Significance
Influenza A viruses cause a contagious human respiratory disease that can result in high mortality rates. Here we identify a new antiviral activity of the cellular ZAPL protein. This activity involves its C-terminal PARP domain and is directed against influenza A virus. The ZAPL PARP domain binds the viral PA and PB2 proteins, leading to their degradation by the proteasome. This degradation pathway involves two posttranslational modifications of PA and PB2, poly(ADP-ribosylation) and ubiquitination. The viral PB1 protein counteracts ZAPL antiviral activity by binding close to the PARP domain, causing PA and PB2 to dissociate from ZAPL and escape degradation, indicating that the PB1 interface with ZAPL is a potential target for antiviral development.
Keywords: ZAPL PARP domain, influenza A virus polymerase protein subunits, proteasomal degradation, ubiquitination, poly(ADP-ribosylation)
Abstract
Previous studies showed that ZAPL (PARP-13.1) exerts its antiviral activity via its N-terminal zinc fingers that bind the mRNAs of some viruses, leading to mRNA degradation. Here we identify a different antiviral activity of ZAPL that is directed against influenza A virus. This ZAPL antiviral activity involves its C-terminal PARP domain, which binds the viral PB2 and PA polymerase proteins, leading to their proteasomal degradation. After the PB2 and PA proteins are poly(ADP-ribosylated), they are associated with the region of ZAPL that includes both the PARP domain and the adjacent WWE domain that is known to bind poly(ADP-ribose) chains. These ZAPL-associated PB2 and PA proteins are then ubiquitinated, followed by proteasomal degradation. This antiviral activity is counteracted by the viral PB1 polymerase protein, which binds close to the PARP domain and causes PB2 and PA to dissociate from ZAPL and escape degradation, explaining why ZAPL only moderately inhibits influenza A virus replication. Hence influenza A virus has partially won the battle against this newly identified ZAPL antiviral activity. Eliminating PB1 binding to ZAPL would be expected to substantially increase the inhibition of influenza A virus replication, so that the PB1 interface with ZAPL is a potential target for antiviral development.
Influenza A viruses cause an annual contagious respiratory disease in humans and are responsible for pandemics that result in higher mortality rates (1). The influenza A virus genome is composed of eight segments of negative sense viral RNAs (2). The three largest segments encode the three subunits (PA, PB1, PB2) of the viral polymerase that is responsible for viral RNA synthesis in infected cells (3, 4). The smallest segment encodes the NS1 protein, a multifunctional nonstructural protein that counters host antiviral responses and regulates other cellular and viral functions (5). A major function of the NS1 protein of most influenza A virus strains is the binding of a cellular pre-mRNA 3′-end processing factor, CPSF30, thereby inhibiting the production of cellular mRNAs, including IFN mRNAs (6).
A major goal of influenza A virus research is to identify specific host proteins that either facilitate or inhibit virus replication. To identify host proteins associated with the NS1 protein that is bound to CPSF30, we purified CPSF30-NS1 complexes from infected cells by sequential affinity selection of CPSF30 and NS1 (7). This purification should yield primarily those cellular and viral proteins that interact with CPSF30 and/or NS1, either directly or indirectly. Because many cellular proteins bind to the NS1 protein, and some of these interactions with NS1 may be mutually exclusive (5), it is likely that there are multiple CPSF30-NS1 complexes that differ with respect to the cellular protein(s) that are bound to the NS1 protein. We have already demonstrated that one cellular protein that is associated with purified CPSF30-NS1 complexes, the DDX21 RNA helicase, is a regulator of influenza A virus gene expression (7).
The cellular ZAPL antiviral protein is also associated with purified CPSF30-NS1 complexes (ref. 7 and the present study). ZAPL is a 902-amino-acid-long PARP protein that contains an N-terminal domain with four zinc fingers, internal TPH and WWE domains, and a C-terminal PARP domain (8). Previous studies showed that ZAPL antiviral activity is mediated by its N-terminal zinc fingers, which bind to mRNAs of several viruses and promote degradation of these mRNAs by recruiting the exosome (8–14). This ZAPL antiviral activity has not been reported to act against influenza A virus. The ZAPS protein, which lacks the PARP domain, also promotes the degradation of specific viral mRNAs.
Much less is known about the antiviral role of the ZAPL PARP domain, which lacks the poly(ADP-ribosylation) activity of other cytoplasmic PARP proteins (15). ZAPL was shown to have stronger activity than ZAPS in degrading viral mRNAs and inhibiting Semliki Forest virus and Moloney murine leukemia virus, but it was not determined how the PARP domain enhances these activities (8). A subsequent study showed that ZAPL is bound to endo/lysosomes via S-farnesylation of a Cys in the PARP domain and that this binding enhances mRNA degradation and antiviral activity against Sinbis virus (16). It is not known how these ZAPL activities are enhanced by endo/lysosomal association. Another study showed that the three amino acids (triad) in the ZAPL PARP domain, which deviates from the triad required for poly(ADP-ribosylation) activity (15), is necessary for antiviral activity against Sindbis virus (17). It is not known why the alternate triad is required for this antiviral activity.
Here we identify the role of the C-terminal PARP domain of ZAPL in antiviral activity directed against influenza A virus. The ZAPL PARP domain binds the PA and PB2 polymerase proteins, leading to their proteasomal degradation. This antiviral activity is counteracted by the PB1 polymerase protein, which binds to an adjacent region of ZAPL and causes PA and PB2 to dissociate from ZAPL and thus escape degradation. Because PB1 antagonizes this ZAPL antiviral activity, influenza A virus replication is only moderately inhibited (20- to 30-fold) by endogenous ZAPL. These results indicate that influenza A virus has partially won the battle against this newly identified ZAPL antiviral activity and that the PB1 interface with ZAPL is a potential target for antiviral development.
Results
ZAPL, Which Is Associated with Purified Infected Cell CPSF30-NS1 Complexes, Inhibits Influenza A Virus Replication.
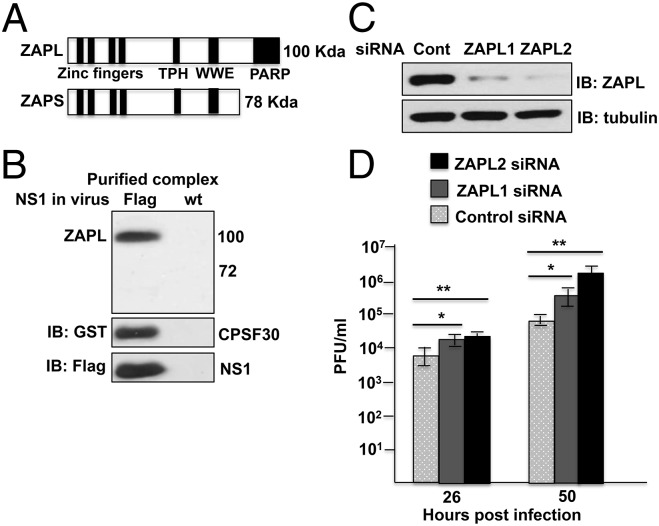
We purified CPSF30-NS1 complexes from influenza A/Udorn/72 (Ud) virus-infected cells as described previously (7). The 293T cells were transfected with a plasmid expressing GST-CPSF30, followed by infection with a recombinant Ud virus that expresses Flag-NS1 or wild-type (WT) NS1 protein as a control. CPSF30-NS1 complexes were purified by sequential affinity selections of GST and Flag. We have already shown that the viral PB1, PB2, PA, NP, and NS1 proteins and the cellular DDX21 RNA helicase are in CPSF30-NS1 complexes (7). In addition, mass spectrometry identified a substantial number of peptides of the cellular ZAP protein only in the proteins of 90- to 110-kDa molecular weight in purified CPSF30-NS1 complexes (7). These results suggested that CPSF30-NS1 complexes probably contain the large ZAPL isoform (100 kDa), but not the small ZAPS isoform (78 kDa) that lacks the C-terminal PARP domain (Fig. 1A). An antibody (Ab) directed against the N-terminal region that is shared by these two isoforms verified that the ZAPL protein, but not the ZAPS protein, is in the purified CPSF30-NS1 complexes and that neither ZAP protein was present in the control sample (Fig. 1B). Both ZAPL and ZAPS are present in virus-infected cells, but ZAPL predominates (Fig. S1A).
Fig. 1.
ZAPL inhibits influenza A virus replication. (A) Domains in human ZAPL and ZAPS. (B) Immunoblot of purified CPSF30-NS1 complexes and control sample with ZAP Ab. (C) HeLa cells were transfected with a ZAPL-specific siRNA, either ZAPL1 or ZAPL2 siRNA or control siRNA, followed by Ud virus infection. (D) Bars show SD of triplicate assays of virus titers at the indicated times after infection. The immunoblot of cell extracts shows the efficiency of ZAPL knockdown. *P < 0.05; **P < 0.005. See also Fig. S1.
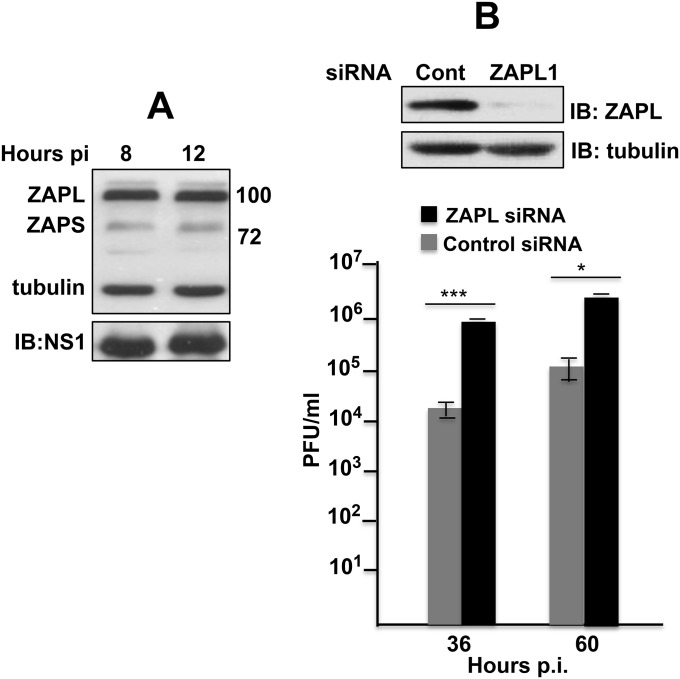
Fig. S1.
ZAPL inhibits influenza A virus replication. (A) Both ZAPL and ZAPS are present in influenza A virus-infected cells. Immunoblot with ZAP Ab of extracts collected at 8 and 12 h after Ud virus infection. (B) A549 cells were transfected with either the ZAPL1 or control siRNA followed by Ud virus infection. Bars show SD of triplicate assays of virus titers at the indicated times after infection. The immunoblot shows the efficiency of ZAPL knockdown. *P < 0.05; ***P < 0.005. This figure is related to Fig. 1.
To establish whether endogenous ZAPL inhibits Ud virus infection, we transfected human HeLa cells with two different ZAPL siRNAs that target the mRNA sequence encoding the ZAPL PARP domain. In this experiment ZAPL2 siRNA was more effective than ZAPL1 in knocking down ZAPL (Fig. 1D). Cells transfected with a ZAPL or control siRNA were infected with Ud virus at a low multiplicity of infection, and virus production was assayed at 26 and 50 h after infection. Replication of Ud virus was enhanced 20- to 30-fold in cells transfected with the ZAPL2 siRNA compared with cells transfected with the control siRNA. Less enhancement of virus replication was observed in cells transfected with the ZAPL1 siRNA that was less effective in ZAPL knockdown. ZAPL knockdown in human A549 cells also enhanced Ud virus replication 20- to 30-fold (Fig. S1B). These results establish that endogenous ZAPL inhibits influenza A virus replication in human cells.
ZAPL Binds the PB1 Protein and Reduces the Levels of the PA and PB2 Proteins.
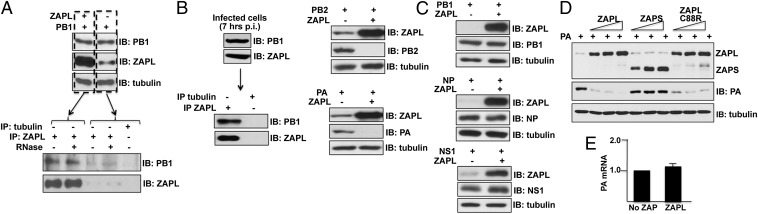
To determine whether ZAPL binds any of the viral proteins previously identified in CPSF30-NS1 complexes, 293T cells were transfected with a plasmid expressing PB1, PB2, PA, NP, or NS1 with or without cotransfection with a plasmid expressing ZAPL. Cell extracts were immunoprecipitated with ZAPL Ab, followed by immunoblots probed with Abs directed against each of the viral proteins. Of these viral proteins, only PB1 bound to ZAPL in a RNase-resistant manner (Fig. 2A), indicating that this binding involves protein–protein interactions. NP also bound to ZAPL, but this binding was eliminated by RNase digestion (Fig. S2A), indicating that this interaction is mediated by RNA. For this reason, we focused on the PB1-ZAPL protein–protein interaction in subsequent experiments. The PB1 protein of another influenza A virus strain also binds to ZAPL in a RNase-resistant manner (Fig. S2B). These results indicate that the binding of ZAPL to PB1 is likely responsible at least in part for the presence of ZAPL in infected cell CPSF30-NS1 complexes. In addition, endogenous ZAPL binds the PB1 protein synthesized during virus infection: PB1 was coimmunoprecipitated from an infected cell extract with ZAPL Ab (Fig. 2B).
Fig. 2.
ZAPL binds the PB1 protein and reduces the levels of the PA and PB2 proteins, but not via RNA-binding activity of its N-terminal zinc fingers. (A) Extracts of 293T cells cotransfected with ZAPL and PB1 plasmids were immunoprecipitated with ZAPL Ab, followed by immunoblots with ZAPL or PB1 Ab as described in SI Materials and Methods. (B) Extracts of 293T cells infected with 0.1 pfu/cell of Ud virus were immunoprecipitated with ZAPL Ab, followed by immunoblots probed with PB1 or ZAPL Ab. (C) Extracts of 293T cells transfected with a NS1, PB1, NP, PB2, or PA plasmid and a ZAPL or empty plasmid were immunoblotted with Ab against the plasmid-expressed ZAPL or viral proteins. (D) Effect of increasing amounts of ZAPL, ZAPS, or ZAPL C88R mutant plasmids on plasmid-driven production of PA protein. (E) Quantitative RT-PCR measurement of PA mRNA in cells cotransfected with a PA plasmid and a ZAPL or empty plasmid was carried out in triplicate as described in SI Materials and Methods. Error bars are shown. See also Fig. S2.
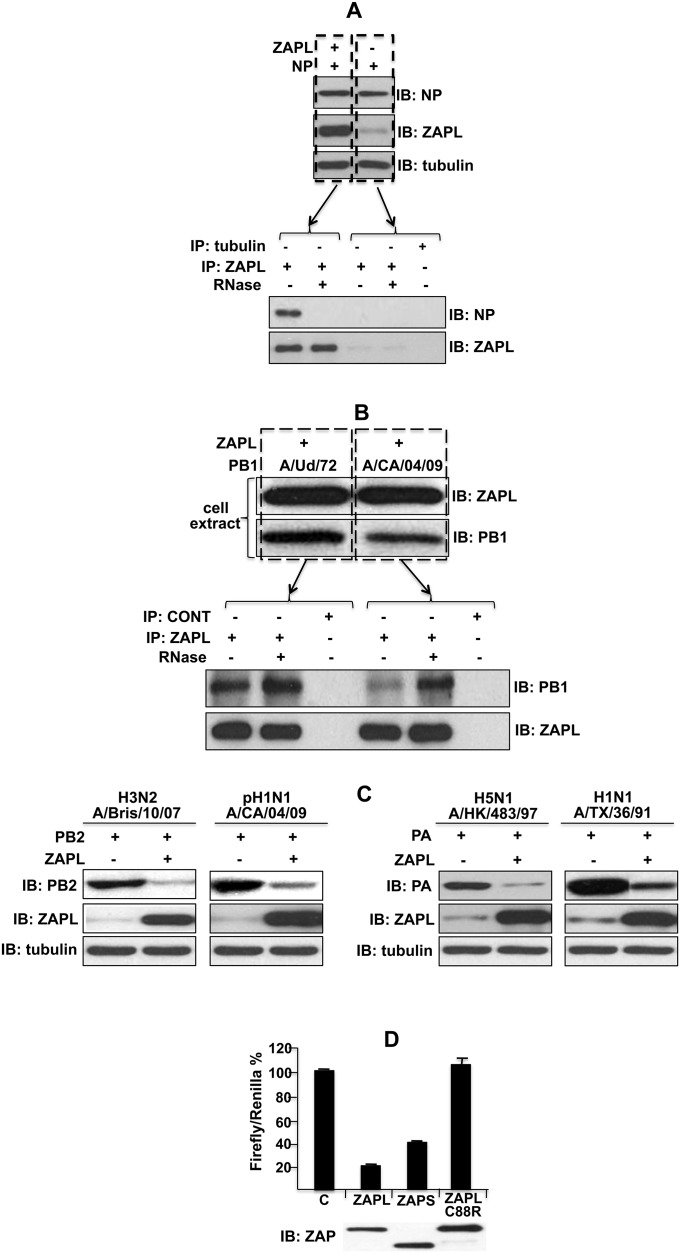
Fig. S2.
NP binding to plasmid-expressed ZAPL was eliminated by RNase digestion; the binding of the PB1 protein of another influenza A virus strain to ZAPL was RNase-resistant; the levels of the PB2 and PA proteins of other influenza A virus strains were reduced by ZAPL and assay of RNA degradation activity of ZAP proteins. (A) The 293T cells were transfected with a ZAPL plasmid and a NP plasmid. Cell extracts were immunoprecipitated with ZAPL or tubulin Ab and were then treated with RNase or not treated. The immunoprecipitates were then immunoblotted with ZAPL or NP Ab. (B) Cells were cotransfected with a ZAPL plasmid and a plasmid expressing the PB1 protein of either pH1N1 influenza A/CA/04/09 or A/Ud/72 virus. Cell extracts were immunoblotted with ZAPL or PB1 Ab. Extracts were immunoprecipitated with ZAPL Ab or tubulin Ab (as a control), with or without RNase treatment, followed by immunoblots with PB1 or ZAPL Ab. pH1N1 is the pandemic 2009 H1N1 virus. (C) Cells were cotransfected with a ZAPL plasmid or an empty plasmid and a plasmid expressing the PB2 or PA protein of the indicated influenza A virus strain. Cell extracts were immunoblotted with PB2, PA, ZAPL, or tubulin Ab. (D) The 293T cells were cotransfected with 50 ng of the LlucSN plasmid and a pcDNA3 plasmid expressing ZAPL (0.5 μg), ZAPS (0.2 μg), ZAPLC88R (1.2 μg), or an empty plasmid (“C” sample). To control for transfection efficiency, 50 ng of a pcDNA3 plasmid expressing Renilla luciferase was included. The total amount of transfected plasmids was equalized by the addition of an empty pcDNA3 plasmid. After 24 h of transfection, cells were collected, and the activities of the two luciferases in cell extracts were determined using a Mithras microplate luminometer. The Firefly/Renilla ratio is shown as a percentage of the control (No ZAP). Transfections were carried out in triplicate, and error bars are shown. The ZAP immunoblot shows that equal amounts of the three ZAP proteins were produced. This figure is related to Fig. 2.
The PB2 and PA proteins did not bind ZAPL in the coimmunoprecipitation experiment described above. Instead, immunoblots of the cell extracts before immunoprecipitation with ZAPL Ab showed that plasmid expression of ZAPL reduced the levels of the PA and PB2 proteins (Fig. 2C). The same result was obtained with the PA and PB2 proteins of other influenza A virus strains (Fig. S2C). In contrast, the levels of the NS1, PB1, and NP proteins were not reduced by plasmid expression of ZAPL (Fig. 2C).
ZAPL-Mediated Reduction of the Level of the PA Protein Does Not Result from Degradation of PA mRNA.
Previous studies of ZAPL showed that it binds to some viral mRNAs via N-terminal zinc fingers and promotes degradation of these mRNAs (8–14). Because a C88R mutation in the second zinc finger of ZAPL eliminates RNA-binding and degradation activities (10), we introduced this mutation into ZAPL and determined whether this mutated ZAPL protein inhibits production of the PA protein. We first verified that, unlike WT ZAPL and ZAPS, the mutated ZAPL lacks mRNA degradation activity, using an assay for ZAP-mediated RNA degradation established by others (8) (Fig. S2D). We transfected cells with the PA plasmid and increasing amounts of plasmids expressing either WT ZAPL, ZAPL with the C88R mutation, or ZAPS (Fig. 2D). The WT ZAPL and the C88R mutant ZAPL effectively reduced the amount of the PA protein to essentially the same extent, indicating that the mRNA degradation activity of ZAPL is not involved in reducing the level of the PA protein. ZAPS, which causes RNA degradation but lacks the PARP domain, did not reduce the level of PA, indicating that the PARP domain is involved in the reduction of PA, which is confirmed below. We verified that mRNA degradation does not have a role in inhibiting PA protein production: quantitative RT-PCR showed that plasmid-expressed ZAPL did not decrease the level of PA mRNA (Fig. 2E).
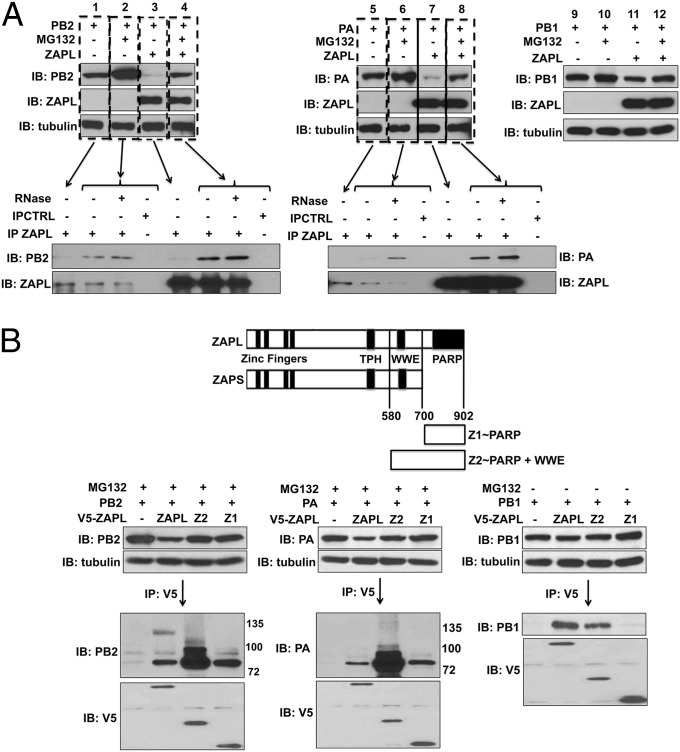
PARP Domain of ZAPL Binds PA and PB2 and Promotes Their Proteasomal Degradation, Whereas PB1 Binds to the Adjacent WWE Region of ZAPL.
Based on the above results, we tested an alternative mechanism for ZAPL action, namely, that plasmid-expressed ZAPL promotes proteasomal degradation of the PA and PB2 proteins. Cells were transfected with the PA or PB2 plasmid with or without the ZAPL plasmid and 18 h later were treated with the MG132 proteasomal inhibitor or DMSO (as a control) for 24 h (Fig. 3A). In cells transfected with the ZAPL plasmid, very little PA or PB2 protein was detected in the absence of MG132 (lanes 3 and 7), and the levels of these two proteins were substantially increased by MG132 treatment (lanes 4 and 8), showing that plasmid-expressed ZAPL results in proteasomal degradation of PA and PB2. In addition, as shown by their coimmunoprecipitation by ZAPL antibody, the PA and PB2 proteins that were rescued from proteasomal degradation by MG132 treatment were bound to ZAPL in a RNase-resistant manner, suggesting that ZAPL binds PB2 and PA to promote their degradation. Plasmid-expressed ZAPL was largely responsible for these effects on the levels of the PB2 and PA proteins because these PB2 and PA levels were substantially lower than the levels in the absence of the ZAPL plasmid, both without and with MG132 treatment. In the absence of the ZAPL plasmid, the levels of the PB2 and PA proteins were increased by MG132 treatment (lanes 1 and 2 and lanes 6 and 7), and the MG132-stabilized PB2 and PA proteins were bound to endogenous ZAPL in a RNase-resistant manner. These results suggest that endogenous ZAPL targets PB2 and PA to promote their proteasomal degradation, which is validated below. In contrast, the level of PB1 was not increased by MG132 treatment in the absence of plasmid-expressed ZAPL and was only slightly increased (∼twofold) by MG132 treatment in cells transfected with the ZAPL plasmid (Fig. 3A, lanes 9–12), in confirmation of the results of Fig. 2C.
Fig. 3.
PB2 and PA bind to the ZAPL PARP domain and undergo proteasomal degradation, whereas PB1 binds to an adjacent WWE region of ZAPL and is not degraded. (A) Cells were treated with MG132 or DMSO after transfection with the indicated plasmids. The levels of PB2, PA, and PA were determined by immunoblots of cell extracts. Cell extracts were immunoprecipitated with ZAPL Ab as described in SI Materials and Methods to determine interaction of PB2 and PA with ZAPL. (B, Top) Diagram of the Z1 and Z2 fragments of ZAPL. The 293T cells were cotransfected with a PB2, PA, or PB1 plasmid and a V5-ZAPL, V5-Z2, V5-Z1, or empty plasmid, followed where indicated by MG132 treatment. Cell extracts were immunoprecipitated with V5 Ab and immunoblotted with the indicated Ab.
To establish whether PA and PB2 bind to the PARP domain of ZAPL, we transfected cells with plasmids expressing different V5-tagged fragments of ZAPL to determine their ability to bind PB2, PA, and PB1 (Fig. 3B). In the PB2- and PA-binding assays, cells were treated with MG132 to minimize PB2 and PA proteasomal degradation by ZAPL. The Z1 ZAPL fragment comprising the PARP domain was sufficient to bind PA and PB2, but not PB1, confirming that the PARP domain is required for binding PB2 and PA, but not PB1. The Z2 fragment comprising both the PARP and WWE domains bound PB1 almost as efficiently as the full-length ZAPL protein, indicating that PB1 probably binds predominantly to the ZAPL region containing the WWE domain. Because a fragment containing only the WWE domain (amino acids 580–700) was very poorly expressed in transfection assays, it was not feasible to use this fragment for binding assays.
In addition, the Z2 fragment bound substantially more PB2 and PA than full-length ZAPL, and some of the Z2-bound PB2 and PA proteins were larger in size, indicating that they contain posttranslational modifications. Because the WWE domain has been shown to bind poly(ADP-ribose) chains (18), it is likely that some of the PB2 and PA proteins associated with the Z2 fragment are poly(ADP-ribosylated), which is verified below.
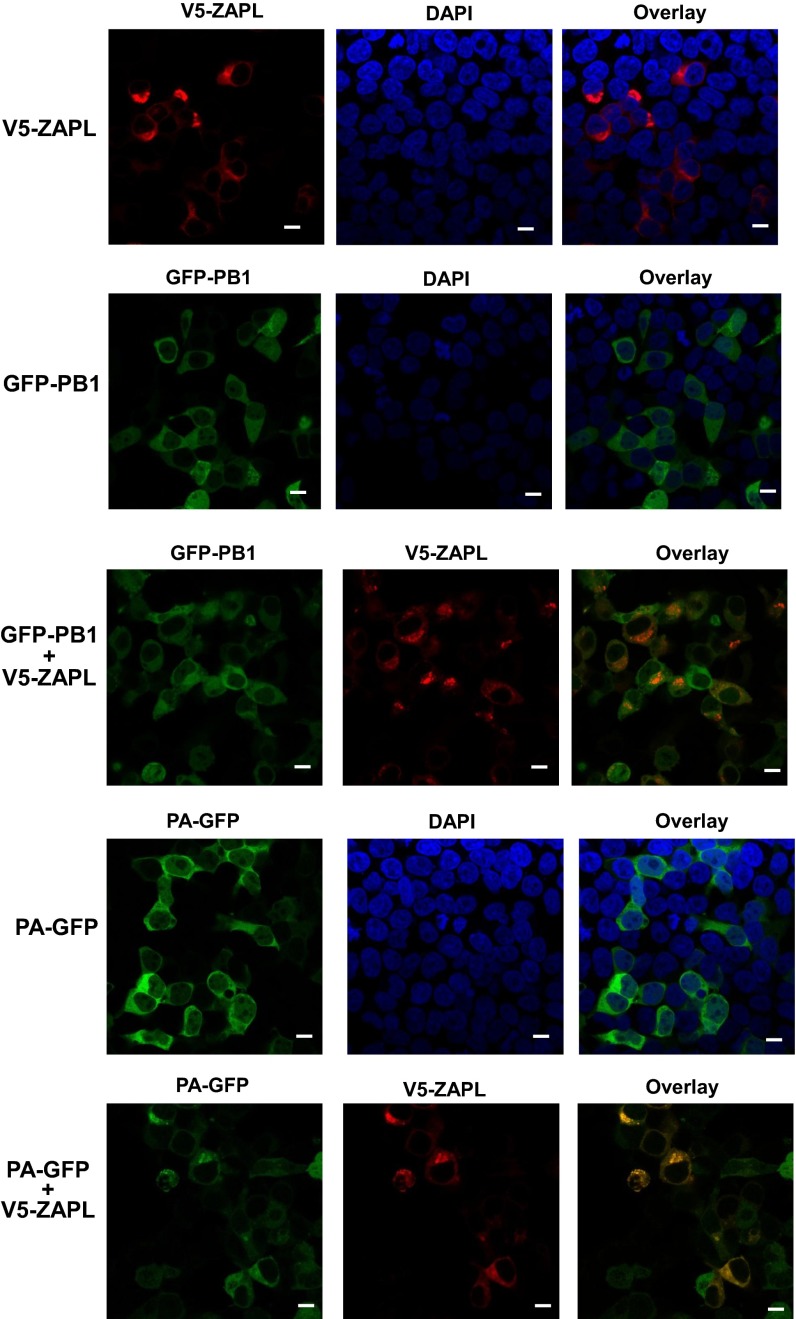
ZAPL, a cytoplasmic PARP protein (16, 19), would be expected to interact with the PA, PB2, and PB1 proteins in the cytoplasm. Individually expressed PA and PB1 proteins are localized primarily in the cytoplasm, whereas PB2 is primarily localized in the nucleus (20). Nonetheless, immunofluorescence analyzed by confocal microscopy of cells cotransfected with a ZAPL plasmid and a PA, PB2, or PB1 plasmid verified that ZAPL interacts in the cytoplasm with PB2 as well as with PA and PB1 (Figs. S3 and S4).
Fig. S3.
ZAPL interacts with PB1 and PA in the cytoplasm. The 293T cells were transfected with plasmids expressing GFP-PB1, PA-GFP, and V5-ZAPL, where indicated, for 36 h. The cells were then fixed with 4% (vol/vol) paraformaldehyde and permeabilized with 0.5% Triton X-100. V5-ZAPL was visualized using mouse anti-V5 antibody and Alexa fluor 546-conjugated goat anti-mouse antibody (Life Technologies). Cell nuclei were stained with DAPI. Images were obtained using a Zeiss LSM 710 confocal microscope, and data were processed with ZEN 2012 software. (Scale bar, 10 μm.)
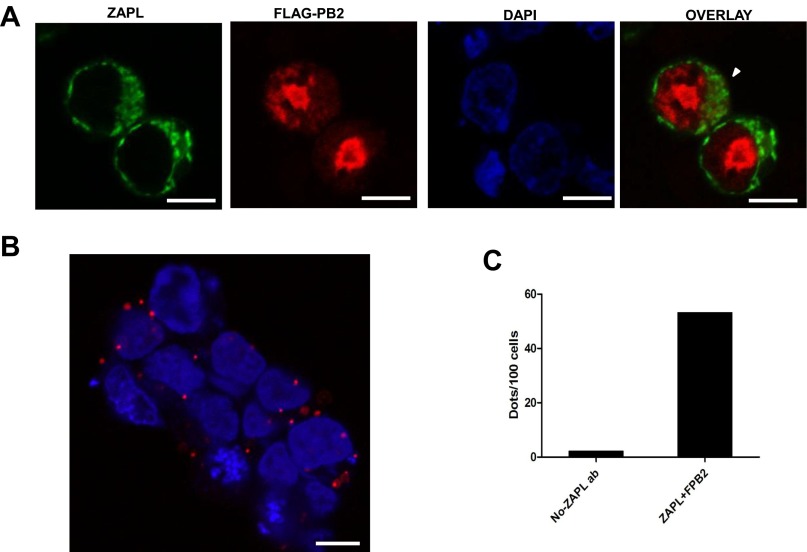
Fig. S4.
ZAPL interacts with PB2 in the cytoplasm. (A) Confocal microscopy. The 293 T cells were transfected with plasmids expressing ZAPL and Flag-PB2 for 24 h, followed by treatment with 5 μM MG132 for 24 h. The cells were fixed with 4% (vol/vol) paraformaldehyde and permeabilized with 0.5% Triton X-100. Flag-PB2 was visualized using mouse monoclonal antibody against Flag and Alexa fluor 546-conjugated goat anti-mouse antibody. ZAPL was visualized by rabbit antibody against ZAPL and Alexa fluor 488-conjugated goat anti-rabbit antibody, respectively. Cell nuclei were stained with DAPI. Partial colocalization of PB2 and ZAPL in the cytoplasm is indicated by the white arrowhead. (Scale bar, 10 μm.) (B) Duolink in situ proximity ligation assay. Cells transfected with ZAPL and Flag-PB2 plasmids were treated as above and incubated with the same primary antibodies, and the cells were then processed for the proximity ligation assay (PLA) (32) as described in the manual of the Duolink PLA start kit. Multiple red dots in cytoplasm were detected, which validated the colocalization of PB2 and ZAPL in the cytoplasm. (Scale bar, 10 μm.) (C) Number of dots per 100 cells was counted with comparison with the negative control in which the ZAPL primary Ab was omitted in the assay. Representative data of three repeats are shown.
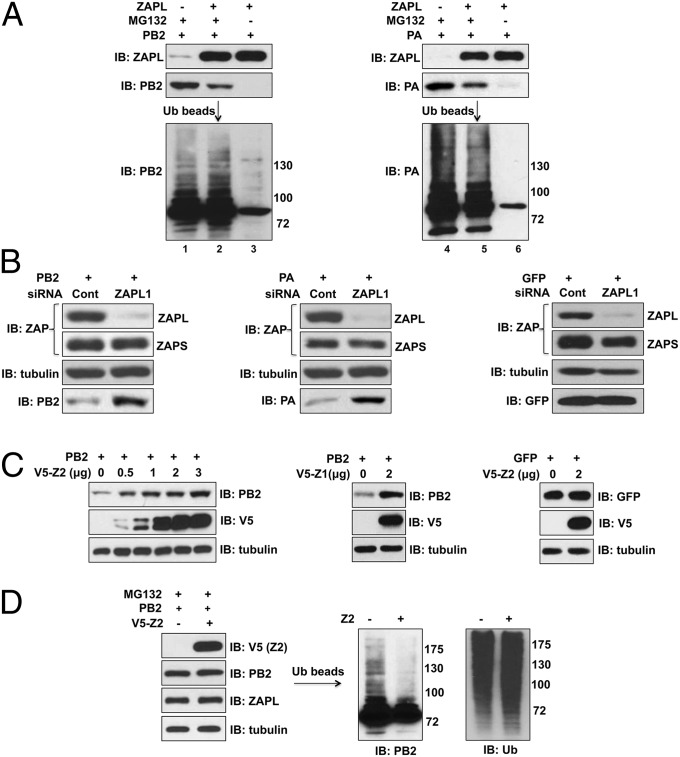
Endogenous ZAPL, Like Plasmid-Expressed ZAPL, Is Required for the Ubiquitination and Degradation of PB2 and PA.
Because the proteasome degrades ubiquitinated proteins, we expected that PA and PB2 would be ubiquitinated before proteasomal degradation. To determine whether ubiquitination of PB2 and PA requires ZAPL, cells were transfected with a ZAPL plasmid and a PB2 or PA plasmid, with and without MG132 treatment. Another set of cells was transfected with only a PA or PB2 plasmid, but not with a ZAPL plasmid, and the cells were then treated with MG132. Ubiquitinated proteins were affinity-selected from cell extracts using beads linked to tandem ubiquitin-binding domains, followed by an immunoblot probed with PA or PB2 Ab. Multiple ubiquitinated proteins larger in size than PB2 or PA were detected in the cells transfected with the ZAPL plasmid (Fig. 4A, lanes 2 and 5). Most of these ubiquitinated proteins were absent when PB2 and PA were not protected against proteasomal degradation by MG132 treatment (lanes 3 and 6), indicating that the observed ubiquitinated proteins are primarily PB2 and PA ubiquitin conjugates.
Fig. 4.
Plasmid-expressed and endogenous ZAPL are required for ubiquitination and degradation of PA and PB2. (A) Extracts of 293T cells transfected with the indicated plasmids, followed, where indicated, with MG132 treatment, were mixed with beads linked to tandem ubiquitin-binding domains to select for ubiquitinated proteins as described in SI Materials and Methods. The selected proteins were immunpoblotted with PB2 or PA Ab. (B) ZAPL was knocked down in 293T cells by ZAPL1 siRNA to determine the effect on the levels of retrovirus-expressed PB2, PA, or GFP. (C) V5-Z2 or Z1 was overexpressed in 293T cells for 48 h before retroviral expression of PB2 or GFP for another 24 h to determine the effect of Z2 and Z1 on the levels of PB2 and GFP. (D) The 293T cells were transfected with a V5-Z2 or empty plasmid for 40 h, followed by infection with a retrovirus expressing PB2 and then MG132 treatment. Ubiquitination of PB2 was determined as described in SI Materials and Methods.
Ubiquitinated PB2 and PA proteins were also detected in the MG132-treated cells in the absence of plasmid-expressed ZAPL (lanes 1 and 4). To determine whether endogenous ZAPL is responsible for this ubiquitination and degradation, we carried out two types of experiments. First, cells were transfected with either a control siRNA or ZAPL1 siRNA that efficiently knocked down endogenous ZAPL (Fig. 4B). Cells were then infected with a retrovirus encoding PB2, PA, or GFP as a control. SiRNA knockdown of endogenous ZAPL effectively rescued PB2 and PA from degradation and did not affect GFP expression. In the second approach, we overexpressed the Z1 or Z2 ZAPL fragment to bind and sequester PB2 away from endogenous ZAPL. Cells were then infected with a retrovirus expressing either PB2 or GFP. Overexpression of either Z1 or Z2 rescued PB2 from degradation by endogenous ZAPL (Fig. 4C). In contrast, Z2 did not affect the level of GFP. These results demonstrate that endogenous ZAPL, like plasmid-expressed ZAPL, is required for the degradation of PB2 and PA. Endogenous ZAPL is also required for ubiquitination: Z2 overexpression substantially reduced the ubiquitination of PB2 that occurs in the absence of plasmid-expressed ZAPL without affecting overall protein ubiquitination (Fig. 4D, right).
Poly(ADP-Ribosylated) PB2 and PA Are Bound to ZAPL and Are Involved in ZAPL-Dependent Proteasomal Degradation.
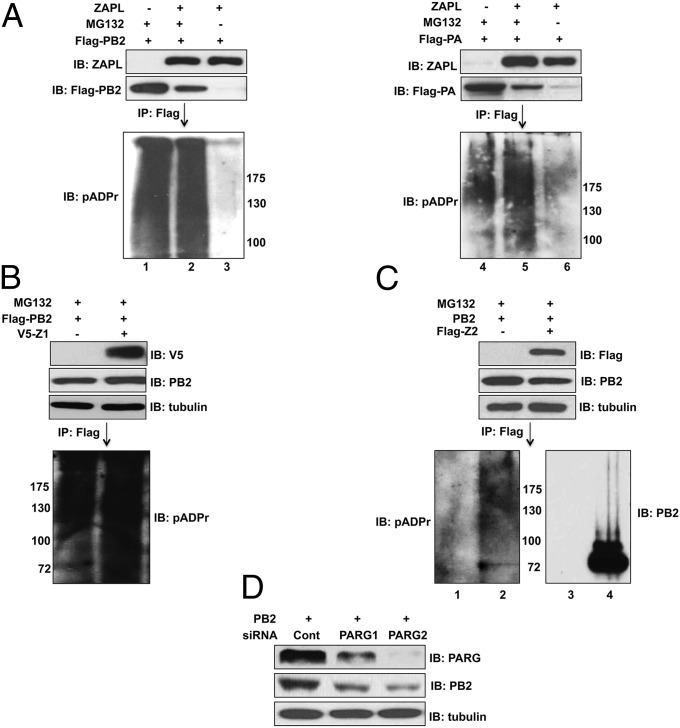
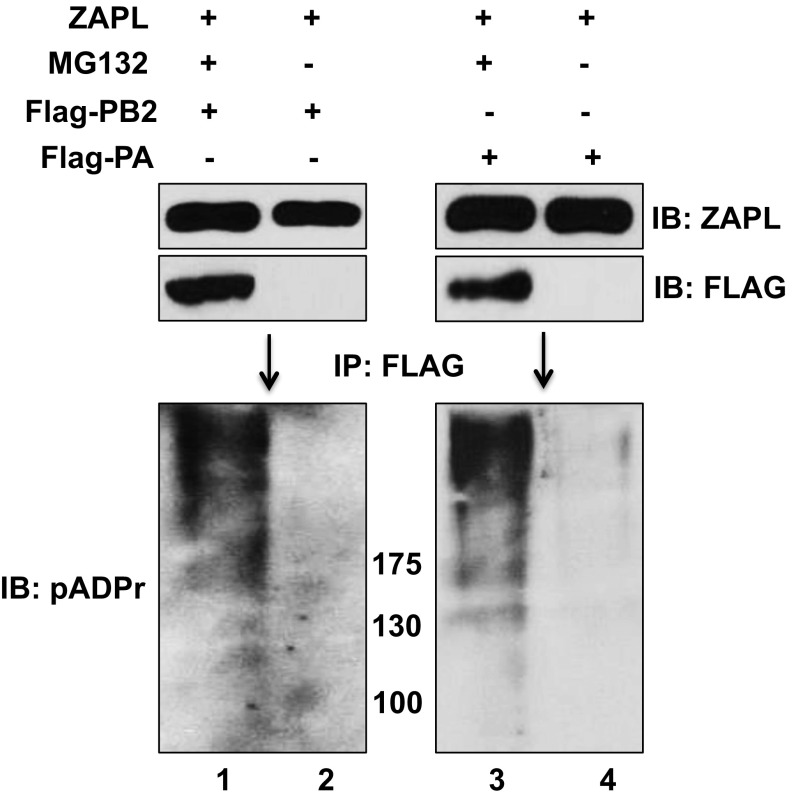
Cytoplasmic PARP-containing proteins can initiate proteasomal degradation by poly(ADP-ribosylating) their target proteins, followed by ubiquitination catalyzed by a E3 ubiquitin ligase that recognizes poly(ADP-ribose) and the PARP protein to which the poly(ADP-ribosylated) target protein is bound (18, 19, 21, 22). However, because the PARP domain of ZAPL lacks the catalytic activity for poly(ADP-ribosylation) (15), it is likely that other PARP proteins would provide this activity, as shown previously for cellular proteins bound to ZAPL (19). To determine whether PA and PB2 are poly(ADP-ribosylated), cells were transfected with a plasmid expressing ZAPL and Flag-PB2 or Flag-PA with and without MG132 treatment. Another set of cells was transfected with only a Flag-PA or Flag-PB2 plasmid with MG132 treatment. Cell extracts were immunoprecipitated with Flag Ab-conjugated beads, followed by an immunoblot probed with a polyclonal Ab generated against poly(ADP-ribose) chains (pADPr Ab) (19). Multiple poly(ADP-ribosylated) protein species larger in size than PB2 and PA were detected in cells transfected with the ZAPL plasmid when proteasome degradation of PB2 and PA was inhibited by MG132 treatment (Fig. 5A, lanes 2 and 5). These poly(ADP-ribosylated) proteins appear as a smear because of the heterogeneity in the length of the poly(ADP-ribose) chains that were added, as previously observed by others (19). Most of these poly(ADP-ribosylated) protein species were absent when PB2 and PA were degraded in the absence of MG132 treatment (lanes 3 and 6), indicating that the observed poly(ADP-ribosylated) proteins (lanes 2 and 5) are primarily poly(ADP-ribosylated) PB2 and PA proteins. To verify that PB2 and PA are poly(ADP-ribosylated), cell extracts were subjected to denaturing conditions to dissociate PB2 and PA from other proteins before immunoprecipitation with Flag Ab-conjugated beads: again poly(ADP-ribosylated) PB2 and PA were detected in cells transfected with the ZAPL plasmid only when proteasome degradation of PB2 and PA was inhibited by MG132 treatment (Fig. S5).
Fig. 5.
PB2 and PA are poly(ADP-ribosylated), but not by ZAPL, and this poly(ADP-ribosylation) is involved in ZAPL-mediated degradation. (A) The 293T cells were transfected with the indicated plasmids and then treated with MG132 where indicated. Cell extracts were immunoprecipitated with Flag Ab, followed by immunoblots probed with pADPr Ab. (B) V5-Z1 was overexpressed in 293T cells followed by retrovirus expression of Flag-PB2 and then MG132 treatment. The level of poly(ADP-ribosylation) of PB2 was determined as described in A. (C) The 293T cells were transfected with a PB2 plasmid and a Flag-Z2 or empty plasmid followed by MG132 treatment. Extracts were immunoprecipitated with Flag Ab, followed by immunoblots probed with pADPr or PB2 Ab. (D) PARG was knocked down in 293T cells by a siRNA to determine the effect on the level of retrovirus-expressed PB2 as described in SI Materials and Methods. See also Fig. S5.
Fig. S5.
Verification that PB2 and PA are poly(ADP-ribosylated). The 293T cells were transfected with the indicated plasmids and then treated with MG132 where indicated. Cell extracts were subjected to denaturing conditions to dissociate PB2 and PA from other proteins. The extracts were then immunoprecipitated with Flag Ab, followed by immunoblots probed with pADPr Ab. This figure is related to Fig. 5.
Poly(ADP-ribosylated) PB2 and PA proteins were also detected in MG132-treated cells in the absence of plasmid-expressed ZAPL (Fig. 5A, lanes 1 and 4). As expected, ZAPL is not required for this poly(ADP-ribosylation): overexpression of the Z1 ZAPL fragment to sequester PB2 away from endogenous ZAPL did not reduce the poly(ADP-ribosylation) of PB2 in the absence of plasmid-expressed ZAPL (Fig. 5B).
A key issue is whether poly(ADP-ribosylated) PB2 and PA are associated with ZAPL and are part of the ZAPL-dependent degradation pathway for PB2 and PA. We have shown that PB2 and PA proteins containing posttranslational modifications are associated with the plasmid-expressed Z2 fragment of ZAPL, which contains the WWE domain as well as the PARP domain (Fig. 3B). To determine whether these posttranslational modifications include the addition of poly(ADP-ribose) chains, cells were transfected with a PB2 plasmid and a Flag-Z2 or empty plasmid, and the cells were treated with MG132 to minimize proteasomal degradation. Cell extracts were affinity-selected with a Flag Ab and immunoblotted with pADPr Ab or PB2 Ab (Fig. 5C). Substantially more poly(ADP-ribosylated) species were detected in cells transfected with the Flag-Z2 plasmid (lane 2) than with the empty plasmid (lane 1), indicating that poly(ADP-ribosylated) PB2 is associated with the Z2 fragment of ZAPL. To demonstrate that poly(ADP-ribosylation) of PB2 is involved in ZAPL-dependent degradation of PB2, poly(ADP-ribosylation) of cytoplasmic proteins was increased by siRNA knockdown of cytoplasmic PARG proteins [poly(ADP-ribose) glycohydrolases] that remove poly(ADP-ribose) modifications from cytoplasmic proteins (23). Two different PARG-specific siRNAs were used. Cells were then infected with a retrovirus expressing PB2. Knockdown of PARG substantially increased PB2 degradation (Fig. 5D). The extent of the siRNA knockdown of PARG correlated with the extent of the increase in PB2 degradation. These results indicate that poly(ADP-ribosylation) of PB2, which is not carried out by ZAPL, is nonetheless involved in ZAPL-mediated degradation of PB2.
Mechanism by Which PB1 Counteracts ZAPL-Dependent Degradation of PA and PB2.
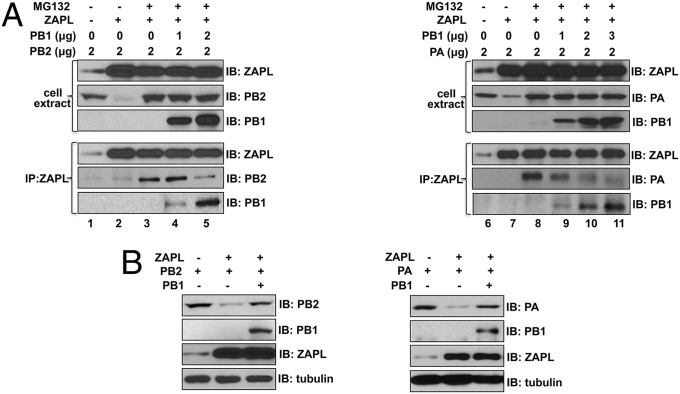
Because the PB1-binding site on ZAPL is relatively close to the PB2- and PA-binding sites (Fig. 3C), we postulated that PB1 binding to ZAPL would lead to the dissociation of PA and PB2 from ZAPL. To test this possibility, we carried out binding assays in cells treated with MG132, conditions under which PB2 and PA are stabilized and remain bound to ZAPL. Plasmid-expressed PA or PB2 that was bound to ZAPL after MG132 treatment was dissociated from ZAPL by the addition of increasing amounts of a PB1 plasmid, concomitant with PB1 binding to ZAPL (Fig. 6A, lanes 4 and 5; lanes 9–11). The dissociation of PA and PB2 from ZAPL would be expected to protect them from ZAPL-mediated degradation. To test this prediction, we determined whether PB1 protects PA and PB2 from ZAPL-mediated degradation in the absence of MG132 treatment (Fig. 6B). The degradation of plasmid-expressed PB2 and PA in cells transfected with the ZAPL plasmid was largely reversed by the transfection of a PB1 plasmid. These results show that PB1 binding to ZAPL dissociates PA and PB2 from ZAPL, thereby protecting PA and PB2 from degradation.
Fig. 6.
PB1 protects PA and PB2 from degradation by binding to ZAPL, leading to dissociation of PA and PB2 from ZAPL. (A) Cells were transfected with the indicated plasmids and then, where indicated, treated with MG132. Cell extracts were immunoprecipitated with ZAPL Ab to determine PB2, PA, and PB1 interactions with ZAPL. (B) Cells were transfected with ZAPL, PB2, PA, and PB1 plasmids where indicated, and cell extracts were immunoblotted with the indicated Ab.
Discussion
Here we identify a new antiviral activity of ZAPL and demonstrate that this activity is directed against influenza A virus. The previously identified ZAPL antiviral activity is mediated by its N-terminal fingers that bind to the mRNAs of several viruses and promote the degradation of these mRNAs (8–14). In contrast, the ZAPL antiviral activity identified in the present study involves its C-terminal PARP domain, which binds the influenza A virus PB2 and PA polymerase proteins. These two viral proteins are poly(ADP-ribosylated), presumably by other cytoplasmic PARP proteins, as shown previously for cellular proteins bound to ZAPL (19). The ZAPL-associated, poly(ADP-ribosylated) PA and PB2 proteins are then ubiquitinated, followed by proteasomal degradation. This ZAPL-dependent mechanism explains why endogenous ZAPL inhibits influenza A virus replication, as we established by a siRNA knockdown experiment.
The PB1 protein counteracts ZAPL antiviral activity by binding to the WWE region adjacent to the PARP domain, causing PB2 and PA to dissociate from ZAPL and escape degradation. This function of the PB1 protein explains why influenza A virus infection is only moderately inhibited (20- to 30-fold) by endogenous ZAPL, indicating that influenza A virus has partially won the battle against this newly identified ZAPL antiviral activity. ZAPL, which is located in the cytoplasm (16, 19), would be expected to interact with the PA, PB2, and PB1 proteins in the cytoplasm before they interact with each other and enter the nucleus. Consistent with this prediction, immunofluorescence of cells cotransfected with a ZAPL and a PA, PB2, or PB1 plasmid verified that ZAPL interacts with PA, PB2, and PB1 in the cytoplasm. The situation in virus-infected cells is more complicated, where cytoplasmic PB1 could potentially interact not only with ZAPL, but also with PA and DDX21 (7, 20), indicating that only some, but not all of the PB1 in the cytoplasm would be available to bind to ZAPL. Nonetheless, we showed that ZAPL binds the PB1 protein in virus-infected cells. Because PB1 binding on ZAPL inhibits ZAPL antiviral activity, eliminating this PB1 binding would be expected to substantially increase the inhibition of influenza A virus replication, so the PB1 interface with ZAPL is a potential target for antiviral development.
ZAPL also has an important cellular function. It binds Argonaute proteins, and both the Argonaute protein and ZAPL itself are poly(ADP-ribosylated) by other cytoplasmic PARP proteins (19). Poly(ADP-ribosylation) of Argonaute increases during stress, leading to the relief of microRNA-mediated silencing of translation and uninhibited translation of cellular mRNAs that are regulated by microRNAs. This cellular function is probably compromised in influenza A virus-infected cells by the diversion of ZAPL molecules to carry out its antiviral function. In other studies, it was shown that poly(ADP-ribosylated) proteins associated with a different PARP-containing protein, tankyrase, are ubiquitinated by an E3 ligase (RNF146) that recognizes poly(ADP-ribose) chains via its WWE domain and also binds to ankyrin repeats in tankyrase itself (22, 24). Hence, tankyrase is an essential scaffold for the E3 ligase that ubiquitinates the poly(ADP-ribosylated) protein bound to tankyrase.
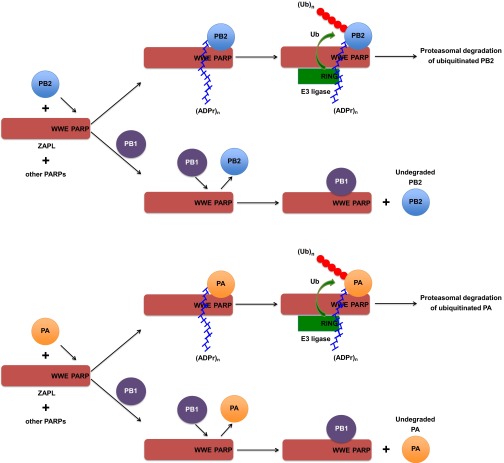
These previous results, coupled with the results that we report in the present paper, provide a working model for the ZAPL-dependent degradation of PB2 and PA (Fig. S6). We showed that PB2 and PA bind to the ZAPL PARP domain and that poly(ADP-ribosylated) PB2 and PA are associated with the region of ZAPL composed of both the PARP and WWE domains. This association can be attributed to the specific binding of the WWE domain to the poly(ADP-ribose) chains attached to PB2 and PA (18). It is likely that the association of poly(ADP-ribosylated) PB2 and PA with this region of ZAPL would be blocked by the binding of the PB1 protein to the WWE region. We found that full-length ZAPL binds substantially less modified PA and PB2 proteins than the Z2 fragment, suggesting that poly(ADP-ribosylated) PB2 and PA proteins in the Z2 region requires ZAPL sequences N-terminal to the WWE domain for further processing, i.e., ubiquitination and degradation. Accordingly, based on the tankyrase model, we propose that poly(ADP-ribosylated) PA and PB2 are ubiquitinated by an E3 ligase that recognizes not only the poly(ADP-ribose) chains on PA and PB2 but also regions of ZAPL that are N-terminal to the WWE domain. It should be noted that in this mechanism ZAPL rather than the E3 ligase provides the WWE domain that binds poly(ADP-ribose) chains, whereas in the tankyrase model the E3 ligase provides the WWE domain (22). The ubiquitinated PB2 and PA proteins are then degraded by the proteasome.
Fig. S6.
Working model for the ZAPL-dependent degradation of PB2 and PA. The mechanism by which PB1 counters ZAPL-dependent degradation of PB2 is also shown. The model is described in Discussion in the main text.
Materials and Methods
The procedures for growth of 293T, HeLa, HEL299, and MDCK cells and for the generation of WT Ud virus and Ud virus encoding a Flag-NS1 protein have been described previously (25, 26). Human ZAPL and ZAPS cDNAs were cloned by RT-PCR of RNA from human HEL299 cells using the primers shown in SI Materials and Methods. Retroviruses expressing PB2 or PA with a C-terminal Flag or GFP were generated as described in SI Materials and Methods. Ubiquitin and poly(ADP-ribosylated) chains on PB2 and PA were identified as described in the legends to Figs. 4 and 5 and in SI Materials and Methods. Complete details of experimental procedures are provided in primers shown in SI Materials and Methods.
SI Materials and Methods
Cells and Viruses.
The procedures for growth of 293T, A549, HeLa, and MDCK cells and for the generation of WT Ud virus and Ud virus encoding a Flag-NS1 protein were described previously (25, 26). Virus stocks were grown in 10-d-old fertilized eggs, and virus titers were determined by plaque assays in MDCK cells. Virus infections were carried out in A549 cells, and transfections were carried out in 293T cells.
Plasmids and Antibodies.
Human ZAPL and ZAPS cDNAs were cloned by RT-PCR of RNA isolated from human HEL299 cells. For ZAPL, the primers were the following: 5′ primer—5′-ATGGCGGACCCGGAGGTG-3′; and 3′ primer—5′-CTAACTAATCACGCAGGC-3′.
For ZAPS, the primers were the following: 5′ primer—5′-ATGGCGGACCCGGAGGTG-3′; and 3′ primer—5′- CTATCTCTTCATCTGCTG-3′. The C88R mutation was introduced into ZAPL using standard oligonucleotide mutagenesis methods. Each cDNA was cloned into the pcDNA3 vector. The LlucSN plasmid was provided by Harmit Malik (8). Rabbit Ab against PB1 was provided by Krister Melen and Ikka Julkunen, Finnish National Institute for Health and Welfare, Helsinki, Finland. Monoclonal Abs against PA and PB2 were provided by Juan Ortin (27). Ab against the major structural proteins (HA, NP, M1) of Ud virus (denoted as Ub Ab) was provided by Robert A. Lamb (28). The Ab against amino acids 300–400 of human ZAPS and ZAPL was from Abcam. Tubulin Ab was from Cell Signaling, and polyclonal poly(ADP-ribosylation) Ab was from BD-Pharmingen.
Retroviruses Expresing PB2, PA, or GFP.
The influenza Ud PB2 or PA sequence with a C-terminal Flag, or untagged GFP sequence was cloned into a pQCXIP retroviral vector kindly provided by Stephen Goff, Columbia University College of Physicians and Surgeons, New York. To generate VSV-G pseudotype retroviruses, 293T cells were cotransfected with the pQCXIP plasmid containing the PB2, PA, or GFP sequence, and pcDNA3-gag-pol and pcDNA3-VSV-G plasmids for 48 h (29). The supernatant containing the released retrovirus was collected and added to 293T cells as indicated in the main text to express PB2, PA, or GFP.
Coimmunoprecipitations.
Cells were cotransfected using the Mirius TransIT-LTR transfection reagent with the plasmids indicated in the text and were extracted with solution A [50 mM Tris⋅HCl pH 7.5, 150 mM NaCl, 5 mM EDTA, 2.5 mM MgCl2, 1% Nonidet P-40, 10% (vol/vol) glycerol, 1 mM PMSF, and 1× Protease inhibitor (Roche)]. An aliquot of the extracts was immunoblotted with the indicated Ab to measure the level of the expressed protein in the cell extracts. The extracts were immunoprecipitated with the Ab against one of the expressed proteins or with tubulin Ab as a negative control, followed by binding to Protein A/G Sepharose beads and RNase (5 μg/mL) treatment, where indicated. After washing the Sepharose beads four times with solution B containing 50 mM Tris⋅HCl pH 7.5, 150 mM NaCl, and 0.5% Nonidet P-40, proteins were eluted by heating the beads at 95 °C in a buffer containing 50 mM Tris⋅HCl 6.8, 2% (vol/vol) SDS, 6% (vol/vol) glycerol, and 2% (vol/vol) β-mercaptoethanol. The eluate was analyzed by immunoblots probed with the indicated Abs.
Ubiquitination Assay.
Cells were cotransfected with the plasmids indicated in the text, and, where indicated, were treated with 40 μM MG132 for 24 h. Cells were extracted with solution A supplemented with the following deubiquitinase inhibitors: 5 mM NEM; 20 μM PR-169; and 5 mM 1,10-phenanthroline (LifeSensors). Cell extracts were incubated with agarose beads linked to tandem ubiquitin-binding domains (TUBE-2, LifeSensors). After washing the beads four times solution B containing 300 mM NaCl, proteins were eluted and immunoblotted as described above for the coimmunoprecipitation experiments.
Poly(ADP-Ribosylation) Assay.
Cells were cotransfected with the plasmids indicated in the text, and, where indicated, were treated with 40 μM MG132. Cells were extracted with pADPr buffer containing 50 mM Hepes pH 7.4, 150 mM NaCl, 1 mM EGTA, 1 mM MgCl2, 1% TX-100, 1 mM DTT, 10 μM nocodazole (Sigma), 10 μg/mL cytochalasin D (Enzo Life Science), 1 μM PARG inhibitor ADP-HPD (EMD millipore), 1 mM NAD+, 1 mM PMSF, and 1× Protease inhibitor (Roche). Cell extracts were incubated with Flag-Ab-conjugated to agarose M2 beads. In some experiments (shown in Fig. S5), cell extracts were subjected to denaturing conditions as described by Levaot et al. (30). The pADPr buffer used to extract infected cells contained 1% SDS, and the extract was incubated at 68 °C for 5 min. The extract was sonicated three times for 15 s and was then diluted twofold to reduce the SDS concentration to 0.5%. After centrifugation to clarify the extract, it was incubated with Flag-Ab–conjugated to agarose M2 beads. After washing the agarose beads four times with the pADPr buffer containing 300 mM NaCl, proteins were eluted by incubating the beads with 500 μg/mL 3× Flag peptides (Sigma). The eluate was heated at 70 °C in a buffer containing 60 mM Tris⋅HCl 6.8, 2% (vol/vol) SDS, 10% (vol/vol) glycerol, and 1.25% (vol/vol) β-mercaptoethanol and was analyzed by immunoblots probed with an Ab against pADPr (LP96-10, BD-Pharmingen) (19, 31) or the Ab against the indicated protein.
Quantitative RT-PCR of PA mRNA.
The 293T cells were cotransfected with the PA plasmid and an empty or ZAPL plasmid. RNA was extracted from cell extracts using the TRIzol reagent, and polyadenylated mRNA was isolated using oligo(dT)-coated beads (Qiagen) and reverse-transcribed using an oligo(dT) primer. PCR amplification of PA mRNA and β-actin mRNA was carried out using the following Ud PA-specific primers: 5′-GAAATGGGGAATGGAGATGA-3′ and 5′-CATGCTCTCGATTTGTTGGA-3′, and β-actin specific primers, respectively, and the FastStart Universal Probe Master Mix (Rox) with Universal Probe Library #134 for Ud PA or #64 for human β-actin. Analysis was carried out using the Perkin-Elmer/Applied Biosystems 7900HT sequence detector. The Ct value for PA mRNA was normalized to the Ct value of β-actin mRNA, and the PA mRNA level in cells tranfected with the ZAPL plasmid was normalized to the PA mRNA level in cells transfected with the empty plasmid.
siRNA Knockdown Experiments.
Knockdown experiment was performed using either a siRNA against target mRNA or control siRNA mixed with X-tremeGENE siRNA Transfection Reagent (Roche) according to the manufacturer’s protocol. To determine the role of endogenous ZAPL in influenza A virus infection, 40 nM of ZAPL-specific siRNA—ZAPL1 (5′-AAAUUUAUCCA-GGAGCUCUGAGUUC-3′), ZAPL2 (5′-CAAGCCGUGCCUAUGUGGAAUCUAU-3′), or a control siRNA (5′-UUGAUGUUGAGCAAUUCACGUUCAU-3′)—was transfected into HeLa or A549 cells for 36 h, followed by infection with 0.01 (HeLa cells) or 0.05 (A549 cells) pfu/cell of Ud virus. Virus production at the indicated hours after infection was determined by plaque assays in MDCK cells. To knock down ZAPL or PARG in 293T cells, 80 nM ZAPL1 siRNA, or PARG-specific siRNA—PARG1 (5′-GCGGUGAAGUUAGAUUA-CAUUUCCA-3′), PARG2 (5′-GAAAUAUUU-CUAGCCUGAAUGUAGA-3′), or the control siRNA—was transfected into the cells for 40 h, followed by infection for 24 h with a retrovirus expressing PA, PB2, or GFP. Cell extracts were immunoblotted with the indicated Ab to determine the level of PA, PB2, or GFP.
Acknowledgments
This work was supported by National Institutes of Health Grant R01-AI11772 (to R.M.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509745112/-/DCSupplemental.
References
- 1.Wright P, Neumann G, Kawaoka Y. 2012. Orthomyxoviruses. Fields Virology, eds Knipe DM, Howley, PM (Lippincott Williams & Wilkins, Philadelphia), 6th Ed, pp 1186–1243.
- 2.Krug RM, Fodor E. 2013. The virus genome and its replication. Influenza Textbook, ed Webster RG, Monto AS, Braciale TJ, Lamb RA (Wiley-Blackwell, Hoboken, NJ), 2nd Ed, pp 57–66.
- 3.Resa-Infante P, Jorba N, Coloma R, Ortin J. The influenza virus RNA synthesis machine: Advances in its structure and function. RNA Biol. 2011;8(2):207–215. doi: 10.4161/rna.8.2.14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pflug A, Guilligay D, Reich S, Cusack S. Structure of influenza A polymerase bound to the viral RNA promoter. Nature. 2014;516(7531):355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- 5.Krug RM, Garcia-Sastre A. The NS1 protein: A master regulator of host and viral functions. In: Webster RG, Monto AS, Braciale TJ, Lamb RA, editors. Influenza Textbook. 2nd Ed. Wiley-Blackwell; Hoboken, NJ: 2013. pp. 114–132. [Google Scholar]
- 6.Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3'end formation of cellular pre-mRNAs. Mol Cell. 1998;1(7):991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 7.Chen G, Liu CH, Zhou L, Krug RM. Cellular DDX21 RNA helicase inhibits influenza A virus replication but is counteracted by the viral NS1 protein. Cell Host Microbe. 2014;15(4):484–493. doi: 10.1016/j.chom.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerns JA, Emerman M, Malik HS. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet. 2008;4(1):e21. doi: 10.1371/journal.pgen.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao G, Guo X, Goff SP. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science. 2002;297(5587):1703–1706. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 10.Guo X, Carroll JW, Macdonald MR, Goff SP, Gao G. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J Virol. 2004;78(23):12781–12787. doi: 10.1128/JVI.78.23.12781-12787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo X, Ma J, Sun J, Gao G. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc Natl Acad Sci USA. 2007;104(1):151–156. doi: 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bick MJ, et al. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol. 2003;77(21):11555–11562. doi: 10.1128/JVI.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Müller S, et al. Inhibition of filovirus replication by the zinc finger antiviral protein. J Virol. 2007;81(5):2391–2400. doi: 10.1128/JVI.01601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu Y, et al. Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc Natl Acad Sci USA. 2011;108(38):15834–15839. doi: 10.1073/pnas.1101676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleine H, et al. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32(1):57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Charron G, Li MM, MacDonald MR, Hang HC. Prenylome profiling reveals S-farnesylation is crucial for membrane targeting and antiviral activity of ZAP long-isoform. Proc Natl Acad Sci USA. 2013;110(27):11085–11090. doi: 10.1073/pnas.1302564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gläsker S, Töller M, Kümmerer BM. The alternate triad motif of the poly(ADP-ribose) polymerase-like domain of the human zinc finger antiviral protein is essential for its antiviral activity. J Gen Virol. 2014;95(Pt 4):816–822. doi: 10.1099/vir.0.060988-0. [DOI] [PubMed] [Google Scholar]
- 18.Wang Z, et al. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 2012;26(3):235–240. doi: 10.1101/gad.182618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leung AK, et al. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell. 2011;42(4):489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fodor E, Smith M. The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex. J Virol. 2004;78(17):9144–9153. doi: 10.1128/JVI.78.17.9144-9153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, et al. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol. 2011;13(5):623–629. doi: 10.1038/ncb2222. [DOI] [PubMed] [Google Scholar]
- 22.DaRosa PA, et al. Allosteric activation of the RNF146 ubiquitin ligase by a poly(ADP-ribosyl)ation signal. Nature. 2015;517(7533):223–226. doi: 10.1038/nature13826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer-Ficca ML, Meyer RG, Coyle DL, Jacobson EL, Jacobson MK. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp Cell Res. 2004;297(2):521–532. doi: 10.1016/j.yexcr.2004.03.050. [DOI] [PubMed] [Google Scholar]
- 24.Guettler S, et al. Structural basis and sequence rules for substrate recognition by Tankyrase explain the basis for cherubism disease. Cell. 2011;147(6):1340–1354. doi: 10.1016/j.cell.2011.10.046. [DOI] [PubMed] [Google Scholar]
- 25.Twu KY, Noah DL, Rao P, Kuo RL, Krug RM. The CPSF30 binding site on the NS1A protein of influenza A virus is a potential antiviral target. J Virol. 2006;80(8):3957–3965. doi: 10.1128/JVI.80.8.3957-3965.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao C, Denison C, Huibregtse JM, Gygi S, Krug RM. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc Natl Acad Sci USA. 2005;102(29):10200–10205. doi: 10.1073/pnas.0504754102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ochoa M, et al. Epitope mapping of cross-reactive monoclonal antibodies specific for the influenza A virus PA and PB2 polypeptides. Virus Res. 1995;37(3):305–315. doi: 10.1016/0168-1702(95)00039-s. [DOI] [PubMed] [Google Scholar]
- 28.Chen BJ, Leser GP, Morita E, Lamb RA. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J Virol. 2007;81(13):7111–7123. doi: 10.1128/JVI.00361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naldini L, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272(5259):263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 30.Levaot N, et al. Loss of Tankyrase-mediated destruction of 3BP2 is the underlying pathogenic mechanism of cherubism. Cell. 2011;147(6):1324–1339. doi: 10.1016/j.cell.2011.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seo GJ, et al. Reciprocal inhibition between intracellular antiviral signaling and the RNAi machinery in mammalian cells. Cell Host Microbe. 2013;14(4):435–445. doi: 10.1016/j.chom.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Söderberg O, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3(12):995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]