Abstract
Recent genetic data on schizophrenia (SCZ) have suggested that proteins of the postsynaptic density of excitatory synapses have a role in its etiology. Mutations in the three SHANK genes encoding for postsynaptic scaffolding proteins have been shown to represent risk factors for autism spectrum disorders and other neurodevelopmental disorders. To address if SHANK2 variants are associated with SCZ, we sequenced SHANK2 in 481 patients and 659 unaffected individuals. We identified a significant increase in the number of rare (minor allele frequency<1%) SHANK2 missense variants in SCZ individuals (6.9%) compared with controls (3.9%, P=0.039). Four out of fifteen non-synonymous variants identified in the SCZ cohort (S610Y, R958S, P1119T and A1731S) were selected for functional analysis. Overexpression and knockdown-rescue experiments were carried out in cultured primary hippocampal neurons with a major focus on the analysis of morphological changes. Furthermore, the effect on actin polymerization in fibroblast cell lines was investigated. All four variants revealed functional impairment to various degrees, as a consequence of alterations in spine volume and clustering at synapses and an overall loss of presynaptic contacts. The A1731S variant was identified in four unrelated SCZ patients (0.83%) but not in any of the sequenced controls and public databases (P=4.6 × 10−5). Patients with the A1731S variant share an early prodromal phase with an insidious onset of psychiatric symptoms. A1731S overexpression strongly decreased the SHANK2-Bassoon-positive synapse number and diminished the F/G-actin ratio. Our results strongly suggest a causative role of rare SHANK2 variants in SCZ and underline the contribution of SHANK2 gene mutations in a variety of neuropsychiatric disorders.
Introduction
Schizophrenia (SCZ) is a complex mental disorder. According to the DSM (Diagnostic and Statistical Manual of Mental Disorders) it is characterized by delusions, hallucinations, disorganized speech, grossly disorganized or catatonic behavior, and negative symptoms. Its life-time prevalence worldwide is about 1%.1 Genetically, SCZ is a heterogeneous heritable disorder.2 Genome-wide association studies,3, 4, 5, 6, 7, 8 copy number variation (CNV) analysis9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20 and exome-sequencing studies21, 22, 23, 24, 25, 26, 27 have pinpointed to distinct loci and genes associated with SCZ. A large fraction of the associated genes are involved in excitatory synapse formation, function and plasticity.
The SHANK protein family is composed of three multidomain scaffolding proteins residing at the postsynaptic density of excitatory glutamatergic synapses.28, 29 SHANK proteins are known to form large homomeric and heteromeric complexes. By numerous specific protein–protein interactions, SHANKs are either directly or indirectly linked to other structural proteins, cell adhesion molecules, receptors, ion channels and to actin-interacting proteins at the postsynaptic density.30 Mutations of all three SHANK family members, SHANK1–3, have already been associated to neurodevelopmental disorders like autism spectrum disorders (ASDs) or intellectual disability (ID).31, 32, 33, 34
There is recent evidence for a genetic as well as a biological overlap between ASD and SCZ. In addition, further overlaps with other neuropsychiatric disorders may exist depending on the genetic background and exposure to environmental risk factors.35 For ASD and SCZ, the same pathways and networks appear to be affected, which are converging to excitatory synapses, possibly affecting synaptic plasticity. As several synaptic proteins like neurexins and neuroligins are interaction partners of the SHANKs and associate with both disorders, the question arises if variants in the SHANK genes themselves can be associated with SCZ.
So far, common variants in the SHANK1 gene were reported to be associated with reduced auditory working memory in SCZ36 and a rare de novo loss-of-function mutation affecting SHANK1 was recently found in a large-scale exome-sequencing study.21 In addition, two de novo SHANK3 mutations were identified in a cohort of 185 SCZ individuals (R536W, R1117X).37 Overexpression of the R1117X variant in hippocampal neurons led to an accumulation of mutated protein within the nucleus of the neuron, altering the transcription of several SCZ risk genes such as Synaptotagmin 1 and LRRTM1.38 These findings provided first evidence that SHANK genes might indeed contribute to the etiology of SCZ.
In this study, we focused on SHANK2, the only SHANK family member for which no association with SCZ was reported so far. SHANK2 mutations were identified in ID and ASD patients.32, 34, 39 Two different SHANK2 knock-out mouse models are available that show alterations in N-methyl-D-aspartate glutamate receptor (NMDAR) function40, 41 and there is good evidence that NMDAR signaling might be impaired in SCZ.42 In addition, recent CNV analysis and exome-sequencing studies in SCZ patients have reported an enrichment of de novo mutations in members of the NMDAR signaling complex.14, 21
Therefore, the aim of our study was to identify variants in the SHANK2 gene in a SCZ patient cohort, analyze their impact on a functional level and determine a possible association with SCZ. To that end, we sequenced the SHANK2 gene in 481 SCZ patients and in 659 controls. Based on the comparison to unaffected controls, on mutation frequency rates and on in silico prediction results, we selected four missense variants for further functional testing.
Materials and methods
Ethics statement
The study was approved by the Ethics Committees of the Universities of Heidelberg and Bonn, Germany.
Patients
The sample comprised 481 cases (275 males and 206 females; mean age=32.6+10.4 years) drawn from consecutive admissions to inpatient psychiatric units in Germany. A total of 403 cases fulfilled DSM-IV (DSM, 4th ed.) diagnostic criteria for SCZ, 71 for schizoaffective disorder and 7 for schizophreniform disorder. Diagnostic assessment involved a best estimate approach and the Interviews for Psychiatric Genetic Studies (IPGS), a comprehensive inventory for phenotype characterization.43 The IPGS comprises: (i) the Structured Clinical Interview for DSM-IV Disorders; (ii) the Operational Criteria Checklist for Psychotic Illness program;44 (iii) a review of medical records and (iv) family history assessment. Our cohort comprised purely schizophrenic patients (according to the diagnostic criteria of the DSM-IV that the disturbance is not due to a general medical condition). Patients with mental retardation, which could have confounded the diagnosis of SCZ, were excluded from the cohort. All subjects were of German descent. All participants were informed about the study and provided written informed consent before study inclusion. Samples of the four individuals with the A1731S variant were subjected to genome-wide genotyping as part of a larger study. Using these data, genome-wide identity-by-state scores were calculated. The respective pair wise identity-by-state scores are <1.65, indicating that no close biological family relationship exists between any two of the individuals. Short clinical reports for these individuals are available in Supplementary Information 1. As a control, DNA from unaffected individuals of European ancestry was used.32
Sequencing
Sequencing primers that cover the SHANK2-SH3 isoform were described previously (Supplementary Table 1).32 PCR amplifications were performed with Paq5000 polymerase (Stratagene, La Jolla, CA, USA) or with Expand High Fidelity PCR System (Roche, Mannheim, Germany). Generated products were analyzed on agarose gels, purified and sequenced directly using the DYEnamic ET Terminator Cycle Sequencing Kit (GE Healthcare, Munich, Germany) and the MegaBACE 1000 DNA Analysis System (GE Healthcare).
Data analysis
Simple burden test for rare SHANK2 missense variants with minor allele frequency<1% was performed, by comparing their accumulated frequency in SCZ cases and controls. Data were analyzed with the statistics program BIAS for Windows, using a two-sided χ2 test with Yates correction (University of Frankfurt/Mainz, Mainz, Germany). In addition, a set-based test implemented in VEGAS45 was applied to the SCZ2 data set of the Psychiatric Genomics Consortium (full single-nucleotide polymorphism (SNP) results, http://www.med.unc.edu/pgc/files/resultfiles/scz2.snp.results.txt.gz%20%20). The test investigates evidence for association on a per gene basis by considering the P-values of all SNPs for each gene including 50 kb from the 5' and 3' UTR, while taking into account the number of SNPs per gene and linkage disequilibrium between markers. All SHANK2 missense variants identified in SCZ patients were analyzed with different prediction programs (MutationTaster, PolyPhen2, SIFT; Supplementary Table 2, December 2011). Prediction programs changed over time and the outcome was slightly different in March 2014 (Supplementary Table 3). Variant A1731S was assessed for potential splicing alterations with the Alamut Visual Software (Interactive Software, Rouen, France), which contains the algorithms SpliceSiteFinder, MaxEntScan, NNSPLICE, GeneSplicer, Human Splicing Finder, ESE-Finder and RESCUE-ESE.
Plasmid constructs
All SHANK2 constructs were cloned in pENTR vectors (Invitrogen, Carlsbad, CA, USA). pENTR2B-SHANK2-SH3 WT was generated in our laboratory.46 pENTR2B-SHANK2-SH3 S610Y, R958S, P1119T, and A1731S were generated using the QuikChange Lightning Site-directed Mutagenesis Kit (Stratagene). The coding sequence of each clone was sequenced to confirm the introduced mutation and to rule out additional unwanted mutations. The described SHANK2E isoform clones were created by modifying the corresponding pENTR2B-SHANK2-SH3 plasmid using the SLiCE cloning system.47 All PCR amplified regions in the constructs were sequenced. Shank3 was subcloned from pGW1-HA-Shank3 expression construct (gift from Dr C Sala) into pENTR3C.48 SHANK expression constructs were generated by transfer to the corresponding DEST vectors using the Gateway recombinant technology (Invitrogen). Detailed description of the cloning procedures and complete list of all used DEST vectors is presented within Supplementary Information 2. The small hairpin RNA constructs were previously described.46 The pGFP(C3)-Vinculin construct was generated in the K. Hahn lab (Addgene, Cambridge, MA, USA, Plasmid 30312). RFP-actin construct was a generous gift from Dr U Engel (NIC, Heidelberg, Germany).
Cell culture and transfection
COS-7 and HEK293 cells were maintained in DMEM media (GIBCO, Carlsbad, CA, USA) supplemented with 10% fetal calf serum, 1 mM glutamine, 100 U ml−1 penicillin and streptomycin. Cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's guidelines.
Protein, antibodies and western blots
Protein isolation and western blot analysis were performed using standard protocols. We used the following primary antibodies: mouse anti-Shank2 (UC Davis/NIH NeuroMab Facility, Davis, CA, USA), rabbit anti-actin (Cytoskeleton, Denver, CO, USA) and rabbit anti-GAPDH (Abcam, Cambridge, UK). IRDye 800CW and IRDye 680 (LI-COR Biosciences, Lincoln, NE, USA) served as secondary antibodies. All western blot band quantification procedures were performed using Image Studio Lite 3.1 software (LI-COR Biosciences).
Imaging and quantitative analyses of spine morphology and dendritic branching patterns
Dissociated primary hippocampal neurons from rat were prepared and transfected as described.46 To determine the effect on spine volume, the mCherry-SHANK2-SH3 constructs were co-transfected with GFP at 10 days in vitro (DIV) and processed for confocal microscopy at 18 DIV. High-resolution z-stack images of GFP-positive neurons were taken with the × 63 objective of the confocal laser scanning microscope (Carl Zeiss, Jena, Germany). The imaging was performed by a second independent person who was blind to the case–control status of samples. Neurons displaying pyramidal morphology and positive for the respective fluorescence markers were chosen randomly and spine volumes were subsequently analyzed with the Fiji software. Signal intensity levels of each construct were similar, excluding the possibility of effects of transfection efficiencies. To investigate the dendritic branching patterns, we took fluorescent images of randomly selected neurons with upright Ni-E microscope (Nikon, Tokyo, Japan), equipped with Nikon Plan Apo λ x20 objective and DS-Qi1Mc camera (Nikon). For digitalization and analysis of the dendrite morphology, we used the ‘Bonfire' program according to its manual.49
Quantitative analysis of rat hippocampal neurons for synaptic density analysis
Primary hippocampal neurons from embryonic day 18/19 (E18/19; Sprague–Dawley rats, Animal Facility, Ulm University or JANVIER LABS, France) were prepared and cultured as described elsewhere.40 After 14 days in vitro (DIV14), neurons were transfected with 1 μg DNA per well (30 000 cells per well). The cells were fixed at DIV15 with 4% paraformaldehyde, washed with phosphate-buffered saline and treated with Triton X-100 and fetal calf serum (alternatively horse serum and BSA) for permeabilization and blocking, respectively. Immunostaining was performed with anti-Bassoon antibody (1:500–1:800) and a respective second antibody. 4′,6-Diamidino-2-phenylindole was used for nuclei staining. Images of randomly chosen transfected neurons with pyramidal cell morphology and an intact nucleus were taken by a second person with a fluorescent microscope at x40 magnification and cells were analyzed with Axiovision software (Carl Zeiss). For SHANK2 synaptic clustering, we counted SHANK2 puncta co-localizing with Bassoon along >1 dendrite. For presynaptic density, we counted Bassoon puncta along >1 dendrite. Significances were evaluated via Student's t-test with 10–17 cells per condition from two to four preparations. P-values <0.05 were stated significant (*P<0,05, **P<0,01, ***P<0,001).
Live cell imaging
We used four-well Lab-Tek II Chambers (Sigma-Aldrich, St Louis, MO, USA) filled with Opti-MEM media (GIBCO) supplemented with 10% fetal calf serum and 1 mM glutamine to seed COS-7 cells at densities of 15 000 cells per well. On the next day, cells were transfected with Lipofectamine 2000. After 24 h of incubation, image acquisition was performed with an inverted Ti microscope (Nikon, Düsseldorf, Germany) equipped with widefield fluorescence and objective total internal reflection fluorescence (TIRF) illumination. During imaging, cells were kept in an on-stage incubation chamber (TokaiHit, Shizuoka-ken, Japan) with control over temperature, CO2 concentration and humidity. We used Nikon Apo TIRF x60 numerical aperture 1.49 objective (Nikon), adjustable for thickness/temperature and Andor iXon3 DU-897 single photon detection EMCCD camera (Andor Technology, Belfast, UK) or Ocra AG camera (Hamamatsu Photonics, Shizuoka, Japan).
F- to G-actin ratio
The F- to G-actin ratio was measured with an Actin Polymerization Assay kit (BK037, Cytoskeleton) according to the provided manual. HEK293 cells were co-transfected with SHANK2 constructs in tag-free pCS2-DEST vector and RFP–actin. After 2 days of incubation, cells were resuspended in F-actin stabilization buffer containing protease inhibitor mix and 1 mM ATP. F-actin was pelleted by centrifugation at 100 000 g/37 °C for 60 min (Optima LE-80 k Ultracentrifuge, SW55Ti rotor, Beckman Coulter, Brea, CA, USA). Actin quantification was performed by western blot using the provided rabbit anti-actin antibody (Cytoskeleton) and secondary anti-Rabbit IRDye 800CW antibody (LI-COR Biosciences). The F- and G-actin band intensities were determined using an Odyssey infrared imaging system (LI-COR Biosciences). F- and G-actin values for SHANK2E WT and SHANK2E A1731S were normalized to the SHANK2 expression level using data from additional western blot experiment with anti-Shank2 and anti-GAPDH antibodies.
Results
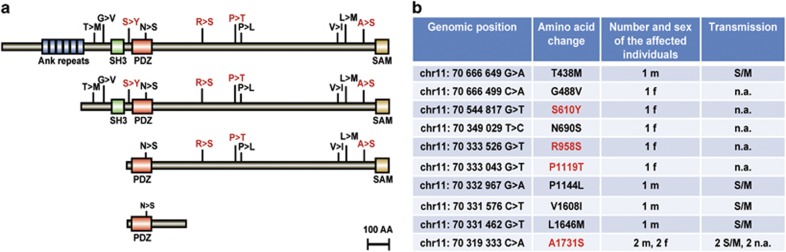
The SHANK2 protein comprises different functional domains including an N-terminal ankyrin repeat, a Src homology 3 (SH3), a PSD95/DLG/ZO1 (PDZ) and a C-terminal sterile alpha motif domain, which exist in different combinations in different isoforms (Figure 1a). Four different isoforms have been described: SHANK2E (AB208025), SHANK2-SH3 (AB208026), SHANK2-PDZ (AB208027) and SHANK2-Short (AF141901).27
Figure 1.
SHANK2 missense variants identified in schizophrenia (SCZ) individuals. (a) Four different human SHANK2 isoforms SHANK2E (AB208025), SHANK2-SH3 (AB208026), SHANK2-PDZ (AB208027) and SHANK2-Short (AF141901) are shown with positions of the missense mutations found in patients with SCZ. The different protein domains are indicated, starting with the N-terminal ankyrin repeats, followed by the SH3 (Src homology 3), the PDZ (postsynaptic density 95/discs large/zona occludens-1) and the C-terminal SAM (sterile alpha motif) domain. The variants selected for functional analysis are labeled in red; AA, amino acid. (b) SHANK2 missense variants found only in SCZ patients. Genomic positions are given according to GRCh37/hg19 assembly and the AA positions correspond to NP_036441.2 sequence. F, female, m, male, n.a., no information available. S/M, transmission from mother to son.
To identify genetic variants in SHANK2, we sequenced the exons including the exon–intron boundaries of the SHANK2-SH3 isoform using genomic DNA isolated from 481 SCZ patients (from 177 individuals parental DNA and parental clinical information were available) and 659 unaffected controls. In total, we identified 10 intronic, 12 synonymous and 15 non-synonymous variants (Supplementary Table 3). The total number of missense variants in the SCZ and control group was comparable (Supplementary Table 4). When we analyzed the frequency of rare missense variants (minor allele frequency<1%) in SCZ and control individuals, 6.9% of SCZ patients (33/481) and 3.9% of controls (26/659) were identified as carriers, pointing to a higher frequency of rare SHANK2 missense variants in SCZ (P=0.039, χ2 test with Yates correction; Supplementary Table 5). SCZ individuals from our study also had a higher frequency (3.6%, 17/466) of rare probably deleterious variants (PolyPhen2) than controls (2.5%, 27/1063).
When we tested the association of the SHANK2 gene in the largest SCZ genome-wide association study to date,3 a significant P-value (P=0.012) resulted, indicating also the contribution of common SHANK2 variants to SCZ etiology. In total, 244 SNPs were considered for the analysis. The SNP with the best P-value (rs11236491, P=0.00012) resided in an intron of the gene.
Ten of the fifteen missense variants in SCZ individuals exclusively occurred in the patient group but not in any of sequenced controls. Five of the variants were inherited (T438M, P1144L, V1608I, L1646M, A1731S, Figure 1b), all with a transmission from the apparently unaffected mother to the schizophrenic son. The inheritance status of the remaining variants (G488V, S610Y, N690S, R958S, P1119T, Figure 1b) could not be determined due to lack of parent DNA. Altogether, we detected rare novel SHANK2 missense variants in 2.7% of SCZ individuals. The schizophrenic subtype of these individuals with SHANK2 variants varied: six suffer from the paranoid, four from the disorganized, one from the catatonic, one from the residual subtype and one individual was classified with schizoaffective disorder (Supplementary Table 6 and Supplementary Information 1).
The most frequent SHANK2 variant in our study was the amino-acid exchange A1731S, which was identified in four unrelated SCZ patients, the only rare variant identified more than once. It was inherited from apparently healthy parents in two of the four patients. Variant A1731S occurred in 0.83% of SCZ individuals and was not detected in 5338 controls (HEVS n=4300, TGP n=379, sequenced controls n=659). In the Human exome variant server, this region has good sequence coverage (67x) and therefore we consider this set of data as an appropriate control. Comparing the frequencies between SCZ cases and controls, we detected a significant difference (P=4,6 × 10−5; two-sided Fisher's exact test) for A1731S.
To address the functional significance, we analyzed four of the newly discovered SHANK2 variants (S610Y, R958S, P1119T and A1731S). Three of them (S610Y, R958S and P1119T) were selected because they had the highest expected probability for a functional effect by three different in silico prediction tools (MutationTaster, PolyPhen2 and SIFT; Supplementary Table 2). Variant P1144L was not considered, despite a high prediction score, because it resides in proximity to P1119T and the change in physico-chemical properties is less severe. On sequence level, the conservation of an affected amino-acid position is the main parameter influencing the prediction outcome by the used tools. Furthermore, structure-based predictions consider the physico-chemical characteristics of the amino acids and the effect of a given substitution on the protein structure and properties. Even though the A1731S variant was predicted to be benign by several (but not all) prediction programs, it was chosen because of its repeated occurrence in SCZ patients and absence in controls. Splicing was not affected by A1731S as predicted by several splice site prediction algorithms of the Alamut software.
Three of the selected variants (R958S, P1119T, A1731S) reside in the three major SHANK2 isoforms (SHANK2E, SHANK2-SH3 and SHANK2-PDZ) that were already confirmed on protein level (Figure 1a). The S610Y variant is not present in SHANK2-PDZ nor SHANK2-Short isoforms but localizes to a highly conserved region between the SH3 and the PDZ domain and was additionally found in an individual with ID from a previous study.32
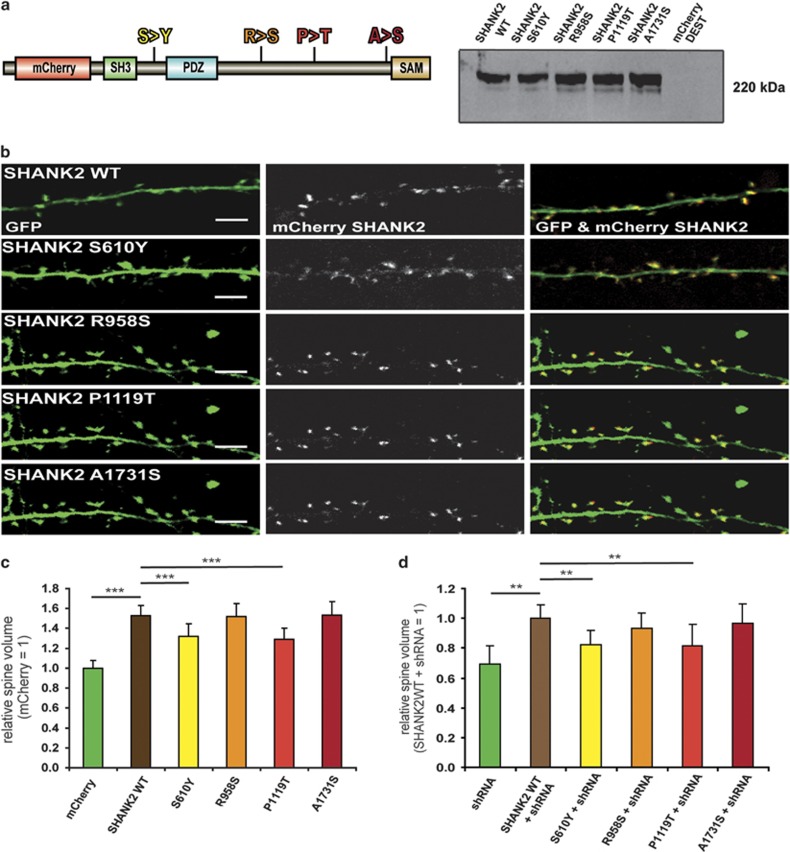
In a first step, we cloned expression constructs for SHANK2 wild type and the variants with N-terminal fusion of mCherry corresponding to the SHANK2-SH3 isoform (Figure 2a). Overexpression of the different constructs in HEK293 cells, followed by western blot analysis with a SHANK2-specific primary antibody, revealed an identical size of the wild-type protein and the mutants (Figure 2a). To study the effects of SHANK2 variants on primary hippocampal neuron morphology, we co-transfected the four variants and the wild-type construct with a GFP-expression vector for whole cell filling (Figure 2b). Laser scanning confocal microscopy was used to obtain z-stack images of mCherry- and GFP-positive neurons to measure the fluorescence intensity of GFP within the two-dimensional projections, which in turn allowed us to calculate the relative spine volumes. Overexpression of SHANK2-SH3 wild type significantly increased the spine volume, compared with neurons transfected with GFP and empty mCherry-vector (Figure 2c). Two of the four variants, S610Y and P1119T, did not increase spine volume to a similar extent as SHANK2 wild type (Figure 2c), pointing to an impairment of protein function. We also measured spine density and the branching of the dendritic arbor. To quantify dendritic arbor complexity, we performed Sholl analysis and analyzed the total number of branching points and terminal points as well as the total neurite length per neuron. All tested four variants showed no significant differences to SHANK2 wild type suggesting that spine density and dendrite branching were not affected (Supplementary Figures 1 and 2).
Figure 2.
Functional analysis of SHANK2 mutations in primary hippocampal neurons. (a) SHANK2 wild type and mutant (S610Y, R958S, P1119T, A1731S) constructs as N-terminal mCherry fusions. Western blot analysis in HEK293 cells displaying the sizes of the different constructs. (b) Representative hippocampal neurons transfected with GFP and mCherry-SHANK2-wild type (WT) or mCherry-mutants. (c) Quantification of the relative dendritic spine volume and the spine density in primary hippocampal neurons overexpressing the different SHANK2 constructs. (d) Rescue of the Shank2 knockdown. Human SHANK2 wild-type protein normalizes spine volume in Shank2 knockdown neurons, whereas the SHANK2-S231Y and the P740T mutant have lost this ability. No effect was seen on spine density. All values are shown as mean±standard deviation. The comparisons of mean differences between groups were made using two-way analysis of variance test including the different conditions and controlling for the replicates followed by Scheffé post hoc test. **P⩽0.01, ***P⩽0.001; (c and d; n=3 experiments, 18 neurons for each condition).
Next we investigated the ability of the SCZ mutants to rescue morphological effects of Shank2 knockdown in primary hippocampal neurons using previously designed and validated small hairpin RNA against rat Shank2.46 The initial Shank2 knockdown experiment revealed a decrease in spine volume as well as a shift of the predominant spine morphology from mature mushroom-like toward immature, thin and filopodia-like. In addition, we observed an increase in dendritic branching but no effect on spine density, as previously described.46 The four different SHANK2 mutants and the wild-type constructs were co-transfected to rescue the Shank2 knockdown phenotype. The S610Y and P1119T mutants showed a significantly reduced ability to rescue the small hairpin RNA-induced decrease in dendritic spine volume compared with the SHANK2 wild type (Figure 2d).
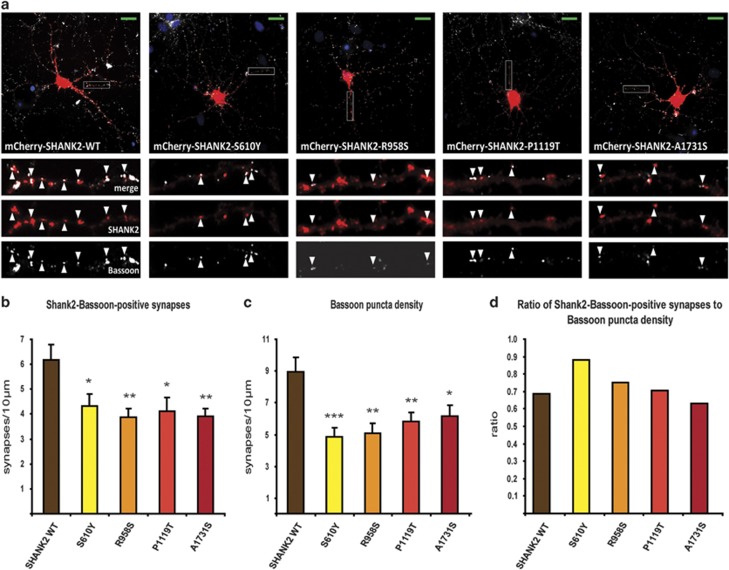
To investigate whether the SHANK2 mutants identified in SCZ individuals have an influence on synaptic clustering and contacts, immune stainings with the presynaptic marker Bassoon were carried out in primary hippocampal neurons (Figure 3a). Subsequently, the co-localization of the Bassoon puncta with the mCherry-SHANK2 wild type and mutants at the synaptic contacts was quantified. Overexpression of the different SHANK2 variants showed a significant reduction of recombinant SHANK2 clustering at synapses for all variants (Figure 3b). We observed an overall loss of presynaptic contacts reaching transfected cells for all four variants, as revealed by a significant reduction of Bassoon puncta along dendrites (Figure 3c). No effect, however, was seen in the ratio SHANK2 containing synapses to Bassoon puncta along dendrites (Figure 3d). The strongest effect with a remarkable reduction in the number of Bassoon-positive synapses along the dendrites was detected for the R958S and A1731S variants (Figure 3b). The strongest loss of presynaptic contacts was seen for the S610Y variant (Figure 3c). Together, these results suggest that overexpression of all four tested SHANK2 variants in primary hippocampal neurons lead to an impairment in synaptic clustering and an overall loss of synaptic contacts.
Figure 3.
SHANK2 mutant constructs lead to a reduction of presynapses and to less mutant protein localized at synapses. (a) DIV14 rat hippocampal neurons were transfected overnight with wild-type and mutant mCherry constructs of ProSAP1/Shank2 (red). Presynapses were immunostained with anti-Bassoon (white), nuclei are visualized with DAPI (blue). Scale bars (green) represent 20 μm. Arrowheads mark co-localizing SHANK2 and Bassoon signals. (b) Analysis of mCherry-SHANK2 Bassoon-positive synapses per 10 μm dendritic length. (b, c) Error bars represent s.e.m. Significance analysis with Student's t-test with following P-values: ***P<0.001, **P<0.01, *P<0.05. n≥10 neurons from ≥2 preparations. (c) Counting of Bassoon puncta reaching dendrites (per 10 μm). (d) Share of mCherry-ProSAP1/Shank2-Bassoon-positive synapses to Bassoon puncta along dendrites (1 is equal to 100%).
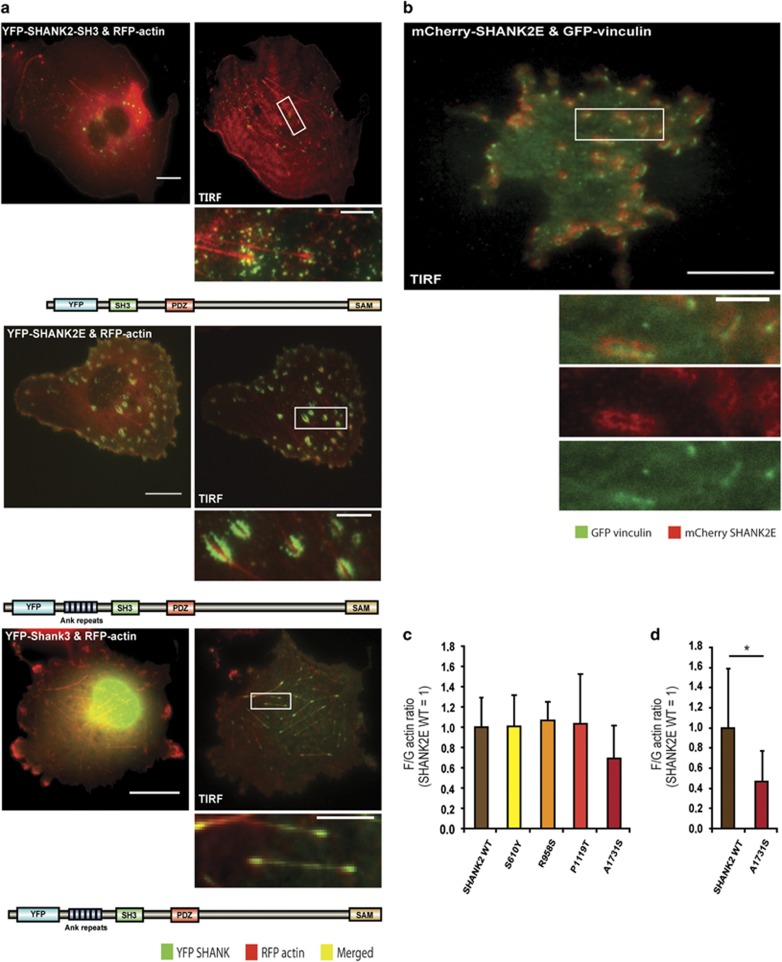
As the SHANK family members and their interaction partners are involved in actin dynamics in neurons, another key point of our study was to investigate whether different SHANK2 variants have an effect on actin structures.50, 51, 52, 53 Therefore, we overexpressed SHANK2-SH3 together with RFP-actin in COS-7 cells and used Shank3 as a control, because it is known to localize to the tip of actin filaments.54 We used live cell TIRF microscopy to observe the localization of used constructs. SHANK2-SH3 protein formed intracellular aggregates of different sizes and did not localize to the actin fiber tips. Taking into consideration, the sequence similarities between the largest SHANK2 isoform SHANK2E and Shank3, we decided to overexpress SHANK2E. In contrast to Shank3, however, we observed compact aggregates of SHANK2E surrounding the tips of the actin fibers (Figure 4a). Furthermore, SHANK2E protein was located in vicinity to the focal adhesion marker vinculin, suggesting a possible interaction with the capping proteins that control the actin filaments growth and filopodia formation (Figure 4b).55 Subsequently, we introduced the four different variants into the SHANK2-SH3 and SHANK2E isoforms (Supplementary Figure 3) but obtained no obvious different localization compared with wild-type proteins. Finally, we performed an actin polymerization assay to quantify the ratio of filamentous (F) to globular (G) actin in HEK293 cells. Interestingly, only the A1731S variant indicated a decreased F/G-actin ratio in our experiments (Figure 4c), suggesting an impairment of actin polymerization. This impairment can probably be linked to the strong reduction of synaptic contacts observed for the A1731S variant.
Figure 4.
Analysis of the effect of SHANK2 wild type and mutants on actin structures and polymerization. (a) YFP-SHANK2E, YFP-SHANK2-SH3 and YFP-Shank3 wild type were co-transfected together with RFP-actin into COS-7 cells. Live cell widefield epifluorescence and TIRF images are presented. SHANK2-SH3 wild type formed intracellular aggregates of different sizes and did not localize to the actin fiber tips. YFP-SHANK2E formed compact aggregates around the actin fiber tips. The same localization was observed with C-terminal SHANK2-SH3-YFP and SHANK2E-YFP constructs (data not shown). YFP-Shank3 was used as a control because it co-localizes to the actin fiber tips. Scale bars: 20 μm (main panels); 5 μm (insets). (b) YFP-SHANK2E wild type was co-transfected together with the focal adhesion marker GFP-vinculin into COS-7 cells. TIRF images are presented. SHANK2E protein accumulated around the focal adhesions but did not co-localize with vinculin. Scale bars: 20 μm (main panels); 5 μm (insets). (c) F/G-actin ratio measurements for all variants tested and SHANK2E-WT in HEK293 cells. Each condition was tested in three independent transfections with two replicates each. (d) A1731S variant and SHANK2E-WT were additionally analyzed in three independent transfections with three replicates each. The A1731S/WT results were normalized to the SHANK2 protein amounts via second western blot with anti-SHANK2 and anti-GAPDH antibodies. All values are shown as mean+standard deviation. Wild-type value is normalized to 1. *P≤0.05 (two-tailed paired t-test).
Discussion
In this study, we analyzed the sequence of the SHANK2 gene in a cohort of 481 SCZ patients and revealed an increased burden of rare SHANK2 missense variants in these individuals compared with controls. The difference in frequency of rare predicted deleterious variants between patients and controls did not reach statistical significance but a trend was noted. The assessment of rare deleterious variant frequencies may be limited by the size of our studied cohort (500 SCZ individuals) and the accuracy of the prediction tools.
Overall, 2.7% of patients harbor novel missense SHANK2 variants that were not found in our sequenced controls. These 10 rare variants are to our knowledge the first identified SHANK2 variants in patients with SCZ. As SHANK2 mutations have been previously identified in patients with ID and ASD, we first compared the variants between the different disorders.39, 46 Previous sequencing of SHANK2 coding exons and exon–intron boundaries in ID and ASD cohorts of 851 patients yielded 1 de novo stop mutation (R462X), 1 inherited micro-duplication of 6 nucleotides and 13 rare missense variants, which were not found in 1090 controls. 27, 32 All the reported ASD missense variants were either shown to be inherited or no parental DNA was available for sequencing, similar to the findings in our SCZ cohort. One of our four functionally tested SCZ variants, S610Y, was also previously identified in an individual with ID suggesting that distinct SHANK2 mutations can contribute to the etiology of more than one brain disorder. Copy number screenings also have identified several SHANK2 de novo deletions in ASD patients32, 39 but no CNVs of this chromosomal region have been reported in SCZ individuals so far. The SCZ patients participated also in a genome-wide CNV screen.56 No SCZ-associated CNVs were found in cases harboring candidate SHANK2 variants.
All identified SHANK2 variants in SCZ patients were heterozygous and either inherited from apparently unaffected parents or are of unknown origin, suggesting variable penetrance (in two-thirds of the patients, parent information was not available). In all five individuals with inherited mutation, the transmission was from mother to son, similar to previous findings in ASD patients. The reasons for this gender preference are not clear.
Demonstrating the causality of rare missense variants is a common challenge in targeted gene mutation screenings. To assess the potential impact of the variants on protein function, we performed various in vitro assays using primary hippocampal neurons and cell lines (COS-7, HEK293) as a model. The four tested variants (S610Y, R958S, P1119T and A1731S) were initially selected based on in silico predictions, frequency and absence in the 1000 Genome project and Human exome variant server databases. Three of them, S610Y, R958S and P1119T, are affecting highly conserved amino-acid positions in SHANK2 with high predicted probabilities to impair protein function (Supplementary Tables 2 and 7). In contrast, the A1731S variant does not locate to a very highly conserved region and was considered to be benign by several prediction softwares (Supplementary Tables 2 and 7). A reduced number of SHANK2-Bassoon-positive synapses was measured for variants S610Y, R958S and P1119T, supporting the hypothesis that conservation of a given affected amino acid is a good indicator for a functional output.39 Two of the variants, S610Y and P1119T, also show a reduced ability to increase dendritic spine volume. Reduction in the potential for spine enlargement was observed before for isolated SHANK2 variants in ASD and ID.32
The A1731S variant was found four times in 481 SCZ patients and has not been detected in 5338 controls, which makes it a potent risk factor for SCZ (P=4,6 × 10−5, two-sided Fisher's exact test). To reach this number of unaffected controls, we integrated our control group of German individuals with European American individuals from the Human Exome Variant Server and from the 1000 Genome project. However, A1731S was also not present in individuals with different ethnic origins in these databases. Such high frequency of detection is unique for a SHANK2 mutation found in individuals with neuropsychiatric disorders so far.
Another interesting point is the apparent overlap in the clinical picture of the four patients sharing the A1731S mutation. All four carriers share an early prodromal phase with an insidious onset of psychiatric symptoms. In addition, all these patients displayed a substantial ego disturbance (depersonalization), persecutory delusions and auditory hallucinations. Parental age at birth was high and varied between 36 and 54 years in fathers and 33 and 39 years in mothers. These findings will be interesting to replicate in a larger patient cohort. High parental age, especially high paternal age, increases the probability for additional de novo mutations, which may further contribute to the SCZ manifestation in these individuals.57, 58, 59
To our surprise, the functional assays also underlined A1731S as a variant with strong effect among the functionally tested SCZ variants. In overexpression experiments, mCherry-A1731S showed reduced number of Shank2-Basoon-positive synapses. Another interesting finding is that A1731S was the only variant analyzed that showed a significant effect on the F/G-actin ratio in HEK293 cells. This parameter attracts attention since synapse structure and function are known to take advantage of actin as stable, but yet dynamic structural components. A diminished actin polymerization could be the explanation for an impairment in synapse formation and maintenance, which is indicated by a reduction of synaptic clustering of the overexpressed SHANK2 mutant and the reduced number of presynaptic contacts. The absence of a dendritic spine phenotype is surprising but actin also contributes to morphologically undetectable functions. Alterations in the organization of the junction scaffold components or assisting the trafficking of the whole synaptic machinery can lead to functional disturbances in neurotransmission without visible effect on spine structure.60 Combined with its frequency in patient samples, variant A1731S sets apart from the other missense SHANK2 variants identified in different disorders.39, 46 The in silico analysis excluded possible splice site disruptions, suggesting that the effect of this variant in patients is likely due to the amino-acid exchange. Its location in near proximity to the sterile alpha motif domain proposes a possible effect on the dendritic localization and the self-multimerization of the mutant protein, two features of SHANK2 that are highly dependant on intact C-termini.61, 62
Regardless of its strong functional effect, A1731S resides at a position with low conservation and was predicted as benign by most prediction tools (Supplementary Table 3). The idea that SCZ pathogenesis may occur in evolutionary recent neural cell types and circuits may be an explanation for the presence of serine as a wild-type allele in some species. Comparison of functionally analyzed missense SHANK2 variants (from both our and a previous study)39 to the corresponding in silico predictions indicates significant rates of false predictions (Supplementary Table 8). This lead us to the conclusion that analysis based only on prediction results may lose valuable information and associated rare SHANK2 missense variants should be functionally assessed independent of their prediction status.
In summary, the accumulated evidence strongly suggests that the identified variants represent risk factors for SCZ. Combined with previous data of SHANK1 and 3, our work underlines the SHANK gene family and their allelic variants to appear in key neuronal pathways and circuits in the postsynaptic density of excitatory neurons and, when perturbing their function causing disease (for example, ID, ASD and SCZ). The exact molecular mechanisms and networks affected by SHANK2 mutations in SCZ remain unclear at this point but it is likely that alteration in NMDAR function is involved. This hypothesis is supported by two SHANK2 mouse models that carry different deletions, comparable to those identified in ASD patients.40, 41 Further useful model systems to address these questions may include induced pluripotent stem cell lines reprogrammed into neurons to study the underlying molecular and cellular mechanisms.
Acknowledgments
We thank R Röth, M Geiger and M Bencun for help with the sequencing, R Sprengel and V Endris for experimental support, K Hahn, U Engel, S Wiemann and C Sala for the provided plasmids, Y Zhang and W Edelmann for the PPY strain, and A de Sena Cortabitarte for reading the manuscript. The TIRF imaging was performed at the Nikon Imaging Center at the University of Heidelberg with support from U Engel. This work was supported by the Medical Faculty Heidelberg and EcTop5 grant from CellNetworks. S Peykov is a member of the Hartmut Hoffmann-Berling International Graduate School of Molecular and Cellular Biology (HBIGS) and was funded by an HBIGS fellowship. MM Nöthen and M Rietschel received support for this work from the German Federal Ministry of Education and Research (BMBF) through the Integrated Network IntegraMent (Integrated Understanding of Causes and Mechanisms in Mental Disorders), under the auspices of the e:Med research and funding concept (grants #01ZX1314A and #01ZX1314A). MM Nöthen is a member of the DFG-funded Excellence Cluster ImmunoSensation. Franziska Degenhardt received support from the BONFOR Programme of the University of Bonn, Germany. G Rappold is a member of CellNetworks Cluster of Excellence.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- 1van Os J, Kapur S. Schizophrenia. Lancet 2009; 374: 635–645. [DOI] [PubMed] [Google Scholar]
- 2Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet 2012; 13: 537–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014; 511: 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460: 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe'er I et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature 2009; 460: 753–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D et al. Common variants conferring risk of schizophrenia. Nature 2009; 460: 744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet 2013; 45: 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Schizophrenia Psychiatric Genome-Wide Association Study (GWAS) Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet 2011; 43: 969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S et al. Large recurrent microdeletions associated with schizophrenia. Nature 2008; 455: 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Levinson DF, Duan J, Oh S, Wang K, Sanders AR, Shi J et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry 2011; 168: 302–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell 2012; 148: 1223–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Rees E, Walters JT, Georgieva L, Isles AR, Chambert KD, Richards AL et al. Analysis of copy number variations at 15 schizophrenia-associated loci. Br J Psychiatry 2014; 204: 108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Kirov G, Rees E, Walters JT, Escott-Price V, Georgieva L, Richards AL et al. The penetrance of copy number variations for schizophrenia and developmental delay. Biol Psychiatry 2014; 75: 378–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry 2012; 17: 142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Szatkiewicz JP, O'Dushlaine C, Chen G, Chambert K, Moran JL, Neale BM et al. Copy number variation in schizophrenia in Sweden. Mol Psychiatry 2014; 19: 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Rees E, Moskvina V, Owen MJ, O'Donovan MC, Kirov G. De novo rates and selection of schizophrenia-associated copy number variants. Biol Psychiatry 2011; 70: 1109–1114. [DOI] [PubMed] [Google Scholar]
- 17Raychaudhuri S, Korn JM, McCarroll SA International Schizophrenia C Altshuler D, Sklar P et al. Accurately assessing the risk of schizophrenia conferred by rare copy-number variation affecting genes with brain function. PLoS Genet 2010; 6: e1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Glessner JT, Reilly MP, Kim CE, Takahashi N, Albano A, Hou C et al. Strong synaptic transmission impact by copy number variations in schizophrenia. Proc Natl Acad Sci USA 2010; 107: 10584–10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19International Schizophrenia C. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 2008; 455: 237–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 2008; 320: 539–543. [DOI] [PubMed] [Google Scholar]
- 21Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P et al. De novo mutations in schizophrenia implicate synaptic networks. Nature 2014; 506: 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 2014; 506: 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Xu B, Ionita-Laza I, Roos JL, Boone B, Woodrick S, Sun Y et al. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet 2012; 44: 1365–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Girard SL, Gauthier J, Noreau A, Xiong L, Zhou S, Jouan L et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet 2011; 43: 860–863. [DOI] [PubMed] [Google Scholar]
- 25Kenny EM, Cormican P, Furlong S, Heron E, Kenny G, Fahey C et al. Excess of rare novel loss-of-function variants in synaptic genes in schizophrenia and autism spectrum disorders. Mol Psychiatry 2014; 19: 872–879. [DOI] [PubMed] [Google Scholar]
- 26Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, Levy S et al. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet 2011; 43: 864–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27McCarthy SE, Gillis J, Kramer M, Lihm J, Yoon S, Berstein Y et al. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol Psychiatry 2014; 19: 652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28Sheng M, Kim E. The Shank family of scaffold proteins. J Cell Sci 2000; 113: 1851–1856. [DOI] [PubMed] [Google Scholar]
- 29Boeckers TM, Bockmann J, Kreutz MR, Gundelfinger ED. ProSAP/Shank proteins - a family of higher order organizing molecules of the postsynaptic density with an emerging role in human neurological disease. J Neurochem 2002; 81: 903–910. [DOI] [PubMed] [Google Scholar]
- 30Kreienkamp HJ. Scaffolding proteins at the postsynaptic density: shank as the architectural framework. Handb Exp Pharmacol 2008; 186: 365–380. [DOI] [PubMed] [Google Scholar]
- 31Sato D, Lionel AC, Leblond CS, Prasad A, Pinto D, Walker S et al. SHANK1 deletions in males with autism spectrum disorder. Am J Hum Genet 2012; 90: 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Berkel S, Marshall CR, Weiss B, Howe J, Roeth R, Moog U et al. Mutations in the SHANK2 synaptic scaffolding gene in autism spectrum disorder and mental retardation. Nat Genet 2010; 42: 489–491. [DOI] [PubMed] [Google Scholar]
- 33Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet 2007; 39: 25–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature 2010; 466: 368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35de Lacy N, King BH. Revisiting the relationship between autism and schizophrenia: toward an integrated neurobiology. Annu Rev Clin Psychol 2013; 9: 555–587. [DOI] [PubMed] [Google Scholar]
- 36Lennertz L, Wagner M, Wolwer W, Schuhmacher A, Frommann I, Berning J et al. A promoter variant of SHANK1 affects auditory working memory in schizophrenia patients and in subjects clinically at risk for psychosis. Eur Arch Psychiatry Clin Neurosci 2012; 262: 117–124. [DOI] [PubMed] [Google Scholar]
- 37Gauthier J, Champagne N, Lafreniere RG, Xiong L, Spiegelman D, Brustein E et al. De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proc Natl Acad Sci USA 2010; 107: 7863–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Grabrucker S, Proepper C, Mangus K, Eckert M, Chhabra R, Schmeisser MJ et al. The PSD protein ProSAP2/Shank3 displays synapto-nuclear shuttling which is deregulated in a schizophrenia-associated mutation. Exp Neurol 2014; 253: 126–137. [DOI] [PubMed] [Google Scholar]
- 39Leblond CS, Heinrich J, Delorme R, Proepper C, Betancur C, Huguet G et al. Genetic and functional analyses of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS Genet 2012; 8: e1002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40Schmeisser MJ, Ey E, Wegener S, Bockmann J, Stempel AV, Kuebler A et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature 2012; 486: 256–260. [DOI] [PubMed] [Google Scholar]
- 41Won H, Lee HR, Gee HY, Mah W, Kim JI, Lee J et al. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature 2012; 486: 261–265. [DOI] [PubMed] [Google Scholar]
- 42Javitt DC. Glutamatergic theories of schizophrenia. Isr J Psychiatry Relat Sci 2010; 47: 4–16. [PubMed] [Google Scholar]
- 43Fangerau H, Ohlraun S, Granath RO, Nothen MM, Rietschel M, Schulze TG. Computer-assisted phenotype characterization for genetic research in psychiatry. Hum Hered 2004; 58: 122–130. [DOI] [PubMed] [Google Scholar]
- 44McGuffin P, Farmer A, Harvey I. A polydiagnostic application of operational criteria in studies of psychotic illness. Development and reliability of the OPCRIT system. Arch Gen Psychiatry 1991; 48: 764–770. [DOI] [PubMed] [Google Scholar]
- 45Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM et al. A versatile gene-based test for genome-wide association studies. Am J Hum Genet 2010; 87: 139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46Berkel S, Tang W, Trevino M, Vogt M, Obenhaus HA, Gass P et al. Inherited and de novo SHANK2 variants associated with autism spectrum disorder impair neuronal morphogenesis and physiology. Hum Mol Genet 2012; 21: 344–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47Zhang Y, Werling U, Edelmann W. SLiCE: a novel bacterial cell extract-based DNA cloning method. Nucleic Acids Res 2012; 40: e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J et al. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron 1999; 23: 569–582. [DOI] [PubMed] [Google Scholar]
- 49Kutzing MK, Langhammer CG, Luo V, Lakdawala H, Firestein BL. Automated Sholl analysis of digitized neuronal morphology at multiple scales. J Vis Exp 2010; 77: 1160–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50Bockers TM, Mameza MG, Kreutz MR, Bockmann J, Weise C, Buck F et al. Synaptic scaffolding proteins in rat brain. Ankyrin repeats of the multidomain Shank protein family interact with the cytoskeletal protein alpha-fodrin. J Biol Chem 2001; 276: 40104–40112. [DOI] [PubMed] [Google Scholar]
- 51Lim S, Sala C, Yoon J, Park S, Kuroda S, Sheng M et al. Sharpin, a novel postsynaptic density protein that directly interacts with the shank family of proteins. Mol Cell Neurosci 2001; 17: 385–397. [DOI] [PubMed] [Google Scholar]
- 52Qualmann B, Boeckers TM, Jeromin M, Gundelfinger ED, Kessels MM. Linkage of the actin cytoskeleton to the postsynaptic density via direct interactions of Abp1 with the ProSAP/Shank family. J Neurosci 2004; 24: 2481–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53Hering H, Sheng M. Activity-dependent redistribution and essential role of cortactin in dendritic spine morphogenesis. J Neurosci 2003; 23: 11759–11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54Durand CM, Perroy J, Loll F, Perrais D, Fagni L, Bourgeron T et al. SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Mol Psychiatry 2012; 17: 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, Borisy GG. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell 2004; 118: 363–373. [DOI] [PubMed] [Google Scholar]
- 56Priebe L, Degenhardt F, Strohmaier J, Breuer R, Herms S, Witt SH et al. Copy number variants in German patients with schizophrenia. PLoS One 2013; 8: e64035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G et al. Rate of de novo mutations and the importance of father's age to disease risk. Nature 2012; 488: 471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58Malaspina D, Corcoran C, Fahim C, Berman A, Harkavy-Friedman J, Yale S et al. Paternal age and sporadic schizophrenia: evidence for de novo mutations. Am J Med Genet 2002; 114: 299–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59Jaffe AE, Eaton WW, Straub RE, Marenco S, Weinberger DR. Paternal age, de novo mutations and schizophrenia. Mol Psychiatry 2014; 19: 274–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60Cingolani LA, Goda Y. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat Rev Neurosci 2008; 9: 344–356. [DOI] [PubMed] [Google Scholar]
- 61Boeckers TM, Liedtke T, Spilker C, Dresbach T, Bockmann J, Kreutz MR et al. C-terminal synaptic targeting elements for postsynaptic density proteins ProSAP1/Shank2 and ProSAP2/Shank3. J Neurochem 2005; 92: 519–524. [DOI] [PubMed] [Google Scholar]
- 62Baron MK, Boeckers TM, Vaida B, Faham S, Gingery M, Sawaya MR et al. An architectural framework that may lie at the core of the postsynaptic density. Science 2006; 311: 531–535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.