Significance
Bacteria have a tremendous capacity to rapidly adapt their gene expression profiles and metabolic rates through global regulatory responses. Toxin–antitoxin complexes regulate their own expression under exponential growth but inhibit energy-demanding processes like protein synthesis during stress. A majority of toxins display exquisite endonucleolytic specificity for mRNAs but only in the context of the ribosome. The molecular basis for this selectivity is unclear given their simple microbial RNase architecture. Here, we demonstrate the mechanistic determinants for host inhibition of growth B (HigB) toxin selection of mRNA substrates. Moreover, we propose that ribosome-dependent toxins recognize their mRNA substrates primarily through identification of the third nucleotide of the codon, contrary to how tRNAs and other translation factors also recognize the A site.
Keywords: toxin–antitoxin systems, protein synthesis, RNases, stringent response, ribosome
Abstract
Bacteria contain multiple type II toxins that selectively degrade mRNAs bound to the ribosome to regulate translation and growth and facilitate survival during the stringent response. Ribosome-dependent toxins recognize a variety of three-nucleotide codons within the aminoacyl (A) site, but how these endonucleases achieve substrate specificity remains poorly understood. Here, we identify the critical features for how the host inhibition of growth B (HigB) toxin recognizes each of the three A-site nucleotides for cleavage. X-ray crystal structures of HigB bound to two different codons on the ribosome illustrate how HigB uses a microbial RNase-like nucleotide recognition loop to recognize either cytosine or adenosine at the second A-site position. Strikingly, a single HigB residue and 16S rRNA residue C1054 form an adenosine-specific pocket at the third A-site nucleotide, in contrast to how tRNAs decode mRNA. Our results demonstrate that the most important determinant for mRNA cleavage by ribosome-dependent toxins is interaction with the third A-site nucleotide.
Bacteria live in dynamic environments and as a consequence have developed robust stress responses to survive harsh conditions including temperature fluctuations, nutrient deprivation, and oxidative stress (1, 2). Lack of nutrients activates the stringent response and the synthesis of the “magic spot” alarmone, guanosine penta/tetraphosphate [(p)ppGpp]. (p)ppGpp serves as a global signaling molecule and facilitates transcription of prosurvival genes and activation of downstream proteolysis of select substrates that inhibit replication and translation (1, 3, 4). This rapid inhibitory switch suppresses metabolite consumption and temporarily halts cell growth to promote bacterial survival until nutrients are readily available. Among the prosurvival genes regulated by (p)ppGpp production are toxin–antitoxin modules, which have additional roles in antibiotic resistance and tolerance, biofilm and persister cell formation, and niche-specific colonization (5–11). The critical roles toxin–antitoxin pairs play in bacterial physiology underscore the importance of understanding their molecular targets and modes of action.
There are five different classes (I to V) of toxin–antitoxin systems defined by how the antitoxin represses toxin function (1). Type II toxin–antitoxin pairs form protein–protein complexes during exponential growth that serve two purposes: inhibition of toxin activity by antitoxin binding and transcriptional autorepression to limit toxin expression (12). Antitoxins are proteolytically degraded after (p)ppGpp accumulation, leading to derepression at the toxin–antitoxin promoter (8, 12). Liberated toxin proteins inhibit the replication or translation machinery by targeting DNA gyrase, initiator tRNAfMet, glutamyl-tRNA synthetase, EF-Tu, free mRNA, ribosome-bound mRNA, and the ribosome itself (13–20).
Ribosome-dependent toxins cleave mRNAs on the ribosome between the second and third nucleotides of the aminoacyl (A)-site codon (21–23). Although collectively Escherichia coli ribosome-dependent toxins target a diverse range of codons, each individual toxin appears to have a strong codon preference and cleaves at defined positions along the mRNA (24–26). RelE cleaves at UAG stop codons and the CAG sense codon (all codons denoted in the 5′–3′ direction); YoeB cleaves at codons following a translational AUG start site and at the UAA stop codon; and YafQ cleaves a single AAA sense codon (16, 24, 27–29). In contrast, Proteus vulgaris host inhibition of growth B (HigB) toxin degrades multiple codons encoding for different amino acids, with the defining codon signature being a single adenosine (30). Furthermore, recent studies have suggested that a number of toxins cleave a spectrum of codons irrespective of codon family (i.e., codons coding for the same amino acid) (16, 25, 27). This loose specificity exhibited by HigB and other toxins strongly suggests that canonical codon identity may play little to no role in defining a toxin mRNA substrate and, as an extension of this, that toxins recognize A-site codons in a manner distinct from tRNAs and release factors. However, because all ribosome-dependent toxins adopt a conserved microbial RNase fold (31–34) and cleave the mRNA using the same mechanism of in-line attack on the scissile phosphate (21–23), the structural features of each toxin that define different nucleotide preferences in the context of the ribosome remain elusive.
Here, we elucidate the molecular basis for translation inhibition and nucleotide selectivity by the HigB toxin. We demonstrate that HigB recognizes each A-site nucleotide position distinctly. Because ribosome-dependent toxins contain a conserved protein architecture, these results provide a molecular framework for rationalizing the specificity of this enzyme family. Our structures reveal that HigB recognizes mRNA using hydrogen-bonding capability to select for the second A-site nucleotide, whereas an adenosine-specific binding pocket is formed by both HigB and 16S rRNA residues to confer specificity at the third A-site position. Lastly, a single HigB residue modulates adenosine selectively at this third A-site position by a mode that is distinct from other ribosome-dependent toxins (21, 22). Because toxin proteins play prominent roles in persister cell formation and represent novel antimicrobial targets (8, 11, 35, 36), determining the molecular basis of mRNA substrate specificity may provide insights to subvert toxin activity.
Results and Discussion
Structural Determination of Ribosome–HigB Complexes.
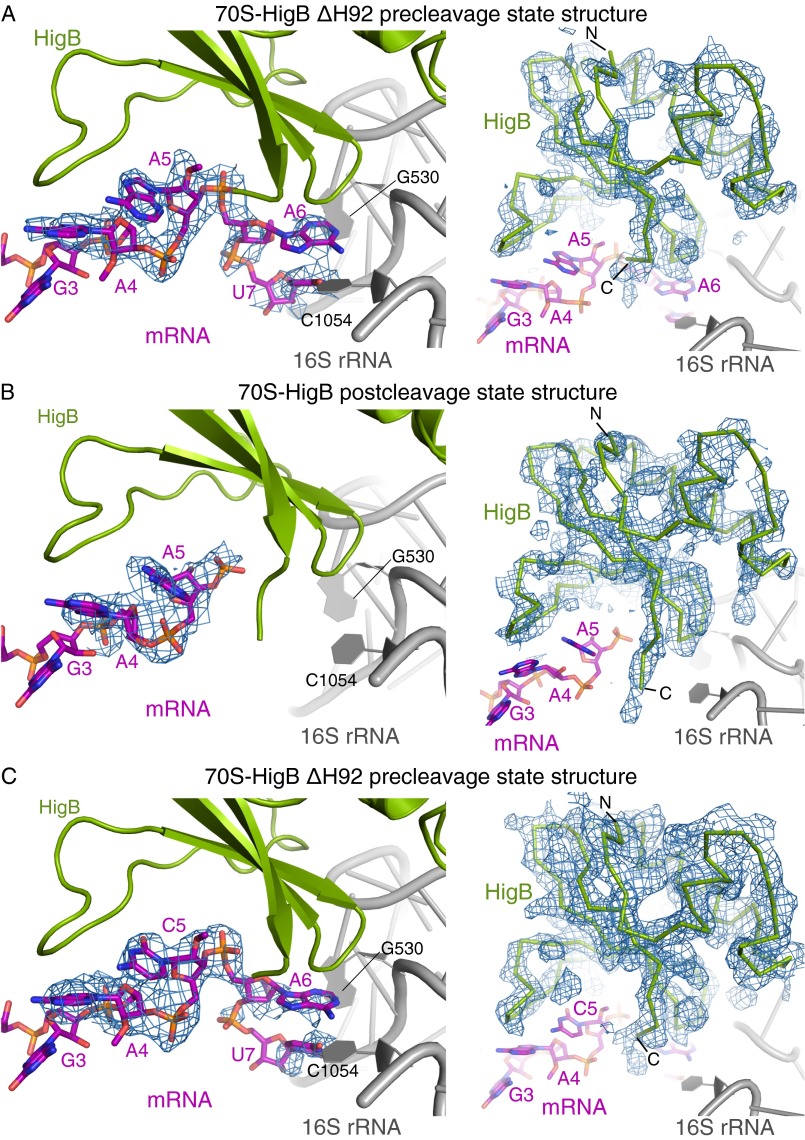
To determine the molecular basis for substrate recognition by HigB, we solved three X-ray crystal structures of HigB bound to the Thermus thermophilus (Tth) 70S containing either an AAA or ACA codon in the A site in pre- or postcleavage states and a high-resolution structure of HigB (Tables S1 and S2). The first precleavage state 70S structure was trapped by using a catalytically inactive HigB variant (ΔH92) (30) and mRNA containing a 2′-OCH3 modification at all three A-site nucleotides (AmAmAm Lys codon). This structure was determined to 3.4 Å (I/σ = 1.8) (Fig. 1 A and B, Fig. S1A, and Table S1). The second structure is a postcleavage state that contains wild-type HigB bound to unmodified mRNA and was solved to 3.3-Å resolution (I/σ = 1.8) (Fig. 1C, Fig. S1B, and Table S1). Additionally, a 70S-HigB ΔH92 precleavage state bound to an AmCmAm codon was solved to 3.1-Å resolution (I/σ = 1.9) (Fig. S1C and Table S1). In all three structures, Fo-Fc difference electron density maps allowed unambiguous placement of P-site tRNAfMet, mRNA, and A site-bound HigB (Fig. S1 A–C). Lastly, a 1.25-Å X-ray structure of HigB was used as a starting model to confidently build HigB into lower-resolution (3.1–3.4 Å) 70S electron density maps (Table S2).
Table S1.
70S-HigB structures
| A-site codon state (HigB/A-site codon) | Precleavage (ΔH92/AmAmAm) | Postcleavage (WT/AAA) | Precleavage (ΔH92/AmCmAm) |
| Data collection | |||
| Space group | P 21 21 21 | P 21 21 21 | P 21 21 21 |
| Cell dimensions | |||
| a, b, c (Å) | 214.1, 453.9, 607.6 | 214.1, 453.9, 607.6 | 214.2, 453.3, 609.9 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution, Å | 35–3.4 (3.52–3.4) | 35–3.3 (3.42–3.3) | 35–3.1 (3.21–3.10) |
| Rmeas,* % | 24 (113) | 19 (88) | 23 (82) |
| Rpim, % | 9.6 (45) | 8.9 (41) | 11 (39) |
| I/σ | 8.2 (1.8) | 8.9 (1.8) | 5.6 (1.9) |
| Completeness, % | 98.9 (99.6) | 94.3 (94.4) | 99.4 (95.9) |
| Redundancy | 6.0 (6.0) | 4.1 (3.9) | 4.4 (4.0) |
| Refinement | |||
| Resolution, Å | 35–3.4 | 35–3.3 | 35–3.1 |
| Total reflections | 4,743,702 | 3,393,142 | 4,584,941 |
| Unique reflections | 793,673 | 827,408 | 1,053,398 |
| Rwork/Rfree, % | 21.5/24.8 | 21.0/23.8 | 20.3/23.8 |
| No. of atoms | 297,549 | 297,400 | 294,445 |
| Protein/RNA | 296,237 | 296,156 | 293,095 |
| Ligand/ion | 1,312 | 1,244 | 1,350 |
| Water | 0 | 0 | 0 |
| B-factors | 91.4 | 94.7 | 75.3 |
| Protein/RNA | 91.7 | 95.0 | 75.5 |
| Ligand/ion | 35.2 | 45.2 | 41.3 |
| rms deviations | |||
| Bond lengths, Å | 0.005 | 0.004 | 0.004 |
| Bond angles, ° | 0.84 | 0.83 | 0.86 |
| PDB ID code | 4YPB | 4W4G | 4YZV |
Values in parentheses are for the highest resolution shell.
Table S2.
HigB structure
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 57.3, 31.3, 50.9 |
| α, β, γ (°) | 90, 112.2, 90 |
| Resolution (Å) | 15.7−1.25 (1.29−1.25) |
| Rmeas (%)* | 5.4 (65) |
| Rpim (%) | 2.0 (24) |
| I / σ | 19.3 (3.2) |
| Completeness (%) | 99.9 (99.9) |
| Redundancy | 7.2 (7.0) |
| Refinement | |
| Resolution (Å) | 15.7−1.25 |
| Total reflections | 168,822 |
| Unique reflections | 23,296 |
| Rwork/Rfree (%) | 15.4/18.9 |
| No. atoms | 1033 |
| Protein | 907 |
| Ion | 4 |
| Water | 122 |
| Mean B-factors (Å2) | 17.7 |
| Protein (Å2) | 16.1 |
| Ion (Å2) | 29.7 |
| Water (Å2) | 29.7 |
| rms deviations | |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 1.49 |
| PDB ID code | 4PX8 |
Values in parentheses are for the highest resolution shell.
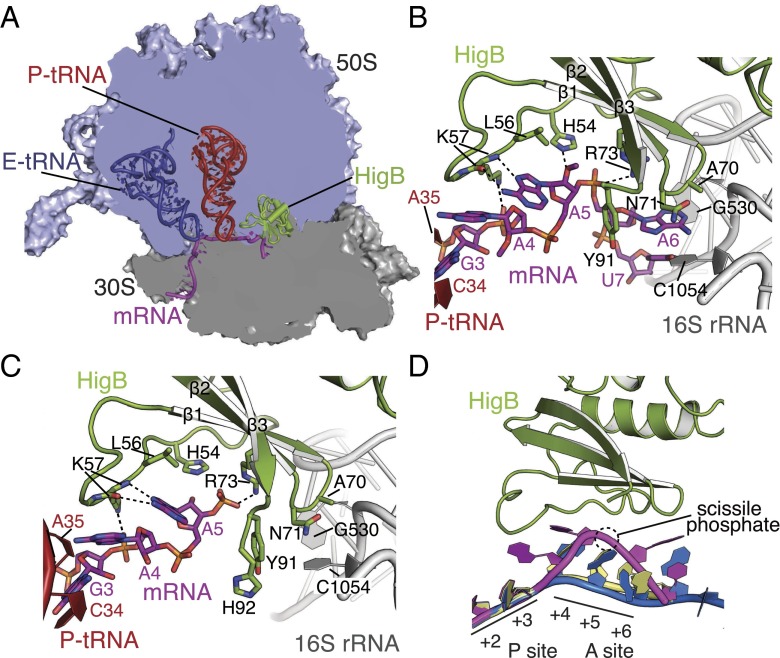
Fig. 1.
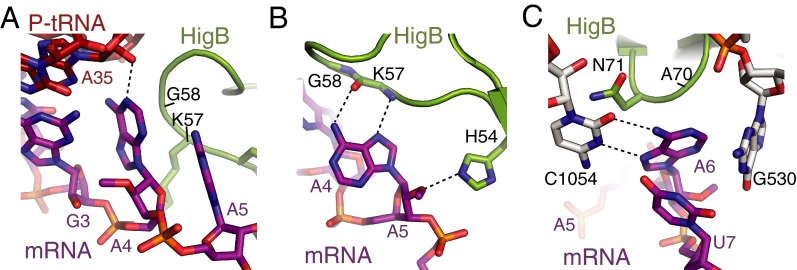
Structural basis for HigB recognition of mRNA on the 70S ribosome. (A) Overview of the structure of the 70S-HigB toxin complex containing A-site HigB showing the 30S and 50S subunits, P-site tRNAfMet, E-site tRNAfMet, and an A-site AAA lysine codon. The X-ray crystal structures of the precleavage (B) and postcleavage (C) states reveal how HigB recognizes an AAA A-site lysine codon (precleavage state A-site codon contains 2′-OCH3 modifications to prevent mRNA cleavage). (D) Comparison of the mRNA path in the A-site when bound to HigB (mRNA is in magenta), tRNA [Protein Data Bank (PDB) ID code 4V51; yellow] or an empty A site (PDB ID code 4V6G; blue), emphasizing the large mRNA movement once HigB binds. P-site mRNA nucleotides (+2 and +3), A-site mRNA nucleotides (+4, +5 and +6), and the location of the scissile phosphate during HigB cleavage are shown.
Fig. S1.
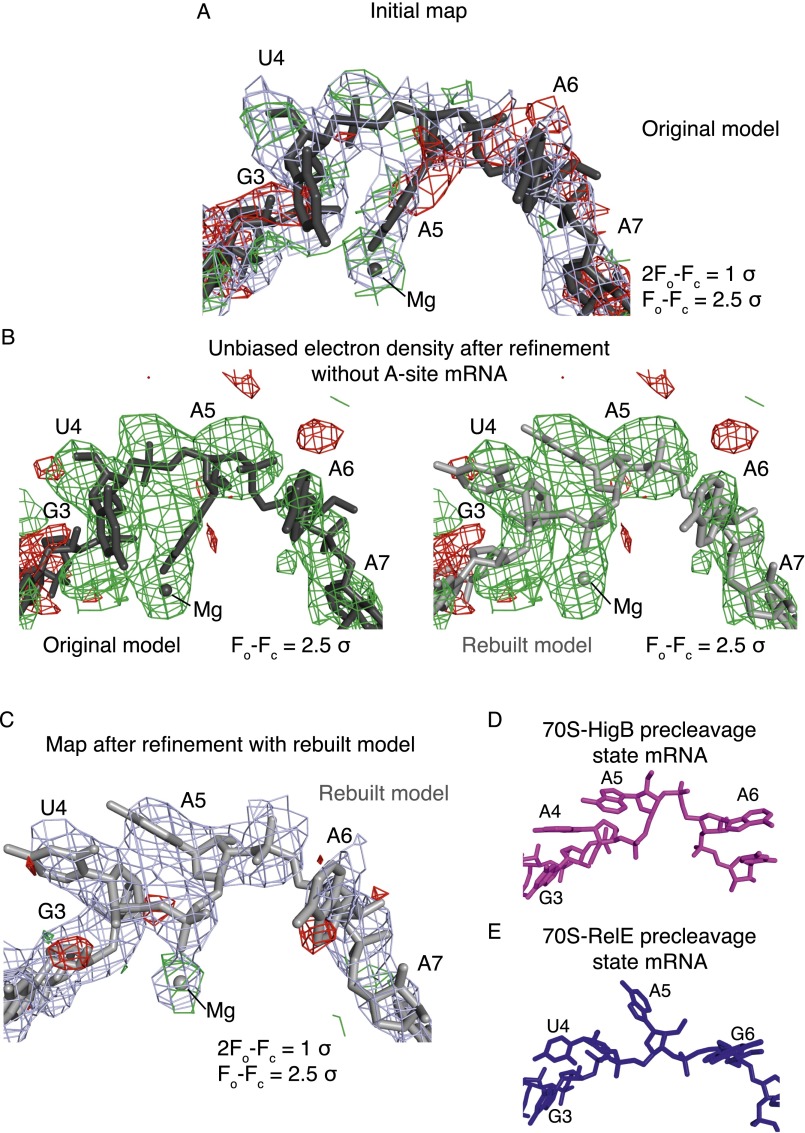
Quality of 70S-HigB bound maps and models. Unbiased Fo-Fc difference electron density (contoured to 2σ unless noted otherwise) for both the mRNA (Left) and HigB (Right) for 70S ribosome structures bound to HigB. (A) A 3.4-Å X-ray crystal structure of the 70S-HigB ΔH92 precleavage state containing an A-site AmAmAm lysine codon. (B) A 3.3-Å X-ray crystal structure of the 70S-HigB postcleavage state containing an A-site AAA lysine codon. (C) A 3.1-Å X-ray crystal structure of the 70S-HigB ΔH92 precleavage state containing an A-site AmCmAm lysine codon. Here, unbiased Fo-Fc difference electron density is contoured to 1.5σ and 2σ the mRNA (Left) and HigB (Right), respectively.
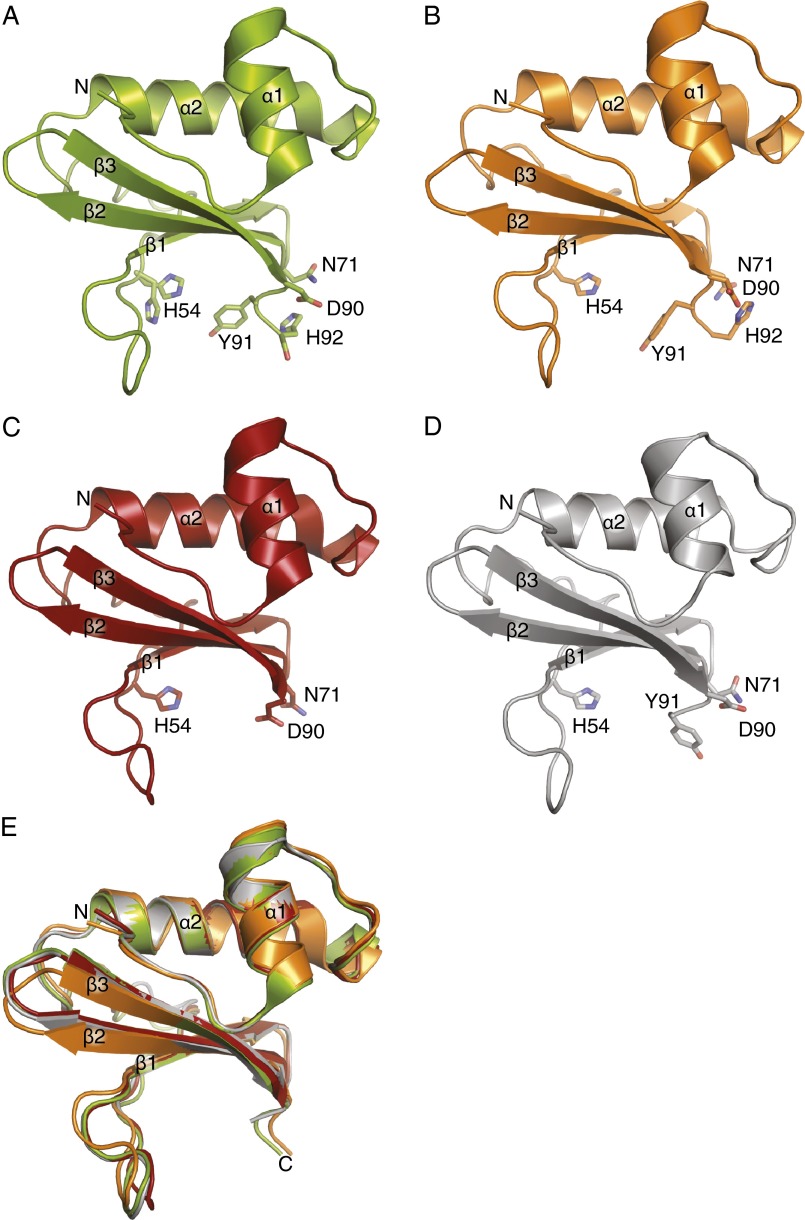
The 1.25-Å structure of free HigB reveals that it adopts a microbial RNase fold consisting of a three-stranded, β-sheet appended by two N-terminal α-helices (Fig. S2A). Comparison of this structure, HigB from the HigBA complex (33), and HigB in the 70S bound structures (discussed in the next section) shows a similar overall HigB fold and concave cleft, the likely location of the active site (rmsd of 0.4–1.0 Å for residues 1–92) (Fig. S2). Although other toxins are proposed to undergo allosteric regulation after release from the antitoxin (32, 37, 38), our data suggest that HigB has a preformed tertiary structure primed for interaction with the ribosome.
Fig. S2.
Comparison of HigB in different states. (A) A 1.25-Å X-ray crystal structure of free HigB with active site residues shown as sticks. (B) A 2.8-Å X-ray crystal structure of the HigBA complex was solved but only HigB is depicted (PDB code 4MCT). (C) Structure of HigB from the 2.1-Å X-ray crystal structure of the HigBA complex (PDB code 4MCX). (D) A 3.4-Å X-ray crystal structure of HigB ΔH92 bound to the 70S ribosome in the precleavage state. (E) Overlay of all four HigB structures demonstrating their overall similar fold (rmsd 0.4–1.0 Å for residues 1–92).
Recognition of the A Site by HigB Involves Distortion of the mRNA.
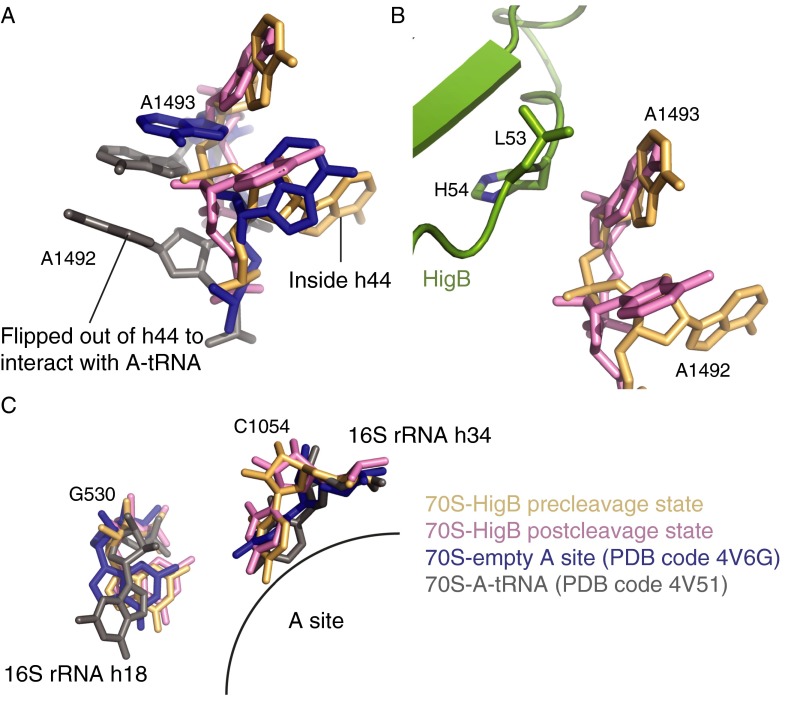
HigB binds the A site between the head and body of the small subunit, similar to where tRNA interacts with mRNA (Fig. 1A) (39). The selection of correct tRNAs results from the ribosome monitoring the Watson–Crick base pairing between the tRNA anticodon stem loop and the A-site codon; 16S rRNA nucleotides A1492/A1493 flip from an internal loop of helix 44 (h44), whereas G530 changes from a syn to an anti conformation to inspect the minor groove of the codon–anticodon interaction (39). In the two 70S-HigB precleavage state structures, A1492 remains within h44, whereas A1493 adopts an intermediate state between its fully extended flipped position, seen when a cognate mRNA-tRNA pair is present in the A site and its position inside of h44 (Fig. S3A). In the postcleavage state structure, both A1492 and A1493 occupy an intermediate state, with A1493 close to 23S rRNA nucleotide A1913 (Helix 69) and HigB residues L53 and H54 (Fig. S3 A and B). In all structures, G530 adopts a syn conformation, resembling an empty A-site 70S complex (40) (Fig. S3C).
Fig. S3.
Orientation of A-site decoding center nucleotides upon HigB binding to the 70S. (A) Comparison of 16S rRNA residues A1492 and A1493 when HigB binds to the 70S trapped in a precleavage state (tan), in a postcleavage state (pink), when no A-site ligand is bound (PDB code 4V6G; blue) and when cognate A-site tRNA is bound (PDB code 4V51; gray). (B) Location of A1492 and A1493 with respect to HigB in a precleavage state and in a postcleavage state and their proximity to loop 3 of HigB. (C) Orientation of 16S rRNA residues G530 and C1054 in precleavage and postcleavage state structures of 70S-HigB compared with when the ribosomal A site contains cognate tRNA (gray) or is empty (blue).
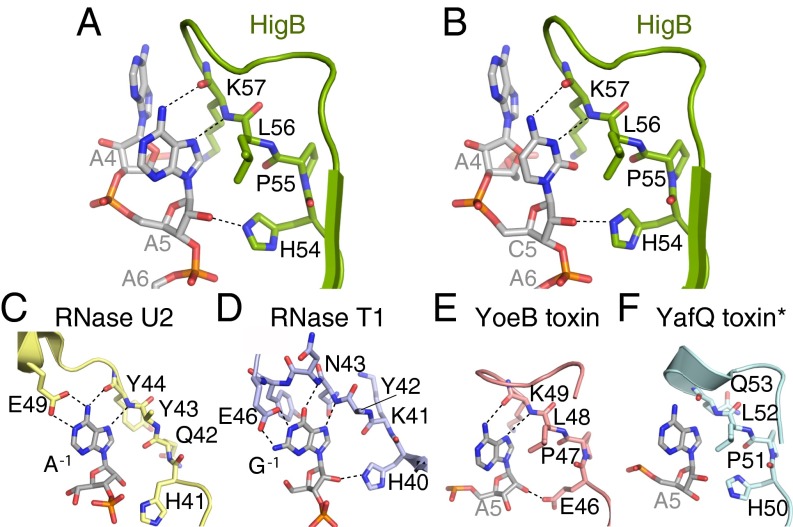
In both the pre- and postcleavage state 70S structures, HigB binding causes a distortion of the mRNA backbone with the position of the A6 nucleotide changing the most dramatically (5- and 9-Å movement of the C1′ and phosphate atoms, respectively) (Fig. 1D). The structures of HigB bound to the 70S reveal that HigB interacts with each of the three A-site nucleotides in distinct ways. Although a potential hydrogen bond between the nucleobase of A4 with the 2′-OH of the adjacent P-site tRNA nucleotide 35 may be present, no other direct interactions between HigB and the A4 nucleotide are observed, indicating any nucleotide would be recognized by HigB at this position (Fig. 2A). In contrast, HigB residues H54, K57, A70, N71, R73, Y91, and H92 flank the +5 and +6 mRNA positions (Figs. 1 B and C). HigB loop residues H54 and K57 (located between α2 and β1) directly interact with the +5 position; the Hoogsteen face of the A5 nucleotide forms two hydrogen bonds with the backbone of HigB lysine residue (K57) (Fig. 2B). The only other nucleotide that can fulfill this same hydrogen-bonding pattern is cytosine in an anti conformation (Fig. S4). An additional contact includes the nitrogen ε2 atom of the imidazole side chain of H54 with the 2′-OH of A5 (a 2′-OCH3 in the precleavage structure) (Fig. 2B).
Fig. 2.
HigB recognition of A-site mRNA nucleotides. (A) HigB forms no interactions with the A4 mRNA nucleobase or ribose to specify nucleotide identity. (B) Hydrogen bonds between the nucleobase of A5 with the backbone of HigB drive nucleotide specificity. (C) HigB residue N71 and 16S rRNA residue C1054 form an adenosine-specific binding pocket at the +6 mRNA position.
Fig. S4.
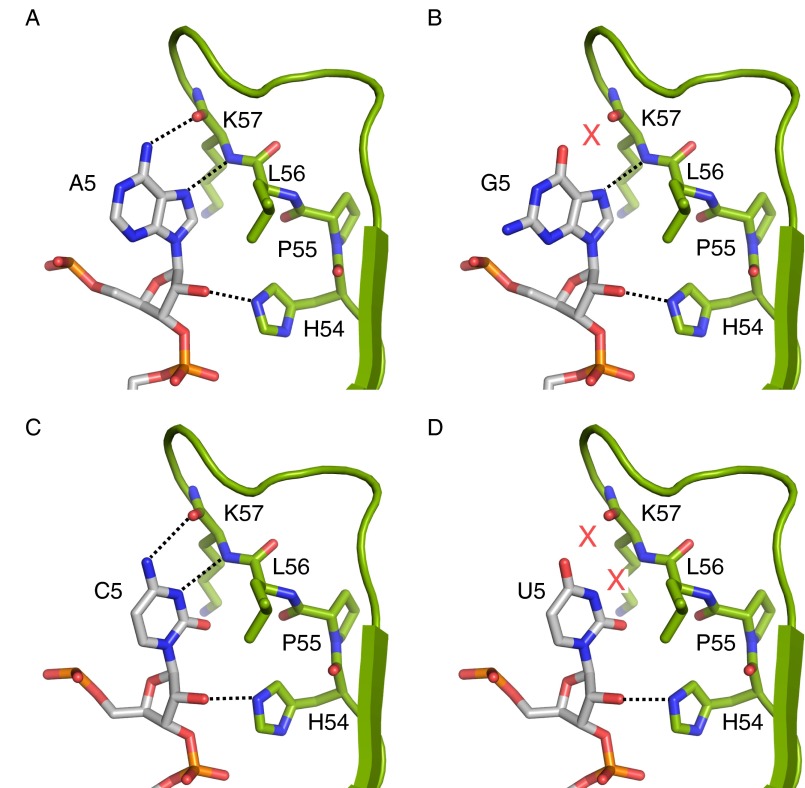
Model for the +5 nucleotide recognition by HigB that predicts G5 and U5 are incompatible. (A) A5 forms hydrogen bonds with the backbone of HigB, as seen in our 70S-HigB precleavage state structure. (B) Model for how G5 interacts with the HigB backbone. Note the guanosine O6 atom and the backbone carbonyl of K57 are unable to form a hydrogen bond (red X). (C) C5 forms hydrogen bonds with the backbone of HigB, as seen in our 70S-HigB precleavage state structure. (D) Model for how U5 interacts with the HigB backbone. Note the uracil O4 and N3 atoms and the backbone carbonyl of K57 are unable to form hydrogen bonds (red Xs).
Surprisingly, C-terminal HigB residues A70, N71, R73, and Y91 and 16S rRNA residues surround the nucleotide of A6, forming a nucleotide-specific pocket (Fig. 2C). HigB R73 interacts with the A6 phosphate (the scissile phosphate), and HigB residues A70 and N71 flank one face of the nucleobase of A6 whereas 16S rRNA residues G530 and C1054 frame the opposite side. HigB residue N71 stacks with 16S rRNA nucleotide C1054, orienting the Watson–Crick face of C1054 for interaction with the Hoogsteen edge of A6. The interaction of A6 with the conserved C1054 provides one mechanism by which HigB selects for adenosines at the +6 position. HigB recognition of the +6 nucleotide is most similar to how release factors 1 or 2 (RF1/2) recognize stop codons, with one significant difference being the +6 nucleotide base stacks with decoding center nucleotide G530 upon RF binding, an interaction absent in HigB-mRNA recognition (41, 42).
A-Site Nucleotide Requirements for HigB Cleavage.
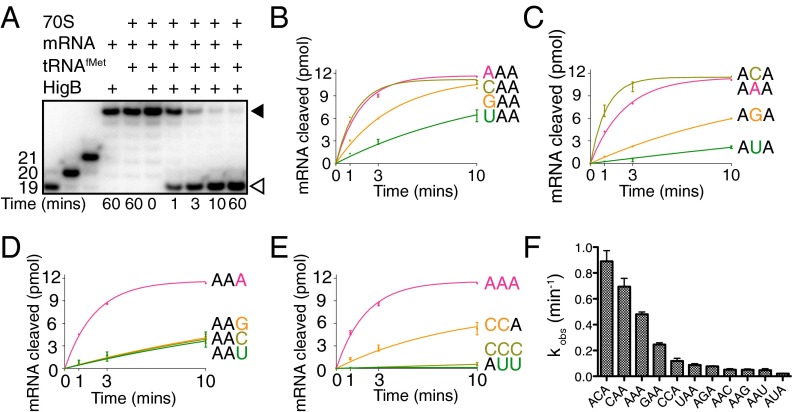
Primer-extension analysis of five transcripts cleaved upon HigB overexpression identified a preference for adenosine-rich codons (codons cleaved in the preference of AAA > GAA > CAA > AAC) but also codons containing a single adenosine (e.g., GCA > CCA > CAU) (30). We confirmed in vitro that HigB efficiently cleaves an AAA codon in a ribosome-dependent manner (Fig. 3A). Our structures reveal that HigB interacts with each of the three A-site nucleotides in distinct ways providing initial evidence that toxins may not have a strict three-nucleotide codon requirement. To further test this hypothesis, we performed in vitro cleavage assays in which each A-site nucleotide position was varied in the context of the preferred AAA codon. Similar results were seen when an adenosine, guanosine, or cytosine was located at the +4 position [observed rate constants (kobs) of 0.57, 0.25, and 0.69 min−1, respectively], whereas an uridine substitution was cleaved less efficiently (0.088 min−1) (Fig. 3B and Fig. S5A). Together, these data strongly suggest that there is no strong nucleotide preference at the +4 position.
Fig. 3.
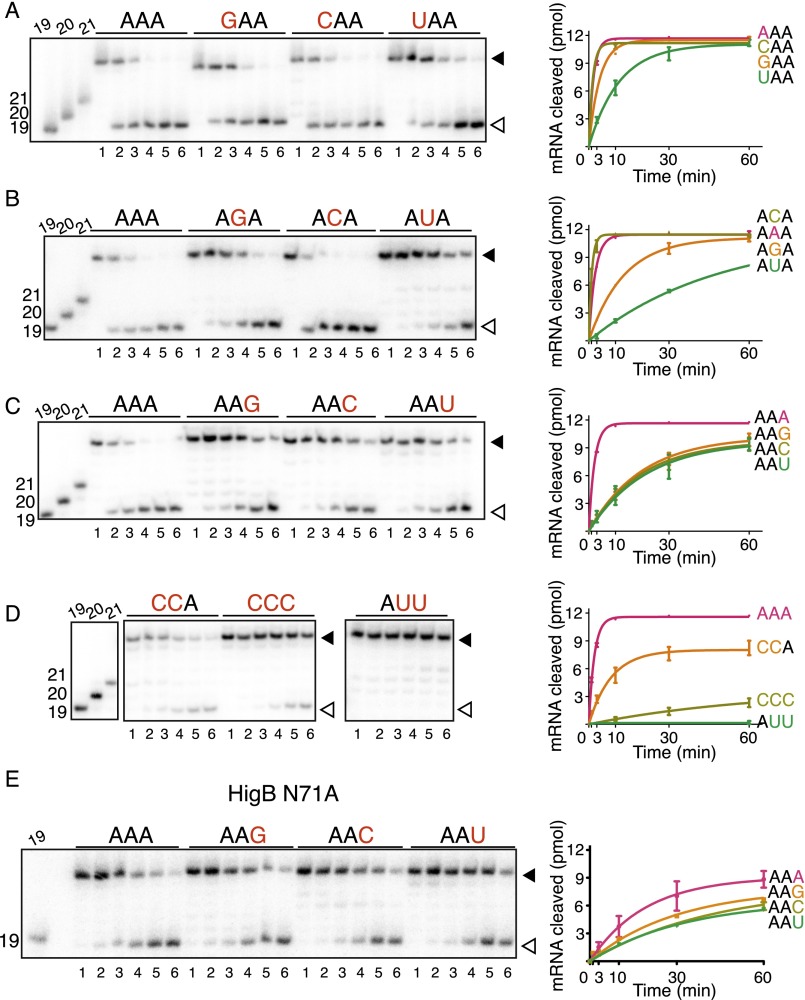
HigB demonstrates a clear preference at the third A-site nucleotide. (A) E. coli 70S ribosomes were programmed with 5′-[32P]mRNA containing an A-site AAA lysine codon, P-site tRNAfMet, and HigB. The reaction was followed for 60 min, and the amount of mRNA cleaved (open arrow) compared with uncleaved (closed arrow) was determined by denaturing RNA gels (Fig. S5). A-site mRNA was varied at the +4 nucleotide position (B), the +5 nucleotide position (C), the +6 nucleotide position (D), and +4, +5 and +6 combinations (E). Each assay was performed for two technical replicates with the mean value ± SEM (F) kobs values (min−1), as calculated from assays in B–E.
Fig. S5.
In vitro analyses of ribosome-dependent mRNA cleavage by HigB. E. coli 70S ribosomes were programmed with 5′-[32P]mRNA containing an A-site AAA lysine codon, P-site tRNAfMet, and HigB. The reaction was followed for 60 min, and the amount of mRNA cleaved (open arrow) compared with uncleaved (closed arrow) was determined by denaturing RNA gels (Left); 19-, 20-, and 21-mer standards are shown at the left of each gel. For wild-type HigB experiments, A-site mRNA was varied at the +4 nucleotide A-site position (A), the +5 nucleotide A-site position (B), the +6 nucleotide A-site position (C), and +4, +5 and +6 combinations (D). Lanes 1–6 represent 0, 1, 3, 10, 30, and 60 min time points for each codon tested in A–D. (E) For the HigB N71A variant, the +6 nucleotide A-site position was varied. Lanes 1–6 represent 1, 3, 10, 30, 60, and 120 min time points. The results were quantified using Image Quant, and product progression curves of the RNA gels are plotted on the (mean values ± SEM are displayed from at least two replicates) (Right).
Our structures of HigB bound to the ribosome reveal A5 makes specific interactions with the HigB backbone, with cytosine being the only other nucleotide capable of forming similar interactions (Fig. S4). We tested the nucleotide preference at the +5 position, and our results demonstrate similar observed cleavage rates for AAA and ACA codons (kobs = 0.57 and 0.89 min−1, respectively), whereas AGA and AUA codons were cleaved 7- and 27-fold less efficiently (0.077 and 0.021 min−1, respectively; Fig. 3C and Fig. S5B), further supporting our predictions based upon our structural work.
To directly test the possibility that an ACA codon forms a similar hydrogen-bonding network as the AAA codon, we solved a 3.1-Å X-ray crystal structure of 70S-HigB ΔH92 bound to the AmCmAm codon in a precleavage state (Table S1). The first and third A-site nucleotides (A4 and A6) are in identical positions as previously seen in our 70S-HigB bound to the AAA A-site codon structure (Fig. 4 A and B and Fig. S1). Although C5 occupies a similar local position as A5, it adopts an anti conformation to form an analogous hydrogen-bonding pattern between A5 and the HigB backbone (Fig. 4 A and B). Specifically, the N3 amine and the N4 amino groups of C5 form hydrogen bonds with the backbone amino and carbonyl groups of K57, respectively. Additionally, the nitrogen ε2 atom of the imidazole side chain of H54 is within ∼3 Å from the 2′-OH of C5 (Fig. 4B). The requirement of an adenosine or cytosine at the +5 position suggests that HigB catalytic efficiency is controlled by hydrogen-bonding complementarity for proton abstraction and in-line attack on the scissile phosphate.
Fig. 4.
Structural basis for toxin specificity at the +5 nucleotide position and similarities to general RNases. (A and B) The 3.4-Å X-ray crystal structure of the 70S-HigB ΔH92 precleavage state (AmAmAm A-site codon) compared with the 3.1-Å X-ray crystal structure of the 70S-HigB ΔH92 precleavage state (AmCmAm A-site codon). Despite the differences at the +5 positions, similar hydrogen bonds are formed between A5 and the HigB backbone and C5 and the HigB backbone. Structures of the nucleotide recognition loop of RNase U2 (PDB ID code 3AGN) (C), RNase T1 (PDB ID code 1RGA) (D), and ribosome-dependent toxins YoeB (PDB ID code 4V8X) (E) and YafQ (PDB ID code 4ML2) (F) highlight the similarities for recognition of the nucleobase preceding the scissile phosphate. The side chains of RNase U2 and T1 residues Y43 and Y42, respectively, stack with the −1 position but were removed for clarity. The 70S-YoeB mRNA was rebuilt using structure factors from the PDB (Fig. S6). The backbone carbonyl and amino groups of YoeB K49 and YafQ Q53 are proposed to interact with A5 similar to HigB K57. The YafQ toxin structure was solved in the absence of the 70S ribosome, and its interactions with A5 are based upon modeling in the ribosomal A site (denoted by a star).
Fig. S6.
Rebuild of the 70S-YoeB structure. (A) Examination of the electron density of the 70S-YoeB precleavage state structure (PDB ID code 4V8X) suggests an alternative modeling of the first two A-site nucleotides. Original mRNA model (dark gray) with electron density shown. (B) Original model (dark gray) (Left), rebuilt model (light gray) (Right), and difference electron density from refinement of 4V8X without mRNA nucleotides +4 to +8. (C) Rebuilt mRNA model and electron density map resulting from refinement. (D) mRNA paths when HigB binds to the A site (this study) and when RelE (E) binds the A site (PDB ID code 4V7J), revealing similar paths when ribosome-dependent toxins YoeB, RelE, and HigB bind the 70S.
Next, we tested the nucleotide requirements of HigB at the +6 position. Our results show AAG/C/U codons were cleaved 9- to 10-fold less efficiently than the optimal AAA codon (0.051, 0.051, and 0.047 min−1, respectively; Fig. 3D and Fig. S5C), demonstrating a clear preference for an adenosine. Taken together, these data support a model where HigB nucleotide specificity increases 5′ to 3′ of the A-site codon, with the identity of the +4 nucleotide playing little to no role in HigB specificity. Conversely, tRNAs and RFs have vastly different recognition requirements. During tRNA selection, nucleotide identity and specifically a Watson–Crick interaction between the anticodon and codon is essential at the +4 and +5 positions, whereas variation at the +6 or wobble position allows for degeneracy in genetic code. During termination by RFs, the identify of all three A-site nucleotides is important. The differential recognition patterns of the three A-site nucleotides by HigB is surprising given what is known about tRNA and RF A-site recognition of mRNA.
Microbial endonucleases RNases T1 and U2 contain a recognition loop that interacts with the purine nucleobase 5′ of the scissile phosphate in a similar manner as toxins probe the identity of the +5 nucleotide (this is also the nucleotide 5′ of the scissile phosphate) (Fig. 4 C and D) (43, 44). Moreover, RNase T1 and U2 binding forces the preceding purine to adopt a syn conformation similar to what we observe with A5 in the 70S-HigB structures. This syn purine conformation allows for recognition of the Hoogsteen face of the nucleobase by the peptide backbone and recognition of the Watson–Crick face by a conserved glutamate residue. A conserved histidine in both RNase T1 and U2 is within hydrogen-bonding distance to the 2′-OH. This histidine likely functions as a general base to initiate the in-line attack reaction, similar to a possible role of HigB residue H54 adjacent to the 2′-OH. One key difference between microbial RNases and HigB is that HigB does not interact with the Watson-Crick face of the nucleotide preceding the scissile phosphate (i.e., +5 position) (Fig. 4A). One possible reason for this lack of inspection is that the HigB recognition loop is shortened as compared to either RNase T1 or U2 (8 or 11 residues, respectively). One consequence of this is that HigB, and possibly other ribosome-dependent toxins, discriminates irrespective of base size but, rather, by complementary surfaces between the mRNA and toxin.
Cross-Talk Between A-Site Nucleotides Drives Efficient HigB Recognition of mRNA.
To more completely define HigB sequence recognition requirements, we next varied more than one nucleotide position in the preferred AAA codon to test the combinatorial effects of adding together two single substitutions that are either cleaved efficiently or inefficiently. A CCA codon should, in theory, be efficiently cleaved by HigB because of C5 and A6. Compared with the CAA and ACA codons, a CCA codon is less efficiently cleaved (six- to sevenfold reduction, respectively; 0.12 min−1) (Fig. 3E and Fig. S5D). Consistent with the importance of a codon containing an adenosine at the +6 position, a CCC codon is cleaved by HigB with extremely low efficiency (Fig. 3E). To define the effects of combining two substitutions that are cleaved inefficiently (AUA and AAU, which each results in a ∼27- and 10-fold reduction of mRNA cleavage compared with AAA, respectively), we tested HigB cleavage of the AUU codon (Fig. 3E). Uridines at the +5 and +6 positions (AUU) completely ablated HigB activity. These data imply that HigB cleavage is context-dependent with communication occuring between the +5 and +6 nucleotides.
These cleavage assays define the codon signatures recognized by HigB and demonstrate that HigB selects for a spectrum of A-site codons containing specific nucleotides at the +5 and +6 nucleotide positions. Based on these studies, we propose that upon entering the A site, HigB probes the nucleotide sequence at the +5 and +6 positions via hydrogen-bonding with the +5 nucleobase while also confirming an adenosine at the +6 position. If both requirements are met, the mRNA is optimally positioned for efficient HigB cleavage. Moreover, although we found the identity at the +4 position plays little role in HigB recognition when adenosines are present at the +5 and +6 A-site positions, we determined that the +4 and +5 nucleotides have a combinatorial effect on HigB activity (Fig. 3E). Although the codon specificities of only RelE and HigB have been quantitated with defined in vitro assays, we predict YafQ and YoeB are also likely to cleave a spectrum of codons as observed for HigB and RelE (24, 27, 28).
A Single HigB Residue Modulates Codon Selectivity.
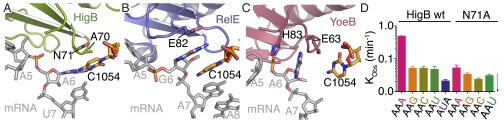
HigB selects for an adenosine at the +6 position through a trans Watson–Crick-Hoogsteen interaction with 16S rRNA C1054, and additionally N71 stacks with C1054 (Fig. 5A). Asparagine 71 is highly conserved in HigB homologs (>87% sequence identity), and to test the effect of N71 substitution, we assayed the HigB N71A variant for its ability to cleave +6 nucleotide substitutions in the context of the AAA lysine codon. Our results show that HigB N71A cleaves the AAA codon ∼10-fold less efficiently than wild-type HigB, but, surprisingly, HigB N71A cleaves AAG/C/U codons only ∼twofold less efficiently than wild-type HigB (Fig. 5D and Fig. S5E). These results strongly suggest that the N71A variant corrupts the A6-binding pocket, allowing for nucleotide promiscuity. The productive interaction between HigB N71 and C1054 likely promotes efficient cleavage of codons containing adenosines at the +6 positions, rather than discriminating against other nucleotides. In support of this, all codons tested for HigB N71A cleavage show similar low levels of mRNA degradation, demonstrating that HigB specificity is conferred by a single amino acid.
Fig. 5.
A single conserved HigB residue drives sequence specificity at the +6 position. Structural comparison of how ribosome-dependent toxins and 16S rRNA residues select for nucleotides at the +6 mRNA position by the formation of stacking and electrostatic interactions. (A) A trans Watson–Crick-Hoogsteen interaction with A6 is formed by 16S rRNA C1054, whereas HigB residue N71 stacks with C1054. (B) In contrast, a continuous stack between RelE residue E82, G6, and C1054 (PDB ID code 4V7J) forms. (C) YoeB residues H83 and E63 stack around A6 with C1054, playing little to no role in nucleotide selection (PDB ID code 4V8X). All precleavage state structures of 70S bound to ribosome-dependent toxins were solved using uncleavable mRNA. (D) HigB N71A mRNA kobs values were determined upon substitution at the +6 nucleotide position. Wild-type HigB cleavage assays were described in Fig. 3 and Fig. S5. HigB N71A cleavage assays were performed with two biological replicates with the mean values ± SEM reported.
Conclusions
We report a comprehensive molecular analysis of the bacterial toxin HigB, a type II ribosome-dependent mRNA endonuclease. The X-ray structures of HigB and HigB bound in a pre- and postcleavage states reveal insights into mRNA specificity by toxins. Recent studies demonstrate that type II toxins play important roles in bacteria, such as in persister cell formation (8, 11, 36). Therefore, determining the molecular basis for mRNA degradation provides significant insights into toxin function. Because many bacterial genomes encode for multiple ribosome-dependent toxins that upon overexpression inhibit translation and cell growth, whether all ribosome-dependent toxins function similarly during stress to simply impair translation, or whether each has a defined role in response to stress, is an unresolved, fundamental question.
Comparison of ribosome-dependent toxins reveals similarities in how each defines a spectrum of codons for degradation but also reveals a number of striking distinctions. HigB, YoeB, and YafQ all contain a 4-amino acid motif (H/E-P-L-X) in the recognition loop that directly contacts the +5 position of the A-site mRNA (Fig. 4 A, E, and F). The HigB recognition loop specifies an A or a C at the +5 nucleotide position, with YoeB similarly recognizing an A5 in a syn conformation (22). Based on our biochemical assays of HigB and the structural analyses of other toxins bound to the 70S (21, 22), we predict that YafQ and YoeB toxins may efficiently cleave codons containing +5 cytosines, along with their well-characterized ability to cleave +5 adenosines (24, 27–29). In contrast, RelE preferentially recognizes an A5 nucleotide that adopts an anti conformation. This unique mode of binding suggests RelE uses a different mechanism for recognition of a purine at the +5 position (16, 21).
The most striking difference in the mechanism of mRNA selection by ribosome-dependent toxins is in how the +6 nucleotide is engaged by each toxin. Our results clearly show a strict HigB requirement for an adenosine at the +6 position, where A6 forms a trans Watson–Crick-Hoogsteen interaction with C1054 (Fig. 5A). However, in the 70S-RelE structure, the nucleobase edge of G6 does not interact with C1054 to enforce specificity, but, rather, G6 and C1054 stack consistent with a RelE preference for a purine at this position (Fig. 5B) (16, 21). In the 70S-YoeB structure, the only contact with A6 is with YoeB itself (Fig. 5C) (22). These differences in how ribosome-dependent toxins interact with the +6 nucleotide define the spectrum of different codons selected.
In a complementary set of experiments, we identified HigB residue N71 as an important determinant of mRNA selection, suggesting a mechanism by which specificity for an adenosine is enforced at the +6 position. Our characterization of the HigB N71A variant reveals that the mutant protein is functional in our cleavage assays but, surprisingly, has an altered nucleotide selectivity and exhibits enzyme promiscuity. The identification of a single HigB residue that controls mRNA specificity implies that sequence-specific targeting has an adaptive advantage for HigB, and other ribosome-dependent toxins may be similarly manipulated. Although N71 is highly conserved among HigB homologs (87%), in some cases, a glutamine or proline substitutes (4% and 5%, respectively). In these cases, it is likely that either residue could similarly form a +6 adenosine-specific pocket by maintaining stacking interactions with C1054. In the other diverse HigB homologs that do not contain an asparagine, glutamine or proline amino acid at position 71 (∼4%), we predict altered mRNA selectivity. Future experiments to determine the codon specificity of YafQ and YoeB will be important to identify the degree of functional overlap among toxin family members.
What is the biological consequence of multiple ribosome-dependent toxins that degrade a spectrum of codons? One possibility is that the codon substrate and catalytic rate may be used to tune the global cellular rate of translation where mRNA cleavage is dependent on enzyme rate and codon frequencies that vary among different bacteria. An interesting additional possibility is that ribosome-dependent toxins active during translation elongation will have a higher probability of degrading the most highly translated mRNA, that is, mRNA with the highest ribosome occupancy. It remains unclear why some toxins target the translation initiation and translation termination steps. These steps are the slowest during protein synthesis, perhaps allowing more time for toxins to efficiently recognize their preferred codons. Alternatively, differential degradation of mRNA could lead to a selective translational program, as seen for the ribosome-independent toxin MazF (14). Lastly, toxins may have a role in halting translation via ribosomal stalling to protect the translational machinery during the stringent response that would allow for rapid resumption of translation upon removal of the stress. We hypothesize that functional distinctions among ribosome-dependent toxins impact bacterial physiology by fine-tuning translation to modulate key cellular pathways.
Materials and Methods
See SI Materials and Methods for strains and plasmids and detailed descriptions of experimental conditions, sequence and structural alignments, structural determination of HigB and 70S-HigB complexes, and mRNA cleavage assays.
SI Materials and Methods
Strains and Plasmids.
E. coli BW25113 [Δ(araD-araB)567 ΔlacZ4787(::rrnB-4) lacIp-400(lacIQ)λ- rpoS396(Am) rph-1 Δ(rhaD-rhaB)568 rrnB-4 hsdR514] cells were used for all bacterial growth assays and wild-type HigB(His)6, HigBΔH92(His)6, and HigBN71A(His)6 protein expression (45). Plasmids pBAD/Myc-HisA-HigB(His)6 and pBAD/Myc-HigA-HigB ΔH92(His)6 were generous gifts from Nancy A. Woychik, Rutgers University, Piscataway, NJ. All point mutations were introduced by site-directed mutagenesis, and sequences were verified by DNA sequencing (Genewiz).
Sequence and Structural Alignments.
HigB homolog sequences were identified using the P. vulgaris HigB sequence as a search model in the National Center for Biotechnology Information protein Basic Local Alignment Search Tool using an e value cutoff of 10−4 (46). All structural comparisons were aligned using the least squares function in Coot (47). For comparisons of the mRNA paths, 70S structures with A, P, and E site-bound tRNA (PDB ID code 4V51) or 70S structures containing P- and E-site tRNAs, but with an empty A site (PDB ID code 4V6G), were each aligned to the P-site mRNA of the 70S-HigB precleavage state structure (AmAmAm codon). Residues 1–90 of HigB in the 70S-HigB precleavage state structure were aligned to HigB free residues 1–90. For comparison of the +6 interactions, the 30S head domains of the 70S-RelE and 70S-YoeB structures were aligned to that of the 70S-HigB structure.
Wild-Type HigB, HigB ΔH92, and HigB N71A Expression and Purification.
E. coli BW25113 cells transformed with either pBAD/Myc-HisA-HigB(His)6, pBAD/Myc-HisA-HigBΔH92(His)6, or pBAD/Myc-HisA-HigBN71A(His)6 were grown at 37 °C in M9 minimal medium [48 mM Na2HPO4, 22 mM KH2PO4, 8.6 mM NH4Cl, 18.7 mM NaCl, 1 mM MgSO4, 0.1 mM CaCl2 and 0.2% (wt/vol) casamino acids] supplemented with 0.21% (vol/vol) glycerol. Protein expression was induced at 0.7 OD600 with 0.04% wt/vol arabinose. After 3 h of protein expression, cells were harvested and frozen at −20 °C, and protein purification was carried out as described for the P. vulgaris HigBA complex except that a S75 10/300 column (GE Healthcare) was used (33).
Structural Determination of HigB.
Trypsinized wild-type HigB crystals (20 mg mL−1) were grown in 30–40% (wt/vol) polyethylene glycol (PEG) monomethylether (MME) 2000 and 0.15 M KBr to dimensions of 50 × 400 × 400 μm in 1 d. Crystals were cryoprotected in the crystallization mother liquor supplemented with 10–30% ethylene glycol and decreasing PEG MME 2000 from 30% to 10% in stepwise additions. Diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22BM beamline at the Advanced Photon Source (APS) (Argonne, IL) and processed to 1.25 Å with the X-ray Detector Software (XDS) package (48). The structure was solved by molecular replacement using Phenix AutoMR (49) with the P. vulgaris HigB-HigA coordinates (PDB ID code 4MCT) (33). Iterative refinement using the xyz coordinates, occupancies and B-factors (anisotropically for proteins and isotropically for waters), and model building was performed in Phenix and Coot, respectively (47), to final R/Rfree values of 15.4/18.9%.
Structure Determination of 70S-HigB Complexes.
Tth 70S ribosomes were purified as previously described (50). Purified E. coli tRNAfMet was purchased from Chemical Block, unmodified RNA was purchased from IDT, and modified RNA was purchased from Thermo Fisher Dharmacon (mRNAs are listed in Table S3). The 70S-HigB ΔH92 precleavage state (AmAmAm codon) and 70S-HigB postcleavage state structures (AAA codon) were obtained by cocomplexing the Tth 70S with unmodified or modified mRNA, tRNAfMet, and HigB ΔH92 protein or wild-type HigB, respectively. Crystal growth was performed as previously described (50); 70S ribosomes at a final concentration of 4.4 μM were first incubated with a twofold molar excess of either modified or unmodified mRNA for 6 min at 55 °C, then a fourfold molar excess of tRNAfMet for 30 min at 55 °C, and a fivefold molar excess of wild-type HigB or HigB ΔH92 at 37 °C for 1 h. A final concentration of 2.4 mM detergent Deoxy Big Chap was added last before crystallization. Crystals grew to 70 × 70 × 400 μm in 1–2 wk and were serially cryoprotected by increasing the concentration of PEG 550 MME along with the addition of 25 μM HigB or HigB ΔH92 in the final cryo before flash-freezing in liquid nitrogen.
Table S3.
mRNAs used for structural and biochemical analysis
| mRNAs, 5′–3′ | A-site codon | Assay(s) used |
| GGCAAGGAGGUAAAAAUGAAAUAGU | AAA | mRNA cleavage assay WT HigB postcleavage structure |
| GGCAAGGAGGUAAAAAUGAmAmAmUAGU | AmAmAm | 70S-HigB ΔH92 precleavage sate structure |
| GGCAAGGAGGUAAAAAUGGAAUAG | GAA | mRNA cleavage assay |
| GGCAAGGAGGUAAAAAUGCAAUAGU | CAA | mRNA cleavage assay |
| GGCAAGGAGGUAAAAAUGUAACAGA | UAA | mRNA cleavage assay |
| GGCAAGGAGGUAAAAAUGAGAUAGU | AGA | mRNA cleavage assay |
| GGCAAGGAGGUAAAAAUGACAUAGU | ACA | mRNA cleavage assay |
| GGCAAGGAGGUAAAAAUGAUAUAGU | AUA | mRNA cleavage assay |
| GGCAAGGAGGUAAAAAUGAAGUAGU | AAG | mRNA cleavage assay |
| GGCAAGGAGGUAAAAAUGAACUAGU | AAC | mRNA cleavage assay |
| GGCAAGGAGGUAAAAAUGAAUCAGA | AAU | mRNA cleavage assay |
| GGCAAGGAGGUAAAAAUGCCAGAAA | CCA | mRNA cleavage assay |
| GGCAAGGAGGUAAAAAUGCCCGAAA | CCC | mRNA cleavage assay |
| GGCAAGGAGGUAAAAAUGAUUUAGU | AUU | mRNA cleavage assay |
| GGCAAGGAGGUAAAAAUGAmCmAmUUU | AmCmAm | HigB ΔH92 precleavage state structure |
Nucleotides followed by “m” contain a 2′-OCH3 group used to prevent mRNA cleavage during crystallization.
For the 70S-HigB ΔH92 precleavage state structure containing the A-site codon AmCmAm, HigB was soaked into preformed Tth 70S empty A-site crystals. Formation of the complex was performed as previously described, except a fivefold molar excess of CC-puromycin (Dharmacon) was first incubated with ribosomes for 30 min at 55 °C. Before cryoprotecting, 100 μM HigBΔH92 was incubated with crystals for 17 h. Crystals were cryoprotected and frozen as described above.
Data were collected by either continuous vector scanning at the Northeast Collaborative Access Team (NE-CAT) ID-24C and ID-24E beamlines or through 4° wedges at the SER-CAT 22ID beamline. For all 70S data collections, at least 90° of data were collected using between 0.1° and 0.2° oscillations. Data were processed by XDS (48) and solved by molecular replacement using two copies of the 70S initiation model (40) (PDB ID code 4V6G) using 30S and 50S and their corresponding Mg2+ ions as rigid body groups in Phenix (49). Initial Fo-Fc difference electron density indicated that mRNA, tRNAfMet, and HigB were bound. tRNAfMet and mRNA 5′ of the A site were modeled in Coot, and individual site refinement was performed in Phenix using base pair restraints and translation/libration/screw parameterization. Modeling of HigB was next performed. The best HigB density was in the 70S-HigB ΔH92 precleavage state structure (AmCmAm codon; 3.1 Å), and this model was used as a starting model for all other 70S structures. Residues 1–90 of the 1.25 Å HigB(His)6 model was placed into Fo-Fc electron density, built in Coot, and refined in Phenix. In all 70S-HigB precleavage state structures, the electron density allowed for the placement of three nucleotides in the A site and one nucleotide 3′ toward the mRNA entry tunnel, whereas in the 70S postcleavage state structure, only density for two A-site nucleotides and a 3′ phosphate was visible, indicating HigB cleavage of the mRNA.
mRNA Cleavage Assays.
E. coli 70S ribosomes were purified as previously described (51). A final concentration of 1.2 μM ribosomes was programmed at 37 °C with 0.6 μM 5′-[32P]mRNA for 6 min and 3 μM E. coli tRNAfMet for 30 min (Table S3). HigB protein at a final concentration of 0.9 μM was added to programmed ribosomes, and aliquots were taken at 1, 3, 10, 30, and 60 min after HigB addition; the reaction was quenched with an equal volume of 2× formamide dye [98% formamide, 10 mM EDTA (pH 8.0) and 0.2 mg mL−1 bromophenol blue], heated at 70 °C for two mins, and stored on ice or at −20 °C. Cleaved mRNA product was separated from full length on an 8M urea, 1× Tris/Borate/EDTA buffer, 18% polyacrylamide sequencing gel for 1.5 h. Quantitation of the cleaved product was performed by exposure of the dried gel to a GE phosphor screen imaging on a Typhoon FLA 7000 gel imager (GE Healthcare), with band density counted by ImageQuant software. Plots of the amount of product versus time were fit by GraphPad Prism 5 (GraphPad) using the single exponential equation:
where Pmax is the mRNA cleavage product plateau (pmol mRNA cleaved), k is the observed rate constant (min−1), and t is the time the reaction progressed (min).
Remodeling of the 70S-YoeB mRNA A-Site mRNA.
Electron density maps for the 70S-YoeB precleavage state structure were generated in Phenix (49) from the downloaded structure factors and coordinates (PDB ID code 4V8X). A-site mRNA nucleotides +4 to +8 were removed from the model, and this new model was refined against structure factors. The mRNA path was modeled in Coot (47), and individual site refinement (mRNA only) and grouped B factor refinement were performed in Phenix using base pair restraints.
Acknowledgments
We thank F. M. Murphy IV and staff members of the Northeast Collaborative Access Team (NE-CAT) beamlines for assistance during data collection, and C.M.D. laboratory members C. Fagan, E. Hoffer, S. Subramanian, A. Ruangprasert, and D. West and Profs. G. Conn and A. Corbett for critical reading of the manuscript. This work was supported by National Science Foundation Faculty Early Career Development Program (CAREER) Award Molecular and Cellular Biosciences 0953714 (to C.M.D.), National Institutes of Health (NIH) Biochemistry, Cellular and Molecular Biology Graduate Training Grant 5T32GM8367, and NIH National Research Service Award F31 Fellowship GM108351 (to M.A.S.). C.M.D. is a Pew Scholar in the Biomedical Sciences. This work is based on research conducted at the Advanced Photon Source (APS) on the NE-CAT beamlines, which is supported by National Center for Research Resources NIH Award RR-15301, and at the Southeast Regional Collaborative Access Team beamline. Use of the APS, an Office of Science User Facility operated for the US Department of Energy Office of Science by Argonne National Laboratory, was supported under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.H.D.C. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4PX8, 4YPB, 4W4G, and 4YZV).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512959112/-/DCSupplemental.
References
- 1.Maisonneuve E, Gerdes K. Molecular mechanisms underlying bacterial persisters. Cell. 2014;157(3):539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 2.Boutte CC, Crosson S. Bacterial lifestyle shapes stringent response activation. Trends Microbiol. 2013;21(4):174–180. doi: 10.1016/j.tim.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Potrykus K, Cashel M. (p)ppGpp: Still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 4.Magnusson LU, Farewell A, Nyström T. ppGpp: A global regulator in Escherichia coli. Trends Microbiol. 2005;13(5):236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Harrison JJ, et al. The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm. Antimicrob Agents Chemother. 2009;53(6):2253–2258. doi: 10.1128/AAC.00043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim Y, Wood TK. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem Biophys Res Commun. 2010;391(1):209–213. doi: 10.1016/j.bbrc.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ren D, Bedzyk LA, Thomas SM, Ye RW, Wood TK. Gene expression in Escherichia coli biofilms. Appl Microbiol Biotechnol. 2004;64(4):515–524. doi: 10.1007/s00253-003-1517-y. [DOI] [PubMed] [Google Scholar]
- 8.Maisonneuve E, Castro-Camargo M, Gerdes K. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell. 2013;154(5):1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 9.Norton JP, Mulvey MA. Toxin-antitoxin systems are important for niche-specific colonization and stress resistance of uropathogenic Escherichia coli. PLoS Pathog. 2012;8(10):e1002954. doi: 10.1371/journal.ppat.1002954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Wood TK. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl Environ Microbiol. 2011;77(16):5577–5583. doi: 10.1128/AEM.05068-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helaine S, et al. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 2014;343(6167):204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerdes K, Christensen SK, Løbner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat Rev Microbiol. 2005;3(5):371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 13.Bernard P, Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J Mol Biol. 1992;226(3):735–745. doi: 10.1016/0022-2836(92)90629-x. [DOI] [PubMed] [Google Scholar]
- 14.Vesper O, et al. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell. 2011;147(1):147–157. doi: 10.1016/j.cell.2011.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winther KS, Gerdes K. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc Natl Acad Sci USA. 2011;108(18):7403–7407. doi: 10.1073/pnas.1019587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedersen K, et al. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell. 2003;112(1):131–140. doi: 10.1016/s0092-8674(02)01248-5. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, et al. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol Cell. 2003;12(4):913–923. doi: 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 18.Castro-Roa D, et al. The Fic protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat Chem Biol. 2013;9(12):811–817. doi: 10.1038/nchembio.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaspy I, et al. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat Commun. 2013;4:3001. doi: 10.1038/ncomms4001. [DOI] [PubMed] [Google Scholar]
- 20.Schifano JM, et al. Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal A site. Proc Natl Acad Sci USA. 2013;110(21):8501–8506. doi: 10.1073/pnas.1222031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neubauer C, et al. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell. 2009;139(6):1084–1095. doi: 10.1016/j.cell.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng S, et al. YoeB-ribosome structure: A canonical RNase that requires the ribosome for its specific activity. Nucleic Acids Res. 2013;41(20):9549–9556. doi: 10.1093/nar/gkt742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maehigashi T, Ruangprasert A, Miles SJ, Dunham CM. Molecular basis of ribosome recognition and mRNA hydrolysis by the E. coli YafQ toxin. Nucleic Acids Res. 2015;43(16):8002–8012. doi: 10.1093/nar/gkv791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Inouye M. The inhibitory mechanism of protein synthesis by YoeB, an Escherichia coli toxin. J Biol Chem. 2009;284(11):6627–6638. doi: 10.1074/jbc.M808779200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurley JM, Cruz JW, Ouyang M, Woychik NA. Bacterial toxin RelE mediates frequent codon-independent mRNA cleavage from the 5′ end of coding regions in vivo. J Biol Chem. 2011;286(17):14770–14778. doi: 10.1074/jbc.M110.108969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshizumi S, et al. Staphylococcus aureus YoeB homologues inhibit translation initiation. J Bacteriol. 2009;191(18):5868–5872. doi: 10.1128/JB.00623-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christensen SK, et al. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: Involvement of the yefM-yoeB toxin-antitoxin system. Mol Microbiol. 2004;51(6):1705–1717. doi: 10.1046/j.1365-2958.2003.03941.x. [DOI] [PubMed] [Google Scholar]
- 28.Christensen-Dalsgaard M, Gerdes K. Translation affects YoeB and MazF messenger RNA interferase activities by different mechanisms. Nucleic Acids Res. 2008;36(20):6472–6481. doi: 10.1093/nar/gkn667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prysak MH, et al. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol Microbiol. 2009;71(5):1071–1087. doi: 10.1111/j.1365-2958.2008.06572.x. [DOI] [PubMed] [Google Scholar]
- 30.Hurley JM, Woychik NA. Bacterial toxin HigB associates with ribosomes and mediates translation-dependent mRNA cleavage at A-rich sites. J Biol Chem. 2009;284(28):18605–18613. doi: 10.1074/jbc.M109.008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takagi H, et al. Crystal structure of archaeal toxin-antitoxin RelE-RelB complex with implications for toxin activity and antitoxin effects. Nat Struct Mol Biol. 2005;12(4):327–331. doi: 10.1038/nsmb911. [DOI] [PubMed] [Google Scholar]
- 32.Kamada K, Hanaoka F. Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol Cell. 2005;19(4):497–509. doi: 10.1016/j.molcel.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 33.Schureck MA, et al. Structure of the Proteus vulgaris HigB-(HigA)2-HigB toxin-antitoxin complex. J Biol Chem. 2014;289(2):1060–1070. doi: 10.1074/jbc.M113.512095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruangprasert A, et al. Mechanisms of toxin inhibition and transcriptional repression by Escherichia coli DinJ-YafQ. J Biol Chem. 2014;289(30):20559–20569. doi: 10.1074/jbc.M114.573006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams JJ, Hergenrother PJ. Exposing plasmids as the Achilles’ heel of drug-resistant bacteria. Curr Opin Chem Biol. 2008;12(4):389–399. doi: 10.1016/j.cbpa.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maisonneuve E, Shakespeare LJ, Jørgensen MG, Gerdes K. Bacterial persistence by RNA endonucleases. Proc Natl Acad Sci USA. 2011;108(32):13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Li GY, Zhang Y, Inouye M, Ikura M. Inhibitory mechanism of Escherichia coli RelE-RelB toxin-antitoxin module involves a helix displacement near an mRNA interferase active site. J Biol Chem. 2009;284(21):14628–14636. doi: 10.1074/jbc.M809656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bøggild A, et al. The crystal structure of the intact E. coli RelBE toxin-antitoxin complex provides the structural basis for conditional cooperativity. Structure. 2012;20(10):1641–1648. doi: 10.1016/j.str.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogle JM, et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292(5518):897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 40.Jenner L, Demeshkina N, Yusupova G, Yusupov M. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat Struct Mol Biol. 2010;17(9):1072–1078. doi: 10.1038/nsmb.1880. [DOI] [PubMed] [Google Scholar]
- 41.Weixlbaumer A, et al. Insights into translational termination from the structure of RF2 bound to the ribosome. Science. 2008;322(5903):953–956. doi: 10.1126/science.1164840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laurberg M, et al. Structural basis for translation termination on the 70S ribosome. Nature. 2008;454(7206):852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 43.Zegers I, Haikal AF, Palmer R, Wyns L. Crystal structure of RNase T1 with 3′-guanylic acid and guanosine. J Biol Chem. 1994;269(1):127–133. [PubMed] [Google Scholar]
- 44.Noguchi S. Isomerization mechanism of aspartate to isoaspartate implied by structures of Ustilago sphaerogena ribonuclease U2 complexed with adenosine 3′-monophosphate. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 7):843–849. doi: 10.1107/S0907444910019621. [DOI] [PubMed] [Google Scholar]
- 45.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313(5795):1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 51.Powers T, Noller HF. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991;10(8):2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]