesis, rcf3 mutants were transformed with two HYL1 phospho-mimics in which all potentially phosphorylated serines were mutated to aspartic acid (S > D) or alanine (S > A), to mimic either a hyper- or hypophosphorylated HYL1, respectively (3). Supporting a role of RCF3 in HYL1 phosphorylation, the morphological rcf3 mutant defects were suppressed only in plants transformed with the hypophosphomimic (S > A) (Fig. 6 B and C). This rescue of the rosette leaf phenotype accords with our observation that RCF3 is highly expressed in the vegetative apex, where leaf shape is determined in leaf primordia.
Discussion
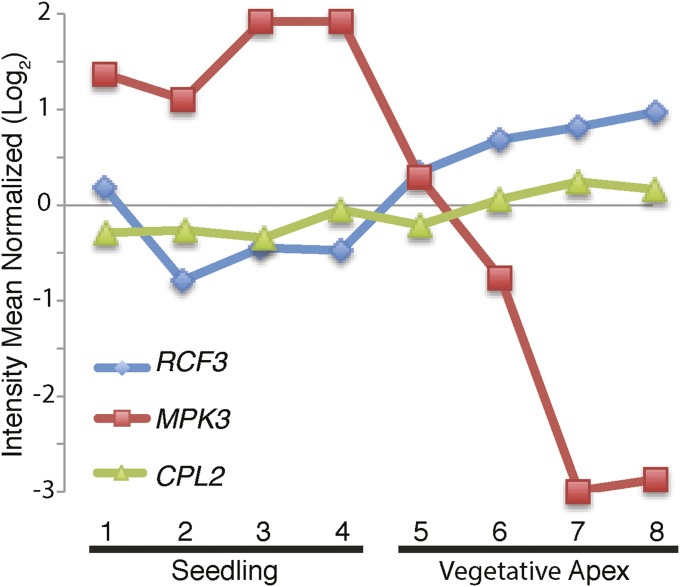
In animals, a series of proteins with specialized roles in miRNA biogenesis have been identified (10). In contrast, the differential contributions of individual proteins to miRNA biogenesis in plants are less well-understood even though mutations in these cofactors often cause distinct developmental and physiological defects. We have identified the KH domain protein RCF3 as a miRNA biogenesis cofactor that acts preferentially at the vegetative and reproductive apices. RCF3 promotes dephosphorylation of HYL1 (Fig. 6), likely through interaction with CPL1 and its homolog CPL2. In the vegetative apex, rcf3 mutations seem to have a stronger effect on HYL1 phosphorylation than cpl1 single mutations. This phenomenon might be explained by both CPL1 and CPL2 being subject to RCF3 action in this tissue, thus with rcf3 mimicking cpl1/cpl2 double mutants. A comparison of the HYL1 phosphorylation status detected in Fig. 6A, using young tissues, and in a previous report (3), where fully expanded leaves were used, suggests tissue-specific and possibly developmental stage-specific regulation of HYL1 activity by changes in its phosphorylation profile. An expression analysis using transcriptome datasets (25) revealed that, whereas CPL2 is accumulated rather evenly across tissues (CPL1 is not included in the analyzed datasets), RCF3 is expressed most strongly in apices (Fig. S8). In a recent study, the protein kinase MPK3 was reported to trigger HYL1 phosphorylation, potentially antagonizing CPL1/2 function (28). Because MPK3 transcripts are low in vegetative apices compared with other tissues, opposite to RCF3 (Fig. S8), the shoot apex might be a niche where HYL1 activity, and therefore miRNA biogenesis, is particularly high because of high RCF3, but low MPK3 activity.
Fig. S8.
RCF3, CPL2, and MPK3 expression profiles. mRNA abundance, quantified by Affymetrix microarrays, was extracted from the AtGenExpress database (25) and expressed as the mean of the normalized reads. Points 1 and 2, 8-d-old seedling; 3 and 4, 21-d-old seedling; 5, 7-d-old apex + young leaves; 6, 7-d-old apices; 7, 14-d-old apices before bolting; 8, 21-d-old apices after bolting.
CPL1 and RCF3 negatively regulate the mRNA capping of several genes and play a role in splicing (22). Links between miRNA biogenesis, mRNA splicing, and capping activity have been established earlier for SE and the cap-binding complex (5, 29). These observations possibly position the CPL1–RCF3 complex close to SE and the cap-binding complex, suggesting that CPL1 and RCF3 might be recruited to pri-miRNA molecules early during their transcription. Even though RCF3 can interact with CPL1 and CPL2 (this work and refs. 30–34), its overall effects are more limited, at least partially because of the more restricted expression of RCF3. Simultaneous loss of CPL1 and CPL2 causes embryonic lethality whereas rcf3-null mutants have only minor developmental defects.
Integrating our data into the larger picture of miRNA biogenesis, we propose a hypothetical mode of action for RCF3 in the context of CPL1/2 and HYL1. Recently, it has been shown that DCL1 is recruited to pri-miRNAs during transcription (35). Due to their role in splicing, it is possible that additionally also SE and CPL1 are, like DCL1, among the first components to be recruited to the pri-miRNA during transcription. In the absence of CPL1, likely CPL2 will be incorporated instead (3). After this initial complex is formed, HYL1 and RCF3 might join the complex, a step dependent on both proteins’ phosphorylation status, and anchor the precursors by specific RNA and protein interactions (3, 26). Both HYL1 and RCF3 are recruited to the complex in their hypophosphorylated isoforms: states that are reached due to the CPL1/2 phosphatase activity and antagonized by MPK3 (3, 26, 28). Localization experiments revealed that CPL1 activity is required for the correct localization of RCF3 and HYL1 (3, 21). The RCF3 mechanism of action, once the protein is recruited to the complex, remains unclear. We found that RCF3 affects the CPL1/2-mediated desphosphorylation of HYL1, but the specifics of this action are still unresolved. In this sense, rcf3 mutants present normal CPL1 nuclear speckle localization (21), excluding the positivity of RCF3 being necessary for the CPL1 recruitment to the complex. One possibility is that RCF3 directly stimulates the enzymatic activity of CPL1/2 or that it serves as a bridge between CPL1 and HYL1. In any case, an extensive biochemical analysis will be required to dissect these possibilities.
Materials and Methods
Plant Material.
A. thaliana accession Columbia (Col-0) or Nicotiana benthamiana seeds were surface sterilized with 10% (vol/vol) bleach and 0.5% SDS and stratified for 2–3 d at 4 °C. Plants were grown at 23 °C either on soil or on Murashige–Skoog (MS) plates (1/2 MS, 0.8% agar, pH 5.7) in long days (16 h light: 8 h dark). hyl1-2 (N564863, SALK_064863), cpl1-7, and rcf3-4 mutants have been described (3, 4, 22).
Transgenes.
RCF3 and CPL2 coding regions were amplified by RT-PCR from RNA isolated from 10-days-old seedlings and cloned into pCR8GWTOPO. These constructs were used for Gateway-mediated recombination with ProQuest (Life Technologies) compatible vectors and pGREEN vectors. A detailed list of all constructs can be found in Table S1. The miRNA activity reporter (throughout the manuscript named “reporter”), the CPL1-fusion proteins, and the phosphomimic constructs have been described (3). As a positive luciferase control, we mutated the amiRNA:luc target site in the luciferase cDNA to make it insensitive to the amiRNA-mediated silencing (PRO35S:rLUC).
Table S1.
Plasmids
| Name | ID | Description |
| 35S:rLuc | pPM118 | Pro35S driving artificial miRNA targeting luciferase expressed from the same vector. The luciferase cDNA was mutated to escape the amiRNA silencing. |
| miRNA-activity reporter | pPM085 | Pro35S driving artificial miRNA targeting luciferase expressed from the same vector |
| gRCF3 | pFRK007 | Genomic RCF3 |
| cRCF3 | pPM347 | RCF3 cDNA |
| cRCF3 w/o stop | pPM348 | RCF3 cDNA without stop codon |
| RCF3:eGFP | pPM350 | Genomic RCF3 without stop fusion to C-terminal citrine |
| DCL1:mCherry | pPM115 | Pro35S driving mutated Cherry (mCherry) N-terminal-fusion to DCL1 |
| SE:mCherry | pPM050 | Pro35S driving mutated Cherry (mCherry) N-terminal-fusion to SE |
| CPL1:mCherry | pPL015 | Pro35S driving mutated Cherry (mCherry) N-terminal-fusion to CPL1 |
| GAL4AD:RCF3 | pPM384 | GAL4 activation domain fusion to RCF3 |
| GAL4BD:RCF3 | pPM385 | GAL4 binding domain fusion to RCF3 |
| GAL4AD:CPL1 | pPM358 | GAL4 activation domain fusion to CPL1 |
| GAL4BD:CPL1 | pPM359 | GAL4 binding domain fusion to CPL1 |
| GAL4AD:DCL1 | pPM258 | GAL4 activation domain fusion to DCL1 |
| GAL4AD:DCL4 | pPM261 | GAL4 activation domain fusion to DCL4 |
| GAL4AD:SE3 | pPM275 | GAL4 activation domain fusion to SE3 |
| GAL4AD:HYL1 | pPM273 | GAL4 activation domain fusion to HYL1 |
| GAL4AD:HEN1 | pPM271 | GAL4 activation domain fusion to HEN1 |
| GAL4AD:HASTY | pPM272 | GAL4 activation domain fusion to HASTY |
| GAL4AD:ABH1 | pPM274 | GAL4 activation domain fusion to ABH1 |
| GAL4AD:CPL2 | pPL004 | GAL4 activation domain fusion to CPL2 |
| GAL4AD:AGO1 | pPM262 | GAL4 activation domain fusion to AGO1 |
| GAL4AD:AGO7 | pPM268 | GAL4 activation domain fusion to AGO7 |
| GAL4AD:AGO9 | pPM269 | GAL4 activation domain fusion to AGO9 |
| GAL4AD:AGO10 | pPM270 | GAL4 activation domain fusion to AGO10 |
| GAL4BD:CPL2 | pPL003 | GAL4 binding domain fusion to CPL2 |
| Pro35S:mHYL1 | pPM444 | Pro35S driving mutated HYL1 phosphomimic Ser > Asp (last Ser > Glu) |
| Pro35S:mHYL1 | pPM445 | Pro35S driving mutated HYL1 phosphomimic Ser > Ala |
| cRCF3 | pPL036 | RCF3 (713–1959) fragment in pGEM |
| Pro35S:CPL2:N Citridine | pPL022 | BiFC CPL2 construct |
| Pro35S:CPL2:C Citridine | pPL023 | BiFC CPL2 construct |
| Pro35S:CPL1:N Citridine | pPM362 | BiFC CPL1 construct |
| Pro35S:RCF3:C Citridine | pPM388 | BiFC RCF3 construct |
| Pro35S:RCF3:N Citridine | pPM389 | BiFC RCF3 construct |
| Pro35S: AT2G29210:C Citridine | pPM375 | BiFC “Empty Vector” construct |
| Pro35S: AT2G29210:N Citridine | pPM376 | BiFC “Empty Vector” construct |
| ProRCF3:GUS | pPM378 | RCF3 promoter:GUS fusion |
Constructs for plant transformation are based on pGREEN and confer either Basta or kanamycin resistance in plants.
Mutant Screen and Luciferase Visualization.
rcf3 mutants were isolated and genetically mapped as described (3). For luminescence analysis, plants were sprayed with 1 mM d-Luciferin-K-Salt (PJK GmbH) twice within 24 h, and imaged with an Orca 2-BT cooled CCD camera (Hamamatsu Photonics). To quantify the bioluminescence intensity (Fig. S4B) of apices versus leaves in mutant plants, 10 plants for each genotype were imaged as described. To avoid saturated spots, unified settings of exposure, gain, and contrast were determined and applied to all images. Luminescence intensity was measured in unprocessed gray-scale pictures using ImageJ. Values were normalized by the measured area size.
RNA Analysis.
Total RNA was extracted using TRIZOL reagent (Life Technologies). Reverse transcription was performed on 1–10 μg of total RNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). Quantitative RT-PCR on mature miRNAs, miRNA precursors, and miRNA targets was executed with biological duplicates and technical triplicates, using BETA-TUBULIN2 (At5g62690) or ACTIN2/8 (At3g18780/ At1g49240) as reference. RNA blots were performed as previously described (3). In situ hybridization was performed as described (36). The probe spans nucleotides 713 and 1959 of the RCF3 cDNA. Sequences of oligonucleotides used for RT-qPCR experiments and RNA blots can be found in Table S2.
Table S2.
DNA oligonucleotide primers and probes
| Gene | Sequence | Purpose |
| RCF3 genomics | F:GGAGGTTAGGACTGCCACGTA | Cloning |
| R:GTACAAGAGGATGGACCGTGA | ||
| RCF3 cDNA | F:ATGGAGAGATCTAGATCCAAGAG | Cloning |
| R:GAGCATACAAGAGGATGGACCGTGA | ||
| RCF3 cDNA | F:TCGGATTCTTCCAAGAGAAAG | Splicing detection |
| R:GACGAGATGATACAATGGCTAAA | ||
| RCF3 cDNA w/o stop | R:GAGCATACAAGAGGATGGACCG | Cloning |
| CPL2 cDNA | F:ATGAATCGTTTGGGTCATAAAT | Cloning |
| R:TATGAAACCTTGCACCCAAGGCT | ||
| miR160 | TGGCATACAGGGAGCCAGGCA | RNA blot |
| miR171 | GATATTGGCGCGGCTCAATC | RNA blot |
| U6 | GCTAATCTTCTCTGTATCGTTCC | RNA blot |
| miR156c | F:GCGGCGGTGACAGAAGAGAGT | qRT-PCR |
| miR159 | F:GCGGCGTTTGGATTGAAGGGA | qRT-PCR |
| miR319 | F:CGTCGTTGGACTGAAGGGAG | qRT-PCR |
| miR394 | F:CGCCATGTTGGCATTCTGTCC | qRT-PCR |
| miRNA RT | R:GTGCAGGGTCCGAGGT | qRT-PCR |
| pri-miR156c | F:ACTCCAACACCTTCAAAGTCTGC | qRT-PCR |
| R:GAGAGAGAAAGTGAGAGATGGGAAC | ||
| pri-miR164a | F:CCCTCATGTGCTTGGAAATG | qRT-PCR |
| R:GCAAATGAGACGGATTTCGTG | ||
| SPL3 | F:ACGCTTAGCTGGACACAACGAGAGAAG | qRT-PCR |
| R:TGGAGAAACAGACAGAGACACAGAGGA | ||
| CUC1 | F:GAAGAGTTGTTGGGTCATGC | qRT-PCR |
| R:CGAAATCAATCTGTCCCGATG | ||
| TCP4 | F:CAACCGATACAGGAAACGGAG | qRT-PCR |
| R:CTGGTATGCGAAAACCCGAAG | ||
| LCR | F:CTATCCACAGCACACTTTAC | qRT-PCR |
| R:CACAGCATTGTAGGTTATCAG | ||
| BETA-TUBULIN2 | F:GAGCCTTACAACGCTACTCTGTCTGTC | qRT-PCR |
| R:ACACCAGACATAGTAGCAGAAATCAAG | ||
| RCF3 | F:CCTAAGCTCGTTACGAAATCAAGA | qRT-PCR |
| R:TCACGGTCCATCCTCTTGTATGCTC | ||
| RCF3 | F:TGAAAAACGCTTTAGCCATTG | In situ probe |
Small RNA Sequencing and Analysis.
RNA for two biological replicates was extracted using TRIZOL reagent for each genotype. Sequencing libraries were prepared with the NEBNext (Set 1) and the TruSeq Small RNA Library Prep Set for Illumina (V2) kit. Raw 50-bp reads were sorted by barcode and adapter and quality trimmed using SHORE v0.9.0 (37). Only reads with a 3′ adapter sequence and trimmed size of 17–25 nt were aligned with bwa v0.7.12 and zero mismatches to the TAIR10 genome (Athaliana_167; phytozome.jgi.doe.gov/pz/portal.html), or the A. thaliana miRBase hairpin and mature miRNA sequences (miRBase.org, release 21).
Perfectly matched reads for each miRNA and miRNA* were counted and normalized to the total number of mapped reads per library. Small RNA hierarchical clustering, at each miRNA locus, was performed by normalized coverage of 18- to 24-nt-long small RNA reads in windows extending 20 bp on both sides of the mature miRNA sequence. The sRNA-seq datasets were deposited in the European Nucleotide Archive (ENA) under accession number PRJEB10589.
Protein Analyses.
For subcellular localization and bimolecular fluorescence complementation (BiFC) assays, N. benthamiana leaves were harvested 3 d after transient transformation and imaged with a TCS SP2 (Leica) or an FV1000 (Olympus) confocal microscope. For BiFC, a nonrelated nuclear protein coding sequence (AT2G29210) was cloned into the corresponding vectors and used as an empty vector (EV) counterpart. Detection of phosphoisoforms was done as previously described (3). Yeast two-hybrid assays were performed using the ProQuest Two-Hybrid System (Life Technologies). For reduction of CPL1 autoactivation, the selection medium was supplemented with 25–150 mM 3-amino-1,2,4-triazole (3-AT).
Note.
During the revision of this article, a second report independently confirmed our observation of RCF3 acting in the miRNA biogenesis, reporting aberrant overaccumulation of miRNA*s in 12-d-old rcf3 seedlings (26).
Supplementary Material
Acknowledgments
We thank Detlef Weigel and Rebecca Schwab for fruitful discussions and input during this project. This work was supported by the Max Planck Society, the Human Frontier Science Program, the International Centre for Genetic Engineering and Biotechnology, and grants from the German Research Foundation. P.A.M. is a member of Consejo Nacional de Investigaciones Científicas y Técnicas. We thank the Deutscher Akademischer Austauschdienst for a short-term fellowship (to P.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the European Nucleotide Archive (ENA) (accession no. PRJEB10589).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512865112/-/DCSupplemental.
References
- 1.Francisco-Mangilet AG, et al. THO2, a core member of the THO/TREX complex, is required for microRNA production in Arabidopsis. Plant J. 2015;82(6):1018–1029. doi: 10.1111/tpj.12874. [DOI] [PubMed] [Google Scholar]
- 2.Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA. 2004;101(34):12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manavella PA, et al. Fast-forward genetics identifies plant CPL phosphatases as regulators of miRNA processing factor HYL1. Cell. 2012;151(4):859–870. doi: 10.1016/j.cell.2012.09.039. [DOI] [PubMed] [Google Scholar]
- 4.Vazquez F, Gasciolli V, Crété P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol. 2004;14(4):346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 5.Laubinger S, et al. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105(25):8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136(4):669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 7.Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J. SERRATE: A new player on the plant microRNA scene. EMBO Rep. 2006;7(10):1052–1058. doi: 10.1038/sj.embor.7400806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schauer SE, Jacobsen SE, Meinke DW, Ray A. DICER-LIKE1: Blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002;7(11):487–491. doi: 10.1016/s1360-1385(02)02355-5. [DOI] [PubMed] [Google Scholar]
- 9.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005;15(16):1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38(3):323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 11.Ben Chaabane S, et al. STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis. Nucleic Acids Res. 2013;41(3):1984–1997. doi: 10.1093/nar/gks1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren G, et al. Regulation of miRNA abundance by RNA binding protein TOUGH in Arabidopsis. Proc Natl Acad Sci USA. 2012;109(31):12817–12821. doi: 10.1073/pnas.1204915109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Speth C, Willing EM, Rausch S, Schneeberger K, Laubinger S. RACK1 scaffold proteins influence miRNA abundance in Arabidopsis. Plant J. 2013;76(3):433–445. doi: 10.1111/tpj.12308. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, et al. NOT2 proteins promote polymerase II-dependent transcription and interact with multiple MicroRNA biogenesis factors in Arabidopsis. Plant Cell. 2013;25(2):715–727. doi: 10.1105/tpc.112.105882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu X, et al. A role for the RNA-binding protein MOS2 in microRNA maturation in Arabidopsis. Cell Res. 2013;23(5):645–657. doi: 10.1038/cr.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu B, et al. The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci USA. 2008;105(29):10073–10078. doi: 10.1073/pnas.0804218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang CI, et al. Importin subunit alpha-2 is identified as a potential biomarker for non-small cell lung cancer by integration of the cancer cell secretome and tissue transcriptome. Int J Cancer. 2011;128(10):2364–2372. doi: 10.1002/ijc.25568. [DOI] [PubMed] [Google Scholar]
- 18.Jauvion V, Elmayan T, Vaucheret H. The conserved RNA trafficking proteins HPR1 and TEX1 are involved in the production of endogenous and exogenous small interfering RNA in Arabidopsis. Plant Cell. 2010;22(8):2697–2709. doi: 10.1105/tpc.110.076638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brodersen P, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320(5880):1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 20.Lorković ZJ, Barta A. Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 2002;30(3):623–635. doi: 10.1093/nar/30.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen T, et al. A KH-domain RNA-binding protein interacts with FIERY2/CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis. PLoS Genet. 2013;9(10):e1003875. doi: 10.1371/journal.pgen.1003875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang J, et al. The arabidopsis RNA binding protein with K homology motifs, SHINY1, interacts with the C-terminal domain phosphatase-like 1 (CPL1) to repress stress-inducible gene expression. PLoS Genet. 2013;9(7):e1003625. doi: 10.1371/journal.pgen.1003625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guan Q, Wen C, Zeng H, Zhu J. A KH domain-containing putative RNA-binding protein is critical for heat stress-responsive gene regulation and thermotolerance in Arabidopsis. Mol Plant. 2013;6(2):386–395. doi: 10.1093/mp/sss119. [DOI] [PubMed] [Google Scholar]
- 24.Schneeberger K, et al. SHOREmap: Simultaneous mapping and mutation identification by deep sequencing. Nat Methods. 2009;6(8):550–551. doi: 10.1038/nmeth0809-550. [DOI] [PubMed] [Google Scholar]
- 25.Schmid M, et al. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37(5):501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 26.Chen T, Cui P, Xiong L. The RNA-binding protein HOS5 and serine/arginine-rich proteins RS40 and RS41 participate in miRNA biogenesis in Arabidopsis. Nucleic Acids Res. 2015;43(17):8283–8298. doi: 10.1093/nar/gkv751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeong IS, et al. Arabidopsis C-terminal domain phosphatase-like 1 functions in miRNA accumulation and DNA methylation. PLoS One. 2013;8(9):e74739. doi: 10.1371/journal.pone.0074739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raghuram B, Sheikh AH, Rustagi Y, Sinha AK. MicroRNA biogenesis factor DRB1 is a phosphorylation target of mitogen activated protein kinase MPK3 in both rice and Arabidopsis. FEBS J. 2015;282(3):521–536. doi: 10.1111/febs.13159. [DOI] [PubMed] [Google Scholar]
- 29.Gregory BD, et al. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell. 2008;14(6):854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 30.Koiwa H, et al. Arabidopsis C-terminal domain phosphatase-like 1 and 2 are essential Ser-5-specific C-terminal domain phosphatases. Proc Natl Acad Sci USA. 2004;101(40):14539–14544. doi: 10.1073/pnas.0403174101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiong L, et al. Repression of stress-responsive genes by FIERY2, a novel transcriptional regulator in Arabidopsis. Proc Natl Acad Sci USA. 2002;99(16):10899–10904. doi: 10.1073/pnas.162111599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuda O, Sakamoto H, Nakao Y, Oda K, Iba K. CTD phosphatases in the attenuation of wound-induced transcription of jasmonic acid biosynthetic genes in Arabidopsis. Plant J. 2009;57(1):96–108. doi: 10.1111/j.1365-313X.2008.03663.x. [DOI] [PubMed] [Google Scholar]
- 33.Ueda A, et al. The Arabidopsis thaliana carboxyl-terminal domain phosphatase-like 2 regulates plant growth, stress and auxin responses. Plant Mol Biol. 2008;67(6):683–697. doi: 10.1007/s11103-008-9348-y. [DOI] [PubMed] [Google Scholar]
- 34.Koiwa H, et al. C-terminal domain phosphatase-like family members (AtCPLs) differentially regulate Arabidopsis thaliana abiotic stress signaling, growth, and development. Proc Natl Acad Sci USA. 2002;99(16):10893–10898. doi: 10.1073/pnas.112276199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang X, Cui Y, Li Y, Qi Y. Transcription and processing of primary microRNAs are coupled by Elongator complex in Arabidopsis. Nature Plants. 2015;1:15075. doi: 10.1038/nplants.2015.75. [DOI] [PubMed] [Google Scholar]
- 36.Weigel D, Glazebrook J. 2002. Arabidopsis: A Laboratory Manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY), pp 354.
- 37.Ossowski S, et al. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;18(12):2024–2033. doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126(6):1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.