Significance
Systemic lupus erythematosus is a complex autoimmune disease where genetic and environmental factors play equally important roles. Recent research has identified that upregulation of the innate immune Toll-like receptor 7 (TLR7) is fundamental for the development of severe disease. In our Sle1Tg7 model system, we show that increased expression of TLR7 in dendritic cells (DCs) is important for the development of kidney disease. Furthermore, we identify conventional CD11b+ DCs infiltrating the glomeruli of diseased mice. In a resting state, human myeloid DCs express negligible levels of TLR7. However, exposure to multiple stimuli results in a dramatic increase in expression. Together, these findings suggest a role for myeloid DC-TLR7 in clinical disease and that TLR7 expression in this cellular compartment may represent a novel target for therapeutic investigation.
Keywords: TLR7, dendritic cells, SLE, nephritis, autoimmunity
Abstract
Glomerulonephritis is a common and debilitating feature of systemic lupus erythematosus (SLE). The precise immune mechanisms that drive the progression from benign autoimmunity to glomerulonephritis are largely unknown. Previous investigations have shown that a moderate increase of the innate Toll-like receptor 7 (TLR7) is sufficient for the development of nephritis. In these systems normalization of B-cell TLR7 expression or temporal depletion of plasmacytoid dendritic cells (pDCs) slow progression; however, the critical cell that is responsible for driving full immunopathology remains unidentified. In this investigation we have shown that conventional DC expression of TLR7 is essential for severe autoimmunity in the Sle1Tg7 model of SLE. We show that a novel expanding CD11b+ conventional DC subpopulation dominates the infiltrating renal inflammatory milieu, localizing to the glomeruli. Moreover, exposure of human myeloid DCs to IFN-α or Flu increases TLR7 expression, suggesting they may have a role in self-RNA recognition pathways in clinical disease. To our knowledge, this study is the first to highlight the importance of conventional DC-TLR7 expression for kidney pathogenesis in a murine model of SLE.
The innate pathogen recognition receptor Toll-like receptor 7 (TLR7) has recently been implicated in the development of autoimmunity. This receptor, along with TLR8 and TLR9, was originally described by Beutler and colleagues (1), with ligand identification and functional characterization by Akira and colleagues just over a year later (2). TLR7 recognizes single-stranded RNA (ssRNA), inducing downstream activation of signaling molecules, including Jnk and NF-κB through a myeloid differentiation primary response gene 88 (MyD88)-dependent cascade (3). This process is central to host defense against invading viruses; however, TLR7 hyperactivity can also drive the initiation and progression of autoimmunity.
Systemic lupus erythematosus (SLE) is a complex autoimmune disease characterized by the presence of antinuclear autoantibodies (ANA) (4). The ANAs form immune complexes with host-derived nuclear material, which accumulate as deposits within the tissues and organs. The ensuing inflammation can lead to organ failure, particularly glomerulonephritis (GN) (5). Prospective studies by Harley and colleagues (6) demonstrated that ANAs are detectable ∼6 years before clinical presentation of SLE, with only a small percentage of these individuals who have ANAs progressing to pathogenic autoimmunity. These data, together with murine studies, suggest that defects in multiple pathways contribute to the initiation and progression of systemic autoimmunity (7).
Multiple investigations have shown that TLR7 and the MyD88 signaling pathway are critical for the initiation of autoimmunity and development of self-reactivity, because genetic ablation of either TLR7 or MyD88 prevents the development of both ANAs and subsequent immune-pathology (8–10). Furthermore, this signaling pathway is specifically required within B cells (11, 12). In TLR7-sufficient autoimmune prone systems, additional immune alterations, such as lpr (lymphoproliferation), sle3, or yaa (Y-linked autoimmune-accelerating locus) can lead to severe autoimmunity. The yaa murine susceptibility locus is a region comprising 16 genes, including TLR7, which translocated from the X chromosome to the Y (13, 14). The yaa-associated increase in TLR7 expression and function was determined to be critical for the development of severe disease, independent of the SLE-model (15–17). More recently, we demonstrated that a twofold increase in TLR7 alone on the Sle1 background is sufficient to drive disease in an almost identical manner to the addition of the yaa susceptibility locus (18). Normalization of B-cell TLR7 did not affect GN, suggesting that the increase in other cells drives severe pathology. Additional studies have suggested that although plasmacytoid dendritic cells (pDCs) contribute to pathology of a yaa-associated model, other cell types are critical for full disease pathogenesis (19). Determining these critical cell types will clarify the mechanisms of progression for therapeutic targeting.
In this study we investigated the contribution of DCs to TLR7-mediated SLE-associated GN. Previous data has presented conflicting evidence on their role in lupus models. Although B-cell proliferation and antibody production can be enhanced by DC activation and genetic ablation diminishes disease progression (20, 21), DCs also play a protective role in regulatory T-cell development and their elimination can result in autoimmunity (22, 23). The overall response in each system may depend on the genetic background or incomplete ablation of DC subsets [discussed by Platt and Randolph (24)].
We bred conditional SLE mice overexpressing TLR7 (Sle1Tg7) with a CD11cCre reporter strain (25) and demonstrated that the increase in TLR7 within CD11c+ cells was essential for all severe autoimmune traits, including splenomegaly, T- and B-cell activation, and GN. Purification of DCs and other leukocyte subsets confirmed normalization of TLR7 mRNA in CD11b+ conventional (c)DCs and pDCs. We identified and characterized a novel CD11b+ cDC infiltrating diseased kidneys, which was absent, together with all other pathological traits, upon restoration of TLR7. Taken together, our data suggest that the CD11b+ cDC, rather than the pDC, is playing the principle role in driving end organ disease. Finally, we determined that primary human BDCA1+ DCs or monocyte-derived DCs express extremely low levels of TLR7 mRNA, but this can be rapidly up-regulated on exposure to virus or IFN-α. This work has uncovered a key role for DCs in one of the most clinically important phases of SLE, the progression to lupus nephritis.
Results
Overexpression of TLR7 in DCs Is Required for the Development of Severe Autoimmunity in SLE Mice.
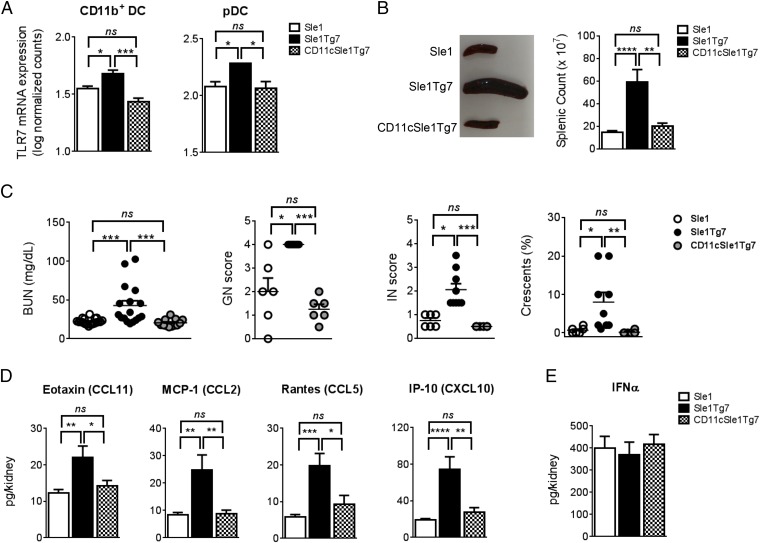
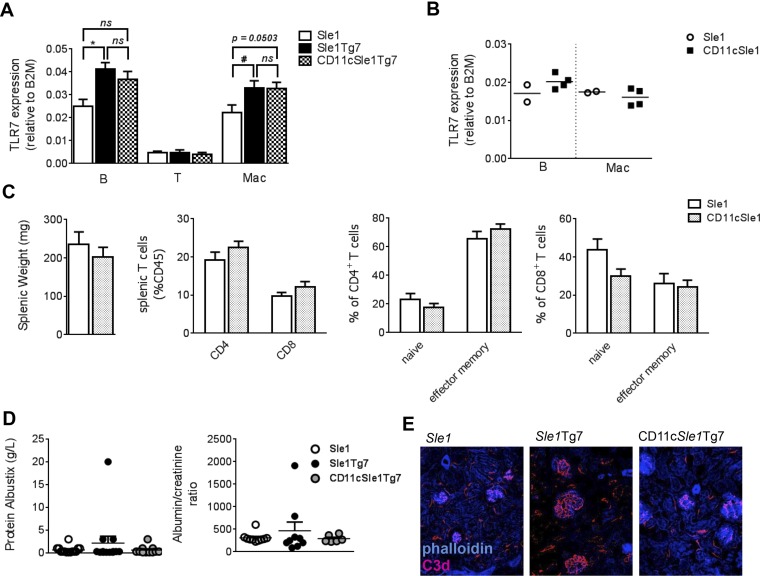
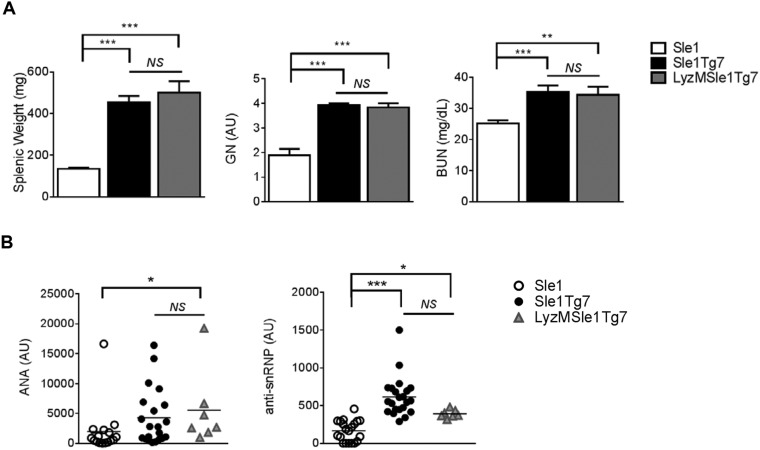
Previous studies have demonstrated that B cells are not the critical cell type driving TLR7-dependent kidney disease in the Sle1Tg7 murine model (18). These findings, together with the conflicting data on the role of DCs in autoimmunity, led us to examine the contribution of TLR7 in this compartment. CD11cCre-mediated normalization of TLR7 mRNA expression in CD11b+ cDCs and pDCs was confirmed using Nanostring technology (Fig. 1A). No significant differences in TLR7 expression were observed in splenic B or peritoneal macrophages, whereas TLR7 expression in T cells was low or undetectable (Fig. S1A). Additionally, the presence of the CD11cCre did not affect either TLR7 mRNA expression levels (Fig. S1B), splenomegaly, T-cell ratios, or T-cell activation in Sle1 mice (Fig. S1C). Five- to 7.5-mo-old female CD11cCreSle1Tg7 (CD11cSle1Tg7) mice, together with Sle1 and Sle1Tg7 controls, were examined for the development of autoimmune disease. Splenomegaly is a characteristic feature of lupus-prone strains and this was prevented in CD11cSle1Tg7 mice (Fig. 1B). Functional assessment of kidney disease showed normal levels of blood urea nitrogen (BUN) in CD11cSle1Tg7 mice, similar to Sle1 controls (Fig. 1C). Proteinuria did not differ between strains (Fig. S1D). Furthermore, there was no evidence of kidney disease by microscopy in either Sle1 or CD11cSle1Tg7 mice, in contrast to the expected nephritis exhibited by the Sle1Tg7 strain (Fig. 1C). Immunofluorescent staining of kidneys from Sle1Tg7 mice revealed bright glomerular complement (C3d) staining, whereas lower levels were observed in CD11cSle1Tg7 and Sle1 mice (Fig. S1E). Analysis of renal supernatants from Sle1Tg7 mice revealed significantly higher levels of chemokines associated with renal infiltration and GN, including Rantes, MCP-1, and IP-10 (Fig. 1D and Table S1) (18, 26, 27). These were normalized in the CD11cSle1Tg7 strain. IFN-α, which is often associated with pDC activation, was similar in all three strains (Fig. 1E). In contrast to the CD11cCre effects, there was no reduction in disease pathogenesis upon normalization of TLR7 using the M-lysozymeCre (LyzMCre) reporter strain, showing that higher levels of TLR7 in macrophages and neutrophils were not central in driving severe autoimmune traits (28) (Fig. S2).
Fig. 1.
Prevention of severe autoimmune pathology with CD11c-Cre-normalization of TLR7. (A) TLR7 mRNA expression in pDCs and CD11b+ cDCs assessed by Nanostring in 6- to 8-wk-old mice (n = 4–5 or n = 3 per strain, respectively, for cDC and pDC). Five- to 7.5-mo-old Sle1, Sle1Tg7, and CD11cSle1Tg7 mice were analyzed for: (B) splenic size (n = 12–13 per strain); (C) concentrations of BUN (n = 13–19 per strain), GN, and interstitial renal disease (IN) scored from 0 to 4 and quantification of glomerular crescent formation (n = 6–9 mice per strain). (D) Analysis of chemokine levels in renal supernatants (n = 12–13 per strain) and (E) IFN-α levels in kidney supernatants by ELISA (n = 11–13 per strain). Bars represent mean + SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, and ns, not significant (one-way ANOVA or Kruskal–Wallis test).
Fig. S1.
TLR7 mRNA expression levels in lymphoid and non-DC myeloid lineages and autoimmune pathology. (A) TLR7 mRNA levels were assessed by quantitative RT-PCR in B- and T-lymphoid cells and macrophages from CD11cSle1Tg7 and control mouse spleens at 1–4 mo of age (n = 4–6 per strain for B cells and n = 4–5 mice per strain for T cells and macrophages). (B) B-cell and macrophage TLR7 mRNA levels were assessed by quantitative RT-PCR at 11 mo of age in Sle1 (n =2) and CD11cSle1 mice (n = 4). (C) Splenic weight and analysis of T-cell subsets by flow cytometry in Sle1 (n =12) and CD11cSle1 mice (n = 8) at a median of 11 mo of age (all mice aged between 8 and 13 mo). (D) Proteinuria in Sle1, Sle1Tg7 and CD11cSle1Tg7 mice (Albustix, n = 11–14 mice per strain, albumin/creatinine ratio n = 6–10 per strain). (E) Frozen kidney sections were assessed for complement activation by staining with anti-C3d antibody. C3d deposition is evident in the enlarged glomeruli of Sle1Tg7 mice and to a lesser extent in the glomeruli of Sle1 and CD11cSle1Tg7 mice. Microscopy magnification: 200×, images are representative of two to three mice per strain. Bars represent mean + SEM. *P < 0.05 (Kruskal–Wallis test with post-hoc Dunn’s multiple correction test); #P < 0.05 (multiple t test corrected for multiple comparisons using the Holm–Sidak method); ns, not significant.
Table S1.
Kidney supernatant analysis by Luminex assay
| Chemokines and cytokines | Sle1 (n = 13) | Sle1Tg7 (n = 12) | CD11cSle1Tg7 (n = 11) |
| Mean ± SEM (pg/kidney) | Mean ± SEM (pg/kidney) | Mean ± SEM (pg/kidney) | |
| Eotaxin | 12.27 ± 0.9962 | 22.03 ± 3.154 | 13.39 ± 1.216 |
| G-CSF | 10.23 ± 2.221 | 24.32 ± 7.513 | 9.7 ± 1.264 |
| GM-CSF | 4.446 ± 2.083 | 4.358 ± 1.735 | 6.582 ± 2.101 |
| IFNg | 5.662 ± 1.073 | 4.65 ± 1.565 | 3.891 ± 1.08 |
| IL-10 | 8.258 ± 1.186 | 5.882 ± 1.361 | 8.145 ± 1.775 |
| IL-12p40 | 26.99 ± 4.156 | 21.65 ± 5.169 | 22.35 ± 3.257 |
| IL-12p70 | 25.88 ± 3.014 | 19.17 ± 2.546 | 18.44 ± 3.634 |
| MCP-1 | 8.354 ± 0.8829 | 24.78 ± 5.536 | 8.873 ± 1.247 |
| IP-10 | 19.15 ± 1.507 | 74.44 ± 13.59 | 27.59 ± 5.012 |
| LIF | 2.015 ± 0.281 | 63.18 ± 17.94 | 4.991 ± 0.9669 |
| MIG | 264.6 ± 22.76 | 784.4 ± 158.5 | 370.3 ± 61.87 |
| Rantes | 5.792 ± 0.7022 | 19.77 ± 3.366 | 9.3 ± 2.416 |
| IL-1a | 40.32 ± 6.746 | 33.07 ± 7.838 | 41.7 ± 8.666 |
| IL-1b | 19.9 ± 2.407 | 26.59 ± 6.167 | 28.89 ± 4.421 |
| MIP1a | 3.346 ± 0.7272 | 9.317 ± 2.376 | 3.6 ± 0.897 |
| IL-15 | 20.8 ± 1.6 | 17.6 ± 2.4 | 20.6 ± 2.4 |
| IL-2 | 9.6 ± 1.2 | 7.6 ± 1.6 | 9.8 ± 1.5 |
| IL-7 | 4.2 ± 0.4 | 4.0 ± 0.5 | 3.7 ± 0.5 |
| IL-9 | 272.6 ± 19.8 | 264.8 ± 23.5 | 257.2 ± 17.0 |
| KC | 28.2 ± 1.9 | 30.7 ± 4.2 | 26.6 ± 2.3 |
| M-CSF | 3.0 ± 0.4 | 6.0 ± 1.0 | 3.6 ± 0.3 |
Kidney supernatants from 5- to 7.5-mo-old mice were assayed for the presence of chemokines and cytokines. IL-3, IL-4, IL-5, IL-6, MIP-1b, MIP-2, TNFa, VEGF, and IL-13 were all undetectable.
Fig. S2.
M-lysozyme-Cre-normalization of TLR7 does not ameliorate severe autoimmune disease pathogenesis. (A) Splenic weight, GN score, and BUN in Sle1, Sle1Tg7, and LyzMSle1Tg7 mice (6-mo-old mice, n ≥ 12 per strain). (B) LyzM-Cre-normalization does not lead to significant changes in ANAs and anti-snRNP levels as determined by ELISA (n ≥ 7 mice per strain). Bars represent mean + SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 (one-way ANOVA); ns, not significant.
Overexpression of TLR7 in DCs Is Required for the Production of Specific Subtypes of ANA in SLE Mice.
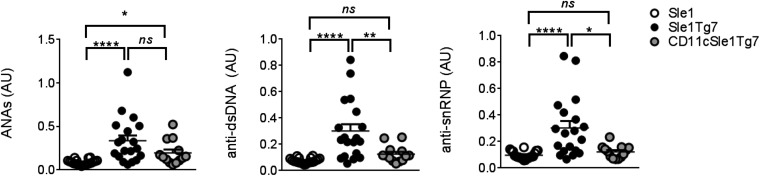
We have previously described an increase in a variety of autoreactive antibodies in the Sle1 strain, which are augmented by the introduction of either yaa or the Tg7-BAC (18). The predominant IgG autoantibodies are reactive to histone, chromatin, and double-stranded (ds)DNA. Despite the reduction in kidney pathology, there was no change detected in the total ANA titer of CD11cSle1Tg7 compared with Sle1Tg7 mice (Fig. 2, Left). However, both anti-dsDNA anti-snRNP levels were normalized to Sle1 levels in CD11cSle1Tg7 mice (Fig. 2, Center and Right). These data show that although DC-TLR7 overexpression is not responsible for the amount of autoantibodies produced in this SLE model, it does affect the extent of recognition of some self-antigens.
Fig. 2.
DC-normalization of TLR7 changes ANA specificity. Sera from 5- to 7.5-mo-old female mice were analyzed for ANA levels by ELISA: IgG dsDNA/histone/chromatin autoantibodies (ANAs), IgG anti-dsDNA Abs (anti-dsDNA), or IgG anti-snRNP Abs (anti-snRNP) (n = 13–20 mice per strain) (AU, absorbance units). Bars represent mean + SEM. *P < 0.05, **P < 0.01, ****P < 0.0001; and ns, not significant (one-way ANOVA or Kruskal–Wallis test).
Overexpression of TLR7 in DCs Is Required for Splenic Autoimmune Traits in SLE Mice.
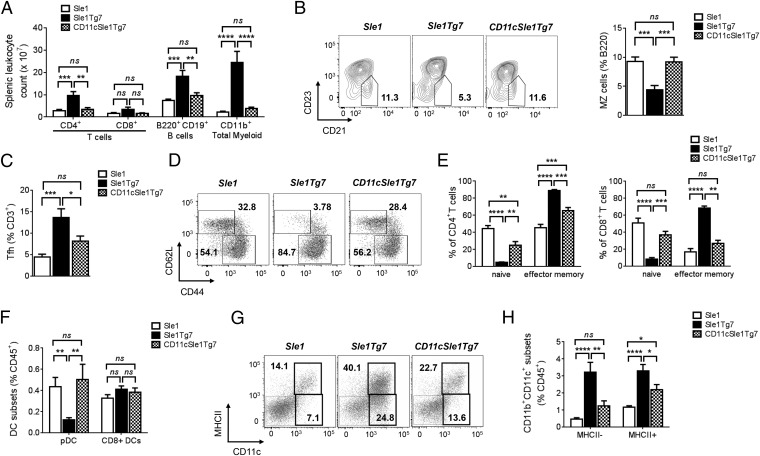
Alongside kidney disease and the production of ANAs, both yaa and Tg7-driven SLE models develop splenomegaly characterized by leukocyte expansion, including myelopoiesis. Aged Sle1Tg7 mice demonstrated significant increases in absolute numbers of CD4+ T cells, B220+ B cells, and CD11b+ myeloid cells within the spleen, compared with Sle1 mice (Fig. 3A). Normalization of DC-TLR7 expression prevented this expansion, with leukocyte numbers comparable to those of Sle1 mice. Furthermore, CD4+ and CD8+ T cells from Sle1Tg7 mouse spleens expressed high levels of the activation markers PD-1, ICOS, and CD69, and the T-cell population was characterized by an increase of effector memory cells (CD62LloCD44hi) (29, 30) concomitant with a reduction in the naïve population (Fig. 3 D and E and Table S2). These phenomena were either reduced in CD11cSle1Tg7 mice or comparable to Sle1 mice (Fig. 3E and Table S2). Additionally, yaa and TLR7-driven murine lupus models are associated with an expansion of the T-follicular helper (Tfh) cell population and a marginal zone B-cell defect: both were detected in the Sle1Tg7 strain and not in CD11cSle1Tg7 mice (Fig. 3 B and C) (13, 14, 18). Similar trends were observed in expanded populations of plasmablasts, plasma cells and germinal-center B cells (Table S2).
Fig. 3.
DC-normalization of TLR7 prevents yaa-associated splenic autoimmune traits. Sle1, Sle1Tg7, and CD11cSle1Tg7 mice were analyzed for: (A) Splenic leukocyte counts; major splenic subsets using CD4 and CD8 for T cells, B220, and CD19 for B cells and CD11b for the myeloid lineage. (B) Marginal zone (MZ) B-cell subsets were identified through CD21 and CD23 expression on B220+CD138− cells (18). (C) Tfh were identified as CD4+, CXCR5+, and ICOS+. (D) Naïve and effector memory CD4+ T cells were gated using CD62L and CD44 expression with (E) the frequencies of CD4+ (Left) and CD8+ T cells (Right). (F) Flow cytometric analysis of splenic pDC and CD8+ DC subsets. (G) Myeloid CD11c+ populations (CD19/CD3/Gr-1−CD11b+), were subgated through MHCII and CD11c into CD11c+MHCII+ (CD11b+ cDCs) and CD11c+MHCII− cells. (H) Quantification of data from G. Data shown is cumulative from three independent aging cohorts, total n = 9–10 mice per strain (A–H). Plots are representative of one mouse per strain (B, D, and G). Bars represent mean + SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; and ns, not significant (one-way ANOVA or Kruskal–Wallis test).
Table S2.
Splenic cell counts and expression of activation markers in 5- to 7-mo-old female mice assessed by flow cytometry
| Cells | Sle1 (n = 10) | Sle1Tg7 (n = 9) | CD11cSle1Tg7 (n = 9) |
| Mean ± SEM | Mean ± SEM | Mean ± SEM | |
| Plasma (B220−CD138+) (106) | 1.8 ± 0.1 | 15.7 ± 4.6** | 3.9 ± 1.0ns |
| Plasmablast (B220+CD138+) (106) | 2.3 ± 0.5 | 18.2 ± 11.2* | 4.4 ± 1.1ns |
| B220+CD19+ B cells (107) | 7.5 ± 0.6 | 18.1 ± 2.8*** | 9.7 ± 1.2ns |
| GC (Fas+GL7+) (106) | 3.0 ± 0.3 | 11.6 ± 2.7** | 3.4 ± 0.6ns |
| MZ (106) | 6.8 ± 0.7 | 7.5 ± 2.0ns | 7.8 ± 1.1ns |
| CD4+ T cells (107) | 2.8 ± 0.5 | 8.8 ± 1.8*** | 3.2 ± 0.8ns |
| PD-1 (MFI) | 88.1 ± 9.8 | 276 ± 34.4**** | 142.4 ± 24.6* |
| ICOS1 (MFI) | 891 ± 85.9 | 2771.1 ± 380.1**** | 1513.2 ± 217.5* |
| CD69+ (% CD4+) | 3.1 ± 0.3 | 8.3 ± 2.0** | 4.7 ± 0.9ns |
| Effector memory CD4+ T (CD62LloCD44hi) (106) | 12.3 ± 2.1 | 77.2 ± 15.7**** | 22.2 ± 5.6ns |
| Naïve CD4+ T (CD62LhiCD44lo) (106) | 12.6 ± 2.7 | 4.0 ± 0.7** | 6.3 ± 1.3ns |
| Tfh (106) | 1.1 ± 0.2 | 11.7 ± 3.2*** | 2.5 ± 0.5ns |
| CD8+ T cells (107) | 1.7 ± 0.3 | 3.2 ± 0.9ns | 1.5 ± 0.3ns |
| PD-1 (MFI) | 66.6 ± 9.4 | 135.8 ± 16.3**** | 71.0 ± 14.8ns |
| ICOS1 (MFI) | 578.1 ± 35.6 | 644.4 ± 131.4ns | 593.0 ± 67.5ns |
| CD69+ (% CD8+) | 2.1 ± 0.2 | 4.0 ± 0.7** | 2.9 ± 0.4ns |
| Effector memory CD8+ T (CD62LloCD44hi) (106) | 2.2 ± 0.3 | 22.3 ± 7.2**** | 4.3 ± 0.8ns |
| Naïve CD8+ T (CD62LhiCD44lo) (106) | 9.7 ± 2.5 | 2.4 ± 0.5* | 5.5 ± 1.3ns |
| CD11b+ total myeloid (107) | 2.3 ± 0.4 | 24.5 ± 4.9**** | 3.7 ± 0.7ns |
| Eos (106) | 1.7 ± 0.4 | 22.3 ± 7.2**** | 3.2 ± 0.5ns |
| PMN (106) | 5.8 ± 1.3 | 36.8 ± 8.9*** | 9.1 ± 2.4ns |
| Gr1+ monocytes (106) | 2.4 ± 0.3 | 39.6 ± 7.9**** | 4.7 ± 0.7ns |
| CD11b+CD11c+MHCII+ DC (106) | 2.2 ± 0.31 | 29.3 ± 6.9*** | 4.8 ± 1.1ns |
| CD8+ DC (106) | 0.7 ± 0.16 | 3.4 ± 0.7*** | 0.8 ± 0.2ns |
| pDC (106) | 0.7 ± 1.5 | 0.7 ± 1.1ns | 0.8 ± 1.5ns |
One-way ANOVA was used to analyze data as described in Materials and Methods. Significant differences between Sle1 and Sle1Tg7 are shown in the “Sle1Tg7” column and significant differences between Sle1 and CD11cSle1Tg7 are shown in the “CD11cSle1Tg7” column: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant. Eos, eosinophils; GC, germinal center B cells; MFI, mean fluorescence intensity; MZ, marginal zone B cells; Tfh, T follicular helper cells; PMN, polymorphonuclear leukocytes. Indented entries are gated on the populations highlighted in bold immediately above.
Characterization of CD11b+ populations in the spleen using flow cytometry indicated that the cell numbers of CD11cSle1Tg7 polymorphonuclear leukocytes (PMNs), Gr1+ monocytes, and eosinophils were normalized to Sle1 levels (Table S2).
The remaining Gr1−CD11b+ cells were subdivided according to CD11c and MHCII expression (Fig. 3G). Higher frequencies of CD11c+MHCII+ DCs and CD11c+MHCII− cells were present in Sle1Tg7 mice, in contrast to CD8+ DCs, which were unchanged (Fig. 3 F and H). This expansion of CD11b+c DCs was prevented with DC-normalization of TLR7 expression (Fig. 3H and Table S2). The frequency of pDCs was found to be lower in Sle1Tg7 mice compared with either Sle1 or Sle1Tg7CD11c (Fig. 3F); however, cell counts reveal that there are no differences in the actual cell numbers across the different strains, indicating that the pDC are not an expanding population in these mice (Table S2).
Overexpression of TLR7 in DCs Is Required for the Infiltration of a Novel CD11c+ Subset into Diseased Kidneys of SLE Mice.
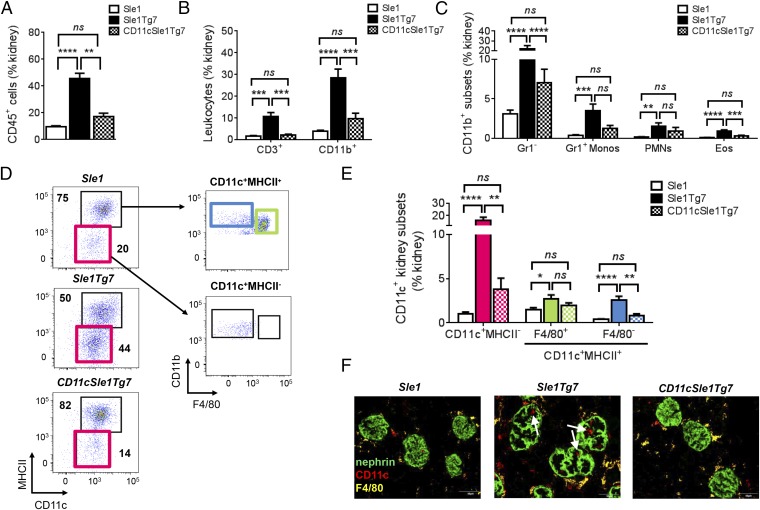
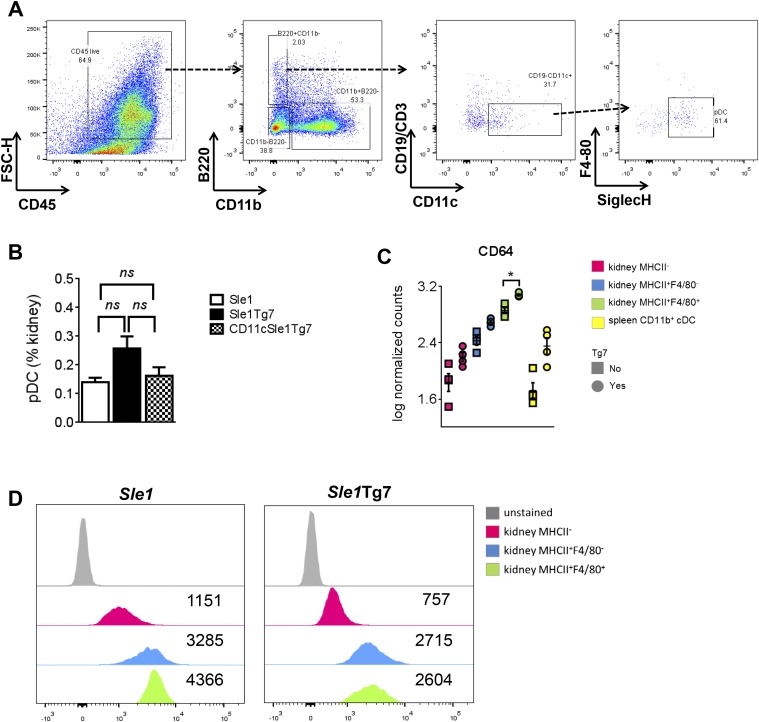
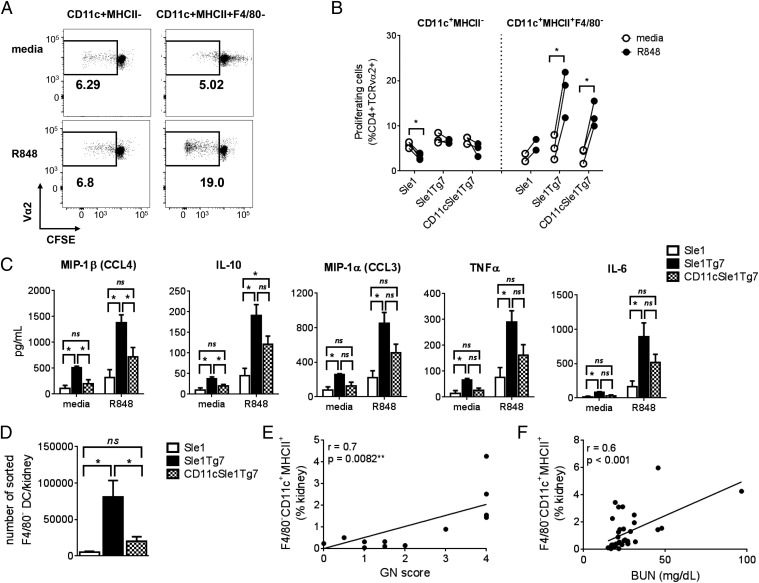
Renal pathology in SLE mice is associated with an infiltration of CD45+ leukocytes, comprising primarily CD3+ T cells, as well as several myeloid populations (18). Normalization of TLR7 expression levels in CD11c+ DCs prevented this infiltration (Fig. 4 A and B). The frequency of pDCs, which were identified using the gating strategy shown, was not found to be higher in diseased mice (Fig. S3 A and B). Analysis of myeloid cell frequencies in the kidney revealed that CD11cSle1Tg7 mice possessed a significantly lower proportion of eosinophils and Gr1− myeloid cells within the leukocyte population, compared with their Sle1Tg7 counterparts (Fig. 4C). To identify the CD11c+ subpopulations present, we characterized the CD45+CD19/3−CD11b+Gr1− subsets in terms of CD11c and MHCII expression (Fig. 4D). We identified two CD11b+CD11c+ populations in Sle1Tg7 mice that were significantly more abundant than in Sle1 and DC-TLR7 normalized mice (Fig. 4E). These populations were F4/80− and either MHCII− or MHCII+. Immunofluorescent imaging of the glomerular leukocyte infiltrate confirmed CD11c+F4/80− cells were markedly more abundant in Sle1Tg7 mice (Fig. 4F). In contrast, the abundance of the F4/80+ myeloid subset was not affected by DC-TLR7 normalization (Fig. 4E). Taken together, these data indicate that overexpression of TLR7 in CD11c-expressing DC drives accumulation of two novel myeloid CD11c+F4/80− subsets in the diseased murine kidney.
Fig. 4.
CD11cCre-normalization of TLR7 prevents renal leukocyte infiltration. (A) Five- to 7.5-mo-old female Sle1, Sle1Tg7, and CD11cSle1Tg7 mice were analyzed for (A) CD45+ renal leukocytes (n = 12–13 mice per strain, four independent aging cohorts). (B) Major leukocyte subtypes infiltrating the kidneys using CD3 for identification of T cells (n = 6 mice per strain) and CD11b for the total myeloid population (n = 12–13 mice per strain). (C) Analysis of CD11b+ PMNs, eosinophils, inflammatory Gr1+ monocytes, and remaining Gr1− myeloid cells (n = 12–13 mice per strain). (D) Gating strategy identifying CD11b+CD11c+ populations in the kidney based on CD11c, MHCII and F4/80 expression. Pink gate represents CD11c+MHCII− cells, blue gate represents CD11c+MHCII+F4/80− cells and the green gate identifies MHCII+F4/80+ cells. (E) Frequencies of CD11b+CD11c+ kidney subsets (n = 12–13 mice per strain). (F) Confocal microscopy showing CD11c+F4/80− cells infiltrating the inflamed, enlarged glomeruli. Microscopy magnification: 400×. (Scale bars, 50 µm.) Micrographs are representative of three mice per strain. Bars represent mean + SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; and ns, not significant (one-way ANOVA or Kruskal–Wallis test).
Fig. S3.
Quantification of kidney pDCs and CD64 expression in sorted renal cell subsets. (A) To identify pDCs, CD45+ singlets which did not express CD11b were gated for B220 expression. These cells were subsequently gated as CD19−CD11c+ and finally pDCs identified as the population of F480−SiglecH+ cells. (B) Flow cytometry was used to assess the frequency of pDCs in the kidneys of Sle1, Sle1Tg7, and CD11cSle1Tg7 mice (n = 6 mice per strain). (C) mRNA expression levels assessed by Nanostring as described. (D) Surface CD64 expression levels assessed by flow cytometry. Representative histograms with median fluorescence intensities are shown. *P < 0.05 (computed as described under Nanostring Gene-Expression Analysis); ns, not significant (one-way ANOVA).
Gene Expression Profiles Reveal Distinct Properties of Disease-Associated DC Subsets in SLE Mice.
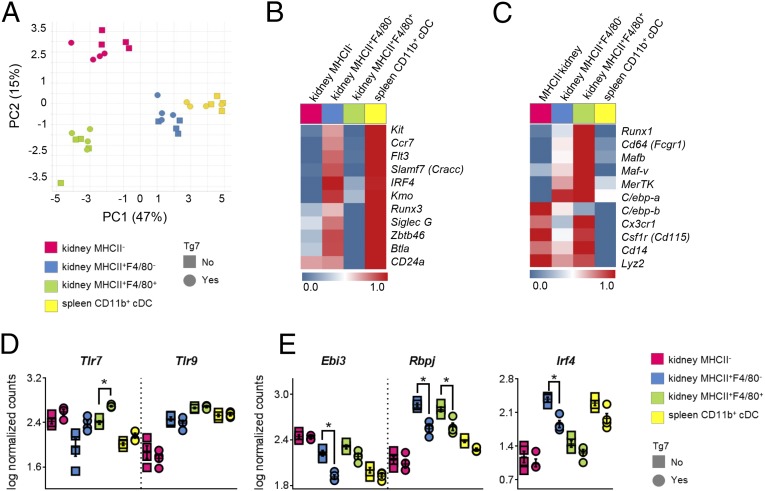
It is unclear whether the CD11c+F4/80+ cells present in all strains are either macrophages or DCs. They were originally described as a kidney resident sentinel DC (31), but more recently they were reclassified as macrophages based on their morphological and functional features (32, 33). Therefore, to confirm the ontogeny of these and our novel infiltrating CD11c+F4/80− populations, we purified the three populations identified in Fig. 4 and analyzed their transcriptional profiles in diseased Sle1Tg7 mice and Sle1 controls. We sorted splenic CD11b+ cDC from the same mice for validation purposes. Nanostring technology was used to accurately measure the abundance of mRNAs of selected genes associated with DC or macrophage ontogeny, as identified previously (34–37). Additionally, we quantified mRNAs of selected inflammatory genes and several that have been associated with the development of murine lupus (Table S3). A two-way ANOVA of normalized data showed that the expressed genes were differentially expressed among the four populations, and we therefore subjected them to principal component analysis (PCA) (Fig. 5A and Table S3). PCA showed that infiltrating kidney F4/80−MHCII+ cells clustered together with splenic CD11b+ cDCs, whereas kidney-resident F4/80+MHCII+ myeloid cells and infiltrating MHCII− cells formed two separate clusters (Fig. 5A). Devolution of these clusters demonstrated that renal F4/80−MHCII+ cells expressed core DC-ontogeny genes (including Slamf7, Zbtb46, Kmo, and Btla) (34) and Irf4, an important transcription factor in nonlymphoid tissue CD11b+ DCs (35) (Fig. 5B). Nanostring and flow cytometric analyses of the renal cDCs demonstrated that they expressed CD64 (Fig. S3 C and D), suggesting that they are monocyte-derived (38, 39). This cDC gene signature was absent from the F4/80+MHCII+ cells, which expressed a typical array of tissue macrophage-related genes such as Cd64, Mafb, MerTK, C/ebp-a, and Cd14 (36) (Fig. 5C). The infiltrating CD11c+MHCII− population, which was particularly abundant in the nephritic kidneys, clustered as a separate population and did not express typical cDC or macrophage signature genes (Fig. 5 A–C). However, the expression of monocyte/macrophage-related genes Csfr1r, Lyz2, C/ebp-b, Cd14, and Cx3cr1 points toward a monocyte-derived cell type or a mixed population (36, 40). In summary, these findings indicate that the resident F4/80+ cells are macrophages and the infiltrating F4/80−MHCII+ cells are CD11b+cDCs.
Table S3.
Complete list of probes used for Nanostring gene-expression analysis
| Gene | Cell Type | Tg7 |
| Adjusted P value | ||
| DC and macrophage ontogeny | ||
| Cd64 (FcGR1) | 4.75E-11 | 3.30E-05 |
| Rbpj | 1.64E-13 | 3.30E-05 |
| Irf4 | 2.70E-12 | 0.0004899 |
| Kmo | 1.64E-13 | 0.001432 |
| C/ebp-b | 5.67E-12 | 0.003363 |
| Esam | 1.93E-06 | 0.003809 |
| Zbtb46 | 2.15E-18 | 0.004737 |
| Batf3 | 2.12E-16 | P > 0.05 |
| Btla | 4.76E-13 | P > 0.05 |
| C/ebp-a | 4.13E-10 | P > 0.05 |
| Ccr7 | 1.97E-11 | P > 0.05 |
| Cd14 | 4.65E-12 | P > 0.05 |
| Cd24a | 1.23E-04 | P > 0.05 |
| Csf1 | 4.70E-02 | P > 0.05 |
| Csf1r(Mcsfr/Cd115) | 9.63E-12 | P > 0.05 |
| Cx3cr1 | 2.72E-09 | P > 0.05 |
| Flt3 | 1.43E-16 | P > 0.05 |
| Gmcsfr (Csf2ra) | 4.05E-07 | P > 0.05 |
| Id2 | 3.54E-12 | P > 0.05 |
| Ikzf1 | 1.20E-10 | P > 0.05 |
| Irf2 | 5.28E-04 | P > 0.05 |
| Irf8 | 3.48E-07 | P > 0.05 |
| Kit | 8.59E-09 | P > 0.05 |
| Klf4 | 1.39E-12 | P > 0.05 |
| Lyz2 | 2.11E-07 | P > 0.05 |
| Mafb | 2.34E-08 | P > 0.05 |
| Maf-v | 1.90E-06 | P > 0.05 |
| Mertk | 7.62E-08 | P > 0.05 |
| Pu.1(Sfpi1) | 1.97E-11 | P > 0.05 |
| RelB | 1.74E-13 | P > 0.05 |
| Runx1 | 2.11E-07 | P > 0.05 |
| Runx2 | 1.93E-06 | P > 0.05 |
| Runx3 | 1.07E-07 | P > 0.05 |
| Spib | 1.60E-11 | P > 0.05 |
| Tcf4(E2.2) | 3.28E-08 | P > 0.05 |
| Tcf7 | 1.78E-07 | P > 0.05 |
| Tlr5 | 7.95E-05 | P > 0.05 |
| Traf1 | 3.25E-13 | P > 0.05 |
| Il34 | ||
| Gmcsf (Csf2) | ||
| Inflammation | ||
| Ebi3 | 4.64E-13 | 3.30E-05 |
| Fcer1g | 2.48E-08 | 0.002286 |
| Il1b | 1.23E-03 | 0.00401 |
| Ccl2(Mcp1) | 1.07E-07 | 0.004239 |
| Ccl5(Rantes) | 1.01E-07 | P > 0.05 |
| Il10 | 3.97E-08 | P > 0.05 |
| Il12p40 (Il2b) | 3.80E-05 | P > 0.05 |
| Il15 | 1.15E-03 | P > 0.05 |
| Il15ra | 8.26E-05 | P > 0.05 |
| Il18 | 1.03E-06 | P > 0.05 |
| Ly6C | 2.27E-03 | P > 0.05 |
| Tgfb1 | 2.73E-06 | P > 0.05 |
| Tnf | 1.94E-05 | P > 0.05 |
| Il12p35(IL12a) | ||
| Il6 | ||
| Il17a | ||
| Il23p19(Il23a) | ||
| Il9 | ||
| Kc | ||
| Ifna1 | ||
| Ifna4 | ||
| Ifna5 | ||
| Ifnb | ||
| Lab/SLE interest | ||
| Tlr7 | 4.01E-07 | 0.000273 |
| Ccr9 | 1.28E-05 | P > 0.05 |
| Cd229 (Slamf3) | 1.26E-02 | P > 0.05 |
| Cd244 | 1.07E-07 | P > 0.05 |
| Cd48 | 8.91E-07 | P > 0.05 |
| CD84_v1 | 3.52E-11 | P > 0.05 |
| Cd84_v2 | 6.01E-09 | P > 0.05 |
| Cd9 | 3.79E-14 | P > 0.05 |
| Dap12(Tyrobp) | 9.63E-06 | P > 0.05 |
| Dr6 | 3.82E-05 | P > 0.05 |
| SiglecE | 4.42E-07 | P > 0.05 |
| SiglecG | 1.43E-16 | P > 0.05 |
| Slamf7 (Cracc) | 2.97E-15 | P > 0.05 |
| Slamf9 | 2.72E-09 | P > 0.05 |
| Tlr9 | 5.21E-13 | P > 0.05 |
| SiglecH | ||
| Housekeeping genes | B2m, Gapdh, Cltc, Tbp, Tubb4, Eefg1 | |
Genes in boldface were not expressed above background levels in the cell populations analyzed. Two-way ANOVA analysis of the Nanostring expression data across all four populations and adjusted P values based on comparison between cell types or between normal or increased TLR7 copy number (Tg7).
Fig. 5.
RNA profile of purified CD11b+CD11c+ populations from the kidney. Cells were sorted from aged Sle1 and Sle1Tg7 mice, mRNA purified and analyzed using Nanostring. (A) PCA of gene expression by the indicated kidney and splenic populations (n = 4 per strain, age 7–9 mo). Characterization of CD11b+CD11c+ subpopulations using genes associated with DC (B) or macrophage (C) ontogeny. Heat maps represent the average gene expression level (n = 8) normalized from the lowest (0, dark blue) to the highest expression (1, bright red). Data visualization was performed using TIBCO Spotfire. (D and E) Differences in expression levels of indicated genes between Sle1 (squares, n = 4) and Sle1Tg7 (circles, n = 4) in the indicated cell subsets *P (adjusted) < 0.05 (see Materials and Methods for detailed statistical analysis).
All myeloid subsets expressed Tlr7, with the expected increase exhibited by Sle1Tg7 cells compared with Sle1 controls (Fig. 5D). Tlr9 was also detected but levels were unaffected by Sle1Tg7-associated disease (Fig. 5D). Further analysis of renal MHCII+F4/80− DCs showed a significant down-regulation in Ebi3, Slamf9, Irf4, Rbpj, Batf3, and Kmo in diseased Sle1Tg7 mice (Fig. 5E and Table S4). Analysis of renal macrophages indicated higher levels of several signaling lymphocyte-activating molecule immunoreceptor family (SLAMF) members in Sle1Tg7 mice: Cd84, Cracc, Cd229, Cd48, and Cd244 (Table S4). Additionally, macrophages expressed the highest amounts of Ccl2 (Mcp-1) transcripts, which correlates with the increased soluble levels in Sle1Tg7 kidneys (Fig. 1D and Table S4). These gene-expression changes suggest that renal macrophages in diseased mice contribute to the disease progression initiated by the DC.
Table S4.
Nanostring expression data of all of the genes analyzed in Sle1 vs. Sle1Tg7 kidney DCs and Sle1 vs, Sle1Tg7 kidney macrophages
| Symbol | Sle1 | Sle1Tg7 | Adjusted P value |
| Average ± SD | Average ± SD | ||
| Kidney DCs (CD11c+MHCII+F4/80−) | |||
| Ebi3 | 2.22 ± 0.07 | 1.91 ± 0.05 | 0.023 |
| Slamf9 | 2.70 ± 0.07 | 2.39 ± 0.06 | 0.023 |
| Irf4 | 2.38 ± 0.08 | 1.86 ± 0.15 | 0.035 |
| Rbpj | 2.86 ± 0.04 | 2.55 ± 0.08 | 0.035 |
| Batf3 | 2.15 ± 0.09 | 1.86 ± 0.09 | 0.042 |
| Kmo | 2.63 ± 0.11 | 2.29 ± 0.10 | 0.042 |
| Btla | 2.36 ± 0.14 | 1.93 ± 0.17 | 0.071 |
| Cd84_v2 | 1.25 ± 0.12 | 1.60 ± 0.03 | 0.071 |
| Zbtb46 | 2.41 ± 0.10 | 2.12 ± 0.12 | 0.071 |
| SiglecG | 2.18 ± 0.13 | 1.86 ± 0.12 | 0.071 |
| Flt3 | 2.52 ± 0.08 | 2.14 ± 0.17 | 0.073 |
| Tcf4(E2.2) | 2.06 ± 0.05 | 1.87 ± 0.09 | 0.073 |
| Il18 | 2.13 ± 0.06 | 1.91 ± 0.10 | 0.080 |
| Cd64(Fcgr1) | 2.43 ± 0.13 | 2.69 ± 0.05 | 0.092 |
| Cd84_v1 | 1.89 ± 0.15 | 2.19 ± 0.05 | 0.092 |
| Runx3 | 1.54 ± 0.11 | 1.79 ± 0.12 | 0.097 |
| C/ebp-b | 2.79 ± 0.24 | 3.23 ± 0.06 | 0.113 |
| Slamf7(Cracc) | 2.67 ± 0.07 | 2.51 ± 0.10 | 0.151 |
| Cd229 | 2.10 ± 0.16 | 2.35 ± 0.11 | 0.158 |
| Cd48 | 2.72 ± 0.05 | 2.63 ± 0.05 | 0.158 |
| Cd9 | 2.63 ± 0.08 | 2.76 ± 0.07 | 0.158 |
| SiglecE | 2.01 ± 0.11 | 1.84 ± 0.04 | 0.158 |
| Tlr5 | 1.12 ± 0.10 | 1.29 ± 0.11 | 0.158 |
| Esam | 0.85 ± 0.25 | 0.49 ± 0.17 | 0.162 |
| Klf4 | 2.68 ± 0.12 | 2.44 ± 0.16 | 0.162 |
| Mafb | 2.42 ± 0.16 | 2.63 ± 0.06 | 0.162 |
| RelB | 2.48 ± 0.14 | 2.67 ± 0.08 | 0.162 |
| Tlr7 | 1.95 ± 0.31 | 2.38 ± 0.12 | 0.162 |
| Traf1 | 2.44 ± 0.13 | 2.62 ± 0.07 | 0.162 |
| Lyz2 | 3.22 ± 0.13 | 3.39 ± 0.05 | 0.165 |
| Ly6c | 1.97 ± 0.16 | 2.27 ± 0.23 | 0.171 |
| Fcer1g | 3.37 ± 0.07 | 3.45 ± 0.04 | 0.197 |
| Cd14 | 2.27 ± 0.16 | 2.54 ± 0.23 | 0.206 |
| Il15 | 1.57 ± 0.15 | 1.27 ± 0.26 | 0.206 |
| Gmcsfr(Csf2ra) | 3.01 ± 0.01 | 2.95 ± 0.05 | 0.228 |
| Irf2 | 2.44 ± 0.08 | 2.35 ± 0.07 | 0.254 |
| Ccl2(Mcp1) | 1.56 ± 0.30 | 1.92 ± 0.32 | 0.274 |
| Csf1r(Mcsfr/Cd115) | 3.10 ± 0.13 | 3.22 ± 0.02 | 0.274 |
| Id2 | 2.95 ± 0.15 | 3.09 ± 0.03 | 0.274 |
| C/ebp-a | 2.59 ± 0.03 | 2.54 ± 0.04 | 0.279 |
| Pu.1(Sfpi1) | 3.11 ± 0.08 | 3.18 ± 0.02 | 0.279 |
| Tcf7 | −0.21 ± 0.92 | 0.54 ± 0.10 | 0.323 |
| Il10 | 0.47 ± 0.42 | 0.81 ± 0.24 | 0.342 |
| Runx1 | 2.28 ± 0.07 | 2.21 ± 0.08 | 0.343 |
| Ccr7 | 1.85 ± 0.30 | 2.06 ± 0.13 | 0.373 |
| Tlr9 | 2.47 ± 0.07 | 2.39 ± 0.10 | 0.373 |
| Cx3cr1 | 2.70 ± 0.11 | 2.62 ± 0.05 | 0.390 |
| Il1b | 2.91 ± 0.25 | 3.08 ± 0.12 | 0.390 |
| Runx2 | 1.68 ± 0.08 | 1.77 ± 0.12 | 0.390 |
| Spib | 2.03 ± 0.13 | 1.92 ± 0.13 | 0.401 |
| Mertk | 1.94 ± 0.35 | 2.15 ± 0.07 | 0.401 |
| Ikzf1 | 2.10 ± 0.05 | 2.13 ± 0.05 | 0.431 |
| Kit | 1.62 ± 0.15 | 1.71 ± 0.15 | 0.510 |
| Ccr9 | 0.87 ± 0.17 | 0.75 ± 0.23 | 0.565 |
| Il12p40(Il12b) | 1.05 ± 0.20 | 1.16 ± 0.23 | 0.597 |
| Il15ra | 1.32 ± 0.17 | 1.39 ± 0.11 | 0.597 |
| Ccl5(Rantes) | 2.26 ± 0.23 | 2.16 ± 0.16 | 0.600 |
| Tgfb1 | 3.00 ± 0.08 | 3.02 ± 0.05 | 0.688 |
| Cd24a | 1.39 ± 0.14 | 1.33 ± 0.17 | 0.707 |
| Csf1 | 1.09 ± 0.10 | 1.17 ± 0.36 | 0.761 |
| Dr6 | 2.11 ± 0.08 | 2.09 ± 0.09 | 0.825 |
| Dap12(Tyrobp) | 3.58 ± 0.06 | 3.57 ± 0.04 | 0.827 |
| Irf8 | 2.15 ± 0.14 | 2.16 ± 0.06 | 0.947 |
| Maf-v | 1.67 ± 0.11 | 1.67 ± 0.14 | 0.952 |
| Tnf | 2.01 ± 0.21 | 2.01 ± 0.08 | 0.952 |
| Cd244 | 1.14 ± 0.17 | 1.15 ± 0.31 | 0.963 |
| Kidney macrophages (CD11c+MHCII+F4/80+) | |||
| Tlr7 | 2.40 ± 0.06 | 2.70 ± 0.02 | 0.0331 |
| Ccl2(Mcp1) | 1.83 ± 0.12 | 2.32 ± 0.10 | 0.0331 |
| Lyz2 | 3.25 ± 0.07 | 3.50 ± 0.07 | 0.0452 |
| Gmcsfr(Csf2ra) | 3.01 ± 0.05 | 2.84 ± 0.04 | 0.0461 |
| Rbpj | 2.80 ± 0.06 | 2.57 ± 0.10 | 0.0499 |
| Cd64(Fcgr1) | 2.86 ± 0.08 | 3.08 ± 0.03 | 0.0499 |
| Cd84_v1 | 2.18 ± 0.05 | 2.31 ± 0.03 | 0.0499 |
| Slamf7(Cracc) | 1.71 ± 0.14 | 2.14 ± 0.02 | 0.0499 |
| Cd229 | 1.96 ± 0.12 | 2.31 ± 0.11 | 0.0499 |
| Cd48 | 2.49 ± 0.03 | 2.58 ± 0.03 | 0.0499 |
| Fcer1g | 3.32 ± 0.04 | 3.44 ± 0.05 | 0.0499 |
| Cd14 | 3.13 ± 0.02 | 2.86 ± 0.08 | 0.0499 |
| Dr6 | 1.69 ± 0.11 | 1.95 ± 0.06 | 0.0499 |
| Cd244 | 0.38 ± 0.15 | 0.81 ± 0.15 | 0.0499 |
| IL15ra | 1.11 ± 0.10 | 1.34 ± 0.09 | 0.0581 |
| Relb | 1.98 ± 0.05 | 2.13 ± 0.08 | 0.0895 |
| Ebi3 | 2.31 ± 0.05 | 2.18 ± 0.07 | 0.1030 |
| Id2 | 2.35 ± 0.13 | 2.58 ± 0.06 | 0.1133 |
| C/ebp-b | 2.37 ± 0.24 | 2.77 ± 0.16 | 0.1235 |
| MerTK | 2.19 ± 0.12 | 2.42 ± 0.11 | 0.1235 |
| Csf1r(Mcsfr/Cd115) | 3.48 ± 0.05 | 3.40 ± 0.04 | 0.1327 |
| Irf8 | 2.43 ± 0.08 | 2.28 ± 0.09 | 0.1401 |
| Il1b | 2.82 ± 0.12 | 3.00 ± 0.07 | 0.1520 |
| Maf-v | 1.92 ± 0.07 | 2.04 ± 0.06 | 0.1653 |
| Cx3cr1 | 3.09 ± 0.05 | 3.01 ± 0.05 | 0.1731 |
| Il12p40(Il12b) | 0.85 ± 0.11 | 0.49 ± 0.27 | 0.1731 |
| Tlr5 | 1.08 ± 0.07 | 0.83 ± 0.21 | 0.2191 |
| Slamf9 | 3.01 ± 0.04 | 2.91 ± 0.09 | 0.2390 |
| Irf4 | 1.45 ± 0.14 | 1.26 ± 0.14 | 0.2390 |
| Csf1 | 0.85 ± 0.22 | 1.12 ± 0.18 | 0.2391 |
| Batf3 | 2.11 ± 0.08 | 1.99 ± 0.10 | 0.2457 |
| Tcf7 | 0.30 ± 0.13 | 0.14 ± 0.15 | 0.3230 |
| Mafb | 2.95 ± 0.09 | 2.87 ± 0.02 | 0.3434 |
| Traf1 | 1.99 ± 0.03 | 1.94 ± 0.06 | 0.3434 |
| Pu.1(Sfpi1) | 2.87 ± 0.03 | 2.91 ± 0.03 | 0.3434 |
| CCL5(Rantes) | 1.35 ± 0.40 | 1.68 ± 0.09 | 0.3434 |
| Tgfb1 | 2.79 ± 0.04 | 2.83 ± 0.04 | 0.3434 |
| Ly6c | 0.70 ± 1.14 | 1.58 ± 0.17 | 0.3730 |
| Runx2 | 0.48 ± 0.99 | 1.26 ± 0.28 | 0.3730 |
| Runx1 | 2.47 ± 0.08 | 2.38 ± 0.11 | 0.3758 |
| Kmo | 1.70 ± 0.14 | 1.81 ± 0.12 | 0.4385 |
| Spib | −0.32 ± 0.79 | 0.18 ± 0.30 | 0.4731 |
| Btla | −0.14 ± 0.59 | 0.23 ± 0.25 | 0.4733 |
| Il10 | 0.52 ± 0.40 | 0.77 ± 0.21 | 0.4733 |
| Runx3 | 0.88 ± 0.16 | 0.36 ± 0.92 | 0.5027 |
| Tlr9 | 2.66 ± 0.03 | 2.68 ± 0.03 | 0.5096 |
| C/ebp-a | 2.54 ± 0.11 | 2.61 ± 0.09 | 0.5105 |
| Tnf | 1.91 ± 0.21 | 1.74 ± 0.29 | 0.5241 |
| Cd9 | 2.43 ± 0.09 | 2.35 ± 0.14 | 0.5427 |
| Il18 | 2.03 ± 0.08 | 1.99 ± 0.03 | 0.5849 |
| CD24a | 0.01 ± 0.71 | 0.51 ± 1.00 | 0.5849 |
| Dap12(Tyrobp) | 3.44 ± 0.06 | 3.46 ± 0.03 | 0.6564 |
| Tcf4(E2.2) | 2.15 ± 0.06 | 2.12 ± 0.08 | 0.6614 |
| SiglecE | 2.52 ± 0.06 | 2.49 ± 0.03 | 0.6614 |
| Zbtb46 | 0.65 ± 0.15 | 0.72 ± 0.15 | 0.6692 |
| SiglecG | 0.23 ± 0.19 | 0.32 ± 0.28 | 0.7057 |
| Esam | 0.77 ± 0.30 | 0.68 ± 0.16 | 0.7057 |
| Ccr7 | 0.32 ± 0.31 | 0.42 ± 0.38 | 0.8093 |
| Flt3 | 0.76 ± 0.22 | 0.72 ± 0.22 | 0.8642 |
| Cd84_v2 | 1.63 ± 0.08 | 1.62 ± 0.11 | 0.9159 |
| Ccr9 | 0.34 ± 0.26 | 0.32 ± 0.14 | 0.9588 |
| Irf2 | 2.24 ± 0.03 | 2.23 ± 0.09 | 0.9740 |
| Kit | −0.02 ± 0.66 | 0.02 ± 0.69 | 0.9740 |
| Klf4 | 1.71 ± 0.15 | 1.71 ± 0.16 | 0.9931 |
| IL15 | 1.70 ± 0.08 | 1.70 ± 0.08 | 0.9931 |
| Ikzf1 | 1.82 ± 0.08 | 1.82 ± 0.07 | 0.9974 |
Average ± SD values are shown (n = 4 per strain). Genes that are significantly differentially expressed are in boldface (based on adjusted P values).
Isolated Sle1Tg7-Derived Renal DCs Are Functionally Active Cells.
To ascertain the functional role of the renal cDCs, we isolated the infiltrating CD11c+MHCII− and MHCII+F4/80− subsets (Fig. 4 D and E) and assessed their ability to present antigen to CD4+ T cells using the ovalbumin/OT II system. Because the candidate genes for the Sle1 region are homotypic SLAMF members, we generated Sle1OTII mice for these studies. We determined that purified CD11c+MHCII+F4/80− renal DCs stimulated with TLR7 ligand (R848) from all three strains induced T-cell proliferation, whereas the R848-stimulated purified CD11c+MHCII− populations were unable to induce proliferation above levels of unstimulated cells (Fig. 6 A and B). Dose–response curves to the TLR7 ligand were not possible because of the low numbers retrieved from the kidney.
Fig. 6.
Kidney CD11c+MHCII+F4/80− DCs are functional and their numbers correlate with kidney disease. (A) Indicated kidney cell subsets were sorted from 5- to 7.5-mo-old mice, stimulated overnight with R848 (or media), pulsed with 50 μg/mL ovalbumin and cocultured with CFSE-stained Sle1.OT-II splenocytes for 5 d. The percentages of CD4+TCRVα2+ T-cell proliferation were assessed with flow cytometry by CFSE dilution. (B) Cumulative data from A. Each circle represents kidney cells sorted from an individual mouse (n = 3–4), except for Sle1, where CD11c+MHCII+F4/80− cells were isolated and pooled from two mice each (total of four mice). A paired t test was used to assess statistical significance. (C) Supernatants from R848 (or media) stimulated CD11c+MHCII+F4/80− DCs were analyzed by Luminex. (D) Numbers of sorted cells retrieved from kidneys of each strain (n = 5 for Sle1Tg7 and CD11cSle1Tg7, for Sle1 7 mice were pooled into three groups). Bars represent mean + SEM. *P < 0.05; ns, not significant (one-way ANOVA). Graphs showing the correlation between GN score (E) or BUN (F) and proportion of CD11c+MHCII+F4/80− DCs. The P value and r were computed by nonparametric Spearman correlation.
The secreted cytokines and chemokines following overnight exposure to R848 were measured using Luminex. Isolated renal DCs from Sle1Tg7 mice secreted MIP-1β, IL-10, MIP-1α, TNF-α, and IL-6 under resting conditions, which were increased further on R848 stimulation (Fig. 6C). Normalization of DC-TLR7 expression lowered basal secretion for IL-10 and MIP-1β, with similar trends being observed for the remaining mediators. The responses following R848 appeared to be intermediate to Sle1 and Sle1Tg7; however, this was not significant. These midway functional responses by Sle1Tg7CD11c cells suggests that other cell types expressing the TLR7 transgene may still be impacting the DC activation state before isolation.
Although the Sle1Tg7CD11c DCs still possess a capacity for antigen presentation and cytokine/chemokine production following R848 stimulation, there are significantly fewer cells infiltrating the kidney compared with Sle1Tg7 mice, with levels similar to Sle1 mice (Fig. 6D). Furthermore, the proportion of DCs in the kidney correlates with GN and BUN, which are established measures of kidney disease (Fig. 6 E and F).
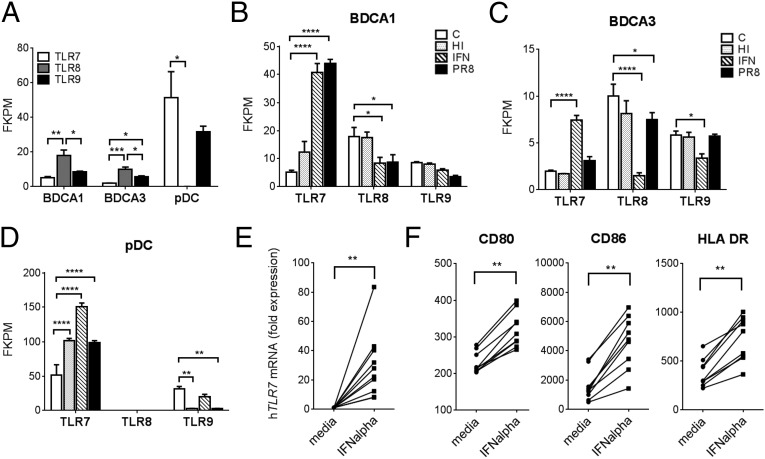
Primary Human BDCA1 and BDCA3 DCs Express TLR7 and Up-Regulate Its Transcription in Response to IFN-α.
Previous reports provide conflicting data on the expression of TLR7 in human in vitro monocyte-derived DCs (mono-DCs) (41, 42). Therefore, we set out to determine the levels of TLR7 in the two subsets of myeloid DCs that have been described in human peripheral blood: BDCA1+ and BDCA3+. DCs were enriched, sorted, then pooled from nine donors and the expression of TLR7, -8, and -9 mRNAs was assessed using RNAseq. Both BDCA1+ and BDCA3+ DC subsets expressed TLR7, but at levels 10- and 20-fold lower, respectively, than pDCs (Fig. 7A). TLR8 was expressed by both myeloid DC subsets, which was consistent with an earlier report comparing TLR7, -8, and -9 expression in BDCA1+ DCs and pDCs using two-step real-time PCR (43). Interestingly, the increased sensitivity of our approach revealed that both BDCA1+ and BDCA3+ DCs also expressed TLR9, albeit at low levels. Previous studies have shown that TLR7, -8, and -9 expression may be modulated by type-I IFN, which is a central mediator in the host response to viruses and has been associated with the development of clinical SLE (44–46). Therefore, the purified DC subsets were exposed to IFN-α, heat inactivated (HI) influenza virus (Flu), or active PR8-Flu. TLR7 mRNA expression significantly increased above control levels in BDCA1+ DCs following treatment with either IFN-α or PR8, with a concomitant decrease in TLR8 expression (Fig. 7B). BDCA3+ DCs showed minimal responses to either HI or active Flu, but increased TLR7 and decreased TLR8 and TLR9 expression in response to IFN-α (Fig. 7C). Exposure of pDCs to all three challenges resulted in significantly increased TLR7 expression compared with controls and a concomitant decrease in TLR9 following treatment with either active or inactive Flu (Fig. 7D). We then generated mono-DCs from 10 healthy human donors and stimulated them overnight with IFN-α. The cultured GM-CSF/IL-4 mono-DCs were then analyzed for TLR7 mRNA expression. We found a significant up-regulation in TLR7 mRNA following IFN-α exposure (Fig. 7E). An earlier report has shown that the addition of IFNα to monocytes cultured with autologous serum induces DC maturation. In a similar manner, we detected an increased maturation of the mono-DCs, as demonstrated by the up-regulation of CD80, CD86, and MHC II (Fig. 7F) (47).
Fig. 7.
Human cDC subsets express low levels of TLR7 and can up-regulate its expression. Expression of TLR7, -8, and -9 mRNA assessed by RNAseq in indicated human DC subsets under: (A) basal conditions and following in vitro stimulation with Live (PR8) or HI Flu or IFN-α compared with a media control, “C”, in BDCA1+ DCs (B), BDCA3+ DCs (C), and pDCs (D) (n = 3 from nine pooled individuals). (E) GM/IL-4-mono-DCs were stimulated with IFN-α or left untreated (media). TLR7 expression was measured using RT-PCR and normalized to β-actin expression and the untreated (media) for each donor (n = 10). (F) Flow cytometric analysis of indicated surface markers (n = 9). Bars represent mean + SEM. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 (two-way ANOVA followed by Dunnett's multiple comparisons test). FPKM, fragments per kilobase of exon per million mapped reads. (E and F) P values were computed by a Wilcoxon matched-pairs signed rank test, **P < 0.01.
In summary, these data highlight a particular responsiveness of primary human and in vitro-derived DCs, which express low basal levels of TLR7 that can be dramatically increased following viral exposure or treatment with IFN-α, a cytokine often implicated in the development of SLE (48).
Discussion
In these studies we have shown the central role of the CD11b+ cDC in initiating a severe autoimmune response in SLE-prone mice. We have shown that a moderate increase in TLR7 expression in DCs creates a new infiltrating population of CD11b+ cDCs that are localized to the glomeruli of nephritic kidneys, creating an inflammatory environment. Moreover, we have established that primary human myeloid DCs and in vitro generated mo-DCs express extremely low levels of TLR7, but may up-regulate expression in response to ex vivo viral or IFN-α exposure. These studies show a central role for CD11b+ cDCs in the transition from benign autoimmunity to severe pathogenesis in the Sle1Tg7 model system, and suggest this as a valid mechanism in human disease.
To investigate the cellular role of TLR7, we used a conditional low-copy BAC transgenic TLR7, Tg7, murine strain (18). In combination with the sle1 lupus susceptibility region, Sle1Tg7 mice develop severe lupus nephritis comparable to other yaa-containing lupus strains, including B6.Sle1Yaa, B6.Nba2Yaa, and B6.Yaa FcγRIIB−/− (13, 14, 17, 18). Using the Cre-lox system, we normalized expression of TLR7 in CD11c+ DCs and completely prevented the development of nephritis and all other disease traits associated with this model, including splenomegaly, myelopoesis, B- and T-cell activation, and Tfh expansion. Although CD11cCre deletion normalized expression in both CD11b+ cDCs and pDCs, the absence of IFN-α in diseased kidneys, together with a lack of pDC expansion in Sle1Tg7 mice, suggests that CD11b+ cDCs play the more dominant role in driving the inflammatory events leading to nephritis. Consistent with this hypothesis, temporal depletion of pDCs in BXSB mice ameliorates, but does not eliminate all aspects of disease (19).
We have previously described a multistep pathway to the development of severe autoimmunity (7). The first step is characterized by a loss of self-tolerance and the presence of ANAs. This stage is a prerequisite for immune complex deposition, tissue inflammation, and subsequent nephritis. Known genes within step 1 include the MHC alleles, the sle1 lupus susceptibility region, and SLAMF members. Previous data by a number of groups has shown that the presence of TLR7 is also a requirement for ANA production (9, 11, 12, 49, 50). Moreover, MyD88 signaling, specifically within B cells, is necessary for the development of ANAs and downstream pathology (10, 12). Furthermore, Reizis and colleagues (51) have recently shown that pDCs can have a critical role in this initial stage because reduction of E2-2 alters pDC function to a more tolerogenic phenotype, which eliminates ANAs altogether and therefore downstream autoimmune pathogenesis. In the second step of disease progression, known modifiers of pathogenesis include sle3, lpr, and TLR expression variations: yaa and Tg7, which have amplified TLR7. These immune alterations alone do not confer end-organ pathology but, in combination with pathway 1, result in severe disease. In these studies, we have now shown that a moderate up-regulation of TLR7 within CD11b+ cDCs is necessary to drive the inflammatory events leading to nephritis in the Sle1Tg7 murine lupus model. Thus, TLR7 plays two major roles in a multistep model of disease progression: first, in ANA production, and second, in promoting DC-mediated inflammatory events that lead to pathogenesis. In a transgenic murine system where TLR7 is increased by 8- to 32-fold, additional genes for step 1 are not required because TLR7 itself leads to the loss of tolerance (15). As expected, disruption of the humoral immune response using SAP- or CD40L-deficient systems eliminates germinal center formation, the production of ANAs, and therefore the development of GN (52). Interestingly, this high-expressing TLR7 transgenic also exhibits a large expansion of CD11b+ cDCs (15) that may contribute to kidney pathogenesis. Our floxed BAC Tg7 mice do not develop B-cell autoimmunity in the absence of the sle1 region (18). Furthermore, although modest TLR7 up-regulation contributes to leukocyte activation and ANA production in Sle1Tg7 mice, B-cell normalization does not eliminate pathogenesis (18).
Elimination of MyD88 within DCs in the MRLlpr strain had a lower impact on the prevention of disease compared with the central role they appear to play here (10). The presence of the TLR7 and MyD88 is a prerequisite for ANA production in step 1 in our model of autoimmune progression, and this has to occur before progression to step 2 and severe disease (53). Furthermore, in the MRLlpr model, the Fas mutation (lpr) disrupts the normal processes of apoptosis in T cells required for tolerance; the immune system becomes overwhelmed with activated autoreactive T cells, and the role of DCs in directing T-cell activation for pathogenesis is perhaps less significant than in other SLE models.
Thus, different murine models have specific dependencies on their cellular role, depending on the genes driving the disease progression. Stronger perturbations of the immune system may not be reflective of the human disease, directing the inflammation more toward one molecule or cell type compared with another. Milder alterations involve multiple pathways and cell types and are harder to decipher, but perhaps are more reflective of clinical disease. Taking these data together, however, we can understand the multiple pathways involved in clinical SLE and reconcile with the heterogeneity of the clinical presentation.
We identified three major myeloid CD11b+CD11c+ populations in the diseased murine kidney. MHCII+F4/80+ macrophages were the most abundant population in nondiseased kidneys. These were originally described as a tissue-resident sentinel DC population; however, our data strongly support their reclassification as macrophages based on gene-expression profiling (31, 54, 55). Examination of diseased kidneys from Sle1Tg7 mice identified the presence of CD11c+F4/80− cells specifically within the glomeruli of diseased mice, suggesting that these cells are critical for the development of GN. Analysis of ontogeny gene mRNA expression confirmed that the infiltrating CD11c+F4/80−MHCII+ subpopulation were CD11b+cDCs. However, CD11c+F4/80−MHCII− cells did not possess either a typical DC or macrophage gene signature, and probably represent a mixed myeloid-infiltrating population, contrary to an earlier investigation that classified these cells as DCs (33).
In vitro analyses showed that purified renal DCs were efficient at presenting antigen and secreted a number of chemokines under resting conditions, with higher levels following R848 stimulation. Some of the functional phenotypes suggest that the responses by CD11cSle1Tg7 were intermediate to Sle1 and Sle1Tg7. Because these cells were isolated from the kidney, they have been exposed to an environment where other non-DCs, including the kidney cells, may influence DC activation, maturation, and the subsequent response to inflammatory stimuli.
Further characterization of the CD11b+CD11c+MHCII+ DCs by flow cytometry revealed the presence of surface CD64, suggesting that these are monocyte-derived rather than arising directly from a bone marrow precursor (Fig. S3C) (38, 39). An earlier investigation by Carlin et al. (56) previously demonstrated that TLR7 enhances the migration of monocytes into the kidney. We propose a mechanism whereby the damage to the kidney caused by the immune complex deposition results in chemokines being produced by epithelial and endothelial cells. These chemokines recruit monocytes into the tissue, where the inflammatory milieu directs differentiation toward a DC phenotype. We have shown that these Sle1Tg7 DCs actively secrete more inflammatory mediators compared with their Sle1 and Sle1Tg7CD11c counterparts and their increased presence correlates with established methods of assessing kidney damage (GN and BUN levels), indicating their primary role in disease progression.
Although the underlying molecular mechanisms driving pathology mediated by DC TLR7 expression are a subject for future study, several additional changes that we have found in the gene-expression profile of renal DCs in Sle1Tg7 mice may provide some clues: the decreased expression of several myeloid developmental genes may suggest a difference in maturation or activation of the cells (34, 35, 37). Furthermore, there is a lower expression level of Ebi3 (Epstein–Barr virus-induced gene 3), which encodes the EBI3 subunit of IL-27 and IL-35. Deficiency in the EBI3 subunit has been associated with the development of lupus nephritis and consistent with these findings, an increase was protective (57, 58).
Existing data on the role of TLR7 in human myeloid DCs suggests a minor role in inflammation: they express extremely low levels of TLR7 and respond to Flu challenge primarily through TLR8 (recognizing ssRNA) and cytoplasmic receptors, such as RIG-I (recognizing both ssRNA and dsRNA) (59). However, we found that primary human BDCA1+ DCs have the capacity to markedly up-regulate TLR7 expression in response to ex vivo viral exposure. In support of these findings, monocyte-derived DCs showed a remarkable capacity to up-regulate TLR7 following overnight exposure to IFN-α, a cytokine known to be important in clinical SLE (48). Therefore, under these conditions, both TLR7 and TLR8 may act as RNA sensors, each contributing to the inflammatory responses by these cells.
In summary, our results demonstrate that up-regulation of TLR7 within the CD11c+ DC population is required for the development of severe autoimmunity in Sle1Tg7 mice. Restoration of TLR7 expression to normal levels in CD11c+ cells completely prevented the development of the major autoimmune traits associated with the model, including splenomegaly with myelopoesis and GN. Furthermore, we have shown that human DCs express low levels of TLR7 and have the capacity to up-regulate expression in response to viral stimuli. Moreover, mono-DCs derived from healthy donors up-regulate TLR7 expression in response to IFN-α. These data support our original multistep hypothesis of SLE progression, where loss of tolerance to self (susceptibility sle1 locus) combined with additional immune alteration (moderate TLR7 up-regulation) culminates in severe kidney disease. Taken together, these findings also suggest that targeting myeloid DC-TLR7 expression may be a productive avenue of research leading to novel therapeutic options for human SLE.
Materials and Methods
Mice.
Mice were bred at either the University of Texas Southwestern Medical Center or in the Biomedical Resource Centre, Singapore. Breeding pairs for C57BL/6J (B6), CD11cCre, and M-LysozymeCre mice were originally obtained from the Jackson Laboratory (25, 28). The derivation of Sle1 and the generation of the conditional BAC Tg7 have been previously described (18, 60). The sle1 region is flanked by D1Mit17 and D1Mit202 and is NZM2410-derived. For pathological and immunological analysis, female mice were aged in three to four separate cohorts of ≥3 mice per strain for 5–7.5 mo. Additional cohorts were set up for purification of leukocyte subsets from 6- to 8-wk-old mice.
Study Approval.
The care and use of laboratory animals conformed to the National Institutes of Health guidelines and all experimental procedures were conducted according to an Institutional Animal Care and Use Committee-approved animal protocol. Written informed consent was received from healthy donors before inclusion in the study. Ethical approval was obtained from the National University of Singapore (NUS) Institutional Review Board (NUS-IRB 10-250) and Health Sciences Authority HAS-IRB-201306-5 (Singapore).
Assessment of Renal Disease.
BUN was assessed using the QuantiChrom Urea Assay Kit (BioAssay Systems). To detect GN, kidneys were fixed in formalin and embedded in paraffin for blinded analysis by an independent pathologist according to World Health Organization specifications, as previously described (16).
Kidney Supernatant Analysis.
Kidneys supernatants were prepared as described previously (18). Briefly, whole kidneys were minced and resuspended into 0.5 mL PBS. Cells were spun down and the supernatants were stored at −80 °C. For IFN-α analysis, supernatants were assayed using a Mouse IFN-alpha Platinum ELISA (Affymetrix) as per the manufacturer’s instructions. The presence of cytokines and chemokines was determined by Luminex multiplexing technology using the MCYTOMAG-70K MILLIPLEX MAP Mouse Cytokine/Chemokine Magnetic Bead Panel Immunology Multiplex Assays (Merck Millipore) as per the manufacturer’s instructions.
Serology.
Serum auto-antibodies were measured using ELISAs detecting anti-histone and anti-dsDNA, anti-U1snRNP, or anti-dsDNA IgG, as described previously (16).
Imaging.
Kidneys sections from frozen OCT embedded tissues were fixed with 1:1 acetone/methanol and stained with antinephrin (AF488), anti-CD11c (PE), and anti-F4/80 (AF647) antibodies. Images were captured with a FV-100 confocal system comprising an Olympus IX81 microscope and FV1000 scan head and processed with Olympus FV10-ASW Fluoview v2.0b.
Flow Cytometry and Cell Sorting of Murine Cells.
A single-cell suspension was prepared from dissected kidneys, as described previously (16). Splenocytes or kidney cells were resuspended in PBS with 1% (vol/vol) FCS and labeled with a combination of up to 12 directly conjugated antibodies. Red blood cell (RBC) lysis was achieved using BD FACS Lysing solution as per manufacturer’s instructions. To identify murine splenic DCs, we gated on CD45 expression then analyzed B220 and CD11b expression, excluding cells expressing CD19 and CD3 (17A2) before taking into account subtype-specific markers: pDCs were defined as B220+CD11b−CD19−CD11cloSiglecH+Gr1+; CD8+ DCs were B220−CD11b−CD8+CD19/3−CD11c+MHCII+; and total myeloid/conventional DCs were CD11b+B220−Gr1−Side-scatterloCD11c+MHCII+. Renal myeloid subsets were CD45+CD11b+CD11c+ and CD3/19−Gr1−. Further gating identified MHCII− and MHCII+ populations and the MHCII+ population was subdivided based on F4/80 expression. Acquisition and analysis was completed using a BD LSR II with Flowjo 7.6 for Windows (Treestar). For sorting, RBC lysis was achieved with ACK lysing buffer (Lonza) and live DAPI− cells were sorted using a BD FACSAria system. Antibodies were purchased either from BD Biosciences, eBioscience, BioLegend, or Life Technologies.
Nanostring Gene-Expression Analysis.
The nCounter Analysis System by Nanostring allows absolute quantification of very small amounts of mRNA without an amplification step, which can introduce variability and bias (61). Following fluorescent labeling procedures described above, murine kidney cells or splenocytes were sorted on a BD FACSAria in the SIgN Flow Cytometry Core Facility according to the gating strategy described above and shown in the Results. Fifteen-thousand cells in 5 μL of RLT buffer (Qiagen) with 1% (vol/vol) 2-mercaptoethanol were used for the Gene Expression Assay using the Nanostring nCounter system, as per the manufacturer’s recommended protocol. Probes were designed by Nanostring (standard assay) with the gene-expression list shown in Table S3. The housekeeping genes B2m, Cltc, and Tbp were used to normalize for RNA loading differences. A two-way ANOVA with post hoc Tukey’s test was used for each gene with the cell type and the Tg7 transgene status as the factors together with their interaction term. Student’s t test with Welch correction was used to determine significant differences in the expression of each gene between transgenic versus nontransgenic within each cell type. Multiple testing correction was done using the method of Benjamini and Hochberg with P < 0.05 postcorrection being considered significant. All of the statistical analysis was conducted using the R statistical language version 2.15.2. Data visualization was performed using TIBCO Spotfire.
Human DC Isolation and Stimulation.
Peripheral blood mononuclear cells (PBMCs) were obtained from blood of healthy donors by Ficoll-Hypaque centrifugation (Amersham Pharmacia Biotech). Primary human DCs were enriched from PBMCs, using a Dynabead Human DC Kit (Life Technologies). Enriched DCs were further purified by FACS: BDCA1 subset (Lin1−HLADR+CD11c+BDCA1+) BDCA3+ (Lin1−HLADR+CD11c+BDCA3+), and pDCs (Lin1−HLADR+CD123+CD11c−). Cells were then seeded in a 96-well plate to give a minimum cell number of 1 × 104 cells per well. DC subsets from individual donors were stimulated separately with either IFN-a2b (1000 U; Miltenyi Biotec), live PR8 (0.512:1 HAU; Charles River), heat-inactivated PR8 (0.512:1 HAU) or were not stimulated, for 8 h. Cells were subsequently lysed in mirVana buffer and stored at –80 °C until RNA isolation and RNAseq analysis.
RNA Isolation and RNA-Seq.
Human DCs were isolated from nine donors as described above. Total RNA was isolated from the DC pellets using a mirVana miRNA Isolation kit, according to the manufacture’s guidelines, and analyzed on Agilent Bioanalzyer for quality assessment. Samples were pooled into three groups; cDNA libraries were prepared using 24–30 ng of total RNA and 2 μL of a 3:5,000 dilution of External RNA Controls Consortium (ERCC) RNA Spike in Controls (Ambion). The fragmented mRNA samples were subjected to cDNA synthesis using Illumina TruSeq RNA sample preparation kit version 1 (low-throughput protocol) according to the manufacturer’s protocol, with the following modifications: (i) use of 14 PCR cycles, and (ii) two additional rounds of Agencourt Ampure XP SPRI beads (Beckman Courter) to remove >600 bp double-stranded cDNA.
RNAseq Analysis.
RNAseq fastq paired end data were first quality checked with fastqc. (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Data were then mapped to an ERCC spikein database with Bowtie using default parameters except max-insert-size set to 500 (www.thermofisher.com/order/catalog/product/4456740) (62). Picard tools (picard.sourceforge.net/) were used to determine the mean mate distance and SD for each sample. The data were mapped to HG19 with TopHat2 (tophat.cbcb.umd.edu (63) using default parameters, except for the mate information, Gencode V16 annotation as a guide (www.gencodegenes.org/), with no novel junctions. A custom script based on the R subreads package was then used to generate raw counts per exon using the gencode V16 annotation, accumulated at the gene level. Counts were normalized by fragments per kilobase of exon per million mapped reads. edgeR (64) was used to determine differential gene expression on a pairwise basis. All of this methodology was written within the Pipeline Pilot frame work (accelrys.com). Statistical significance was determined using a two-way ANOVA followed by Dunnett’s multiple comparisons test in GraphPad Prism 6.05.
Antigen Presentation.
Kidney CD11c+MHCII− and CD11c+MHCII+F4/80− cells were sorted as described, resuspended in complete media, and seeded in round-bottom 96-well plates at 104cells per well. Where indicated, cells sorted from multiple mice were pooled to achieve sufficient cell numbers. Cells were stimulated in overnight cultures with 1 µg/mL R848 (InvivoGen) or left untreated, and then incubated with 50 µg/mL ovalbumin (Sigma-Aldrich). After 4 h, the media was replaced with a suspension of CFSE-stained splenocytes from Sle1.transgenic OT-II T-cell receptor mice at 2 × 105 cells per well. After 5 d of incubation, the proliferation of T cells was assessed by CFSE dilution using flow cytometry. Supernatants from overnight stimulations (media and R848) were collected and immediately frozen at −80°. The presence of cytokines and chemokines was assessed by Luminex as described for kidney supernatants.
Monocyte-Derived Dendritic Cell Isolation and Stimulation.
PBMCs were obtained from healthy donor blood as described above and CD14+ monocytes were purified using a CD14-microbead kit (Miltenyi Biotec). Cells were cultured in a six-well plate at 2 × 105 cells per well, 2 mL per well in RPMI complete medium (RPMI CM; RPMI-1640 containing 10% (vol/vol) heat-inactivated FCS, 10 mM Hepes, 2 mM l-glutamine, 100 IU/mL penicillin, and 100 µg/mL streptomycin) with GM-CSF (1,000 U/mL) and IL-4 (1,000 U/mL). Fresh medium and cytokines were added to the culture on day 4. Mono-DCs were harvested and seeded on day 6 in a 96-well U-bottom plate at 1 × 105 cells per well, before stimulation with IFN-a2b (1,000 U), or left unstimulated, for 16 h. Cells were then collected by centrifugation and an aliquot was stained for flow cytometric analysis with antibodies directed to CD80, CD86, and HLADR (BD Pharmingen). A near IR Live Dead Dye (Life Technologies) was used to gate out dead cells. The rest of the cells were resuspended in RLT buffer (Qiagen), homogenized by syringing and immediately frozen on dry ice. Cell lysates were stored at −80 °C until RNA isolation.
Quantitative Real-Time PCR.
RNA was extracted using the RNeasy Micro Kit (Qiagen) and concentrations were measured using Nanodrop 2000 spectrophotometer (Thermo Scientific). RNA was reverse-transcribed and RT-PCR was performed using the Power SYBR Green PCR master mix (Applied Biosystems) on the 7900H fast real-time PCR system (Applied Biosystems). Primers for human TLR7 were from Invivogen (catalog #rtp-htlr7) and primers for β-actin were as described previously (65). The results were analyzed using SDS 2.4, Microsoft Excel for Windows, and GraphPad Prism v6.05.
Statistics.
Nanostring and RefSeq data were analyzed as described. Data are expressed as the arithmetic mean + SEM and were statistically analyzed using GraphPad Prism 6.05. When comparing two groups, multiple t tests were used, corrected for multiple comparisons with the Holm–Sidak method. Where indicated, a paired t test or Wilcoxon matched-pairs signed rank test was used to compute P values for paired data. When three groups were compared, parametric data were assessed by ordinary one-way ANOVA with post hoc Bonferroni’s multiple comparison test and nonparametric data were assessed with the Kruskal–Wallis test with post hoc Dunn’s multiple comparisons test. Correlation was assessed by Spearman’s rank correlation test. Where there is no notation of significance on a figure, there was no significant difference found.
SI Materials and Methods
Proteinuria.
Proteinuria was assessed using Albustix (Bayer). Albumin levels in urine were assayed using an Albumin Mouse ELISA Kit (Abcam) according to the manufacturer’s instructions; samples were assayed at a dilution of 1:400. Samples were normalized for creatinine using a Creatinine (urinary) Colorimetric Assay Kit (Cayman Chemical) according to the manufacturer’s instructions; initial sample dilution of 1:10.
Cell Sorting, RNA Isolation, and RT-PCR.
Splenic B cells were sorted as live CD45+Gr1−B220+CD19+, splenic T cells as live CD45+Gr1−CD3+CD5+ and peritoneal macrophages as live CD45+Gr1−CD11bhiF4/80hi. Sorted cells were centrifuged, resuspended in TRIzol (Life Technologies) and stored at −80°. RNA was extracted by TRIzol/chloroform and purified with the Qiagen RNeasy Mini purification kit according to the manufacturer's instructions (Qiagen). Concentration and purity were measured using a Nanodrop spectrophotometer (Thermo Fisher Scientific). A TaqMan RNA-to-CT 1-Step Kit (Applied Biosystems) was used to perform the reverse transcription and quantitative PCR reactions according to the manufacturer’s instructions using TaqMan gene expression assays (Applied Biosystems) to either Tlr7 (Mm00446590) or the B2m housekeeping gene (Mm00437762). Real-time PCR was performed on the 7900H fast real-time PCR system and analyzed using SDS 2.4 (Applied Biosystems). Relative mRNA expression was calculated using the comparative Ct method.
Imaging.
Kidney sections from OCT embedded tissue were fixed with 4% paraformaldehyde before permeabilization with acetone and stained with Phalloidin (AF647) and anti-CD3d (unlabeled Ab followed by secondary staining with donkey anti-goat Dylight 550). Images were captured with a Zeiss LSM800 microscope at 200× magnification and processed with Zen imaging software.
Acknowledgments
We thank Dr. Lucy Robinson of Insight Editing London for critical review and assistance in manuscript preparation; Florent Ginhoux for discussions on myeloid ontogeny; and the Flow Cytometry and Functional Genomic cores at the Singapore Immunology Network, A*STAR, Singapore, and the flow cytometry core at the University of Texas Southwestern Medical Center, Dallas, TX. This work was supported by core funding from the Singapore Immunology Network and Institute of Cell and Molecular Biology at A*STAR, Singapore (to A.-M.F. and J.C.), and grants from the National Institutes of Health and the Alliance for Lupus Research (to E.K.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.M.-R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507052112/-/DCSupplemental.
References
- 1.Du X, Poltorak A, Wei Y, Beutler B. Three novel mammalian toll-like receptors: Gene structure, expression, and evolution. Eur Cytokine Netw. 2000;11(3):362–371. [PubMed] [Google Scholar]
- 2.Hemmi H, et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol. 2002;3(2):196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 3.Barton GM, Medzhitov R. Toll-like receptor signaling pathways. Science. 2003;300(5625):1524–1525. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- 4.Tan EM, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25(11):1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 5.Maroz N, Segal MS. Lupus nephritis and end-stage kidney disease. Am J Med Sci. 2013;346(4):319–323. doi: 10.1097/MAJ.0b013e31827f4ee3. [DOI] [PubMed] [Google Scholar]
- 6.Arbuckle MR, et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003;349(16):1526–1533. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- 7.Fairhurst AM, Wandstrat AE, Wakeland EK. Systemic lupus erythematosus: Multiple immunological phenotypes in a complex genetic disease. Adv Immunol. 2006;92:1–69. doi: 10.1016/S0065-2776(06)92001-X. [DOI] [PubMed] [Google Scholar]
- 8.Nickerson KM, et al. TLR9 regulates TLR7- and MyD88-dependent autoantibody production and disease in a murine model of lupus. J Immunol. 2010;184(4):1840–1848. doi: 10.4049/jimmunol.0902592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santiago-Raber ML, et al. Critical role of TLR7 in the acceleration of systemic lupus erythematosus in TLR9-deficient mice. J Autoimmun. 2010;34(4):339–348. doi: 10.1016/j.jaut.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Teichmann LL, Schenten D, Medzhitov R, Kashgarian M, Shlomchik MJ. Signals via the adaptor MyD88 in B cells and DCs make distinct and synergistic contributions to immune activation and tissue damage in lupus. Immunity. 2013;38(3):528–540. doi: 10.1016/j.immuni.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berland R, et al. Toll-like receptor 7-dependent loss of B cell tolerance in pathogenic autoantibody knockin mice. Immunity. 2006;25(3):429–440. doi: 10.1016/j.immuni.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 12.Soni C, et al. B cell-intrinsic TLR7 signaling is essential for the development of spontaneous germinal centers. J Immunol. 2014;193(9):4400–4414. doi: 10.4049/jimmunol.1401720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pisitkun P, et al. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312(5780):1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- 14.Subramanian S, et al. A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc Natl Acad Sci USA. 2006;103(26):9970–9975. doi: 10.1073/pnas.0603912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deane JA, et al. Control of toll-like receptor 7 expression is essential to restrict autoimmunity and dendritic cell proliferation. Immunity. 2007;27(5):801–810. doi: 10.1016/j.immuni.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fairhurst AM, et al. Yaa autoimmune phenotypes are conferred by overexpression of TLR7. Eur J Immunol. 2008;38(7):1971–1978. doi: 10.1002/eji.200838138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santiago-Raber ML, et al. Evidence for genes in addition to Tlr7 in the Yiaa translocation linked with acceleration of systemic lupus erythematosus. J Immunol. 2008;181(2):1556–1562. doi: 10.4049/jimmunol.181.2.1556. [DOI] [PubMed] [Google Scholar]
- 18.Hwang SH, et al. B cell TLR7 expression drives anti-RNA autoantibody production and exacerbates disease in systemic lupus erythematosus-prone mice. J Immunol. 2012;189(12):5786–5796. doi: 10.4049/jimmunol.1202195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowland SL, et al. Early, transient depletion of plasmacytoid dendritic cells ameliorates autoimmunity in a lupus model. J Exp Med. 2014;211(10):1977–1991. doi: 10.1084/jem.20132620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sang A, et al. Dysregulated cytokine production by dendritic cells modulates B cell responses in the NZM2410 mouse model of lupus. PLoS One. 2014;9(8):e102151. doi: 10.1371/journal.pone.0102151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teichmann LL, et al. Dendritic cells in lupus are not required for activation of T and B cells but promote their expansion, resulting in tissue damage. Immunity. 2010;33(6):967–978. doi: 10.1016/j.immuni.2010.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birnberg T, et al. Lack of conventional dendritic cells is compatible with normal development and T cell homeostasis, but causes myeloid proliferative syndrome. Immunity. 2008;29(6):986–997. doi: 10.1016/j.immuni.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 23.Ohnmacht C, et al. Constitutive ablation of dendritic cells breaks self-tolerance of CD4 T cells and results in spontaneous fatal autoimmunity. J Exp Med. 2009;206(3):549–559. doi: 10.1084/jem.20082394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Platt AM, Randolph GJ. Does deleting dendritic cells delete autoimmunity? Immunity. 2010;33(6):840–842. doi: 10.1016/j.immuni.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8− dendritic cells in the spleen. J Exp Med. 2007;204(7):1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haberstroh U, et al. Expression of the chemokines MCP-1/CCL2 and RANTES/CCL5 is differentially regulated by infiltrating inflammatory cells. Kidney Int. 2002;62(4):1264–1276. doi: 10.1111/j.1523-1755.2002.kid572.x. [DOI] [PubMed] [Google Scholar]
- 27.Romagnani P, et al. IP-10 and Mig production by glomerular cells in human proliferative glomerulonephritis and regulation by nitric oxide. J Am Soc Nephrol. 2002;13(1):53–64. doi: 10.1681/ASN.V13153. [DOI] [PubMed] [Google Scholar]
- 28.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8(4):265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 29.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401(6754):708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 30.Wherry EJ, et al. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol. 2003;4(3):225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- 31.Soos TJ, et al. CX3CR1+ interstitial dendritic cells form a contiguous network throughout the entire kidney. Kidney Int. 2006;70(3):591–596. doi: 10.1038/sj.ki.5001567. [DOI] [PubMed] [Google Scholar]
- 32.Lionakis MS, et al. CX3CR1-dependent renal macrophage survival promotes Candida control and host survival. J Clin Invest. 2013;123(12):5035–5051. doi: 10.1172/JCI71307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sahu R, Bethunaickan R, Singh S, Davidson A. Structure and function of renal macrophages and dendritic cells from lupus-prone mice. Arthritis Rheumatol. 2014;66(6):1596–1607. doi: 10.1002/art.38410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller JC, et al. Immunological Genome Consortium Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol. 2012;13(9):888–899. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlitzer A, et al. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. 2013;38(5):970–983. doi: 10.1016/j.immuni.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gautier EL, et al. Immunological Genome Consortium Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13(11):1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guilliams M, et al. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol. 2014;14(8):571–578. doi: 10.1038/nri3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langlet C, et al. CD64 expression distinguishes monocyte-derived and conventional dendritic cells and reveals their distinct role during intramuscular immunization. J Immunol. 2012;188(4):1751–1760. doi: 10.4049/jimmunol.1102744. [DOI] [PubMed] [Google Scholar]
- 39.Plantinga M, et al. Conventional and monocyte-derived CD11b(+) dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity. 2013;38(2):322–335. doi: 10.1016/j.immuni.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 40.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19(1):71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- 41.Larangé A, Antonios D, Pallardy M, Kerdine-Römer S. TLR7 and TLR8 agonists trigger different signaling pathways for human dendritic cell maturation. J Leukoc Biol. 2009;85(4):673–683. doi: 10.1189/jlb.0808504. [DOI] [PubMed] [Google Scholar]
- 42.Rissoan MC, et al. Reciprocal control of T helper cell and dendritic cell differentiation. Science. 1999;283(5405):1183–1186. doi: 10.1126/science.283.5405.1183. [DOI] [PubMed] [Google Scholar]
- 43.Guiducci C, et al. RNA recognition by human TLR8 can lead to autoimmune inflammation. J Exp Med. 2013;210(13):2903–2919. doi: 10.1084/jem.20131044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Derkow K, et al. Multiple sclerosis: Modulation of Toll-like receptor (TLR) expression by interferon-β includes upregulation of TLR7 in plasmacytoid dendritic cells. PLoS One. 2013;8(8):e70626. doi: 10.1371/journal.pone.0070626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bekeredjian-Ding IB, et al. Plasmacytoid dendritic cells control TLR7 sensitivity of naive B cells via type I IFN. J Immunol. 2005;174(7):4043–4050. doi: 10.4049/jimmunol.174.7.4043. [DOI] [PubMed] [Google Scholar]
- 46.Rönnblom L, Pascual V. The innate immune system in SLE: Type I interferons and dendritic cells. Lupus. 2008;17(5):394–399. doi: 10.1177/0961203308090020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blanco P, Palucka AK, Gill M, Pascual V, Banchereau J. Induction of dendritic cell differentiation by IFN-alpha in systemic lupus erythematosus. Science. 2001;294(5546):1540–1543. doi: 10.1126/science.1064890. [DOI] [PubMed] [Google Scholar]
- 48.Pascual V, Farkas L, Banchereau J. Systemic lupus erythematosus: All roads lead to type I interferons. Curr Opin Immunol. 2006;18(6):676–682. doi: 10.1016/j.coi.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 49.Christensen SR, et al. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25(3):417–428. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 50.Groom JR, et al. BAFF and MyD88 signals promote a lupuslike disease independent of T cells. J Exp Med. 2007;204(8):1959–1971. doi: 10.1084/jem.20062567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sisirak V, et al. Genetic evidence for the role of plasmacytoid dendritic cells in systemic lupus erythematosus. J Exp Med. 2014;211(10):1969–1976. doi: 10.1084/jem.20132522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walsh ER, et al. Dual signaling by innate and adaptive immune receptors is required for TLR7-induced B-cell-mediated autoimmunity. Proc Natl Acad Sci USA. 2012;109(40):16276–16281. doi: 10.1073/pnas.1209372109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Celhar T, Magalhães R, Fairhurst AM. TLR7 and TLR9 in SLE: When sensing self goes wrong. Immunol Res. 2012;53(1-3):58–77. doi: 10.1007/s12026-012-8270-1. [DOI] [PubMed] [Google Scholar]
- 54.Krüger T, et al. Identification and functional characterization of dendritic cells in the healthy murine kidney and in experimental glomerulonephritis. J Am Soc Nephrol. 2004;15(3):613–621. doi: 10.1097/01.asn.0000114553.36258.91. [DOI] [PubMed] [Google Scholar]
- 55.Taylor PR, et al. Macrophage receptors and immune recognition. Annu Rev Immunol. 2005;23:901–944. doi: 10.1146/annurev.immunol.23.021704.115816. [DOI] [PubMed] [Google Scholar]
- 56.Carlin LM, et al. Nr4a1-dependent Ly6C(low) monocytes monitor endothelial cells and orchestrate their disposal. Cell. 2013;153(2):362–375. doi: 10.1016/j.cell.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Igawa T, et al. Deficiency in EBV-induced gene 3 (EBI3) in MRL/lpr mice results in pathological alteration of autoimmune glomerulonephritis and sialadenitis. Mod Rheumatol. 2009;19(1):33–41. doi: 10.1007/s10165-008-0117-1. [DOI] [PubMed] [Google Scholar]
- 58.Shinsuke N, Hiroshi I. Overexpression of Epstein-Barr virus-induced gene 3 protein (EBI3) in MRL/lpr mice suppresses their lupus nephritis by activating regulatory T cells. Autoimmunity. 2013;46(7):446–454. doi: 10.3109/08916934.2013.809422. [DOI] [PubMed] [Google Scholar]
- 59.Jensen S, Thomsen AR. Sensing of RNA viruses: A review of innate immune receptors involved in recognizing RNA virus invasion. J Virol. 2012;86(6):2900–2910. doi: 10.1128/JVI.05738-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morel L, et al. Functional dissection of systemic lupus erythematosus using congenic mouse strains. J Immunol. 1997;158(12):6019–6028. [PubMed] [Google Scholar]
- 61.Kulkarni MM. 2011. Digital multiplexed gene expression analysis using the NanoString nCounter system. Curr Protoc Mol Biol Chapter 25:Unit25B.
- 62.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kokkinopoulos I, Jordan WJ, Ritter MA. Toll-like receptor mRNA expression patterns in human dendritic cells and monocytes. Mol Immunol. 2005;42(8):957–968. doi: 10.1016/j.molimm.2004.09.037. [DOI] [PubMed] [Google Scholar]