Significance
Eukaryotic genomes are compacted into chromosomes, in which heterochromatin is generally considered to be distinct from euchromatin in chromosomal packaging levels and locations. In Drosophila, heterochromatin is mainly found in pericentric and telomeric regions. In this study, we show that heterochromatin landscapes that interspersed in euchromatic arms are counteracted by CDK12, a major RNA polymerase II C-terminal domain kinase. After the depletion of CDK12, heterochromatin enrichment can be observed on euchromatic arms, especially on the X chromosome, which leads to transcriptional attenuation in targeted genes and defects in neuronal functions. Our findings provide insights into the regulation of heterochromatin domain in the natural chromosomal context and suggest a chromatin regulatory role of CDK12 in neuronal functions.
Keywords: epigenetic transition, HP1, RNAPII C-terminal domain kinase, histone H3 on Lys9 methylation, neuronal function
Abstract
Dynamic regulation of chromatin structure is required to modulate the transcription of genes in eukaryotes. However, the factors that contribute to the plasticity of heterochromatin structure are elusive. Here, we report that cyclin-dependent kinase 12 (CDK12), a transcription elongation-associated RNA polymerase II (RNAPII) kinase, antagonizes heterochromatin enrichment in Drosophila chromosomes. Notably, loss of CDK12 induces the ectopic accumulation of heterochromatin protein 1 (HP1) on euchromatic arms, with a prominent enrichment on the X chromosome. Furthermore, ChIP and sequencing analysis reveals that the heterochromatin enrichment on the X chromosome mainly occurs within long genes involved in neuronal functions. Consequently, heterochromatin enrichment reduces the transcription of neuronal genes in the adult brain and results in a defect in Drosophila courtship learning. Taken together, these results define a previously unidentified role of CDK12 in controlling the epigenetic transition between euchromatin and heterochromatin and suggest a chromatin regulatory mechanism in neuronal behaviors.
Appropriate regulation of gene transcription is essential throughout the life of an organism. In eukaryotes, DNA and histone octamers are assembled into nucleosomes and further compacted into higher-order structures (1). Dynamic changes in the chromatin architecture affect gene transcription in many aspects of developmental and physiological processes, such as neural plasticity, memory formation, and cognition (2, 3). Eukaryotic genomes are composed of two basic forms, euchromatin and heterochromatin, which are originally characterized by their cytological chromatin packaging levels. Euchromatic regions are generally associated with a relatively open chromatin configuration and mainly contain transcriptionally active genes, whereas heterochromatin is highly compacted and less accessible to the transcriptional machinery (4). Drosophila heterochromatin is mainly localized at the pericentric and subtelomeric regions and enriched for methylation of histone H3 on Lys9 (H3K9me), which provides a docking site for heterochromatin protein 1 (HP1) (5, 6), a highly conserved protein involved in heterochromatin formation (7). Previous studies also show that HP1 and H3K9me associate with a subset of loci on euchromatic regions, and they presumably serve to fine tune the level of gene transcription (8–10). One significant question that comes up is how the chromatin structure is dynamically regulated to impact expression of genes in complex neuronal processes, such as learning and memory. Our earlier study on chromatin domain mapping of chromosome 4 suggests that heterochromatic domains may be regulated by the activities of RNA polymerase II (RNAPII) complexes, which form a “barrier” to prevent heterochromatin spreading and transcriptional gene silencing (11). However, the mechanism underlying such a mode of regulation and the consequence on reprogrammed transcription remain to be investigated.
Results
Cyclin-Dependent Kinase 12 Counteracts Heterochromatin Enrichment on Euchromatic Regions.
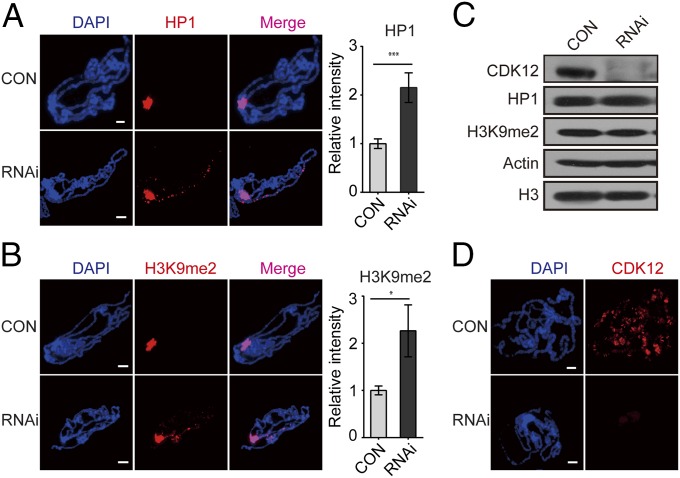
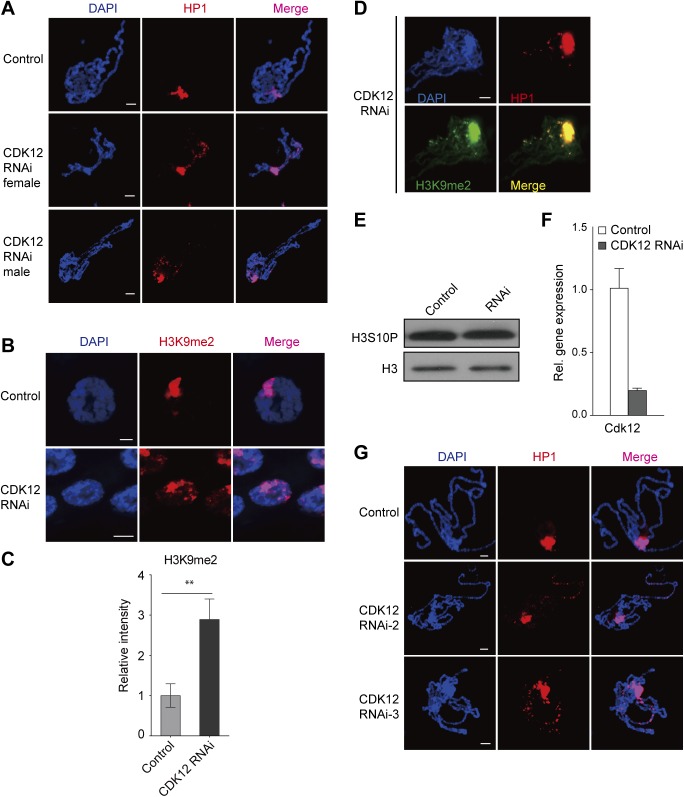
To understand how heterochromatin domains are regulated on chromosomes, we intended to screen for factors that affect HP1 distribution in Drosophila polytene chromosomes. Because kinases play important roles in modulating the cellular localization of their substrates and kinase JIL-1 has been reported to regulate HP1 binding on chromosomes (12), we performed a pilot transgenic RNAi screen focusing on kinases. The “visible” and consistent cytological localization of HP1 at the chromocenter on giant polytene chromosomes from the third-instar larvae allows for evaluation of the switch between the heterochromatic and euchromatic domains through immunofluorescence assays. We screened transgenic RNAi lines against protein kinases in Drosophila using a salivary gland-specific (SG) GAL4 (SG-GAL4). Surprisingly, we found substantial accumulation of HP1 in euchromatic regions in a cyclin-dependent kinase 12 (CDK12) RNAi line (TH00344.N) (Fig. 1A). In addition, similar HP1 binding patterns were observed in male and female flies after CDK12 knockdown (Fig. S1A), suggesting that this phenomenon is sex-independent. We next studied whether the localization of H3K9me2 was also changed along with the distribution of HP1 after CDK12 knockdown, because these two heterochromatin components are often coregulated, and both are essential for heterochromatin formation (7). Indeed, H3K9me2 also accumulated on euchromatic arms after depletion of CDK12 (Fig. 1B and Fig. S1 B and C), with a pattern indistinguishable from that of HP1 (Fig. S1D), which suggests that CDK12 counteracts heterochromatin enrichment on euchromatic arms. To examine whether CDK12 depletion affects the overall levels of HP1 and H3K9me2, we performed Western blot analysis. There was no obvious change (Fig. 1C), indicating that heterochromatin enrichment on euchromatic arms is not caused by increased levels of HP1 and H3K9me2.
Fig. 1.
Depletion of CDK12 triggers heterochromatin enrichment on euchromatic arms. (A and B) HP1 and H3K9me2 spread to euchromatic arms after CDK12 knockdown. Polytene chromosomes were squashed and stained with (A) anti-HP1 and (B) anti-H3K9me2. The relative fluorescence intensities of HP1 and H3K9me2 on the euchromatic arms based on raw data are shown in Right. Error bars indicate SD (n = 3). *P < 0.05; ***P < 0.001. (C) Western blot results showing that CDK12 depletion has no obvious effect on the level of HP1 or H3K9me2. Whole-tissue extracts were prepared from third-instar larvae. Antibodies specific for CDK12, HP1, and H3K9me2 were used. Antibodies against Actin and histone H3 were used as loading controls. Three independent replicates were performed. (D) The efficiency of CDK12 depletion was validated by immunostaining. Polytene chromosomes were labeled with anti-CDK12 (red). CON, control. (Scale bar: 10 μm.)
Fig. S1.
Heterochromatin enrichment occurs in both males and females after CDK12 knockdown. (A) CDK12 depletion induces ectopic accumulation of HP1 on euchromatic arms in both male and female Drosophila. Polytene chromosomes obtained from CDK12-depleted larvae and control were stained with anti-HP1 (red) and DAPI (blue). (B and C) H3K9me2 spreads to euchromatic arms after CDK12 knockdown in the whole-salivary gland immunostaining. RNAi-induced knockdown was driven by SG-GAL4. The relative fluorescence intensity of H3K9me2 on the euchromatic arms based on raw data is shown in C. Error bars indicate SD (n = 3). **P < 0.01. (D) Colocalization of HP1 (red) and H3K9me2 (green) on polytene chromosomes isolated from CDK12-depleted larvae. (E) CDK12 depletion has no effect on the level of phosphorylated H3S10. Protein extracts were prepared from salivary glands. (F) The RNAi efficiency of CDK12 in third-instar larvae was detected by qRT-PCR. Female UAS-CdkRNAi transgenic flies were crossed with males carrying actin-GAL4; w1118 was used as a WT control line, and rp49 was used as an internal control gene. Error bars indicate SD (n = 3). (G) The HP1 binding pattern in two independent CDK12 RNAi strains is shown. Polytene chromosomes were isolated from CDK12 RNAi-2 and CDK12 RNAi-3 and stained with anti-HP1 (C1A9; red). (Scale bar: 10 μm.)
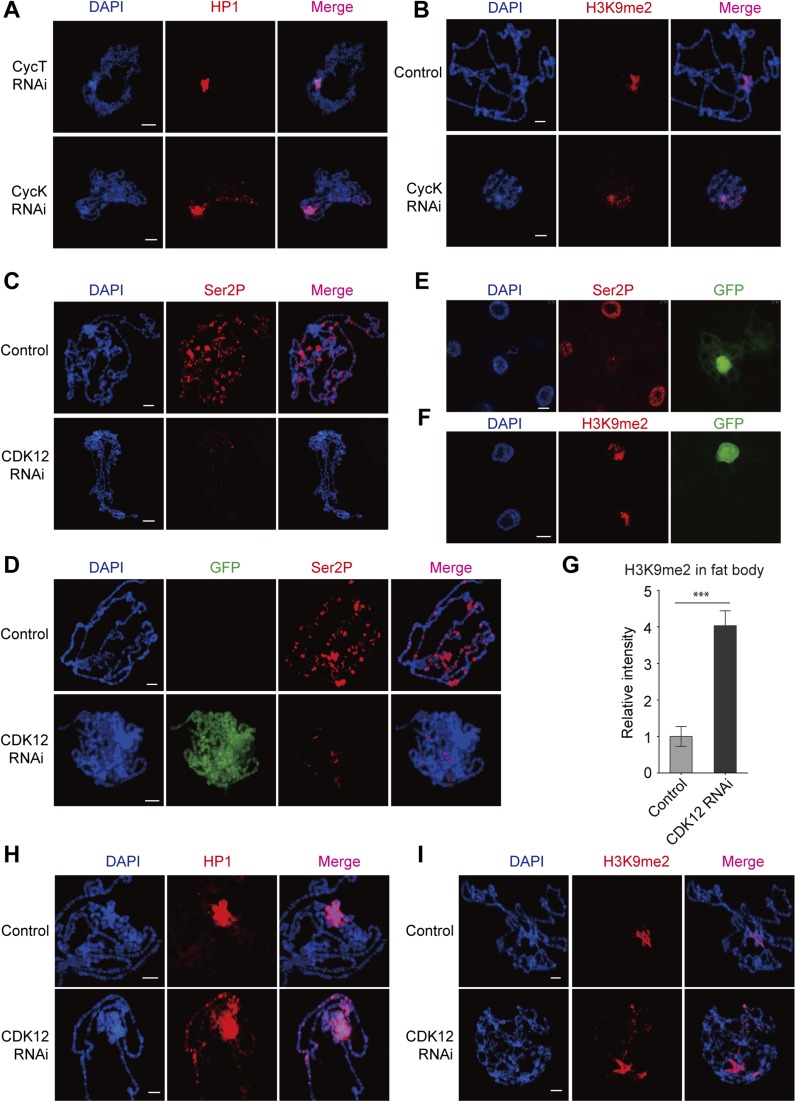
The CDK12 RNAi efficiency was validated by quantitative RT-PCR (qRT-PCR), Western blot analysis, and immunostaining. Our results showed that both the transcription level and the protein level of CDK12 were greatly reduced in third-instar larvae driven by actin-GAL4 (Fig. 1C and Fig. S1F). By immunostaining with a CDK12-specific antibody, we found abundant CDK12 signals in control chromosomes as previously reported. However, after CDK12 knockdown, these signals were efficiently eliminated in the polytene chromosomes (Fig. 1D). Furthermore, we tested two other independent CDK12 RNAi lines and found a similar HP1 binding pattern to that observed with the original RNAi line (TH00344.N) (Fig. S1G). In addition, we found that depletion of cyclin K (CycK), a known functional partner of CDK12 (13–15), induced heterochromatin enrichment on the chromosomes (Fig. S2 A and B), supporting that CDK12 is involved in counteracting heterochromatin enrichment. Recently, CDK12 has been identified as an RNAPII C-terminal domain (CTD) kinase that catalyzes the bulk of Ser2 phosphorylation (Ser2P) in Drosophila (13). Immunostaining of polytene chromosomes showed a dramatic decrease of Ser2P after CDK12 knockdown (Fig. S2C). Consistently, clonal assays in the salivary gland and fat body also showed reduced Ser2P levels in CDK12 knockdown cells compared with those in the neighboring control cells (Fig. S2 D and E). Moreover, we observed heterochromatin enrichment in fat body cells as indicated by H3K9me2 on CDK12 depletion (Fig. S2 F and G), suggesting that this phenomenon is preserved between different cell types. Taken together, these results show that CDK12 is a previously unidentified regulator for counteracting heterochromatin enrichment on euchromatic arms in Drosophila.
Fig. S2.
Depletion of CDK12/CycK induces heterochromatin ectopic accumulation. (A) Polytene chromosomes from CycK-depleted and CycT-depleted third-instar larvae were squashed and stained with anti-HP1 (C1A9; red). RNAi-induced knockdown was driven by SG-GAL4. (B) Chromosomes isolated from control and CycK-depleted larvae were stained with anti-H3K9me2 (red). (C) Ser2P was strongly decreased after CDK12 knockdown. Immunolocalization of Ser2P in polytene chromosomes isolated from WT control and CDK12-depleted larvae driven by SG-GAL4. (D–G) Ser2P was strongly decreased after CDK12 knockdown in clonal assay. (D) The polytene chromosome and (E and F) the fat body were dissected from third-instar larvae driven by the flip-out system and stained with Ser2P (red) in D and E and anti-H3K9me2 (red) in F. CDK12-depleted cells were detected by an antibody recognizing GFP, and cells without GFP staining served as a control. The relative fluorescence intensity of H3K9me2 on the euchromatic arms in the fat body cell is shown in G. Error bars indicate SD (n = 3). ***P < 0.001. (H and I) Heterochromatin enrichment occurs on euchromatic arms after CDK12 knockdown in the GAL80 system. The ectopic accumulations of (H) HP1 and (I) H3K9me2 on polytene chromosomes are shown. (Scale bar: 10 μm.)
Heterochromatin Enrichment Triggered by Loss of CDK12 Primarily Occurs on the X Chromosome.
Consistent with previous reports that the X chromosome is a preferential target for heterochromatin enrichment (12, 16), the accumulation of HP1 induced by CDK12 depletion was prominent on the X chromosome marked by acetylation of histone H4 on Lys16 in males (Fig. 2A). However, the loss of CDK12 driven by SG-GAL4 remarkably reduced the size of the salivary gland, increasing the difficulty in precisely mapping the distribution of HP1 on the X chromosome. Thus, we introduced tub-GAL80ts, a temperature-sensitive inhibitor of GAL4, for controlling GAL4 activity. At 25 °C, GAL80 activity is partially inhibited and allows a low level of GAL4/UAS activation, and the size of the salivary gland was found to be comparable with that in control. We found that HP1 exhibited a prominent distribution pattern on the X chromosome (Fig. 2B and Fig. S2H). A similar binding pattern was observed for H3K9me2 (Fig. S2I). We then mapped HP1 signals to multiple regions on the X chromosome and found that HP1 mainly associated with band regions rather than interbands (Table S1). Band regions are shown by the deep staining of DAPI for DNA and generally considered to be transcriptionally inactive. In summary, our observations reveal that the depletion of CDK12 induces ectopic HP1 accumulation in transcription-inactive regions on the X chromosome.
Fig. 2.
Heterochromatin enrichment mainly occurs on the X chromosome after CDK12 knockdown. (A) HP1 spreads to the X chromosome in male flies after CDK12 depletion. Polytene chromosomes isolated from male CDK12-depleted and control larvae were stained with anti-HP1 (red), antiacetylation of histone H4 on Lys16 (anti-H4K16ac; green), and DAPI (blue). The relative fluorescence intensities of HP1 on the X chromosome based on raw data are shown. Error bars indicate SD (n = 3). **P < 0.01. (B) HP1 enrichment occurs in band regions on the X chromosome; tub-GAL80ts was combined with the SG-GAL4 to allow low levels of GAL4 activity at 25 °C. The presented chromosome is assembled from three immunostained X chromosomes. The photographic map of the X chromosome is reproduced from ref. 40, with permission from Elsevier. CON, control. (Scale bar: 10 μm.)
Table S1.
HP1 distribution on the X chromosome after CDK12 knockdown
| X chromosome | Binding intensity | X chromosome | Binding intensity | X chromosome | Binding intensity |
| 1A 1–5* | +++ | 8A | + | 12F 4–5 | + |
| 1B 9–10* | +++ | 8B | + | 13A | + |
| 1E 1–3 | + | 8C 1–3 | + | 13B | + |
| 3A 2 | + | 8E | + | 13C | + |
| 3C 3–9* | +++ | 9A | + | 14B3* | ++++ |
| 4A 1 | ++ | 9F | + | 14A 1–3* | ++++ |
| 4B | ++ | 10A | + | 14D* | ++++ |
| 4C 2* | +++ | 10B | + | 16F 3–6* | ++++ |
| 4D | ++ | 10E | + | 17A* | ++++ |
| 4F 2–9* | +++ | 11A 1–7* | +++++ | 17C* | ++++ |
| 5B | + | 11B 1 | + | 18A | + |
| 5C | + | 11C | + | 18C | + |
| 5D | + | 11D 4 | + | 18E | + |
| 5E | 11F 1 | + | 19A* | ++++ | |
| 6A | ++ | 12A 1–2 | ++ | 19C* | ++++ |
| 6F 1 | ++ | 12C 4–5 | + | 19E* | ++++ |
| 7A | + | 12D 1–3* | ++++ | 19F* | ++++ |
| 7B | + | 12E 1–4* | ++++ | 20A* | ++++ |
| 7C | + | 12F 1–3* | ++++ | 20F* | ++++ |
The position of the HP1-enriched banded regions on the X chromosome and the binding intensity are shown; +, ++, +++, ++++, and +++++ represent increasing intensity levels.
Indicate regions with strong enrichment of HP1 after CDK12 depletion.
Heterochromatin Enrichment Predominantly Occurs in Long Genes After CDK12 Depletion.
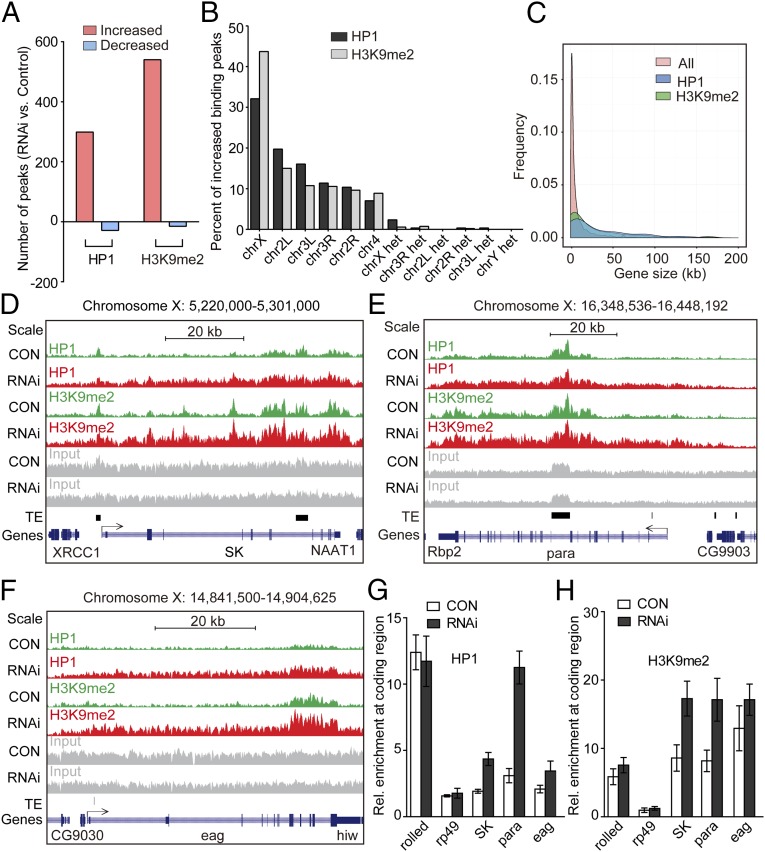
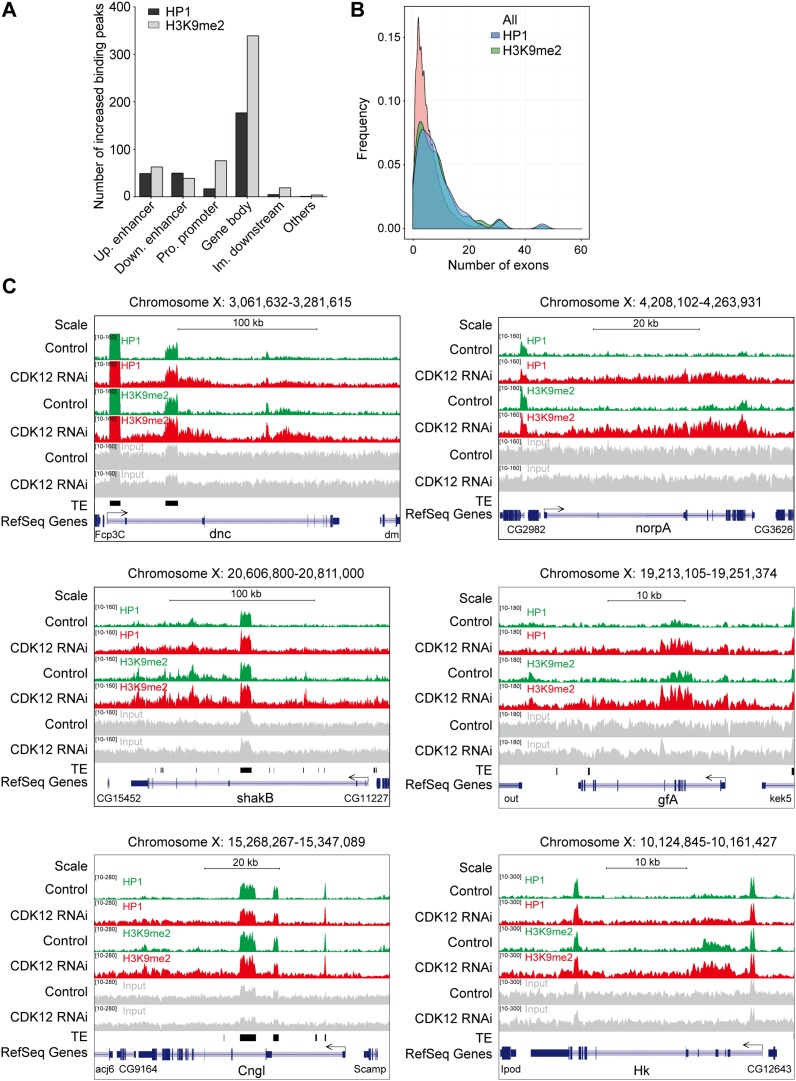
To further determine the target regions of HP1 accumulation on the X chromosome, we performed ChIP and sequencing (ChIP-seq) using tissues from control and CDK12-depleted larvae. Compared with those in control, we found that 299 chromatin peaks showed an increase in HP1 binding, whereas only 29 peaks showed a decrease in HP1 binding after CDK12 depletion (Fig. 3A and Dataset S1). Similarly, a global increase in the binding of H3K9me2 was also evident, with 540 peaks exhibiting increased binding and only a few peaks showing decreased binding (Fig. 3A and Dataset S1), validating that the loss of CDK12 resulted in heterochromatic territories. In addition, among the euchromatic peaks with increased association of HP1, more than 30% of the peaks were located on the X chromosome (Fig. 3B), which is consistent with the preferential binding pattern of HP1 on the X chromosome as indicated by the cytological mapping after CDK12 knockdown. Additional analyses indicated that the enrichment of both HP1 and H3K9me2 occurred mainly in the gene bodies after CDK12 depletion, counting for ∼60% of the total binding detected (Fig. S3A). In accordance with previous reports that HP1 prefers to bind with long genes in euchromatic regions (for example, >10 kb) (17), we found that the increased association of HP1 was biased toward long genes with multiple exons after CDK12 knockdown (Fig. 3C and Fig. S3B).
Fig. 3.
ChIP-seq analysis of heterochromatin enrichment after CDK12 knockdown. (A) The binding of HP1 and H3K9me2 is increased genome-wide after CDK12 depletion. The numbers of increased (red) and decreased (blue) binding peaks of H3K9me2 or HP1 are indicated. (B) HP1 and H3K9me2 preferentially associate with the X chromosome. The percentages of increased binding peaks of HP1 (black) or H3K9me2 (gray) on different chromosomes and heterochromatic domains are indicated. (C) HP1 and H3K9me2 preferentially associate with long genes. The comparisons of all Drosophila genes and genes with increased HP1 and H3K9me2 bindings with regard to the gene size are shown. The data for the gene length were obtained from the University of California at Santa Cruz Genome Browser. (D–F) The distribution of HP1 and H3K9me2 at three long genes on the X chromosome, (D) SK, (E) para, and (F) eag, in control (green) and CDK12 depletion (red) larvae. ChIP-seq input (gray), transposable elements (TEs; black), and the reference genes (blue) are illustrated. All annotated TE insertions on the X chromosome were obtained from FlyBase. The direction of gene transcription is indicated by arrows. The height range for all peak panels in D and F is 10–215, and the height range for all peak panels in E is 10–410. (G and H) ChIP assay and quantitative PCR analysis validate the increased binding of HP1 and H3K9me2 at gene bodies on the X chromosome after CDK12 depletion. The bars show enrichment of HP1 or H3K9me2 normalized to input and the internal control gene GS, which shows no binding of HP1 and H3K9me2 in the ChIP-seq analysis; rolled serves as a positive control, whereas rp49 serves as a negative control. The primers were designed based on sequences within the transcribed regions of these genes. Error bars indicate SD (n = 3). CON, control.
Fig. S3.
CDK12 depletion induces heterochromatin enrichment on the gene body of euchromatic genes with multiple exons. (A) The distribution of increased binding peaks of HP1 (black) or H3K9me2 (gray) at different regulatory regions of genes is shown. (B) The comparison of all Drosophila genes and genes with increased HP1 or H3K9me2 binding with regard to the exon numbers is shown. The data for the exon numbers were obtained from the University of California at Santa Cruz Genome Browser. (C) ChIP-seq analysis reveals the ectopic accumulation pattern of HP1 and H3K9me2 at the individual gene level in both controls (green) and CDK12-depleted (red) larvae. Heterochromatin landscape in six genes on the X chromosome is displayed. The relative positions of transposable elements (TEs; black) in the reference genes (blue) are also shown. The direction of gene transcription is indicated by the arrows.
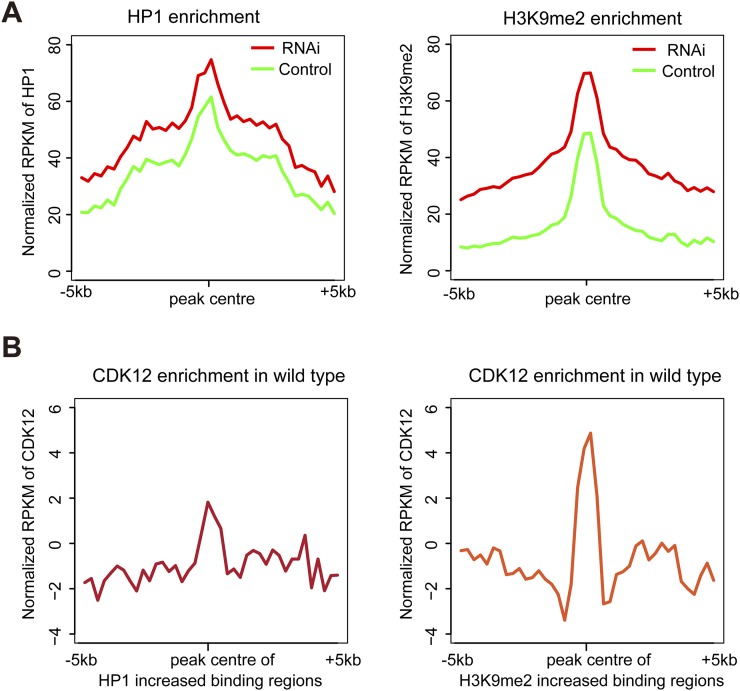
Furthermore, we examined the distribution patterns of heterochromatin enrichment at single-gene resolution by focusing on a group of 10 X-chromosomal genes with highly increased HP1 and H3K9me2 bindings after loss of CDK12. Consistent with a previous report (9), low levels of HP1 and H3K9me2 bindings exist in transcribed regions in 10 chosen genes in control. Interestingly, after CDK12 depletion, abundant levels of HP1 and H3K9me2 bind the gene bodies of these loci (Fig. 3 D–F and Fig. S3C). The increased levels of HP1 and H3K9me2 in the X-chromosomal genes were also confirmed by quantitative PCR with another independent ChIP preparation (Fig. 3 G and H). Because we noticed that these X-chromosomal genes were initially covered with low levels of HP1 and H3K9me2, we continued to analyze whether this is a general phenomenon in the whole genome. We found that, in the average profile, the regions with increased heterochromatin enrichment are originally associated with low levels of HP1 and H3K9me2 in the control (Fig. S4A). Moreover, to test whether CDK12 initially associates with these regions, we performed ChIP-seq analysis. The results suggest a certain level of CDK12 association in these regions (Fig. S4B). Taken together, our ChIP-seq analysis supports that heterochromatin enrichment preferentially occurs on the X chromosome after CDK12 knockdown and that the increased association of HP1 predominantly locates in long genes, which were initially covered with a low level of HP1.
Fig. S4.
The average enrichment levels of HP1, H3K9me2, and CDK12 in the peak regions with increased heterochromatin accumulation after CDK12 depletion. (A) The average enrichment levels of HP1 and H3K9me2 around the peak regions in the whole genome were calculated in both control and CDK12 knockdown. The peak regions are where the binding levels of HP1 or H3K9me2 have increased after CDK12 depletion. (B) The average enrichment of CDK12 was calculated in the regions of the genome where the binding peaks of HP1 or H3K9me2 were increased after CDK12 depletion.
Heterochromatin Enrichment by CDK12 Depletion Impairs Neuronal Gene Activation.
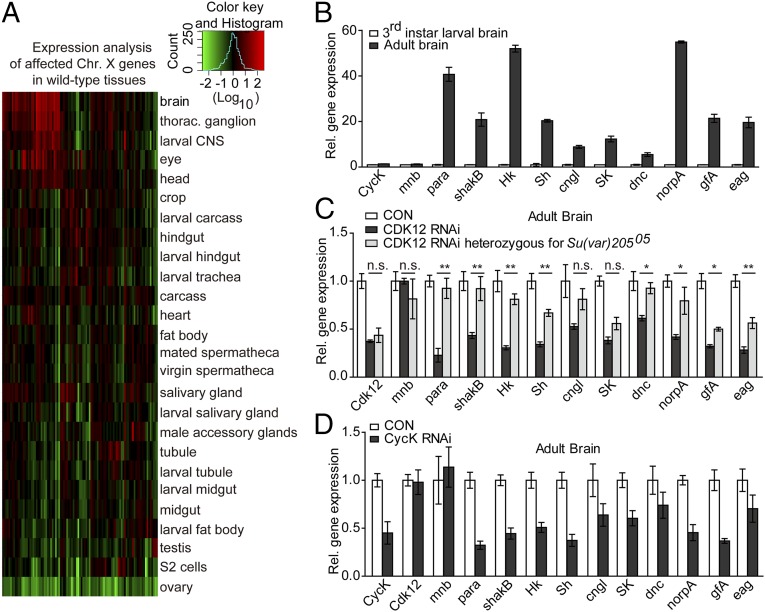
To further characterize the developmental functions of the targeted genes with heterochromatin accumulation after CDK12 knockdown, we selected the genes with increased HP1 binding on the X chromosome and analyzed their expression patterns in various wild type (WT) tissues using the public FlyAtlas database (18). Interestingly, about one-half of the affected X-chromosomal genes on CDK12 knockdown are preferentially expressed in the brain and ganglion of WT flies (Fig. 4A and Table S2), especially during the adult stage. A similar expression pattern was observed for genes with increased H3K9me2 binding (Fig. S5A and Table S2), indicating a major role of CDK12 in antagonizing heterochromatin enrichment on neuronal genes. Based on neuronal functions, these genes are mainly divided into three categories. The first group of genes encodes for voltage-gated and mechanosensitive ion channels, including Sh, Hk, eag, and para. The second group of genes encodes for ligand-gated channels/receptors, including SK, Cngl, and gfA. The third group of genes includes norpA, a phospholipase C, and dnc, a cAMP phosphodiesterase that is expressed in Drosophila mushroom body (MB) neuropil and regulates learning acquisition in flies (19).
Fig. 4.
Heterochromatin enrichment on CDK12 knockdown impairs neuronal gene activation. (A) Heat map shows that the affected X-chromosome genes are highly expressed in the brain of WT Drosophila; 52 X-chromosome genes with increased HP1 binding after CDK12 depletion are selected. Expression data were obtained from the FlyAtlas database (18). The relative expression levels were determined using a log10 scale and normalized to the whole-fly genome-wide average. (B) The neuronal genes are highly activated in the adult brain. The relative expression levels of neuronal genes in the third-instar larval brain and the adult brain in WT were monitored by qRT-PCR and normalized to rp49 mRNA; mnb showed no association with HP1 or H3K9me2 in our ChIP-seq analysis and served as a negative control. Error bars indicate SD (n = 3). (C and D) The depletion of CDK12 reduced the transcription of neuronal genes, whereas introducing Su(var)20505 restored the expression level. The relative expression levels of neuronal genes in the adult brain after (C) CDK12 depletion or CDK12 depletion heterozygous for Su(var)20505 or (D) CycK depletion were monitored by qRT-PCR as described in B. Significance was analyzed by ordinary one-way ANOVA in GraphPad Prism. Error bars indicate SD (n = 3). CON, control; n.s., not significant. *P < 0.05; **P < 0.01.
Table S2.
List of the affected genes on the X chromosome (increased HP1 or H3K9me2 binding genes after CDK12 depletion) that are highly expressed in the WT brain (ratio > 2)
| Gene symbol | Brain mean | Whole-fly mean | Ratio |
| Increased HP1 binding after CDK12 depletion | |||
| AlstR | 143.3 | 2.9 | 49.4 |
| Sh | 355.3 | 7.6 | 46.6 |
| mmd | 84.1 | 2.1 | 41.0 |
| CG9170 | 774.2 | 21.7 | 35.7 |
| rab3-GEF | 1,005.3 | 33.5 | 30.0 |
| SK | 262.8 | 10.4 | 25.4 |
| CG15478 | 110.6 | 5.0 | 22.1 |
| stnB | 1963.5 | 90.6 | 21.7 |
| X11Lβ | 199.6 | 9.4 | 21.2 |
| para | 135.5 | 7.9 | 17.2 |
| CG1504 | 15.2 | 0.9 | 16.9 |
| inaE | 186.5 | 17.0 | 11.0 |
| shakB | 56.7 | 6.3 | 8.9 |
| kirre | 61.3 | 8.6 | 7.1 |
| Hers | 500.0 | 73.8 | 6.8 |
| CG12496 | 3.2 | 0.5 | 6.4 |
| Scgδ | 37.0 | 6.4 | 5.8 |
| sdt | 88.7 | 16.9 | 5.3 |
| dnc | 294.2 | 72.0 | 4.1 |
| CG42340 | 19.3 | 4.9 | 3.9 |
| Ste:CG33236 | 31.5 | 8.2 | 3.8 |
| CG33496 | 15.9 | 4.2 | 3.8 |
| rdgA | 34.4 | 9.9 | 3.5 |
| Fas2 | 197.1 | 59.2 | 3.3 |
| CG12531 | 10.0 | 3.0 | 3.3 |
| eag | 12.7 | 4.2 | 3.0 |
| disco | 61.2 | 20.8 | 2.9 |
| α-Man-I | 166.8 | 58.0 | 2.9 |
| Increased H3K9me2 binding after CDK12 depletion | |||
| gfA | 195.7 | 2.5 | 78.3 |
| D2R | 222.8 | 4.1 | 54.3 |
| CG42492 | 744.4 | 13.9 | 53.7 |
| AlstR | 143.3 | 2.9 | 49.4 |
| CG6123 | 114.3 | 2.4 | 47.6 |
| Sh | 309.3 | 6.7 | 46.1 |
| mmd | 84.1 | 2.1 | 41.0 |
| Syt12 | 241.1 | 6.4 | 37.7 |
| CG9170 | 774.2 | 21.7 | 35.7 |
| nAcRα-7E | 120.9 | 3.6 | 33.6 |
| CG8909 | 897.8 | 28.4 | 31.6 |
| CG7135 | 202.8 | 6.7 | 30.3 |
| rab3-GEF | 1,005.3 | 33.5 | 30.0 |
| Vsx1 | 39.7 | 1.4 | 28.4 |
| Rph | 390.2 | 14.0 | 27.9 |
| cngl | 86.5 | 3.4 | 25.6 |
| SK | 262.8 | 10.4 | 25.4 |
| stnB | 1,963.5 | 90.6 | 21.7 |
| NetB | 628.3 | 31.0 | 20.3 |
| Hk | 289.7 | 16.8 | 17.2 |
| Para | 135.5 | 7.9 | 17.2 |
| CG1504 | 15.2 | 0.9 | 16.9 |
| Nep3 | 387.1 | 24.0 | 16.1 |
| CG43902 | 111.2 | 8.2 | 13.6 |
| CG42450 | 24.3 | 1.9 | 13.1 |
| Tty | 60.8 | 4.7 | 12.9 |
| CG32653 | 74.2 | 5.9 | 12.6 |
| acj6 | 84.8 | 7.0 | 12.1 |
| CG11085 | 14.4 | 1.2 | 12.0 |
| Hiw | 322.2 | 27.8 | 11.6 |
| tomosyn | 1,683.5 | 145.9 | 11.5 |
| CG32532 | 51.8 | 4.5 | 11.5 |
| Rg | 385.4 | 33.7 | 11.5 |
| CG34411 | 14.5 | 1.5 | 9.7 |
| shakB | 56.7 | 6.3 | 8.9 |
| CanA-14F | 176.9 | 20.4 | 8.7 |
| CG32791 | 69.0 | 9.0 | 7.7 |
| Hers | 500.0 | 73.8 | 6.8 |
| Fend | 426.1 | 62.9 | 6.8 |
| Or13a | 18.2 | 2.8 | 6.5 |
| mamo | 36.0 | 6.2 | 5.9 |
| DIP1 | 1,183.1 | 214.4 | 5.5 |
| Tyn | 87.7 | 16.2 | 5.4 |
| Sdt | 88.7 | 16.9 | 5.3 |
| Pde9 | 73.0 | 14.9 | 4.9 |
| Ca-α1T | 22.0 | 4.5 | 4.9 |
| CG11294 | 34.4 | 7.3 | 4.7 |
| CG14431 | 17.5 | 3.9 | 4.5 |
| CG11071 | 5.8 | 1.3 | 4.5 |
| norpA | 326.5 | 73.4 | 4.4 |
| CG32521 | 524.6 | 118.2 | 4.4 |
| Br | 45.7 | 10.4 | 4.4 |
| Proc-R | 3.4 | 0.8 | 4.3 |
| Dnc | 294.2 | 72.0 | 4.1 |
| CG42340 | 19.3 | 4.9 | 3.9 |
| Hexo2 | 805.0 | 205.1 | 3.9 |
| CG33491 | 15.9 | 4.2 | 3.8 |
| CG42594 | 14.4 | 3.8 | 3.8 |
| CG33639 | 15.8 | 4.4 | 3.6 |
| rdgA | 34.4 | 9.9 | 3.5 |
| Fas2 | 197.1 | 59.2 | 3.3 |
| CG42343 | 7.4 | 2.3 | 3.3 |
| Eag | 12.7 | 4.2 | 3.0 |
| Mgl | 180.0 | 60.6 | 3.0 |
| dlg1 | 132.4 | 50.9 | 2.6 |
| Sbr | 550.2 | 223.5 | 2.5 |
| kek5 | 44.6 | 18.9 | 2.4 |
| dmrt11E | 2.1 | 0.9 | 2.3 |
| CG6847 | 41.1 | 18.3 | 2.2 |
| CG43154 | 153.4 | 70.2 | 2.2 |
Expression data were obtained from the FlyAtlas database. The relative expression ratios were determined by log10 scale and normalized to the whole-fly, genome-wide mean. Ratio > 2 was taken as highly expressed genes in WT brain.
Fig. S5.
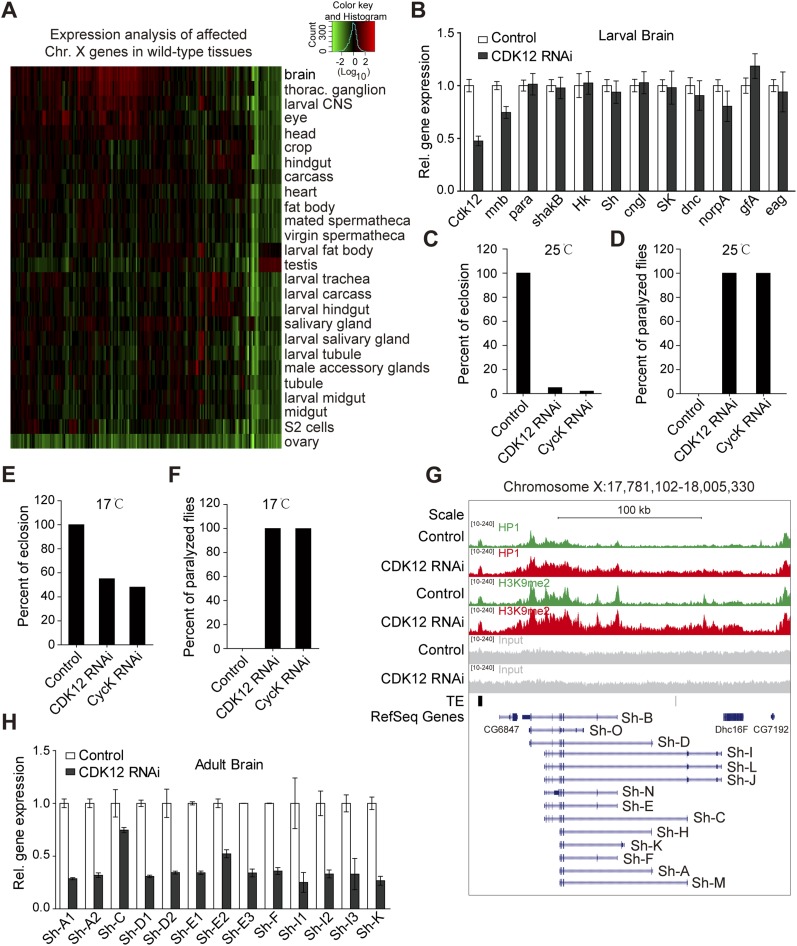
CDK12 does not affect neuronal gene expression in larval stage, and depletion of CDK12 induces locomotion defect in adult stage. (A) Heat map shows that the affected X-chromosome genes are highly expressed in the brain of WT Drosophila; 141 X-chromosome genes with increased H3K9me2 binding after CDK12 depletion are selected. Expression data in different tissues (mainly from the larval and adult stages) were obtained from the FlyAtlas database. The relative expression levels were determined using a log10 scale and normalized to the whole-fly genome-wide average. (B) The relative expression level of neuronal genes in third-instar larval brain after CDK12 depletion was monitored by qRT-PCR and normalized to rp49 mRNA; mnb showed no association with HP1 or H3K9me2 in our ChIP-seq analysis and served as a negative control. Error bars indicate SD (n = 3). (C) The percentage of eclosion from puparia was calculated in control, CDK12-depleted, and CycK-depleted flies. The CDK12 or CycK knockdown in the adult brain was driven by elav-GAL4 at 25 °C. (D) The percentage of flies showing paralyzed phenotype was calculated after CDK12 or CycK depletion. The CDK12 or CycK knockdown in the adult brain was driven by elav-GAL4 at 25 °C. (E) The percentage of eclosion from puparia was calculated as in C. The CDK12 or CycK knockdown in the adult brain was driven by elav-GAL4 at 17 °C. (F) The percentage of flies showing paralyzed phenotype was calculated as in D. The CDK12 or CycK knockdown in the adult brain was driven by elav-GAL4 at 17 °C. (G) The distribution pattern of HP1 and H3K9me2 within the Sh gene in CDK12-depleted (red) larvae and controls (green) is shown. Input (gray), transposable element (TE) locations (black), and reference genes (blue) are indicated. The isoforms of the Sh gene are shown at the bottom. The direction of transcription of the Sh gene is from right to left. (H) The relative expression levels of transcripts encoding different Sh isoforms were analyzed by qRT-PCR and normalized to rp49 mRNA. Primers were designed to target these isoforms, and some primers mapped more than one isoform of Sh. Error bars indicate SD (n = 3).
To detect whether the heterochromatinization on the X chromosome after CDK12 depletion could result in transcription down-regulation, we performed qRT-PCR analysis with brain tissues on 10 selected neuronal genes. By using a panneuronal driver elav-GAL4 to conduct neuron-specific knockdown of CDK12, we found no obvious change in the transcription of these genes in the larval brain after CDK12 depletion (Fig. S5B), which may be because of the relatively low expression levels of these genes in the larval brain compared with those in the adult brain as indicated by gene expression analysis using the public FlyAtlas database (Fig. 4A). These neuronal genes may be developmentally regulated, because insects, like Drosophila, go through metamorphosis in which the nervous system is remarkably remodeled from larva stage to adult stage (20). Therefore, we compared expression levels of 10 genes between third-instar larvae and 1-d adult in the brain. qRT-PCR results showed that expression levels of these genes were 3- to 60-fold higher in the adult brain compared with those in the larval brain (Fig. 4B).
Next, we wondered whether the depletion of CDK12 in the adult brain would cause neuronal defects. Notably, we found that flies failed to eclose from puparia after CDK12 knockdown driven by elav-GAL4 at 25 °C (Fig. S5C) and that these flies exhibited a severely paralyzed phenotype if dissected out of puparia (Fig. S5D). A similar phenotype was also observed after CycK knockdown (Fig. S5 C and D). When the culture temperature was shifted to 18 °C, ∼50% of the flies were able to eclose from puparia, but all of them still displayed a paralyzed phenotype (Fig. S5 E and F). This paralyzed phenotype may be induced by the decreased expression of para, because a previous report showed that para mutants exhibit a paralyzed phenotype (21).
We next assessed the transcription levels of these neuronal genes in the brains of 1-d adults. The qRT-PCR results showed two- to threefold reduction in their transcription after CDK12 knockdown compared with those in control (Fig. 4C). Consistently, the depletion of CycK also resulted in a decrease in the transcription of the tested genes in the adult brain (Fig. 4D). To further address whether CDK12 affects the expression of these neuronal genes through modulating heterochromatin, we introduced HP1 loss-of-function allele Su(var)20505 into CDK12 transgenic RNAi stock. The qRT-PCR results showed that the expression levels of eight neuronal genes were significantly rescued (Fig. 4C). Moreover, we found that the paralyzed phenotype was clearly suppressed in CDK12 knockdown flies that are heterozygous for Su(var)20505 (Movies S1–S3). In addition, to detect whether CDK12 affects alternative splicing, we focused on the Sh gene, which is processed into multiple RNA transcripts (Fig. S5G). The qRT-PCR results showed that the different isoforms of Sh were uniformly decreased in the adult brain (Fig. S5H), suggesting that CDK12 is not associated with the alternative splicing of the Sh gene. Overall, these results indicate that the heterochromatinization on the X chromosome after CDK12 depletion impairs the transcriptional activation of neuronal genes during the adult stage.
Heterochromatin Enrichment on CDK12 Depletion Affects Drosophila Courtship Learning.
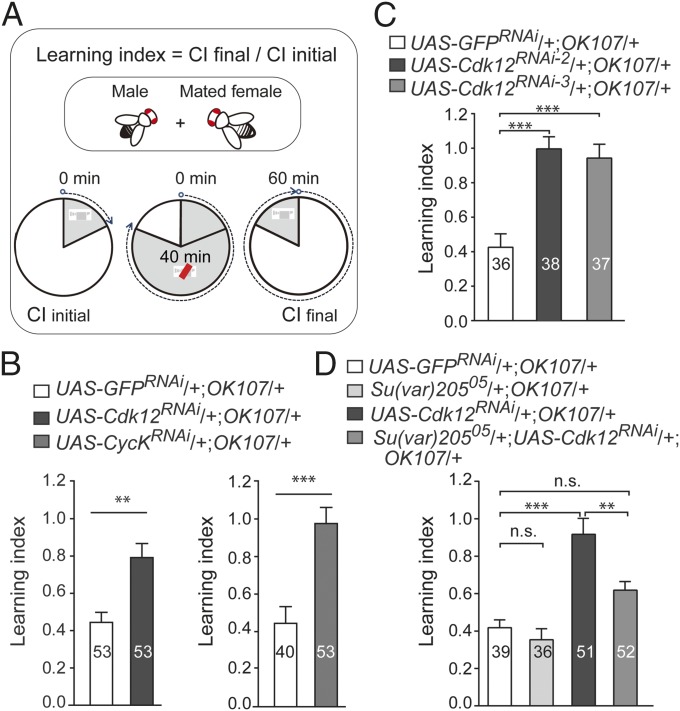
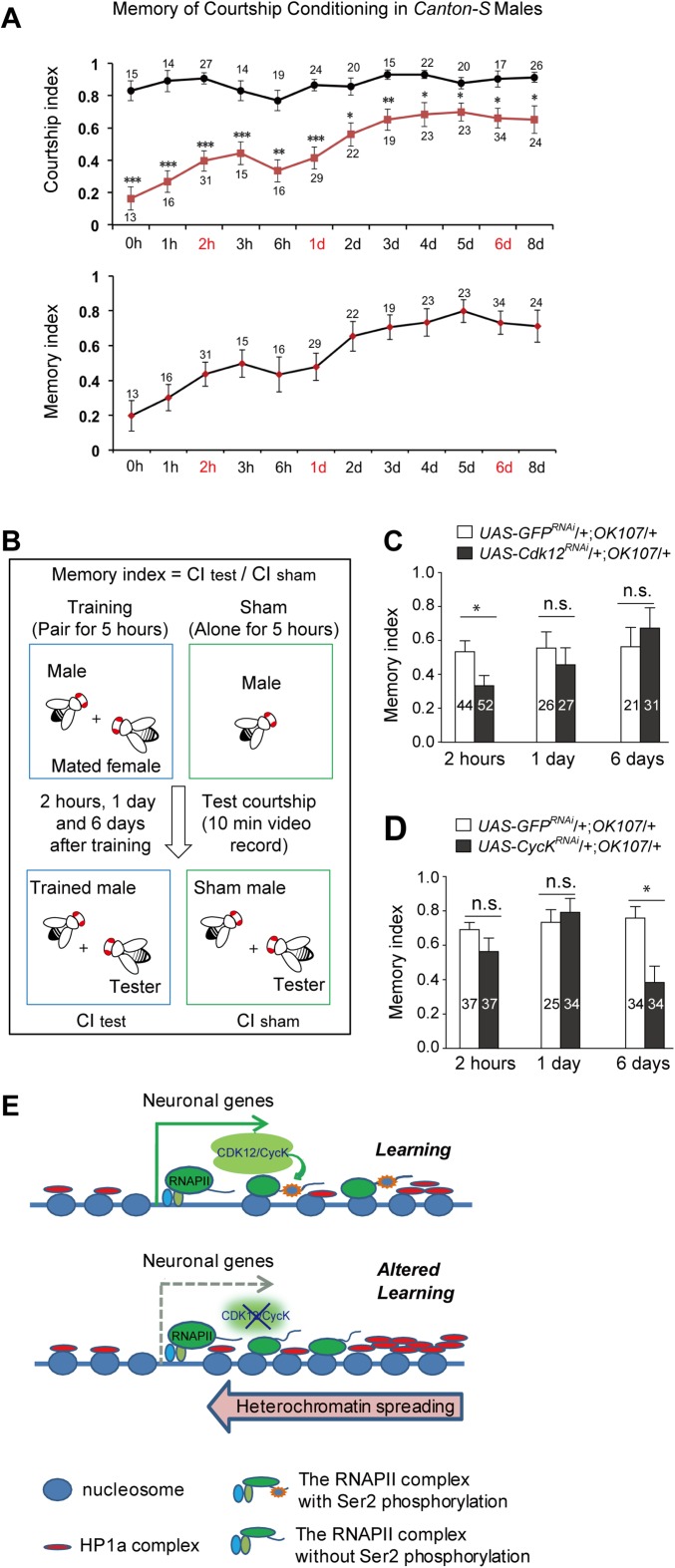
The decreased expression of neuronal genes in the adult brain prompted us to test whether the loss of CDK12 or CycK would have any effect on neuronal behaviors. Because the potassium channel genes Sh, Hk, eag, and SK and the cAMP phosphodiesterase dnc are all known to be involved in Drosophila learning and memory (19, 22–25), we performed a courtship conditioning test using an OK107-GAL4 driver, which is mainly active in the MBs (putative learning and memory centers in the Drosophila brain). Unsuccessful courtship reduces the subsequent courtship behavior of male flies toward virgin females, and this courtship conditioning behavior is known as one of the major learning and memory paradigms in Drosophila. Our results showed that knockdown of CDK12 in the MBs induced a severe courtship learning defect (Fig. 5 A and B and Movies S4 and S5). Furthermore, we tested two other independent CDK12 RNAi lines and found a consistent courtship learning defect (Fig. 5C). We then tested the courtship memory behaviors of the control and the CDK12 RNAi flies at 2 h, 1 d, and 6 d after a 5-h training period based on the memory curves of Canton-S flies (Fig. S6A) and found no obvious change in memory indexes between the control and CDK12-depleted lines at these different time points (Fig. S6 B and C). Consistently, we also observed that the depletion of CycK resulted in similar defects in courtship learning but not in courtship memory (Fig. 5B, Fig. S6D, and Movie S6). To further address whether CDK12 affects courtship learning through modulating heterochromatin status, we performed a learning test on CDK12 knockdown flies that were also heterozygous for Su(var)20505 (Fig. 5D and Movie S7). We found that the learning index of these flies had decreased significantly compared with that of CDK12 knockdown flies, which shows that heterozygous HP1 mutation suppresses the learning defect induced by CDK12 knockdown in the MB. Overall, our results suggest that CDK12 regulates Drosophila courtship learning by antagonizing heterochromatin enrichment on neuronal genes.
Fig. 5.
Heterochromatin enrichment upon CDK12 depletion affects Drosophila courtship learning. (A) The schematics illustrating procedures used to analyze Drosophila courtship learning. The learning index was determined as the ratio of the courtship level during the final 10 min of training [courtship index final (CIfinal)] to that of the initial 10 min [courtship index initial (CIinitial)]. If the learning index is ≥1, it indicates that there is no learning. (B) Courtship learning indexes were measured in (Left) CDK12-depleted and (Right) CycK-depleted flies. Significance was estimated using Student's t test. (C and D) Heterozygous Su(var)20505 mutation suppresses the learning defects induced by CDK12 knockdown. Courtship learning indexes were measured in (C) two independent CDK12-depleted flies and (D) CDK12 RNAi flies heterozygous for Su(var)20505. Significance was analyzed by ordinary one-way ANOVA in GraphPad Prism. The genotypes of tested flies are listed. Error bars indicate SEM. n.s., not significant. **P < 0.01; ***P < 0.001.
Fig. S6.
A model depicting the possible mechanism of how CDK12 contributes to courtship learning in Drosophila. (A) Memory of courtship conditioning in Canton-S males. After a 5-h training with a mated female, the CI was determined during 10 min of testing with a tester; right after training; 1, 2, 3, or 6 h later; or 1, 2, 3, 4, 5, 6, or 8 d later. Squares and circles represent the mean CIs of trained males and age-matched shams, respectively. Error bars indicate SEM. The MI was calculated by dividing the mean of CItest by the mean of CIsham. (B) The schematics illustrating procedures used to analyze Drosophila courtship memory. The MI was calculated by dividing the CItest by the mean of the CIsham. If MI is ≥1, it indicates that there is no memory. (C and D) Courtship MIs were measured in (C) CDK12-depleted and (D) CycK-depleted flies. Significance was estimated using the Student’s t test. The CDK12 and CycK knockdowns in the MBs of adult flies were driven by OK107-GAL4. (E) A model depicting the mechanism by which the CDK12/CycK complex counteracts heterochromatin enrichment and regulates the expression of neuronal genes. In the presence of CDK12/CycK, the Ser2 of the CTD in RNAPII is phosphorylated and maintains the processes of transcriptional elongation. The RNAPII elongation complex together with other histone remodeling factors, including histone acetyltransferase, prevent the ectopic accumulations of H3K9me2/HP1 from the enriched 3′ region toward the promoter region. In the absence of CDK12/CycK, RNAPII loses the phosphorylation at Ser2, which may lead to a reduction in its elongating activity and further increases the levels of H3K9me2 and HP1. The heterochromatinization caused by the ectopic accumulations of H3K9me2 and HP1 results in the transcriptional attenuation of specific genes, such as neuronal genes, when they are required to be highly activated at specific developmental stages. n.s., Not significant. *P < 0.05; **P < 0.001; ***P < 0.0001.
Discussion
In Drosophila, heterochromatin is featured by strong enrichment of “silencing marks” HP1 and H3K9me2 (9). In this study, we have shown that the depletion of CDK12 induces heterochromatin accumulation within euchromatic arms, particularly on the X chromosome. Previous reports show that overexpression of Su(var)3–7 induces ectopic heterochromatin formation on the euchromatic arms, with preferential enrichment of HP1 and H3K9me2 in the male X chromosome (16). This pattern is associated with the dosage compensation complex, and it is distinct from CDK12 depletion-triggered heterochromatin enrichment. In addition, mutation of the histone H3S10 kinase JIL-1 results in heterochromatin spreading to the euchromatic arms, especially on the X chromosome in both male and female flies (12). However, there was no obvious change in the level of H3S10 phosphorylation (H3S10P) after CDK12 knockdown (Fig. S1E). Furthermore, we noticed that the overall levels of HP1 and H3K9me2 remain unchanged after CDK12 knockdown (Fig. 1C), which resembles what has been observed in the JIL null mutant (12). A similar pattern was reported in the spindle-E mutant, where HP1 was highly enriched on the entire chromosome, whereas the overall level of HP1 was not affected (26). There are two possibilities of why the HP1 level remained unchanged while an enrichment on the euchromatic regions was observed. The first possibility is that HP1 may be relocated in the nucleus. HP1 is also found in the nucleoplasmic fraction (27), and its association with chromatin is dynamic (28). After CDK12 depletion, there may be more HP1 becoming associated with chromatin from the nucleoplasm. A second possibility is that HP1 is redistributed on the chromosome. In certain heterochromatin regions, HP1 is highly enriched and may be saturated. The current research methods may not be sensitive enough to detect the slight decrease in HP1 levels in the heterochromatin regions.
Intriguingly, the X chromosome is shown to be a preferential target for heterochromatin enrichment in this study and previous reports. This phenomenon may be partly explained by modENCODE data in Drosophila, which show that the X chromosome contains large “facultative heterochromatin” domains with moderate levels of H3K9me2/me3 (9). These domains, thus, may serve as seed regions for ectopic heterochromatinization. Indeed, our ChIP-seq analysis on the X chromosome showed that genes with heterochromatin enrichment were initially covered with low levels of HP1 and H3K9me2 without CDK12 depletion.
RNAPII CTD serves as a scaffold for orchestrating chromatin modifiers and transcription apparatus during eukaryotic gene expression (29). Here, we provide the first evidence, to our knowledge, that an RNAPII CTD kinase functions as a heterochromatin antagonizer on euchromatic arms in Drosophila. Previous reports show that Ctk1, the yeast ortholog of CDK12, directed RNAPII CTD Ser2P to provide a docking site for the H3K36me3 methyltransferase (30), whereas HP1 promotes H3K36me3 demethylation through interaction with dKDM4A in Drosophila (31), which indicates a potential counteractive interplay between HP1 and the transcription elongation machinery. Moreover, our earlier study on chromatin domain mapping of chromosome 4 suggests that the transcriptional activity of the RNAPII complexes prevents heterochromatin spreading by forming a barrier (11). Because CTD Ser2 is the only known substrate for CDK12 (13), we speculate that CTD Ser2P catalyzed by CDK12 provides a platform for counteracting heterochromatin enrichment (Fig. S6E). Neverthless, emerging evidence has shown that CDK12 contains the arginine/serine domain and is involved in RNA processing (32, 33). In addition, CDK12 has been implicated in mRNA splicing (15, 34, 35). However, from our data, we cannot exclude the possibility that the arginine/serine domain of CDK12 also contributes to heterochromatin remodeling. Additional research is needed to better understand the mechanism of heterochromatin enrichment on CDK12 depletion.
The mechanism of epigenetic regulation on neuronal and behavioral plasticity remains an intriguing issue in neuroscience. So far, ∼30 chromatin regulators have been implicated in mental retardation and psychiatric disorders (36). Moreover, the pathology of Alzheimer’s disease is linked to alterations in heterochromatin status (37). In this study, we provide evidence that heterochromatin enrichment induced by CDK12 knockdown hinders the transcriptional activation of neuronal genes, leading to a defect in courtship learning (Fig. S6E). Previous reports show that large facultative heterochromatin domains are prominent in the Drosophila BG3 neuronal cell line (9), and our data show that heterochromatin enrichment mainly occurs on neuronal genes, especially on long genes encoding ion channel subunits. A partial explanation for these observations is that the facultative heterochromatin domain acts in a dynamic equilibrium to fine tune the transcription elongation rate of RNAPII to ensure RNA processing on long neuronal genes with multiple exons (38). However, after heterochromatin accumulates, the transcription efficiency is greatly impeded, especially for genes that need to be expressed at high levels for specific neuronal processes. Therefore, after CDK12 depletion, those neuronal genes that are robustly up-regulated in the adult brain are the ones affected the most in our study. Because evidence shows that CDK12 affects the expression of long genes in mammals (14), it will be interesting to determine whether heterochromatin enrichment on long genes triggered by CDK12 depletion is conserved in vertebrates and affects important neuronal processes, such as learning or memory.
Materials and Methods
For total larval chromatin preparation, ∼3,500 third-instar larvae were collected. Larvae extracts were treated as described previously with some modifications (39). The details for fly strains, antibodies, immunostaining, ChIP-seq analysis, qRT-PCR, Western blot analysis, and courtship learning tests are described in SI Materials and Methods.
SI Materials and Methods
Fly Stocks.
All of the stocks were raised on corn-agar medium at 25 °C unless otherwise indicated. SG-GAL4 (1824) was from the Bloomington Stock Center. The following strains were obtained from the TsingHua Fly Center: UAS-Cdk12RNAi (TH00344.N), UAS-CycKRNAi (THU1386 and HMS01003), UAS-GFPRNAi (TH00781.N), 7018 tub-GAL80ts, and actin-GAL4. Balancer stock U7 (yw;Sco/CyO;Sb/TM6B) was a gift from Jose Carlos Pastor-Pareja, School of Life Sciences, Tsinghua University, Beijing. The following strains were obtained from Yi Zhong, School of Life Sciences, Tsinghua University, Beijing: elav-GAL4, OK107-GAL4, and flip-out strain (hsp70-flp; actin < y+< GAL4, UAS-GFP/CyO). HP1 loss-of-function allele Su(var)20505 was described previously (41) and provided by Lori Wallrath, University of Iowa, Iowa City, IA. UAS-GFPRNAi and w1118 were used as controls.
Immunostaining of the Polytene Chromosomes.
The salivary glands were rapidly dissected from third-instar larvae in PBS (pH 7.2). For nonhistone protein immunostaining, the chromosomes were incubated in fixation solution containing 1.85% (vol/vol) formaldehyde and 45% (vol/vol) acetic acid in double-distilled water (ddH2O) for 15 min; for immunostaining of the histone modifications, the chromosomes were fixed in 3.7% (vol/vol) formaldehyde and 1% Triton X-100 in PBS for 6 min and then transferred to a solution containing 3.7% formaldehyde and 50% (vol/vol) acetic acid in ddH2O for 10 min. Alternatively, the chromosomes were fixed in 1% citric acid, 6 mM MgCl2, and 1% Triton X-100 for 2 min as described in a previous report (42). The squashed chromosomes were labeled with the following antibodies: monoclonal mouse C1A9 anti-HP1 (1:100), mouse anti-H3K9me2 (1:50; ab1220), rabbit anti-H3K9me2 (1:50; 07–441; Upstate), rabbit antiacetylation of histone H4 on Lys16 (1:50; 07–329; Upstate), monoclonal mouse anti-Ser2 phosphorylated CTD of RNAPII (1:50; H5; Covance), and rabbit anti-CDK12 (1:400; gift from Arno Greenleaf, Duke University Medical Center, Durham, NC). The chromosomes were then incubated with secondary antibodies (1:200; Jackson ImmunoResearch Laboratories) and stained with DAPI at 0.1 μg/mL. The chromosomes were examined with a Leica Microscope using a 60× oil lens (Leica DMI 4000B), and the images were processed with Adobe Photoshop CS3 software.
To compare the levels of HP1 and H3K9me2 on the polytene chromosomes between the control and the CDK12 RNAi flies, we captured the images with identical exposure times and quantified the results of three representative images under the same conditions using ImageJ. To quantify the staining, we first selected the area of interest (with immunofluorescence signal) using the freeform tool and measured the area to get the integrated density readings. We then selected the regions without any immunofluorescence signal and got the mean background readings. Finally, we used the following formula to calculate the corrected immunofluorescence intensity (CIFI):
Average CIFIs in the control and the CDK12 RNAi results were recorded. The relative fluorescence intensities were calculated by normalizing CDK12 RNAi readings to control values. Error bars denote SD. Significance was estimated using a Student’s two-tailed t test assuming equal variance.
Flip-Out Analysis and Whole-Salivary Gland Immunostaining.
Random clones in the fat body and the salivary gland were generated using the flip-out system. For clones in the fat body, embryos (2–6 h after egg laying) were heat-shocked at 37 °C for 1 h; for clones in the salivary gland, first-instar larvae were heat-shocked at 37 °C for 1 h. Third-instar larvae that expressed GFP were selected in cold PBS. For polytene chromosome immunostaining, the salivary glands were squashed and immunostained as described above.
For fat body immunostaining, the fat bodies were dissected and fixed in 3.7% (vol/vol) formaldehyde in PBS solution with 0.3% Triton X-100 (PBST) for 20 min at room temperature. After rinsing three times in PBST, the fat bodies were incubated in blocking solution [5% (vol/vol) goat serum in PBST] for 1 h at room temperature followed by incubation with primary antibodies at 4 °C overnight and secondary antibodies for 2 h at room temperature.
For whole-salivary gland immunostaining, salivary glands were dissected and fixed in 3.7% (vol/vol) formaldehyde in PBS containing 1% Triton X-100 for 15 min at room temperature. After rinsing three times in PBS containing 0.1% Triton X-100, the glands were permeabilized in PBS containing 1% Triton X-100 for 1 h. Then, the salivary glands were incubated in primary antibodies [diluted in PBS containing 5% (vol/vol) goat serum and 0.1% Triton X-100] at 4 °C overnight and secondary antibodies for 2 h at room temperature.
The following primary antibodies were used: mouse anti-H3K9me2 (1:50; ab1220), monoclonal mouse anti-Ser2 phosphorylated CTD of RNAPII (1:50; H5; Covance), and rabbit anti-GFP (1:400; 598; MBL). The images were captured by confocal microscopy (Leica SP5) and processed with Adobe Photoshop CS3 software.
ChIP-Seq.
CDK12 RNAi virgin females were crossed with actin-GAL4 males to drive the ubiquitous knockdown and raised at 18 °C. The w1118 strain was crossed with the same GAL4 strain as a control. For total larval chromatin preparation, ∼3,500 third-instar larvae were collected, frozen in liquid nitrogen, and stored at −80 °C. Larvae extracts were treated as described previously with some modifications (39). At room temperature, the homogenized cell suspensions were cross-linked with 1% formaldehyde (F8775; Sigma) for 20 min, quenched with 125 mM glycine for 5 min, and filtered through Miracloth (475855; CalbioChem) to remove debris. After sonication in a Branson Digital Sonifer with 25% output for 10 min (2 s on and 4 s off), the chromatin was sheared into fragments with a length of 150–500 bp (with peaks at ∼200–300 bp).
Twelve independent immunoprecipitations were conducted for each antibody in a biological replicate. For each immunoprecipitation, 4 μL HP1 polyclonal antibody (anti-rabbit), 4 μL CDK12 polyclonal antibody (anti-rabbit; gift from Arno Greenleaf), or 3 μL H3 antibody (ab1791) and the H3K9me2 antibody (ab1220) were used. Protein A Sepharose Fast Flow (GE Healthcare) was preblocked and prepared as a 50% suspension; 30 μL bead suspension was added to each immunoprecipitation. The immune complexes were washed, and the beads were collected into one tube and eluted in freshly prepared elution buffer. After reverse cross-linking, the input DNA and the immunoprecipitation DNA were purified with a PCR Purification Kit (Qiagen). To validate the ChIP enrichment, quantitative real-time PCR was performed using a Bio-Rad Detection System (iQ5). Three replicates were prepared in a 20 μL final volume with SYBR Green Mix (Takara). To compare the HP1 enrichment on target genes in different samples, rp49 was used as an internal reference gene. The ChIP signal for each immunoprecipitation was calculated relative to the input and further normalized to rp49. The H3K9me2 ChIP signal was normalized to the H3 ChIP signal and rp49. To validate the ChIP enrichment of CDK12, Actin and rp49 served as positive control genes. Primer sequences are available on request.
For construction of the sequencing library, the recovered DNA fragments from both the input and immunoprecipitation samples were analyzed on an Agilent 2100 Bioanalyzer to determine the size and concentration. Library construction was performed according to the Illumina DNA Library Construction protocol, and sequencing was performed on HiSeq1500 and HiSeq2500.
Reads Alignment and ChIP-Seq Peak Calling.
The ChIP-seq reads were mapped to the Drosophila melanogaster genome (dm3) using the BWA software with default parameters. Two biological replicates were merged in compressed binary version of the sequence alignment/map (BAM) format, which then was transformed to Browser Extensible Data (BED) format for peak calling. The peak calling was performed using MACS (version 1.4.2). In addition to the default parameters, some advanced parameters were set in MACS considering that HP1 and H3K9me2 associated with repetitive elements, which are not modeled. Additionally, shift size = 73, and the maximum number of duplicate tags at the same position = 5. Enrichment of the HP1 and H3K9me2 signal was calculated relative to the input. The complete dataset is available at www.ncbi.nlm.nih.gov/geo/ (accession no. GSE63011). The occupancy profiles of HP1 and H3K9me2 were recorded into files in wiggle track format for analysis of the data in the University of California at Santa Cruz Genome Browser.
For analyzing increased or decreased HP1/H3K9me2 binding peaks after CDK12 depletion, nonoverlapping peaks between samples were chosen as candidate differential binding regions. The tag count of these regions in each sample was normalized to total mapped reads and their corresponding input reads. Then, each candidate peak was assigned a fold change of normalized tag count between samples. Peaks with a fold change larger than 1.2 were considered as differential binding regions. We defined unique peaks in CDK12-depleted samples as increased binding peaks and unique peaks in control samples as decreased binding peaks.
To analyze the average enrichment of HP1 and H3K9me2 in the control and CDK12-depleted samples, we focused on the peak regions in the genome where the binding levels of HP1 or H3K9me2 had increased after CDK12 depletion and conducted a plot analysis in the regions 5 kb upstream and downstream from the peak center. Each 10-kb window was divided into 40 bins, the average read per kilobase of bin per million signals in each bin was calculated, and a curve graph was drawn in R. The same method was used in analyzing the average enrichment of CDK12 in the targeted regions in the control where the binding levels of HP1 or H3K9me2 had increased after CDK12 depletion.
Real-Time qRT-PCR.
RNA from larvae and adult brains was isolated using TRIzol Reagent according to the manufacturer’s instructions (Invitrogen). The RNA concentration was quantitated with a NanoDrop 2000c Spectrophotometer (Thermo Scientific). A sample of 2 μg total RNA was used to prepare 20 μL cDNA with oligod(T)18 primer (Takara) and M-MLV Reverse Transcriptase (Promega). Real-time quantitative PCR amplification reactions were performed on a Bio-Rad Detection System (iQ5). Three replicates were prepared in a 20 μL final volume with SYBR Green Mix (Takara). The primer sequences are available on request. To compare the gene expression in different samples, rp49 was used as an internal reference gene. The target gene expression levels in the RNAi samples were calculated relative to the control and further normalized to rp49.
Western Blot Analysis.
Sixty third-instar larvae were homogenized in 240 μL 2× SDS loading buffer [250 mM Tris⋅HCl, pH 6.8, 10% (wt/vol) SDS, 50% (vol/vol) glycerol, 0.5 M DTT, 0.1% bromophenol blue] and boiled for 10 min followed by centrifugation at 13,000 × g at 4 °C for 15 min. The supernatants were transferred to a new tube for the subsequent experiments. For Western blotting using salivary glands, 120 pairs of salivary glands from CDK12 RNAi and control larvae were dissected in ice-cold PBS, homogenized in 200 μL 4% (wt/vol) SDS, and centrifuged in a QIAshredder Column (QIAGEN). The protein concentration was determined using the BCA Protein Assay Kit (Novagen). The lysate was boiled in loading buffer for 10 min. Samples containing 20 μg protein were loaded per well in 12% (wt/vol) running SDS/PAGE gel, and the proteins were transferred to PVDF membranes (Millipore). The following antibodies were used: rabbit anti-CDK12 (1:1,000; gift from Arno Greenleaf), monoclonal mouse C1A9 anti-HP1 (1:1,000), mouse anti-H3K9me2 (1:2,000; ab1220), mouse anti-H3 (1:4,000; ab1791), mouse anti-Actin (1:2,000; 5A7; abmart), and rabbit anti-H3S10P (1:1,000; ab32107).
FlyAtlas Data Analysis.
The tissue expression data were from the FlyAtlas database. Log10 of the expression ratios was taken and normalized to the whole-fly genome-wide mean; 52 and 141 genes were shown to have increased HP1 and H3K9me2 association, respectively, on the X chromosome after CDK12 depletion. Heat maps were generated by clustering these genes with the hclust and dist functions in R with default parameters.
Courtship Learning and Memory.
Flies were raised at 25 °C and 60–70% humidity under a 12-h/12-h light/dark cycle and collected within 4 h after eclosion. The males were collected and isolated in small food vials individually (1.5-mL Eppendorf tubes with food). The females were collected and stored in groups in glass tubes containing food (∼30 animals per tube). Flies were aged for ∼4–5 d until sexually mature. On the day before the courtship suppression training, a virgin female was placed with a naive male overnight, and we then collected the mated females.
For the learning assay, male flies were placed in each cell (diameter of 15 mm and depth of 8 mm) of the eight-cell wheel chamber using a mouth aspirator and allowed to adapt to the chamber for ∼3–5 min. A mated female was introduced to each cell and paired with the male for 1 h. Video was recorded of the initial 10 min and the final 10 min of the 1-h training period (43).
For the long-term memory training, a male was paired with a mated female in a small food vial (1.5-mL Eppendorf tubes with cotton plugs) for 5 h. After training, the males were transferred into fresh food vials until testing. The memory of the males was tested by memory-pairing each fly with a freeze-killed virgin female (tester, freeze 1 min at −80 °C immediately before the test) in the chamber. Video was recorded for 10 min. As a control (sham training), each male was isolated in a food vial for 5 h and then submitted to the same testing.
For each 10-min recording, we calculated the courtship index (CI) of the males, CIinitial, CIfinal, CItest, and CIsham, which is the fraction of time that a male showed courtship behavior (orientation, wing vibration, licking, and attempted copulation) during the 10-min recording. If copulation occurred during training or if the CIinitial was too low (<0.1), the data were eliminated from additional analysis. The learning index (LI) was determined as the ratio of the courtship level in the final 10 min of the training (CIfinal) to that of the initial 10 min (CIinitial). The memory index (MI) was calculated by dividing CItest by the mean of CIsham (43). If LI or MI is ≥1, it indicates that there is no learning or memory:
Supplementary Material
Acknowledgments
We thank Dr. Arno L. Greenleaf for providing the CDK12 antibody and Dr. Lori Wallrath for the Su(var)20505 fly stock. We thank Dr. Jose Carlos Pastor-Pareja for Balancer stock. We thank Dr. Yi Zhong for sharing equipment and members of his laboratory for constructive suggestions. We thank Dr. Haitao Li for insights and helpful discussions. We thank the TsingHua Fly Center and the Bloomington Drosophila Stock Center for fly strains. This work is supported by Ministry of Science and Technology of the People’s Republic of China Grants 2011CB965300, 2013CB835100, 2013CB35102, and 2015BAI09B03; National Natural Science Foundation of China Grants 31330043, 31371496, 31371489, and 91419304; Projects of the Science and Technology Program of Yunnan Province Grant 2013GA003; the Shanghai Pujiang Program 13PJ1408100; the 1000 Talents Youth Program; and the Tsinghua-Peking Center for Life Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE63011).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1502943112/-/DCSupplemental.
References
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 3.Day JJ, Sweatt JD. Epigenetic mechanisms in cognition. Neuron. 2011;70(5):813–829. doi: 10.1016/j.neuron.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elgin SC, Grewal SI. Heterochromatin: Silence is golden. Curr Biol. 2003;13(23):R895–R898. doi: 10.1016/j.cub.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410(6824):120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 6.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410(6824):116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 7.Eissenberg JC, Elgin SC. HP1a: A structural chromosomal protein regulating transcription. Trends Genet. 2014;30(3):103–110. doi: 10.1016/j.tig.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu LP, Ni JQ, Shi YD, Oakeley EJ, Sun FL. Sex-specific role of Drosophila melanogaster HP1 in regulating chromatin structure and gene transcription. Nat Genet. 2005;37(12):1361–1366. doi: 10.1038/ng1662. [DOI] [PubMed] [Google Scholar]
- 9.Riddle NC, et al. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 2011;21(2):147–163. doi: 10.1101/gr.110098.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cryderman DE, et al. Role of Drosophila HP1 in euchromatic gene expression. Dev Dyn. 2005;232(3):767–774. doi: 10.1002/dvdy.20310. [DOI] [PubMed] [Google Scholar]
- 11.Sun FL, et al. cis-Acting determinants of heterochromatin formation on Drosophila melanogaster chromosome four. Mol Cell Biol. 2004;24(18):8210–8220. doi: 10.1128/MCB.24.18.8210-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W, et al. The JIL-1 histone H3S10 kinase regulates dimethyl H3K9 modifications and heterochromatic spreading in Drosophila. Development. 2006;133(2):229–235. doi: 10.1242/dev.02199. [DOI] [PubMed] [Google Scholar]
- 13.Bartkowiak B, et al. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010;24(20):2303–2316. doi: 10.1101/gad.1968210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blazek D, et al. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011;25(20):2158–2172. doi: 10.1101/gad.16962311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng SW, et al. Interaction of cyclin-dependent kinase 12/CrkRS with cyclin K1 is required for the phosphorylation of the C-terminal domain of RNA polymerase II. Mol Cell Biol. 2012;32(22):4691–4704. doi: 10.1128/MCB.06267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delattre M, Spierer A, Jaquet Y, Spierer P. Increased expression of Drosophila Su(var)3-7 triggers Su(var)3-9-dependent heterochromatin formation. J Cell Sci. 2004;117(Pt 25):6239–6247. doi: 10.1242/jcs.01549. [DOI] [PubMed] [Google Scholar]
- 17.de Wit E, Greil F, van Steensel B. Genome-wide HP1 binding in Drosophila: Developmental plasticity and genomic targeting signals. Genome Res. 2005;15(9):1265–1273. doi: 10.1101/gr.3198905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39(6):715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 19.Dudai Y, Jan YN, Byers D, Quinn WG, Benzer S. dunce, a mutant of Drosophila deficient in learning. Proc Natl Acad Sci USA. 1976;73(5):1684–1688. doi: 10.1073/pnas.73.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veverytsa L, Allan DW. Subtype-specific neuronal remodeling during Drosophila metamorphosis. Fly (Austin) 2013;7(2):78–86. doi: 10.4161/fly.23969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki DT, Grigliatti T, Williamson R. Temperature-sensitive mutations in Drosophila melanogaster. VII. A mutation (para-ts) causing reversible adult paralysis. Proc Natl Acad Sci USA. 1971;68(5):890–893. doi: 10.1073/pnas.68.5.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bushey D, Huber R, Tononi G, Cirelli C. Drosophila Hyperkinetic mutants have reduced sleep and impaired memory. J Neurosci. 2007;27(20):5384–5393. doi: 10.1523/JNEUROSCI.0108-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abou Tayoun AN, Pikielny C, Dolph PJ. Roles of the Drosophila SK channel (dSK) in courtship memory. PLoS One. 2012;7(4):e34665. doi: 10.1371/journal.pone.0034665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cowan TM, Siegel RW. Drosophila mutations that alter ionic conduction disrupt acquisition and retention of a conditioned odor avoidance response. J Neurogenet. 1986;3(4):187–201. doi: 10.3109/01677068609106849. [DOI] [PubMed] [Google Scholar]
- 25.Griffith LC, Wang J, Zhong Y, Wu CF, Greenspan RJ. Calcium/calmodulin-dependent protein kinase II and potassium channel subunit eag similarly affect plasticity in Drosophila. Proc Natl Acad Sci USA. 1994;91(21):10044–10048. doi: 10.1073/pnas.91.21.10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pal-Bhadra M, et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303(5658):669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 27.Piacentini L, et al. Heterochromatin protein 1 (HP1a) positively regulates euchromatic gene expression through RNA transcript association and interaction with hnRNPs in Drosophila. PLoS Genet. 2009;5(10):e1000670. doi: 10.1371/journal.pgen.1000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheutin T, et al. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299(5607):721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 29.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20(21):2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 30.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128(4):707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 31.Lin CH, et al. Heterochromatin protein 1a stimulates histone H3 lysine 36 demethylation by the Drosophila KDM4A demethylase. Mol Cell. 2008;32(5):696–706. doi: 10.1016/j.molcel.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eifler TT, et al. Cyclin-dependent kinase 12 increases 3′ end processing of growth factor-induced c-FOS transcripts. Mol Cell Biol. 2015;35(2):468–478. doi: 10.1128/MCB.01157-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davidson L, Muniz L, West S. 3′ end formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes Dev. 2014;28(4):342–356. doi: 10.1101/gad.231274.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Even Y, et al. CDC2L5, a Cdk-like kinase with RS domain, interacts with the ASF/SF2-associated protein p32 and affects splicing in vivo. J Cell Biochem. 2006;99(3):890–904. doi: 10.1002/jcb.20986. [DOI] [PubMed] [Google Scholar]
- 35.Ko TK, Kelly E, Pines J. CrkRS: A novel conserved Cdc2-related protein kinase that colocalises with SC35 speckles. J Cell Sci. 2001;114(Pt 14):2591–2603. doi: 10.1242/jcs.114.14.2591. [DOI] [PubMed] [Google Scholar]
- 36.Ronan JL, Wu W, Crabtree GR. From neural development to cognition: Unexpected roles for chromatin. Nat Rev Genet. 2013;14(5):347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frost B, Hemberg M, Lewis J, Feany MB. Tau promotes neurodegeneration through global chromatin relaxation. Nat Neurosci. 2014;17(3):357–366. doi: 10.1038/nn.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alló M, et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol. 2009;16(7):717–724. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- 39.Larschan E, et al. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell. 2007;28(1):121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 40.Lefevre G. A photographic representation and interpretation of the polytene chromosomes of Drosophila melanogaster salivary glands. In: Ashburner M, Novitski E, editors. The Genetics and Biology of Drosophila. Academic; New York: 1976. pp. 31–66. [Google Scholar]
- 41.Eissenberg JC, Morris GD, Reuter G, Hartnett T. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics. 1992;131(2):345–352. doi: 10.1093/genetics/131.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Srinivasan S, Dorighi KM, Tamkun JW. Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS Genet. 2008;4(10):e1000217. doi: 10.1371/journal.pgen.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ejima A, Griffith LC. Assay for courtship suppression in Drosophila. Cold Spring Harb Protoc. 2011;2011(2):t5575. doi: 10.1101/pdb.prot5575. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.