Abstract
Motivation: The accurate discovery and annotation of regulatory elements remains a challenging problem. The growing number of sequenced genomes creates new opportunities for comparative approaches to motif discovery. Putative binding sites are then considered to be functional if they are conserved in orthologous promoter sequences of multiple related species. Existing methods for comparative motif discovery usually rely on pregenerated multiple sequence alignments, which are difficult to obtain for more diverged species such as plants. As a consequence, misaligned regulatory elements often remain undetected.
Results: We present a novel algorithm that supports both alignment-free and alignment-based motif discovery in the promoter sequences of related species. Putative motifs are exhaustively enumerated as words over the IUPAC alphabet and screened for conservation using the branch length score. Additionally, a confidence score is established in a genome-wide fashion. In order to take advantage of a cloud computing infrastructure, the MapReduce programming model is adopted. The method is applied to four monocotyledon plant species and it is shown that high-scoring motifs are significantly enriched for open chromatin regions in Oryza sativa and for transcription factor binding sites inferred through protein-binding microarrays in O.sativa and Zea mays. Furthermore, the method is shown to recover experimentally profiled ga2ox1-like KN1 binding sites in Z.mays.
Availability and implementation: BLSSpeller was written in Java. Source code and manual are available at http://bioinformatics.intec.ugent.be/blsspeller
Contact: Klaas.Vandepoele@psb.vib-ugent.be or jan.fostier@intec.ugent.be
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
One of the major challenges in systems biology is gaining a full understanding of gene transcriptional regulation. Transcription factors, for which the binding sites are usually hidden in the promoter sequence of the gene, are in this respect of particular importance. Computational approaches for de novo motif discovery can be classified in (i) methods to identify binding sites in promoter sequences of co-regulated genes within a single genome and (ii) comparative approaches using homologous sequences from multiple related species (Das and Dai, 2007).
The first category uses clusters of co-expressed genes, which are assumed to be regulated by the same set of transcription factors. A drawback of these methods is that the relationship between co-expression and co-regulation relies on complex regulatory mechanisms, making it difficult to assemble reliable datasets since co-expression does not necessarily imply that there is a common binding site involved. Two different algorithmic approaches coexist: the statistical (Bailey et al., 2009; Hughes et al., 2000; Liu et al., 2001; Thijs et al., 2002; Wei and Yu, 2007) and the exhaustive, word-based algorithms. The latter contain graph-based approaches (Eskin and Pevzner, 2002; Liang et al., 2004; Satya and Mukherjee, 2004) and methods based on index structures (Marsan and Sagot, 2000; Marschall and Rahmann, 2009; Pavesi et al., 2001).
Due to the growing availability of genome sequences, a second category of algorithms based on phylogenetic footprinting emerged (Blanchette and Tompa, 2002): orthologous regulatory regions from multiple species are compared with the underlying assumption that functional elements evolve at a much slower pace, compared to the non-functional part of the genome, due to selective pressure (Berezikov et al., 2004). Most comparative motif discovery approaches rely in some way on multiple sequence alignments, in which regulatory signals are expected to be well-aligned. Pioneering algorithms in this category are Conreal (Berezikov et al., 2004), Phylonet (Wang, 2005) and Phyloscan (Carmack et al., 2007). More recent algorithms relying on alignments are used to study mosquitoes (Sieglaff et al., 2009), Fusarium (Kumar et al., 2010), vertebrates (Ettwiller et al., 2005) and mammals (Xie et al., 2005).
It has, however, been shown that known regulatory elements are not always correctly aligned (Siggia, 2005), an issue that is further complicated by the different alignments produced by various alignment programs (Pollard et al., 2004). Transcription factor (TF) binding sites are short, flexible against certain mutations and even mobile which explains why they are sometimes misaligned. Mechanisms have been observed that allow the modification of regulatory sequences without altering their function: divergence driving words and binding site turnover. Regulatory sequences can diverge freely if the divergence driving words, which are specific short words in the non-coding DNA, are not altered (Bradley et al., 2010). Since a TF can often bind to multiple similar sites, mutations turning one site into another should not affect regulation. Binding site turnover, on the other hand, is the mechanism where the gain of a redundant binding site allows the loss of a previously functional site (Venkataram and Fay, 2010). The corresponding TF can then bind to the new site, maintaining the regulatory interaction. This allows binding sites to relocate within the regulatory sequence, making it difficult for alignment algorithms to correctly align them.
Binding site discovery, especially in plants, has to deal with large divergence times and complex diversification mechanisms such as genome duplications. This makes approaches based on whole genome alignments, often used in de novo algorithms, impractical. Some of these problems have been addressed in earlier studies. Stark et al. (2007) used a mixed approach in a study with 12 Drosophila species, starting from whole genome alignments but allowing for limited motif movement within an alignment. Elemento and Tavazoie (2005) designed an alignment-free algorithm to discover overrepresented k-mers over the exact ACGT alphabet in pairs of related genomes. Finally, MDOS (Wu et al., 2008) is a new version of this algorithm with improved statistics.
In this article, four monocotyledonous plant species are studied using a phylogenetic footprinting approach: Oryza sativa ssp. indica (osa), Brachypodium distachyon (bdi), Sorghum bicolor (sbi) and Zea mays (zma). We adopt a gene-centric approach, where the promoter sequences of orthologous genes are grouped into gene families. A word-based discovery algorithm was designed to exhaustively report all genome-wide conserved motifs. The term conserved relates to the occurrence of the motif in multiple promoter sequences of a particular gene family. Genome-wide conservation relates to the fact that this conservation occurs in more gene families than what is expected by chance. Motifs are modeled as words (k-mers) over an alphabet that contains the four bases (ACGT) and (optionally) additional degenerate characters from the IUPAC alphabet (Cornish-Bowden, 1985). This degeneracy allows a motif to model a collection of binding sites. The algorithm can be run in both alignment-free or alignment-based mode. In case of alignment-free discovery, the conservation of a motif is scored irrespective of its orientation or position within a promoter sequence. This relaxed definition of conservation was previously used by Gordân et al. (2010) and is especially relevant when studying more diverged species for which accurate multiple sequence alignments are difficult to generate. Alignment-based discovery adds the constraint that motifs must be aligned, i.e. occur at the same position in the multiple sequence alignment.
Robust algorithms for comparative genomics are expected to gain in power when more related species are added. Most studies so far only consider motifs that are conserved within all organisms. The branch length score (BLS) was developed to quantify motif conservation in a biologically meaningful manner and ranges from 0% (not conserved) to 100% (conserved in all sequences). The BLS takes the phylogenetic relationships between the species into account by representing a relative evolutionary distance over which a candidate binding site is conserved within a gene family. The BLS was first used in a comparative study with 12 Drosophila genomes (Stark et al., 2007) and allows studying motifs only conserved in subsets of the organisms.
Whereas most current algorithms avoid exploring the full motif space by using greedy algorithms, our method is unique in the sense that it is exhaustive. MDOS (Wu et al., 2008) only processes promising k-mers and gradually adds degeneracy if this improves the conservation score. Kellis et al. (2003) and Stark et al. (2007) use the mini-motifs approach (van Helden et al., 2000) only processing promising trinucleotide duos before adding degeneracy. Here, every word that occurs in one of the input sequences, including their degenerate variants, is considered as a candidate motif. The only imposed restrictions are a prespecified minimum and maximum length and a maximum number of degenerate IUPAC characters. The advantage of such exhaustive approach is that the method yields globally optimal results. In order to strongly reduce the runtime and avoid excessive memory requirements, the MapReduce programming model (Dean and Ghemawat, 2004) was adopted as a means to take advantage of a parallel, distributed-memory cloud computing environment. By enabling disk I/O to store intermediate results, the current MapReduce implementation overcomes the memory bottleneck in a prototype implementation of this software that relied on the Message Passing Interface (MPI) for parallelization (De Witte et al., 2013).
2 Methods
2.1 Generation of gene families
The orthology relationships between the genes of the four different monocot plant species were inferred using the ‘integrative orthology viewer’ in the PLAZA 2.5 platform (Proost et al., 2009; Van Bel et al., 2012). Homologous (i.e. orthologous and paralogous) genes were grouped in gene families and their promoter sequences 2 kbp upstream from the translation start site were extracted. In its most simple form, a family consists of four orthologous genes: one from each organism. In that case, the phylogenetic tree by Reineke et al. (2011) is used. For gene families that comprise one or more paralogs, gene family-specific phylogenetic trees can be constructed that take into account the specific order in which the duplications and speciation events occurred. For simplicity, we assume that all paralogous gene duplications occurred recently. This is modeled by adding a bifurcation with a branch length of zero to the phylogenetic tree which means that only conservation between different species contributes to the branch length score. Note that besides promoter regions, additional homologous sequences of interest (e.g. intronic regions) could be added to the input dataset.
2.2 Intrafamily step: conservation within a gene family
For all gene families individually, all words with a length between and characters that occur in any of the sequences are exhaustively enumerated and their degree of conservation within that family is quantified. Words are spelled in the IUPAC alphabet or a subset thereof. Up to degenerate (i.e. non-ACGT) characters are allowed per word. The intrafamily phase can operate in alignment-free or alignment-based mode.
In the alignment-free approach, a generalized suffix tree (GST) is constructed (Giegerich et al., 1999) from the promoter sequences and their reverse complements in the gene family. Using Sagot’s Speller algorithm (Marsan and Sagot, 2000), the GST is used to efficiently and exhaustively report all words in the IUPAC alphabet along with the sequences in which they occur. Additional algorithmic details and runtime information are described in Supplementary Methods 1.1.
The alignment-based mode requires a pregenerated multiple sequence alignment (MSA) of the orthologous promoters in a gene family. Dialign-TX (Subramanian et al., 2008) was chosen to create these MSAs in view of good results on a non-coding alignment benchmark (Pollard et al., 2004). For every position in the alignment, a small GST is generated containing only the suffixes of the sequences that start at that position. The same Speller algorithm is run to report all words and the sequences in which they occur at aligned positions, again using the IUPAC alphabet.
For every word, the degree of conservation in each gene family is quantified using the branch length score (BLS). Given the sequences in which the word occurs, the BLS can be calculated by finding the minimum spanning tree that connects these sequences in the phylogenetic tree. The sum of the weights of the horizontal branches in the minimum spanning tree then represents the BLS (Stark et al., 2007). In alignment-based mode, the same motif can occur at multiple aligned positions within a single family; in that case only the highest BLS value is used. Only words for which the BLS exceeds a prespecified threshold T are retained. Such words are said to be conserved within the gene family.
2.3 Interfamily step: genome-wide conservation
The conserved words of all gene families are sorted according to base content and partitioned into permutation groups whose elements are permutations of each other. All words in a permutation group hence have the same length, base content and degeneracy. For example, the words AWTC, WTAC and CAWT belong to the same permutation group.
The number of occurrences for each distinct word within a permutation group is counted. This number corresponds to the number of gene families in which that word is conserved with a and is referred to as the conserved family count F(T). Genome-wide conserved motifs are selected based on the fact that they have a conserved family count F(T) that is (much) higher than the median conserved family count of the member instances of their permutation group. This median value, denoted as (bg = background) represents the expected conserved family count for a word in that permutation group. is approximated by randomly generating a large number (default = 1000) of instances of the permutation group, i.e. random words with the same length and base content and computing the median value for the conserved family count. Note that some of those random instances can have a conserved family count equal to zero.
A confidence score C, adopted from (Stark et al., 2007), is obtained for each word in the permutation group by comparing F(T) and as follows:
Words for which and are considered genome-wide conserved motifs and are retained by the method where and denote user-defined thresholds. The output of the method consists of an exhaustive list of motifs which satisfy these thresholds, along with the F(T) and C(T) metrics. Similar to Stark et al. (2007), rather than using a single threshold T, multiple BLS thresholds Ti can be used in a single run. The confidence score is then computed for all thresholds Ti individually, i.e. . Here, denotes the number of families in which the motif is conserved with a BLS higher than the threshold Ti. Similarly, is the corresponding value for the background model. Words for which and for any of the BLS thresholds Ti are retained.
2.4 MapReduce implementation
The method was implemented using the MapReduce (Dean and Ghemawat, 2004) programming model. The map phase corresponds to the intrafamily phase in which the gene families are processed in parallel by the different mappers. The reduce phase corresponds to the interfamily phase in which the permutations groups are processed in parallel by the different reducers. In between the map and reduce step, the candidate motifs are sorted according to length and base content in order to create the permutation groups.
3 Results and discussion
3.1 BLSSpeller algorithm
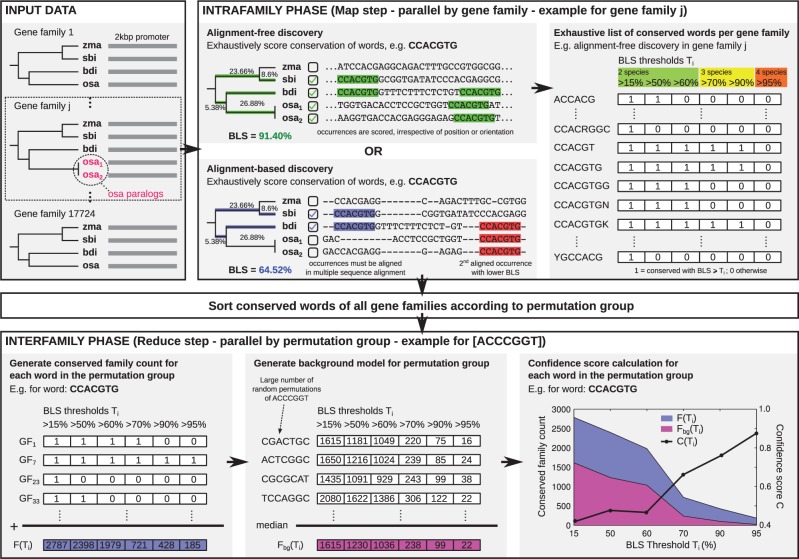
The workflow of BLSSpeller is illustrated in Figure 1. The input consists of gene families containing homologous promoter sequences from related species. The algorithm consists of an intrafamily and an interfamily step with a sorting step in between.
Fig. 1.
Overview of BLSSpeller. The input consists of homologous promoter sequences grouped into gene families. During the intrafamily phase, conserved words are exhaustively enumerated for each gene family individually. A word is considered to be conserved in a gene family if its branch length score (BLS) exceeds threshold T. Multiple BLS thresholds Ti can be used in a single run. In the alignment-free mode, the BLS of a word is computed irrespective of its orientation or relative position within the promoter sequences. Alternatively, in the alignment-based mode, words must appear aligned in the multiple sequence alignment. During the sorting phase, conserved words of all gene families are sorted according to permutation group, i.e. words with the same length and base content are grouped together. In the interfamily phase, permutation groups are handled individually. First, for each word, the conserved family count , i.e. the number of gene families in which the word is conserved with BLS , is established for all BLS thresholds Ti. Next, a background model is created by selecting the median value of the conserved family count of a large number of randomly generated instances of the permutation group, again for each threshold Ti. Finally, a confidence score is computed for each Ti. Words for which and for any threshold Ti are considered to be genome-wide conserved motifs and are retained
3.1.1 Intrafamily step
In the intrafamily step, for each gene family individually, all words with a length between and characters that occur in the promoter sequences of that gene family are exhaustively enumerated. Words are spelled in the restricted IUPAC alphabet that consists of 11 characters: 4 base pairs (ACGT), 6 twofold-degenerate characters (RYSWKM) and the ‘any’ character (N). A maximum of degenerate characters are allowed per candidate motif. The degree of conservation of a word within the gene family is scored using the branch length score (BLS). The intrafamily step can operate in either alignment-free (AF) or alignment-based (AB) mode. In case of AF discovery, the BLS of a word is scored irrespective of its orientation or relative position within the promoter sequences. AB discovery adds the constraint that the words must be aligned in the multiple sequence alignment of the promoter sequences. Words for which the BLS exceeds threshold T are considered to be conserved within the gene family and retained for further processing. Six BLS thresholds Ti (i.e. 15, 50, 60, 70, 90 and 95%) were used in this study. At the end of this phase an exhaustive list of conserved words has been generated for each gene family individually.
3.1.2 Interfamily step
Using the data from the intrafamily step, for each word, the conserved family count , i.e. the number of gene families in which the word is conserved with a BLS , is counted for each BLS threshold Ti. Next, a confidence score , adopted from Stark et al. (2007), is established for each candidate motif (see Section 2). Two thresholds apply: motifs are only retained when and for any of the BLS thresholds Ti. Here, represents a threshold on the conserved family count and is used to eliminate words that are conserved in only few gene families and hence typically do not correspond to TF binding sites. Additionally, ensures that the candidate motif is conserved in a much higher number of gene families than what is expected for such a word (i.e. a word with the same length, base composition and degeneracy) and can hence be considered a potentially functional element. Motifs that satisfy both thresholds are considered to be genome-wide conserved motifs.
Note that the branch length score thresholds Ti on the one hand and conserved family count threshold and confidence score threshold on the other hand are independent. The former provides information about the degree of conservation within a single gene family whereas the latter are indicative of the degree of genome-wide conservation. Certain motifs only show up as being genome-wide conserved for high BLS thresholds. This is typically the case for short and/or highly degenerate motifs, where also permutations of that motif are conserved with a moderate BLS in a rather large number of families, resulting in a low confidence score C. Conversely, a lower BLS threshold allows for the detection of longer motifs with genome-wide conservation in only a subset of the species. Using only a single BLS threshold would therefore limit the sensitivity of the method.
3.2 Exhaustive motif discovery in four monocot species
BLSSpeller was applied to four monocot species: O.sativa ssp. indica (osa), B.distachyon (bdi), S.bicolor (sbi) and Z.mays (zma). Based on conserved gene content and genome organization, these grass species are considered to be a single genetic system (Benntzin and Freeling, 1993), making a comparative motif discovery approach feasible. The dataset consists of 17 724 gene families each containing four orthologous genes (one from each organism). Additionally, 10 636 paralogs are taken into account. Hence, a total of 163 064 regulatory sequences (forward and reverse strands) with a length of 2 kbp each, were analyzed.
BLSSpeller was run on this dataset using both the alignment-free (AF) and the alignment-based (AB) discovery mode on the Amazon Web Services (Elastic MapReduce) cloud infrastructure using 20 nodes of the type m1.xlarge. On every node, 7 map tasks and 2 reduce tasks were run in parallel. The computational requirements are listed in Supplementary Results 2.1. Based on the Amazon pricing of 2014, the financial cost for performing these simulations amounted to 1080$ and 278$ for the AF and AB cases, respectively.
After the intrafamily step and using the AF discovery mode, an aggregated number of 537 billion words were found with a BLS (i.e. conservation in at least two species) over all 17 724 gene families. Note that these words are not necessarily unique as the same word can be conserved in multiple gene families. Using the AB discovery mode, only 82 billion words were found with a BLS . This is because the AB discovery mode imposes the additional constraint that words should appear aligned in the multiple sequence alignment. After the interfamily step and using and , the number of genome-wide conserved motifs amounted to 6.62 and 6.26 billion unique motifs, for the AF and AB discovery mode respectively.
The reason why the number of motifs is high is twofold. First, very relaxed thresholds and were used. It is computationally cheap to further filter this list using more stringent (and biologically meaningful) thresholds (see below). A second reason is the exhaustive, word-based nature of BLSSpeller. If a word is found to be genome-wide conserved, a large number of redundant, highly similar (e.g. slightly more degenerate) variants of that word may also appear in the final output of the method.
3.3 Estimation of the false discovery rate
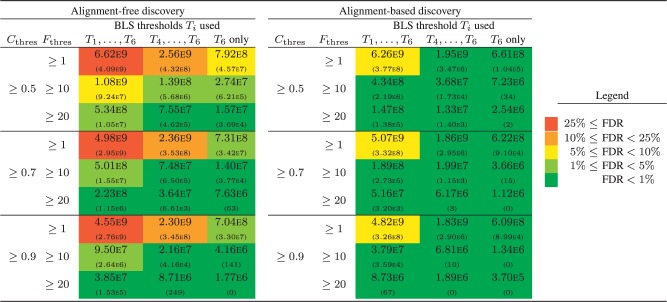
The output of BLSSpeller consists of a list of motifs, along with the conserved family count and conservation score for the six different BLS thresholds Ti. This list was filtered using more stringent thresholds for (i.e. 1, 10 and 20) and (i.e. 0.5, 0.7 and 0.9). Additionally, the list can be filtered by considering only a (stricter) subset of the BLS thresholds Ti (i.e. all six thresholds , three thresholds corresponding to conservation in at least three species, a single threshold T6 corresponding to conservation in all four species). The number of genome-wide conserved motifs for all 27 parameter combinations is shown in Figure 2 for both AF and AB discovery. Clearly, each of the parameters has a strong influence on the final number of motifs in both the AF and AB discovery.
Fig. 2.
Number of genome-wide conserved motifs for both alignment-based and alignment-free discovery for different values of and and different subsets of the six BLS thresholds Ti ( and ). Top number: real Monocot dataset; bottom number between brackets: random dataset (zeroth-order Markov model). The colors represent the false discovery rate (see legend)
In order to assess the specificity of the method for the different parameter combinations, we estimate the false discovery rate (FDR) in an empirical fashion by running BLSSpeller on a random dataset generated using a zeroth-order Markov model (preservation of mononucleotide frequencies) as provided by RSAT (Thomas-Chollier et al., 2008). A more detailed version of Figure 2 is available as Supplementary Figure S4. Additional discussion of the limitations of the FDR analysis, higher-order Markov models and FDR analysis as a function of motif length and degeneracy is provided in Supplementary Results 2.2.
A number of observations can be made. First, for comparable parameter settings, AB discovery has a lower FDR compared to AF discovery. The multiple sequence alignment method increases the specificity for AB discovery as relatively few words will be aligned in random data purely by chance. Second, low values of result in a poor FDR. The reason for this is that in such case, the output consists of a large number of words that are conserved in only a single gene family. If these words are long and/or have low degeneracy, most random permutations of that word will not be conserved in any gene family, resulting in a confidence score . We therefore recommend to impose a certain threshold on the conserved family count. As functional transcription factors typically target multiple genes, this appears to be a biologically reasonable approach. Third, a reasonable threshold on the confidence score should be applied. Applying this threshold filters words for which their random permutations are conserved in a comparable number of gene families. This comprises low-complexity motifs and/or highly degenerate motifs. Finally, a more stringent definition of conservation results in an improved FDR. This can be obtained by imposing higher BLS thresholds Ti.
Even though there is a clear correlation between each of the parameters and the FDR, the exact FDR is hard to predict up front and likely also depends on the dataset that is used. We therefore recommend to run BLSSpeller with relaxed parameter settings on both real and random data, and to filter this output using more stringent parameters until a reasonable FDR is obtained.
For reasonably stringent parameter settings where the FDR , the AF discovery mode reports 3.1–6.8 times more motifs compared to the AB discovery. At first glance, this may seem to be a trivial consequence of the relaxed definition of conservation in the AF methodology. Indeed, a word that is found to be conserved in a gene family with BLS using the AB discovery will also be conserved in the AF method. Therefore, for each word. However, in order to establish the confidence score C(T), the conserved family count F(T) is compared to the corresponding median value of the background distribution (see Section 2). As is also computed using the relaxed, alignment-free definition of conservation, it holds that . Therefore, there is no reason to assume a priori that the AF mode will pick up more motifs than its AB counterpart, as can indeed be observed in Figure 2 for a few parameter combinations, e.g. and BLS thresholds . The reason that we do find more genome-wide conserved motifs for most parameter combinations (including those with good FDR) is because we found a significant number of known motif instances to be misaligned in this relatively highly diverged Monocot dataset. This is exemplified in Section 3.5.
3.4 Motif instance predictions correlate with experimental cis-regulatory datasets
The genome-wide conserved motifs discovered by BLSSpeller are highly redundant. High-scoring, motifs (AF discovery; BLS ; 38 462 976 motifs in total) were mapped back to the promoter sequences and were found to cluster around specific genomic regions (see Supplementary Figs. S8 and S9). Certain loci are covered by thousands of highly similar motif variants. Nevertheless, the high-scoring motifs delineate distinct conserved genomic intervals on the promoter sequences. For these conserved regions, we investigated the accessibility for transcription factor binding in the promoter sequences of rice genes. DNase I hypersensitive sites are associated with regions of open chromatin where the DNA is accessible and as such provide a global perspective on possible protein-binding to the genome. Such regions were recently characterized by Zhang et al. (2012). We performed overlap analysis between conserved genomic regions (as determined by BLSSpeller) and open chromatin regions (see Supplementary Methods 1.2). We found a significant enrichment (3.005 fold) of conserved regions for open chromatin regions (P-value < 0.001) (see Table 1). For a stricter subset of motifs (AF discovery; BLS ; 1 769 963 motifs in total), the fold enrichment increased to 3.796.
Table 1.
Overlap between conserved genomic regions as identified by BLSSpeller and experimentally profiled open chromatin regions in rice and transcription factor binding sites inferred through protein-binding microarrays in rice and maize
| Overlap with experimentally profiled open chromatin regions (OCR) in O.sativa | |||||
|---|---|---|---|---|---|
| BLSSpeller | No. of conserved | No. of OCR regions | No. of conserved regions | No. of rand. conserved regions | enrichment |
| thresholds | regions | within OCR regions | within OCR regions | fold | |
| BLS 15%, 0.9, 20 | 754 205 | 77 247 | 121 026 | 40 277 | 3.005 |
| BLS 95%, 0.9, 20 | 464 229 | 77 247 | 98 681 | 25 996 | 3.796 |
|
Overlap with experimentally profiled TF binding sites (TBS) in O.sativa | |||||
|---|---|---|---|---|---|
| BLSSpeller | No. of conserved | No. of TBS regions | No. of TBS regions within | No. of TBS regions within | enrichment |
| thresholds | regions | conserved regions | rand. conserved regions | fold | |
| BLS 15%, 0.9, 20 | 754 205 | 442 506 | 159 542 | 42 522 | 3.752 |
| BLS 95%, 0.9, 20 | 464 229 | 442 506 | 37 093 | 5 689 | 6.520 |
|
Overlap with experimentally profiled TF binding sites (TBS) in Z.mays | |||||
|---|---|---|---|---|---|
| BLSSpeller | No. of conserved | No. of TBS regions | No. of TBS regions within | No. of TBS regions within | enrichment |
| thresholds | regions | conserved regions | rand. conserved regions | fold | |
| BLS 15%, 0.9, 20 | 828 400 | 482 317 | 156 929 | 66 564 | 2.358 |
| BLS 95%, 0.9, 20 | 454 221 | 482 317 | 35 710 | 10 755 | 3.320 |
Regions are required to fully overlap in order to be scored.
Additionally, we investigated the enrichment of TF binding sites determined in vitro (Weirauch et al., 2014) towards conserved genomic regions in rice and maize. Transcription factor DNA binding specificities are the primary mechanism by which transcription factors recognize genomic features and regulate genes. Recently, a dataset containing a large number of these binding specificities was generated using protein-binding microarrays (PBM) (Weirauch et al., 2014). From this database, PWMs were downloaded for 481 TFs in rice and for 615 TFs in maize. These were mapped onto the respective rice and maize promoters and overlap analysis was performed (see Supplementary Methods 1.2). In rice, of the 754 205 constrained genomic regions (BLS ), 159 542 contain a PBM-based TF binding site, leading to 3.752 fold enrichment (P-value < 0.001). Again, for the stricter subset of conserved motifs (BLS ), fold enrichment increased to 6.520. Maize showed a fold enrichment of 2.358 and 3.320 (P-value < 0.001) respectively. Overall, these analyses revealed that a large part of the conserved non-coding sequences can be accessed by DNA binding proteins and as such can act as functional transcription factor binding sites, and that these conserved non-coding sequences show enrichment for the binding sites of a large number of TFs inferred using PBMs.
3.5 Conservation of the ga2ox1-like KN1 binding site
KNOTTED1 (KN1) transcription factors are involved in the establishment and maintenance of plant meristems and are thought to be conserved among the family of grasses (Bolduc and Hake, 2009). Bolduc et al. (2012) profiled KN1 binding sites in Z.mays using ChIP-seq experiments. The overlapping loci in two samples of immature ears were retained and assigned to the nearest gene within a range of 10 kbp. The ChIP-Seq peaks were found to be mainly situated in the 5′ en 3′ regions extending from the gene but also occur in introns and exons. Thus, a set of candidate KN1-regulated maize genes were identified. For approximately 7% of these genes, a binding site reminiscent of the intronic KN1 binding site in ga2ox1, was identified. For these so-called ga2ox1-like KN1 binding sites, a Position Weight Matrix (PWM) was derived by Bolduc et al. (2012). Translated to the IUPAC alphabet, this PWM corresponds to TGAYNGAYDGAY.
We investigate whether BLSSpeller is able to discover the ga2ox1-like KN1 motifs and binding sites through a comparative study of the four monocot species. From the BLSSpeller output, all genome-wide conserved motifs of length 12 that match the ga2ox1-like KN1 PWM identified by Bolduc et al. (2012) were retained. Using alignment-free discovery, and using and (FDR , see Fig. 2), 51 genome-wide conserved motif variants are identified. In total, these motifs are conserved in 165 gene families with a BLS (i.e. conservation in at least two species). From the 51 identified motif variants, only 19 are required to explain the conservation in all 165 gene families. These essential motifs are listed in Table 2 along with their respective metrics. In turn, these gene families contain 213 maize genes in total, 51 of which were also identified in Bolduc et al. (2012). These results were compared to those obtained by Fastcompare (Elemento and Tavazoie, 2005; see Supplementary Results 2.3), a method that also performs motif discovery in an alignment-free and exhaustive manner. However, Fastcompare is limited to the exact ACGT alphabet and pairwise species comparisons. Because of these limitations, Fastcompare could identify only 36 maize gene targets, 10 of which were also identified by Bolduc et al. (2012).
Table 2.
List of genome-wide conserved ga2ox1-like KN1 motif variants identified by BLSSpeller using both AF and AB discovery
|
Alignment-free discovery |
Alignment-based discovery |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| KN1 motif variant | KN1 motif variant | ||||||||
| TGATNGATKGAY | 59 | 0.93 | 75 | 24 | TGATNGAYGGAY | 11 | 0.91 | 10 | 3 |
| TGATNGAYKGAT | 59 | 0.93 | 74 | 20 | TGATNGATKGAY | 11 | 0.82 | 11 | 3 |
| TGAYNGATKGAT | 54 | 0.93 | 68 | 21 | TGAYNGACKGAC | 10 | 0.90 | 11 | 3 |
| TGATNGAYWGAT | 40 | 0.88 | 50 | 11 | TGAYGGAYGGAY | 9 | 1.00 | 9 | 3 |
| TGAYNGAYTGAT | 36 | 0.89 | 48 | 11 | TGATNGAYRGAT | 9 | 0.89 | 10 | 3 |
| TGAYTGAYTGAY | 33 | 0.97 | 42 | 9 | TGAYNGAYTGAC | 8 | 0.88 | 9 | 2 |
| TGATNGAYTGAY | 32 | 0.88 | 40 | 7 | TGACNGAYTGAY | 8 | 0.88 | 10 | 3 |
| TGAYNGATWGAT | 31 | 0.84 | 42 | 12 | TGACNGACWGAY | 7 | 0.86 | 7 | 2 |
| TGATNGATWGAY | 30 | 0.83 | 36 | 9 | TGACAGAYRGAY | 3 | 1.00 | 4 | 0 |
| TGATNGATRGAY | 29 | 0.86 | 39 | 9 | |||||
| TGAYNGATRGAT | 27 | 0.85 | 37 | 9 | |||||
| TGATNGAYRGAT | 26 | 0.85 | 35 | 8 | |||||
| TGAYNGATTGAY | 25 | 0.84 | 34 | 7 | |||||
| TGAYNGATGGAY | 24 | 0.88 | 35 | 9 | |||||
| TGATNGAYGGAY | 24 | 0.88 | 31 | 8 | |||||
| TGAYTGAYWGAT | 22 | 0.91 | 27 | 6 | |||||
| TGAYNGACTGAY | 22 | 0.91 | 28 | 9 | |||||
| TGAYNGAYTGAC | 21 | 0.90 | 27 | 8 | |||||
| TGAYNGACKGAC | 20 | 0.90 | 25 | 10 | |||||
| Union (all variants) | 165 | – | 213 | 51 | Union (all variants) | 37 | – | 41 | 10 |
denotes the number of gene families in which the motif is conserved with while denotes the corresponding confidence score. denotes the number of maize genes contained in the gene families while denotes the intersection with experimentally profiled maize genes.
Similarly, using BLSSpeller’s alignment-based discovery mode, conservation with a BLS is observed in only 37 gene families, even with very relaxed thresholds ( and ) (FDR ). The nine essential motif variants required to explain this conservation are listed in Table 2. The 37 gene families contain 41 maize genes, 10 of which are also reported in Bolduc et al. (2012). Inspection of the promoter sequence alignments of the gene families reveals that the ga2ox1-like KN1 variants are often not aligned, either because the motif instances in the different species are located at entirely different positions in the promoter sequences or because they appear on different strands (see Supplementary Table 3). Therefore, alignment-based motif discovery approaches such as BLSSpeller in AB mode or the ‘mini motifs’ approach as used by Stark et al. (2007) suffer from reduced sensitivity on diverged datasets.
4 Conclusion
A novel phylogenetic footprinting approach was developed for the sensitive discovery of conserved cis-regulatory elements even in diverged sequences. Using IUPAC strings as motif model and using the MapReduce programming model to enable distributed computing, it was shown that it is feasible to compute all genome-wide conserved words in a large dataset in an exhaustive manner. For a given false discovery rate, it was demonstrated that an alignment-free approach detects more conserved words than an alignment-based approach. Even though millions of genome-wide conserved motifs were identified by our method, mapping of these motifs to the promoter sequences results in constrained conserved genomic regions. It was shown that these conserved regions were significantly enriched for experimentally profiled open chromatin regions in rice and for TF binding sites inferred through protein-binding microarrays in rice and maize. Finally, it was shown that the alignment-free approach shows an improved recovery of the ga2ox1-like KN1 binding site, compared to the alignment-based approach or competing methods.
Supplementary Material
Acknowledgements
We acknowledge the support of Ghent University (Multidisciplinary Research Partnership ‘Bioinformatics: From Nucleotides to Networks’) and Dries Vaneechoutte, Kenneth Hoste, Ewan Higgs and Stijn De Weirdt for technical assistance. Part of the computational resources (Stevin Supercomputer Infrastructure) and services used in this work were provided by the VSC (Flemish Supercomputer Center), funded by Ghent University, the Hercules Foundation and the Flemish Government department EWI.
Funding
J.V.D.V. is supported by the Institute for the Promotion of Innovation by Science and Technology in Flanders (IWT-Vlaanderen).
Conflict of Interest: none declared.
References
- Bailey T.L., et al. (2009) MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. , 37, W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benntzin J., Freeling M. (1993) Grasses as a single genetic system: genome composition, collinearity and compatibility. Trends Genet. , 9, 259–261. [DOI] [PubMed] [Google Scholar]
- Berezikov E., et al. (2004) CONREAL: conserved regulatory elements anchored alignment algorithm for identification of transcription factor binding sites by phylogenetic footprinting. Genome Res. , 14, 170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M., Tompa M. (2002) Discovery of regulatory elements by a computational method for phylogenetic footprinting. Genome Res. , 12, 739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N., Hake S. (2009) The maize transcription factor knotted1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell , 21, 1647–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc N., et al. (2012) Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. , 26, 1685–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley R.K., et al. (2010) Binding site turnover produces pervasive quantitative changes in transcription factor binding between closely related Drosophila species. PLoS Biol. , 8, e1000343+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmack C.S., et al. (2007) PhyloScan: identification of transcription factor binding sites using cross-species evidence. Algorithms Mol. Biol. , 2, 1+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish-Bowden A. (1985) Nomenclature for incompletely specified bases in nucleic acid sequences: recommendations 1984. Nucleic Acids Res. , 13, 3021–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M., Dai H.K. (2007) A survey of DNA motif finding algorithms. BMC Bioinformatics , 8, S21+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Witte D., et al. (2013) A parallel, distributed-memory framework for comparative motif discovery. Parallel Process. Appl. Math. , 8385, 268–277. [Google Scholar]
- Dean J., Ghemawat S. (2004) MapReduce: simplified data processing on large clusters. Operat. Syst. Des. Implement. , 53, 137–150. [Google Scholar]
- Elemento O., Tavazoie S. (2005) Fast and systematic genome-wide discovery of conserved regulatory elements using a non-alignment based approach. Genome Biol. , 6, R18+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskin E., Pevzner P.A. (2002) Finding composite regulatory patterns in DNA sequences. Bioinformatics (Oxford, England) , 18, 354–363. [DOI] [PubMed] [Google Scholar]
- Ettwiller L., et al. (2005) The discovery, positioning and verification of a set of transcription-associated motifs in vertebrates. Genome Biol. , 6, R104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegerich R., et al. (1999) Efficient implementation of lazy suffix trees. In: Vitter J.S., Zaroliagis C. (eds) International Workshop on Algorithm Engineering , pp. 30–42. Springer-Verlag, London, UK. [Google Scholar]
- Gordân R., et al. (2010) Finding regulatory DNA motifs using alignment-free evolutionary conservation information. Nucleic Acids Res. , 38, e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J.D., et al. (2000) Computational identification of Cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. , 296, 1205–1214. [DOI] [PubMed] [Google Scholar]
- Kellis M., et al. (2003) Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature , 423, 241–254. [DOI] [PubMed] [Google Scholar]
- Kumar L., et al. (2010) Systematic discovery of regulatory motifs in Fusarium graminearum by comparing four Fusarium genomes. BMC Genomics , 11, 208+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S., et al. (2004) cWINNOWER algorithm for finding fuzzy dna motifs. J. Bioinform. Comput. Biol. , 2, 47–60. [DOI] [PubMed] [Google Scholar]
- Liu X., et al. (2001) BioProspector: discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. , 6, 127–138. [PubMed] [Google Scholar]
- Marsan L., Sagot M.F. (2000) Algorithms for extracting structured motifs using a suffix tree with an application to promoter and regulatory site consensus identification. J. Comput. Biol. , 7, 345–362. [DOI] [PubMed] [Google Scholar]
- Marschall T., Rahmann S. (2009) Efficient exact motif discovery. Bioinformatics (Oxford, England) , 25, 356–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi G., et al. (2001) An algorithm for finding signals of unknown length in DNA sequences. Bioinformatics (Oxford, England) , 17, S207–S214. [DOI] [PubMed] [Google Scholar]
- Pollard D.A., et al. (2004) Benchmarking tools for the alignment of functional noncoding DNA. BMC Bioinformatics , 5, 6+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proost S., et al. (2009) PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell Online , 21, 3718–3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke A.R., et al. (2011) Evolutionary divergence and limits of conserved non-coding sequence detection in plant genomes. Nucleic Acids Res. , 39, 6029–6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satya R.V., Mukherjee A. (2004) Pruner: algorithms for finding monad patterns in DNA sequences. In: CSB , pp. 662–665. IEEE Computer Society. [Google Scholar]
- Sieglaff D.H., et al. (2009) Comparative genomics allows the discovery of cis-regulatory elements in mosquitoes. Proc. Natl. Acad. Sci. , 106, 3053–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggia E.D. (2005) Computational methods for transcriptional regulation. Curr. Opin. Genet. Dev. , 15, 214–221. [DOI] [PubMed] [Google Scholar]
- Stark A., et al. (2007) Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature , 450, 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A.R., et al. (2008) DIALIGN-TX: greedy and progressive approaches for segment-based multiple sequence alignment. Algorithms Mol. Biol. AMB , 3, 6+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thijs G., et al. (2002) A Gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J. Comput. Biol. , 9, 447–464. [DOI] [PubMed] [Google Scholar]
- Thomas-Chollier M., et al. (2008) RSAT: regulatory sequence analysis tools. Nucleic Acids Res. , 36, W119–W127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bel M., et al. (2012) Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. , 158, 590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helden J., et al. (2000) Discovering regulatory elements in non-coding sequences by analysis of spaced dyads. Nucleic Acids Res. , 28, 1808–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataram S., Fay J.C. (2010) Is transcription factor binding site turnover a sufficient explanation for cis-regulatory sequence divergence? Genome Biol . Evol. , 2, 851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. (2005) Identifying the conserved network of cis-regulatory sites of a eukaryotic genome. Proc. Natl. Acad. Sci. , 102, 17400–17405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Yu X. (2007) Comparative analysis of regulatory motif discovery tools for transcription factor binding sites. Genomics Proteomics Bioinf. , 5, 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch M.T., et al. (2014) Determination and inference of eukaryotic transcription factor sequence specificity. Cell , 158, 1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., et al. (2008) Discovering regulatory motifs in the Plasmodium genome using comparative genomics. Bioinformatics (Oxford, England) , 24, 1843–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., et al. (2005) Systematic discovery of regulatory motifs in human promoters and 3[prime] UTRs by comparison of several mammals. Nature , 434, 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., et al. (2012) High-resolution mapping of open chromatin in the rice genome. Genome Res. , 22, 151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.