Significance
The field of microbiome research is moving from 16S rDNA gene sequencing to metagenomic sequencing of complete communities, which clearly gives a more comprehensive genomic and functional representation of the organisms present. Here we describe, quantify, and compare biases associated with four currently available next-generation sequencing library preparation methods using a synthetic DNA mock community and an extraction spike-in control of microbial cells. Our study highlights a critical need for consistency in protocols and data analysis procedures, especially when attempting to interpret human microbiome data for human health.
Keywords: microbiome, genomics, sequencing
Abstract
Observations from human microbiome studies are often conflicting or inconclusive. Many factors likely contribute to these issues including small cohort sizes, sample collection, and handling and processing differences. The field of microbiome research is moving from 16S rDNA gene sequencing to a more comprehensive genomic and functional representation through whole-genome sequencing (WGS) of complete communities. Here we performed quantitative and qualitative analyses comparing WGS metagenomic data from human stool specimens using the Illumina Nextera XT and Illumina TruSeq DNA PCR-free kits, and the KAPA Biosystems Hyper Prep PCR and PCR-free systems. Significant differences in taxonomy are observed among the four different next-generation sequencing library preparations using a DNA mock community and a cell control of known concentration. We also revealed biases in error profiles, duplication rates, and loss of reads representing organisms that have a high %G+C content that can significantly impact results. As with all methods, the use of benchmarking controls has revealed critical differences among methods that impact sequencing results and later would impact study interpretation. We recommend that the community adopt PCR-free–based approaches to reduce PCR bias that affects calculations of abundance and to improve assemblies for accurate taxonomic assignment. Furthermore, the inclusion of a known-input cell spike-in control provides accurate quantitation of organisms in clinical samples.
Next-generation sequencing (NGS) of microbial genomes and metagenomes is now widely used in various applications including forensic genetics, clinical diagnostics, pathogen outbreaks, and infectious disease surveillance. Since the publication of the first human microbiome study in 2006 (1), there has been an explosion of human microbiome studies for both healthy and disease conditions; the great majority of these studies now routinely use NGS technologies. Several recent articles have raised alarm about a lack of data robustness and reproducibility among published 16S rDNA and whole-genome sequencing (WGS) metagenomic studies between sequencing runs and laboratory cores (2–5). Finucane et al. (6), for example, compared several obesity microbiome publications and suggested that no simple taxonomic signature could be found and that several groups did not agree about the microbiome association with body mass index. A similar picture evolves in the analysis of microbiome samples of nonalcoholic fatty liver disease, for which conflicting microbial signatures exist (7). In addition, experimental variation remains a significant hurdle in many published studies. For example, researchers recently published evidence of variation in 16S rDNA gene profiling from microbiome specimens processed using several nucleic acid extraction protocols (8, 9). Franzosa et al. (10, 11) further revealed the impact of sample collection on the stability of the metagenome and metatranscriptome of stool specimens.
Improvements and development of novel chemistries and sequencing technologies have provided the scientific community with tools to obtain high-resolution measurements from microbiome samples, with advances in NGS technology resulting in several library preparation products currently available on the market. Rapid development of new library chemistries and approaches provide novel low-cycle PCR and PCR-free tools, as well as chemical and physical shearing approaches, that can be used to analyze the microbiome. However, these tools may introduce unanticipated artifacts in the data. In the current study we focus on this major essential upstream step of human microbiome analysis—library preparation. We comprehensively assess NGS library preparation platforms by comparing the three major platforms, Illumina TruSeq DNA PCR-free (TSF), Illumina Nextera XT (XT), and Kapa Hyper Prep (KF) [both Kapa Hyper Prep PCR (KP) and PCR-free (KF)] in a controlled setting of identical samples, equipment, and handlers. In addition, we further assessed the three major platforms using a set of longitudinal stool specimens collected following amoxicillin treatment. Analysis of these samples on the aforementioned platforms provides additional insight into potential bias of primary specimens compared with a synthetic mock community.
Results
Sequencing, Assembly, and Annotation Statistics of Mock Community and Stool Samples.
To determine the impact of the XT, KP, KF, and TSF library preparation methods on microbiome community quantitation, we generated two independent libraries from each of five DNA samples (the mock community and four clinical samples) for each library chemistry. To maintain unbiased comparison among different libraries, postsequencing quality analysis and annotation were based on equivalent numbers of raw reads. For taxonomy profiling, 4.85 million paired-end reads (minimal yield among the samples) were used for all the samples. For assembly, ORF prediction, and functional annotation, where additional depth was desired, 11 million paired-end raw reads were used for the analysis (Dataset S1, Table S1). Two stool sample libraries generated by TSF did not reach sufficient coverage. The two technical replicates for these two samples were pooled together to achieve 11 million raw reads. XT libraries generated an average of 28% low-quality reads, a duplication rate of 1.38%, and a contig N50 length of 10,727 bp. Sequence data generated from KP libraries had an average of 6.41% low-quality reads, a duplication rate of 1.29%, and a contig N50 length of 30,814 bp. Libraries generated using the KF approach resulted in sequence data with an average of 15.39% low-quality reads, a duplication rate of 0.07%, and an N50 of 32,864 bp. Last, sequence data produced from TSF libraries resulted in an average of 15.41% low-quality reads, a duplication rate of 0.04%, and an N50 of 45,707 bp (Dataset S1, Table S1). In summary, the TSF libraries produced the longest contigs; KP produced the largest total contig length; and XT libraries resulted in significantly shorter individual contig length and total contig length (Fig. S1 and Dataset S1, Table S1).
Impact of the Library Preparation Method on Taxonomic Abundance and Functional Predictions.
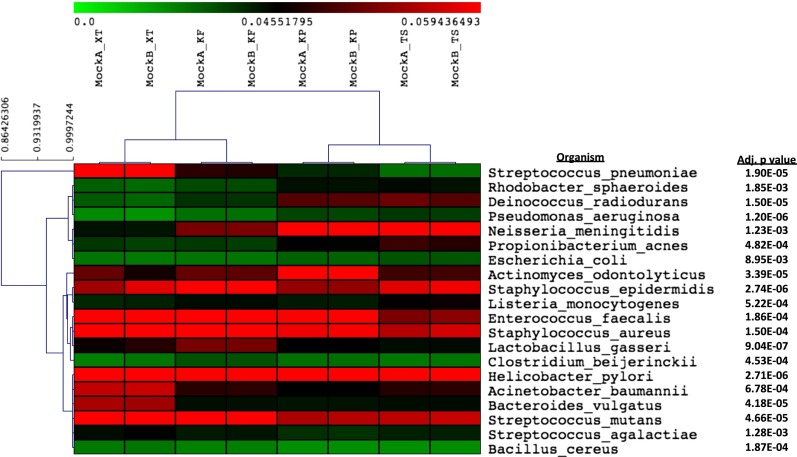
To assess the impact of the library preparation method on metagenomic shotgun sequencing data, measurements of taxonomic relative abundance were calculated for the mock DNA community [Biodefense and Emerging Infections Research Resources Repository (BEI Resources) HM-276D] (Dataset S1, Table S2) across the four different protocols. Hierarchical clustering of the four libraries based on the relative genome abundance (RGA) profiles of the constituent microbes revealed two major groups: XT and KF in one group and KP and TSF in the other (Fig. 1). Unexpectedly, cluster analysis indicates that KF matches more closely with XT than with TSF, pairing a PCR-free sample more closely to a PCR-amplified sample than to another PCR-free sample and suggesting that the low-cycle PCR amplification step did not result in any bias. Cluster analysis also illustrated closer grouping between platforms rather than within platforms, as demonstrated closer clustering between XT and KF and between KP and TSF, rather than between XT and TSF or between KF and KP, indicating that library chemistry does not equate with clustering similarity. Using one-way ANOVA analysis to compare the relative abundance measurements of the members of the mock community across the four mock-community libraries revealed significant variation based on library preparation (Dataset S1, Table S3). Among the largest variations were the relative abundances of Helicobacter pylori, Streptococcus pneumoniae, Deinococcus radiodurans, and Pseudomonas aeruginosa with SDs from 1.16–1.61% or ∼4,000–6,000 reads. Furthermore, there were no correlations across library protocols between organism abundance in the mock community and variation based on RGA.
Fig. 1.
One-way ANOVA analysis across library preparation methods. Relative abundance measurements were calculated for the mock community across the four different protocols and analyzed for consistency between library preparations from both technical replicates. Shading in the heat map indicates relative abundance in the mock-community DNA mixture from low (green) to high (red) abundance. Adjusted P values were calculated based on a maximum P value of 0.01. Samples and organisms were clustered based on an uncentered Pearson complete linkage analysis. The letters “A” and “B” indicate technical replicates for each sample preparation.
To examine how library preparation impacts functional annotation, we analyzed genes that can be recovered from the assembled sequences, which is the basis for predicting the function and pathway landscape of metagenomes (Dataset S1, Table S4). Here, two ORFs sets, predicted ORFs from the assembled scaffolds and true ORFs called from the complete mock reference genomes, are compared using cd-hit-2d at ≥98% sequence identity over 95% of the length of predicted ORFs to find matched ORFs between these two sets (Fig. S2). Because predicted ORFs tend to be more fragmented, multiple predicted ORFs may be aligned to a single true ORF. Overall, around 96% of true ORFs were recovered by 95% of predicted ORFs for the 20 strains in the mock community across four libraries; XT showed slightly but notably worse performance than the other libraries. XT data also resulted in more fragmented ORFs than did KP, KF, and TSF. Most of the 20 individual strains demonstrate a pattern very similar to the combined data from the 20 strains. Among the 20 mock genomes, Clostridium beijerinckii exhibited more significant differences between XT and other libraries: XT resulted in approximately fivefold more unmapped ORFs (840 vs. ∼140), approximately threefold more missing genes (237 vs. ∼60), and a 20% higher fragmentation ratio (1.23% vs. 1.03%) than the other libraries. This analysis agrees with our observations regarding the assembly status and genome coverage. We did not identify significant differences in functional annotation between KP, KF, and TSF in all 20 mock genomes. For the remaining 19 community organisms, XT performed as well as KP, KF, and TSF.
Impact of Library Procedure on Quantitative Assessment of Clinical Stool Samples.
RGA measurements for a longitudinal study were analyzed to examine the impact of the selected library preparation approaches on clinical stool samples. The clinical samples originated from an ongoing research project to determine the impact of antibiotic selection on the microbiome. Samples were collected from the participant before (day 0) and following amoxicillin treatment (days 3 and 7 and week 8). Because genomic libraries prepared from real human microbiome samples contain greater microbial complexity than the mock community, the inclusion of these samples may help determine the degree to which the four library protocols impact any downstream analyses. Libraries were prepared in duplicate from the extracted stool DNAs using the XT, TSF, KF, and KP protocols as described above.
Precision analysis for clinical stool sample replicates demonstrate little variation between technical replicates, with an average R2 = 0.999 (Dataset S1, Table S5). The RGA profiles, based on rank order analysis, show a high degree of correlation among library preparations, with an average R2 = ∼0.97 (Dataset S1, Table S6). To determine if specific library preparation methods introduce quantitative variation based on RGA measurements at the species/strain level, a one-way ANOVA analysis was performed on the day 0 clinical stool sample. Data revealed significant variation in the representation of the abundance of 25 microbial species/strains (P value of 0.01; Dataset S1, Table S7). A one-way ANOVA analysis based on a P value of 0.01 with standard Bonferroni correction was applied to the data from the day 3, day 7, and week 8 postantibiotic time points to assess quantitative variation introduced by kit-specific library preparations. The data revealed statistically significant variation in the representation of the abundance of 27 organisms in the day 3 sample, 8 organisms in the day 7 sample, and 36 organisms in the week 8 sample. To examine if a significant difference in measured sample diversity exists based on library preparation protocol, we performed a Shannon index analysis of the clinical specimens before and after antibiotic treatment (Fig. S3) (12). There was no significant impact in measured diversity.
Impact of Library Procedure on Qualitative Assessment of Clinical Stool Samples.
To examine whether the observed quantitative variation introduced by individual library preparation methods impacts the qualitative assessment of the microbiome, we performed a Bayesian temporal analysis for all time points within individual library preparations (13). We observed changes in response to the antibiotic treatment in 233 microbial species/strains based on a P value of 0.01 in the XT libraries following antibiotic administration (Dataset S1, Table S8). We observed similar changes following antibiotic administration in 234 microbiome species/strains in the KP libraries, 235 microbial species/strains in the KF libraries, and 243 microbial species/strains in the TSF libraries (Dataset S1, Tables S9–S11). Venn diagram analysis was used to identify common microbial species/strains that were significantly modulated following the administration of antibiotic. Our analysis revealed 217 microbial species/strains were identified in all library preparations as significantly modulated following antibiotic selection (Fig. S4). Species/strains not uniformly identified across all of the four library preparation approaches tended to represent organisms with a percentage RGA of <0.00001.
Percent G+C Bin and Genome Coverage Mapping Analysis.
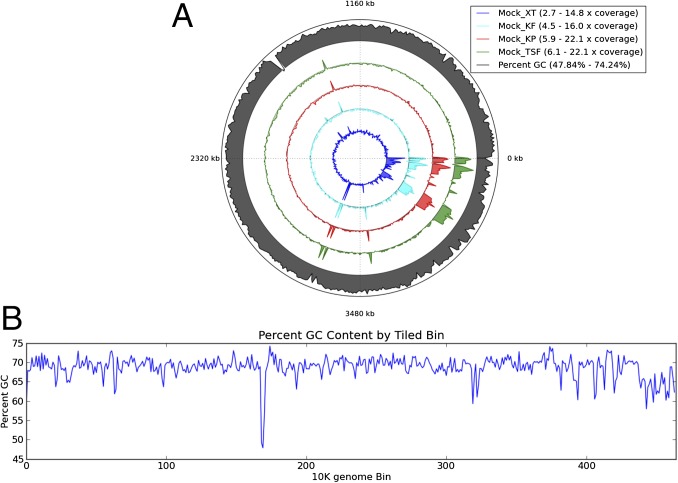
Previously published studies report a potential bias in the %G+C in XT libraries sequenced on the Roche 454 Titanium platform (14) and on the MiSeq platform (15). Using the mock community, we analyzed the relative measurements of species abundance for each library protocol with respect to %G+C. Consistent with the cluster analysis of the mock community, XT and KF have a similar pattern of higher representation of organisms with a low %G+C and a corresponding lower representation of organisms with a high %G+C, as marked by yellow and red boxes, respectively, in Fig. S5. To examine the possible relationship between read coverage and genome %G+C by method, we calculated both the %G+C of the bin and the mean read-depth coverage (from nonoverlapping 10-kbp windows across the genome) (Fig. 2B). Coverage patterns are very comparable across methods, with equivalent region-specific depth spikes and valleys. Examination of sequences underlying the most extreme spikes per genome revealed repetitive genomic elements. This analysis is consistent with recent observations that the annotated 16S rRNA gene copy number of many organisms is incorrect (16). Of note, the Rhodobacter sphaeroides coverage plots show large stretches of increased coverage at the end of the circle plots (Fig. 2A). These regions correspond to the boundaries and regions of plasmids found in this organism, suggesting that these plasmids are multicopy. Similar evaluations were made for Escherichia coli and C. beijerinckii (Figs. S6 and S7). To examine read depth coverage biases, we simulated the sequencing of each genome to an average read depth ranging from 0.1–20 and calculated the expected proportion of the genome expected to have at least one read when sequenced at that depth. Although the specific effect seems to vary slightly from genome to genome, there is a general trend for TSF, KP, and KF preparation methods to behave nearly identically, whereas XT requires a deeper average sequencing depth to capture an equivalent fraction of the genome with reads.
Fig. 2.
Map of mean GC content and mean relative sequencing depth by library prep method across the genome for R. sphaeroides. (A) The complete genome for the organisms is used, including any known plasmids, and (B) sub division of the genome into 10-kb bins for mean analysis. Outer grey ring depicts the delta from 50% GC content for a sequence bin. The inner 4 colored rings depict the delta of the average sequencing depth for the bin from the average sequencing depth of the whole genome. Maximum and minimum values per ring are given in the legend.
qPCR Determination of XT and KP Bias After Library Preparation.
qPCR was used to quantitate accurately the 20 species within the mock-community DNA sample. Three genes unique to each of the 20 members of the mock community were identified and used to develop organism-specific qPCR assays (Dataset S1, Table S12). Genomic DNAs were used to optimize reaction conditions and to establish a standard curve for quantitation of strain-specific DNA concentrations within the mock-community DNA and to serve as a positive control. When possible we used at least two independent qPCR measurements and corresponding primer-specific standard curves to determine percent composition for each organism in the mock community. The percent delta between RGA from the sequencing data generated from the four library preparations and the average qPCR quantification measurements demonstrates the impact of library strategy and sequencing on the accurate representation of individual mock-community organisms (Table 1). We observed over- and underrepresentation of strain DNAs, with a deviation of 2.0% or more or of −2.0% or less, compared with qPCR measurements, for mock-community organisms in all the library strategies tested based on RGA (10/20 XT; 12/20 KF; 7/20 KP; and 7/20 TSF). To determine if the bias should be attributed to library preparation or downstream clustering/sequencing, qPCR was performed on the XT and KP library preparations using strain-specific primers for representative organisms in the mock community. Direct comparison of qPCR measurements of the mock-community DNA and the XT library revealed an underrepresentation of ≥1.9% in two (D. radiodurans and R. sphaeroides) of the three organisms with high (≥60%) G+C content, and in the KP library R. sphaeroides and Propionibacterium acnes were underrepresented by ∼4% (Table 2). To determine which library protocol had the greatest similarity to the starting DNA composition of the mock community, we computed a Pearson correlation matrix between samples (Dataset S1, Table S13). We observed that the qPCR measurements of the mock-community DNA were more similar to the generated libraries than to the resulting sequencing data. Specifically, the mock-community DNA had greater similarity with the XT library (0.58) than did the KP library (0.20).
Table 1.
Comparison of RGA and initial DNA input measured by qPCR
| Organism | Mock community by PCR, % | XT RGA, % | KF RGA, % | KP RGA, % | TSF RGA, % |
| Helicobacter pylori | 5.92 | 15.61 | 13.50 | 13.09 | 14.78 |
| Lactobacillus gasseri | 2.84 | 4.69 | 5.21 | 4.55 | 4.35 |
| Streptococcus mutans | 2.21 | 6.27 | 6.11 | 5.52 | 5.60 |
| Streptococcus pneumoniae | 2.55 | 6.36 | 4.77 | 3.83 | 2.54 |
| Streptococcus agalactiae | 2.67 | 4.39 | 4.14 | 3.69 | 3.97 |
| Neisseria meningitidis | 2.42 | 4.18 | 5.22 | 5.95 | 6.10 |
| Actinomyces odontolyticus | 4.78 | 4.88 | 5.09 | 6.43 | 4.91 |
| Propionibacterium acnes | 7.08 | 4.32 | 4.29 | 5.54 | 5.86 |
| Staphylococcus epidermidis | 5.94 | 5.33 | 5.82 | 4.97 | 5.40 |
| Enterococcus faecalis | 7.14 | 6.97 | 7.11 | 6.42 | 5.29 |
| Staphylococcus aureus | 5.90 | 6.37 | 6.89 | 5.84 | 5.58 |
| Listeria monocytogenes | 3.76 | 4.02 | 4.49 | 4.21 | 4.74 |
| Deinococcus radiodurans | 5.80 | 2.82 | 3.65 | 5.02 | 5.08 |
| Escherichia coli | 7.40 | 2.46 | 2.47 | 2.80 | 3.06 |
| Rhodobacter sphaeroides | 6.42 | 2.74 | 3.22 | 4.26 | 4.31 |
| Acinetobacter baumannii | 4.43 | 5.59 | 4.75 | 4.50 | 4.72 |
| Bacteroides vulgatus | 4.84 | 5.45 | 4.30 | 4.18 | 4.34 |
| Bacillus cereus | 6.01 | 2.44 | 2.26 | 2.00 | 1.99 |
| Pseudomonas aeruginosa | 6.38 | 2.75 | 3.61 | 4.64 | 4.96 |
| Clostridium beijerinckii | 5.50 | 2.36 | 3.11 | 2.55 | 2.42 |
qPCR quantitation of the 20 species within the mock-community DNA sample. To ensure accuracy of species abundance, we quantitated the mock-community DNA to determine the exact proportions of each organism in the community.
Table 2.
Comparison of initial DNA input and postlibrary generation measured by qPCR
| Strain | qPCR of mock community, % | XT library qPCR, % | KP library qPCR, % | XT RGA, % | KF RGA, % | KP RGA, % | TSF RGA, % |
| R. sphaeroides | 5.99 | 1.79 | 1.63 | 2.74 | 3.22 | 4.26 | 4.31 |
| C. beijerinckii | 5.13 | 4.23 | 4.17 | 2.36 | 3.11 | 2.55 | 2.42 |
| L. gasseri | 2.65 | 2.99 | 2.12 | 4.69 | 5.21 | 4.55 | 4.35 |
| E. coli | 6.90 | 8.28 | 5.28 | 2.46 | 2.47 | 2.80 | 3.06 |
| P. acnes | 6.59 | 6.63 | 2.17 | 4.32 | 4.29 | 5.54 | 5.86 |
| D. radiodurans | 5.41 | 3.52 | 7.60 | 2.82 | 3.65 | 5.02 | 5.08 |
Quantitative impact assessed by qPCR of KP and XT on mock-community DNA. We performed qPCR on the two PCR-based library protocols, XT and KP, to determine if bias was introduced at the library preparation stage or sequencing.
qPCR Determination of Raw Mapped Reads in Relation to Genomic Units.
To evaluate further the accuracy of the XT approach and to calculate the correlation between raw mapped reads, RGA, and genomic units (GU), qPCR was performed on a stool sample spiked with cells of Shewanella oneidensis, as described above. Cells were spiked into three stool samples at a known concentration and processed through our specimen total DNA extraction pipeline; libraries were prepared using the XT approach and were sequenced to determine the correlation to DNA input, mapped reads, RGA estimates, and GU. The total number of mapped 125-nt reads generated in each of the spiked and control stool specimens ranged from 11.7 × 106 to 12.4 × 106. In parallel, total DNA extracted from each spiked stool specimen was processed by qPCR to quantitate the amount of S. oneidensis DNA in the sample before library preparation and sequencing. Data revealed that S. oneidensis represented 2.21–3.14% of the total extracted DNA in samples as determined by the equation: number of copies = [amount of DNA (in nanograms) * 6.022 × 1023] / [length (in base pairs) * 1 × 109 * 650] (Table 3). qPCR analysis indicates that S. oneidensis DNA is present at ∼5 × 103 GU per nanogram of total extracted spiked specimen DNA. The percentage of reads uniquely mapped to S. oneidensis from the total mapped reads strongly agreed with the calculated percent RGA. We calculated the number of mapped reads that correlate to one GU for a 4.9-Mb genome, based on calculated GU, total mapped reads, unique reads mapped to S. oneidensis, and the amount of library loaded for clustering. We estimate that 33–40 125-bp reads represent one S. oneidensis GU as determined by the equation: reads to GU = [(number of S. oneidensis mapped reads per GU) * percent of sequenced library]. We propose, based on these calculations and calibration standards, an estimation of approximately eight 125-nt mapped reads per genome size (in million base pairs) per 10 × 106 reads, which equates to 1 GU.
Table 3.
Comparison of initial DNA input and postlibrary generation measured by qPCR
| Sample | S. oneidensis DNA measured by qPCR, ng | Calculated number of S. oneidensis GU in samples | S. oneidensis DNA going into library preparation, % | Library sample clustered, % | Number of reads mapped to S. oneidensis by XT | Reads mapped to S. oneidensis by XT, % | % S. oneidensis RGA, XT | Total number of mapped reads by XT | Calculated S. oneidensis reads to GU | Estimated reads per megabase per GU |
| HLI1264 | 0.0265 | 5.01E+03 | 3.14 | 96.22 | 206,073 | 1.76 | 1.70 | 11,727,217 | 40 | 8 |
| HLI1270 | 0.0292 | 5.52E+03 | 2.45 | 72.28 | 255,385 | 2.12 | 2.80 | 12,056,364 | 33 | 7 |
| HLI1271 | 0.0278 | 5.25E+03 | 2.21 | 55.26 | 373,061 | 3.00 | 3.00 | 12,423,257 | 39 | 8 |
Correlation of raw mapped reads to GU. To correlate mapped sequence reads to GUs, we compared the qPCR measurement of DNA from S. oneidensis cells as a control spiked into three stool samples to the RGA generated from sequence analysis. qPCR was performed on three stool specimens spiked with S. oneidensis cells. The cell-spiked specimen was processed through the HLI specimen extraction pipeline. qPCR was used to quantitate the amount of S. oneidensis DNA present in the total extracted specimen DNA. We calculated an estimated number of reads to genomic units per million base pairs based on genome length and number of mapped reads.
Discussion
At the beginning of the NIH Roadmap Initiative Human Microbiome Project (HMP), there were no standards for human sampling, sample handling, DNA extraction, DNA sequencing, or data analysis. Eight years since the beginning of the HMP, and despite efforts by many investigators to set standards for many aspects of microbiome research, great variability in approach, methodology, results, and interpretation are routinely reported. These inconsistencies have resulted in significant misinterpretation of data and confusion in the field. DNA sequence generation is fundamental to all metagenome projects; however, although there has been benchmarking for 16S rDNA sequencing, there has been no benchmarking of NGS platforms or library preparation methods for metagenomic sequencing using established controls. Here we quantified the biases associated with four available library preparation methods for the Illumina HiSeq 2500 (Version 4 reagents) platform using a publicly available synthetic DNA mock community (BEI Resources HM-276D) and a DNA extraction spike-in control of S. oneidensis cells. The Illumina HiSeq platform was selected because the technology is estimated to account for the majority of all microbiome WGS studies that currently are under way. We also performed a qualitative comparison of a WGS metagenomic analysis of stool specimens using several library methods. Two of the methods were PCR-free: TruSeq DNA PCR-Free (TSF) and Kapa PCR Free (KF), and two include PCR amplification steps: Nextera XT (XT) and Kapa PCR (KP). Quantitative PCR (qPCR) was also used to validate the mock-community and library preparations.
From our analysis it is evident that each method has advantages and disadvantages. Both PCR-free methods generated very long contig N50s (150–178 kb) for the mock-community DNA, low duplication rates, and low numbers of low-quality reads. The PCR-based systems had much higher duplication and error rates, but the KF libraries also had a high contig N50 (∼142 kb) for the mock-community DNA. The XT libraries had the poorest performance, with 28% low-quality reads, 1.4% duplication rate, and a contig N50 of 35 kb for the mock-community DNA. Comparison of contig N50s for the clinical specimens showed that the TSF had the longest average contig N50s: 12.9 kb compared with 4.2 kb for XT, 3.5 kb for KF, and 2.9 kb for KP. RGA calculations of the mock-community control demonstrated an average RGA SD of <1% across all library preparations in 16 of 20 (80%) of mock-community organisms (Table 1). Direct comparison of RGA data with the qPCR measurements of the input mock-community DNA did demonstrate over- and underrepresentation of strain DNAs with a deviation of 2.0% or more or 2.0% or less compared with qPCR measurements for mock-community organisms in all the library strategies tested based on RGA (10/20 for XT; 12/20 for KF; 7/20 for KP; 7/20 for TSF). Direct comparison by qPCR of the XT and KP mock-community libraries with the pure mock-community DNA demonstrates that the XT library has greater similarity to the original starting sample than the KP library (Dataset S1, Table S13). The level of quantitative variability of RGA compared with qPCR measurements is extremely interesting and alarming. Numerous publications report a change in abundance of ∼0.1% as significant. Although statistically significant based on study design, the biological significance/relevance of a reported ∼0.1% change in abundance in microbiome studies may be questioned. Our observations and other reports indicate that multiple factors can influence sequencing accuracy and quantitation and also account for chemistry-to-chemistry variability using the same template DNA molecule. Previous reports have demonstrated the introduction of significant bias based on genome amplification enzymes (17). Furthermore, the level of genome fragmentation can bias flowcell clustering efficiency, with smaller DNA fragments clustering more efficiently than longer fragments. This difference in clustering efficiency may explain the RGA deviation between XT tagmentation- and Covaris fragmentation-based libraries. Furthermore, the variability in the efficiency of platform-specific polymerase enzymes and in enzymatic performance in DNA with high %G+C content is well known. Given the multiple influential variables (i.e., DNA fragmentation, library amplification, and size selection, among others), we strongly suggest the inclusion of a calibration control for microbiome studies.
To correlate mapped sequence reads to GUs, we compared the qPCR measurement of DNA from S. oneidensis cells as a control spiked into three stool samples to the RGA generated from sequence analysis. Our analysis revealed a strong correlation between the percent of S. oneidensis DNA in the total extracted stool sample (2.21%, 2.45%, and 3.14% in technical replicates) and the corresponding RGA (3%, 2.8%, and 1.7%). We are able to estimate the number of raw mapped reads equating to a GU. The calculated number of S. oneidensis GUs in the DNA mixture is ∼5 × 103. S. oneidensis DNA represents ∼2–3% of the total extracted DNA, based on qPCR measurements. Using the same ratio of S. oneidensis DNA to total extracted DNA, we calculate that in stool DNA [assuming an average genome size of 1.5–4 Mb (18, 19)] there are ∼2.32 × 105 to 6.18 × 105 GU per nanogram of DNA. We calculate a range of 41–71 (average ∼43) 125-nt reads equate to one GU, based on the total number of reads and number of mapped reads. Assuming average genome sizes of 1.5, 3.9 Mb, 4.9 Mb, and 5.5 Mb, we estimate that 19, 50, 62 and 73 125-nt reads correlate to one GU, respectively. With these calculations we can begin to estimate the actual number of GUs per nanogram of DNA per milligram of specimen and establish an individual’s personalized microbiome baseline per organism. Our analysis indicates that both PCR and PCR-free based systems can introduce bias into the downstream analysis. PCR-free systems offer the ability to increase contig length without the potential bias that PCR-based systems may introduce. The KF systems appear to be the best solution for specimens in which ∼500 ng of high-quality DNA is available for library preparation. For metagenomic samples, a PCR-free system potentially would give the user the ability to measure considerably more potential GUs [∼1.68 × 108 GUs; GU number of copies = (1,000 ng * 6.022 × 1023)/(5.5 Mb * 1 × 109 * 650)].
By comparing four different NGS library preparations using a DNA mock community and a cell spike-in control of known concentration, we have revealed biases in error profiles, duplication rates, and loss of reads in organisms with a high %G+C; these biases can impact metagenomic results significantly. We also have identified methods that can provide high contig N50s that may better predict single genome assemblies within metagenomic samples. As with all methods, the use of benchmarking controls has revealed critical differences between methods that impact sequencing results and later would impact study interpretation. Standardization to one system is not feasible, because technology continues to advance, amounts and availability of microbiome samples are highly variable, and scientific budgets are constrained. We propose standardization based on sample type and material abundance and the inclusion of a cell spike-in for DNA extraction controls. In the majority of cases, stool specimens provide sufficient material (i.e., ∼250–500 ng of DNA or more) to use in a PCR-free system. We propose that the community use PCR-free–based approaches (such as Kapa Hyper Prep PCR-free and TruSeq DNA PCR-free) to reduce PCR bias in calculations of abundance and to improve assemblies for accurate taxonomic assignment. Furthermore, the inclusion of a cell spike-in control will permit a more accurate quantitation of organisms based on a known input value. In the case of precious, low-abundance material, we propose the use of a low-cycle (three to five cycles) PCR-based approach in parallel with the cell spike-in control. These findings suggest that, until standardized platforms and practices are adopted, the microbiome community will be at risk of over-interpreting datasets, generating conflicting results, and hindering progress in the field.
Materials and Methods
Mock-Community DNA and Cell Lysis Controls.
A mock microbial community DNA sample, composed of a mixture 20 microbial genomic DNAs that vary in genome size (1.6–6.2 Mb) and G+C content (32–69%), was obtained from BEI Resources (HM-276D; even, high-concentration v5.1H) (Dataset S1, Table S2). For cell lysis control, cells of the S. oneidensis strain MR-1 were grown from a single colony for 18 h in Lysogeny Broth (10 g tryptone, 10 NaCl, 10 g yeast extract per liter) at 37 °C; then the culture was adjusted to an OD600 of 1.0, which equals 6.6 × 109 cells/mL. Cell counts per OD600 were determined previously by quadruplicate counts of independent cultures using a Petroff–Hausser cell-counting chamber.
Human Subject Enrollment and Sampling.
A 22-y-old healthy female gave informed consent for the study. The subject received a 7-d course of amoxicillin (day 1 through day 7 of the study) and donated fecal samples on days 0 (before antibiotic therapy), 3, and 7 and at week 8 after receiving the antibiotic. Specimens were immediately frozen at −80 °C until use [as described in Human Microbiome Project v12.0 protocol (20)]. This study was approved by the Institutional Review Board of the University of California, San Diego.
Sample Preparation Platforms.
Nextera XT libraries were prepared manually following the manufacturer’s protocol (15031942) (Illumina). Briefly, samples were normalized to 0.2 ng/µL DNA material per library using a Quant-iT PicoGreen assay system (Q33120; Life Technologies) on an AF2200 plate reader (Eppendorf) and then were fragmented and tagged via tagmentation. Amplification was performed by Veriti 96-well PCR (Applied Biosystems) followed by AMPure XP bead cleanup (A63880l; Beckman Coulter). Two technical replicates were generated for each biological sample, resulting in 10 XT-generated libraries. Kapa Hyper Prep libraries were prepared manually following the manufacturer’s protocol (KK8504l; Kapa Biosystems). Samples were normalized to either 1 µg or 1 ng, and DNA was sheared by sonication with a Covaris LE220. Adapters were ligated, and double solid-phase reversible immobilization (SPRI) size selection was performed using SPRI beads from Beckman Coulter (B23318). Samples starting with 1 ng of input DNA were amplified with a 12-cycle PCR and cleaned up with AMPure beads. Samples with a starting input of 1 µg of DNA were processed without PCR amplification. Two technical replicates were generated for each biological sample, resulting in 10 KP and 10 KF libraries, i.e., a total of 20 Kapa libraries. Illumina TruSeq DNA PCR-Free libraries were prepared manually following the manufacturer’s protocol for Illumina TruSeq DNA PCR-Free (15036187; Illumina) with minor modifications. Briefly, samples were normalized to 1 µg DNA and sheared by sonication with a Covaris LE220. AMPure XP beads were used for cleanup and size selection, and adapters then were ligated. Fragment sizes for all libraries were measured using a Labchip GX Touch Hi Sens, and qPCR was performed on a QuantsStudio 6 Flex Real-Time PCR System (Applied Biosystems) with the Kapa library quantification kit. Two technical replicates were generated for each biological sample, resulting in 10 TSF libraries. Sequencing was performed on an Illumina HiSeq 2500 using V4 reagents.
qPCR Primers and Quantitation Assays.
qPCR plates were prepared using 1:10 dilutions of a combined stock of forward and reverse primers at a final concentration of 1.25 μM (Invitrogen) (Dataset S1, Table S12). Three species-specific primers were designed for each of the 20 mock-community organisms. The final reaction concentration of all primers was 0.125 μM. Individual pure genomic DNAs that were component members of the mock community were used to create a standard curve to quantify the strain-specific makeup of the mock community and to establish a melting-curve profile. All qPCR reactions using purified or mock-community DNAs were interrogated by melting-curve analysis for amplification artifacts. Purified S. oneidensis genomic DNA also was used to quantify the composition of S. oneidensis in spiked specimens. Organism-specific, specimen, and mock-community genomic DNAs were diluted to 2 ng/µL and then were serially diluted to 0.02 pg/μl. The designated amount of each DNA sample then was combined with 5.5 μL of 2× SYBR Green Master Mix (Kapa Biosystems) to a total reaction volume of 11 μL per well. Thermal cycling conditions used were 96 °C for 3 min followed by 35 cycles of 96 °C for 30 s, 58 °C for 30 s, and 72 °C for 20 s. A final melting-curve cycle was performed starting at 95° for 15 s, 58° for 1 min, and 95 °C for 15 s. All qPCR reactions were run on the Applied Biosystems QuantStudio 6 system. Median threshold cycle (Ct) values for qPCR duplicate reactions were calculated. Organisms with multiple unique targets were analyzed for quantitative correlation. Calculations were performed using the Applied Biosystems QuantStudio 6 Flex software (Applied Biosystems).
Supplementary Material
Acknowledgments
We thank Dr. Orianna Bretschger (J. Craig Venter Institute) for her kind donation of S. oneidensis bacteria.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information BioProject database, www.ncbi.nlm.nih.gov/bioproject (project ID PRJNA298489).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1519288112/-/DCSupplemental.
References
- 1.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flores R, et al. Assessment of the human faecal microbiota: II. Reproducibility and associations of 16S rRNA pyrosequences. Eur J Clin Invest. 2012;42(8):855–863. doi: 10.1111/j.1365-2362.2012.02659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim M, Yu Z. Variations in 16S rRNA-based microbiome profiling between pyrosequencing runs and between pyrosequencing facilities. J Microbiol. 2014;52(5):355–365. doi: 10.1007/s12275-014-3443-3. [DOI] [PubMed] [Google Scholar]
- 4.Ravel J, Wommack KE. All hail reproducibility in microbiome research. Microbiome. 2014;2(1):8. doi: 10.1186/2049-2618-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt TS, Matias Rodrigues JF, von Mering C. Limits to robustness and reproducibility in the demarcation of operational taxonomic units. Environ Microbiol. 2015;17(5):1689–1706. doi: 10.1111/1462-2920.12610. [DOI] [PubMed] [Google Scholar]
- 6.Finucane MM, Sharpton TJ, Laurent TJ, Pollard KS. A taxonomic signature of obesity in the microbiome? Getting to the guts of the matter. PLoS One. 2014;9(1):e84689. doi: 10.1371/journal.pone.0084689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Best N, Jansen PL, Rensen SS. The gut microbiota of nonalcoholic fatty liver disease: Current methods and their interpretation. Hepatol Int. 2015;9(3):406–415. doi: 10.1007/s12072-015-9640-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner Mackenzie B, Waite DW, Taylor MW. Evaluating variation in human gut microbiota profiles due to DNA extraction method and inter-subject differences. Front Microbiol. 2015;6:130. doi: 10.3389/fmicb.2015.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeSantis TZ, Stone CE, Murray SR, Moberg JP, Andersen GL. Rapid quantification and taxonomic classification of environmental DNA from both prokaryotic and eukaryotic origins using a microarray. FEMS Microbiol Lett. 2005;245(2):271–278. doi: 10.1016/j.femsle.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 10.Franzosa EA, et al. Sequencing and beyond: Integrating molecular ‘omics’ for microbial community profiling. Nat Rev Microbiol. 2015;13(6):360–372. doi: 10.1038/nrmicro3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franzosa EA, et al. Relating the metatranscriptome and metagenome of the human gut. Proc Natl Acad Sci USA. 2014;111(22):E2329–E2338. doi: 10.1073/pnas.1319284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eichner CA, Erb RW, Timmis KN, Wagner-Döbler I. Thermal gradient gel electrophoresis analysis of bioprotection from pollutant shocks in the activated sludge microbial community. Appl Environ Microbiol. 1999;65(1):102–109. doi: 10.1128/aem.65.1.102-109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aryee MJ, Gutiérrez-Pabello JA, Kramnik I, Maiti T, Quackenbush J. An improved empirical bayes approach to estimating differential gene expression in microarray time-course data: BETR (Bayesian Estimation of Temporal Regulation) BMC Bioinformatics. 2009;10:409. doi: 10.1186/1471-2105-10-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeAngelis KM, et al. PCR amplification-independent methods for detection of microbial communities by the high-density microarray PhyloChip. Appl Environ Microbiol. 2011;77(18):6313–6322. doi: 10.1128/AEM.05262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia LC, Cram JA, Chen T, Fuhrman JA, Sun F. Accurate genome relative abundance estimation based on shotgun metagenomic reads. PLoS One. 2011;6(12):e27992. doi: 10.1371/journal.pone.0027992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brooks JP, et al. Vaginal Microbiome Consortium The truth about metagenomics: Quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol. 2015;15:66. doi: 10.1186/s12866-015-0351-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Probst AJ, Weinmaier T, DeSantis TZ, Santo Domingo JW, Ashbolt N. New perspectives on microbial community distortion after whole-genome amplification. PLoS One. 2015;10(5):e0124158. doi: 10.1371/journal.pone.0124158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nayfach S, Pollard KS. Average genome size estimation improves comparative metagenomics and sheds light on the functional ecology of the human microbiome. Genome Biol. 2015;16:51. doi: 10.1186/s13059-015-0611-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manor O, Borenstein E. MUSiCC: A marker genes based framework for metagenomic normalization and accurate profiling of gene abundances in the microbiome. Genome Biol. 2015;16:53. doi: 10.1186/s13059-015-0610-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Human Microbiome Project Consortium A framework for human microbiome research. Nature. 2012;486(7402):215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.