Significance
A recent puzzle in the big data scientific literature is that an increase in explanatory variables found to be significantly correlated with an outcome variable does not necessarily lead to improvements in prediction. This problem occurs in both simple and complex data. We offer explanations and statistical insights into why higher significance does not automatically imply stronger predictivity and why variables with strong predictivity sometimes fail to be significant. We suggest shifting the research agenda toward searching for a criterion to locate highly predictive variables rather than highly significant variables. We offer an alternative approach, the partition retention method, which was effective in reducing prediction error from 30% to 8% on a long-studied breast cancer data set.
Keywords: statistical significance, prediction, high-dimensional data, variable selection classification
Abstract
Thus far, genome-wide association studies (GWAS) have been disappointing in the inability of investigators to use the results of identified, statistically significant variants in complex diseases to make predictions useful for personalized medicine. Why are significant variables not leading to good prediction of outcomes? We point out that this problem is prevalent in simple as well as complex data, in the sciences as well as the social sciences. We offer a brief explanation and some statistical insights on why higher significance cannot automatically imply stronger predictivity and illustrate through simulations and a real breast cancer example. We also demonstrate that highly predictive variables do not necessarily appear as highly significant, thus evading the researcher using significance-based methods. We point out that what makes variables good for prediction versus significance depends on different properties of the underlying distributions. If prediction is the goal, we must lay aside significance as the only selection standard. We suggest that progress in prediction requires efforts toward a new research agenda of searching for a novel criterion to retrieve highly predictive variables rather than highly significant variables. We offer an alternative approach that was not designed for significance, the partition retention method, which was very effective predicting on a long-studied breast cancer data set, by reducing the classification error rate from 30% to 8%.
An early 2013 Nature Genetics article (1), “Predicting the influence of common variants,” identified prediction as an important goal for current genome-wide association studies (GWAS). However, a puzzle that has recently arisen in the GWAS-related literature is that an increase in newly identified variants (variables) does not necessarily seem to lead to improvements in current predictive models. Although intuitively it would seem that the addition of information (more statistically significant variants) should increase predictive powers, in recent models of prediction the power is not increased when adding more significant variants to classical significance test-based approaches (2–5). [We refer to “statistically significant” variables throughout this paper as simply “significant.”]
A typical GWAS study collects data on a sample of subjects: cases, who have a disease, and controls, who are disease-free. A very large list of single-nucleotide polymorphisms (SNPs) is evaluated for each individual where each SNP corresponds to a given locus on the genome, and can take on the value 0, 1, or 2 depending on how many copies of the “minor” allele show up. The SNPs are distributed over the whole genome. Typically the researcher wants to select a subgroup of the SNPs that is associated with the disease, so that she can study how the disease works. She may also be interested in predicting whether a new individual has the disease by analyzing the individual’s selected SNPs.
Whether or not an individual has the disease is regarded as the dependent variable. [Here we focus on discrete outcomes, as is common in GWAS studies that are case-control.] The SNP values are the explanatory variables. In a typical study there may be several thousand subjects and hundreds of thousands of SNPs. From the scientist’s point of view there are two basic problems, complicated by the large size of the data set. These are variable selection and prediction. For variable selection, we wish to find a relatively small set of SNPs associated with the disease. For prediction we wish to find how a small set of such variables can be used to predict whether the subject has the disease. The size of the data set is such that the typical approach to variable selection has been to see how well correlated each SNP value is with the disease, and to keep only those for which the statistical significance was very high. Only recently has there been serious consideration of the possible interactions among two or more SNPs by some investigators. The prediction problem has typically been approached by using some variation of linear regression based on the limited number of SNPs from the variable selection stage.
If predictivity is measured by how well the method works on the (training) data used to derive the predictions, we are almost bound to get overoptimistic results. Methods of cross-validation will result in more accurate estimates. Alternatively one may use a separate test sample, independent of the data used to produce the prediction model. Much of our discussion is also relevant to large data sets in other fields of study. Indeed, this problem is not unique to genetic data; we find cases of similar problems in the social sciences. For instance, significant explanatory variables for civil wars serve nearly negligible input for predicting civil wars (6). Likewise, variables found to be significant for fluctuations in the stock market index carry no predictive power (7). This phenomenon is pervasive across different types of data as well as different sample sizes. Thus, the goal of this paper is to offer theoretical insight and illuminating examples to demonstrate precisely how finding highly significant variables is different from finding highly predictive ones—regardless of data type. For illustrative purposes however, we use the lens of prediction for genetic data throughout.
One might ask why one method of variable selection that works perfectly well for a significance-based research question might not work so well for a classification-based research question. Fundamentally, the main difference is that what constitutes a good variable for classification and what constitutes a good variable for significance depend on different properties of the underlying distributions. The test for significance is a test of the null hypothesis that the distributions of X under the two states are the same, whereas the classification error is a test of whether X belongs to one state or the other. Different properties of the distributions are involved. The tests used also may or may not be efficient. In fact, significance was not originally designed for the purposes of prediction.
Some might also comment that perhaps it is clear and intuitive why it is that some significant variables do not appear as highly predictive. After all, variables may be significantly associated with the outcome simply for a small group of individuals in the population, thereby leading to poor prediction on the population. This is true to an extent. However, there is still a fair amount of research using significant variables to predict, perhaps because of a lack of obvious alternative options for variable selection. For instance, currently, prediction-oriented GWAS research uses genetic variants for constructing additive prediction models for estimating disease risk. A recent New England Journal of Medicine article illustrates one example of such an approach, whereby researchers constructed a model based on five genetic variants from GWAS results on prostate cancer; the researchers report that the variants do not increase predictive power (8). Likewise, Gränsbo et al. show that chromosome 9p21, although significantly associated with cardiovascular disease, does not improve risk prediction (9).
In addition, whereas the intuition behind significant variables not appearing predictive might be reasonably obvious, the fact that highly predictive variables do not appear necessarily as highly significant is perhaps less so. We discuss and then demonstrate this phenomenon with both a theoretical explanation and a series of examples. Finally, whereas superficially we might reason that indeed, significance cannot be the same as predictivity, why this is precisely so and what makes for their differences is also not quite so obvious.
With this in mind, we provide a short theoretical explanation for the differences between highly significant and highly predictive variables. We then demonstrate, with a series of artificial examples of increasing relevance, how and why seeking significance and prediction can lead to very different decisions in variable selection. These examples are artificial, partly because they assume that the underlying probabilities are known, whereas the scientist can only infer these from the data. In these examples we compare significance and prediction, and show how the relatively simple I score, defined in Materials and Methods, which we have used in our partition retention (PR) approach to variable selection (10–13), seems to correlate well with predictivity. We offer the I score as one possible useful tool in the study of increasing predictivity. We show a highly successful real application of the PR approach for increasing predictivity in the analysis of a longstanding data set on breast cancer, for which we show some results. Finally, some conclusions are offered to aid in the study of improving predictivity in GWAS research.
There is a long-established literature in statistics on classification with major applications to biology. In recent years the fields of pattern recognition, machine learning, and computer science became heavily involved, often with different terminology and new ideas adapted to the increasing size of the relevant data sets. In the Supporting Information, we present a very brief description of some of the techniques, approaches, and terminology.
Highly Significant vs. Highly Predictive Variables
Data has substantially grown in recent years with both exponential increases in the number of variables and, in many cases, increases in sample sizes as well. This has served as stimulation for a large number of applications via the novel retooling of well-known concepts. Two popular concepts, statistical significance and prediction (including classification), serve as the focus of this article. Historically, significance has played a larger role in statistical inference whereas prediction has served more in identifying future data behavior. The retooling of significance has found a role in data dimension reduction for prediction, that of guiding the feature selection/variable selection step (14). We evaluate this retooling and consider how significance and predictivity are related in the goal of good prediction.
We have mentioned that a key difference between what makes a variable highly significant versus highly predictive lies in different properties of their underlying distributions. We elaborate on this point a bit more here.
Suppose a statistician is given a variable set denoted by X. It is assumed that among control observations X follows a distribution and among cases X follows a distribution . The statistician wishes to test the null hypothesis that against the alternative hypothesis that , where is not specified, using observed data, and assess the statistical significance of the observed data with respect to the null hypothesis. He also wishes to evaluate how strong a predictor based on this variable set could be in predicting the case/control label of future data. Particularly, in a case-control study, he is interested in whether case samples (from ) are significantly differently from control samples (from ).
To carry out a test between and based on variable set x, the statistician chooses a test statistic and, based on the observed values x of X for the n cases and n controls, calculates . Then one can claim that and are significantly different if the probability , which we call the P value, is sufficiently small.
To decide whether x, the observed value of X for a single individual, comes from the distribution or from , when the costs of false positives and false negatives are equal and both possibilities are equally likely, the appropriate Bayes decision rule is to decide in favor of the larger of and . Then the corresponding error rates are and . The average of these two is which, together with , add to 1. [We note that the prediction rate can be seen as equal to 1 minus the average error rate. For continuous distributions, Eq. 1 would be written with integrals rather than summations.] Thus, we may write
| [1] |
Here x represents the possibly multivariate observation that can assume a finite number of values; and are its probability distributions, under case and control, respectively. Eq. 1 defined above requires the knowledge of the true probability distributions, whereas, in practice, the statistician can only infer such knowledge from the data.
The key difference between finding a subset of variables to be highly significant versus finding it to be highly predictive is that the former uses assumptions on, but no knowledge of, the exact distributions of the variables, whereas the latter, as shown in Eq. 1, requires knowledge of both and .
Should the statistician still wish to pursue the significance route to identify variables that are highly predictive, he might wish to compare two subsets of explanatory variables, x and , for their usefulness in the prediction problem. Here has distributions and . It is a current practice to carry out the comparison by testing the null hypotheses and and seeing which has a smaller P value. Because of his limited knowledge on the underlying distributions he is restricted to use tests that are not necessarily powerful enough. Often he is reduced to using a χ2 technique, recommended, for example, in the studies of complex diseases, which is not very powerful for the multiple variable cases. The suboptimality of the test procedure makes the significance level an unreliable basis for comparing subsets of variables and for the usefulness in prediction. It is no surprise that searching for variables based on significance level and based on correct prediction rate can lead us in conflicting directions.
The statistician’s P value for the test is a random variable and here we have assigned the significance value to be the median of the P values, which we may calculate, knowing the probability distributions. The statistician sees only the P value. To make his prediction using x, in the case of equal sample sizes and equal costs of error, he can select for each observed value x, either D or H depending on whether there are more cases or controls in his samples corresponding to x. A naive estimate of the correct prediction rate, the training prediction rate, is obtained by simply using this method on the observed samples. It tends to be overoptimistic. Many sampling properties, such as the significance, the expected training prediction rate, and the median of the I score, can often be calculated conveniently by simulation.
Our next section uses artificial examples to illustrate how highly significant variables and highly predictive variables might differ.
Three Examples
Although we are concerned with large data, our first few examples use only a few observations to cleanly illustrate the issues. The three examples are followed by comparisons, based on a set of 546 more relevant and related examples, each involving 6 SNPs and many observations as summarized as example 4. These examples will show how and why significance and predictivity can differ and that the I score can serve as a useful sign of predictivity. They also show that the problems we run into in prioritizing significance instead of predictivity in our variable selection stage can grow with the complexity of the data. The comparisons in the last example require many simulations and are meant to demonstrate a complicated data scenario, more akin to a GWAS.
Example 1.
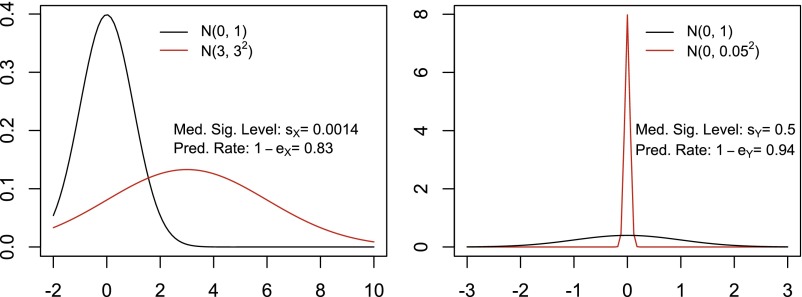
For example 1, there is a single observation X, the distribution of which is normal with mean 0 and SD 1 under a hypothesis H, which can be thought of as health. But, there is an alternative hypothesis K, under which X has a normal distribution with mean 3 and SD 3. We wish to use X to determine whether H or K is the correct hypothesis. Our problem can be thought of as predicting or classifying the state of an individual yielding the observation X. It is a standard problem of testing the hypothesis H and we may regard large values of X as favoring K and suggesting rejection of H.
Statistical theory tells us that the optimal test of H consists of rejecting H when the likelihood ratio is large. For any choice c of what constitutes large enough, we have two error probabilities, and , which are the probabilities of making the wrong decision under H and K, respectively. Notice that if c increases it becomes harder to reject H and decreases while increases. It is possible to calculate the value of c which minimizes the average of and and to call this minimal value , the minimal average error probability associated with X.
For this problem a plausible, if slightly suboptimal, test is to reject H when X is sufficiently large. For each possible value x of X, there is a probability , under H, that X will be as large as x or larger. Then is called the P value when X is observed. Before observing X, we know that X and the P value are random variables. Under H, is uniformly distributed between 0 and 1, but under K, will have a different distribution. If X is very good at discriminating between H and K, should be very small with large probability under K. We label the median value of under K as the significance associated with X. In this case and . Note that is an optimal error rate, but we calculated based on a suboptimal test that a researcher, not knowing the underlying probability distributions, could reasonably have decided to use. In that sense the significance was treated unfairly (Fig. 1). Note also that predictivity, measured by , is associated with a test of the hypothesis H against the alternative K, and is related to the classification problem of deciding which of several (in this case two) situations applies. Thus, prediction, classification, and hypothesis testing are different names for the same problem.
Fig. 1.
Simple example of reversals.
Now suppose that there is another variable Y which is also normally distributed with mean 0 and SD 1 under H, but normally distributed with mean 0 and SD 0.05 under K. Here we calculate and if we insist on using the silly test of rejecting H when Y is large, we obtain . (Surprisingly, in this strange case a much better test would consist of rejecting H when the absolute value of Y is too small.) Forgetting for the moment how silly the test is, let us consider the dilemma of the scientist who must decide, based on these numbers, whether to observe X or Y. He prefers Y if he decides on the basis of error rate or predictivity and X if the decision is based on significance. We refer to this situation where the preferred choice between X and Y depends on the use of significance or predictivity as a reversal.
There are several explanations for the reversal. One is that there was some arbitrariness in our choices of measures of predictability and significance (measures and ). Another is that even though the two choices are aimed at measuring the force of inference, they depend on different properties of the probability distributions involved. Another important point is that because we know the probability distributions in this admittedly artificial example, we used that knowledge to calculate the ideal average error probabilities. On the other hand we did not use the optimal test procedure based on the likelihood ratio for calculating the significance. This may be important because for real data sets we have to use the data to calculate significance levels and predictability. Our estimates may depend as much on the limited capability of our methods of analysis as on the unknown probabilities.
The following two examples, illustrated in Fig. 2, are more relevant and show the same sort of reversal under considerably more reasonable circumstances. They are also more conventional examples of obtaining significance for the test of a null hypothesis.
Fig. 2.
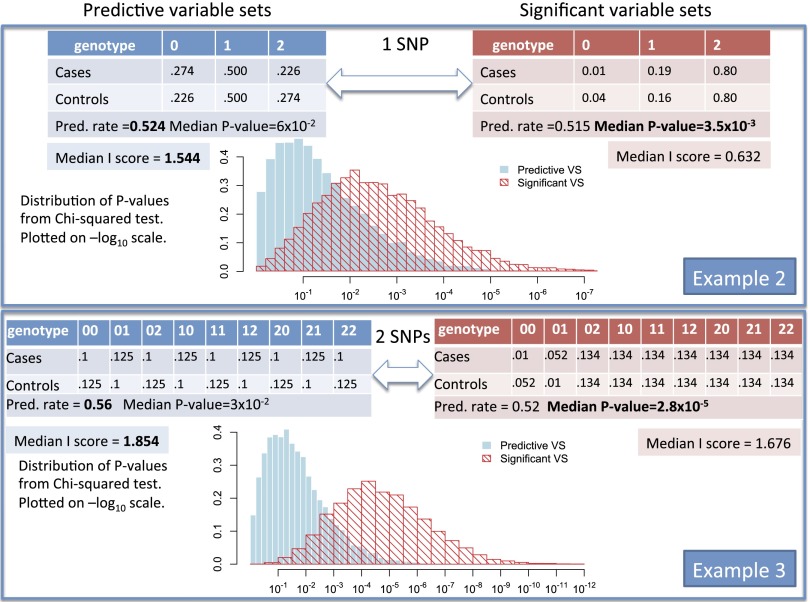
Reversals of predictive and significant variable sets in SNP examples. Example 2 has one explanatory variable (1 SNP) for which the probabilities under cases and controls are listed in the tables. Example 3 has two explanatory variables (2 SNPs) for which the probabilities under cases and controls are listed in the tables. Left-hand-side tables (in blue) are for more predictive variable sets, whereas right-hand-side tables (in red) are for more significant variable sets. The prediction rate (proportion of correct predictions) of each variable set (of size 1 or 2) can be directly computed using the genotype frequencies specified. Using sample sizes of 500 cases and 500 controls, we simulate random case-control data sets by simulating genotype counts among cases and controls using the genotype frequencies specified. I score and the χ2 test statistic were computed for each simulated data set. Simulation details can be found in the Supporting Information.
Example 2.
In example 2 the outcome variable is case or control status. The explanatory variable X is the reading on one SNP for each of 500 cases and 500 controls, for which the probabilities under cases and controls are listed in the blue table in Fig. 2. In this case the minor allele frequency (MAF) is 0.5 and the odds ratio is close to 1 for each of the three possible observations 0, 1, and 2. For Y, based on the other SNP described in the red table in Fig. 2, the MAF is between 0.1 and 0.2 depending on what proportion of the population is healthy. For Y, the odds ratio varies from 4 to 1. In this example we have (prediction rate ) and (prediction rate ). We calculate the significance level using the standard χ2 test for the null hypothesis that the two distributions for case and control are the same. This yields and . Once more we have a reversal because the smaller average error rate is not accompanied by the smaller median P value. The figure also lists the median I score for both X and Y, which favors X as does the prediction rate.
Example 3.
Example 3 is also presented in Fig. 2. Here the variable X in the blue table consists of the outcome of two SNPs (two-way interaction effect). This outcome can fall in one of the cells . Again there is a reversal and the median I score favors X as opposed to Y (in the red table) as does the prediction rate. Whereas the prediction rates are comparable, the median P values are wildly different. Note in both plots of distributions of the predictive variable sets (predictive VS) and significant variable sets (significant VS) in examples 1 and 2, there is overlapping between variable sets but large portions of predictive variable sets are not significant and vice versa. In addition, in both examples the I score follows the preferred prediction rate and not the significance (median P values).
Comparing Significance Tests with the I Score
Before drawing conclusions from the three examples, we present a more complex data simulation for example 4, which consists of a comparison of 546 related, more relevant cases with large numbers of subjects.
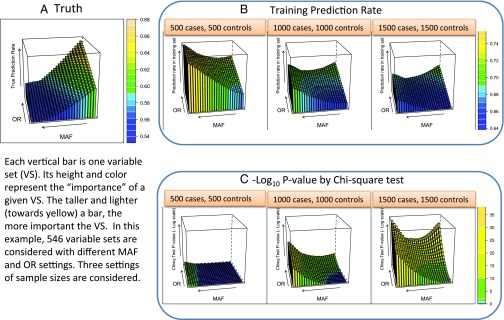
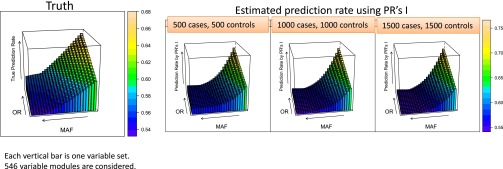
In these cases we deal with six independent but similar SNPs (encapsulating six-way interaction effects), and the observation for a given subject falls into one of cells. The 546 levels of disease are controlled by 26 MAFs and 21 odds ratios (ORs). The results in Fig. 3 present truth, training prediction rate, and significance. Truth is the ideal prediction rate given the MAF and OR. The training prediction rate is the overoptimistic rate based on deciding according to the observed number of cases and controls in each of the 729 cells. The significance level depends on the use of the χ2 test. The latter two are medians of measures based on observed data and their calculation requires extensive simulations. The graphs show how poorly these correlate with truth until the number of subjects becomes very large. Whereas the I score and its median are also based on the data, Fig. 4 shows that it is very well correlated with the truth for modest sample sizes; at large sample sizes I is still better correlated with truth than are the training prediction rate and χ2 test.
Fig. 3.
Disconnect between true prediction power of a variable set and its empirical training set prediction rate and test-based significance. We use 546 variable sets of 6 SNPs with varying levels of disease information (both MAFs and ORs). This results in a partition of 729 cells, each corresponding to a genotype combination on the 6 SNPs represented by this variable set. Three levels of sample size are considered, 500 cases and 500 controls, 1,000 cases and 1,000 controls, and 1,500 cases and 1,500 controls. For each variable set, the theoretical Bayes rate is computed based on the population frequencies and odds ratios. Two thousand independent simulations under each variable sets—given a sample size specification—were used to evaluate the average training prediction error, P value from the χ2 test, and the I-score prediction rate. A depicts the true prediction rate for each of the 546 variable sets for the varying OR and MAF levels. B shows the corresponding training prediction rate as the sample size increases from 500 cases and 500 controls up to 1,500 cases and 1,500 controls. C depicts the corresponding χ2 test P value for each of the variable sets across the three sample sizes. Simulation details can be found in the Supporting Information.
Fig. 4.
Proposed estimated prediction rate based on I scores correlates well with the truth.
Applying the I Score to Real Breast Cancer Data
To reinforce the previous section we turn to a brief examination of real disease data. As noted before, our research team has made heavy use of the I measure in a variable selection method called “partition retention.” This method, applied to real disease data, has not only been quite successful in finding possibly interacting influential variable sets but has also resulted in variable sets that are very predictive and do not necessarily show up as significant through traditional significance testing (10, 15, 16). Here “predictive” refers to both high in I score as well as having high correct prediction rates as determined by k-fold cross-validation. We present examples of some discovered variable sets found to be highly predictive for a real data set on breast cancer (17) that are not highly significant. When using these newly found variable sets, the team was able to reduce the error rate on prediction from the literature standard of to . These results are found from the analysis and data used in ref. 15.
In Table 1 we investigate the top five-variable module (subset of interacting variables) in the breast cancer data found to be predictive through both top I score and performance in prediction in cross-validation and an independent testing set in ref. 15. To find how significant these variables are, we calculate the individual, marginal association of each variable in the marginal P value. When testing 1,000 variables having no effect, it is likely that some will have P values of around 0.001. Here, we have 4,918 variables and therefore desire a P value of , the familywise threshold, to announce significance. None of these variables show up as statistically significant. Measuring the joint influence of all five variables does not have a P value that is significant either.
Table 1.
Real breast cancer example: Five genes in the top returned predictive variable set from van’t Veer data
| Systematic name | Gene name | Marginal P value |
| Contig45347_RC | KIAA1683 | 0.008 |
| NM_005145 | GNG7 | 0.54 |
| Z34893 | ICAP-1A | 0.15 |
| NM_006121 | KRT1 | 0.9 |
| NM_004701 | CCNB2 | 0.003 |
Joint I score: 2.89; joint P value: 0.005; familywise threshold: .
Comments and Conclusion
In our exposition of the differences between highly predictive versus highly significant variable sets, we use artificial examples. We need to know the true relevant underlying probability distributions to treat the problem as one of testing a simple hypothesis against a known alternative for which statistical theory can calculate optimal tests and predictive rates. Our four simulated examples can demonstrate with clarity the reversals we see in choosing significant versus predictive variable sets. Real examples are more difficult because the researcher must rely on a limited number of individuals to infer the relevant distributions and the number of possible variables is huge. However, to demonstrate the potential usefulness of our proposed measure, we additionally provided the highly promising results of applying the I score to the real and well-known van’t Veer breast cancer data set (ccb.nki.nl/data/).
One may wonder whether the shortcoming of using significance is due to the custom of using marginal significance and not taking into account the possible interaction effects of groups of variables. In our examples the problem of reversals seems to increase when using significance-based measures on routine tests when dealing with groups of interacting variables. In example 4, six-way interactions are considered and traditional significance approaches do not capture predictive variable sets. However, using the PR approach based on the measure I for the variable selection stage does well for prediction. Finally, even when we can capture joint effects that are highly predictive, as in the case of the captured variable sets in the van’t Veer example, these groups of variables were not significant. Seeking highly predictive groups of variables through significance alone would not have retrieved these variable sets.
If that is the case, how did we manage to get good results in the breast cancer problem? We used the PR approach, relying heavily on the I score for the variable selection aspect. For reasons we only partly understand, the I score seems to correlate well with predictivity. Having selected the relatively small number of candidate “influential” variables, an intensive use of a variety of known techniques in classification was applied. These were more sophisticated than simple linear regressions.
The issue of obtaining high predictivity from large data demands study. We encourage exploration away from significance-based methodologies and toward prediction-oriented ones. We propose the I score and the PR method of variable selection as candidate tools for the latter.
Materials and Methods
The PR approach to variable selection depends heavily on the I score applied to small groups of explanatory variables. Suppose we have n observations on a disease phenotype Y. When dealing with a small group of m SNPs, each individual is represented by a value Y of the dependent variable and one of possible cells into which the m variables fall. Then the value of I is given by
where corresponds to the ith individual, is the mean of all n Y values, s is the SD of all n Y values, is the mean of the Y values in cell j, is the number of individuals in cell j, and n is the total number of individuals. The measure I is a statistic which may be calculated from the observed data, and does not involve knowing the underlying distributions, as did truth in example 4.
The I score has several desirable properties. First it does not require specification of a model for the joint effect of the m SNPs on Y. It is designed to capture the discrepancy between the conditional means of Y given the values of the SNPs and the overall mean of Y. Unlike ORs as a measure of effect in assessing simple tables, I captures and aggregates all discrepancy (signals) from all cells and forms a flexible measure. It can be used as a measure to assess joint influence or effect sizes, and, importantly, is well-correlated with predictivity.
Second, under the null hypothesis that the subset has no effect on Y, the expected value of I remains nonincreasing when dropping variables from the subset. In other words, the I score is robust to changes to the number of SNPs, m. And, I has the property that adjoining to the group another variable which is independent of Y will tend to decrease I; the PR method is based on selecting a group at random and sequentially eliminating those variables which diminish I the most, and retaining those for which I can no longer be diminished. Those variables, that are retained most often from many randomly chosen groups are candidates for variable selection. The fact that I does not automatically increase as more variables are added to the group being measured is a good property of the I score.
Finally, under the null hypothesis of no effect I acts like a weighted average of independent χ2s with one degree of freedom. Therefore, I values substantially larger than 1 are worth noting.
Online Supporting Materials I
Simulation Details for Fig. 2 (Examples 2 and 3).
The prediction rate (proportion of correct predictions) of each variable set (of size 1 or 2) can be directly computed using the genotype frequencies specified.
Using sample sizes of 500 cases and 500 controls, we simulate B = 1,000 random case-control data sets by simulating genotype counts among cases and genotype counts among controls using the genotype frequencies specified. For each simulated data set for each variable set considered, we compute the statistic of the χ2 test of independence. For each variable set, we summarize these test statistics using histograms shown in Fig. 2, with a vertical bar indicating the corresponding 5% significance level.
Simulation Details for Figs. 3 and 4 (Example 4).
We generate VSs of size 6 . Each represents a genotype of an SNP (0,1, or 2) with MAF . The population frequencies of the genotypes are then and , respectively. Given the values at the 6 variables in a given VS, we have a partition of 729 cells, each corresponding to a genotype combination on the 6 SNPs represented by this VS. Population frequencies of these partition cells are decided by the s. In this simulation, we set for all simulated scenarios.
To simulate case-control data of a complex disease, we generated a fixed base-level vector of ORs (denoted by ) for the 729 partition cells. It remains fixed and was used in all our simulation scenarios. It was generated at the beginning of our simulation. Specifically, 10% of the genotypes of the highest population frequencies were selected to be disease associated (OR 1), then assigned logarithm of odds ratios drawn from , corresponding to having 95% of ORs between 0.55 and 1.82. The log ORs were sorted from the highest to lowest according to their absolute values before being assigned to genotypes sorted by population frequencies. To generate VSs with different levels of predictive information, we created 26 levels of MAF between 0.15 and 0.40 for (denoted as “MAF” in Figs. 3 and 4). The OR amplifying factor γ is between 1 and 2. For a given simulated VS, the actual OR for the partition cells is . Twenty-one levels of γ between 1 and 2 were considered (denoted as “OR” in Figs. 3 and 4). In total, there were 26 21 = 546 variable sets. Three levels of sample sizes were considered: 500 cases and 500 controls, 1,000 cases and 1,000 controls, and 1,500 cases and 1,500 controls.
For each specified VS, we first computed the theoretical Bayes rate, based on the population frequencies and ORs (Fig. 3). Using 2,000 independent simulations under each VS, given a sample size specification, we evaluated (i) the average training prediction error, (ii) P value from χ2 test of independence, and (iii) our proposed estimated prediction rate using PR’s I. (i) and (ii) are commonly used approaches in the current literature.
Online Supporting Materials II
Variable selection or feature selection refers to the approach of selecting a subset of an original group of variables to construct a model. Often feature selection is used on data of large dimensionality with modest sample sizes (18). In the context of high-dimensional data, such as GWAS, with perhaps redundant or irrelevant information, this dimensionality reduction can be a very important step. Unlike projection- or compression-based approaches (such as principal component analysis or use of information theory), feature selection methods do not change the variables themselves.
The types of approaches and tools developed for feature selection are both diverse and varying in degrees of complexity; however, there is general agreement that three broad categories of feature selection methods exist. These are filter, wrapper, and embedded methods. Filter approaches tend to select variables through ranking them by various measures (correlation coefficients, entropy, information gains, χ2, etc.). Wrapper methods use black-box learning machines to ascertain the predictivity of groups of variables; because wrapper methods train similar predictive models for each subset of variables, they can be computationally intensive. Embedded techniques search for optimal sets of variables via a built-in classifier construction. A popular example of an embedded approach is the least absolute shrinkage and selection operator (LASSO) method for constructing a linear model, which penalizes the regression coefficients, shrinking many to zero. Often cross-validation is used to evaluate the prediction rates. For a more comprehensive survey of the feature selection literature see, among others, refs. 14, 18–20.
Although a spectrum of feature selection approaches exists, many scientists have taken the approach of tackling prediction through the use of important and hard-to-discover influential variables found to be statistically significant in previous studies. In the context of high-dimensional data and in the spirit of further investigating variables known to be influential, it is reasonable to hope that these same variables can prove useful for predictive purposes as well. This approach is in some ways most similar to a univariate filter method, as it is independent of the classifier and has no cross-validation or prediction step for variable selection. Our purpose here is to address this popular approach of variable selection via statistical significance. We illustrate how and why the popular filter approach of variable selection through statistical significance might be different from variable selection through predictivity.
Acknowledgments
This research is supported by National Science Foundation Grant DMS-1513408.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518285112/-/DCSupplemental.
References
- 1.Anonymous Predicting the influence of common variants. Nat Genet. 2013;45(4):339. doi: 10.1038/ng.2605. [DOI] [PubMed] [Google Scholar]
- 2.Clayton DG. Prediction and interaction in complex disease genetics: Experience in type 1 diabetes. PLoS Genet. 2009;5(7):e1000540. doi: 10.1371/journal.pgen.1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de los Campos G, Gianola D, Allison DB. Predicting genetic predisposition in humans: The promise of whole-genome markers. Nat Rev Genet. 2010;11(12):880–886. doi: 10.1038/nrg2898. [DOI] [PubMed] [Google Scholar]
- 4.Jakobsdottir J, Gorin MB, Conley YP, Ferrell RE, Weeks DE. Interpretation of genetic association studies: Markers with replicated highly significant odds ratios may be poor classifiers. PLoS Genet. 2009;5(2):e1000337. doi: 10.1371/journal.pgen.1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janssens AC, van Duijn CM. Genome-based prediction of common diseases: advances and prospects. Hum Mol Genet. 2008;17(R2):R166–R173. doi: 10.1093/hmg/ddn250. [DOI] [PubMed] [Google Scholar]
- 6.Ward MD, Greenhill BD, Bakke KM. The perils of policy by p-value: Predicting civil conflicts. J Peace Res. 2010;47(4):363–375. [Google Scholar]
- 7.Welch I, Goyal A. A comprehensive look at the empirical performance of equity premium prediction. Rev Financ Stud. 2008;21:1455–1508. [Google Scholar]
- 8.Zheng SL, et al. Cumulative association of five genetic variants with prostate cancer. N Engl J Med. 2008;358(9):910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]
- 9.Gränsbo K, et al. Chromosome 9p21 genetic variation explains 13% of cardiovascular disease incidence but does not improve risk prediction. J Intern Med. 2013;274(3):233–240. doi: 10.1111/joim.12063. [DOI] [PubMed] [Google Scholar]
- 10.Chernoff H, Lo SH, Zheng T. Discovering influential variables: A method of partitions. Ann Appl Stat. 2009;3(4):1335–1369. [Google Scholar]
- 11.Lo SH, Zheng T. A demonstration and findings of a statistical approach through reanalysis of inflammatory bowel disease data. Proc Natl Acad Sci USA. 2004;101(28):10386–10391. doi: 10.1073/pnas.0403662101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng T, Chernoff H, Hu I, Ionita-Laza I, Lo SH. In: Handbook of Computational Statistics: Statistical Bioinformatics. Lu HHS, Scholkopf B, Zhao H, editors. Springer; New York: 2010. [Google Scholar]
- 13.Zheng T, Wang H, Lo SH. Backward genotype-trait association (BGTA)-based dissection of complex traits in case-control designs. Hum Hered. 2006;62(4):196–212. doi: 10.1159/000096995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guyon I, Elisseeff A. An introduction to variable and feature selection. J Mach Learn Res. 2003;3:1157–1182. [Google Scholar]
- 15.Wang H, Lo SH, Zheng T, Hu I. Interaction-based feature selection and classification for high-dimensional biological data. Bioinformatics. 2012;28(21):2834–2842. doi: 10.1093/bioinformatics/bts531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lo SH, Chernoff H, Cong L, Ding Y, Zheng T. Discovering interactions among BRCA1 and other candidate genes associated with sporadic breast cancer. Proc Natl Acad Sci USA. 2008;105(34):12387–12392. doi: 10.1073/pnas.0805242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van ’t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 18.Saeys Y, Inza I, Larrañaga P. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23(19):2507–2517. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]
- 19.Hua J, Tembe WD, Dougherty ER. Performance of feature-selection methods in the classification of high-dimension data. Pattern Recognit. 2009;42(3):409–424. [Google Scholar]
- 20.Bolón-Canedo V, Sánchez-Maroño N, Alonso-Betanzos A. A review of feature selection methods on synthetic data. Knowl Inf Syst. 2013;34(3):483–519. [Google Scholar]