Significance
Assigning biological relevance and molecular function to large catalogues of mutated genes in cancer is a major challenge. Likewise, pinpointing drivers among thousands of transcriptionally or epigenetically dysregulated genes within a cancer is complex and limited by the lack of tools for high-throughput functional cancer genomic analyses. We show here for the first time, to our knowledge, application of the CRISPR/Cas9 genome engineering system for simultaneous (multiplexed) mutagenesis of large gene sets in adult mice, allowing high-throughput discovery and validation of cancer genes. We characterized applications of CRISPR/Cas9 multiplexing, resulting tumor phenotypes, and limitations of the methodology. By using defined genetic or environmental predisposing conditions, we also developed, to our knowledge, the first mouse models of CRISPR/Cas9-induced hepatocellular carcinoma and show how multiplexed CRISPR/Cas9 can facilitate functional genomic analyses of hepatobiliary cancers.
Keywords: in vivo CRISPR/Cas9, somatic multiplex-mutagenesis, hepatocellular carcinoma, intrahepatic cholangiocarcinoma, chromosome engineering
Abstract
Here, we show CRISPR/Cas9-based targeted somatic multiplex-mutagenesis and its application for high-throughput analysis of gene function in mice. Using hepatic single guide RNA (sgRNA) delivery, we targeted large gene sets to induce hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma (ICC). We observed Darwinian selection of target genes, which suppress tumorigenesis in the respective cellular/tissue context, such as Pten or Cdkn2a, and conversely found low frequency of Brca1/2 alterations, explaining mutational spectra in human ICC/HCC. Our studies show that multiplexed CRISPR/Cas9 can be used for recessive genetic screening or high-throughput cancer gene validation in mice. The analysis of CRISPR/Cas9-induced tumors provided support for a major role of chromatin modifiers in hepatobiliary tumorigenesis, including that of ARID family proteins, which have recently been reported to be mutated in ICC/HCC. We have also comprehensively characterized the frequency and size of chromosomal alterations induced by combinatorial sgRNA delivery and describe related limitations of CRISPR/Cas9 multiplexing, as well as opportunities for chromosome engineering in the context of hepatobiliary tumorigenesis. Our study describes novel approaches to model and study cancer in a high-throughput multiplexed format that will facilitate the functional annotation of cancer genomes.
For decades, a major bottleneck in cancer research has been our limited ability to identify genetic alterations in cancer. The revolution in array-based and sequencing technologies and the recent development of insertional mutagenesis tools in animal models enable the discovery of cancer-associated genetic alterations on a genome-wide scale in a high-throughput manner. Next-generation sequencing (NGS) of cancer genomes and transposon-based genetic screening in mice, for example, are currently creating large catalogs of putative cancer genes for principally all cancer types (1–3). A challenge for the next decades will be to validate the causative cancer relevance of these large gene sets (to distinguish drivers from passengers) and to understand their biological function. Moreover, pinpointing downstream targets of mutated cancer genes or drivers among the thousands of transcriptionally or epigenetically dysregulated genes within individual cancers is complex and limited by the lack of tools for high-throughput functional cancer genomic analyses.
The development of technologies for targeted manipulation of the mouse germ line has opened tremendous opportunities to study gene function (4, 5). Mouse models recapitulate the extensive biological complexity of human cancer and have given insights into many fundamental aspects of the disease that can be studied only at an organismal level (6). However, the speed and efficiency of such studies is limited by the long time frames needed to genetically engineer, intercross, and breed mouse cancer models.
The prokaryotic clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated protein 9 (Cas9) system has been recently adapted for genetic engineering in mammalian cells (7–13). Using 20-bp single guide RNA sequences (sgRNAs), the endonuclease Cas9 can be directed to desired genomic positions to cause a double strand break. This break is repaired by nonhomologous end joining, which commonly leaves a short insertion or deletion (indel), allowing homozygous disruption of the targeted gene. Recent studies showed that CRISPR/Cas9 is functional in germ cells and somatic cells of mice and can be used for gene editing and cancer induction in the lung and the biliary compartment (14–20). Targeting of Pten and p53 in the liver was reported to induce intrahepatic cholangiocarcinoma (ICC) (16), but CRISPR/Cas9-based modeling of hepatocellular carcinoma (HCC) (which accounts for 90% of liver cancers) has not been achieved so far, nor has complex combinatorial gene targeting. We therefore developed CRISPR/Cas9 hepatic mutagenesis approaches in multiplexed formats for high-throughput in vivo applications.
Results and Discussion
Inducing HCC and ICC by Hepatic Delivery of Multiplexed CRISPR/Cas9 in Adult Mice.
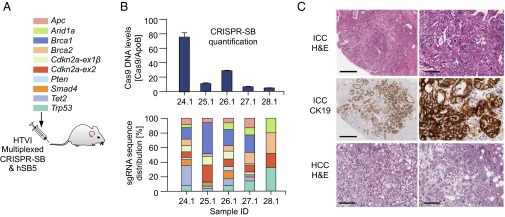
To deliver CRISPR/Cas9 to hepatocytes, we used hydrodynamic tail vain injection (HTVI) (21). We generated a vector (CRISPR-SB) carrying sgRNA and Cas9 expression cassettes (11) flanked by Sleeping Beauty (SB) inverted repeats (SI Appendix, Fig. S1). HTVI of CRISPR-SB and an SB-transposase vector (hSB5) enables, in principle, both transient CRISPR/Cas9 expression from episomal plasmids and long-term expression from SB-mobilized/genome-integrated vectors. Using HTVI of two different vectors followed by fluorescence-based detection of their cellular delivery, we found that multiple plasmids can enter a cell (SI Appendix, Fig. S2 and Supplementary Methods), providing a rationale for combinatorial CRISPR/Cas9-based tumor suppressor gene (TSG) targeting.
NGS recently discovered many putative novel ICC/HCC cancer genes (22–29), but their functional validation is largely lacking. Based on a literature search (SI Appendix, Table S1), we have chosen to target (i) bona fide TSGs that are often mutated, deleted, or epigenetically silenced in ICC/HCC (e.g., Trp53, Smad4, Pten, Cdkn2a, and Apc), (ii) TSG Arid1a, a novel commonly mutated chromatin modifier in ICC (and less frequently in HCC), and (iii) TSG Tet2, a putative (negatively regulated) downstream target of the ICC oncogenes Idh1/Idh2. We also targeted the TSGs Brca1/2, which are not or only rarely altered in ICC/HCC. Targeting efficiencies of multiple sgRNAs per gene were validated in vitro using T7E1 assays before choosing best performing sgRNAs for HTVI (SI Appendix, Fig. S3). A dominant pathway activated in ICC/HCC is Ras/MAPK signaling (30, 31). We have therefore used oncogenic KrasG12D to accelerate tumorigenesis.
We coinjected hSB5 transposase plasmid and 10 CRISPR-SB vectors and confirmed their successful delivery 2 wk later: real time quantitative PCR (qPCR) showed a random distribution pattern of the 10 sgRNAs in most animals (Fig. 1 A and B). We euthanized eight mice 20–30 wk post-HTVI and collected 21 macroscopic liver tumors (Fig. 1C). At this stage, mice typically had one to three small tumors (1–3 mm), occasionally more. We found both ICCs and HCCs (Fig. 1C and SI Appendix, Figs. S4 and S5). Conventional type ICCs showed CK19 positivity, reflecting biliary differentiation, and featured a Collagen-4–positive stromal reaction like the human disease (Fig. 1C and SI Appendix, Fig. S5). These early onset cancers were triggered by CRISPR/Cas9 because KrasG12D alone induces only low-penetrance late-onset tumors: We observed no ICCs/HCCs in a control cohort of 53 Alb-Cre;KrasLSL-G12D/+ mice aged up to 38 wk. Furthermore, we didn’t observe ICCs/HCCs in Alb-Cre;KrasLSL-G12D/+ control cohorts injected with hSB5 and Cas9-only expressing CRISPR-SB (n = 8).
Fig. 1.
Hepatic delivery of multiplexed CRISPR/Cas9 for somatic mutagenesis in mice. (A) Genes targeted simultaneously upon hydrodynamic tail vein injection (HTVI). For details of the two-vector system, see SI Appendix, Fig. S1. (B) Analysis of healthy livers 2 wk post-HTVI. (Upper) Quantification of hSpCas9 DNA copies using qPCR; Error bars, SEM from triplicate determinations. (Lower) Quantitative analysis of sgRNA distribution using qPCR with guide-specific forward primers and a generic reverse primer (color code from A). (C) Microscopic images of CRISPR/Cas9-induced tumors in Alb-Cre;KrasLSL-G12D/+ mice. ICC, intrahepatic cholangiocarcinoma. (Top) H&E staining. (Middle) Cytokeratin 19 (CK19) IHC staining. HCC, hepatocellular carcinoma. (Bottom) Two H&E-stained HCCs. (Scale bars: Left panel in Top and Middle row, 50 µm; Right panel in Top and Middle row, 400 µm; both images in Bottom row, 100 µm.)
Quantitative Analysis of Target Site Mutations in Healthy Livers And Cancers.
We performed NGS of PCR-amplified target sites in tumors and related healthy livers (Fig. 2). Because sequence reads with large deletions are often filtered out during mapping using standard bioinformatics tools, we used manually inspected/mapped capillary sequencing data of cloned PCR products (SI Appendix, Fig. S6) to optimize the algorithms for NGS-based high-throughput indel detection. Whereas Cas9-only injected control mice had no mutations at the CRISPR/Cas9 target sites, we found a total of 167 indels in the 21 tumors (Fig. 2 A and B). The majority were small and located at the position of the Cas9 double strand break insertion [3 bp upstream of the protospacer adjacent motif (PAM)]. Large deletions (>50 bp) were rare (Fig. 2 and SI Appendix, Figs. S7 and S8).
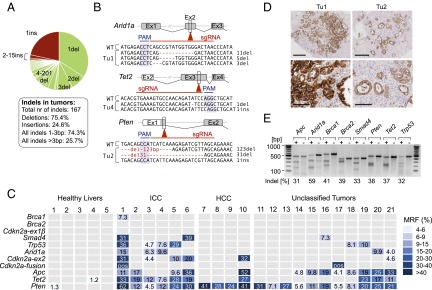
Fig. 2.
Analysis of CRISPR/Cas9 target site mutations in healthy liver and liver tumors. (A) Pie chart showing the type (deletion or insertion) and size of all indels derived from liver tumors (n = 21) of Alb-Cre;KrasLSL-G12D/+ mice (n = 8) 20–30 wk after HTVI of hSpCas9 and ten sgRNAs. (B) Examples of indel sequence context in selected tumors for Arid1a/Tet2/Pten. PAM, protospacer adjacent motif. WT, WT sequence. (C) Mutant read frequencies (MRFs) at individual target sites as determined by amplicon-based next-generation sequencing. Multiple indels at individual target sites (shown in detail in SI Appendix, Fig. S9) are presented here as combined MRFs. Frame-shift causing indels with MRFs >1% and 4% are shown for healthy livers and tumors, respectively. Pos, tumors with Cdkn2a-ex1β/Cdkn2a-ex2 fusions. Small macroscopic tumors (<1 mm) were used entirely for genomic analyses (unclassified; no histology available). (D) Comparison of cancer cell to nonneoplastic cell contents in Tu1 and Tu2. CK19 staining marks ICC. (Scale bars: Upper, 400 µm; Lower, 100 µm.) (E) Surveyor assays to assess CRISPR/Cas9 efficiency upon transient transfection of the mouse pancreatic cancer cell line PPT-53631. Indel frequencies are indicated. Cell lines were transfected with the target sgRNA (+) or a control sgRNA (−). Because PPT-53631 has homozygous Cdkn2a deletions, Cdkn2a sgRNAs were tested in a different cell line (PPT-4072) (SI Appendix, Fig. S10).
We next compared the frequency of CRISPR/Cas9-induced frame shifts causing indels at target sites in tumors and healthy livers from the same mice (Fig. 2C and detailed view in SI Appendix, Fig. S9). In-frame deletions <10 bp are less likely to have functional consequences and are therefore shown only in SI Appendix, Fig. S7 and Table S2. Normal liver samples from tumor-bearing mice exhibited no or only few mutations with low mutant read frequencies (MRFs) (fraction of mutant-reads/all-reads at individual target sites) (Fig. 2C). In contrast, all tumors had several mutations above the 4% MRF threshold, which was used to exclude their origin in healthy tissue (Fig. 2C). In Tu1, for example, MRFs reached up to 62% for individual target loci, reflecting clonal expansion of mutations. Further details about the type and frequency of mutations at individual positions are shown in SI Appendix, Fig. S9 and Table S2.
Differences of MRFs between tumors can at least partly be explained by the varying content of nonneoplastic cells. Tu2, for example, which generally had lower MRFs at mutated target sites than Tu1, also had a significantly smaller tumor/normal cell ratio (Fig. 2D and SI Appendix, Fig. S5). In contrast, extensive differences between MRFs at different target sites within one tumor could reflect intratumor heterogeneity, as shown later.
Cancer-Relevant Mutations Undergo Positive Selection.
A global comparison of the incidence of CRISPR/Cas9-induced target site mutations across tumors showed a nonrandom distribution (P = 2.2 × 10−15; χ2 test). Pten, for example, was mutated in all 21 tumors whereas Brca1 or Brca2 mutations were largely absent (only one low-frequency Brca1 mutation in Tu1). This distribution suggests that biologically relevant mutations are selected for in vivo. The high incidence of Pten mutations can indeed be explained by the key importance of PI3K signaling in hepatobiliary tumorigenesis in humans and mice (32–34). Likewise, the lack of Brca1/2 mutations reflects their extremely rare alteration in human ICC/HCC (SI Appendix, Table S1). Overall, several genes were targeted significantly more frequently than Brca1/2, including Pten (P = 6.4 × 10−15), Apc (P = 9.3 × 10−7), Tet2 (P = 6.6 × 105), Cdkn2a-ex2 (P = 0.0007), p53 (P = 0.007), and Arid1a (P = 0.02; Fisher's exact test).
The possibility of technical problems underlying the low incidence of Brca1/2 mutations in tumors can be excluded because (i) surveyor assays in vitro confirmed similar efficiencies of Brca1/2 targeting to other loci (Fig. 2E), and (ii) the “background” Brca1/2 mutation rate in healthy livers was similar to other target genes (SI Appendix, Table S2). We therefore conclude that Darwinian selection of indels with pathogenetic relevance in the specific tissue context drives tumorigenesis in our model.
Another level of evidence for in vivo selection comes from the comparison of the two Cdkn2a sgRNAs that we used: one targeting exon-1β to inactivate p19Arf and the second directed against exon-2 to disrupt both p19Arf and p16Ink4a. Whereas Cdkn2a-ex2 was mutated in 33% (7/21) of tumors, no mutations above the “background” mutation rate in healthy liver were found in Cdkn2a-ex1β (P = 0.009; Fisher's Exact test) (Fig. 2C). This observation suggests selective pressure for the double-mutant and also reflects the predominant CDKN2A inactivation pattern in human hepatobiliary cancers. To confirm that sgRNAs against both exons are in fact functional, we performed surveyor assays, which showed similar efficiencies of Cdkn2a-ex2 and Cdkn2a-ex1β targeting (SI Appendix, Fig. S10).
The pathogenic relevance of TSGs like Pten or Trp53 in ICC/HCC has been shown in vivo (35, 36). For Arid1a, which was recently discovered to be recurrently mutated in ICC/HCC (23, 24, 26, 27), such biological information is lacking. We found Arid1a alterations in 24% of tumors (Fig. 2C). In addition, 80% (11/14) of hepatobiliary cancers (3/3 ICCs and 8/11 HCCs) induced in a second HTVI approach targeting a larger set of genes (described below) had CRISPR/Cas9-induced mutations of Arid1a and/or Arid1b, another chromatin modifier that was recently discovered to be frequently mutated in ICC/HCC. These observations strongly support a role of chromatin modifying enzymes in hepatobiliary tumorigenesis.
CRISPR/Cas9 has been recently adapted for genetic screening in vitro (37–39) and also in a transplantation model (40). We show that somatic mutagenesis and cancer gene discovery are also feasible directly in vivo. A surprising finding was the high frequency of CRISPR/Cas9-induced mutations in Tet2 (particularly in ICCs) (Fig. 2C). Its tumor suppressive function might be linked to IDH1/IDH2, which carry oncogenic mutations in >10% of ICCs (23, 24, 41), leading to dioxigenase inhibition by 2-hydroxyglutarate production (42, 43). Among the 70 2OG-dependent dioxigenases, TET2 is considered a promising cancer-relevant target: TET2 and IDH1/2 mutations induce similar hypermethylation phenotypes (41, 44) and are mutually exclusive in AML, suggesting similar effects on cellular transformation (45). TET2 is not mutated in human ICC, but IDH1/2 alterations are associated with impaired TET2 function (41). Our data support TET2's pathogenetic relevance in ICC and exemplify how genetic screening can pinpoint cancer genes that are not mutated, but dysregulated by other means.
Intratumor Heterogeneity in a Small Subset of CRISPR/Cas9-Induced Cancers.
In some cancers (e.g., Tu1, -4, -5, and -21), MRFs differed extensively between individual target sites, and often more than two mutations at individual sites existed within a tumor (Fig. 2 and SI Appendix, Fig. S9). One explanation for this observation could be that some mutations occur in the transfected founder cell whereas others happen only after the first cell division in subsequent daughter cells. To explore this possibility, we compared three different regions in Tu1 (SI Appendix, Fig. S11): the large area R1 and the small microdissected areas R2 (with a well-differentiated tubular growth pattern) and R3 (showing poor differentiation and more solid growth). Target sites sequencing revealed that, even within R2/R3, many MRFs were low, suggesting additional intraregional minority clones and a complex subclonal structure, which is only partly resolved. The only mutation with consistently high MRFs in all three regions was Cdkn2a-ex2, suggesting its position at the trunk of a phylogenetic tree. R2/R3 comparison revealed substantial differences regarding driver mutations in dominant clones (SI Appendix, Fig. S11C), with Smad4-1del defining the dominant clone in R2 and Pten-1del-b in R3, suggesting that genetic heterogeneity underlies phenotypic intratumor diversity. The possibility of R1/R2/R3 being independent tumors is highly unlikely because of (i) the presence of specific high-frequency founder Cdkn2a mutations in all three regions (including a single base deletion and an 18-kb CRISPR/Cas9-induced deletion) (SI Appendix, Fig. S9 and Table S2), and (ii) the small size (3 mm) of this solitary tumor in an otherwise healthy liver.
Chromosomal Rearrangements Induced by Combinatorial CRISPR/Cas9 Targeting.
One potential limitation of multiplexed CRISPR/Cas9 mutagenesis is that, in principle, combinatorial sgRNA targeting could lead to undesired large chromosomal rearrangements (18, 20). To examine this possibility, we performed PCR-based screening for all possible deletions at chromosomes that were targeted by multiple sgRNAs (SI Appendix, Fig. S12). Out of the 105 possible deletions in 21 tumors, we found evidence for fusion products between the Cdkn2a-ex1β and Cdkn2a-ex2 sgRNA target sites in two cancers (Fig. 2C). In both cases the resulting deletion of ∼18 kb led to inactivation of both p16Ink4a and p19Arf (SI Appendix, Fig. S12). Because small indels in exon-2 mediated by a single Cdkn2a-ex2 sgRNA also inactivates both p16Ink4a and p19Arf, there is no selective pressure beyond exon-2 mutations for the 17.7-kb deletion to occur. It therefore seems that this relatively small deletion of 17.7 kb is a fairly efficient process.
We therefore next studied such potentially undesired effects of CRISPR/Cas9 multiplexing in a scenario of higher level multiplexing (18 sgRNAs targeting known or putative hepatobiliary cancer genes). Furthermore, to examine whether ICCs/HCCs can be induced by CRISPR/Cas9 multiplexing in environmental cancer-predisposing contexts, we have used not only the Kras-mutant background but also a CCl4-induced liver injury model. We have analyzed a total of 41 tumors collected in these experiments. All cancers induced in the CCl4 context (n = 35) were HCCs whereas in Alb-Cre;KrasLSL-G12D mice, we found both ICCs and HCCs. Detailed information about tumor incidences is provided in SI Appendix, Table S3. The general conclusions drawn from target site mutation sequencing were in concordance with our observations made in the 10 sgRNA studies: For example, the incidence of Brca1, Brca2, or isolated Cdkn2a-ex1β mutations was very low (20%, 10%, or 7%) whereas Pten or epigenetic regulators (Arid1a and/or Arid1b) were hit in 93% and 78% of cancers, respectively, further confirming that pathogenetically relevant mutations are selected for in vivo.
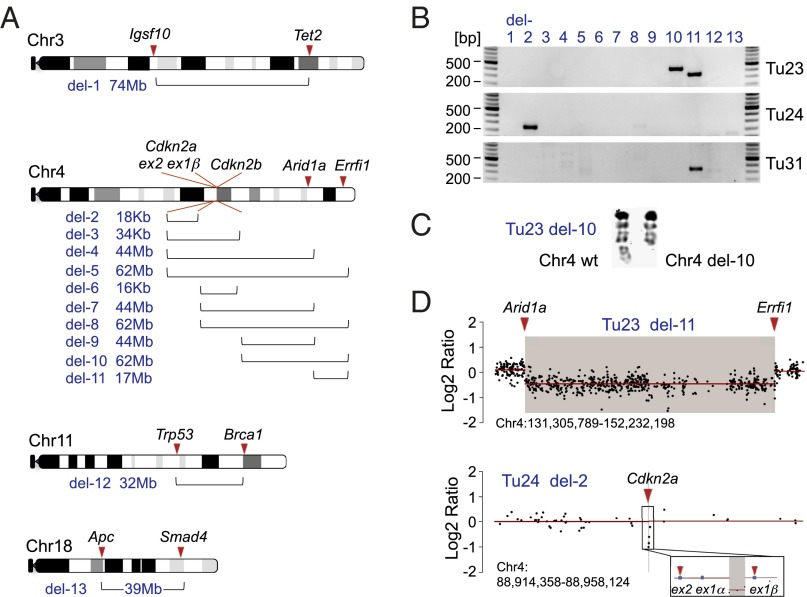
With respect to CRISPR/Cas9-induced rearrangements, we screened for all 533 possible large intrachromosomal deletion/fusion events in the 41 tumors using PCR and in a subset of tumors also by comparative genomic hybridization (CGH) and multicolor fluorescence in situ hybridization (M-FISH) (Fig. 3 and SI Appendix, Figs. S13 and S14). We identified four deletions in three cancers: an 18-kb deletion at the Cdkn2a locus (Tu24), a 62-Mb deletion between TSGs Cdkn2b and Errfi1 (Tu23), and 17-Mb deletions between Arid1a and Errfi1 (Tu23 and Tu31). The 62-Mb deletion identified in Tu23 by fusion-PCR was “silent” in CGH because of its subclonal occurrence. It was, however, detectable by FISH (1/7 metaphases positive for the deletion) (Fig. 3C). There were no interchromosomal translocations in the cell lines analyzed by M-FISH (n = 2) (SI Appendix, Fig. S14). Because stable integration of CRISPR/Cas9 vectors was very rare in our cancers (integrations identified by PCR-based detection of CRISPR-SB vectors in only 3 out of 62 tumors), we conclude that transient expression of multiplexed CRISPR/Cas9 can be sufficient to induce one or more intrachromosomal rearrangements within a cell in vivo.
Fig. 3.
Intrachromosomal fusions induced by combinatorial CRISPR/Cas9 targeting. (A) Scheme of chromosomes with two or more CRISPR/Cas9 target sites in the 18 sgRNA multiplexing experiments. Brackets indicate the predicted sizes of possible deletions (del-1 to del-13), ranging from 16 kb to 74 Mb. (B) PCR screening for all 533 possible intrachromosomal fusions in 41 liver tumors revealed four large deletions in three tumors: an 18-kb deletion (del-2, Tu24; Cdkn2a-ex1β/Cdkn2a-ex2 fusion), two large deletions in Tu23 (del-10, Cdkn2b/Errfi1, 62Mb; del-11, Arid1a/Errfi1, 17 Mb), and another Arid1a/Errfi1 fusion in Tu31. (C) DAPI staining of metaphase spreads of Tu23 cell line confirms the large deletion (del-10) in chromosome 4. (D) Del-2 in Tu24 and del-11 in Tu23 are detected as copy number losses by array CGH.
One implication of these results is that the extent of multiplexing will have limitations. Either it will require careful selection/combination of target sites or the possibility of undesired chromosomal damage occurring will need to be tested for. These findings are also relevant for genome-wide in vitro CRISPR/Cas9 screening, particularly in experimental settings where multiple sgRNAs are delivered to a cell. On the other hand, the observation that chromosome engineering is feasible somatically in the context of liver cancer offers great opportunities. GWAS and whole genome sequencing studies are currently identifying hundreds of ICC/HCC variant hot spot regions, many of which are located in genomic deserts, coinciding with putative regulatory regions, such as enhancers (www.genome.gov/encode). Our results suggest that these regions can be systematically targeted using multiplexed CRISPR/Cas9 to study their biological role in cancer.
No Off-Target Effects in CRISPR/Cas9-Induced Liver Tumors.
We have screened eight tumors for undesired off-target effects by amplicon-based NGS of each sgRNA's top five off-targets (at least three exonic off-targets). We found no indels at off-target sites with a mutant read frequency of 0.2% or higher (a cutoff used to exclude sequencing errors for both on- and off-target site analyses). We also screened CGH data from six tumors for 266,778 potential intrachromosomal deletions resulting from combinations of potential off-target cleavage events (1,010 and 1,550 off-target sites for 10 sgRNAs and 18 sgRNAs, respectively) (SI Appendix, Fig. S13). Off-target sites were defined to be potentially causative if they were within a distance of 500,000 bp (and 20 probes or fewer) to an aberration detected by CGH. These analyses did not identify chromosomal deletions attributable to off-target effects.
CRISPR/Cas9-Induced Mutations Are Predominantly Biallelic.
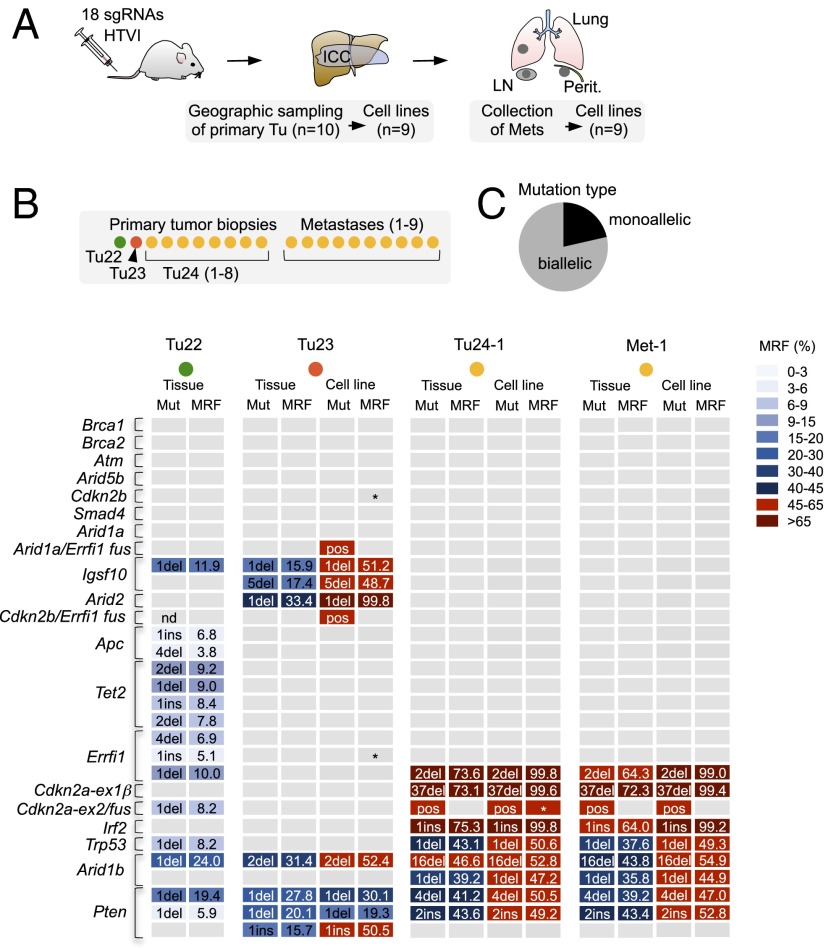
To assess the incidence of biallelic vs. monoallelic target gene mutations, we next analyzed cancer cell lines isolated from an aggressive ICC induced by 18-sgRNA multiplexing (Fig. 4 and SI Appendix, Fig. S15 show that these cell lines are transplantable). In contrast to all other tumors analyzed in this study (which were identified early by regular MRI screening and were therefore small), one animal had an early onset large (>2 cm) tumor mass and numerous metastases to lymph nodes, peritoneum, and lungs (SI Appendix, Fig. S16). Extensive geographical sampling of the tumor mass (n = 10) and subsequent target site sequencing revealed three independent primary cancers (Tu22, Tu23, and Tu24), with Tu24 being predominant (8/10 samples). The analysis of CRISPR/Cas9-induced indel patterns also allowed phylogenetic tracking of metastatic clones: All metastases (n = 9) originated from Tu24 (Fig. 4B and SI Appendix, Table S4).
Fig. 4.
Allelic frequencies of target site mutations and phylogenetic tracking of CRISPR/Cas9-induced metastatic ICC. (A) A 2-cm tumor mass (ICC) and numerous metastases (Mets) in lungs, lymph nodes (LN), and peritoneum in a mouse 20 wk after HTVI of 18 sgRNAs. (B) CRISPR/Cas9 target sites underwent NGS in a total of 35 tumor/metastasis tissues and cell lines. Indel patterns revealed three independent primary tumors. All metastases originate from Tu24. Frame-shift causing indels with a cumulative mutant read frequency (MRF) >4% per target site are shown for representative samples. Note that MRFs are underestimated in cancer tissue (because of healthy stromal components) but are accurately reflected in cell lines. Tumors with indicated fusion products are marked as positive (pos). Asterisks indicate a lack of WT sequence. (C) Allelic frequency of mutations in cell lines of Tu23 and Tu24 as determined by a combined quantitative analysis of indel frequencies, presence/absence of large fusions, and the presence/absence of WT reads.
Comparative indel analysis of primary tumor tissue and corresponding cell lines showed that accurate estimation of MRFs is difficult in primary cancer tissue due to stromal components (Fig. 4B and SI Appendix, Table S4). A combined quantitative analysis of (i) indel frequencies, (ii) the presence or absence of large deletions (fusions), and (iii) the frequency of WT reads at target sites in these cell lines revealed that 79% of mutated target loci have biallelic inactivation (Fig. 4C and SI Appendix, Table S5), despite the fact that none of these tumors had stably integrated CRISPR/Cas9. The predominant homozygous inactivation underlines the potential of CRISPR/Cas9 for recessive genetic screening and gene function analysis.
Hepatic loss-of-function screening has been performed using RNAi-based gene knock-down in transplantation models (e.g., intrasplenic implantation of bipotent liver progenitor cells) (46) or by HTVI-based/transposon-mediated genome integration of shRNAs (47). Our results show that RNAi and CRISPR/Cas9 are complementary tools with unique beneficial characteristics, depending on the experimental context. CRISPR/Cas9-induced homozygous gene knockout is a major advance for recessive genetic screening whereas RNAi-based knockdown (which is typically only partial) has advantages for the study of dosage effects or reversible phenotypes. Likewise, the ability to perform chromosome engineering by CRISPR/Cas9 is an important novel technological innovation but can be disadvantageous if such effects are not desired.
Concluding Remarks
Our work describes novel approaches to model and study cancer in mice. We provide, to our knowledge, the first demonstration and characterization of highly multiplexed direct in vivo CRISPR/Cas9 mutagenesis, including (i) the description of proof-of-principle applications (genetic screening for cancer gene validation/discovery), (ii) a characterization of tumor phenotypes at the genetic level (tumor heterogeneity, allelic mutation frequency, phylogenetic metastasis tracking, single cell cloning), and (iii) a thorough analysis/discovery of possible caveats (frequency/size/extent of chromosomal rearrangements). This multilayered characterization gives comprehensive insights into the potential and limitations of in vivo CRISPR/Cas9 multiplexing and thus guidance for its appropriate use. In defined genetic (KrasG12D) and liver damage models (CCl4), we also show for the first time, to our knowledge, that CRISPR/Cas9 somatic gene targeting can be used to induce HCC, one of the leading causes of cancer-related death worldwide, and we provide support for the emerging role of chromatin modifiers in hepatobiliary tumorigenesis. Multiplexing CRISPR/Cas9 will enhance the speed and efficiency of assigning biological function to DNA sequence, one of the big scientific challenges in the postgenomic era.
Methods
A detailed description of experimental procedures is available in SI Appendix. Briefly, CRISPR/Cas9 cleavage efficiencies were tested in vitro using T7E1 or Surveyor assays. Hepatic delivery of CRISPR/Cas9 vectors was performed by HTVI, as described earlier (21). All animal studies were conducted in compliance with European guidelines for the care and use of laboratory animals and were approved by the Institutional Animal Care and Use Committees (IACUC) of Technische Universität München, Regierung von Oberbayern, and the UK Home Office. CRISPR/Cas9 target site mutations were identified using amplicon-based NGS. Liver tumors were characterized by immunohistochemistry (IHC), CGH, and M-FISH.
Supplementary Material
Acknowledgments
We thank Olga Seelbach, Ruth Hillermann, Teresa Stauber, and Daniel Kull for excellent technical assistance. J.d.l.R. is recipient of a Federation of European Biochemical Societies (FEBS) fellowship. The work was supported by the German Cancer Consortium Joint Funding Program and the Helmholtz Gemeinschaft (Preclinical Comprehensive Cancer Center).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512392112/-/DCSupplemental.
References
- 1.Stratton MR. Exploring the genomes of cancer cells: Progress and promise. Science. 2011;331(6024):1553–1558. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 2.Copeland NG, Jenkins NA. Harnessing transposons for cancer gene discovery. Nat Rev Cancer. 2010;10(10):696–706. doi: 10.1038/nrc2916. [DOI] [PubMed] [Google Scholar]
- 3.Rad R, et al. PiggyBac transposon mutagenesis: A tool for cancer gene discovery in mice. Science. 2010;330(6007):1104–1107. doi: 10.1126/science.1193004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradley A, Evans M, Kaufman MH, Robertson E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature. 1984;309(5965):255–256. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- 5.Doetschman T, et al. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330(6148):576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- 6.van Miltenburg MH, Jonkers J. Using genetically engineered mouse models to validate candidate cancer genes and test new therapeutic approaches. Curr Opin Genet Dev. 2012;22(1):21–27. doi: 10.1016/j.gde.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 7.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(5962):167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 8.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468(7320):67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 9.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482(7385):331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 11.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32(4):347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin H, et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32(6):551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xue W, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature. 2014;514(7522):380–384. doi: 10.1038/nature13589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Platt RJ, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159(2):440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maddalo D, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516(7531):423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sánchez-Rivera FJ, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014;516(7531):428–431. doi: 10.1038/nature13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blasco RB, et al. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Reports. 2014;9(4):1219–1227. doi: 10.1016/j.celrep.2014.10.051. [DOI] [PubMed] [Google Scholar]
- 21.Yant SR, et al. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat Genet. 2000;25(1):35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- 22.Ong CK, et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat Genet. 2012;44(6):690–693. doi: 10.1038/ng.2273. [DOI] [PubMed] [Google Scholar]
- 23.Jiao Y, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45(12):1470–1473. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan-On W, et al. Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat Genet. 2013;45(12):1474–1478. doi: 10.1038/ng.2806. [DOI] [PubMed] [Google Scholar]
- 25.Ross JS, et al. New routes to targeted therapy of intrahepatic cholangiocarcinomas revealed by next-generation sequencing. Oncologist. 2014;19(3):235–242. doi: 10.1634/theoncologist.2013-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujimoto A, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44(7):760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 27.Guichard C, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44(6):694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang J, et al. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet. 2012;44(10):1117–1121. doi: 10.1038/ng.2391. [DOI] [PubMed] [Google Scholar]
- 29.Schulze K, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47(5):505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delire B, Stärkel P. The Ras/MAPK pathway and hepatocarcinoma: Pathogenesis and therapeutic implications. Eur J Clin Invest. 2015;45(6):609–623. doi: 10.1111/eci.12441. [DOI] [PubMed] [Google Scholar]
- 31.Sia D, et al. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology. 2013;144(4):829–840. doi: 10.1053/j.gastro.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horie Y, et al. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J Clin Invest. 2004;113(12):1774–1783. doi: 10.1172/JCI20513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simbolo M, et al. Multigene mutational profiling of cholangiocarcinomas identifies actionable molecular subgroups. Oncotarget. 2014;5(9):2839–2852. doi: 10.18632/oncotarget.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyault S, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45(1):42–52. doi: 10.1002/hep.21467. [DOI] [PubMed] [Google Scholar]
- 35.Xu X, et al. Induction of intrahepatic cholangiocellular carcinoma by liver-specific disruption of Smad4 and Pten in mice. J Clin Invest. 2006;116(7):1843–1852. doi: 10.1172/JCI27282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Dell MR, et al. Kras(G12D) and p53 mutation cause primary intrahepatic cholangiocarcinoma. Cancer Res. 2012;72(6):1557–1567. doi: 10.1158/0008-5472.CAN-11-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343(6166):80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343(6166):84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera MdelC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32(3):267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 40.Chen S, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160(6):1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang P, et al. Mutations in isocitrate dehydrogenase 1 and 2 occur frequently in intrahepatic cholangiocarcinomas and share hypermethylation targets with glioblastomas. Oncogene. 2013;32(25):3091–3100. doi: 10.1038/onc.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2010;465(7300):966. doi: 10.1038/nature09132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ward PS, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guilhamon P, et al. Meta-analysis of IDH-mutant cancers identifies EBF1 as an interaction partner for TET2. Nat Commun. 2013;4:2166. doi: 10.1038/ncomms3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zender L, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135(5):852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xue W, et al. A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc Natl Acad Sci USA. 2012;109(21):8212–8217. doi: 10.1073/pnas.1206062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.