Significance
Hair cells, the sensory receptors of the inner ear, underlie our ability to hear and maintain balance. In mammals, these cells are formed by birth, and they cannot be restored through regeneration. Mammals therefore lack the capacity to recover hearing and balance after the loss of hair cells. By assessing gene expression during inner ear development in mice, we identified several genes that are downregulated at the cessation of hair cell production. We demonstrated that two of these genes—Sox4 and Sox11—are necessary and sufficient for the production of hair cells in the sensory epithelia of the inner ear. Our data suggest that Sox4 and Sox11 represent targets in the development of therapies for deafness and disequilibrium.
Keywords: auditory system, cochlea, hair cell, utricle, vestibular system
Abstract
Hair cells, the mechanosensory receptors of the inner ear, underlie the senses of hearing and balance. Adult mammals cannot adequately replenish lost hair cells, whose loss often results in deafness or balance disorders. To determine the molecular basis of this deficiency, we investigated the development of a murine vestibular organ, the utricle. Here we show that two members of the SoxC family of transcription factors, Sox4 and Sox11, are down-regulated after the epoch of hair cell development. Conditional ablation of SoxC genes in vivo results in stunted sensory organs of the inner ear and loss of hair cells. Enhanced expression of SoxC genes in vitro conversely restores supporting cell proliferation and the production of new hair cells in adult sensory epithelia. These results imply that SoxC genes govern hair cell production and thus advance these genes as targets for the restoration of hearing and balance.
Unlike mammals, nonmammalian vertebrates can regenerate hair cells effectively throughout life and thus recover from hearing and balance deficits (1). The discovery of the ear's regenerative potential in avian species (2–4) initiated a wave of studies directed toward understanding the molecular basis of hair cell regeneration and the deficiency of this process in mammals. Two distinct mechanisms of regeneration have emerged (5). The first involves the production of hair cells by the transdifferentiation of supporting cells, which are the epithelial cells that separate and provide metabolic support for hair cells (6–8). A rudimentary form of this process occurs in mammals (9, 10). A limitation of this pathway, however, is that transdifferentiation depletes the population of supporting cells and thereby interferes with the ability of sensory organs to function properly (11). The second mode of regeneration involves supporting cell proliferation, which restores both hair cells and supporting cells. Prevalent in the auditory sensory epithelia of nonmammalian species, this mechanism allows functional recovery (5). The corresponding mechanism is absent in mammals, however, and little is known about the molecular events involved (12, 13).
In the sensory epithelia of the mammalian inner ear, the ability to restore hair cells after trauma declines late in development, largely as a result of the diminished proliferative capacity of supporting cells (10). Reasoning that this transition should be reflected by differences in the expression of genes involved in proliferation, differentiation, and regeneration, we investigated the genes expressed late in the development of the murine utricle. With a simple architecture and just under 4,000 hair cells in an adult animal (14), the sensory epithelium of the utricle—the macula—represents a useful model system. Although gene expression has been characterized in early otic development (15, 16) and in the neonatal organ of Corti (17, 18), corresponding data are lacking for the developing utricle.
Results
Chronology of Diminishing Proliferative Capacity in the Ear.
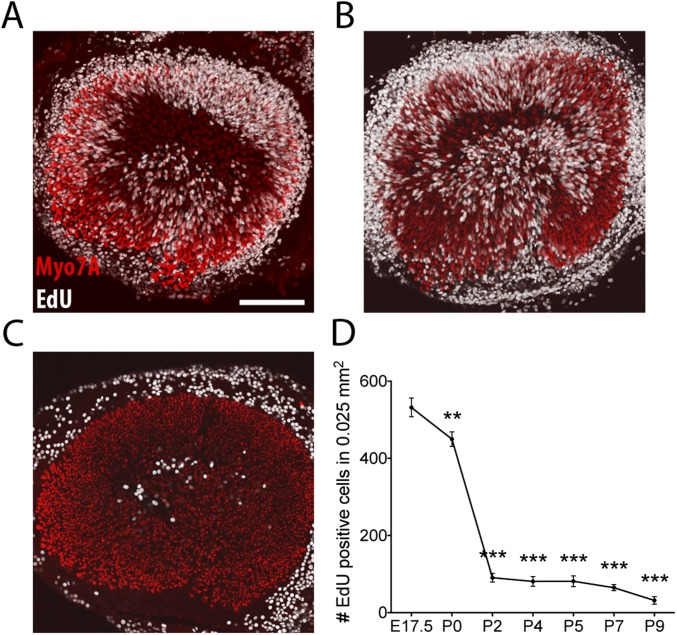
To assess the time course of the ear's decline in proliferative ability, we dissected utricles from mice at embryonic day 17.5 (E17.5) through postnatal day 9 (P9). By culturing the organs in medium supplemented with 5-ethynyl-2′-deoxyuridine (EdU), we labeled the proliferating cells at each stage. In accord with previous results (14), we observed proliferating cells at the periphery of the utricular macula and in the striola, the organ's central region (Fig. S1 A–C). In keeping with previous reports, we term these proliferating cells “supporting cells”; however, we cannot exclude the possibility that proliferation was confined to sensory-progenitor cells or a subset of the morphologically defined supporting cells. The number of proliferating cells decreased by 80% at P2 and by 93% at the end of the first week of life (Fig. S1D). These results indicate that the proliferative potential of supporting cells in the utricle is largely lost within 48 h after birth.
Fig. S1.
Decrease in the proliferative capacity of the utricle. (A) In an E17.5 utricle after 24 h of EdU labeling in culture, proliferating cells (white) occur in the macula, which is marked by hair cells immunolabeled for Myo7A (red). Additional proliferation transpires in the nonsensory epithelium surrounding the macula. (Scale bar, 100 μm.) (B) In an identical preparation of a P0 utricle the results are similar. (C) In an identical preparation of a P9 utricle numerous proliferating cells occur in the nonsensory epithelium but very few occur in the macula. (D) The number of dividing cells in the utricular macula decreases significantly between E17.5 and P9. **P < 0.001, ***P < 0.0001; n = 8, 10, 10, 12, 8, 12, 6 on days indicated.
Genes Involved in the Development of the Utricular Macula.
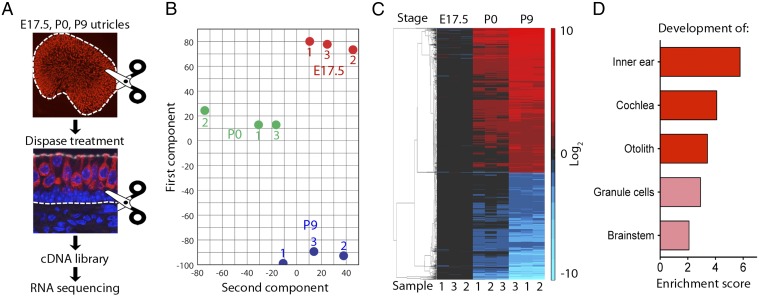
To identify the transcriptional changes responsible for silencing proliferation and hair cell production, we conducted RNA sequencing to compare the genes expressed before and after the cessation of cellular proliferation and hair cell differentiation. We developed a method for isolating utricular sensory epithelia and prepared sequencing libraries at E17.5, P0, and P9 (Fig. 1A). To determine the consistency of our results, we analyzed three independent samples at each developmental stage. Principal-component analysis demonstrated a high degree of similarity among the samples at each stage but a significant difference between stages (Fig. 1B). These distinctions also were reflected upon hierarchical clustering of the differentially expressed genes (Fig. 1C), which revealed that groups of genes were up- or down-regulated to similar levels in all three samples at each stage and that the samples from P0 exhibited expression patterns intermediate between those from E17.5 and P9 mice. Gene-ontology analysis revealed that the greatest enrichment occurred for transcripts known to be involved in inner ear development (Fig. 1D).
Fig. 1.
Transcriptional changes in late utricular development. (A) In a schematic representation of the protocol for isolation of the utricular macula and preparation of cDNA libraries, the white dashed lines represent the borders between the macular epithelium and the nonsensory tissues that were removed by microdissection and enzymatic digestion. Hair cells are immunolabeled for Myo7A (red) and nuclei are stained blue. (B) Principal-component analysis of the RNA-sequencing results for nine samples reveals that the first two components capture 56% (ordinate) and 14% (abscissa) of the variance in the set of samples from E17.5, P0, and P9 mice. (C) In a hierarchical-clustering analysis of transcriptional changes in the developing utricle, the vertical rows represent the nine samples in B. Each horizontal stripe represents a gene whose level of expression changed at least twofold between E17.5 and P9 (P < 0.05). Each expression level is normalized to the average value for that gene at E17.5. Increased expression is indicated in shades of red and decreased expression in blue. (D) According to gene-ontology analysis, the principal categories up-regulated between E17.5 and P9 are associated with the ear's development. P < 0.05 for the top three categories.
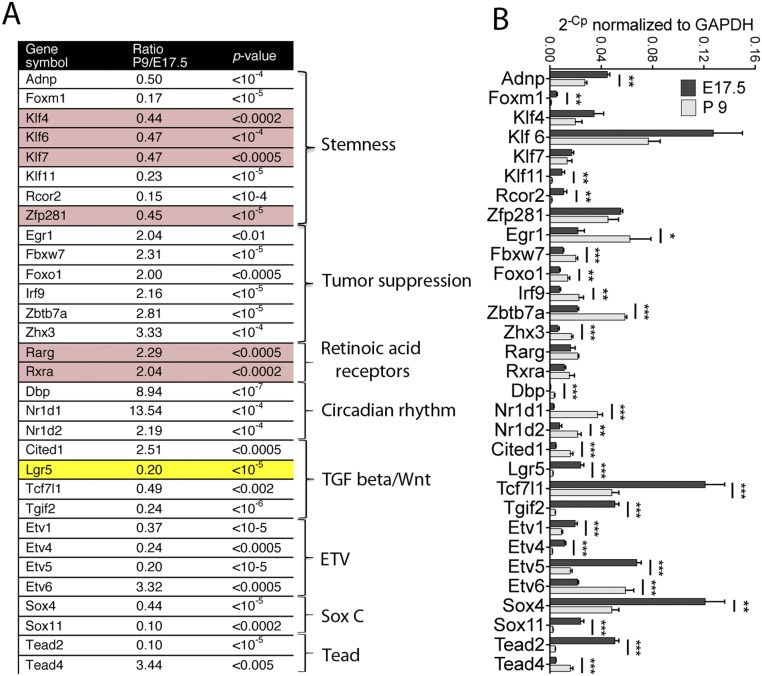
Many of the genes that showed at least a twofold change in expression between E17.5 and P9 already have been found to play roles in hair cell development. The cognate proteins include constituents of the Notch pathway such as Hey1 and Hes6 (19, 20) and members of the Wnt cascade such as the G-protein–coupled receptor Lgr5 (21, 22). On the basis of gene function—stem cell maintenance, cell-cycle progression, tumor suppression, and neurogenesis—we selected 31 transcription factors that might be expected to influence the regenerative capacity of sensory epithelia (Fig. S2A). To validate the RNA-sequencing data, we performed quantitative PCR (qPCR) for selected genes with RNA isolated from E17.5 and P9 mice. Because of its declining expression in developing sensory epithelia (21, 22), we chose Lgr5 as a positive control for our validation. qPCR confirmed changes in the level of expression for 25 of the transcription factors, suggesting that they participate in inner ear development (Fig. S2B).
Fig. S2.
Gene expression in the developing utricle. (A) A table of transcription factors displays the ratio between the expression of the cognate genes at P9 and at E17.5. The right column displays the pathways in which the transcription factors participate. The genes whose changes in expression are not significant by the qPCR analysis in B are highlighted in red. Lgr5, highlighted in yellow, serves as a positive control. (B) qPCR confirms the changes in gene expression between E17.5 and P9 shown in A. *P < 0.05, **P < 0.001, ***P = 0.0001; n = 6 in each instance.
Expression of Sox4 and Sox11 in the Mammalian Inner Ear.
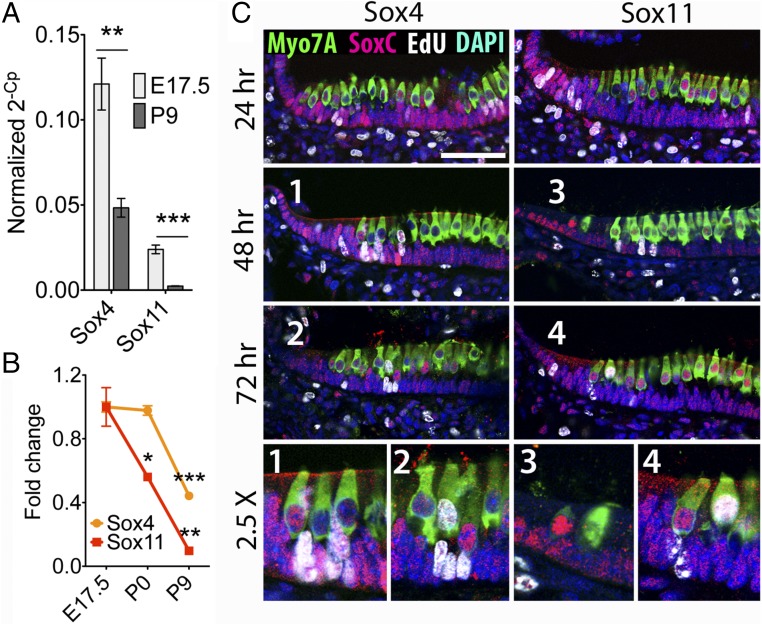
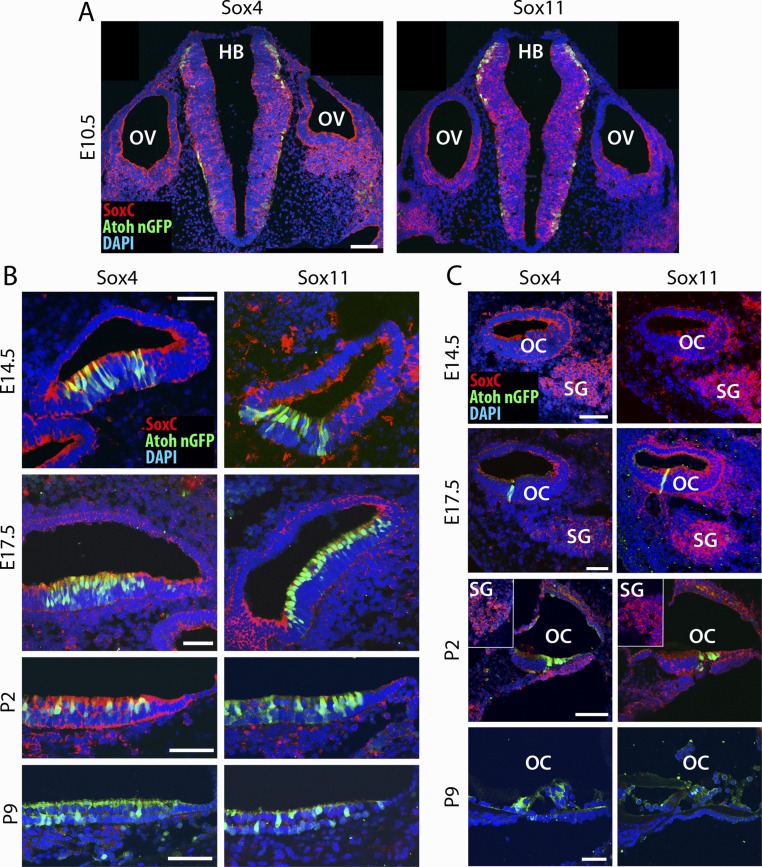
As shown by RNA sequencing and confirmed by qPCR analysis, two SoxC genes were highly expressed in the developing utricular macula at E17.5. Their expression was strongly down-regulated by P9, Sox4 by 50% and Sox11 by 90% (Fig. 2A). In situ hybridization demonstrated a similar pattern of expression for both genes. At E10.5, Sox4 and Sox11 were present throughout the prosensory domains of the otic vesicle and in the hindbrain (Fig. S3A). The intense expression also was detected at E14.5 in all the developing sensory organs of the inner ear and in the spiral ganglion (Fig. S3 B and C). At E17.5 the intense signals were restricted to the actively growing edges of the vestibular maculae (Fig. S3B). The cochlea manifested strong expression of SoxC genes throughout the organ of Corti and in the spiral ganglion (Fig. S3C). Although Sox4 expression remained robust until P2, Sox11 expression was markedly down-regulated in the utricular macula and the organ of Corti. Both genes remained highly expressed in the spiral ganglion during this period. The difference in the timing of down-regulation for Sox4 and Sox11 was corroborated by RNA sequencing: When normalized to E17.5 expression levels, Sox4 expression remained unchanged at P0, whereas Sox11 expression declined significantly by 50% (Fig. 2B). By P9 we could no longer detect RNA for either gene by in situ hybridization.
Fig. 2.
SoxC expression in the developing sensory organs of the inner ear. (A) qPCR confirms the decrease in expression of SoxC genes between E17.5 and P9. The significance of the change in expression is **P = 0.0012 for Sox4 (n = 6) and ***P = 0.0001 for Sox11 (n = 6). (B) Normalized to the RNA-sequencing data from E17.5 mice, the fold changes in the number of reads per kilobase of exon per million fragments mapped emphasize the delay in the down-regulation of Sox4 relative to Sox11. The change is significant at P0 for Sox11 (*P = 0.023, n = 3) and at P9 for both Sox4 (***P = 0.0001, n = 3) and Sox11 (**P = 0.0017, n = 3). (C) Twenty-four hours after injection, EdU labeling and antibody labeling for Sox4 and Sox11 (red) demonstrate that these proteins occur in actively proliferating supporting cells (white) at the periphery of each utricular macula. Some labeled hair cells (green) are newly formed by the criteria of EdU labeling, peripheral localization, absence of mature hair bundles, and weak immunolabeling for Myo7A. Representative SoxC-expressing hair cells are shown in the bottom images at a further enlargement of 2.5×. Nuclei are labeled blue. (Scale bar, 50 μm.)
Fig. S3.
SoxC expression in the developing sensory organs of the inner ear. (A) FISH on sections of the utricles from Atoh1-nGFP mice reveals the locations of Sox4 and Sox11 RNAs. At E10.5 the reaction product for each gene (red) occurs throughout the prosensory domains of the otic vesicle (OV). Strong expression also is observed in the hindbrain (HB). (B) At E14.5 robust labeling for SoxC RNAs occurs throughout the sensory epithelium of the developing utricle. At E17.5 the reaction product for each gene (red) occurs at the periphery of the utricular macula and in hair cells immunolabeled for GFP (green). By P2 Sox4 expression remains apparent, but Sox11 RNA cannot be detected; neither transcript is present in utricular maculae on P9. Nuclei are shown in blue. (Scale bars, 50 μm.) (C) A similar preparation reveals that the reaction product for each gene (red) also occurs in the organ of Corti (OC) and spiral ganglion (SG). A similar pattern of expression emerges at E14.5, E17.5, and P2. In a preparation at P9 neither Sox4 nor Sox11 transcripts are present. Hair cells are immunolabeled for GFP (green); nuclei are shown in blue. (Scale bars, 50 μm.)
To investigate the relationship between the expression of Sox4 and Sox11 and the production of new hair cells, we injected pregnant mice with EdU at E16.5 and analyzed the inner ears of progeny 24, 48, and 72 h later. Using antibody labeling for SoxC proteins, we found that 24 h after EdU injection the periphery of the macula and the striolar region contained actively proliferating progenitor cells that robustly expressed both Sox4 and Sox11 (Fig. 2C). Young EdU+ hair cells, which also strongly expressed the two transcription factors, appeared by 48 h after EdU injection. The expression of SoxC proteins was down-regulated in mature hair cells. In light of the neurogenic role of SoxC genes (23, 24), these findings suggest that Sox4 and Sox11 are involved in the regulation of cell proliferation and hair cell formation in the developing sensory organs of the mammalian inner ear.
Vestibular Dysfunction and Deafness in SoxC-Deficient Mice.
The overlapping expression patterns and functional redundancy of SoxC genes allow some members of the gene family to compensate in part for deficiencies in others (25). We therefore generated mice with inner ear-specific conditional knockout (CKO) of both Sox4 and Sox11 genes by crossing Pax2-Cre mice with Sox4fl/fl Sox11fl/fl Sox12−/− animals (26–28). The phenotypic effects of Sox12 are known to be weak (29), and our RNA-sequencing data showed an insignificant expression of Sox12 in the sensory epithelia of the inner ear. We therefore disregarded the Sox12 genotype in our subsequent analysis.
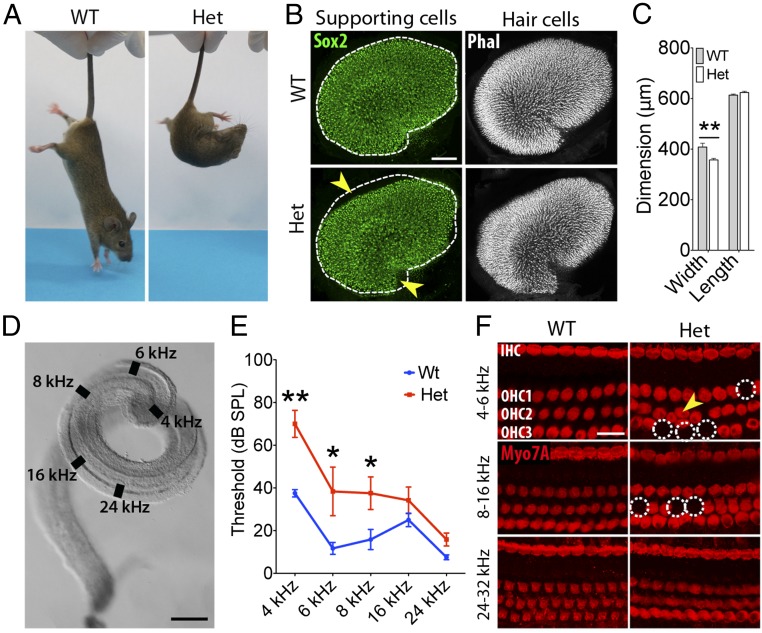
By 4 wk of age, heterozygous Sox4 Sox11 double-CKO animals repetitively demonstrated signs of vestibular dysfunction, including circling, head tossing and bobbing, and loss of balance (Movie S1). To characterize this phenotype further, we performed a battery of vestibular and hearing tests (30, 31). In tail-suspension tests we observed frequent trunk curling and propelling behavior in the heterozygous CKO animals but not in their WT littermates (Fig. 3A and Movie S2). This result suggests that there are defects in the utricle and saccule, the organs that detect static position. In contrast, we detected no difference in contact righting; the organs that sense rotation, the cristae of the semicircular canals, apparently remained functional. Although the gross appearance of the inner ear was relatively normal in the heterozygous CKO animals, we noticed an aberrant shape and a minor but statistically significant reduction in the size of the vestibular organs (Fig. 3B). The utricles of heterozygous CKO mice were 6% smaller in area than those of WT littermates, a difference caused by a consistent reduction in organ width (Fig. 3C).
Fig. 3.
Vestibular dysfunction and deafness in heterozygous SoxC CKO mice. (A) In the tail-suspension test, a 4-wk-old WT mouse reaches downward for the substrate, whereas a heterozygous SoxC CKO animal (Het) displays ataxic curling. (B) In utricles from 4-wk-old WT and Het animals, supporting cells are immunolabeled with anti-Sox2 (green), and hair bundles are revealed by phalloidin labeling (white). Fitting the outline of the WT macula (dashed line) to the mutant's macula demonstrates a decrease in the organ’s width, which was quantified by measuring the distance across the organ at the notch on its neural edge and perpendicular to its long axis. (Scale bar, 100 μm.) (C) Measurements confirm that the difference observed in B is statistically significant and corresponds to a 13% decrease in the organ’s width (**P = 0.0076) (WT n = 5, Het n = 6). (D) A dissected murine cochlear duct shows the anatomical positions of the hair cells most sensitive to the frequencies at which auditory brainstem responses were measured. (Scale bar, 250 μm.) (E) Heterozygous SoxC CKO mice (n = 6) display a significant elevation in their thresholds for auditory brainstem responses with respect to WT animals (n = 6). *P < 0.05, **P < 0.001. (F) Immunolabeling of a WT ear for Myo7A (red) reveals a regular hair cell array with one row of inner hair cells (IHC) and three rows of outer hair cells (OHC1, OHC2, and OHC3) in the apical (4–6 kHz and 8–16 kHz) and middle (16–24 kHz) cochlear turns. In a heterozygous SoxC CKO animal, in contrast, some hair cells in the apical turn are missing (dotted circles), and others are misaligned (yellow arrowhead). (Scale bar, 25 μm.)
We evaluated the hearing of heterozygous CKO mice by recording auditory brainstem responses to stimulation at 4–24 kHz, frequencies represented in the apex and middle turn of the organ of Corti (Fig. 3D) (30). The mutant mice displayed a significant 30-dB elevation of hearing thresholds for low-frequency sounds (Fig. 3E), implying that they could detect only sounds more than 30-fold louder than the faintest stimuli perceived by their WT littermates. The difference in threshold was smaller at 8 kHz and was insignificant for frequencies of 16–24 kHz. The increased hearing thresholds in the low-frequency range were mirrored by aberrations in the cochlear structure of SoxC-deficient mice: Many hair cells in the apical turn were misaligned or absent, but the defects were less severe in the region subserving 8–16 kHz and were absent from the basal organ of Corti (Fig. 3F).
Role of Sox4 and Sox11 During Inner-Ear Development.
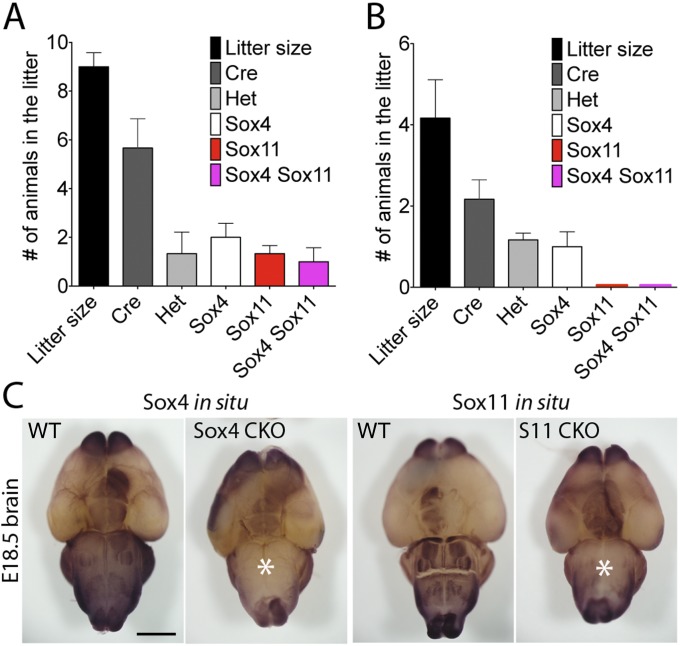
We obtained CKO mice homozygously deficient for various combinations of SoxC genes by crossing Pax2-Cre Sox4fl/+ Sox11fl/+ Sox12−/+ and Sox4fl/fl Sox11fl/fl Sox12−/− animals (Fig. S4 A and B). Homozygous Sox4 or Sox11 CKO animals exhibited respiratory distress, became cyanotic, and died shortly after birth. In accord with the reported pattern of Cre activity in Pax2-Cre mice (26), in situ hybridization demonstrated an absence of Sox4 and Sox11 expression in most of the hindbrain and midbrain (Fig. S4C). Because the relevant nuclei are located in these brain regions, cardiopulmonary dysfunction likely underlay the neonatal mortality of the homozygous Sox4 or Sox11 CKO mutants.
Fig. S4.
Early lethality in homozygous SoxC CKO mice. (A) A graphic representation of the genotype distribution seen at E18.5 shows nearly Mendelian ratios. (B) The corresponding distribution at P0 lacks Sox11 CKO and Sox4 Sox11 CKO animals, suggesting late fetal or early neonatal lethality for these mutants. (C) In situ hybridization with digoxigenin-labeled Sox4 and Sox11 antisense probes demonstrates that the expression of these SoxC genes is largely absent from the midbrains and hindbrains (white asterisks) of the mutant animals. (Scale bar, 1 μm.)
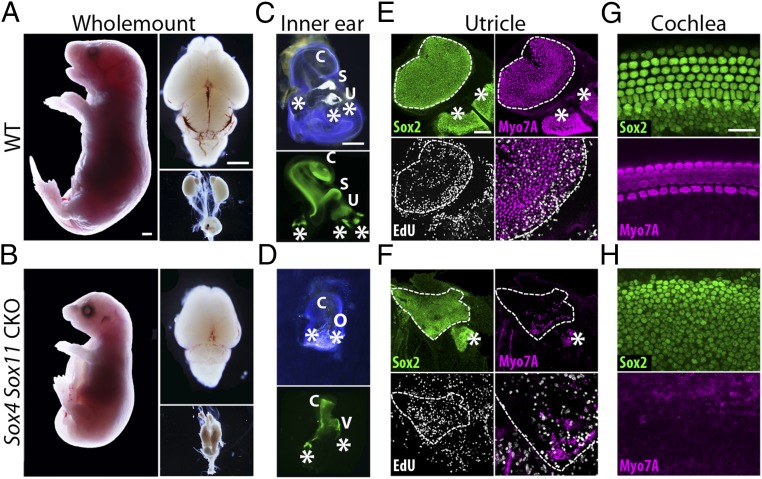
Homozygous Sox4 or Sox11 CKO mice were small but appeared anatomically normal (Fig. S5 A and B). The Pax2-Cre–expressing organs of these mutants—the kidney, midbrain, and hindbrain—were grossly intact. Sox4 Sox11 double-CKO mice were born dead. Much smaller than WT mice, these animals displayed severe defects including microcephaly and gross anatomical malformations of the brain (Fig. 4 A and B). They also manifested severe edema, likely caused by greatly reduced kidneys.
Fig. S5.
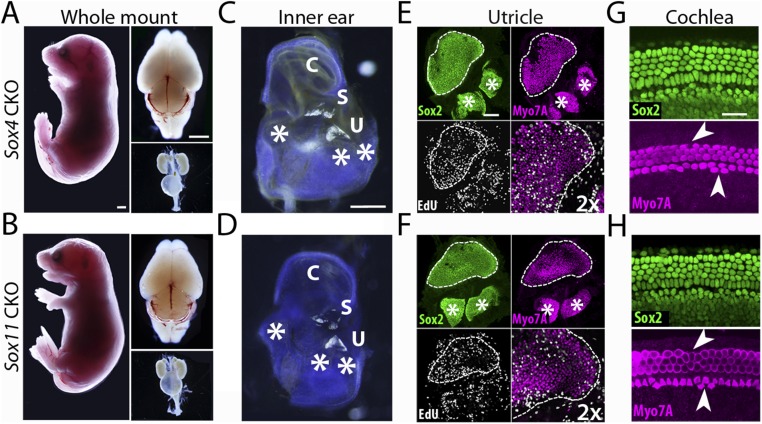
Anatomical defects in homozygous Sox4 or Sox11 CKO mice. (A) An E17.5 Sox4 CKO embryo (Left) displays a relatively normal brain (Upper Right) and kidneys and bladder (Lower Right). (Scale bars, 1 μm.) (B) The same organs are shown for an E17.5 Sox11 CKO embryo, which also is grossly normal. (C) A whole-mount preparation of the inner ear from a Sox4 CKO mouse shows the cochlea (C), sacculus (S), utricle (U), and ampullae (white asterisks) of the posterior (left asterisk), horizontal (center asterisk), and anterior (right asterisk) semicircular canals. (Scale bar, 500 μm.) (D) A preparation of the inner ear from a Sox11 CKO mouse reveals similar structures. (E, Upper Left) Supporting cells in the utricular macula (white outline) and cristae (white asterisks) of the anterior and horizontal semicircular canals are immunolabeled for Sox2 (green). (Upper Right) Hair cells are immunolabeled for Myo7A (magenta). The proliferating supporting cells at the edge of the utricle are labeled with EdU (Lower Left, white) and are enlarged twofold in the image at the Lower Right. (Scale bar, 100 μm.) (F) Identical labeling of the utricle and cristae in a Sox11 CKO animal reveals similar structures. (G, Upper) In the organ of Corti from an E17.5 Sox4 CKO embryo, supporting cells are immunolabeled for Sox2 (green). (Lower) The four rows of hair cells are labeled for Myo7A (magenta). The supporting cells are misaligned; some outer hair cells are absent (upper arrowhead), and ectopic inner hair cells are present (lower arrowhead). (Scale bar, 50 μm.) (H) The cochlea of an identically labeled Sox11 CKO embryo of the same age demonstrates similar defects.
Fig. 4.
Anatomical defects in homozygous Sox4 Sox11 double-CKO mice. (A) An E17.5 WT embryo (Left), its brain (Upper Right), and its kidneys and bladder (Lower Right) are presented for reference. (Scale bars, 1 μm.) (B) An E17.5 Sox4 Sox11 double-CKO embryo is substantially smaller and displays microcephaly as well as malformations of the brain and kidneys. (C, Upper) A whole-mount preparation of the inner ear from a WT mouse shows the cochlea (C), saccule (S), utricle (U), and ampullae (white asterisks) of the posterior (left asterisk), horizontal (middle asterisk), and anterior (right asterisk) semicircular canals. (Lower) Immunolabeling for Sox2 (green) reveals the size and position of the sensory epithelia within these organs. (Scale bar, 500 μm.) (D) Identical preparations of the inner ear of a Sox4 Sox11 double-CKO mouse highlight the underdevelopment of the cochlea (C), a single otolithic organ (O), and only two ampullae (white asterisks). (E, Upper Left) Supporting cells in a WT utricular macula (white outline) and cristae of the anterior and horizontal semicircular canals (white asterisks) are immunolabeled for Sox2 (green). (Upper Right) Hair cells are immunolabeled for Myo7A (magenta). (Lower Left) The proliferating supporting cells labeled for EdU (white) include a prominent band at the abneural edge of the utricle, shown in 2× enlargement at Lower Right. (Scale bar, 100 μm.) (F) Identical labeling of the single otolith organ (white outline) and rudimentary crista of a semicircular canal (white asterisk) in a Sox4 Sox11 double-CKO animal reveals that proliferating supporting cells are not concentrated at the organ's periphery and that hair cells are nearly absent. (G) In the organ of Corti of an E17.5 WT embryo, supporting cells are immunolabeled for Sox2 (green), and the four rows of hair cells are immunolabeled for Myo7A (magenta). (Scale bar, 50 μm.) (H) The cochlea of an identically labeled Sox4 Sox11 double-CKO embryo of the same age contains a field of disordered supporting cells and no hair cells.
We next assessed the development of the inner-ear structures in each variety of CKO mouse. At E18.5 the inner ear of a homozygous Sox4 or Sox11 CKO embryo had a relatively normal structure. The cochlea, utricle, saccule, and all three cristae of the semicircular canals were present, but the shapes of the utricle and saccule and the patterns of their otoconia were altered (Fig. 4C and Fig. S5 C and D). The inner ear of a homozygous Sox4 Sox11 double-CKO animal exhibited severe malformations. The size of the cochlea was markedly reduced in comparison with that of a WT animal, all three semicircular canals were absent, and there were no otoconia in either the utricle or the saccule (Fig. 4 C and D). Sox2 labeling revealed further irregularities: The organ of Corti was short and broad, only a single sensory patch was seen at the expected location of the utricle and saccule, and the cristae of the anterior and horizontal semicircular canals were replaced by a single rudimentary crista (Fig. 4 E and F). However, the crista of the posterior semicircular canal appeared only slightly smaller than normal (Fig. 4D).
To ascertain whether the morphological abnormalities of the vestibular organs in the SoxC CKO mice arose from deficiencies in supporting cell proliferation, we labeled proliferating cells with EdU and analyzed the inner ears of E18.5 embryos. As shown previously (14), a band of proliferating supporting cells occurred along the convex abneural periphery of a WT utricle (Fig. 4E). In contrast, the utricle of a homozygous Sox4 or Sox11 CKO mice was smaller and had a concave abneural edge with a dearth of proliferating cells (Fig. S5 E and F). The density of hair cells appeared normal. Although the single observable macula of a Sox4 Sox11 double-CKO mouse was not much smaller than the utricle of a homozygous Sox4 or Sox11 CKO animal, the structure of this putative vestibular organ was inconsistent with that of either a utricle or a saccule. In addition, proliferating cells were lacking from the periphery of the organ and hair cells were nearly absent (Fig. 4F).
The structure of the organ of Corti in a homozygous Sox4 or Sox11 CKO animal was relatively normal but less regular than that of a WT control (Fig. S5 G and H). The organ's distinctive supporting cells—pillar cells, Deiters’s cells, and Hensen’s cells—were present but occurred in a disordered array. The hair cells were misaligned, with ectopic hair cells among the inner hair cells and gaps between the outer hair cells. These defects were more prominent in the apical and middle turns and were nearly absent from the basal turn. As in the vestibular macula, the supporting cells were completely disorganized in homozygous Sox4 Sox11 double-CKO animals; no hair cells or distinctive subtypes of supporting cells were detected (Fig. 4 G and H).
Restorative Effect of Sox4 and Sox11 Overexpression.
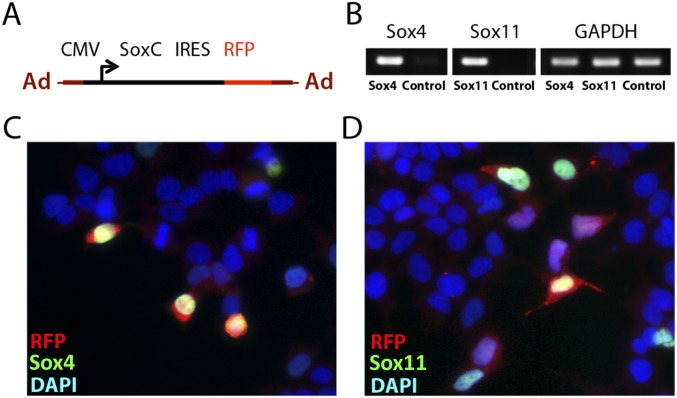
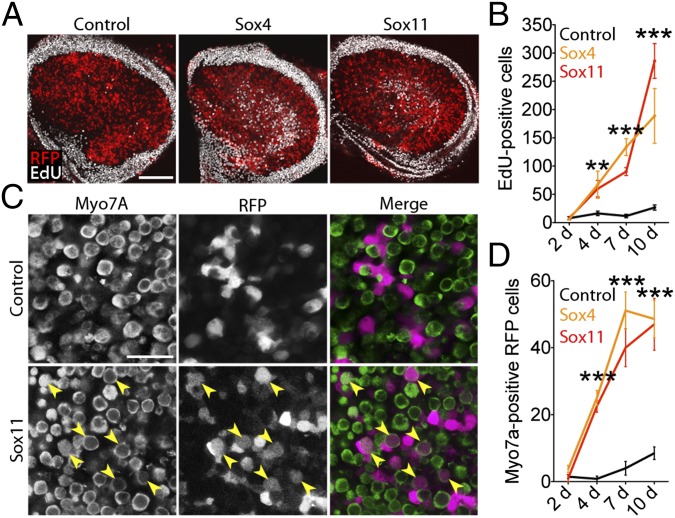
To address the functional relationship between SoxC expression, supporting cell proliferation, and the formation of new hair cells, we overexpressed the two principal SoxC genes in the utricular maculae of young adult mice. We developed adenoviral overexpression vectors carrying Sox4 (Ad-Sox4-RFP) and Sox11 (Ad-Sox11-RFP) and confirmed their efficacy in human embryonic kidney cells (Fig. S6). Cultured utricles of P12–P14 mice were infected with either vector and were compared with those infected with the control construct Ad-RFP (Fig. 5A). After 4 d of culture, the average number of proliferating cells in a macula overexpressing Sox4 or Sox11 was fourfold that in a control preparation (Fig. 5B). After 10 d a Sox4-overexpressing utricle and a Sox11-overexpressing macula showed proliferation seven- and 11-fold higher, respectively, than in a control. A similar 10-fold increase in the number of proliferating supporting cells was observed in the utricles of 6-wk-old mice when Sox4 or Sox11 was overexpressed for 10 d.
Fig. S6.
Characterization of SoxC expression vectors in HEK cells. (A) A schematic representation of an adenoviral expression construct shows the coding sequence for Sox4 or Sox11 driven by a cytomegaloviral promoter (CMV). The construct also includes the coding sequence for RFP following an internal ribosome-entry site (IRES). (B) Gel electrophoresis of Sox4, Sox11, and Gapdh cDNA fragments amplified by PCR demonstrates that Sox4 and Sox11 expression is induced in HEK cells upon transfection with the construct shown in A. The level of Gapdh expression, used as a control, remains constant. (C) Immunolabeling for Sox4 (green) in HEK cells transfected with Ad-Sox4-RFP (red) demonstrates that Sox4 is expressed specifically in transfected cells. All nuclei are labeled blue. (Scale bar, 25 μm.) (D) An identical preparation of HEK cells transfected with Ad-Sox11-RFP (red) demonstrates Sox11 immunolabeling (green) in the transfected cells.
Fig. 5.
Effects of viral overexpression of SoxC genes in adult utricles. (A) In representative images of cultured utricles transfected 7 d previously with control, Ad-Sox4-RFP, or Ad-Sox11-RFP virus, transfected cells are marked by RFP (red) and proliferating cells are labeled by EdU (white). (Scale bar, 100 μm.) (B) The number of EdU+ cells rises significantly in SoxC-transfected utricles between 2 d and 10 d in culture. **P < 0.01 ***P < 0.001. (Control n = 4, 7, 8, 8; Sox4 n = 3, 8, 10, 4; Sox11 n = 3, 3, 10, 4.) (C) In transfected utricles after 7 d in culture, transfected cells are marked by RFP, and hair cells are immunolabeled for Myo7A. Newly formed hair cells displaying both labels occur in the Sox11-overexpressing culture (yellow arrowheads) but not in the control preparation. (Scale bar, 20 μm.) (D) The number of newly formed hair cells, which are doubly positive for RFP and Myo7A, increases significantly during culture (***P < 0.001). (Control n = 5, 6, 5, 5; Sox4 n = 5, 5, 7, 5; Sox11 n = 6, 5, 9, 7.)
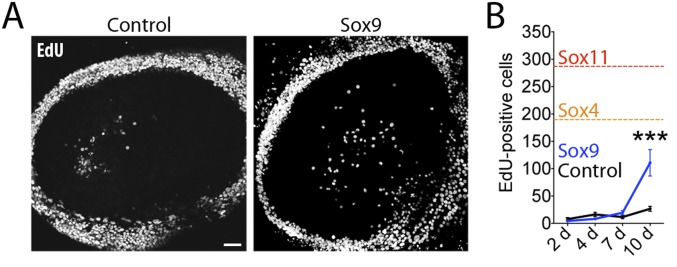
To test whether the mitogenic effect of SoxC proteins was specific, we overexpressed a member of the related SoxE family of transcription factors—Sox9—in P12 utricles in vitro. No effect on supporting cell proliferation was observed 2, 4, or 7 d after the infection (Fig. S7). Although after 10 d Sox9-overexpressing utricles showed slightly elevated levels of proliferation (Fig. S7A), they contained significantly fewer proliferating cells than SoxC-overexpressing cultures (Fig. S7B).
Fig. S7.
Effects of viral overexpression of Sox9 in P12 utricles. (A) In representative images of cultured utricles transfected 10 d previously with control or Ad-Sox9 virus, proliferating cells are labeled for EdU (white). (Scale bar, 100 μm.) (B) The number of EdU+ cells does not rise significantly in Sox9-transfected utricles between 2 d and 7 d in culture (control n = 4, 7, 8; Sox9 n = 4, 4, 4), but the difference is significant (***P < 0.001) at 10 d. (Control n = 8; Sox9 n = 3.) As a reference, the numbers of EdU+ cells in Sox4- and Sox11-overexpressing cultures after 10 d in culture are indicated by dashed lines.
To determine whether overexpression of SoxC genes yielded new hair cells, we also analyzed the transfected utricles for expression of Myo7A, a marker of mature hair cells. Because adenovirus of serotype 5 preferentially infects supporting cells (32), any cell doubly positive for RFP and Myo7A likely represented a hair cell newly derived from a transfected supporting cell. Two days after infection utricles contained virtually no doubly positive cells in cultures exposed to Ad-Sox4-RFP, Ad-Sox11-RFP, or Ad-RFP viruses (Fig. 5C). By 4 d, however, RFP+ hair cells were seen in the Sox4- or Sox11-overexpressing utricles but not in the control preparations. After 7 d the number of doubly positive hair cells increased to 13-fold the control value in Sox4-overexpressing cultures and to 10-fold the control value in Sox11-overexpressing cultures (Fig. 5D).
Discussion
Here we show that SoxC proteins play an integral role in the development of the vestibular and auditory receptor organs. Our results indicate that, by guiding the proliferation of progenitor cells, SoxC proteins are necessary for the growth of the sensory epithelia of the inner ear. Deletion during development of an increasing number of alleles for the cognate genes produced a progressive decrease in the size of the vestibular organs, ranging from a modest reduction in heterozygous CKO animals to diminutive or absent sensory organs in homozygous Sox4 Sox11 double-CKO mutants. The decrease in organ size was accompanied by defects in cell proliferation; conversely, overexpression of SoxC genes restored proliferation in the sensory epithelia of adult utricles.
Our observations on the mitogenic effects of SoxC proteins are congruent with previously known roles for these transcription factors. Sox4 and Sox11 are highly expressed early during murine embryonic development and in the stem cells of adult neural tissues (24, 25). Additionally, the cognate proteins control the survival and proliferation of neural progenitor cells (27). In the hippocampal subgranular zone, for example, Sox4 and Sox11 are strongly expressed in proliferating progenitor cells and newly generated neuroblasts (23). The loss of the two proteins during early neurogenesis results in major cortical defects and a severely stunted or absent hippocampus (23).
Our data additionally suggest that SoxC genes play a prosensory role by directing the differentiation of supporting cells into sensory receptors. In the periphery of the utricular macula, Sox4 and Sox11 were highly expressed in proliferating progenitor cells and in newly generated hair cells, but their activity declined sharply in maturing sensory receptors. Although severely underdeveloped, vestibular and auditory organs nonetheless were present in homozygous Sox4 Sox11 double-CKO mice; however, these organs completely lacked hair cells. We also found that the overexpression of Sox4 or Sox11 in adult murine utricles resulted in hair cell production through the direct differentiation of infected supporting cells.
Previous research has demonstrated a prosensory role for SoxC genes. For example, it has been shown that the activation of SoxC expression in the eye increases the production of retinal ganglion cells (24). SoxC transcription factors also directly foster the expression of neuron-specific genes during neurogenesis in the adult hippocampus (23). SoxC proteins support the flexion of DNA and thereby promote the formation of transcriptional enhancer complexes (25). It is possible that Sox4 and Sox11 proteins function upstream of the transcription factor Atoh1, a master regulator of hair cell differentiation (33). We note the presence of multiple SoxC-binding sites in the 3′ enhancer of Atoh1, the principal regulatory region for this gene (34).
Our findings demonstrate that the proper functioning of SoxC genes is necessary and sufficient for hair cell production in the sensory epithelia of the murine inner ear. Although further investigation of the mechanism of action of SoxC transcription factors in the inner ear is required, these proteins hold great potential as therapeutic targets for hair cell regeneration.
Materials and Methods
Experiments were conducted in accordance with the policies of Rockefeller University’s Institutional Animal Care and Use Committee. After dissection of murine utricles (35) and isolation of total RNA (RNeasy Micro Kit; Qiagen), samples were analyzed by single-end, 100-bp sequencing (Illumina HiSeq 2500). In situ hybridization, immunohistochemistry, and qPCR were conducted by standard protocols. The AdEasy Adenoviral Vector System (36) was used to create adenoviral vectors containing the full-length coding sequence of murine Sox4 or Sox11 under the control of a cytomegalovirus promoter. The experimental procedures are detailed in SI Materials and Methods. The RNA-sequencing data have been deposited under accession code GSE72293 in the Gene Expression Omnibus of the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE72293). The primers were designed with PrimerQuest (Integrated DNA Technologies) and are listed in Table S1.
Table S1.
qPCR primers used for the validation of RNA-sequencing data
| Gene name | Forward primer | Reverse primer |
| Adnp | ACAGTTCATCCGGTTGAGAAA | GTGGACTAGATGCAGAGTGATG |
| Cited1 | AGATGAACTCTCTGGCCTACT | GTTGGCTTTGGCTCCATTTG |
| Dbp | GCTTGACATCTAGGGACACAC | AAGTCTCATGGCCTGGAATG |
| Egr1 | AACAACCCTATGAGCACCTG | GAGTCGTTTGGCTGGGATAA |
| Etv1 | AGAAGCCACAAGTGGGAATG | GGAGTATGAGCTGTGTTTGGAG |
| Etv4 | GGTGATGGAGTGATGGGTTATG | CTTCCTGCTTGATGTCTCCTTC |
| Etv5 | AGTCAGGAGTTCCTGGAGATAG | GGTCATGGTATTCCTGCTTGA |
| Etv6 | TCAGTGTCTCCTGTCGAGAATA | CAGGATCAAAGGGTGCATGA |
| Fbxw7 | GGAGACGAGGAGAACTCAAATC | TGTTGTCATCAGAACCACTAACT |
| Foxm1 | AGGAGGACATAGCACACTTAGA | GGAGTTGGATAGCTCCTCTTTG |
| Foxo1 | CGTGCCCTACTTCAAGGATAAG | GCACTCGAATAAACTTGCTGTG |
| Gapdh | TCAACAGCAACTCCCACTCTTCCA | ACCCTGTTGCTGTAGCCGTATTCA |
| Irf9 | GAGAGAGGTCGTATGGATGTTG | CGCTTGCATGGTGATTTCTG |
| Klf4 | CAGCTTGCAGCAGTAACAAC | TGGGTTAGCGAGTTGGAAAG |
| Klf6 | GGTACATCGGTGCCACTTTA | AACCTTCCCATGAGCATCTG |
| Klf7 | AAGTGCTCATGGGAAGGATG | CTGTCGCAGTGGTTACACTTA |
| Klf11 | TGCTGTTCAGACCTGCTTAC | ACAGGGATCATCTGGCAAAG |
| Lgr5 | CCCGTTTCTGGATGAGATCAA | GCGTGGGTAAAGGATACTAAGG |
| Nr1d1 | CTTCATCCTCCTCCTCCTTCTA | GTAATGTTGCTTGTGCCCTTG |
| Nr1d2 | CCAGCTCAGTGATGAGGAAAT | GCCTCCACTGAGTTGACATT |
| Rarg | CAGCACTAAGGGAGCAGAAA | GGAGTCGTCCTCAAACATCTC |
| Rcor2 | GACCACTGATGAGCAACTTCTA | CACTTGGGTCAGAGTCTTGTT |
| Rxra | GCTAACAGAGCTGGTGTCTAAG | CCTTAGAGTCAGGGTTGAACAG |
| Sox4 | AAGGACAGCGACAAGATTCC | GCCCGACTTCACCTTCTTT |
| Sox11 | CAGGAAGATCATGGAGCAGTC | TCCCTGATGAACGGGATCT |
| Tcf7l1 | GAGTGTACCCTGAAGGAAAGTG | CCGGGCAAGCTCATAGTATTT |
| Tead2 | GCGGCAGATCTACGACAAATT | CCAGAACTTGACGAGGAAGAAG |
| Tead4 | TGGACATCCGCCAAATCTATG | CCCAGAACTTCACAAGGAAGAA |
| Tgif2 | CTCGGTGCTGCAGATATGTAA | GGAGATGGTGAACTGATTAGGG |
| Zbtb7a | CGATTCACCAGACAGGACAA | GTGGTTCTTCAGGTCGTAGTT |
| Zfp281 | CAGGAGTTAGCTAGCCAGATTG | CTGCCCGACTTAAACTGAGAA |
| Zhx3 | TGTGGCAGATATAAGCAGTGAG | GTTCTCAGACAGCTCCGAATAG |
SI Materials and Methods
Animal Care and Strains.
Experiments were conducted in accordance with the policies of Rockefeller University’s Institutional Animal Care and Use Committee. Swiss Webster mice with timed pregnancies were obtained from Charles River Laboratories. To create CKO mice, we mated heterozygous Pax2-Cre animals with homozygous Sox4fl/fl Sox11fl/fl Sox12−/− mice. The F1 Pax2-Cre Sox4fl/+ Sox11fl/+ Sox12−/+ progeny then were bred with Sox4fl/fl Sox11fl/fl Sox12−/− animals to generate homozygous knockouts of Sox4 or Sox11 at an expected frequency of 25% and double knockouts at 12.5% frequency.
Utricle Dissections.
Pregnant mice were killed, and embryos were extracted and placed into ice-cold HBSS (Life Technologies). Utricles were dissected using a standard procedure (35) with two modifications: The organs were approached from the medial surface of the temporal bone, and tissue was disinfected by five rinses of 1–3 min each in ice-cold HBSS supplemented with 105 U/L penicillin and 170 μM streptomycin (Sigma). For RNA-extraction experiments, utricles were treated with 10 mg/L dispase (Life Technologies) for 1 h at 37 °C in a tissue-culture incubator charged with 5% CO2. The sensory epithelia then were separated with a hair knife and collected in ice-cold lysis solution (RNeasy Micro Kit; Qiagen) containing 130 mM β-mercaptoethanol (Sigma).
Organotypic Cultures.
Utricles were cultured on inserts with 0.4-μm-diameter pores (Millipore) in DMEM/F12 medium supplemented with 33 mM D-glucose, 19 mM sodium bicarbonate, 15 mM Hepes, 1 mM glutamine, 1 mM nicotinamide, 29 nM sodium selenite, 20 mg/L EGF, 20 mg/L FGF, 10 mg/L insulin, and 5.5 mg/L transferrin. All reagents were purchased from Sigma-Aldrich.
RNA Sequencing and Analysis.
Total RNA was isolated by a standard protocol (RNeasy Micro Kit; Qiagen). The yield and quality of the product were measured with an Agilent 2100 bioanalyzer; samples with RNA integrity numbers of 8.5–10 were used for library preparation. Approximately 200 ng of total RNA, representing 20–30 utricular maculae, was used to create each cDNA library. Three samples per lane were analyzed by Illumina HiSeq 2500 single-end, 100-bp sequencing. We used the Genomics Suite of Partek software to analyze the data. We evaluated the statistical significances of gene expression within each sample and of differential expression between samples by ANOVA followed by a false-discovery control through the Benjamini–Hochberg test.
In Situ Hybridization.
The entire coding sequences of the Sox4 and Sox11 genes were amplified from a cDNA library made from a P0 murine brain. Amplicons were cloned into the pCRII-TOPO vector (Dual-Promoter TOPO TA Cloning Kit; Invitrogen). Sense and antisense riboprobes were each synthesized from 1 μg of linearized DNA with the SP6/T7 DIG RNA-labeling kit (Roche Applied Science). In situ hybridization was performed on 20-μm frozen sections of inner ears as described (35). Alkaline phosphatase activity was detected with a fluorescent fast red substrate (Roche). After fluorescence in situ labeling of frozen sections from inner ears of Atoh1-nGFP mice, the hair cells were marked by anti-GFP immunoreactivity. Nuclei were counterstained with 3 μM DAPI (Sigma-Aldrich).
Immunohistochemistry.
Utricles and cochleas were dissected in ice-cold HBSS and fixed in 4% (vol/vol) formaldehyde for 1 h at room temperature. Alternatively, inner ears were dissected and fixed in formaldehyde for 18 h at 4 °C, dehydrated in 0.88 M sucrose for 18 h at 4 °C, embedded in Tissue-Tek optimum cutting temperature compound (Sakura), and frozen in liquid-nitrogen vapor. Whole-mount sensory epithelia or 10-μm frozen sections then were blocked with 3% (vol/vol) normal donkey serum (Sigma-Aldrich) in 500 mM NaCl and 20 mM Tris(hydroxymethyl)aminomethane at pH 7.5 (Bio-Rad) containing 0.3% Triton X-100 (Sigma). The following primary antisera were used: goat anti-Sox2 (Santa Cruz), rabbit anti-Myo7A (Proteus BioSciences), guinea pig anti-Sox4, guinea pig anti-Sox11, and rabbit anti-GFP (Torrey Pines Biolabs). Primary antisera were reconstituted in the blocking solution and applied overnight at 4 °C. Samples were washed with PBS solution containing 0.1% Tween 20 (Sigma-Aldrich); Alexa Fluor-labeled secondary antisera (Life Technologies) were applied in the same solution for 1 h at room temperature.
EdU pulse experiments were initiated by single i.p. injections of 50 μg EdU (Life Technologies) per gram of maternal body mass. The animals were killed at the indicated times and the sensory epithelia of the inner ear were analyzed by Click-iT EdU labeling (Life Technologies) to reveal newly formed cells. The nuclei of all cells were counterstained with 3 μM DAPI.
Imaging and Cell Counting.
Confocal imaging was performed with an Olympus IX81 microscope equipped with a Fluoview FV1000 laser-scanning system (Olympus America). To enumerate EdU+ cells, we imaged whole-mounted utricles as Z-stacks and selected representative slices for maximal-intensity Z-projections.
Inner ears for low-magnification whole-mounts were dehydrated in 100% ethanol, cleared with methyl salicylate (Sigma-Aldrich), and imaged with an Olympus SZX7 dissecting microscope and DP71 camera.
The statistical significance of comparisons between cell counts was determined by two-tailed Student’s t tests.
qPCR Analysis.
The RNA for cDNA synthesis was obtained as described above from six to eight utricles for each sample. The primers were designed with PrimerQuest (Integrated DNA Technologies) and are listed in Table S1. Relative gene-expression levels were obtained by normalization to the expression of Gapdh in each sample. qPCR analyses were performed on an Applied Biosystems 7900HT Sequence Detection System with FastStart Universal SYBR Green Master (Roche Applied Science).
Adenoviral Gene Transfer.
The AdEasy Adenoviral Vector System (36) was used to create adenoviral vectors containing the full-length coding sequence of murine Sox4 or Sox11 under the control of a cytomegalovirus promoter. To permit the identification of infected cells, we included sequences for an internal ribosome-entry site and for RFP under the control of the same promoter. Viral particles were amplified in HEK cells and purified by CsCl-gradient centrifugation followed by dialysis (Viral Vector Core Facility, Sanford-Burnham Medical Research Institute). Utricles were dissected at P12–P14 and infected in 200 μL of culture medium with 10 μL of virus at a titer of 1010 pfu/mL. Ad-RFP virus (Vector Biolabs) and Ad-Sox9 (Signagen) at the same titer were used as controls. One day later 2 mL of culture medium was supplemented with 10 μM EdU. Preparations were analyzed after 2, 4, 7, or 10 d of culture.
Supplementary Material
Acknowledgments
We thank Dr. V. Lefebvre for the invaluable Sox4fl/fl Sox11fl/fl Sox12−/− mice, Dr. J. E. Johnson for the Atoh1-nGFP line, Dr. A. K. Groves for the Pax2-Cre animals. Dr. E. Sock provided the anti-Sox4 and anti-Sox11 antisera and Dr. C.-T. Huang prepared the Sox4- and Sox11-overexpression viruses. Dr. T. Iafe and the members of our research group provided comments on the manuscript. K.G. was supported by Howard Hughes Medical Institute, of which A.J.H. is an Investigator.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc (accession no. GSE72293).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1517371112/-/DCSupplemental.
References
- 1.Warchol ME. Sensory regeneration in the vertebrate inner ear: Differences at the levels of cells and species. Hear Res. 2011;273(1-2):72–79. doi: 10.1016/j.heares.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Cruz RM, Lambert PR, Rubel EW. Light microscopic evidence of hair cell regeneration after gentamicin toxicity in chick cochlea. Arch Otolaryngol Head Neck Surg. 1987;113(10):1058–1062. doi: 10.1001/archotol.1987.01860100036017. [DOI] [PubMed] [Google Scholar]
- 3.Ryals BM, Rubel EW. Hair cell regeneration after acoustic trauma in adult Coturnix quail. Science. 1988;240(4860):1774–1776. doi: 10.1126/science.3381101. [DOI] [PubMed] [Google Scholar]
- 4.Corwin JT, Cotanche DA. Regeneration of sensory hair cells after acoustic trauma. Science. 1988;240(4860):1772–1774. doi: 10.1126/science.3381100. [DOI] [PubMed] [Google Scholar]
- 5.Stone JS, Cotanche DA. Hair cell regeneration in the avian auditory epithelium. Int J Dev Biol. 2007;51(6-7):633–647. doi: 10.1387/ijdb.072408js. [DOI] [PubMed] [Google Scholar]
- 6.Kelley MW. Regulation of cell fate in the sensory epithelia of the inner ear. Nat Rev Neurosci. 2006;7(11):837–849. doi: 10.1038/nrn1987. [DOI] [PubMed] [Google Scholar]
- 7.Stone JS, Rubel EW. Delta1 expression during avian hair cell regeneration. Development. 1999;126(5):961–973. doi: 10.1242/dev.126.5.961. [DOI] [PubMed] [Google Scholar]
- 8.Daudet N, et al. Notch regulation of progenitor cell behavior in quiescent and regenerating auditory epithelium of mature birds. Dev Biol. 2009;326(1):86–100. doi: 10.1016/j.ydbio.2008.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawamoto K, Izumikawa M, Beyer LA, Atkin GM, Raphael Y. Spontaneous hair cell regeneration in the mouse utricle following gentamicin ototoxicity. Hear Res. 2009;247(1):17–26. doi: 10.1016/j.heares.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golub JS, et al. Hair cell replacement in adult mouse utricles after targeted ablation of hair cells with diphtheria toxin. J Neurosci. 2012;32(43):15093–15105. doi: 10.1523/JNEUROSCI.1709-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haddon C, et al. Hair cells without supporting cells: Further studies in the ear of the zebrafish mind bomb mutant. J Neurocytol. 1999;28(10-11):837–850. doi: 10.1023/a:1007013904913. [DOI] [PubMed] [Google Scholar]
- 12.Groves AK. The challenge of hair cell regeneration. Exp Biol Med (Maywood) 2010;235(4):434–446. doi: 10.1258/ebm.2009.009281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atkinson PJ, Huarcaya Najarro E, Sayyid ZN, Cheng AG. Sensory hair cell development and regeneration: Similarities and differences. Development. 2015;142(9):1561–1571. doi: 10.1242/dev.114926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burns JC, On D, Baker W, Collado MS, Corwin JT. Over half the hair cells in the mouse utricle first appear after birth, with significant numbers originating from early postnatal mitotic production in peripheral and striolar growth zones. J Assoc Res Otolaryngol. 2012;13(5):609–627. doi: 10.1007/s10162-012-0337-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Powles N, Babbs C, Ficker M, Schimmang T, Maconochie M. Identification and analysis of genes from the mouse otic vesicle and their association with developmental subprocesses through in situ hybridization. Dev Biol. 2004;268(1):24–38. doi: 10.1016/j.ydbio.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 16.Hartman BH, Durruthy-Durruthy R, Laske RD, Losorelli S, Heller S. Identification and characterization of mouse otic sensory lineage genes. Front Cell Neurosci. 2015;9:79. doi: 10.3389/fncel.2015.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waldhaus J, Durruthy-Durruthy R, Heller S. Quantitative high-resolution cellular map of the ogan of Corti. Cell Reports. 2015;11(9):1385–1399. doi: 10.1016/j.celrep.2015.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheffer DI, Shen J, Corey DP, Chen ZY. Gene expression by mouse inner ear hair cells during development. J Neurosci. 2015;35(16):6366–6380. doi: 10.1523/JNEUROSCI.5126-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng JL, Shou J, Guillemot F, Kageyama R, Gao WQ. Hes1 is a negative regulator of inner ear hair cell differentiation. Development. 2000;127(21):4551–4560. doi: 10.1242/dev.127.21.4551. [DOI] [PubMed] [Google Scholar]
- 20.Qian D, et al. Basic helix-loop-helix gene Hes6 delineates the sensory hair cell lineage in the inner ear. Dev Dyn. 2006;235(6):1689–1700. doi: 10.1002/dvdy.20736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi F, Kempfle JS, Edge AS. Wnt-responsive Lgr5-expressing stem cells are hair cell progenitors in the cochlea. J Neurosci. 2012;32(28):9639–9648. doi: 10.1523/JNEUROSCI.1064-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bramhall NF, Shi F, Arnold K, Hochedlinger K, Edge AS. Lgr5-positive supporting cells generate new hair cells in the postnatal cochlea. Stem Cell Rep. 2014;2(3):311–322. doi: 10.1016/j.stemcr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mu L, et al. SoxC transcription factors are required for neuronal differentiation in adult hippocampal neurogenesis. J Neurosci. 2012;32(9):3067–3080. doi: 10.1523/JNEUROSCI.4679-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Y, et al. Transcription factors SOX4 and SOX11 function redundantly to regulate the development of mouse retinal ganglion cells. J Biol Chem. 2013;288(25):18429–18438. doi: 10.1074/jbc.M113.478503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dy P, et al. The three SoxC proteins--Sox4, Sox11 and Sox12--exhibit overlapping expression patterns and molecular properties. Nucleic Acids Res. 2008;36(9):3101–3117. doi: 10.1093/nar/gkn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohyama T, Groves AK. Generation of Pax2-Cre mice by modification of a Pax2 bacterial artificial chromosome. Genesis. 2004;38(4):195–199. doi: 10.1002/gene.20017. [DOI] [PubMed] [Google Scholar]
- 27.Bhattaram P, et al. Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nat Commun. 2010;1(1):1–12. doi: 10.1038/ncomms1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lefebvre V, Dumitriu B, Penzo-Méndez A, Han Y, Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int J Biochem Cell Biol. 2007;39(12):2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoser M, et al. Sox12 deletion in the mouse reveals nonreciprocal redundancy with the related Sox4 and Sox11 transcription factors. Mol Cell Biol. 2008;28(15):4675–4687. doi: 10.1128/MCB.00338-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardisty-Hughes RE, Parker A, Brown SD. A hearing and vestibular phenotyping pipeline to identify mouse mutants with hearing impairment. Nat Protoc. 2010;5(1):177–190. doi: 10.1038/nprot.2009.204. [DOI] [PubMed] [Google Scholar]
- 31.Willott JF. 2006. Measurement of the auditory brainstem response (ABR) to study auditory sensitivity in mice. Curr Protoc Neurosci 34:8.21B:8.21B.1–8.21B.12.
- 32.Brandon CS, Voelkel-Johnson C, May LA, Cunningham LL. Dissection of adult mouse utricle and adenovirus-mediated supporting-cell infection. J Vis Exp. 2012;61:3734. doi: 10.3791/3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen P, Johnson JE, Zoghbi HY, Segil N. The role of Math1 in inner ear development: Uncoupling the establishment of the sensory primordium from hair cell fate determination. Development. 2002;129(10):2495–2505. doi: 10.1242/dev.129.10.2495. [DOI] [PubMed] [Google Scholar]
- 34.Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127(6):1185–1196. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- 35.Cunningham TJ, Chatzi C, Sandell LL, Trainor PA, Duester G. Rdh10 mutants deficient in limb field retinoic acid signaling exhibit normal limb patterning but display interdigital webbing. Dev Dyn. 2011;240(5):1142–1150. doi: 10.1002/dvdy.22583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo J, et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc. 2007;2(5):1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.