Significance
Mitochondrial function is critical for health and longevity. Mutation of the highly conserved Rieske iron–sulfur subunit (ISP-1) of complex III in the respiratory chain results in pleiotropic phenotypes in Caenorhabditis elegans, including delayed development and increased lifespan. We identified intragenic mutations within a conserved 6-aa tether region of ISP-1. These suppressors are capable of suppressing all of the phenotypes associated with the isp-1(qm150) mutation. We further demonstrated that this mutation/suppressor relationship is conserved in the Rieske iron–sulfur protein (Rip1) of yeast complex III. These findings provide insights into conserved features of the structure and function of this protein, and allow us to propose a unique “spring-loaded” mechanism to account for these effects, supported by empirical physicochemical data.
Keywords: mitochondrial iron–sulfur protein, aging, Rip1, isp-1, complex III
Abstract
Mitochondria play an important role in numerous diseases as well as normative aging. Severe reduction in mitochondrial function contributes to childhood disorders such as Leigh Syndrome, whereas mild disruption can extend the lifespan of model organisms. The Caenorhabditis elegans isp-1 gene encodes the Rieske iron–sulfur protein subunit of cytochrome c oxidoreductase (complex III of the electron transport chain). The partial loss of function allele, isp-1(qm150), leads to several pleiotropic phenotypes. To better understand the molecular mechanisms of ISP-1 function, we sought to identify genetic suppressors of the delayed development of isp-1(qm150) animals. Here we report a series of intragenic suppressors, all located within a highly conserved six amino acid tether region of ISP-1. These intragenic mutations suppress all of the evaluated isp-1(qm150) phenotypes, including developmental rate, pharyngeal pumping rate, brood size, body movement, activation of the mitochondrial unfolded protein response reporter, CO2 production, mitochondrial oxidative phosphorylation, and lifespan extension. Furthermore, analogous mutations show a similar effect when engineered into the budding yeast Rieske iron–sulfur protein Rip1, revealing remarkable conservation of the structure–function relationship of these residues across highly divergent species. The focus on a single subunit as causal both in generation and in suppression of diverse pleiotropic phenotypes points to a common underlying molecular mechanism, for which we propose a “spring-loaded” model. These observations provide insights into how gating and control processes influence the function of ISP-1 in mediating pleiotropic phenotypes including developmental rate, movement, sensitivity to stress, and longevity.
Mitochondria are sites for adenosine 5′-triphosphate (ATP) production by oxidative phosphorylation, cellular calcium buffering, iron–sulfur cluster biogenesis, reactive oxygen species (ROS) formation, and regulation of apoptosis. Although inherited defects in mitochondrial function are most often associated with severe childhood disorders, a large number of age-related diseases such as heart disease, cancer, diabetes, obesity, and neurodegeneration have also been linked to mitochondrial dysfunction (1, 2).
In Caenorhabditis elegans, multiple studies have demonstrated that reduced electron transport chain activity (ETC) can lead to increased lifespan. These include mutations in the coenzyme Q biosynthetic gene clk-1, the pyrophosphokinase gene tpk-1, and the Rieske iron–sulfur protein isp-1 (3–7). Following RNAi knockdown of ETC components, several other proteins have been implicated in lifespan extension, including HIF-1, GCN-2, CEP-1, CEH-23, TAF-4, AHA-1, CEH-18, JUN-1, NHR-27, and NHR-49 (8–12). In addition, it was proposed that the mitochondrial unfolded protein response (mtUPR) directly mediated lifespan extension from ETC inhibition (13); however, more recent work has suggested that induction of the mtUPR is neither necessary nor sufficient to extend lifespan in worms (14). A self-consistent model is emerging suggesting that isp-1(qm150) animals have increased levels of ROS, which induces activation of the intrinsic apoptotic pathway to extend lifespan (15).
ISP-1 is an evolutionary conserved, nuclear-encoded iron–sulfur (2Fe-2S) protein that functions within complex III of the electron transport chain (16). The isp-1(qm150) allele, which results in a proline to serine substitution, has been particularly well studied due to its robust positive effect on lifespan (6). In this context, we set out to further explore the biochemical and molecular mechanisms by which the isp-1(qm150) mutation causes pleiotropic phenotypes including delayed development and increased lifespan. Here we report the identification of intragenic suppressors of isp-1(qm150) all located in the highly conserved six amino acid tether (also sometimes referred to as a “hinge”) region of ISP-1. These mutations suppressed all of the phenotypes associated with isp-1(qm150) examined, including a previously unreported sensitivity to hyperoxia. In addition, we show a similar relationship between the isp-1(qm150) mutation and two of the isolated suppressors in the budding yeast Rieske iron–sulfur protein Rip1, demonstrating a striking conservation of the structure–function relationship across widely divergent phyla. Analysis of the extensive literature on physicochemical parameters involving the role of ISP reveals a “spring-loaded” mechanism of action, summarized in the discussion and further supported in the SI Appendix.
Results
Isolation of Mutations That Suppress C. elegans isp-1(qm150) Slow Development.
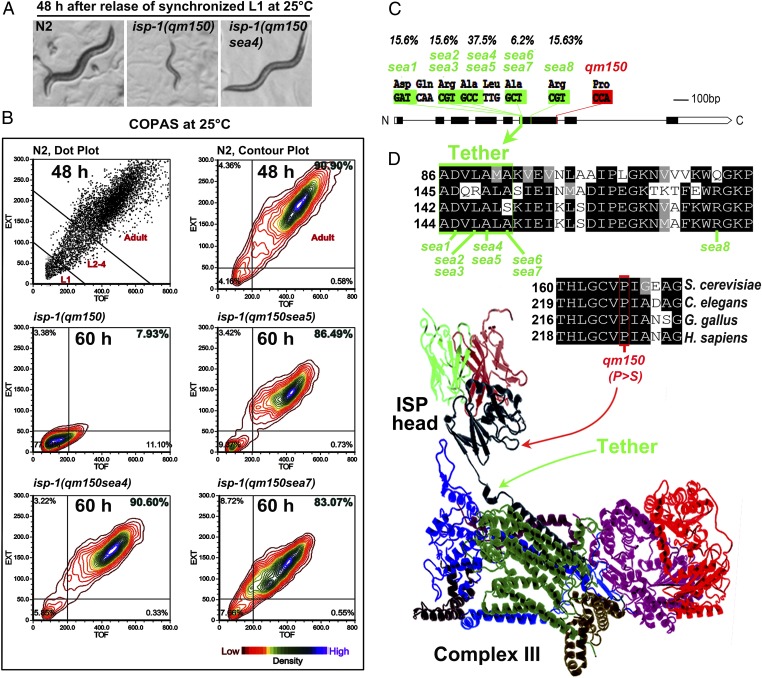
We performed a forward genetic screen to identify suppressors of the isp-1(qm150) slow development phenotype (Fig. 1A). At 25 °C isp-1(qm150) homozygotes require about 5 d to reach adulthood, whereas the N2 (wild type) and the suppressor mutants develop to adulthood in 2–3 d. Using ethyl methanesulfonate/N-ethyl-N-nitrosourea mutagenesis, ∼50,000 haploid genomes were screened and eight unique suppressor mutations within ISP-1 were isolated (Table 1 and SI Appendix, Table S1). These strains were serially backcrossed to isp-1(qm150) three times and then reassessed for their phenotypes.
Fig. 1.
Intragenic suppressors of isp-1(qm150) suppress the slow development and are located in the tether region. (A) Development of N2, isp-1(qm150), and one of the suppressors, 48 h after release of synchronized L1 at 25 °C. (B) Complex Object Parametric Analyzer and Sorter (COPAS) assessment of the size of the individuals in the population cultivated at 25 °C for 48 h for N2, or 60 h for isp-1(qm150), and the suppressor strains (n = 6,000 per strain). N2 were assayed at 48 h to prevent substantial contamination with progeny. (C) Structure of the isp-1 gene showing the positions, residues, and the incidence of the suppressor mutations in green and the P > S mutation in isp-1(qm150) in red. (D) Protein sequences of ISP from S. cerevisiae, C. elegans, Gallus gallus, and Homo sapiens, were aligned using ClustalW and represented with Boxshade. Amino acid numbers corresponding to the first shown residue are to the left of each segment. Functionally identical residues are indicated with black background. The proline that is mutated to serine in isp-1(qm150) is highlighted in red, and suppressor mutations within the tether region are in green background. Arrows in the ribbon structure of the cytochrome bc1 complex show the tether and P > S mutation locations.
Table 1.
Suppressor mutations in C. elegans MQ887 isp-1(qm150) background
| Supressor | Supressor mutations | Nucleotide change | Mutation location |
| isp-1(qm150sea1) | D146N | g > a | Tether |
| isp-1(qm150sea2) | R148C | c > t | Tether |
| isp-1(qm150sea3) | R148H | g > a | Tether |
| isp-1(qm150sea4) | A149T | g > a | Tether |
| isp-1(qm150sea5) | A149V | c > t | Tether |
| isp-1(qm150sea6) | A151D | c > a | Tether |
| isp-1(qm150sea7) | A151T | g > a | Tether |
| isp-1(qm150sea8) | R171C | c > t | Close to tether |
To determine the relative effect of each suppressor on developmental growth rate, we quantified the size of N2, isp-1(qm150), and suppressor strains at 25 °C using flow cytometry (Fig. 1B). L1 synchronized N2 animals reach adulthood about 40 h after plating on food, whereas isp-1(qm150) animals need about 100 h to develop to adulthood. The suppressor mutants were each able to reach to adult stage in 55–75 h after L1 release.
Seven out of the eight unique intragenic suppressor mutations were clustered in a six amino acid region (DQRALA) tether region of ISP-1 located 73 residues upstream of the P-to-S mutation in the isp-1(qm150) strain (Fig. 1C). Mutations at position 149 (alanine) were the most prevalent intragenic suppressor mutation found in our screen (Fig. 1C and SI Appendix, Table S1). The only intragenic suppressor outside the tether was located 20 amino acids downstream of the tether region, sea8, R171 (Fig. 1 C and D). The tether region of the ISP protein is highly conserved (17) (Fig. 1D and SI Appendix, Fig. S1). Because alanine residues in the ISP-1 tether region are highly conserved, and because all of the suppressor mutants grew at relatively similar rates, we focused our further analyses on A149T/V (sea4 and sea5 for comparison in the same residue) and A151T (sea7 for comparison in another residue).
Intragenic Suppressors of the isp-1(qm150) Developmental Delay also Suppress Defects in Fecundity, Motility, and Pumping.
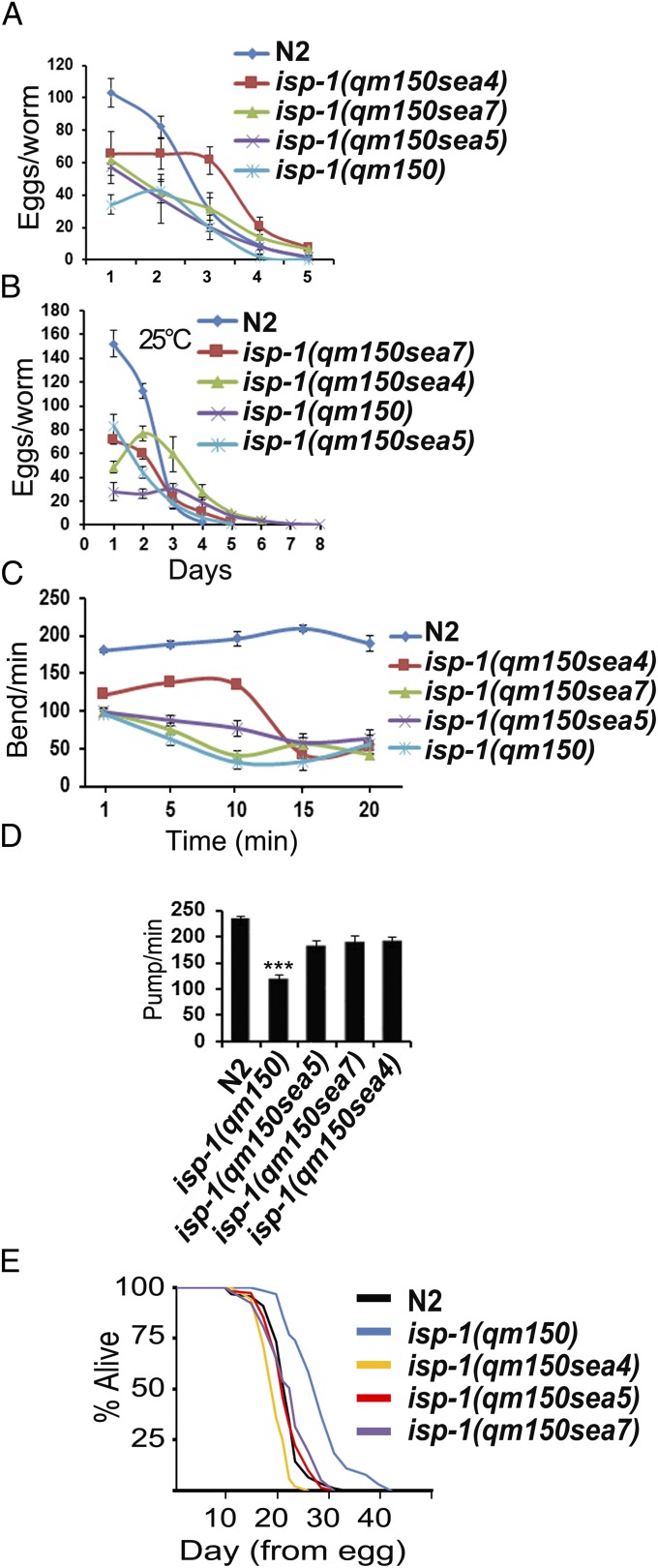
The isp-1(qm150) mutation causes a dramatic decrease in fecundity (brood size) compared with wild type (6). We measured the brood size of isp-1(qm150) animals carrying the sea4, sea5, or sea7 suppressor mutations at 20 °C and 25 °C, and found that all three suppressor strains produced significantly more progeny than isp-1(qm150) animals (Fig. 2 A and B and SI Appendix, Table S2). Likewise, increased motility was observed in sup mutants, as assessed by a “thrashing” (lateral swimming) assay (Fig. 2C and SI Appendix, Table S2). The isp-1(qm150) mutation also results in dramatic reductions in pharyngeal pumping rate and motility, relative to wild-type C. elegans (15). In every case examined, the suppressor mutations resulted in increased pharyngeal pumping rate compared with the isp-1(qm150) animals (Fig. 2D, SI Appendix, and Movies S1–S5).
Fig. 2.
Physiological parameters of the isp-1(qm150) suppressors. (A and B) Brood size of N2, isp-1(qm150), and one of the suppressors at two different temperatures (20 °C and 25 °C), n = 25 for each strain/condition. (C) Thrashing in the course of 20 min. (D) Pharyngeal pumping rates, n = 30 per strain, the two-tailed heteroscedastic t test was performed on two strains at a time in all 10 combinations to establish which differences in pumping rate were statistically significant. (E) Lifespan of the suppressors, at 25 °C. The depicted result is a single typical trial. N2 (n = 90) isp-1(qm150) (n = 65), isp-1(qm150sea4) (n = 53), isp-1(qm150sea5) (n = 158), isp-1(qm150sea7) (n = 198). The graph indicates animals that died during the course of the experiment. Animals that left the agar surface are not considered. No other censoring was performed. Error bars: SEM for highlighted experiments, *P < 0.05; **P < 0.01; ***P < 0.001.
Intragenic Suppressors of the isp-1(qm150) Developmental Delay also Suppress Lifespan Extension and Activation of the Mitochondrial Unfolded Protein Response.
We determined whether suppression of the slow development of the isp-1(qm150) mutant is associated with suppression of the lifespan extension. Animals carrying the developmental suppressor alleles also had reduced life span extension, with the degree of suppression dependent upon temperature. At 25 °C, the lifespan of isp-1(qm150sea5) and isp-1(qm150sea7) were similar to N2, and isp-1(qm150sea4) was short lived; whereas, at 15 °C, all sup mutations tested partially suppressed the lifespan extension of isp-1(qm150) (Fig. 2E and SI Appendix, Table S3).
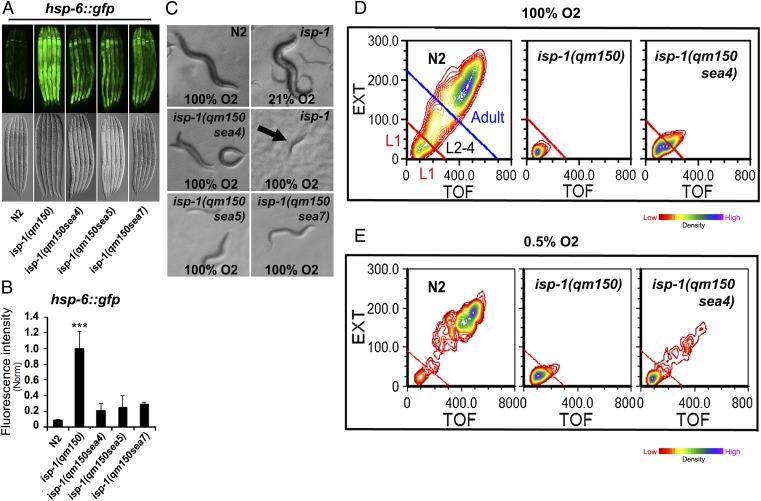
The mitochondrial unfolded protein response (mtUPR) is induced by mitochondrial stress in C. elegans, as assessed by expression of the hsp-6p::gfp reporter (18). We outcrossed the hsp-6p::gfp strain with the isp-1 strain and three of the suppressor strains. All three isp-1(qm150) suppressors reduced hsp-6p::gfp fluorescence in the isp-1(qm150); hsp-6p::gfp strain, indicating that activation of the mtUPR is also attenuated (Fig. 3 A and B).
Fig. 3.
Effects of intragenic suppressors on isp-1(qm150) induction of the mitochondrial unfolded protein response, sensitivity to hyperoxia, and sensitivity to hypoxia. (A) Suppressors attenuated the hsp-6p::gfp expression (mtUPR response). Epifluorescence and DIC Nomarski images of the worms expressing hsp-6p::gfp. (B) hsp-6p::gfp fluorescence intensity of the strains were compared by ImageJ software (50). (C) Intragenic suppressors partially rescued isp-1(qm150) hyperoxia lethality. N2 at 100% O2 produced next generation. isp-1(qm150) at 21% O2 produced next generation. isp-1(qm150) died at L1 stage at 100% O2. Intragenic suppressors reached to L3/4 stage under 100% O2. Images are from day eight from synchronized L1 seeded onto NGM-OP50 at 25 °C. (D and E) COPAS sorted worms in hyperoxia and hypoxia. N2 worms were sorted at day two and other strains at day five at 25 °C. Please note that L1 in N2 are from the next generation. Error bars: SEM for highlighted experiments, *P < 0.05 **P < 0.01 ***P < 0.001.
The isp-1(qm150) Mutation Causes Sensitivity to High and Low Oxygen That Is Suppressed by Mutations in the Tether Region.
Exposure to oxygen concentrations higher or lower than atmospheric levels (21%) can increase ROS production (19, 20). As previously reported (21), we observed that development and reproduction of wild-type animals were not significantly affected by hyperoxia (100% oxygen, 90 kPa). In contrast, a previously unreported defect was observed in hyperoxia when isp-1(qm150) showed a profound sensitivity to hyperoxia and failed to develop, ultimately dying as L1/2 larvae after 3–5 d. The tether region mutations conferred partial suppression of this phenotype, as animals carrying isp-1(qm150) suppressors developed to the L4 stage; however, they never reached the adult reproductive stage (Fig. 3 C and D, Table 2, and SI Appendix, Fig. S2).
Table 2.
Hyperoxia effect on development at 25 °C
| N2* | isp-1(qm150) | isp-1(qm150sea4) | isp-1(qm150sea5) | isp-1(qm150sea7) | ||||||
| Stage | Worms, # | Worms, % | Worms, # | Worms, % | Worms, # | Worms, % | Worms, # | Worms, % | Worms, # | Worms, % |
| L1 | 113 | 11.3 | 300 | 100 | 671 | 67.1 | 772 | 77.2 | 745 | 74.5 |
| L2-4 | 254 | 25.4 | 0 | 0 | 329 | 32.9 | 228 | 22.8 | 255 | 25.5 |
| Adult | 633 | 63.3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 1,000 | 100 | 300 | 100 | 1,000 | 100 | 1,000 | 100 | 1,000 | 100 |
N2 were sorted at day 2 (to avoid F1), and other strains were sorted at day 5.
In addition, hypoxia (1% oxygen; ref. 22) also can exacerbate ROS production in respiratory deficient mutants such as isp-1(qm150) (9, 23), and we observed that isp-1(qm150) animals arrested at the L1/2 developmental stage and died in hypoxia at 25 °C (Fig. 3E and SI Appendix, Fig. S3). Under this condition, tether region mutations allow isp-1(qm150) animals to develop further.
Mutations in the isp-1(qm150) Tether Increase Mitochondrial Function and Stabilize Respirasomes.
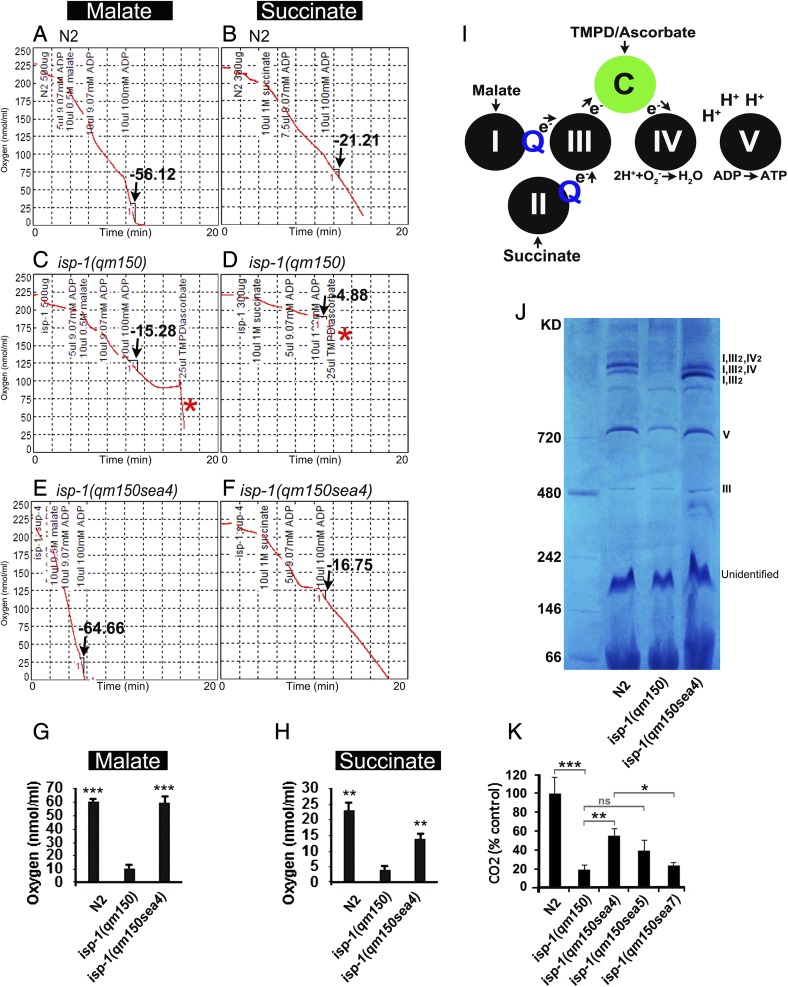
To directly assess the effect of the intragenic suppressors on the capacity for oxidative ATP production, we measured the state 3 (ADP-stimulated) respiration of intact isolated mitochondria. We compared the rates from mitochondria isolated from N2, isp-1(qm150), and one of the suppressors, isp-1(qm150sea4). Mitochondria were isolated from day 1 adults grown in normoxia at 21 ± 2 °C. We found that state 3 respiration in isp-1(qm150) mitochondria is impaired when electrons are donated from either malate or succinate substrates to the respiratory chain upstream of complex III (Fig. 4 A–F), as has been reported by others (24). In contrast, respiration is not affected when electrons are donated via ascorbate plus N,N,N′,N′-tetramethyl-p-phenylenediamine directly to cytochrome c (cyt c) downstream of complex III (Fig. 4 C and D, red asterisks are after adding TMPD/Ascorbate to bypass complex III). Intact respiration of isp-1(qm150) isolated mitochondria, through TMPD/Ascorbate confirms an electron transport defect in isp-1(qm150) at complex III. In isp-1(qm150sea4), state 3 respiration rates are significantly higher, approaching wild-type function, indicating a rescue of complex III (Fig. 4 A–H and SI Appendix, Fig. S4). In addition, isp-1(qm150) has been reported to affect complex I dependent respiration by destabilizing the respirasome supercomplex formed by complexes I, III, and IV (24). In Blue Native Gels, we observed diaphorase activity, commonly attributed to the presence of complex I, in I,III and I,III,IV supercomplex bands from all N2 and all isp-1(qm150; sea4) samples but not in isp-1(qm150) samples (Fig. 4J). We also analyzed the CO2 output of three suppressors (sea4, sea5, and sea7) and compared their CO2 output with N2 and isp-1(qm150). We observed that the isp-1(qm150) mutant produced significantly lower CO2 compared with N2, and the isp-1(qm150sea4) mutant partially restored the CO2 output (Fig. 4K and SI Appendix, Fig. S5 and Table S4).
Fig. 4.
ADP-stimulated respiration of intact isolated mitochondria. Oxygen consumption traces are shown in N2, isp-1(qm150), and isp-1(qm150sea4). (A) State 3 of ADP-stimulation rates (arrow) in N2 is measured by using malate substrate as an upstream (complex I) electron donor. State 3 respiration with 2 mM ADP represents the maximum capacity for oxidative phosphorylation of the mitochondria in N2. (B) Oxygen consumption traces in N2 by using succinate substrate as an upstream (complex II) electron donor. (C–F) comparison of state 3 of ADP-stimulation rates (arrows) in isp-1(qm150) and isp-1(qm150sea4) by using complex III upstream substrates (malate and succinate). In C and D, asterisks (*) show the oxygen consumption in isp-1(qm150) after adding TMPD/ascorbate (electron donor to cytochrome c, downstream of complex III). (G and H) Quantitative results for state 3 measurements of 3 biological replicates are depicted. P values are compared with isp-1(qm150) triplicates. P values in H: N2: 0.000204, isp-1(qm150sea4): 0.000707. P values in I: N2: 0.002405, isp-1(qm150sea4): 0.009303. Respiration of intact isolated mitochondria was measured with a Clark electrode. The traditional unit for the respiration rate, AO/min/mgprotein is equivalent to nmol 1/2O2/min/mgprotein (52). (I) A schematic cartoon depicting electron flow from the electron donor substrates (malate, succinate, or TMPD/ascorbate) through the mitochondrial ETC complexes. ADP stimulates respiration (state 3) because its phosphorylation to ATP by complex V allows proton flow, through complexes I, III, IV (out), and V (in). Note that electron flow from TMPD/ascorbate is independent of functional complex III. (J) Blue Native Gel. Complex I is present in supercomplex bands of N2 and isp-1(qm150sea4) but not in isp-1(qm150). (K) isp-1(qm150) CO2 output compared with the suppressors and N2 (for each strain: n = 1,000 COPAS sorted synchronized young adult animals of identical size (and likely weight) in three technical replicates (3000 worms/strain/condition). Error bars: SEM for highlighted experiments, *P < 0.05 **P < 0.01 ***P < 0.001.
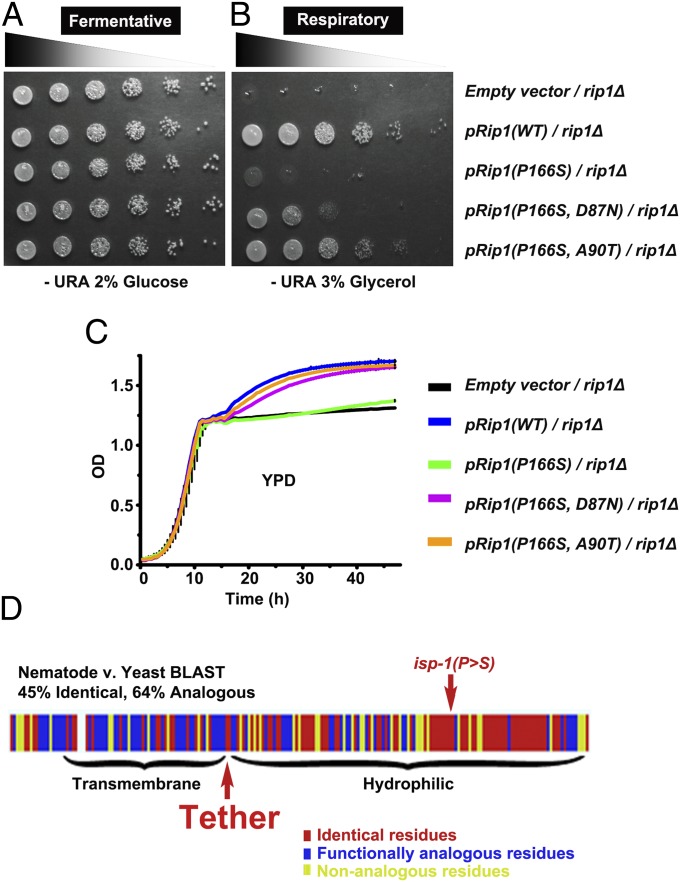
Yeast Expressing the Rip1(P166S) Mutant Protein Display a Respiratory Defect That Is Rescued by Mutations in the Tether Region.
To determine if the structure–function relationship of the isp-1(qm150) mutation and suppressors is conserved across phyla, we introduced analogous mutations into the Saccharomyces cerevisiae Rieske iron–sulfur protein, Rip1. Yeast rip1Δ mutants are capable of fermentative growth (Fig. 5A), but are respiratory deficient (25), as evidenced by a lack of growth on media containing nonfermentable glycerol as the only carbon source (Fig. 5B). To assess the effects of expression of Rip1 with and without suppressors, plasmids containing mutated RIP1 were introduced into haploid yeast lacking the RIP1 gene (rip1Δ). Reintroduction of a wild-type copy of the RIP1 gene rescues the rip1Δ respiratory defect, as expected. Yeast expressing Rip1 containing the P166S mutation, analogous to the worm isp-1(qm150) mutation, exhibited slowed growth under respiratory conditions, indicating this proline to serine mutation strongly diminishes mitochondrial respiratory function in yeast as well as worms. A D87N mutation partially suppressed the respiratory defect caused by the P166S mutation, whereas an A90T mutation rescued this defect more strongly (Fig. 5 A and B). Growth was also assayed under liquid culture conditions, where yeast switch from fermentative to respiratory metabolism upon exhaustion of glucose during a growth phase called diauxic shift (about 12 h of growth in Fig. 5C). Yeast expressing the P166S mutation grew poorly during the diauxic shift, and the suppressing mutations improved this defect. These findings mirror our results in C. elegans, demonstrating that the effects of the isp-1(qm150) mutation and our isolated suppressors are conserved (Fig. 5D).
Fig. 5.
Tether mutations rescue the respiratory defect of S. cerevisiae Rip1(P166S). Yeast lacking the entire RIP1 gene ORF (rip1Δ) were transformed with a plasmid containing empty vector, or plasmids expressing wild-type Rip1(WT), Rip1(P166S), or Rip1(P166S) with D87N (Sea1), or Rip1(P166S) with A90T(Sea4). Respiratory growth of yeast expressing Rip1(P166S) was dramatically impaired, which is rescued by the secondary mutations within the tether region, D87N or A90T. (A and B) Growth was assessed on solid synthetic media containing 2% (wt/vol) glucose (fermentative conditions; A) or 3% (wt/vol) glycerol (respiratory conditions; B). (C) Growth curves were generated from Bioscreen liquid culture experiments in rich media containing 2% (wt/vol) glucose. (D) Comparison of Rip1 conservation with isp-1.
Discussion
In this study, we identified several intragenic suppressors of isp-1(qm150) that not only enhanced the slow developmental rate of isp-1(qm150) animals, but also suppressed other phenotypes including pharyngeal pumping rate, brood size, body movement, mitochondrial unfolded protein response, CO2 production, mitochondrial oxidative phosphorylation (O2 consumption), lifespan extension, and a previously unreported sensitivity to hyperoxia (100% O2). Nearly all of the intragenic isp-1(qm150) suppressor mutations occurred in a small highly conserved tether region of the ISP protein composed of six amino acids DQRALA (D__A_A are conserved in α-proteobacteria, yeast, worm, chicken, and human). Crystal structures of complex III have revealed that the extrinsic mobile domain of ISP containing the 2Fe-2S iron–sulfur cluster can exist either near the electron donor site (Qo-site, catalytic site for ubihydroquinone oxidation) (26), or proximal to the electron acceptor site (cyt c1 position). Because the gap between these electron donors and the acceptor sites is too large to allow the observed rate of electron transfer (17, 27–31), movement of the head unit of ISP was postulated. Multiple studies support the notion that the tether region is a flexible element important in movement of the head region of ISP, with two (17, 32–34) or three (35) main locations for the mobile ISP head domain, either at the electron donor or at the acceptor site, or intermediate. Because the oxidized ISP (ISPox) is a substrate for the rate determining bifurcated reaction that determines the flux through the complex, the pleiotropic phenotypes of the isp-1(qm150) allele and their suppression likely reflect this role.
Previous efforts to identify suppressors of the isp-1(qm150) delayed development phenotype identified a mutation (A170V) in ctb-1, the mitochondrial encoded cytochrome b gene (6). One phenotypic difference between the identified ctb-1 suppressor qm189 and our tether region suppressors is that the lifespan extension of isp-1(qm150) was reported as unchanged by ctb-1(qm189), A170V (6). The reason for this difference is unclear, but may arise from the different subunit locations and from differential effects of these suppressors on complex III and its partner complexes within the respirasome. Suthammarak et al. (24) reported isp-1(qm150) strongly decreased not only complex III but also complex I activity measured in solubilized mitochondria. Suppressor ctb-1(qm189) did not rescue complex III activity but surprisingly improved complex I activity which suggested a rescue mechanism via allosteric effects within the supercomplex. By contrast, our tether region suppressor qm150sea4 restored oxidative phosphorylation of structurally intact mitochondria regardless whether the electrons entered the ETC via complex I or II, indicating rescue of complex III activity. Moreover, normal complex I-dependent respiration in qm150sea4 mitochondria suggested if qm150 caused an additional complex I deficiency, this was rescued too. (Our polarography method did not allow to verify whether a combined complex III and I deficiency actually existed in qm150.)
It is noteworthy that Saccharomyces yeast does not have a true complex I, but instead relies on three single enzyme NADH dehydrogenases (Nde1, Nde2, and Ndi1) to perform this activity. Because the yeast D87N (sea1) and A90T (sea4) mutations were sufficient to rescue the respiratory defect caused by the P166S mutation in Rip1, this further suggests that these tether region mutations are sufficient to restore complex III activity. By homology with the yeast structure, the ctb-1 A170 residue is located in a clamp, contributed by spans from cyt b (both subunits of the dimer) and cyt c1, that holds ISP as it emerges from the membrane at the C-terminal end of the anchoring helix (27). In yeast, the equivalent residue, P174, is adjacent to the ISP tether span -TADV-. Because the clamp constrains the tether, changes may alter control of ISP head movement (6, 35), which could therefore be taken as supporting a model in which a slow conformational change of the isp-1(qm150) head region may account for the reduced ETC function and observed phenotypes (slow development, slow defecation, slow locomotion, etc.).
In yeast, our data illustrate that mutation of proline 166 to serine results in drastically diminished respiratory function, and that secondary intragenic mutations within the tether region (D87N and A90T) can rescue this defect. Another mutation, G175S, located within the same intervening region as P166, has also been reported to reduce the respiratory capacity of yeast, though in a temperature-sensitive manner (36). Due to the location of P166 and the structural rigidity that proline provides, this residue may be important for maintaining the tertiary structure of the loop. Notably, mutation of this proline residue to leucine has previously been found to lower the midpoint redox potential of the iron–sulfur cluster (37), and mutations of the tether region have been shown to raise the midpoint potential of the iron–sulfur cluster (38). However, although these opposing effects correlate with the longevity and suppressor phenomena, studies of the molecular basis of the changes in redox potential, especially from tether mutations, show that they do not involve a direct effect on the cluster, but are mediated by structure–function interactions.
Physicochemical parameters from a wider literature prompted us to examine the interplay between relatively weak binding of the ISP head domain at the Qo-site in the wild type, and forces associated with conformational changes in the tether region, which support an alternative spring-loaded mechanism (39). In brief, we suggest that when complexes of ISP form with Qo site occupants, two sets of counteracting forces are in play, with one set (the forces associated with formation of the complex), pulling on the cluster domain, and the other set (the “spring”), pulling on the tether at the other end of the extrinsic domain. Complex formation is in concert with changes between extended and relaxed conformations of the tether-span spring, with all forces mediated by H-bonding and van der Waals interactions, which are differentially modified on mutation in these regions to give the effects observed.
The ISP head domain is involved in formation of complexes with a variety of occupants of the Qo-site. These include (i) the enzyme–substrate complex (ES-complex) between QH2 and ISPox (the 2Fe-2S cluster oxidized) that initiates the bifurcated reaction; (ii) an enzyme-product (EP-) complex between ISPH (the cluster reduced) and Q; and (iii) complexes between ISPH and inhibitors, exemplified by UHDBT and stigmatellin. (iv) Structures have also been solved for a different type of inhibitors, exemplified by myxothiazol and MOA-type inhibitors, which bind deeper in the Qo-site and do not interact with ISP. Complexes ii and iii involving ISPH can be assayed through their EPR spectra because the reduced cluster is paramagnetic, and interaction induces a new line in the spectrum. On this basis, physicochemical properties involved in formation of these complexes can be determined, as detailed in the SI Appendix. In particular, because formation of a complex pulls the reduced form (ISPH) out of the redox mix, in each of these cases, the redox potential, Em, is raised, and the extent provides a measure of the strength of binding, (SI Appendix, Table S5). The binding of substrates to form the ES-complex can also be assayed, but because the state is metastable, only through kinetic studies. In each of these complexes, a main contribution to the binding energy comes from formation of an H-bond between ISP and the site occupant. The histidine, which ligands the cluster through Nδ, can serve as H-acceptor (-Nε—HO-) when ISPox is involved or as H-donor (-NεH—O=) when ISPH is involved.
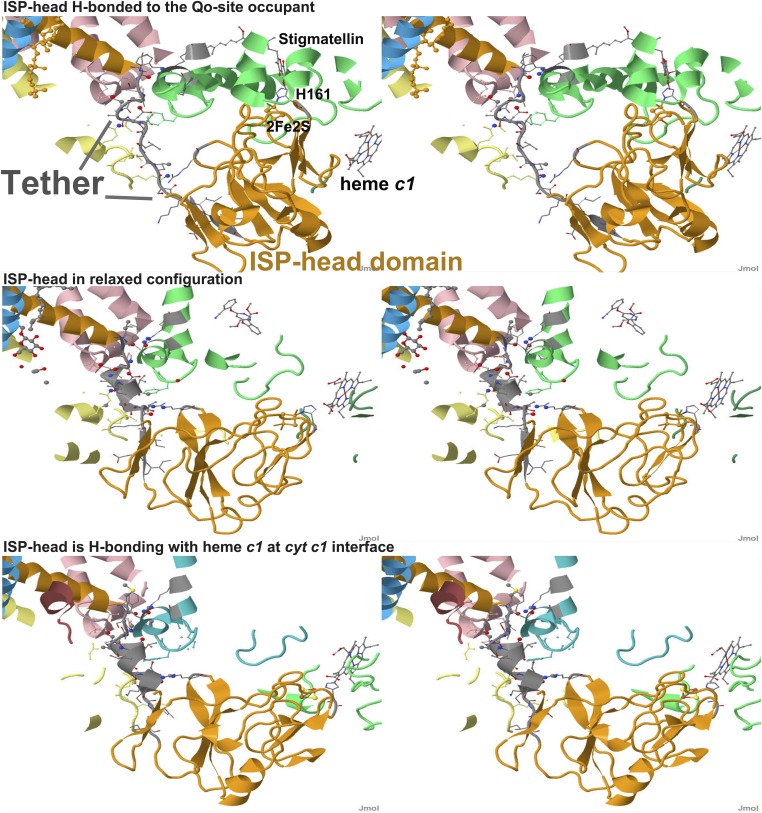
When a complex is formed (17, 27–29, 40), structures show that the ISP head is tightly packed at an interface with cyt b. Otherwise, the ISP head is rotated away, and the histidine makes no H-bond (Fig. 6). These different positions are correlated to changes in the tether span connecting the extrinsic domain to its anchoring helix. When ISP is bound to a Qo-site occupant, the tether is pulled into an extended configuration, but when not, the tether collapses to a helical configuration. From these and similar studies in mutant strains (30, 41, 42), a possible mechanism becomes obvious (39). In normal complex formation, the binding is balanced against a restraining force, mainly from H-bonds in the helix, broken in the extension (SI Appendix, Table S6). When additional residues are inserted to introduce slack, or when the helical configuration is discouraged by replacing residues by helical breakers (glycine or proline), the tether can be extended with less effort, and the apparent strength of the H-bond stabilizing the complex, , increases. When the tether is so modified that the spring loses its pull, the value of is equal to the loss of the balancing force. The physicochemical underpinning of this hypothesis is discussed at length in the SI Appendix.
Fig. 6.
Three configurations of the extrinsic domain of ISP show the spring-loaded mechanism in action. (Top) ISP docked at cyt b interface, H-bonded to the Qo-site occupant [Protein Data Bank (PDB) ID 3H1K, avian complex, stigmatellin bound]. The main features of interest are labeled. (Middle) ISP in relaxed configuration [PDB ID 3L71, avian complex, azoxystrobin (MOA-inhibitor) bound]. (Bottom) ISP H-bonding with heme c1 at cyt c1 interface (PDB ID 1BE3, bovine complex, no Qo-site occupant indicated). In the relaxed configuration (Middle), the cluster ligand (H161 in bovine) makes no H-bonding contacts, so the configuration in the tether region is not under tension. When the H161 H-bonds to the Qo-site occupant (Top), the tether is stretched due to application of a force (mainly from the strength of the H-bond to the occupant) to lengthen the tether and rotate the head. The helical region is pulled into an extended configuration, leading to the breaking of several intra- and interspan H-bonds. When H161 H-bonds to heme c1, the displacements are much smaller, and the tether region retains a H-bonding configuration similar to that of the relaxed state. SI Appendix, Fig. S6 and Table S6 shows details of H-bonding changes between Qo-site docked (stigmatellin) and relaxed (azoxystrobin) configurations. Chains are colored as follows: chain c, cyt b, pink; chain d, cyt c1, yellow; chain e, ISP, orange; chain j, 7.2-kDa protein, cyan; chain p, cyt b, green (or blue-green in 1BE3); chain q, cyt c1, olive (or light green in 1BE3). Gray spans have the cartoon colored by the CPK coloring of the C-atom or C-C bond. Atoms within 7 Å of a neighboring chain are shown as space-filled spheres of 0.2-Å radius. Atoms within 3.5 Å of a neighboring chain are shown as space-filled spheres of 0.4-Å radius. Selected atoms are CPK colored (by atom type). The three structures are shown with the protein scaffold in the same orientation, so that the main difference comes from the configurational changes of the ISP tether and extrinsic domains. Stereoscopic images for crossed-eye viewing.
Similar balancing forces will be in play in formation of the ES-complex, and these are more pertinent to the present discussion since this initiates forward electron transfer. Mutations in the head domain at the interface with cyt b likely inhibit formation of the ES-complex by steric hindrance, decreasing the binding energy available from the H-bond by unfavorable VDW interactions, and thus changing the balance of spring-loading. By limiting ES-complex formation, such mutations, including isp-1(qm150), would restrict the input flux to the Qo-site, and therefore the occupancy of SQo, without affecting the reactions for its removal. This educed occupancy would lower ROS production, perhaps contributing to the longevity-associated effects. In addition, the reduced flux would propagate to the energy metabolism of the cell, lowering the proton gradient, and hence the poise of the ATP synthase, leading to substantial metabolic and physiological effects, which could plausibly account for many pleiotropic phenotypes. Then, all of the suppressor function from tether mutations can be readily explained by an easing of the tension in the spring so that the ES-complex could be formed more easily, and the input flux restored. Additional support for this simple mechanism from earlier mutational studies and a possible role for spring-loading in control and gating are discussed in the SI Appendix.
In humans, specific mutations of cyt b have been reported in patients with severe cardiomyopathy (43) and exercise intolerance (44). Introduction of human counterpart mutations into yeast cyt b results in respiratory defects (45). Multiple suppressors of the respiratory deficient phenotype from cyt b S152P and G291D mutations (both at the docking interface for ISP) were identified within in the tether region of Rip1 or in the region within cyt b located near the Rip1 tether (45). Similar to our findings, many of these suppressors were in alanine residues of the tether, which may be important for flexibility of the tether region. Interestingly, mutation of A90T was among the stronger suppressors of the cyt b G291D respiratory defect, and our data indicate that this mutation (sea4, A149T) was the most potent suppressor identified from our screen in C. elegans. Collectively, these findings highlight the importance of the tether region in modulating the interaction of ISP with cyt b within complex III.
In this study, we have shown that the functional consequences and interactions of mutations in the Rieske iron–sulfur protein of mitochondrial complex III of C. elegans are replicated in the highly evolutionarily diverged unicellular model organism, S. cerevisiae. Our data provide more evidence that intragenic mutations within the tether region can rescue respiratory defects in different organisms. We have provided, to our knowledge for the first time, a framework for discussion of the mechanistic features underlying these effects, which brings a molecular understanding of the role of complex III in mediating effects on metabolic rate, embryonic and post embryonic lethality, developmental delay, defecation cycle, fecundity, stress resistance, and longevity.
Structural mechanistic analysis and discussion on the proposed spring-loaded model is available in the SI Appendix.
Experimental Procedures
Strains and Growth Conditions.
Standard C. elegans procedures were used as described (46–48). Worms were maintained on solid Nematode Growth Medium (NGM) and grown at 20 °C unless otherwise stated. To have a synchronized L1 population, eggs were hatched at room temperature (∼20–22 °C), in M9 overnight. The postembryonic development assay screen was done at 25 °C on solid NGM, seeded with UV arrested OP50 (49). Strains are described in SI Appendix.
EMS/ENU Mutagenesis Screen.
Ten thousand P0 isp-1(qm150) L4 were mutagenized with 50 mM EMS (Sigma M0880-5G) + 1 mM ENU (Sigma N3385-1G) for 6 h. Then the P0 animals were distributed on 10 individual 15-cm plates. F1 animals were grown to adulthood, and their eggs (F2) were obtained by the bleach method. For each plate, 3,000 F2 eggs were seeded onto each new NGM plate. After 72 h at 25 °C, the plates were screened for any fast growing animals. From each P0 population, only one fast growing worm was selected.
Lifespan Analyses.
Lifespan analyses were performed as described (49) (SI Appendix).
Microscopy.
Pictures were taken immediately after slide preparation using a Nikon Eclipse E600 microscope using a QImaging Retigia EX CCD Camera and the fluorescent intensity was analyzed using ImageJ (50).
Hypoxia (0.5% O2) and Hyperoxia (100% O2).
Worms were synchronized by egg-bleach preparation (51) and placed onto NGM into atmospheric chambers at 25 °C in an incubator and raised. Atmospheres were humidified and gas was continually flowed at a rate of one chamber volume per minute. Hypoxic and hyperoxic atmospheres were generated using combinations of compressed air, 100% oxygen and 100% nitrogen mixed with mass flow controllers and analyzed with a paramagnetic oxygen analyzer that was spanned using oxygen standards. Animals were scored dead if they could not move after evoked touch (SI Appendix).
CO2 Production Assay.
One thousand synchronized young adults were sorted by COPAS Biosort (Union Biometrica) in three technical replicates (3,000 worms in total) and placed on agar pads in atmospheric chambers in the absence of food. The atmospheric chambers were flushed with CO2 free air for 15 min then sealed for 15 min. The expired CO2 was measured by LI-COR 6262 CO2 analyzer (SI Appendix).
Brood Size Assay.
Brood size assays were performed at 20 °C by picking 25 L4s onto fresh NGM plates (5 L4 per plate) and transferring them to new plates every 24 h for 4–7 d. Progeny plates were incubated at 20 °C for about 2 d and then scored.
Thrashing Assay.
For each strain, 10 L4s were placed onto an NGM plate seeded with OP50. The thrashing assay was performed by transferring 1 L4 to an unseeded plate containing 1.6 mL of M9 buffer and thrashes were scored with the intervals of 1 min, 5 min, 10 min, 15 min, and 20 min for the duration of 1 min each.
Pharyngeal Pumping Assay.
L4 nematodes were acclimated 20 h on NGM-OP50. The assay was performed within a single day at ∼22 °C (room temperature). Pharyngeal contractions of 30 nematodes for each strain were counted over 10-s intervals in three cycles of 10:10:10:10:10 with and without the aid of reduced speed video, and these measurements were extrapolated into pumps per minute.
Worm Sorting.
The COPAS BIOSORT (Union Biometrica) was used to differentiate the different animals based on size. Time of Flight vs. Extinction were used as gating parameters. The worms were gated to exclude eggs and debris according to COPAS Application Note B-03.
Mitochondrial Fraction Isolation and Respiratory Capacity Assessment.
Measurements of oxidative phosphorylation capacity on freshly isolated mitochondria and Blue Native gel assays were performed as published (52–54) with the modification that animals were grown on 8P plates (55), and nylon woven mesh sheet 30, 40, and 110 microns were used to clean and sort worms based on size.
Yeast Assays.
S. cerevisiae haploid strains were all derived from isogenic BY4741 MATa or BY4742 MATα backgrounds. Yeast lacking rip1 (strain BW998) were obtained from the yeast deletion collection and the presence of the deletion was confirmed by PCR (Open Biosystems). Solid media was CSM-URA supplemented with 2% (wt/vol) glucose and cell number was normalized by equalizing the optical density of each sample. Cells were fivefold serially diluted in water, and 4 µL of each dilution was pipetted onto the indicated solid media containing 2% (wt/vol) agar. For liquid growth experiments, overnight cultures were grown in CSM-URA 2% (wt/vol) glucose liquid media and 5 µL of these overnight cultures were used to inoculate 145 µL of fresh media consisting of 1% (wt/vol) yeast extract, 2% (wt/vol) peptone, and 2% (wt/vol) glucose. Growth rate was assayed using a Bioscreen C MBR instrument (Growth Curves) as described (56, 57).
Supplementary Material
Acknowledgments
Some strains were provided by the Caenorhabditis Genetics Center, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). B.M.W. was supported by NIH Training Grant T32ES007032. J.N.P. was supported by NIH Training Grant T32AG000057. This work was supported by NIH Grant R01AG039390 (to M.K.), the University of Washington Nathan Shock Center of Excellence in the Basic Biology of Aging (NIH P30AG013280), the Healthy Aging and Longevity Research Institute, and an award from the Murdock Charitable Trust (to M.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509416112/-/DCSupplemental.
References
- 1.Wallace DC. Mitochondrial DNA mutations in disease and aging. Environ Mol Mutagen. 2010;51(5):440–450. doi: 10.1002/em.20586. [DOI] [PubMed] [Google Scholar]
- 2.Bratic A, Larsson NG. The role of mitochondria in aging. J Clin Invest. 2013;123(3):951–957. doi: 10.1172/JCI64125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butler JA, Mishur RJ, Bhaskaran S, Rea SL. A metabolic signature for long life in the Caenorhabditis elegans Mit mutants. Aging Cell. 2013;12(1):130–138. doi: 10.1111/acel.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Jong L, Meng Y, Dent J, Hekimi S. Thiamine pyrophosphate biosynthesis and transport in the nematode Caenorhabditis elegans. Genetics. 2004;168(2):845–854. doi: 10.1534/genetics.104.028605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felkai S, et al. CLK-1 controls respiration, behavior and aging in the nematode Caenorhabditis elegans. EMBO J. 1999;18(7):1783–1792. doi: 10.1093/emboj/18.7.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng J, Bussière F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev Cell. 2001;1(5):633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- 7.Lakowski B, Hekimi S. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 1996;272(5264):1010–1013. doi: 10.1126/science.272.5264.1010. [DOI] [PubMed] [Google Scholar]
- 8.Ventura N, et al. p53/CEP-1 increases or decreases lifespan, depending on level of mitochondrial bioenergetic stress. Aging Cell. 2009;8(4):380–393. doi: 10.1111/j.1474-9726.2009.00482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee SJ, Hwang AB, Kenyon C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr Biol. 2010;20(23):2131–2136. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan MH, et al. TAF-4 is required for the life extension of isp-1, clk-1 and tpk-1 Mit mutants. Aging (Albany, NY) 2013;5(10):741–758. doi: 10.18632/aging.100604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walter L, Baruah A, Chang HW, Pace HM, Lee SS. The homeobox protein CEH-23 mediates prolonged longevity in response to impaired mitochondrial electron transport chain in C. elegans. PLoS Biol. 2011;9(6):e1001084. doi: 10.1371/journal.pbio.1001084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker BM, Nargund AM, Sun T, Haynes CM. Protective coupling of mitochondrial function and protein synthesis via the eIF2α kinase GCN-2. PLoS Genet. 2012;8(6):e1002760. doi: 10.1371/journal.pgen.1002760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144(1):79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennett CF, et al. Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans. Nat Commun. 2014;5:3483. doi: 10.1038/ncomms4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yee C, Yang W, Hekimi S. The intrinsic apoptosis pathway mediates the pro-longevity response to mitochondrial ROS in C. elegans. Cell. 2014;157(4):897–909. doi: 10.1016/j.cell.2014.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwata S, Saynovits M, Link TA, Michel H. Structure of a water soluble fragment of the ‘Rieske’ iron-sulfur protein of the bovine heart mitochondrial cytochrome bc1 complex determined by MAD phasing at 1.5 A resolution. Structure. 1996;4(5):567–579. doi: 10.1016/s0969-2126(96)00062-7. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Z, et al. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392(6677):677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 18.Yoneda T, et al. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J Cell Sci. 2004;117(Pt 18):4055–4066. doi: 10.1242/jcs.01275. [DOI] [PubMed] [Google Scholar]
- 19.Mansfield KD, et al. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-alpha activation. Cell Metab. 2005;1(6):393–399. doi: 10.1016/j.cmet.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jamieson D. Oxygen toxicity and reactive oxygen metabolites in mammals. Free Radic Biol Med. 1989;7(1):87–108. doi: 10.1016/0891-5849(89)90103-2. [DOI] [PubMed] [Google Scholar]
- 21.Van Voorhies WA, Ward S. Broad oxygen tolerance in the nematode Caenorhabditis elegans. J Exp Biol. 2000;203(Pt 16):2467–2478. doi: 10.1242/jeb.203.16.2467. [DOI] [PubMed] [Google Scholar]
- 22.Guzy RD, et al. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1(6):401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 23.Guzy RD, Schumacker PT. Oxygen sensing by mitochondria at complex III: The paradox of increased reactive oxygen species during hypoxia. Exp Physiol. 2006;91(5):807–819. doi: 10.1113/expphysiol.2006.033506. [DOI] [PubMed] [Google Scholar]
- 24.Suthammarak W, Morgan PG, Sedensky MM. Mutations in mitochondrial complex III uniquely affect complex I in Caenorhabditis elegans. J Biol Chem. 2010;285(52):40724–40731. doi: 10.1074/jbc.M110.159608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beckmann JD, Ljungdahl PO, Lopez JL, Trumpower BL. Isolation and characterization of the nuclear gene encoding the Rieske iron-sulfur protein (RIP1) from Saccharomyces cerevisiae. J Biol Chem. 1987;262(18):8901–8909. [PubMed] [Google Scholar]
- 26.Brandt U, Haase U, Schägger H, von Jagow G. Significance of the “Rieske” iron-sulfur protein for formation and function of the ubiquinol-oxidation pocket of mitochondrial cytochrome c reductase (bc1 complex) J Biol Chem. 1991;266(30):19958–19964. [PubMed] [Google Scholar]
- 27.Crofts AR, et al. Mechanism of ubiquinol oxidation by the bc(1) complex: Role of the iron sulfur protein and its mobility. Biochemistry. 1999;38(48):15791–15806. doi: 10.1021/bi990961u. [DOI] [PubMed] [Google Scholar]
- 28.Crofts AR, et al. Mechanism of ubiquinol oxidation by the bc(1) complex: Different domains of the quinol binding pocket and their role in the mechanism and binding of inhibitors. Biochemistry. 1999;38(48):15807–15826. doi: 10.1021/bi990962m. [DOI] [PubMed] [Google Scholar]
- 29.Crofts AR, Hong S, Zhang Z, Berry EA. Physicochemical aspects of the movement of the rieske iron sulfur protein during quinol oxidation by the bc(1) complex from mitochondria and photosynthetic bacteria. Biochemistry. 1999;38(48):15827–15839. doi: 10.1021/bi990963e. [DOI] [PubMed] [Google Scholar]
- 30.Darrouzet E, Valkova-Valchanova M, Moser CC, Dutton PL, Daldal F. Uncovering the [2Fe2S] domain movement in cytochrome bc1 and its implications for energy conversion. Proc Natl Acad Sci USA. 2000;97(9):4567–4572. doi: 10.1073/pnas.97.9.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moser CC, Keske JM, Warncke K, Farid RS, Dutton PL. Nature of biological electron transfer. Nature. 1992;355(6363):796–802. doi: 10.1038/355796a0. [DOI] [PubMed] [Google Scholar]
- 32.von Jagow G, Ohnishi T. The chromone inhibitor stigmatellin--binding to the ubiquinol oxidation center at the C-side of the mitochondrial membrane. FEBS Lett. 1985;185(2):311–315. doi: 10.1016/0014-5793(85)80929-7. [DOI] [PubMed] [Google Scholar]
- 33.Matsuura K, Bowyer JR, Ohnishi T, Dutton PL. Inhibition of electron transfer by 3-alkyl-2-hydroxy-1,4-naphthoquinones in the ubiquinol-cytochrome c oxidoreductases of Rhodopseudomonas sphaeroides and mammalian mitochondria. Interaction with a ubiquinone-binding site and the Rieske iron-sulfur cluster. J Biol Chem. 1983;258(3):1571–1579. [PubMed] [Google Scholar]
- 34.Bowyer JR, Dutton PL, Prince RC, Crofts AR. The role of the Rieske iron-sulfur center as the electron donor to ferricytochrome c2 in Rhodopseudomonas sphaeroides. Biochim Biophys Acta Bioenergetics. 1980;592(3):445–460. doi: 10.1016/0005-2728(80)90091-2. [DOI] [PubMed] [Google Scholar]
- 35.Iwata S, et al. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science. 1998;281(5373):64–71. doi: 10.1126/science.281.5373.64. [DOI] [PubMed] [Google Scholar]
- 36.Beckmann JD, Ljungdahl PO, Trumpower BL. Mutational analysis of the mitochondrial Rieske iron-sulfur protein of Saccharomyces cerevisiae. I. Construction of a RIP1 deletion strain and isolation of temperature-sensitive mutants. J Biol Chem. 1989;264(7):3713–3722. [PubMed] [Google Scholar]
- 37.Gatti DL, Meinhardt SW, Ohnishi T, Tzagoloff A. Structure and function of the mitochondrial bc1 complex. A mutational analysis of the yeast Rieske iron-sulfur protein. J Mol Biol. 1989;205(2):421–435. doi: 10.1016/0022-2836(89)90352-5. [DOI] [PubMed] [Google Scholar]
- 38.Darrouzet E, Daldal F. Movement of the iron-sulfur subunit beyond the ef loop of cytochrome b is required for multiple turnovers of the bc1 complex but not for single turnover Qo site catalysis. J Biol Chem. 2002;277(5):3471–3476. doi: 10.1074/jbc.M107974200. [DOI] [PubMed] [Google Scholar]
- 39.Crofts AR, Shinkarev VP, Dikanov SA, Samoilova RI, Kolling D. Interactions of quinone with the iron-sulfur protein of the bc(1) complex: Is the mechanism spring-loaded? Biochim Biophys Acta. 2002;1555(1-3):48–53. doi: 10.1016/s0005-2728(02)00253-0. [DOI] [PubMed] [Google Scholar]
- 40.Esser L, et al. Crystallographic studies of quinol oxidation site inhibitors: A modified classification of inhibitors for the cytochrome bc(1) complex. J Mol Biol. 2004;341(1):281–302. doi: 10.1016/j.jmb.2004.05.065. [DOI] [PubMed] [Google Scholar]
- 41.Darrouzet E, Daldal F. Protein-protein interactions between cytochrome b and the Fe-S protein subunits during QH2 oxidation and large-scale domain movement in the bc1 complex. Biochemistry. 2003;42(6):1499–1507. doi: 10.1021/bi026656h. [DOI] [PubMed] [Google Scholar]
- 42.Darrouzet E, Valkova-Valchanova M, Daldal F. The [2Fe-2S] cluster Em as an indicator of the iron-sulfur subunit position in the ubihydroquinone oxidation site of the cytochrome bc1 complex. J Biol Chem. 2002;277(5):3464–3470. doi: 10.1074/jbc.M107973200. [DOI] [PubMed] [Google Scholar]
- 43.Andreu AL, Checcarelli N, Iwata S, Shanske S, DiMauro S. A missense mutation in the mitochondrial cytochrome b gene in a revisited case with histiocytoid cardiomyopathy. Pediatr Res. 2000;48(3):311–314. doi: 10.1203/00006450-200009000-00008. [DOI] [PubMed] [Google Scholar]
- 44.Dumoulin R, et al. A novel gly290asp mitochondrial cytochrome b mutation linked to a complex III deficiency in progressive exercise intolerance. Mol Cell Probes. 1996;10(5):389–391. doi: 10.1006/mcpr.1996.0053. [DOI] [PubMed] [Google Scholar]
- 45.Fisher N, et al. Human disease-related mutations in cytochrome b studied in yeast. J Biol Chem. 2004;279(13):12951–12958. doi: 10.1074/jbc.M313866200. [DOI] [PubMed] [Google Scholar]
- 46.Kaeberlein TL, et al. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell. 2006;5(6):487–494. doi: 10.1111/j.1474-9726.2006.00238.x. [DOI] [PubMed] [Google Scholar]
- 47.Smith ED, et al. Age- and calorie-independent life span extension from dietary restriction by bacterial deprivation in Caenorhabditis elegans. BMC Dev Biol. 2008;8:49. doi: 10.1186/1471-213X-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sutphin GL, Kaeberlein M. Dietary restriction by bacterial deprivation increases life span in wild-derived nematodes. Exp Gerontol. 2008;43(3):130–135. doi: 10.1016/j.exger.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sutphin GL, Kaeberlein M. Measuring Caenorhabditis elegans life span on solid media. J Vis Exp. 2009;(27):1152. doi: 10.3791/1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stiernagle T. Maintenance of C. elegans. WormBook. 2006 doi: 10.1895/wormbook.1.101.1. Available at www.ncbi.nlm.nih.gov/books/NBK19649/. Accessed February 15, 2014. [DOI] [PMC free article] [PubMed]
- 52.Falk MJ, Kayser EB, Morgan PG, Sedensky MM. Mitochondrial complex I function modulates volatile anesthetic sensitivity in C. elegans. Curr Biol. 2006;16(16):1641–1645. doi: 10.1016/j.cub.2006.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kayser EB, Hoppel CL, Morgan PG, Sedensky MM. A mutation in mitochondrial complex I increases ethanol sensitivity in Caenorhabditis elegans. Alcohol Clin Exp Res. 2003;27(4):584–592. doi: 10.1097/01.ALC.0000060524.62805.D2. [DOI] [PubMed] [Google Scholar]
- 54.Johnson SC, et al. mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science. 2013;342(6165):1524–1528. doi: 10.1126/science.1244360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bianchi L, Driscoll M. Culture of embryonic C. elegans cells for electrophysiological and pharmacological analyses. WormBook. 2006 doi: 10.1895/wormbook.1.122.1. Available at www.ncbi.nlm.nih.gov/books/NBK19713/. Accessed February 15, 2014. [DOI] [PMC free article] [PubMed]
- 56.Olsen B, Murakami CJ, Kaeberlein M. YODA: Software to facilitate high-throughput analysis of chronological life span, growth rate, and survival in budding yeast. BMC Bioinformatics. 2010;11:141. doi: 10.1186/1471-2105-11-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murakami C, Kaeberlein M. Quantifying yeast chronological life span by outgrowth of aged cells. J Vis Exp. 2009;(27):1156. doi: 10.3791/1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.