Significance
This work identifies autophagy as an essential homeostatic process that maintains pancreatic acinar cell function. By preventing endoplasmic reticulum stress, reactive oxygen species accumulation, and DNA damage, basal autophagy preserves the high rates of protein synthesis that characterize the exocrine pancreas. Conversely, loss of autophagy can result in progressive loss of pancreatic function, which leads to development of pancreatitis as well as regenerative responses that may increase the risk of pancreatic cancer.
Keywords: autophagy, ATG7, pancreatitis, protein synthesis
Abstract
Pancreatic acinar cells possess very high protein synthetic rates as they need to produce and secrete large amounts of digestive enzymes. Acinar cell damage and dysfunction cause malnutrition and pancreatitis, and inflammation of the exocrine pancreas that promotes development of pancreatic ductal adenocarcinoma (PDAC), a deadly pancreatic neoplasm. The cellular and molecular mechanisms that maintain acinar cell function and whose dysregulation can lead to tissue damage and chronic pancreatitis are poorly understood. It was suggested that autophagy, the principal cellular degradative pathway, is impaired in pancreatitis, but it is unknown whether impaired autophagy is a cause or a consequence of pancreatitis. To address this question, we generated Atg7Δpan mice that lack the essential autophagy-related protein 7 (ATG7) in pancreatic epithelial cells. Atg7Δpan mice exhibit severe acinar cell degeneration, leading to pancreatic inflammation and extensive fibrosis. Whereas ATG7 loss leads to the expected decrease in autophagic flux, it also results in endoplasmic reticulum (ER) stress, accumulation of dysfunctional mitochondria, oxidative stress, activation of AMPK, and a marked decrease in protein synthetic capacity that is accompanied by loss of rough ER. Atg7Δpan mice also exhibit spontaneous activation of regenerative mechanisms that initiate acinar-to-ductal metaplasia (ADM), a process that replaces damaged acinar cells with duct-like structures.
The pancreatic acinar cell is responsible for production and secretion of numerous digestive enzymes, including amylase, lipase, and various proteases. To cope with the high daily demand for these enzymes, the acinar cell possesses one of the highest protein biosynthetic rates of all cells, together with an extensive rough endoplasmic reticulum (RER) network (1). Due to its high protein synthetic rates, the acinar cell is prone to the accumulation of misfolded proteins and subsequent induction of ER stress (2, 3). ER stress was suggested to be involved in the pathogenesis of pancreatitis, a potentially fatal inflammatory disease of the exocrine pancreas (2, 4). By progressing from acute (sudden onset; duration <6 mo), to recurrent acute (>1 episode of acute pancreatitis), and chronic (duration >6 mo) disease (5), pancreatitis increases the risk of pancreatic ductal adenocarcinoma (PDAC), the fourth deadliest cancer worldwide, with a median survival of 6 mo (6). The molecular mechanisms mediating the progression of pancreatitis from acinar cell damage and inflammation to formation of pancreatic intraepithelial neoplasia (PanIN) and PDAC are not fully understood. Recent studies suggest that in addition to ER stress, insufficient autophagy also contributes to development of pancreatitis (7).
Autophagy is an evolutionarily conserved, catabolic quality control process that maintains cellular homeostasis by degrading damaged organelles, misfolded protein aggregates, and foreign organisms (8). Autophagy is also important for generation of amino acids and other building blocks during starvation (9). There are three classes of autophagy: macroautophagy, microautophagy, and chaperone-mediated autophagy (9). Macroautophagy, the major type of autophagy (hereafter referred to as autophagy), entails formation of double-membrane vesicles (autophagosomes) that sequester damaged organelles and biomolecules and recycle them after transport into lysosomes, where they are degraded. The rate of autophagy is increased in response to diverse stress conditions, including nutrient deprivation, viral infection, and genotoxic stress. In this way, autophagy controls the cross-talk between the intracellular demand for energy, building blocks, and external stimuli (9). Autophagy is critically involved in mammalian development, cell survival, and longevity (10), and its impairment correlates with many pathological conditions (11), including pancreatitis (7). Notably, the mammalian exocrine pancreas exhibits a higher autophagy rate (or autophagic flux) than the liver, kidney, heart, or endocrine pancreas (12), underscoring the likely importance of autophagy in maintaining acinar cell homeostasis and function. Until recently, however, the role of autophagy in pancreatitis has been controversial. On one hand, the genetic inhibition of autophagy [Autophagy Related 5 (Atg5) gene ablation] was found to reduce trypsinogen activation and attenuate pancreatic damage in mice challenged with the pancreatic enzyme secretagogue cerulein (13); but selective and protective autophagy can sequester and degrade potentially deleterious activated zymogens during early pancreatitis (14). More recent findings from experimental models (cerulein-induced pancreatitis), and genetically altered mice (Pdx-Cre; IkkαF/F, also known as IkkαΔpan, and Ptf1a-Cre; Atg5F/F) have demonstrated that insufficient autophagy, at least in mice, can lead to the onset of pancreatitis (15–17). The mechanisms by which disruption of autophagy triggers pancreatitis are poorly understood.
Degradation of long-lived proteins, a major function of autophagy, is impaired during experimental pancreatitis, especially after administration of cerulein, which causes acinar cell vacuolization and excessive trypsinogen activation (18). Moreover, autophagic flux is reduced during pancreatitis due to defective cathepsin-mediated processing of lysosomal proteases (18). Similarly, pancreas-specific ablation of inhibitor of IκB kinase (IKK)α results in acinar damage ranging from vacuole accumulation to chronic pancreatitis (15). IKKα deficiency impairs the completion of autophagy in acinar cells, with accumulation of the chaperon and autophagy substrate ubiquitin-binding protein p62/SQSTM1 as the key pathogenic mechanism (15). However, ablation of ATG5 was reported to either inhibit (13) or promote (16) pancreatitis. In addition, inhibition of autophagy can either accelerate the development of early malignant lesions in mice lacking the transformation-related protein 53 (p53) (19) or cause the death of established pancreatic cancer (20). The latter results gave rise to several ongoing clinical trials (clinicaltrials.gov numbers NCT01494155, NCT01978184, NCT01506973, NCT01128296, and NCT01273805) that intend to evaluate the impact of autophagy inhibitors on human pancreatic cancer. However, a recent commentary has raised concerns about the safety of this therapeutic approach (21). Given all of these questions, we decided to take a closer look at the impact of autophagy inhibition on pancreatic health and function by generating mice that lack autophagy-related protein 7 (ATG7) in pancreatic epithelial cells. These mice, termed Atg7Δpan, exhibit striking acinar cell degeneration, which is followed by pronounced pancreatic inflammation and fibrosis. Whereas loss of ATG7 leads to the expected decrease in autophagic flux, it also results in ER stress, accumulation of dysfunctional mitochondria, oxidative stress, and a marked reduction in protein synthetic ability.
Results
Loss of Pancreatic ATG7 Results in Acinar Cell Damage and Inflammation.
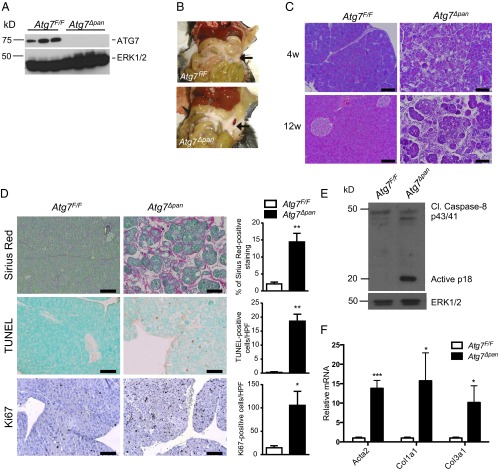
To investigate the role of autophagy in pancreatic physiology and homeostasis we generated Pdx1-Cre; Atg7F/F (Atg7∆pan) mice, which lack the essential autophagy gene Atg7 in all pancreatic epithelial cells (PECs), by crossing Atg7F/F mice (22) with Pdx1-Cre mice. Immunoblot (IB) analysis of whole pancreas lysates confirmed complete loss of ATG7 in pancreata of Atg7∆pan mice compared with Cre-negative controls (Fig. 1A). At 12 wk of age, Atg7∆pan mice exhibited severe pancreatic degeneration as suggested by the whitish, fibrotic appearance of the pancreas (Fig. 1B). Histological analysis of pancreatic tissue from 4- and 12-wk-old Atg7Δpan mice showed absence of healthy morphology and dramatic and progressive tissue damage (Fig. 1C), with enhanced cell death (Fig. 1D) and compensatory proliferation at 12 wk as shown by the increase in Ki67 and PCNA-positive nuclei in the remaining amylase positive cells (Fig. 1D and Fig. S1A). Cell death was accompanied by caspase-8 activation (Fig. 1E), suggesting it may be induced in response to elevated expression of death-promoting cytokines. Sirius red staining, which detects collagen accumulation, showed the onset of fibrosis (Fig. 1D), which was confirmed by up-regulation of α-SMA (Acta2), collagen1A(I) (Col1a1), and collagen3A(I) (Col3a1) mRNAs (Fig. 1F). Atg7-deficient pancreata also displayed up-regulation of numerous cytokine, chemokine, and immune cell marker genes, such as Tnf, Il6, Ccl2, and F4/80 (Fig. S1B).
Fig. 1.
Atg7∆pan mice exhibit pancreatic degeneration, inflammation, and fibrosis. (A) IB analysis of ATG7 in pancreatic lysates of 12-wk-old mice of indicated genotypes. (B) Gross morphology of pancreata in 12-wk-old Atg7F/F and Atg7∆pan mice. (C) H&E staining of pancreatic tissue sections from 4- and 12-wk-old mice. (Scale bars, 100 μm.) (D) Histological analysis (IHC) of pancreatic sections from 12 wk old Atg7F/F and Atg7∆pan mice. Sirius Red, TUNEL, and Ki67 staining, and corresponding quantitation of positive cells per high power (200×) field (HPF). (Scale bars, 100 μm.) (E) IB analysis of caspase 8 in pancreatic lysates from 3-wk-old mice. ERK1/2: loading control. (F) qPCR analysis of α-SMA (Acta2), collagen1A(I) (Col1a1) and collagen 3A(I) (Col3a1) mRNAs in 12-wk-old mice of indicated genotypes. Values in D and F are means ± SEM n = 3–4 mice per condition. *P < 0.05, **P < 0.01, ***P < 0.001.
Fig. S1.
Loss of pancreatic ATG7 results in inflammation, acinar cell death, and compensatory proliferation. (A) PCNA and amylase co-IF in pancreatic sections from 12-wk-old mice of the indicated genotype. (Scale bars, 200 μm.) (B) qPCR analysis of cytokine, chemokine, and immune cell marker mRNAs. Values are means ± SEM n = 3–4 mice per condition. *P < 0.05; ***P < 0.001.
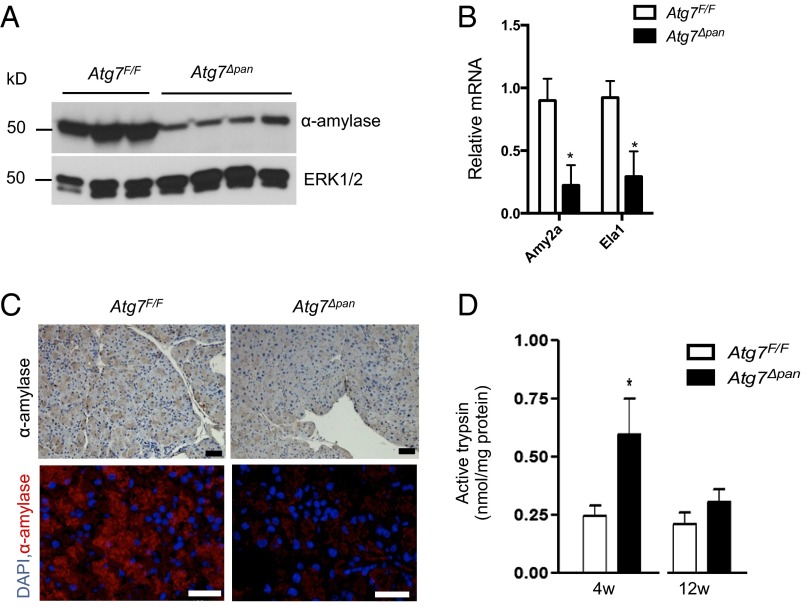
To identify the type of damaged cells in the Atg7∆pan pancreas, we analyzed expression of acinar cell markers and observed reduced protein and mRNA expression of α-amylase and elastase (Fig. 2 A and B). This was accompanied by a decrease in amylase positive cells in Atg7Δpan pancreata (Fig. 2C), highlighting pronounced acinar cell damage and loss. Intrapancreatic trypsin activity was elevated at an early (4 wk) but not at a later (12 wk) time point (Fig. 2D), suggesting that premature, intraacinar, trypsinogen activation is just one of several factors that contribute to this disease (15, 16). Collectively, these results indicate that loss of ATG7 triggers pancreatic atrophy, fibrosis, and chronic pancreatitis.
Fig. 2.
Loss of pancreatic ATG7 results in acinar cell damage. (A–C) Analysis of acinar cell markers in 12-wk-old Atg7F/F and Atg7∆pan mice by IB (A), qPCR (B), IHC (C, Top), or IF (C, Bottom). Results in B are means ± SEM n = 3–4 mice per condition. *P < 0.05. [Scale bars in C, 50 μm (IHC) and 100 μm (IF).] (D) Trypsin activity in pancreata of 4- and 12-wk-old Atg7F/F and Atg7∆pan mice. Results are means ± SEM n = 6 (4 w), n = 3 (12 w). *P = 0.06.
Loss of Pancreatic ATG7 Impairs Autophagy.
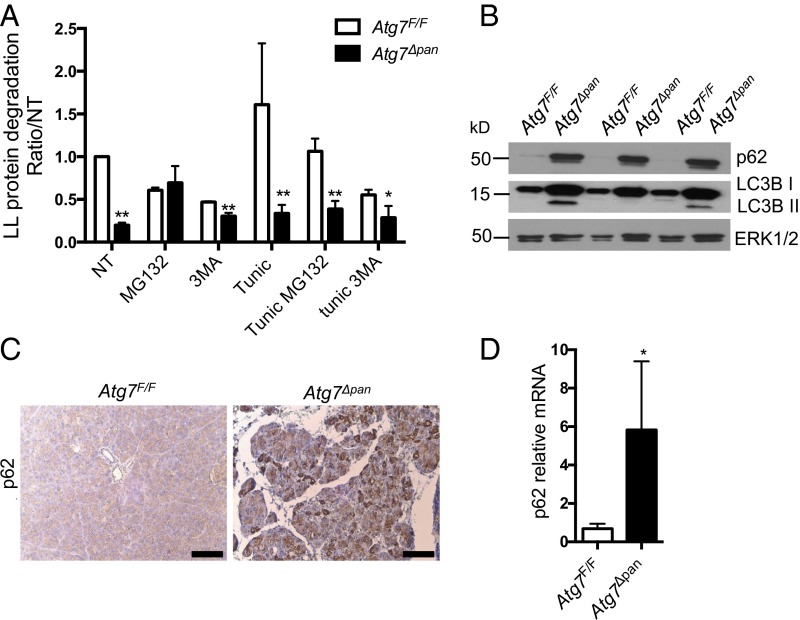
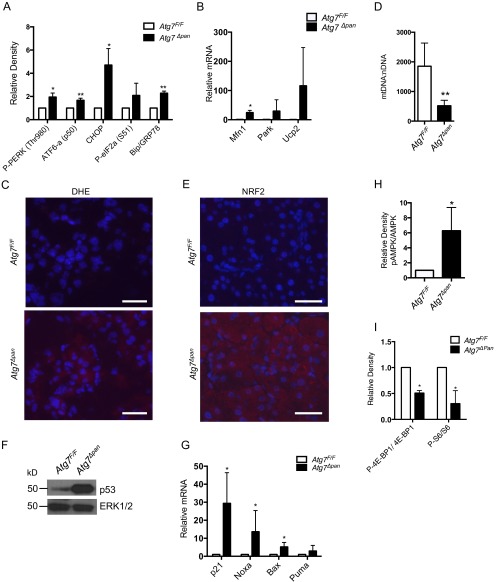
ATG7 is an E1 ligase-like enzyme that activates microtubule-associated protein 1A/1B-light chain 3 (LC3)-I and ATG12 during autophagosome elongation (23). Therefore, its loss should inhibit autophagosome formation and downstream autophagic protein degradation. Indeed, measurement of bulk long-lived protein degradation in Atg7Δpan primary acinar cells in the absence or presence of the autophagy inhibitor 3-methyladenine (3MA) revealed impaired autophagic flux, with a baseline value that was neither further decreased by 3MA treatment, nor induced after stimulation with the ER stress inducer tunicamycin (Fig. 3A). In addition, Atg7Δpan primary acinar cells and pancreata exhibited accumulation of the autophagy substrate and chaperone p62 (Fig. 3 B and C and Fig. S2 A and B) and the unmodified form of LC3 (LC3B-I), although the presence of phosphatidylethanolamine (PE) conjugated LC3 (LC3B-II) (Fig. 3B) indicates residual noncanonical authophagy (24). Whereas expression of key autophagy-related genes was unaffected by Atg7 ablation (Fig. S2C), p62 mRNA was considerably elevated in Atg7Δpan pancreata (Fig. 3D). The increase in p62 mRNA could be due to either activation of a positive feedback loop involving the nuclear factor erythoroid 2 (NRF2) (25) or activation of NF-κB by inflammatory cytokines (26).
Fig. 3.
Impaired autophagy and p62 accumulation in Atg7∆pan pancreata. (A) Autophagic flux in Atg7F/F and Atg7Δpan primary acinar cells treated or not with the ER stress-inducer tunicamycin (10 μg/mL, 4 h) in the absence (not treated, NT) or presence of the autophagy inhibitor 3-MA (10 mM), and MG132 (100 nM). Results are means ± SEM of triplicates. n = 3–4 mice per condition. *P < 0.05. (B) IB analysis of LC3B and p62 in pancreatic lysates from 12-wk-old mice. (C) p62 IHC of pancreatic sections from 12-wk-old mice of the indicated genotype. (Scale bars, 100 μm.) (D) qPCR analysis of p62 mRNA. Results are means ± SEM of triplicates. n = 3–4 mice per condition. *P < 0.05.
Fig. S2.
Impaired autophagy and p62 accumulation in Atg7∆pan pancreata. (A) p62 IB in primary acinar cells. Tubulin loading control, *nonspecific band. (B) IHC of pancreatic sections from 12-wk-old mice of the indicated genotype. (Scale bars, 100 μm.) (C) qPCR analysis of autophagy-related mRNAs. Results are means ± SEM of triplicates. n = 3–4 mice per condition.
Loss of ATG7 Results in ER Stress and Mitochondrial Dysfunction.
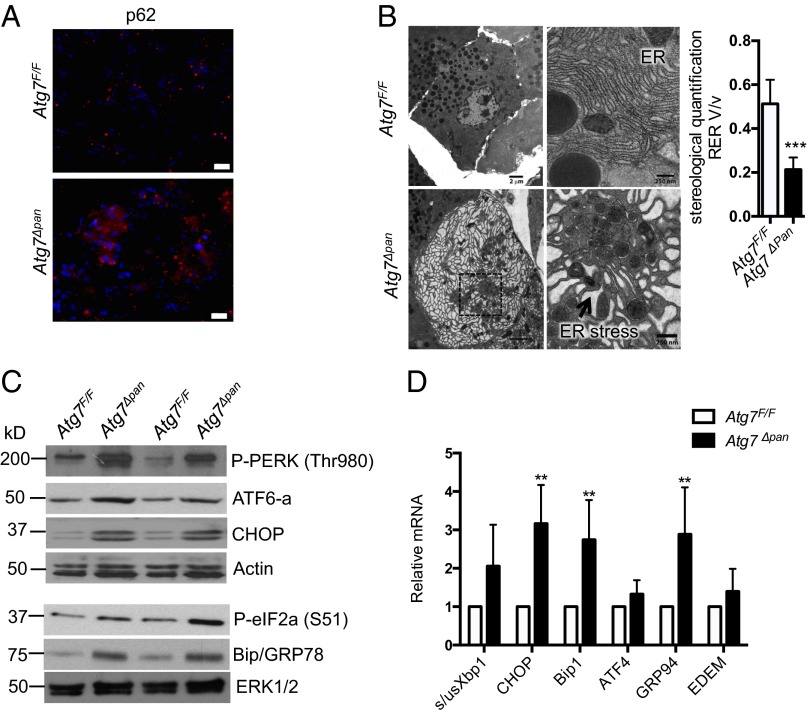
Impaired autophagy can contribute to ER stress by inhibiting degradation of misfolded proteins (15), which undergo ubiquitination and are bound by p62 (27). Indeed, we detected pronounced aggregates/inclusion bodies containing p62 in Atg7Δpan pancreata (Fig. 4A). Fittingly, electron microscopy (EM) revealed extensively distended and dilated ER with nearly complete loss of associated ribosomes in Atg7∆pan pancreata, which was quantified by stereological analysis (Fig. 4B). Furthermore, ER stress activates a complex signaling network in Atg7Δpan pancreata, the unfolded protein response (UPR), to restore homeostasis (28). Indeed, immunoblot analysis revealed increased amounts of cleaved ATF6a p50, phosphorylated PKR-like ER kinase (PERK), and translation initiation factor 2α (eIF2α; Fig. 4C and Fig. S3A), all of which decrease translation initiation. The proteolytic cleavage and activation of ATF6 were confirmed by increased mRNA and protein expression of GRP78/Bip and GRP94 (Fig. 4 C and D). Increased expression of C/EBP-homologous protein (CHOP) which is controlled by the PERK-eIF2α -ATF4 pathway, was also seen (Fig. 4 C and D and Fig. S3A). Moreover, Atg7∆pan pancreata also showed increased XBP1 mRNA splicing, indicating IRE1α activation (Fig. 4D) (28).
Fig. 4.
Loss of ATG7 results in ER stress. (A) IF analysis of p62-containing protein aggregates in Atg7F/F and AtgΔpan pancreata. (Scale bars, 20 μm.) (B) EM images showing dilated ER (arrow) and accumulation of smooth ER, quantified by stereological analysis in 12-wk-old Atg7F/F and Atg7Δpan mice. [Scale bars, 2 μm (Left) and 250 nm (Right).] Results are means ± SEM n = 7–8 micrographs per condition (one mouse for each condition). ***P < 0.001. (C and D) Expression of ER stress markers in pancreata of 12-wk-old Atg7F/F and Atg7Δpan mice. (C) IB analysis of indicated proteins. Loading control: Actin (Top), ERK1/2 (Bottom). (D) Analysis of the indicated mRNAs by qPCR. Results are means ± SEM n = 4 mice per condition. *P < 0.05.
Fig. S3.
ATG7 ablation results in accumulation of aberrant mitochondria, oxidative stress and NRF2 activation. (A) Densitometric analysis of indicated proteins from Fig. 4C, after normalization with loading control. (B) Expression of the indicated pancreatic mRNAs was determined by qPCR. Results are means ± SEM of triplicates. n = 3 mice per condition. *P < 0.05 vs. Atg7F/F. (C) DHE staining of pancreatic sections from 12-wk-old Atg7F/F and Atg7Δpan mice. (Scale bars, 100 μm.) (D) Quantification of mitochondrial DNA (mtDNA) per nuclear DNA (nDNA) in pancreata of 12-wk-old mice. Results are means ± SEM of triplicates. n = 3 mice per condition. **P < 0.01 vs. Atg7F/F. (E) NRF2 IF in pancreatic sections from 12-wk-old Atg7F/F and Atg7Δpan mice. (Scale bars, 100 μm.) (F and G) Activation of p53 and its target genes in pancreata of 12-wk-old mice. Results are means ± SEM of triplicates. n = 3 mice per condition. *P < 0.05 vs. Atg7F/F. (H) Relative amounts of phosphorylated AMPKα from Fig. 5F were determined by densitometry after normalization with ERK1/2. (I) Densitometric analysis of phosphorylated mTORC1 targets from Fig. 5G. An arbitrary value of 1.0 was given to the Atg7F/F specimens. Results in H and I are means ± SEM n = 3 mice per condition. *P < 0.05 vs. Atg7F/F.
ER stress can promote mitochondrial damage (29), and autophagy/mitophagy is responsible for removal and recycling of damaged or aged mitochondria (30). Congruently, EM analysis revealed accumulation of fused, electron-dense, and abnormally looking mitochondria, containing fewer and swollen cristae in Atg7∆pan pancreata (Fig. 5A). These morphological alterations, which were absent in AtgF/F tissue, correlated with increased protein and mRNA expression of parkin, a protein responsible for decorating defective mitochondria with polyubiquitin chains, and mitofusin 1 (Mfn1), a GTPase that regulates mitochondrial fusion (Fig. 5B and Fig. S3B) (30). Damaged mitochondria and ER stress result in accumulation of reactive oxygen species (ROS). This was confirmed by enhanced staining of Atg7Δpan pancreata with dihydroethidium (DHE), which is oxidized by superoxide anions (Fig. S3C). Mitochondrial DNA amounts were decreased (Fig. S3D), further supporting the presence of extensive mitochondrial damage. Atg7∆pan pancreata contained high amounts of Ucp2 mRNA (Fig. S3B), encoding a mitochondrial protein located on the inner mitochondrial membrane, involved in adaptive response to mitochondria-derived ROS and oxidative damage (31).
Fig. 5.
ATG7 ablation results in accumulation of aberrant mitochondria, oxidative stress, and activation of protective and counteractive responses. (A) EM images showing accumulation of abnormally looking mitochondria (arrow) in 12-wk-old mice. [Scale bars, 2 μm (Left), 250 nm (Top Right), and 500 nm (Bottom Right).] Healthy vs. damaged mitochondria were quantified by stereological analysis. Results are means ± SEM n = 7–8 micrographs per condition (one mouse for each condition). ***P < 0.001. (B) Mfn1 and Parkin expression in pancreata of above mice. Protein amounts were determined by densitometry and normalized to actin (Bottom). (C) Protein expression of NRF2 and NQO1 in primary acinar cell (NRF2), and total prancreata samples (NQO1) of 12-wk-old Atg7F/F and Atg7Δpan mice. (D) Expression of the indicated pancreatic mRNAs was determined by qPCR. Results are means ± SEM of triplicates. n = 3 mice per condition. *P < 0.05, **P < 0.01 vs. Atg7F/F. (E) IB analysis of antiapoptotic proteins. (F) IB analysis of total and phosphorylated AMPKα in 12-wk-old pancreata. (G) IB analysis of phosphorylated mTORC1 targets in pancreatic lysates. (H) Protein synthesis in primary acinar cells of Atg7∆pan and Atg7F/F mice, quantified as the ratio of [3H]-leucine incorporated into TCA insoluble material between control and cycloheximide (30 µg/mL) treated cells. *P < 0.05 vs. Atg7F/F. Results are means ± SEM of triplicates. n = 3 mice per condition.
Activation of Protective and Counteractive Responses.
Disruption of autophagy, accumulation of p62, and buildup of ROS also lead to activation of counteractive protective responses. First and foremost, both ROS accumulation and elevated p62 expression result in activation of nuclear factor erythoroid 2 (NRF2), which stimulates the expression of genes encoding antioxidant enzymes (32). As expected, NRF2 was up-regulated in both Atg7Δpan pancreata and primary acinar cells and its up-regulation correlated with elevated expression of numerous NRF2 target genes (Fig. 5 C and D and Fig. S3E).
Immunoblot analysis also revealed accumulation of Bcl-XL and Bcl2, two antiapoptotic proteins that prevent and attenuate apoptosis and necrosis in pancreatitis, and X-linked inhibitor of apoptosis protein (XIAP), which negatively regulates caspase activation (Fig. 5E). Oxidative stress and mitochondrial damage lead to p53 activation (33). Congruently, p53 protein and p53 target genes were elevated in Atg7Δpan pancreata (Fig. S3 F and G).
Reduced ATP production due to mitochondrial damage should lead to activation of AMP-sensitive protein kinase (AMPK), which can stimulate mitochondrial biogenesis and lead to mTORC1 inhibition (34). Indeed, AMPK phosphorylation was markedly elevated in Atg7Δpan tissue (Fig. 5F and Fig. S3H). Fittingly, phosphorylation of the mTORC1 substrate 4E-BP1 was reduced along with phosphorylation of ribosomal protein S6, the substrate of p70S6K, another direct mTORC1 target (Fig. 5G and Fig. S3I). Reduced mTORC1 activity correlated with a decrease in the rate of protein synthesis in Atg7Δpan acinar cells (Fig. 5H). Inhibition of protein synthesis may be responsible for the nearly complete absence of RER in ATG7-deficient pancreata.
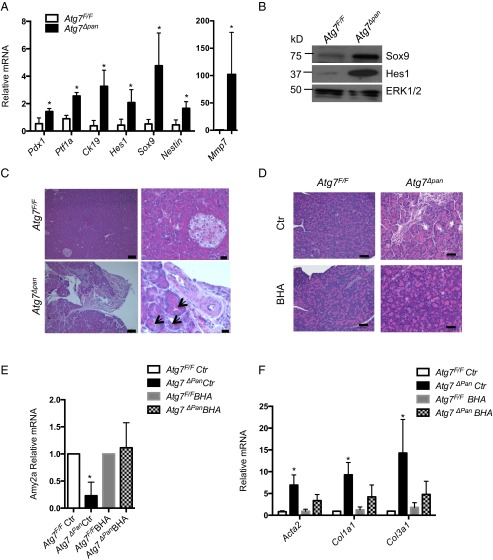
Pancreatic injury also triggers a counteractive regenerative response that restores digestive enzyme-producing acinar cells. This response entails the proliferation of residual acinar tissue and acinar to ductal metaplasia (ADM), a process in which acinar cells dedifferentiate to become ductal progenitors, which eventually restore acinar tissue mass and function (35, 36). Atg7Δpan 12-wk-old pancreata exhibited up-regulation of several pancreatic progenitor and ductal markers, including Ptf1, Sox9, CK19, and Pdx1, as well as the Notch target genes Hes1 and Nestin (Fig. 6A). Protein expression of Sox9 and Hes1 was also elevated (Fig. 6B), as well as the mRNA expression of matrix metalloproteinase 7 (Mmp7; Fig. 6A), a metalloproteinase involved in Notch activation (37). Histological analysis confirmed the presence of structures consistent with ADM in Atg7Δpan pancreata, which were absent in Atg7F/F pancreata (Fig. 6C).
Fig. 6.
Acinar-to-ductal metaplasia and role of oxidative stress in Atg7Δpan pancreatitis. (A) Relative mRNA expression of indicated genes determined by qPCR. Results are means ± SEM of triplicates. n = 3 mice per condition. *P < 0.05 vs. Atg7F/F. (B) Sox9 and Hes1 in total pancreatic lysates of 12-wk-old mice. (C) H&E staining of pancreatic sections from 12-wk-old mice. ADM-like structures indicated by arrows. [Scale bars, 100 μm (Left) and 20 μm (Right).] (D) H&E staining of pancreatic tissue sections from 12-wk-old Atg7F/F and AtgΔpan mice fed with BHA-containing (0.7%) diet (BHA) or normal chow (Ctr) for 4 wk. (Scale bars, 50 μm.) (E and F) mRNA analysis of the indicated genes in total pancreata samples treated as described in D. Results are means ± SEM of triplicates. n = 4 mice per condition. *P < 0.05 vs. Atg7F/FCtr.
Role of p62 and Oxidative Stress in Pancreatic Pathology.
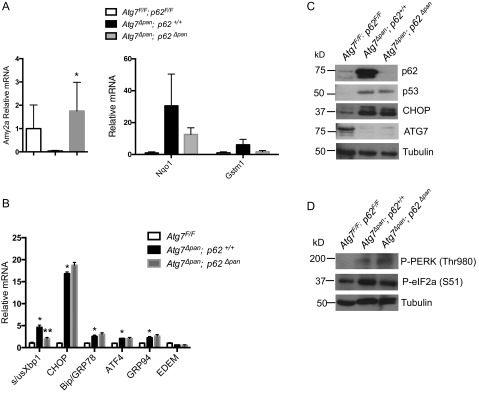
Because p62 is a key pathogenic mediator in another genetic model of chronic pancreatitis (15), we examined its role as a possible mediator of the ATG7-deficient phenotype. The 12-wk-old double knockout mice, deficient in both ATG7 and p62 (Pdx1Cre; Atg7F/F; p62F/F), still exhibited pancreatic damage, inflammation, and ADM formation. Only a partial rescue of the Atg7Δpan phenotype was observed, with a significant increase in acinar gene expression (Amy2A; Fig. S4A, Left), and reduced expression of NRF2 target genes like Gstm1 and Nqo1 (Fig. S4A, Right). However, expression of fibrogenic, progenitor and ductal markers remained unchanged, and genes involved in UPR, with the exception of the down-regulation of XBP1 splicing, remained elevated (Fig. S4B). Immunoblot analysis revealed the persistence of elevated p53, CHOP, and phosphorylated PERK and eiF2a (Fig. S4 C and D), indicating that ER stress, DNA damage, and inhibition of protein synthesis still occur in the absence of p62.
Fig. S4.
The Atg7Δpan phenotype is only partially rescued by additional p62 ablation. (A) Amylase (Amy2a) (Left), Nqo1 and Gstm1 (Right) mRNA amounts in pancreata of indicated genotypes. Results are means ± SEM n = 3 mice per condition. *P < 0.05 vs. Atg7∆pan; p62F/F. (B) mRNA analysis of the indicated genes. Results are means ± SEM n = 3 mice per condition. *P < 0.05 vs. Atg7 F/F; p62F/F. **P < 0.01 vs. Atg7∆pan; p62F/F. (C) IB analysis of total pancreatic lysates from mice of indicated genotypes. Tubulin: loading control. (D) P-PERK and P-eIF2a IB analysis of total pancreatic lysates from mice of indicated genotypes. Tubulin loading control.
Given the extensive oxidative stress in Atg7Δpan mice, we examined whether antioxidants can ameliorate some of the pathology. We fed 2-mo-old mice with chow containing butylated hydroxyanisole (BHA) or normal chow for 4 wk. BHA is a widely used antioxidant and preservative in food and medicines. Histological analysis of pancreatic tissue from Atg7Δpan mice fed with BHA-containing diet showed partial restoration of healthy morphology and less tissue damage than in Atg7Δpan mice fed normal chow (Fig. 6D). The reduction in tissue damage was also highlighted by up-regulation of the acinar marker Amy2a (Fig. 6E). In vitro treatment of primary acinar cells derived from Atg7Δpan and Atg7F/F mice was also protective and increased Amy2a expression (Fig. S5A). Moreover, in vivo BHA treatment reduced fibrosis, as demonstrated by diminished expression of collagen1A(I) (Col1a1) and collagen3A(I) (Col3a1) (Fig. 6F), but failed to attenuate the inflammatory response, characterized by up-regulation of cytokine and chemokine gene transcription (Fig. S5B). Although ER stress and oxidative stress are linked (28), BHA treatment did not prevent p62 accumulation (Fig. S5C) and up-regulation of ER stress markers (Fig. S5 C and D). Phosphorylated eiF2a remained elevated and mTORC1 remained inhibited as indicated by persistence of lower amounts of phospho-S6 (Fig. S5C).
Fig. S5.
Role of oxidative stress in the Atg7Δpan pancreatitis model. (A) Amy2a mRNA analysis in primary acinar cell of the indicated genotype, treated with BHA (5 μM) or vehicle Ctr for 48 h. (B and D) mRNA analysis of the indicated genes in total pancreata from 12-wk-old Atg7F/F and AtgΔpan mice fed with BHA-containing (0.7%) diet (BHA) or normal chow (Ctr) for 4 wk. Results are means ± SEM of triplicates. n = 4 mice per condition. *P < 0.05 vs. Atg7F/FCtr. (C) IB analysis of the indicted proteins in total pancreatic lysates treated as describe in B.
Discussion
The results described above indicate that ATG7-dependent autophagy is absolutely essential in both genders for maintenance of normal pancreatic acinar cell physiology and homeostasis. Unlike previously described IkkαΔpan and Atg5Δpan mice, which exhibit autophagy defects that can be ameliorated by the loss of p62 (15, 16), the more severe pancreatitis and acinar tissue atrophy in Atg7Δpan mice cannot be rescued by additional p62 ablation. These findings also stand in marked contrast to those reported in an earlier study in which Atg5 was ablated using Cre recombinase driven by the elastase promoter (13). In that study, no pancreatic injury was observed and the ATG5 deficiency was even claimed to ameliorate cerulein-induced pancreatic injury (13), a rather odd finding because cerulein-induced pancreatic injury entails inhibition of autophagic protein degradation (18). Notably, a more recent study demonstrated that Ptf1a-Cre–mediated Atg5 ablation does result in the development of pancreatitis (16), albeit not as severe as the pathology seen in Atg7Δpan mice. Although the basis for these differences is not entirely clear, our results strongly suggest that ongoing autophagy is needed for the continuous recycling of misfolded proteins, which spontaneously appear in acinar cells due to their very high protein synthesis rate. Notably, the UPR is also linked to an autophagy-related, proteasome-independent mechanism of protein degradation, known as endoplasmic reticulum-associated degradation, ERAD (2). Therefore, ATG7-dependent autophagy controls the health and survival of acinar cells, by maintaining functional ERAD as well as supporting elevated protein synthesis. ATG7-dependent autophagy leads to clearance of inclusion bodies containing misfolded proteins bound to p62 and also supplies the protein synthetic machinery with amino acids released upon lysosomal proteolysis of misfolded proteins and damaged organelles. Atg7 ablation results in accumulation of inclusion bodies and extensive ER stress, one of whose consequences is inhibition of translation initiation through PERK-mediated phosphorylation of eIF2α (38). Both PERK activation and eIF2α phosphorylation are elevated in Atg7Δpan pancreata. We envision the UPR response, which leads to inhibition of mRNA translation and consequent decrease in ER protein load, as the last opportunity for the stressed acinar cell to fight against chronic damage and restore cellular homeostasis. However, despite UPR activation, the impairment of autophagy-related ERAD prevents the restoration of homeostasis, resulting in other rather severe sequelae, including the perturbation of Ca2+ homeostasis (39), which promotes mitochondrial damage. Damaged mitochondria are cleared via mitophagy, and therefore the ATG7 deficiency leads to extensive accumulation of damaged mitochondria in Atg7Δpan acinar cells, and a compensatory response that attempts to stimulate mitophagy by increasing parkin and mitofusin 1. Mitochondrial damage results in insufficient production of ATP, leading to the activation of AMPK and inhibition of mTORC1, thereby causing further inhibition of protein synthesis due to accumulation of nonphosphorylated 4E-BP1 (33). Although AMPK activation may result in stimulation of mitochondrial biogenesis, this is insufficient for preventing the decrease in mitochondrial DNA in Atg7Δpan cells. Damaged mitochondria, together with aggregated proteins and a chronic ER stress, further enhance ROS accumulation leading to activation of p53 and up-regulation of proapoptotic and antiproliferative genes (33), as well as NRF2 activation (32).
Despite activation of NRF2 and elevated expression of several antioxidant enzymes, the overall metabolic balance in the ATG7-deficient acinar cells is in favor of ROS accumulation and oxidative stress. We tried to ameliorate oxidative stress pharmacologically by feeding Atg7Δpan mice with BHA-containing diet with only partial success. Although BHA feeding prevented loss of acinar cells due to ADM and reduced the extent of fibrosis, it did not prevent ER stress and the inhibition of protein synthesis. Together, ER stress, mitochondrial damage, ATP depletion and oxidative stress promote the death of untreated ATG7-deficient acinar cells, leading to tissue injury and inflammation. Death-promoting cytokines, such as TNF, produced by infiltrating inflammatory cells induce caspase-8 activation in cells with inhibited protein synthesis and further accelerate the death of ATG7-deficient acinar cells. It remains to be determined, however, whether ATG7 deficiency or defects in other essential autophagy mediators underlie some forms of human pancreatitis, or whether the vicious cascade triggered by the ATG7 deficiency represents an extreme version of the pathogenic process triggered by environmental factors, such as alcohol or obesity, which interfere with the initiation or completion of normal autophagy (7).
Materials and Methods
Mice and Treatments.
Atg7F/F and p62F/F mice (22, 40, 41) were intercrossed with B6.FVB-Tg(Ipf1-cre)1Tuv/Nci Pdx1Cre mice (42) to obtain the compound mutants: Pdx1Cre; Atg7F/F (Atg7Δpan), Pdx1Cre; Atg7F/F; p62F/F (Atg7Δpan; p62Δpan). All mice were maintained in filter-topped cages with free access to autoclaved food and water, and experiments were performed in accordance to University of California, San Diego and NIH guidelines and regulations. Cre-negative mice served as controls. Because ATG7 ablation causes the same phenotype in both in males and females, we included both genders in our study, and all experiments were conducted on age- and gender-matched littermates. When indicated, 2-mo-old mice were fed with BHA containing (0.7%) (LabDiet, 5053) or normal chow for 4 wk.
Immunoblot Analysis.
Immunoblot analysis was performed on tissue lysates as described and probed with the antibodies listed in SI Materials and Methods. Densitometric analysis was performed using ImageJ software.
Histology, Immunohistochemistry, Immunofluorescence, and DHE Staining.
Pancreata were dissected and fixed in 10% neutral-buffered formalin and embedded in paraffin. Five-μm sections were prepared and stained with hematoxylin and eosin (H&E) or Sirius red. IHC and DHE were performed as described in SI Materials and Methods.
In situ TUNEL staining was performed using the DeadEnd Colorimetric TUNEL System Kit (Promega). Sirius red-positive area was quantified using ImageJ software. TUNEL- and Ki67-positive cells were counted in at least three random fields (200×) on each slide. All of the quantification results are depicted in the bar graphs next to each panel.
Images were captured on an upright light/fluorescent Imager A2 microscope (Zeiss) equipped with an AxioCam camera.
RNA Extraction and Quantitative PCR.
Total RNA was extracted from pancreata using RNAeasy kit (Qiagen) following manufacturer’s instruction. cDNA was synthesized using iScript cDNA Synthesis kit (Bio-Rad) and qPCR was performed on a CFX-96 Real-Time PCR Detection System (Bio-Rad). Gene expression was calculated as described in SI Materials and Methods.
Autophagic Flux.
Autophagic flux was performed as described (15) (SI Materials and Methods).
Enzyme Assays.
Trypsin activity was measured in pancreatic homogenates as described (15). See SI Materials and Methods for details.
Transmission Electron Microscopy.
Mice were fixed via cardiac perfusion at 37 °C with 2% paraformaldehyde (freshly made), 2.5% glutaraldehyde in pH 7.4, 0.1 M sodium cacodylate buffer with 0.03% calcium chloride. Pancreata were washed five times with ice-cold buffer consisting of 0.1 M sodium cacodylate, 0.03% calcium chloride, and postfixed with ice-cold 1% OsO4, 0.8% KFe(CN)3, 0.03% CaCl2 in 0.1 M sodium cacodylate for 1 h on ice, washed three times with ice-cold distilled water, stained with 2% uranyl acetate at 4 °C for 1 h, dehydrated with graded ethanol solutions, and embedded in Durcupan ACM resin (Fluka). Ultrathin 80-nm-thick sections were made using a Leica Ultracut UCT ultramicrotome and Diatome 45° diamond knife. Sections were poststained with Sato lead before imaging with a FEI Spirit transmission electron microscope operated at 120 kV. Mosaics were collected at 11,000 × g with a Teitz TemCam F224 2k by 2k CCD camera to allow entire cells to be view with their corresponding RER and mitochondria population. Quantification of RER and mitochondrial volume over total cell volume measured as number fraction in EM images was performed using stereology software (IMOD version 4.7, University of Colorado).
mtDNA Quantification.
Mitochondrial DNA (mtDNA) to nuclear DNA (nDNA) ratio was determined by qPCR analysis of the difference in threshold amplification (ΔΔCt method) (43). Primers are listed in SI Materials and Methods.
Measurement of Pancreatic Acinar Cell Protein Synthesis.
Pancreatic acinar cell suspension is prepared as described above, by collagenase digestion, and preincubate in leucine-free DMEM for 2 h supplemented with 50 μM of the proteasome inhibitor MG132, and 30 μg/mL cycloheximide. The cells were labeled with 1 μCi [3H]-leucine (PerkinElmer) for 1 h and washed by centrifugation to remove unincorporated radioactivity and released amino acids. At the end of the incubation, cells were washed twice with PBS. The cells were lysed and proteins were precipitated in ice-cold 10% TCA (Sigma-Aldrich), and then dissolved at 37 °C in 0.2 M NaOH for 2 h. Radioactivity was measured by liquid scintillation counting. Background samples contain NaOH to control for chemiluminiscence. Protein concentration from the TCA precipitated samples was determined using BioRad protein detection reagent.
Statistical Analysis.
Data are expressed as means ± SEM. A two-tailed Student’s t test was used for comparing two groups and results were considered significant at P < 0.05.
SI Materials and Methods
Cell Culture and Treatments.
Isolation of pancreatic acinar cells, were performed as previously described (15). Acinar cells were cultured for 48 h on collagen IV-coated dishes in F12/K medium supplemented with BHA (5 μM; Sigma, W218308) or vehicle as control.
Immunoblot Analysis.
IB analysis was performed on tissue lysates that were separated by SDS/PAGE and transferred to PVDF membranes. Blots were incubated with 5% (wt/vol) nonfat dry milk or 5% (wt/vol) BSA in TBS with 0.05% (wt/vol) Tween 20 (TBST) at room temperature for 1 h to block nonspecific binding, and overnight at 4 °C with primary antibodies in 5% (wt/vol) nonfat dry milk or 5% (wt/vol) BSA in TBST and finally with HRP conjugated secondary antibody in blocking buffer. Blots were developed using an enhanced chemiluminescence detection kit (Clarity Western ECL Substrate, BioRad). The following antibodies were used: rabbit anti-APG7 (H-300; Santa Cruz, sc-33211), ERK1/2 (Cell Signaling, 9102), cleaved caspase 8 (Cell Signaling, 8592), amylase (Sigma, A8273), LC3B (Cell Signaling, 2775), p62 (Progen, GP62-C), mouse monoclonal antiactin (Clone AC-40, Sigma A4700), p-PERK (Thr-981; Santa Cruz, sc-32577), ATF-6α (H-280; Santa Cruz, sc-22799), mouse monoclonal anti-CHOP (GADD153; Thermo Fisher, MA1-250), GRP78 (H-129; Santa Cruz, sc-13968), P-eIF2a (S51; Cell Signaling, 3597), rabbit policlonal Mfn1 (H-65; Santa Cruz, sc-50330), Parkin (H-300; Santa Cruz, sc-30130), NRF2 (C20; Santa Cruz, sc-722), NQO1 (C19, Santa Cruz, sc-16464), Bcl-2 (Cell Signaling, 3498), Bcl-xL (54H6; Cell Signaling, 2764), anti-XIAP (BD transduction, 610717), p53 (1C12; Cell Signaling, 2524), p-AMPK-a (Thr-172; Cell Signaling, 2535), AMPKa1/2 (H-300; Santa Cruz, sc-25792), rabbit anti-phospho-S6 ribosomal protein (Ser235/236; Cell Signaling, 2211), mouse anti-S6 ribosomal protein (54D2; Cell Signaling, 2317S), rabbit anti-4E-BP1 (Cell Signaling, 9452), rabbit antiphospho-4E-BP1 (Thr37/46; Cell Signaling, 2855), tubulin (Sigma, T5168), Sox 9 (H-90; Santa Cruz, sc-20095), and HES1 (D6P2U; Cell Signaling, 11988).
Histology, Immunohistochemistry, and Immunofluorescence.
IHC was performed as follows: after xylene deparaffinization, ethanol dehydration, and antigen retrieval (water bath for 1 h at 98 °C in 10 mM sodium citrate buffer pH 6.0), sections were first blocked with 10% goat serum in 1% BSA/TBST for 15 min and then incubated with primary antibodies diluted in 1% BSA/TBST for 1 h at room temperature, followed by incubation with 1:200 biotinylated secondary antibodies conjugated with HRP for 30 min and 1:500 streptavidin-HRP for 30 min. Bound peroxidase was visualized by 3–5 min incubation in a 3,3′-diaminobenzidine (DAB) solution (Liquid DAB-Plus Substrate kit; Invitrogen 00–2020). Sections were washed, counterstained with hematoxylin, dehydrated, and mounted. For IF staining, frozen/cryostat sections were immediately fixed for 15 min, with 4% formaldehyde diluted in warm 1× PBS, washed three times in PBS for 5 min each, blocked as described above, and incubated overnight with primary antibodies at 4 °C, followed by incubation with secondary antibodies conjugated to Alexa-594, or -488.
The following antibodies were used: Ki67 (GeneTex, GTX16667), p62 (Progen, GP62-C), amylase (Sigma, A8273), PCNA (PC10, Santa Cruz, sc-56), and NRF2 (C20; Santa Cruz, sc-722).
DHE Staining.
Fragments of pancreas were frozen in OCT compound (Sakura Finetek) and frozen sections were prepared using a cryomicrotome. DHE staining was performed as follows: after thawing in room temperature, sections were incubated with 5 μM DHE (Emd Millipore; Biosciences) in PBS solution at 37 °C for 30 min. Sections were washed three times in PBS, mounted using Vectorshield with DAPI (Vectorlab), and images were captured on an upright light/fluorescent Imager A2 microcope (Zeiss) equipped with an AxioCam camera.
Quantitative PCR.
Gene expression was calculated using the 2-ΔΔCt method (15), and normalized relative to cyclophilin or 18S. Primers designed using online primer tools Primer3 or Primer Depot are listed below (forward, reverse):
Tnfα: ACGTCGTAGCAAACCACCAA, GCAGCCTTGTCCCTTGAAGA
Il-1β: AGCTTCCTTGTGCAAGTGTCT, GACAGCCCAGGTCAAAGGTT
Il-6: GGGACTGATGCTGGTGACAA, TTGCCATTGCACAACTCTTTTCT
Tgfb1: TTGCTTCAGCTCCACAGAGA; TGGTTGTAGAGGGCAAGGAC
Ccl2: AGCTGTAGTTTTTGTCACCAAGC, GTGCTGAAGACCTTAGGGCA
Ccl5: CCTCACCATATGGCTCGGAC, ACGACTGCAAGATTGGAGCA
F4/80: CGGGGCTATGGGATGCATAA, TCAGCAACCTCGTGTCCTTG
Cd2: AGGATTCTGGAGAGGGTCTCA, TCGCCTCACACTTGAATGGT
Cd19: GTCATTGCAAGGTCAGCAGTG, GGGTCAGTCATTCGCTTCCT
Lc3: AAGAGTGGAAGATGTCCGGC, TGCAAGCGCCGTCTGATTA
Becn1: GGAAGTAGCTGAAGACCGGG, TTAGACCCCTCCATGCCTCA
Atg12: TGGCCTCGGAACAGTTGTTTA, ATCCCCATGCCTGGGATTTG
Vps34: CCACCTGCGAAGGTATTCCA, CGCTCTCGTCGATCAGACTC
p62: GCTGCCCTATACCCACATCT, CGCCTTCATCCGAGAAAC
Amy2a: TAAAGTGGCTGACAAAGCCC, AGCCTTTTTCAAACTGGTGG
Ela: TATGGAGGATCATGGCACCAC, ACACAGTGAGCAGCGGTCATC
Pnlip: CCGATGCTCAGTTTGTGGAC, CTGGCATTTCGATTCCTCCG
Ppia: ACCAAACACAAACGGTTCCC, TGCCTTCTTTCACCTTCCCAA
Bip/GRP78: CGATACTGGCCGAGACAACA, GACGACGGTTCTGGTCTCAC
Chop: ACCTGAGGAGAGAGTGTTCCA, CAAGGTGAAAGGCAGGGACT
EDEM: CTACCTGCGAAGAGGCCG, GTTCATGAGCTGCCCACTGA
GRP94: AAGAATGAAGGAAAAACAGGACAAAA, CAAATGGAGAAGATTCCGCC
sXBP1: CTGAGTCCGAATCAGGTGCAG, GTCCATGGGAAGATGTTCTGG
tXBP1: TGGCCGGGTCTGCTGAGTCCG, GTCCATGGGAAGATGTTCTGG
usXBP1: CAGCACTCAGACTATGTGCA, GTCCATGGGAAGATGTTCTGG
Alox15: ATCTTGGCCAGCTGGATTGG, TCGTCGCGTCCTTGGTTTTA
Duox1: ATGGGGTTCCACTTAGCTCTG, AGTTCTGAGCTCCCAAAGAAGC
Duox2: CGCTACGACGGCTGGTTTA, TTTGCCCTTAGCGACAGCAT
Nqo1: ATCACAGGTGAGCTGAAGGAC, CAATATCTGGGCTCAGGCGT
Gstm1: TTTGTCCTGCCCACGTTTCT, CGGACGTTCCAGTATCCCAG
Cyp2a5: ACCAAGGACACCAAGTTTCG, AGAGCCCAGCATAGGAAACA
Acta2 (α-SMA): GTCCACCGCAAATGCTTCTA, GAGGCGCTGATCCACAAAAC
Col1a1: GAGAGGTGAACAAGGTCCCG, AAACCTCTCTCGCCTCTTGC
Col1a3: CAGGAGAAAAGGGTCCTCCC, ATACCCCGTATCCCTGGACC
Mfn1: TCTCCAAGCCCAACATCTTC, TCTCTCCATGTGCTGTCTGC
Park2: GCGTAGGTCCTTCTCGACC, ATCGACCTCCACTGGGAAG
Ucp2: ACTTTCCCTCTGGATACCGC, ACGGAGGCAAAGGCTCATCTG
Cpt1a: CCAGGCTACAGTGGGACATT, GAACTTGCCCATGTCCTTGT
Sox9: CGGAACAGACTCACATCTCTCC; GCTTGCACGTCGGTTTTGG
Nestin: CCCTGAAGTCGAGGAGCTG; CTGCTGCACCTCTAAGCGA
Ck19: GGGGGTTCAGTACGCATTGG; GAGGACGAGGTCACGAAGC
Hes1: TCTACACCAGCAACAGTG; TCAAACATCTTTGGCATCAC
Mmp7: CCCGGTACTGTGATGTACCC; AATGGAGGACCCAGTGAGTG
Pdx1 TTAACCTAGGCGTCGCACAAG, TTCCAGAAGTCTGCCAGCATC
Ptf1: GGCACTCTCTTTCCTGGACT, AGTTTTCTGGGGTCCTCTGG
Noxa GAACGCGCCAGTGAACCCAAC, CTTGGGCTCCTCATCCTGCTG
Bax: ATGCGTCCACCAAGAAGCTGA, CCCCAGTTGAAGTTGCCATCAG
Puma: CTGTGAATCCTGTGCTCTGC, GGTCACACGTGCTCTCTCTA.
Enzyme Assays.
Trypsin activity was measured in pancreatic homogenates by a fluorimetric assay (44). Briefly, pancreata were homogenized on ice using a glass-Teflon homogenizer in a buffer containing 5 mM Mes (pH 6.5), 1 mM MgSO4, and 250 mM sucrose. An aliquot of the homogenate was added to the assay buffer containing 50 mM Tris⋅HCl (pH 8.0), 150 mM NaCl, 1 mM CaCl2, and 0.1 mg/mL BSA in a stirred cuvette at 37 °C. The reaction was started by adding a specific substrate, Boc-Gln-Ala-Arg-MCA, which is converted to a fluorescent product that emits fluorescence at 440 nm upon excitation at 380 nm. The increase in fluorescence was linear during the 5-min observation period. Trypsin activity in each sample was determined using a standard curve for purified trypsin (Worthington).
Autophagic Flux.
Dispersed pancreatic acini were cultured for 5 h on collagen IV-coated dishes in leucine-free DMEM (Crystalgen), supplemented with 1 μCi [3H]-leucine (PerkinElmer), and washed by centrifugation to remove unincorporated radioactivity and released amino acids. The cells were then replated and chased for 16 h in fresh F12/K medium containing 2 mM cold leucine, during which time all short-lived proteins were degraded. After that, cells were washed three times by centrifugation in F12/K medium containing 2 mM cold leucine and further incubated for 4 h with 10 μg/mL tunicamycin, or 10 mM 3-methyladenine (3-MA), or 100 nM MG132. At the end of the incubation, proteins were precipitated in ice-cold 10% trichloroacetic acid (TCA; Sigma-Aldrich). Soluble radioactivity, which represents degradation products, was separated from intact proteins by centrifugation at 12,000 × g for 10 min. The pellet was washed with cold 10% (wt/vol) TCA, then dissolved in 0.2 M NaOH for 2 h at 37 °C. Radioactivity was then measured by liquid scintillation counting. The rate of degradation of long-lived proteins was calculated from the ratio of the acid-soluble radioactivity in the medium to that in the acid-precipitated cell fraction.
mtDNA Quantification.
For mitochondrial DNA (mtDNA) to nuclear DNA (nDNA) qPCR the following primers were used:
mtDNA forward primer, CCTATCACCCTTGCCATCAT
mtDNA Reverse primer, GAGGCTGTTGCTTGTGTGAC
nDNA Forward primer, ATGGAAAGCCTGCCATCATG
nDNA Reverse primer, TCCTTGTTGTTCAGCATCAC.
Acknowledgments
We thank Guy Perkins and Tom Deerinck for advice and helpful suggestions and thank Santa Cruz Biotechnology and Cell Signaling for antibody samples. L.A. was supported by a fellowship from Istituto Pasteur–Fondazione Cenci Bolognetti; J.B.F. was supported by postdoctoral fellowships from the Swedish Research Council (524-2011-6817) and the Swedish Society for Medical Research. J.Y.K. was supported by the Superfund Basic Research Project (SBRP; ES010337); J.T. was supported by the Erwin Schroedinger Fellowship from the Austrian Science Fund (J3233) and Univ. Prof. Dr. 17 Matthias M. Müller Fellowship from the Austrian Association for Laboratory Medicine and Clinical Chemistry (ÖGLMKC) and the Austrian Program for Advanced Research and Technology of the Austrian Academy of Sciences; I.G. was supported by NIH Grants AA019730 and DK098108; M.M. and M.H.E. were supported by grants from the International Community Foundation, San Diego, the NIH (GM103412 and GM103426). M.K. is an American Cancer Society Research Professor and the Ben and Wanda Hildyard Chair for Mitochondrial and Metabolic Diseases. Research in his lab was supported by grants from the NIH (CA163798), the SBRP (ES010337), the Lustgarten Foundation (RFP-B-007), and NIH Program Project Grant P01 DK098108.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1519384112/-/DCSupplemental.
References
- 1.Case RM. Synthesis, intracellular transport and discharge of exportable proteins in the pancreatic acinar cell and other cells. Biol Rev Camb Philos Soc. 1978;53(2):211–354. doi: 10.1111/j.1469-185x.1978.tb01437.x. [DOI] [PubMed] [Google Scholar]
- 2.Logsdon CD, Ji B. The role of protein synthesis and digestive enzymes in acinar cell injury. Nat Rev Gastroenterol Hepatol. 2013;10(6):362–370. doi: 10.1038/nrgastro.2013.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang M, Kaufman RJ. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat Rev Cancer. 2014;14(9):581–597. doi: 10.1038/nrc3800. [DOI] [PubMed] [Google Scholar]
- 4.Sah RP, et al. Endoplasmic reticulum stress is chronically activated in chronic pancreatitis. J Biol Chem. 2014;289(40):27551–27561. doi: 10.1074/jbc.M113.528174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LaRusch J, Solomon S, Whitcomb DC. 1993. Pancreatitis Overview, eds Pagon RA, et al. (GeneReviews, University of Washington, Seattle)
- 6.Hausmann S, Kong B, Michalski C, Erkan M, Friess H. The role of inflammation in pancreatic cancer. Adv Exp Med Biol. 2014;816:129–151. doi: 10.1007/978-3-0348-0837-8_6. [DOI] [PubMed] [Google Scholar]
- 7.Gukovsky I, Li N, Todoric J, Gukovskaya A, Karin M. Inflammation, autophagy, and obesity: Common features in the pathogenesis of pancreatitis and pancreatic cancer. Gastroenterology. 2013;144(6):1199–1209 e1194. doi: 10.1053/j.gastro.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reggiori F, Komatsu M, Finley K, Simonsen A. Autophagy: More than a nonselective pathway. Int J Cell Biol. 2012;2012:219625. doi: 10.1155/2012/219625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13(5):495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleming A, Noda T, Yoshimori T, Rubinsztein DC. Chemical modulators of autophagy as biological probes and potential therapeutics. Nat Chem Biol. 2011;7(1):9–17. doi: 10.1038/nchembio.500. [DOI] [PubMed] [Google Scholar]
- 11.Uchiyama Y, Shibata M, Koike M, Yoshimura K, Sasaki M. Autophagy-physiology and pathophysiology. Histochem Cell Biol. 2008;129(4):407–420. doi: 10.1007/s00418-008-0406-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gukovskaya AS, Gukovsky I. Autophagy and pancreatitis. Am J Physiol Gastrointest Liver Physiol. 2012;303(9):G993–G1003. doi: 10.1152/ajpgi.00122.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashimoto D, et al. Involvement of autophagy in trypsinogen activation within the pancreatic acinar cells. J Cell Biol. 2008;181(7):1065–1072. doi: 10.1083/jcb.200712156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grasso D, et al. Zymophagy, a novel selective autophagy pathway mediated by VMP1-USP9x-p62, prevents pancreatic cell death. J Biol Chem. 2011;286(10):8308–8324. doi: 10.1074/jbc.M110.197301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li N, et al. Loss of acinar cell IKKα triggers spontaneous pancreatitis in mice. J Clin Invest. 2013;123(5):2231–2243. doi: 10.1172/JCI64498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diakopoulos KN, et al. Impaired autophagy induces chronic atrophic pancreatitis in mice via sex- and nutrition-dependent processes. Gastroenterology. 2015;148(3):626–638 e617. doi: 10.1053/j.gastro.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Gukovsky I, Gukovskaya AS. Impaired autophagy triggers chronic pancreatitis: Lessons from pancreas-specific atg5 knockout mice. Gastroenterology. 2015;148(3):501–505. doi: 10.1053/j.gastro.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mareninova OA, et al. Impaired autophagic flux mediates acinar cell vacuole formation and trypsinogen activation in rodent models of acute pancreatitis. J Clin Invest. 2009;119(11):3340–3355. doi: 10.1172/JCI38674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenfeldt MT, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504(7479):296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 20.Yang A, et al. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer Discov. 2014;4(8):905–913. doi: 10.1158/2159-8290.CD-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iacobuzio-Donahue CA, Herman JM. Autophagy, p53, and pancreatic cancer. N Engl J Med. 2014;370(14):1352–1353. doi: 10.1056/NEJMcibr1400189. [DOI] [PubMed] [Google Scholar]
- 22.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169(3):425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem. 2001;276(3):1701–1706. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- 24.Codogno P, Mehrpour M, Proikas-Cezanne T. Canonical and non-canonical autophagy: Variations on a common theme of self-eating? Nat Rev Mol Cell Biol. 2012;13(1):7–12. doi: 10.1038/nrm3249. [DOI] [PubMed] [Google Scholar]
- 25.Puissant A, Fenouille N, Auberger P. When autophagy meets cancer through p62/SQSTM1. Am J Cancer Res. 2012;2(4):397–413. [PMC free article] [PubMed] [Google Scholar]
- 26.Fang J, et al. Myeloid malignancies with chromosome 5q deletions acquire a dependency on an intrachromosomal NF-κB gene network. Cell Reports. 2014;8(5):1328–1338. doi: 10.1016/j.celrep.2014.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Komatsu M, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010;12(3):213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 28.Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 2007;18(6):716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhandary B, Marahatta A, Kim HR, Chae HJ. An involvement of oxidative stress in endoplasmic reticulum stress and its associated diseases. Int J Mol Sci. 2012;14(1):434–456. doi: 10.3390/ijms14010434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding WX, Yin XM. Mitophagy: Mechanisms, pathophysiological roles, and analysis. Biol Chem. 2012;393(7):547–564. doi: 10.1515/hsz-2012-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pi J, et al. ROS signaling, oxidative stress and Nrf2 in pancreatic beta-cell function. Toxicol Appl Pharmacol. 2010;244(1):77–83. doi: 10.1016/j.taap.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Itoh K, Tong KI, Yamamoto M. Molecular mechanism activating Nrf2-Keap1 pathway in regulation of adaptive response to electrophiles. Free Radic Biol Med. 2004;36(10):1208–1213. doi: 10.1016/j.freeradbiomed.2004.02.075. [DOI] [PubMed] [Google Scholar]
- 33.Filomeni G, De Zio D, Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 2015;22(3):377–388. doi: 10.1038/cdd.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17(6):596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 35.Murtaugh LC, Keefe MD. Regeneration and repair of the exocrine pancreas. Annu Rev Physiol. 2015;77:229–249. doi: 10.1146/annurev-physiol-021014-071727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ziv O, Glaser B, Dor Y. The plastic pancreas. Dev Cell. 2013;26(1):3–7. doi: 10.1016/j.devcel.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 37.Sawey ET, Johnson JA, Crawford HC. Matrix metalloproteinase 7 controls pancreatic acinar cell transdifferentiation by activating the Notch signaling pathway. Proc Natl Acad Sci USA. 2007;104(49):19327–19332. doi: 10.1073/pnas.0705953104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan W, et al. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc Natl Acad Sci USA. 2002;99(25):15920–15925. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaufman RJ, Malhotra JD. Calcium trafficking integrates endoplasmic reticulum function with mitochondrial bioenergetics. Biochim Biophys Acta. 2014;1843(10):2233–2239. doi: 10.1016/j.bbamcr.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dessimoz J, Bonnard C, Huelsken J, Grapin-Botton A. Pancreas-specific deletion of beta-catenin reveals Wnt-dependent and Wnt-independent functions during development. Curr Biol. 2005;15(18):1677–1683. doi: 10.1016/j.cub.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 41.Müller TD, et al. p62 links β-adrenergic input to mitochondrial function and thermogenesis. J Clin Invest. 2013;123(1):469–478. doi: 10.1172/JCI64209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hingorani SR, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4(6):437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 43.Chen H, et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141(2):280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]