Background: SNAREs mediate membrane fusion and need to be addressed to specific intracellular compartments.
Results: We show that the Q-SNARE SNAP-47 regulates the localization and function of a subset of v-SNAREs.
Conclusion: SNAP-47 localizes mainly in the early secretory pathway where it regulates the transport of selected v-SNAREs.
Significance: SNAP-47 may guide selected v-SNAREs through the early secretory pathway.
Keywords: membrane fusion, membrane trafficking, nuclear translocation, protein secretion, SNARE proteins, vesicles, ERGIC, SNAP-47, VAMP
Abstract
SNAREs constitute the core machinery of intracellular membrane fusion, but vesicular SNAREs localize to specific compartments via largely unknown mechanisms. Here, we identified an interaction between VAMP7 and SNAP-47 using a proteomics approach. We found that SNAP-47 mainly localized to cytoplasm, the endoplasmic reticulum (ER), and ERGIC and could also shuttle between the cytoplasm and the nucleus. SNAP-47 preferentially interacted with the trans-Golgi network VAMP4 and post-Golgi VAMP7 and -8. SNAP-47 also interacted with ER and Golgi syntaxin 5 and with syntaxin 1 in the absence of Munc18a, when syntaxin 1 is retained in the ER. A C-terminally truncated SNAP-47 was impaired in interaction with VAMPs and affected their subcellular distribution. SNAP-47 silencing further shifted the subcellular localization of VAMP4 from the Golgi apparatus to the ER. WT and mutant SNAP-47 overexpression impaired VAMP7 exocytic activity. We conclude that SNAP-47 plays a role in the proper localization and function of a subset of VAMPs likely via regulation of their transport through the early secretory pathway.
Introduction
The secretory pathway is essential for the homeostasis of membrane proteins and cell-cell communication in eukaryotes. Vesicular trafficking is the main mechanism of communication between intracellular membranes along the secretory pathway, and it proceeds through three main steps as follows: budding, transport, and fusion (1). SNAREs are essential for membrane fusion and are thus major players in vesicular trafficking. V-SNARE, on vesicular membrane, and t-SNAREs, on target membrane, form a complex, the so-called SNARE complex or SNAREpin, which drives fusion between two compartments (2). A SNARE complex is composed of three to four different SNAREs, which interact together by their coiled-coil domain or SNARE domain. V-SNAREs are represented by the VAMP family with a central arginine (R) in the SNARE domain, thus being referred to as R-SNAREs. T-SNAREs, also referred as Q-SNAREs due to a central glutamine (Q) in their SNARE domain, fall into two groups: SNAP-25 and syntaxin families (3). How v-SNAREs localize to the membrane compartments where they operate is an essential question to understand their cellular functions.
VAMP7 (also called tetanus neurotoxin-insensitive VAMP, TI-VAMP) is localized in late endosomes, lysosomes, and the trans-Golgi network (TGN)6 (4, 5), and it plays a role in several secretory and endocytic mechanisms (6). It interacts with SNAP-23, SNAP-25, and syntaxins 1, 3, or 4 in exocytosis or syntaxin 7/syntaxin 8/Vti1b in endosomal fusion (7–10). VAMP4 is mainly localized in the TGN and in early endosomes (11). It is involved in TGN-endosome trafficking via the plasma membrane (12) and endosome to TGN transport (13). VAMP8 localizes to early and late endosomes and mediates their homotypic fusion (14). It is also involved in fusion of autophagosomes with endosomes/lysosomes mediated by the SNARE complex also comprising syntaxin 17 and SNAP-29 (15, 16).
VAMPs are C-terminally anchored membrane proteins, synthesized in the cytosol and post-translationally inserted into the ER (17). They are then transported through the secretory pathway and reach the membrane compartments where they operate as v-SNAREs. VAMP7 has a long N-terminal extension, called the Longin domain, which is auto-inhibitory. Its binding to the AP-3 adaptor complex mediates VAMP7 targeting from early to late endosomes and to synaptic vesicles (7, 18). VAMP4 binds to an adaptor complex, AP-1, via a di-leucine motif. This interaction is required for its proper localization (19). We have previously shown that retention of syntaxin 1 in the ER by the absence of Munc18a, a syntaxin 1 partner, regulator, and stabilizer (20), can also affect the subcellular localization of its cognate v-SNAREs VAMP3 and -7 (21). Post-Golgi VAMPs presumably need to be fusion-incompetent while they are in the ER and Golgi to prevent illegitimate fusion until they have reached the membranes where they operate as fusogenic proteins. The molecular mechanisms of their transport from the ER to Golgi are still largely unknown. As of now, only BAP31 has been described to control VAMP3 export from the ER (22).
SNAP-47 is an atypical Q-SNARE, a member of the SNAP-25 family, which has a long N-terminal extension and lacks cysteine palmitoylation-mediated membrane targeting. In vitro, recombinant SNAP-47 substituted for SNAP-25, even though less efficiently, for SNARE complex formation with the neuronal SNAREs syntaxin 1a and VAMP2 and proteoliposome fusion (23). SNAP-47 was recently shown to mediate the secretion of brain-derived neurotrophic factor (BDNF) in neurons (24) and to be crucial for AMPA receptor insertion in long term potentiation (25) and retinoic acid-mediated homeostatic synaptic plasticity (26). These pathways all require VAMP2, but none of these studies characterized the membrane compartments to which SNAP-47 may be targeted in neurons or showed that SNAP-47 operated specifically through VAMP2 binding. Also, the role of SNAP-47 in non-neuronal cells still remains unknown.
Here, we sought to characterize the interactome of VAMP7 in epithelial cells using a proteomics approach. The identification of SNAP-47 as a VAMP7-interacting partner further led us to the characterization of the full spectrum of its VAMP partners. The subcellular localization of SNAP-47 in the ER and ERGIC and the effects of its deleted mutant overexpression and of its inactivation led us to propose a role of SNAP-47 in the early secretory pathway of a subset of post-Golgi v-SNAREs.
Experimental Procedures
Plasmids and Cloning
SNAP47-mcherry WT and ΔCter were cloned in mcherryN1 vector by PCR using set of primers, respectively, 5′-GATAGGAATTCTATGCGCGCGGCTCGC-3′, 3′-GCTGTCGACAAGGTCAGCCTCTTCATCCG-5′ and 5′-GATAGGAATTCTATGCGCGCGGCTCGC-3′, 3′-GCTGTCGACAACTGCAGGTGCAGTGCTGT-5′. SNAP47-mycDDK cDNA was from Origene. GFP-SNAP47 WT and ΔCter were cloned in pEGFPC3 vector in EcoRI/SalI and EcoRI/PstI sites, respectively. SNAP-47mycHis WT and ΔCter were cloned in pcDNA3.1 Myc His B+ vector using BamHI/ApaI and BamHI/XhoI, respectively, for vectors and BglII/ApaI and BglII/SalI, respectively, for inserts. SNAP47-mcherry WT and ΔCter were cloned in mcherryN1 vector in EcoRI/SalI sites. Human GFP-tagged VAMP3 (8), VAMP4 (13), VAMP7 (full length, Longin, ΔLongin) (7), VAMP8 (8), Sec22b (27), and GFP-SNAP23 (28) were previously described and mouse Ykt6 in pEGFP vector obtained from Gene Service Ltd. GFP-syntaxin 1, Munc18a (21), and VAMP7-pHluorin (18) plasmids were previously described. GFP-tagged rat VAMP2 was a generous gift from R. Scheller, (Stanford University). GFP-syntaxins 5 and 13 were generous gifts from L. Johannes (Institut Curie, Paris, France).
Sequence Prediction Analysis
The NCBI human reference sequences used were as follows: NP_444280.2 for SNAP-47; NP_004773.1 for SNAP-29; NP_003816.2 for SNAP-23; and NP_570824.1 for SNAP-25. For prediction of nuclear localization signals (NLS) and nuclear export signals (NES), PSORT II program (29) and NetNES 1.1 server (30) were used, respectively.
Antibodies (Abs)
Primary antibodies used in the study are listed in supplemental Table 2. DaRabbitIRDye800/AlexaFluor 800 and GaMouseIRDye680/AlexaFluor 680 (Rockland Inc.) were used at 1:4000 as Western blotting secondary Abs. GFP-trap beads were from Chromotek.
Cross-linking of Antibodies on Beads
Antibodies used for IP were cross-linked to magnetic beads (Dynabeads, Life Technologies, Inc.). Beads were washed in 0.2 m triethanolamine, pH 8.0, and incubated with antibodies for 4 h at 4 °C. After washes with triethanolamine, antibodies were cross-linked to beads using 20 mm dimethyl pimelimidate (DMP) for 30 min at room temperature and washed with 50% triethanolamine and 50% Tris 50 mm, pH 7.0, solution before use.
Immunoprecipitation (IP) for Proteomics and Western Blot (WB) Assay
GFP, GFP-Longin, and GFP-VAMP7 MDCK cells (31) were washed in PBS and lysed 20 min in TSE (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA) supplemented with 0.5% Triton X-100 and Complete tablets (Roche Applied Science). Lysates were centrifuged 30 min at 13,000 rpm, and supernatant proteins were quantified by the Bradford method. Fluorescent proteins were estimated using a fluorimeter and adjusted accordingly. 30 mg of proteins were immunoprecipitated overnight at 4 °C with rabbit anti-GFP Ab coupled to magnetic beads (Dynabeads) by cross-linking or GFP-trap beads when mentioned. After washing beads with lysis buffer, proteins were eluted using 50 mm glycine, pH 2.5, 2% Triton X-100, equilibrated with 1 m Tris, pH 8.0, and separated by SDS-PAGE using NuPAGE BisTris 4–12% gradient gels (Life Technologies, Inc.). Samples were analyzed using an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) coupled to a nanoLC system (Ultimate 3000, Dionex S.A.). For co-IP assays, IPs and WB were performed as described previously (32) using specific Ab previously cross-linked to magnetic beads (Dynabeads). NEM treatment was done as described by Galli et al. (8).
Liquid Chromatography-MS/MS (LC-MS/MS) and Data Analysis
After IP, proteins were submitted to SDS-PAGE. Excised gel slices were washed, and proteins were reduced with 10 mm DTT prior to alkylation with 55 mm iodoacetamide. After washing and shrinking of the gel pieces with 100% acetonitrile, in-gel digestion was performed using trypsin (Sequencing Grade Modified, Roche Applied Science) overnight in 25 mm ammonium bicarbonate at 30 °C. The extracted peptides were analyzed by nano-LC-MS/MS using an Ultimate3000 system coupled to the LTQ-Orbitrap mass spectrometer. Samples were loaded onto a C18 precolumn (300 μm inner diameter × 5 mm; Dionex) at 20 μl/min in 5% acetonitrile, 0.1% TFA. After 3 min of desalting, the precolumn was switched on line with the analytical C18 column (75 μm inner diameter × 15 cm; C18 PepMapTM, Dionex) equilibrated in 95% solvent A and 5% solvent B (5% acetonitrile, 0.1% formic acid and 80% acetonitrile, 0.085% formic acid). Bound peptides were eluted using a 5–52% gradient of solvent B during 57 min at a 200 nl/min flow rate. Data-dependent acquisition was performed on the LTQ-Orbitrap mass spectrometer in the positive ion mode. Survey MS scans were acquired in the Orbitrap on the 475–1200 m/z range with the resolution set to a value of 60,000. Each scan was recalibrated in real time by coinjecting an internal standard from ambient air into the C-trap (“lock mass option”). The five most intense ions per survey scan were selected for CID fragmentation, and the resulting fragments were analyzed in the linear trap (LTQ). Target ions already selected for MS/MS were dynamically excluded for 180 s. Data were acquired using the Xcalibur software (version 2.0.7), and the resulting spectra were then analyzed via the MASCOTTM software created with Proteome Discoverer (version 1.4, Thermo Scientific) using the SwissProt Canis lupus familiaris database, containing 29,287 protein sequences. Carbamidomethylation of cysteines, oxidation of methionine, and protein N-terminal acetylation were set as variable modifications for all Mascot searches. Specificity of trypsin digestion was set for cleavage after Lys or Arg except before Pro, and two missed trypsin cleavage sites were allowed. The mass tolerances in MS and MS/MS were set to 2 ppm and 0.8 Da, respectively. The resulting Mascot files were further processed using myProMS (33). The estimated false discovery rate of all peptide and protein identifications was set to 1%, by automatically filtering the Mascot score of all peptide identifications. Hits found in both VAMP7 or Longin and control GFP IP samples were excluded.
Yeast Two-hybrid Cloning and Analysis
pB29/pB27, pB66, and pP6 are derived from the original pBTM116 (34), pAS2ΔΔ (36), and pGADGH (35) plasmids, respectively. For SNAP-23, 59 million clones (6-fold the complexity of the library) were screened using a mating approach with Y187 (matα) and L40ΔGal4 (mata) yeast strains as described previously (36). 109 His+ colonies were selected on a medium lacking tryptophan, leucine, and histidine and supplemented with 50 mm 3-aminotriazole to handle bait autoactivation. For SNAP-29, 153 million clones were screened, and 159 His+ colonies were selected on a medium supplemented with 5 mm 3-aminotriazole. The screen with the SNAP-47-LexA (LexA-SNAP-47, respectively) construct yielded 0 His+ (5 respectively) colonies out of 133 (54 respectively) million interactions tested on a medium without 3-aminotriazole. 66 million clones were screened with the Gal4DBD-SNAP-47 construct using the same mating approach with Y187 (matα) and CG1945 (mata) yeast strains. 94 His+ colonies were selected on a medium lacking tryptophan, leucine, and histidine. The prey fragments of the positive clones were amplified by PCR and sequenced at their 5′ and 3′ junctions. The resulting sequences were used to identify the corresponding interacting proteins in the GenBankTM database (NCBI) using a fully automated procedure. The protein interactions from this publication have been submitted to the IMEx consortium through IntAct and were assigned the identifier IM-22287.
Cell Culture and Transfection
Tet-Off MDCK cells expressing GFP, GFP-VAMP7, GFP-ΔLongin, or GFP-Longin were cultured as described (31). L929 cells were cultured like HeLa and COS7 cells as described (32). PC12 cells were cultured in RPMI 1640 medium with 10% horse serum and 5% FCS on collagen-coated plates. For immunofluorescence, cells were plated on coverslips and transfected by Lipofectamine 2000 (Life Technologies, Inc.) according to the manufacturer's instructions, and fixed 16–24 h after transfection. For IP, cells were electroporated using Gene Pulser II (Bio-Rad). After washing with DMEM, 7·106 cells were resuspended in 0.5 ml of Cytomix solution (25 mm HEPES, 120 mm KCl, 10 mm KH2PO4, 0.15 mm CaCl2, 5 mm MgCl2, 2 mm EGTA, pH 7.6). 2 mm ATP, 5 mm glutathione, and 25–50 μg of DNA were added, and cells were electroporated (0.29 kV, 1000 microfarads). IPs were performed 16–24 h after transfection. If required, cells were treated with 5 μm brefeldin A for 45 min or 10 ng/ml leptomycin B for 4 h. Equivalent volumes of DMSO or methanol were used as control, respectively.
In Vitro Binding Assay
HeLa cells were transfected either with GFP-SNAP47 or GFP-SNAP23 and lysed 24 h after transfection in a buffer containing 50 mm Tris HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100. GST-VAMP3, GST-VAMP7, and GST were produced as follows; transformed BL-21 cells were induced by isopropyl 1-thio-β-d-galactopyranoside during 3 h and centrifuged 10 min at 5000 rpm. Bacteria pellets lysed in buffer (50 mm Tris-HCl, pH 8.0, 10% glycerol, 300 mm KCl, 10 mm β-mercaptoethanol) were sonicated 4 min, and Triton X-100 was added at 1% final. After centrifugation for 15 min at 8500 rpm and 1 h at 35,000 rpm, glutathione beads were added to supernatants, incubated overnight, and washed five times with lysis buffer. Lysates from HeLa cells transfected with GFP-SNAP23 and GFP-SNAP47 were incubated with purified GST-VAMPs or GST alone as a control. After five washes, beads were transferred to a 96-well microplate, and fluorescence was measured at 488 nm using a Fluorescence Plate Reader (Wallac 1420 Victor2 Microplate Reader, PerkinElmer Life Sciences).
Immunofluorescence
For SNAP-47 immunofluorescence, HeLa cells were pre-permeabilized with digitonin for 45 s at 4 °C and washed with PBS to remove a soluble pool of SNAP-47 prior to fixation. Cells were fixed with 4% paraformaldehyde for 20 min and incubated for 15 min in 50 mm PBS-NH4Cl. Cells were permeabilized 10 min with PBS 0.1% Triton X-100 and blocked 30 min in PBS 1% BSA or 10% FBS. Primary Abs were incubated for 2 h and secondary and DAPI for 1 h at room temperature in PBS 1% BSA or 3% FBS.
Cell Imaging
For immunostained samples, images were sequentially acquired using a confocal microscope (TCS SP5; Leica Microsystems) and an HCX PL APO ×63/0.6–1.4 NA oil immersion objective or an upright microscope (DMRA2 Leica Microsystems) equipped with a CCD camera CoolSNAP ES (Photometrics, Roper Scientific) and a HCX PL APO ×40/1.25–0.75 NA CS oil immersion Leica objective. For live imaging, HeLa cells were cultured on coverglass and transfected with indicated fluorescent proteins as described under “Cell Culture and Transfection.” Live imaging was carried out at 37 °C in modified Krebs-Ringer buffer (135 mm NaCl, 2.5 mm KCl, 1.2 mm MgCl2, 1 mm CaCl2, 20 mm HEPES, 11.1 mm glucose, pH 7.4) supplemented with 1% FBS. Cells were imaged using an inverted DMI6000B microscope (Leica Microsystems), equipped with a ×63/1.4–0.6 NA Plan-Apochromat oil immersion objective, an EMCCD digital camera (Cascade:512B; Photometrics, Roper Scientific), and controlled by Metamorph software (Roper Scientific, Trenton, NJ). To virtually abrogate latency between two-channel acquisition, illumination was sequentially provided by a 488-nm and a 561-nm diode acousto-optically shuttered lasers whose beam was expanded to fill the field of view (iLas system; Roper Scientific), and a dual-band filter cube optimized for 488/561-nm laser sources (BrightLine; Semrock) was used. Images were acquired every 300 ms (150ms exposure per channel). Environmental temperatures during experimental acquisitions averaged 22 °C. Fiji software was used for bleaching correction (histogram matching plugin) and for montage of movies. Binary mask of particles was generated by applying the wavelet-based spot detector plugin of Icy imaging software (37) to each channel sequence, and an average time-projection of the resulting mask was then created.
Subcellular Distribution Analysis
The subcellular distribution of GFP-tagged VAMPs was estimated using a previously described method (32) with slight modifications. Using ImageJ, a 20-pixel-wide band was designed from the nucleus (defined from the edge of DAPI staining) to the cell leading edge and going through the Golgi apparatus (indicated by TGN46 staining). To normalize the length of the selected band, a binning was applied to obtain a 10-pixel-long line. Fluorescence intensity profile was then measured, expressed as percent of total intensity, and plotted in function of the distance from nucleus to periphery (i.e. 0–100% on the x axis).
Surface Immunostaining of VAMP7-pHluorin
Cells were washed with ice-cold modified Krebs-Ringer buffer and then incubated 30 min at 4 °C with mouse Ab anti-GFP. After extensive washing, cells were fixed and processed for immunofluorescence using goat anti-GFP Ab. This protocol allowed us to distinguish the fraction of VAMP7 expressed at the surface (Cy3 staining) and the total amount of expressed protein (AlexaFluor 488 staining). Quantification of surface/total integrated signals was done on more than 130 VAMP7-pHluorin positive cells coexpressing the SNAP constructs.
siRNA Experiments
siRNAs pool targeting mouse SNAP-47 and control luciferase siRNA (target sequence 5′-CGTACGCGGAATACTTCGA-3′) were purchased from Dharmacon (Thermo Fisher Scientific). Lipofectamine 2000 (Life Technologies, Inc.) was used as follows: 50 pmol of siRNAs were transfected at day 1 and 15 pmol were cotransfected on day 2 with 0.5 μg of GFP-VAMP4 plasmid. Cells were fixed and immunostained at day 4. Colocalizations of VAMP4 with calreticulin and GM130 were quantified with correlation intensity analysis plugin of Fiji software as follows: Pearson's coefficient was measured in whole cell for calreticulin or inside the mask area corresponding to GM130 staining.
Statistical Analyses
Calculations were performed in Microsoft Excel. GraphPad Prism software was used for graph generation and statistical analyses. For each dataset, at least three independent experiments were considered, and all data are shown as mean ± S.E. Student's t test or ANOVA with Bonferroni's, Dunn's, or Tukey's post-tests were applied, as specified in the figure legends (*, p < 0.05; **, p < 0.005; and ***, p < 0.0005; N.S., not significant).
Results
SNAP-47 Is a Partner of VAMP7
To identify new partners of VAMP7, we took advantage of stable MDCK cell lines expressing GFP-VAMP7 and GFP-Longin domain of VAMP7 (31). After immunoprecipitation of GFP-VAMP7, GFP-Longin, and GFP alone (Fig. 1A) and analysis of eluates by mass spectrometry, we were able to identify potential partners of VAMP7. We considered only the proteins that were found in three independent experiments and excluded all the proteins that were present in the GFP control in each of these experiments. We identified proteins from different families, some previously found in our yeast two-hybrid (Y2H) screens of VAMP7 and new ones (Fig. 1B and supplemental Table 1). We did not find AP-3δ (18), Hrb (38), or Varp (39), identified in Y2H screens, suggesting that these partners interact with VAMP7 either transiently or with weak affinity as suggested by structural studies (40, 41) or were not prone to identification by proteomics for technical reasons. First and foremost, we identified SNAREs as VAMP7 partners (Fig. 1B and supplemental Table 1) as follows: SNAP-23 (8), SNAP-29 (18, 42), and syntaxin 10 (18), which are already described partners, whereas SNAP-47 and syntaxin 12/13 appeared as novel interacting proteins. To confirm these interactions, we immunoprecipitated full-length VAMP7, the Longin domain alone, or the protein deleted from this domain (ΔLongin). The extracts were obtained after NEM treatment to inhibit the SNARE complex dissociation by NSF (NEM-sensitive factor) or DTT-inactivated NEM treatment as control as described previously (8). We were able to confirm interactions between VAMP7 (full length and ΔLongin) and STX12/13, SNAP-23, SNAP-29, and SNAP-47 (Fig. 1C). We further confirmed the interaction between SNAP-47 and VAMP7 in both directions using GFP- and Myc-tagged proteins and corresponding anti-tag immunoprecipitations. Because of the low level of SNAP-47 immunoprecipitated by the Myc antibody, we also used the GFP-tagged version of SNAP-47 and RFP-VAMP7 and confirmed their coimmunoprecipitation by the GFP trap method (Fig. 1E). As for the other SNAREs, SNAP-47 binding required VAMP7 SNARE domain because no interaction was detected with the Longin domain alone. Also, SNAP-47 interaction with VAMP7 (Fig. 1, C and D) was increased following NEM treatment compared with NEM + DTT control condition indicating that VAMP7·SNAP-47-containing complexes are likely targeted by NSF for disassembly. We thus identified SNAP-47 as a new partner of VAMP7. This interaction required the VAMP7 SNARE domain, and the two proteins likely participate in the formation of a cognate SNARE complex.
FIGURE 1.
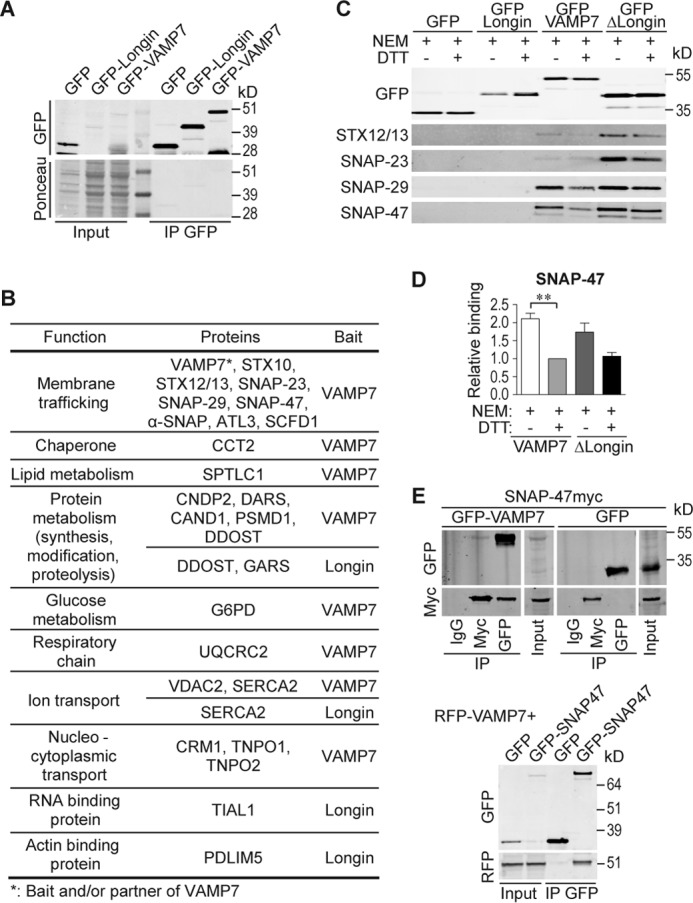
VAMP7 interactome identified by proteomics. A and B, after IP by anti-GFP Ab in transfected MDCK cells, proteins were analyzed by mass spectrometry (LC-MS/MS). A, amount of bait proteins was assessed by Ponceau S stain and the corresponding WB revealed with anti-GFP Ab (input, 50 μg; IP, 1 mg). B, table shows VAMP7- and longin-interacting proteins classified based on their molecular functions. C, GFP-tagged proteins from transfected MDCK cells treated with NEM or NEM + DTT were immunoprecipitated using anti-GFP Ab. The presence of interaction with endogenous STX12/13, SNAP-23, SNAP-29, or SNAP-47 was assessed by WB using corresponding Abs. D, quantification of ratio co-IP/IP (SNAP-47/VAMP7 constructs) from C. SNAP-47/VAMP7 in NEM + DTT condition was arbitrarily set to 1. Column chart displays mean ± S.E. All data are representative of three independent experiments (hereafter n = 3). One-way ANOVA, Bonferroni's post-test. **, p < 0.005. E, interaction between SNAP-47 and VAMP7 demonstrated by IP in COS7 cells expressing either GFP- and Myc-tagged proteins with mouse IgG IPs carried out as control (top panel) or GFP-SNAP47 and RFP-VAMP7 using GFP-trap immunoprecipitation (bottom panel).
SNAP-47 Is Mainly Localized in a Compartment between ER and Golgi Apparatus
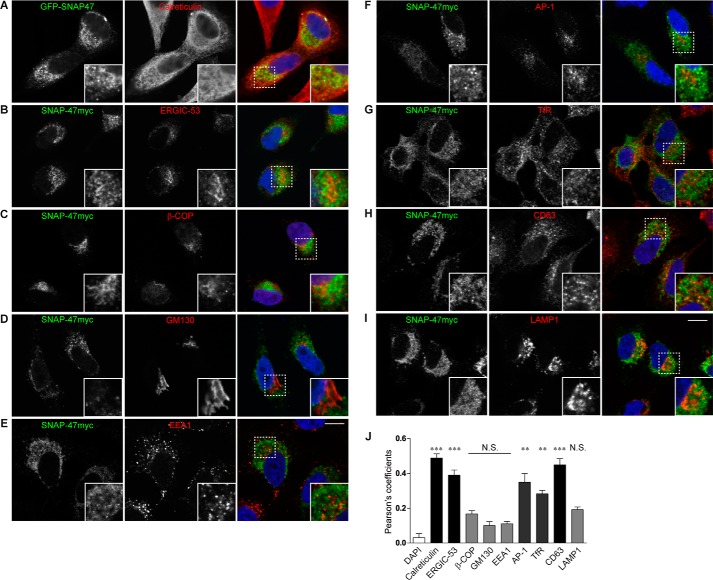
To understand the cellular function of SNAP-47, we performed immunostaining in HeLa cells transfected with SNAP-47myc or GFP-SNAP47 to circumvent low sensitivity of available SNAP-47 antibodies and permeabilized with digitonin prior to fixation to remove the excess cytosolic pool of the protein (Fig. 2, A–I, estimation of colocalizations, Fig. 2J). Immunostaining showed that tagged SNAP-47 partially colocalized with calreticulin, an ER marker (Fig. 2A), and ERGIC-53, an intermediate compartment marker (Fig. 2B). We did not find significant colocalization with β-Cop (Fig. 2C), a COPI coat component involved in a retrograde pathway from Golgi to ER, a cis-Golgi marker, GM130 (Fig. 2D), an early endosomal marker, EEA1 (Fig. 2E), nor the lysosomal marker, LAMP1 (Fig. 2I). However, SNAP-47 also partially colocalized with TGN and endosomal clathrin adaptor AP-1 (Fig. 2F), recycling endosomal transferrin receptor (TfR, Fig. 2G) and late endosomal and secretory lysosomal CD63 (Fig. 2H). Time-lapse imaging experiments further showed that a pool of SNAP-47 is enriched in some poorly motile ERGIC53-positive structures (supplemental Movie 1). Altogether, these data suggested that SNAP-47 localized partially in endosomes and mainly localized to the ER and ERGIC where it may regulate the early secretory pathway of VAMP7.
FIGURE 2.
SNAP-47 localizes mainly to the ER/ERGIC. Immunolocalization of overexpressed SNAP-47 with different compartment markers as follows: calreticulin (A), ERGIC53 (B), β-COP (C), GM130 (D), EEA1 (E), AP-1 (F), transferrin receptor (TfR) (G), CD63 (H), and LAMP1 (I). HeLa cells were transfected with Myc- or GFP-tagged SNAP-47 and processed after 16–24 h for immunofluorescence. Dashed boxes indicate magnified regions displayed in the bottom right corners. Scale bars = 10 μm. Colocalizations were estimated with Pearson's coefficient and compared with the DAPI (J). Column chart displays mean ± S.E. Significance is determined by one-way ANOVA, Dunn's post-test. **, p < 0.005; ***, p < 0.0005; N.S., not significant.
SNAP-47 Interacts with VAMP4, VAMP7, and VAMP8
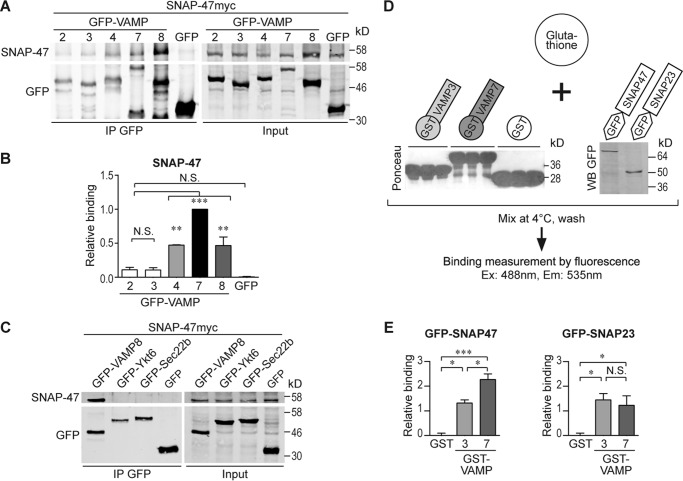
To determine whether or not VAMP7 was the only specific v-SNARE partner of SNAP-47, we immunoprecipitated seven GFP-tagged v-SNAREs and assayed for coprecipitation of Myc-tagged SNAP-47. We found that SNAP-47 interacted with VAMP4, VAMP7, and VAMP8 (Fig. 3, A and B). A weak interaction of SNAP-47 with the brevins VAMP2 and VAMP3 was detectable (Fig. 3, A and B), although we could not detect any significant interaction with the Longins Ykt6 and Sec22b (Fig. 3C). Selective binding of SNAP-47 was confirmed using an in vitro binding assay (Fig. 3, D and E). Indeed, more GFP-SNAP47 was retained by GST-VAMP7 than by GST-VAMP3, although there was no difference in the amount of SNAP-23 retained by these two VAMPs (Fig. 3E). Therefore, SNAP-47 interacted preferentially with RG-SNAREs (43) operating in post-Golgi compartments. Nevertheless, the presence of a longin domain was not critical, as demonstrated by the lack of interaction with the other members of the longin family Ykt6 and Sec22b (Fig. 3C). We then used live imaging of VAMPs with the advantage that exogenous expression allows us to emphasize the newly synthesized pool of proteins. We found that SNAP-47 localized in central and sometimes peripheral poorly motile structures among which some transiently colocalized with VAMP7 and VAMP4. Most colocalization was in perinuclear regions, but a highly dynamic cotransport appeared limited (supplemental Movies 2 and 3). Dynamics studies combined with our biochemical experiments thus suggest that at least some VAMP4 and VAMP7 may interact with SNAP-47 in the ERGIC and/or ER and that VAMPs-positive vesicles may then emerge from these SNAP-47-containing structures.
FIGURE 3.
SNAP-47 interacts mainly with VAMP4, -7, and -8. A–C, COS7 cells were cotransfected with SNAP-47myc and either with GFP-VAMP2–4, -7, and -8 or GFP (A) or either with GFP-VAMP8, -Ykt6, -Sec22b, or GFP (C). GFP-VAMPs were immunoprecipitated using anti-GFP Ab. B, quantification of co-IP/IP (SNAP-47/GFP-tagged proteins) from A. n = 4. SNAP-47/GFP-VAMP7 ratio was arbitrarily set to 1. D and E, in vitro binding assay. Lysates from HeLa cells expressing GFP-SNAP47 or GFP-SNAP23 were incubated with purified control GST, GST-VAMP3 or -7. D, scheme summarizing the assay with Ponceau S stain showing the relative amount of the different GST-tagged proteins coupled to the glutathione beads at the end of the experiment and control GFP immunoblot (E). Quantification of the binding assays (n = 3). Column chart displays mean ± S.E. One-way ANOVA, Tukey's post-test. *, p < 0.05; **, p < 0.005; ***, p < 0.0005; N.S., not significant.
SNAP-47 Interacts with Syntaxin 5 and Syntaxin 1 When Retained in the ER
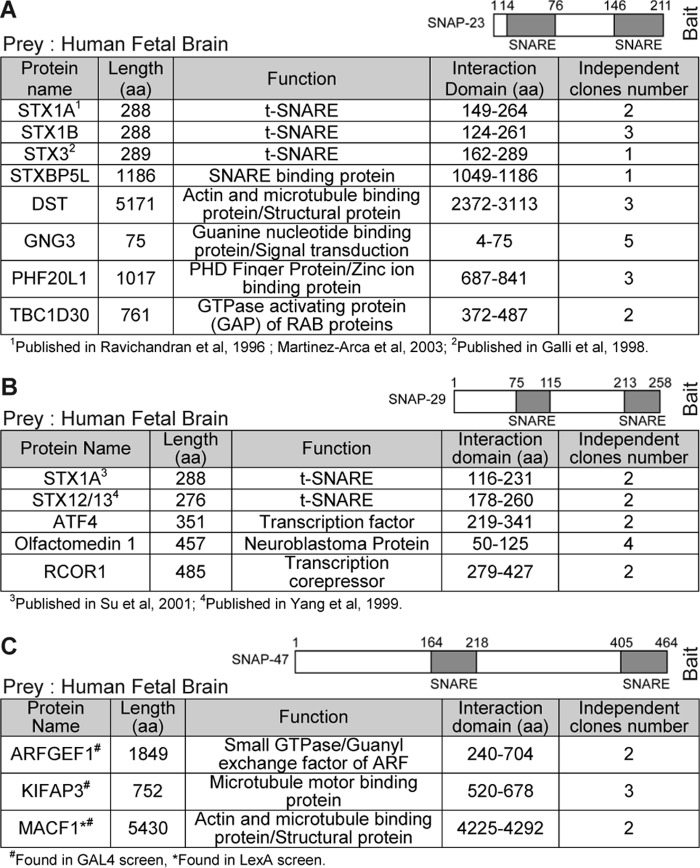
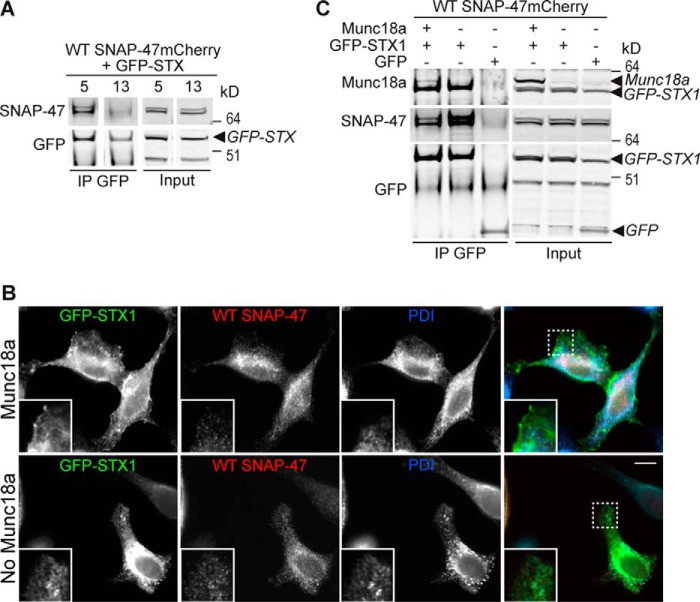
To gain more insights into the molecular properties of SNAP-47 and its partners, we carried out comparative Y2H screens of SNAP-23, SNAP-29, and SNAP-47. Several prey were isolated (Fig. 4), and regarding interaction with t-SNAREs, as expected, syntaxins were found in SNAP-23 and SNAP-29 screens (Fig. 4, A and B): syntaxins from the plasma membrane, i.e. 1A, 1B, and 3 for SNAP-23 and syntaxins 1A and 12/13 for SNAP-29. Nevertheless, while using SNAP-47 as bait (Fig. 4C), we failed to find any SNARE in two independent screens using LexA and Gal4 reporter systems (44), even if its interaction with syntaxin 1 was already described in vitro (23). To go further in SNAP-47 biochemical properties, we tested whether it could effectively interact with t-SNAREs in vivo to participate in the formation of SNARE complexes. As SNAP-47 at least partially localized in the ER and a subset of endosomes (Fig. 2), we tested its interaction with syntaxin 5, which mediates ER to Golgi transport (45), and syntaxin 13, which localizes in tubular early and recycling endosomes (46) and interacts with VAMP7 (Fig. 1, B and C). Using HeLa cell transfections with GFP-tagged syntaxins and SNAP-47 mCherry (Fig. 5), we indeed detected interaction between SNAP-47 and syntaxin 5 but very poor interaction with syntaxin 13 (Fig. 5A). SNAP-47 was already described to interact in vitro with syntaxin 1, which is present at the neuronal plasma membrane (23). Here, we took advantage of the fact that exogenous syntaxin 1 is retained in the ER where it interacts with and retains cognate post-Golgi v-SNAREs VAMP3 and VAMP7 but can reach the plasma membrane when Munc18a is coexpressed in non-neuronal cells (Fig. 5B) (21). We found that SNAP-47 interacted with syntaxin 1 in the absence of Munc18a, i.e. when retained in the ER, and to a much lower extent (57 ± 12% less) in the presence of Munc18a, i.e. when syntaxin 1 is at the plasma membrane (Fig. 5C). Altogether, these data suggest that SNAP-47 is able to participate in the formation of SNARE complexes with syntaxins present in the ER/ERGIC compartments.
FIGURE 4.
SNAP-23, SNAP-29, and SNAP-47 yeast two-hybrid screens results. A–C, Y2H screens using human SNAP-23 (A), -29 (B), and -47 (C) as baits and human fetal brain library as prey. Only high confidence interactions are presented. The detailed proteomics and Y2H screen results, including interactions with a lower confidence score, can be browsed using the PIMRider software.
FIGURE 5.
SNAP-47 interacts with syntaxin 5 and syntaxin 1 when retained in the ER in the absence of Munc18a. A, GFP IP on HeLa cells cotransfected with SNAP-47mcherry and either GFP-syntaxins 5 or 13. B, lysates from HeLa cells expressing SNAP-47mcherry and GFP or GFP-syntaxin 1 in the presence or absence of cotransfected Munc18a were submitted to GFP IP (B). C, control immunostaining of B. PDI was used as an ER marker. Dashed boxes indicate magnified regions displayed in bottom left corners. n = 3. Scale bar = 10 μm.
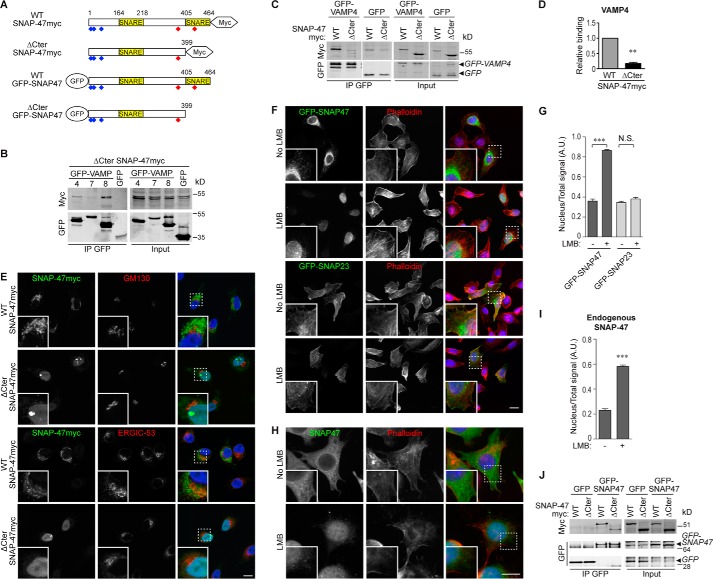
Nuclear Localization and Impaired VAMP Binding Properties of a Truncated SNAP-47 Mutant
In SNAP-47 screens (Fig. 4C), we identified KIFAP3 and MACF1, which are involved in actin and microtubule dynamics (47, 48) like DST in the SNAP-23 screen (Fig. 4A), and ARF-GEF1/BIG1, a guanine nucleotide exchange factor for the small GTPases ARF1 and ARF3 (49). We thus hypothesized that SNAP-47 may also act by recruiting molecules involved in budding (ARF-GEF1) and transport (KIFAP3 and MACF1) for Golgi to ER and post-Golgi transport of SNAREs. If this is indeed the case, affecting SNAP-47 function should lead to defect in the post-Golgi localization and function of its VAMP partners and disturb their functions. We thus generated a truncated form of SNAP-47 by deleting a C-terminal portion (ΔCter SNAP-47) that resulted in loss of the downstream SNARE domain (Fig. 6A). Indeed, in Drosophila with a loss of function in its closest family member, SNAP-29, rescue experiments indicate that both SNARE domains are required for function (50). Interaction of the ΔCter SNAP-47 and v-SNAREs was strongly impaired compared with WT (Fig. 6, B–D). We conclude that removing one coiled-coil domain affects the interactions between SNAP-47 and v-SNAREs. Surprisingly, the deletion mutant localized predominantly in the nucleus (Fig. 6E). We found by sequence prediction analysis several putative NLS in the N-terminal region of SNAP-47 (amino acids 5–70), and two (NES in the C-terminal portion (around amino acids 356 and 426) (Fig. 6A). In HeLa cells treated with leptomycin B (LMB), a CRM1/exportin1-dependent nuclear export inhibitor, a large pool of WT GFP-SNAP47 was further found in the nucleus, suggesting a functional NES in SNAP-47 (Fig. 6, F and G). Furthermore, in ΔCter SNAP-47, only the second NES site had been deleted indicating its requirement for SNAP-47 exit from the nucleus. Interestingly, SNAP-47 is the only member of the human SNAP-25 family (SNAP-23, SNAP-25, SNAP-29, and SNAP-47) to exhibit several predicted NLS sites. Accordingly, LMB had no effect on GFP-SNAP23 localization (Fig. 6, F and quantification in G) demonstrating the specificity of the effect on SNAP-47. We further found that LMB triggered nuclear accumulation of endogenous SNAP-47 in L929 cells (Fig. 6, H and quantification in I).
FIGURE 6.
Nuclear localization and impaired VAMP binding properties of a truncated SNAP-47 mutant. A, GFP- and Myc-tagged constructs of SNAP-47 used in the study. Blue and red diamonds indicate in silico predicted NLS and NES signals, respectively. B, COS7 cells were transfected with ΔCter SNAP-47myc form and GFP-VAMP4, -7, or -8. GFP-tagged VAMPs were immunoprecipitated 16–24 h after transfection (n = 3). C, COS7 cells were transfected with GFP-VAMP4 or GFP alone and SNAP-47myc either WT or ΔCter form. GFP-VAMP4 and GFP were immunoprecipitated 16–24 h after transfection. D, ratios of SNAP-47myc co-IP/GFP-VAMP4 IP were quantified for WT and ΔCter forms. WT SNAP-47 co-IP/GFP-VAMP4 IP ratio was set to 1. n = 3. Significance is determined by Student's t test. E, immunolocalization of SNAP-47myc WT and ΔCter forms in HeLa cells. Cells were transfected with SNAP-47myc and processed after 16–24 h for immunofluorescence. Cells were stained for Myc and costained with GM130 or ERGIC-53 markers. F, immunolocalization of WT GFP-SNAP47 or GFP-SNAP23 after LMB treatment. HeLa cells were transfected with indicated GFP-tagged construct and treated after 16–24 h with LMB or control. Cells were stained with anti-GFP antibody and phalloidin. Dashed boxes indicate magnified insets displayed in bottom left corners. Scale bars = 10 μm. G, quantification of nuclear signal of indicated GFP-tagged protein from F (>29 cells per condition). Significance is determined by one-way ANOVA, Dunn's post-test. H, immunolocalization of endogenous SNAP-47 after LMB treatment in L929 cells. Cells were stained with anti-SNAP-47 antibody and phalloidin. Dashed boxes indicate magnified insets displayed in bottom left corners. Scale bars = 10 μm. I, quantification of nuclear signal of endogenous SNAP-47 from H (>37 cells per condition). Significance is determined by Student's t test. J, HeLa cells were transfected with GFP-SNAP-47 WT or GFP alone and SNAP-47myc either WT or ΔCter forms. GFP-SNAP47 and GFP were immunoprecipitated 16–24 h after transfection. Column charts display mean ± S.E. **, p < 0.005; ***, p < 0.0005; N.S., not significant.
Thus, the C-terminal part of SNAP-47 is involved in interaction with v-SNAREs and accumulation in the cytoplasm by promoting nuclear export. To address the possibility that ΔCter-SNAP-47 expression may affect the function of the endogenous protein in a dominant negative manner, we tested its interaction with the WT form (Fig. 6J). We found both WT and ΔCter-SNAP-47 in the GFP-SNAP47 immunoprecipitates, indicating that SNAP-47 forms multimers and that the C-terminal truncation mutant still interacts with the full-length molecule, albeit to a lesser extent than full length. Therefore, the ΔCter-SNAP-47 mutant could trap SNAP-47 WT and potentially have a dominant negative effect in cells.
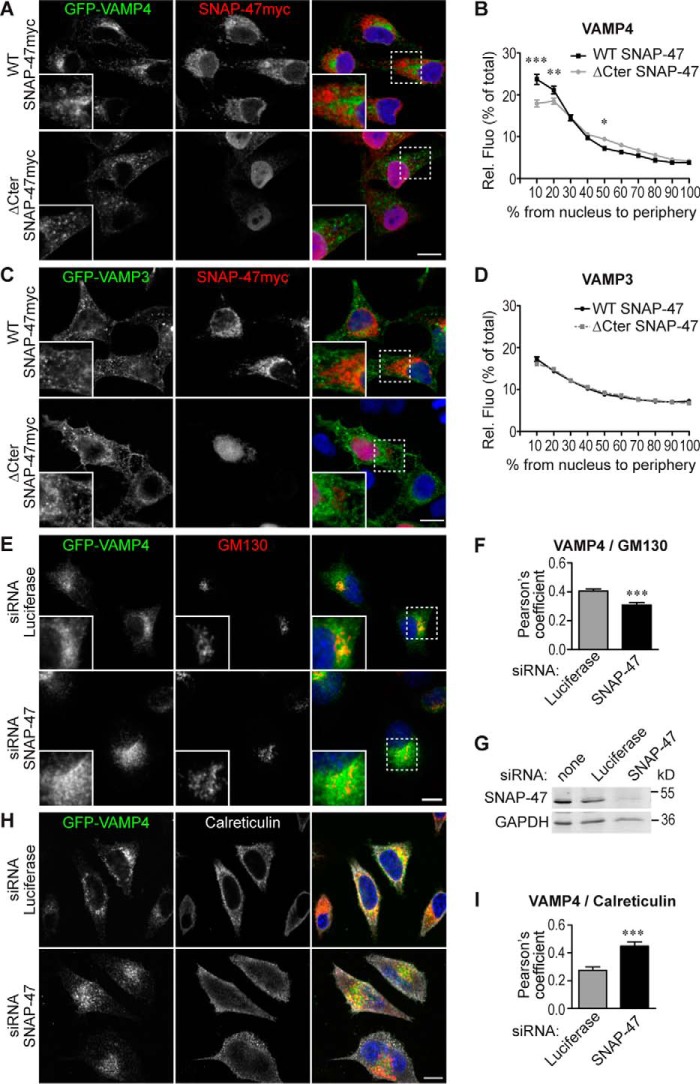
SNAP-47 Regulates VAMP4 Subcellular Localization
Based on the biochemical properties of ΔCter-SNAP-47 and its potential dominant negative role, we sought to test its effect on VAMPs' subcellular localization. To this end, we expressed either full-length SNAP-47myc or the ΔCter-SNAP-47myc with GFP-VAMP4 and GFP-VAMP3 to obtain significant amounts of newly synthesized v-SNAREs (Fig. 7, A and C). We found a partial delocalization of GFP-tagged VAMP4 upon expression of ΔCter-SNAP-47. Indeed, by measuring the distribution of GFP-VAMP4 from the nucleus to periphery, we found that VAMP4 had a more homogeneous distribution in ΔCter-SNAP-47 compared with WT SNAP-47-expressing cells (Fig. 7B). In contrast, we did not find any delocalization of GFP-VAMP3 (Fig. 7D). These results led us to conclude that ΔCter-SNAP-47 has a dominant negative effect on the localization of VAMP4.
FIGURE 7.
Mutation and knockdown of SNAP-47 alter VAMP4 subcellular localization. A–D, HeLa cells were transfected with WT or ΔCter SNAP-47myc and GFP-VAMP4 (A and B) or -3 (C and D). Cells were immunostained (A and C) and relative fluorescence intensity (Rel. Fluo) of VAMP4 (B) and -3 (D) from nucleus to periphery was plotted (see under “Materials and Methods”). Significance is determined by two-way ANOVA, Bonferroni's test (between 28 and 45 cells per condition). E–I, L929 cells were transfected with GFP-VAMP4 and indicated siRNAs (luciferase as control). Cells were immunostained using anti-GFP and anti-GM130 (E) or anti-calreticulin (H) Abs. Scale bars = 10 μm. Dashed boxes indicate magnified regions displayed in bottom left corners. G, efficiency of siRNAs knockdown was checked by WB (GAPDH as loading control). Colocalization between VAMP4 and GM130 (F) or calreticulin (I) was estimated with Pearson's coefficient. Student's t test (>100 cells for F and >16 cells for I). Column charts and line graphs display mean ± S.E. *, p < 0.05; **, p < 0.005; ***, p < 0.0005.
To further define the function of SNAP-47, we silenced its expression in L929 cells, a mouse fibroblast line in which endogenous SNAP-47 was more readily detectable than in HeLa cells using available antibodies, and we assayed for VAMP4 subcellular localization (Fig. 7, E and H, and verification of SNAP-47 knockdown in Fig. 7G). We found that GFP-VAMP4 was less concentrated in the Golgi apparatus (as evidenced by Pearson's coefficient analyses with GM130; Fig. 7, E and F) but more in the ER (Pearson's coefficient analyses with calreticulin; Fig. 7, H and I) in SNAP-47 siRNA-treated compared with control cells. These results led us to conclude that SNAP-47 is required for the proper subcellular localization of VAMP4, likely through a role in the early transport of the v-SNARE in the secretory pathway.
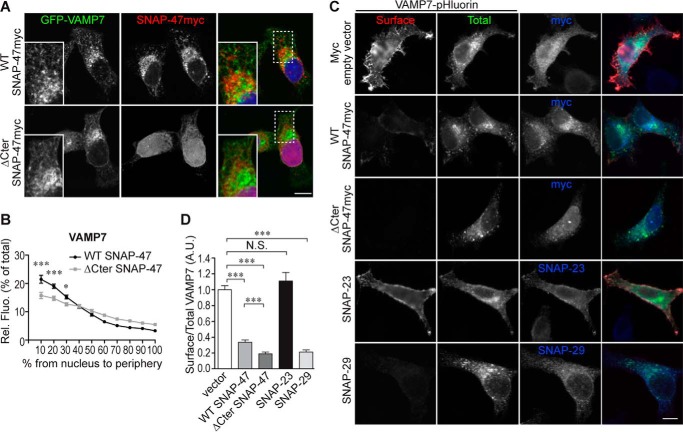
SNAP-47 Regulates VAMP7 Subcellular Localization and Exocytosis
We then sought to determine the role of SNAP-47 on VAMP7 localization and function. To this end, we expressed as above either full-length SNAP-47myc or the ΔCter-SNAP-47myc with GFP-VAMP7 (Fig. 8A). By measuring the distribution of GFP-VAMP7 from the nucleus to periphery, we found that VAMP7 had a more homogeneous distribution in ΔCter-SNAP-47 compared with WT SNAP-47-expressing cells (Fig. 8B), like in the case of VAMP4, suggesting again impaired transport in the early secretory pathway. To go further, we took advantage of the fact that VAMP7 exocytosis can be directly assessed by surface staining of pHluorin-tagged VAMP7 (7, 32). Indeed, when the luminal tag is being exposed to the extracellular medium upon exocytosis of the VAMP7-positive vesicle, it becomes accessible to extracellular anti-GFP antibodies (Fig. 8C). We quantified the amount of VAMP7 at the plasma membrane as a ratio between the surface and the total staining for VAMP7 (Fig. 8D). Expression of ΔCter-SNAP-47 construct had a dramatic effect on VAMP7 exocytosis, probably due to the inefficient transport of VAMP7 to the cell periphery noted above. Surprisingly, SNAP-47 WT overexpression also decreased VAMP7 presence at the cell surface. We then tested whether all the members of the SNAP-25 family induce the same phenotype. Overexpression of the plasma membrane SNAP-23 had no significant effect on surface expression of VAMP7. On the contrary, overexpression of SNAP-29, the closest SNAP-47 member of the family (23) and partner of VAMP7 (Fig. 1C) (50), also had a strong inhibitory effect like SNAP-47. As a conclusion, we demonstrated that SNAP-47 is implicated in the subcellular localization and function of a subset of v-SNAREs likely via a role in their early transport.
FIGURE 8.
Mutation of SNAP-47 alters VAMP7 subcellular localization and exocytosis. A and B, HeLa cells were transfected with GFP-VAMP7 and WT or ΔCter SNAP-47myc. Cells were immunostained (A) and relative fluorescence intensity (Rel. Fluo) of VAMP7 from nucleus to periphery was quantified (B). Dashed boxes indicate magnified regions displayed in bottom left corners. Two-way ANOVA, Bonferroni's post-test (>24 cells per condition). C and D, HeLa cells were transfected with VAMP7-pHluorin and indicated SNAPs plasmid. C, cells were immunostained with anti-Myc, SNAP-23, or -29 and anti-GFP Abs for surface (red) and total (green) VAMP7-pHluorin. D, ratio of surface/total VAMP7-pHluorin fluorescence intensity was quantified (>100 cells) for each condition. One-way ANOVA, Dunn's post test. Column chart and line graph display mean ± S.E. *, p < 0.05; ***, p < 0.0005; N.S., not significant. Scale bars = 10 μm.
Discussion
In this study we report the identification of SNAP-47 as a VAMP7 partner. SNAP-47 interacts with full-length VAMP7 and ΔLongin VAMP7 but not with the longin domain alone, suggesting an interaction through the SNARE domain of VAMP7. We found that SNAP-47 mainly colocalized with ER and ERGIC markers and a pool of SNAP-47 localized in some poorly motile ERGIC53-positive structures in live cells. We also found significant colocalization of SNAP-47 with late and to a lower extent with recycling endosomal markers, suggesting that SNAP-47 could be implicated, as SNAP-29 (50), in several intracellular membrane fusion events. We further found that SNAP-47 not only interacted with VAMP7 but also with VAMP4 and VAMP8. The predominant localization of SNAP-47 in ER and ERGIC and its preferential interaction with the ER/Golgi syntaxin 5 and syntaxin 1 only when the latter is retained in the ER strongly suggest a role in the early secretory pathway. SNAP-47 did not interact with Sec22b and Ykt6, both involved in ER to Golgi and intra-Golgi transports (51–53). Interestingly, even if VAMP7 is predominantly implicated in late endosomal pathways, it has also been shown to be a bona fide functional ER constituent in rat intestines in a complex with syntaxin 5 and involved in the ER to Golgi transport of prechylomicron (54), thus suggesting that in certain conditions and cells, post-Golgi VAMPs could operate also in the early secretory pathway. Altogether, these data suggest that SNAP-47 may interact with VAMPs (4, 7, 8) in cis-complexes being transported in the early secretory pathway until the complexes are disassembled by NSF and fusion competency of the VAMPs is activated (i.e. at the exit of the Golgi apparatus). Alternatively, SNAP-47 may allow VAMPs (4, 7, 8) to mediate fusion events in the early secretory pathway. Further studies are required to definitively sort out these two hypotheses.
To study SNAP-47 function, we generated a C-terminally truncated form of SNAP-47 (ΔCter-SNAP-47), and we found it enriched in the nucleus. In silico study of human SNAP-47 protein sequence unraveled the presence of NLS and allowed us to discover that full-length SNAP-47 shuttles between the nucleus and the cytoplasm by a CRM1/exportin1-dependent pathway, but its potential role in the nucleus will require further study. Importantly, ΔCter-SNAP-47 was shown to interact with WT SNAP-47 suggesting that it could trap endogenous SNAP-47 and act as a dominant negative mutant. Indeed, expression of ΔCter-SNAP-47, which was impaired in VAMP4 interaction, nevertheless affected the subcellular localization of VAMP4. The role of SNAP-47 for the proper subcellular localization of VAMP4 was further demonstrated by the fact that SNAP-47 knockdown resulted in decreased VAMP4 concentration in the Golgi apparatus and retention in the ER. These results likely indicate the occurrence of defective transport of the newly synthesized VAMP4 from the ER to the Golgi when SNAP-47 function is impaired by silencing or expression of a truncated mutant. In good agreement, we further found that expression of ΔCter-SNAP-47 impaired the subcellular localization and transport to the cell surface of VAMP7 thus suggesting that SNAP-47 is required for VAMP7 localization and function. Surprisingly, overexpression of WT SNAP-47 and SNAP-29 but not SNAP-23 also inhibited VAMP7 exocytic function. An inhibitory role of SNAP-29 in plasma membrane fusion has already been described at the apical membrane in Drosophila epithelial cells, although it would have a positive function in the intracellular trafficking route from late endosomes to lysosomes (50). Thus, SNAP-47 and SNAP-29, which are not tightly membrane-associated like palmitoylated SNAP-23, may outcompete SNAP-23 thus preventing exocytic membrane fusion and favor intracellular fusion events at the expense of secretory membrane fusion involving SNAP-23 when they are expressed at a high enough level. This hypothesis will require further in vivo and in vitro investigations.
In addition to its function as a SNARE and potential role in the nucleus, SNAP-47 also likely participates in membrane trafficking via interactions with MACF1, KIFAP3, and ARF-GEF1/BIG1, three molecules that play a role in the ER-Golgi, post-Golgi, and microtubule dynamics, as revealed by Y2H screens. KIFAP3, a subunit of Kinesin 2 motor, is localized in the ER and plays a role in COPI-dependent retrograde transport from Golgi to ER (55). MACF1 is essential for cytoskeletal dynamics and is involved, together with GolginA4, in the transport from the Golgi to the cell periphery of glycosylphosphatidylinositol-anchored proteins and VAMP7 vesicles (32, 56). ARF-GEF1/BIG1 is mainly localized in the TGN and is involved in trafficking between TGN and endosomes (57). Interestingly enough, knockdown of SNAP-47 was found to induce a condensed cis-Golgi, similar to the effect of latrunculin B (58); and knockdown of syntaxin 5 causes a Golgi fragmentation (59) and ER spreading (60). SNAP-47 could thus also recruit factors involved in the regulation of the structural organization of the ER or Golgi-ER and post-Golgi transport possibly through interaction with cytoskeletal elements. From our seminal findings, the precise role of SNAP-47 in connection with non-SNARE partners will now require in-depth investigation.
We conclude that SNAP-47 likely contributes to the proper subcellular localization and function of a subset of VAMPs (4, 7, 8) via the formation of SNARE complexes in the early transport of the VAMPs, and it likely has additional roles, including in the nucleus and together with the cytoskeleton yet to be explored.
Author Contributions
A. K., T. G., and V. P. G. designed the research. A. K., S. N., F. D., B. V., C. G., J. C. B., M. N., T. M., and V. P. G. performed the research. A. K., S. N., and V. P. G. contributed to new reagents or analytic tools. A. K., S. N., D. L., E. F., T. G., and V. P. G. analyzed the data. A. K., S. N., T. G., and V. P. G. wrote the paper.
Supplementary Material
Acknowledgments
We are grateful to N. Sakamoto for help with experiments; T. Binz for SNAP-29 and H. Ben-Tekaya for GFP-ERGIC53 plasmids; and C. Jackson, S. Robine, L. Pintard for discussions. We acknowledge the ImagoSeine Facility, member of the France BioImaging infrastructure supported by the French National Research Agency Grant ANR-10-INSB-04, “Investments of the Future.”
This work was supported by grants from INSERM, CNRS, Association pour la Recherche sur le Cancer, Fondation pour la Recherche Médicale, and Ecole des Neurosciences de Paris, awards from Association Robert Debré pour la Recherche Médicale, French National Research Agency Grant NeuroImmunoSynapse ANR-13-BSV2-0018-02 (to T. G.), GenHomme Network Grant 02490-6088 (to Hybrigenics), and fellowships from Association pour la Recherche sur le Cancer and Who Am I?, and Labex Grant Idex ANR-11-IDEX-0005-01 (to A. K.). M. N, T. M., and E. F. are employees and E. F. is shareholder of Hybrigenics.

This article contains supplemental Movies S1–S3 and Tables S1 and S2.
- TGN
- trans-Golgi network
- ER
- endoplasmic reticulum
- Ab
- antibody
- WB
- Western blot
- MDCK
- Madin-Darby canine kidney cell
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- IP
- immunoprecipitation
- ANOVA
- analysis of variance
- Y2H
- yeast two-hybrid
- LMB
- leptomycin B
- VAMP
- vesicle-associated membrane protein
- NEM
- N-ethylmaleimide
- NLS
- nuclear localization signal
- NES
- nuclear export signal
- NSF
- NEM-sensitive factor
- STX
- syntaxin.
References
- 1.Cai H., Reinisch K., and Ferro-Novick S. (2007) Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell 12, 671–682 [DOI] [PubMed] [Google Scholar]
- 2.Söllner T., Bennett M. K., Whiteheart S. W., Scheller R. H., and Rothman J. E. (1993) A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell 75, 409–418 [DOI] [PubMed] [Google Scholar]
- 3.Südhof T. C., and Rothman J. E. (2009) Membrane fusion: grappling with SNARE and SM proteins. Science 323, 474–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coco S., Raposo G., Martinez S., Fontaine J. J., Takamori S., Zahraoui A., Jahn R., Matteoli M., Louvard D., and Galli T. (1999) Subcellular localization of tetanus neurotoxin-insensitive vesicle-associated membrane protein (VAMP)/VAMP7 in neuronal cells: evidence for a novel membrane compartment. J. Neurosci. 19, 9803–9812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Advani R. J., Yang B., Prekeris R., Lee K. C., Klumperman J., and Scheller R. H. (1999) VAMP-7 mediates vesicular transport from endosomes to lysosomes. J. Cell Biol. 146, 765–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaineau M., Danglot L., and Galli T. (2009) Multiple roles of the vesicular-SNARE TI-VAMP in post-Golgi and endosomal trafficking. FEBS Lett. 583, 3817–3826 [DOI] [PubMed] [Google Scholar]
- 7.Martinez-Arca S., Alberts P., Zahraoui A., Louvard D., and Galli T. (2000) Role of tetanus neurotoxin insensitive vesicle-associated membrane protein (TI-VAMP) in vesicular transport mediating neurite outgrowth. J. Cell Biol. 149, 889–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galli T., Zahraoui A., Vaidyanathan V. V., Raposo G., Tian J. M., Karin M., Niemann H., and Louvard D. (1998) A novel tetanus neurotoxin-insensitive vesicle-associated membrane protein in SNARE complexes of the apical plasma membrane of epithelial cells. Mol. Biol. Cell 9, 1437–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rao S. K., Huynh C., Proux-Gillardeaux V., Galli T., and Andrews N. W (2004) Identification of SNAREs involved in synaptotagmin VII-regulated lysosomal exocytosis. J. Biol. Chem. 279, 20471–20479 [DOI] [PubMed] [Google Scholar]
- 10.Pryor P. R., Mullock B. M., Bright N. A., Lindsay M. R., Gray S. R., Richardson S. C., Stewart A., James D. E., Piper R. C., and Luzio J. P. (2004) Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 5, 590–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Advani R. J., Bae H. R., Bock J. B., Chao D. S., Doung Y. C., Prekeris R., Yoo J. S., and Scheller R. H. (1998) Seven novel mammalian SNARE proteins localize to distinct membrane compartments. J. Biol. Chem. 273, 10317–10324 [DOI] [PubMed] [Google Scholar]
- 12.Tran T. H., Zeng Q., and Hong W. (2007) VAMP4 cycles from the cell surface to the trans-Golgi network via sorting and recycling endosomes. J. Cell Sci. 120, 1028–1041 [DOI] [PubMed] [Google Scholar]
- 13.Mallard F., Tang B. L., Galli T., Tenza D., Saint-Pol A., Yue X., Antony C., Hong W., Goud B., and Johannes L. (2002) Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J. Cell Biol. 156, 653–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antonin W., Holroyd C., Tikkanen R., Höning S., and Jahn R. (2000) The R-SNARE Endobrevin/VAMP-8 mediates homotypic fusion of early endosomes and late endosomes. Mol. Biol. Cell 11, 3289–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itakura E., Kishi-Itakura C., and Mizushima N. (2012) The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151, 1256–1269 [DOI] [PubMed] [Google Scholar]
- 16.Guo B., Liang Q., Li L., Hu Z., Wu F., Zhang P., Ma Y., Zhao B., Kovács A. L., Zhang Z., Feng D., Chen S., and Zhang H. (2014) O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nat. Cell Biol. 16, 1215–1226 [DOI] [PubMed] [Google Scholar]
- 17.Kutay U., Ahnert-Hilger G., Hartmann E., Wiedenmann B., and Rapoport T. A. (1995) Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 14, 217–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez-Arca S., Rudge R., Vacca M., Raposo G., Camonis J., Proux-Gillardeaux V., Daviet L., Formstecher E., Hamburger A., Filippini F., D'Esposito M., and Galli T. (2003) A dual mechanism controlling the localization and function of exocytic v-SNAREs. Proc. Natl. Acad. Sci. U.S.A. 100, 9011–9016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peden A. A., Park G. Y., and Scheller R. H. (2001) The di-leucine motif of vesicle-associated membrane protein 4 is required for its localization and AP-1 binding. J. Biol. Chem. 276, 49183–49187 [DOI] [PubMed] [Google Scholar]
- 20.Toonen R. F., de Vries K. J., Zalm R., Südhof T. C., and Verhage M. (2005) Munc1-1 stabilizes syntaxin 1, but is not essential for syntaxin 1 targeting and SNARE complex formation. J. Neurochem. 93, 1393–1400 [DOI] [PubMed] [Google Scholar]
- 21.Martinez-Arca S., Proux-Gillardeaux V., Alberts P., Louvard D., and Galli T. (2003) Ectopic expression of syntaxin 1 in the ER redirects TI-VAMP- and cellubrevin-containing vesicles. J. Cell Sci. 116, 2805–2816 [DOI] [PubMed] [Google Scholar]
- 22.Annaert W. G., Becker B., Kistner U., Reth M., and Jahn R. (1997) Export of cellubrevin from the endoplasmic reticulum is controlled by BAP31. J. Cell Biol. 139, 1397–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holt M., Varoqueaux F., Wiederhold K., Takamori S., Urlaub H., Fasshauer D., and Jahn R. (2006) Identification of SNAP-47, a novel Qbc-SNARE with ubiquitous expression. J. Biol. Chem. 281, 17076–17083 [DOI] [PubMed] [Google Scholar]
- 24.Shimojo M., Courchet J., Pieraut S., Torabi-Rander N., Sando R. 3rd, Polleux F., and Maximov A. (2015) SNAREs controlling vesicular release of BDNF and development of callosal axons. Cell Rep. 11, 1054–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jurado S., Goswami D., Zhang Y., Molina A. J., Südhof T. C., and Malenka R. C. (2013) LTP requires a unique postsynaptic SNARE fusion machinery. Neuron 77, 542–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arendt K. L., Zhang Y., Jurado S., Malenka R. C., Südhof T. C., and Chen L. (2015) Retinoic acid and LTP recruit postsynaptic AMPA receptors using distinct SNARE-dependent mechanisms. Neuron 86, 442–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pranke I. M., Morello V., Bigay J., Gibson K., Verbavatz J. M., Antonny B., and Jackson C. L. (2011) α-Synuclein and ALPS motifs are membrane curvature sensors whose contrasting chemistry mediates selective vesicle binding. J. Cell Biol. 194, 89–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das V., Nal B., Dujeancourt A., Thoulouze M. I., Galli T., Roux P., Dautry-Varsat A., and Alcover A. (2004) Activation-induced polarized recycling targets T cell antigen receptors to the immunological synapse: Involvement of SNARE complexes. Immunity 20, 577–588 [DOI] [PubMed] [Google Scholar]
- 29.Nakai K., and Horton P. (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24, 34–36 [DOI] [PubMed] [Google Scholar]
- 30.la Cour T., Kiemer L., Mølgaard A., Gupta R., Skriver K., and Brunak S. (2004) Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel, 17, 527–536 [DOI] [PubMed] [Google Scholar]
- 31.Proux-Gillardeaux V., Raposo G., Irinopoulou T., and Galli T. (2007) Expression of the Longin domain of TI-VAMP impairs lysosomal secretion and epithelial cell migration. Biol. Cell 99, 261–271 [DOI] [PubMed] [Google Scholar]
- 32.Burgo A., Proux-Gillardeaux V., Sotirakis E., Bun P., Casano A., Verraes A., Liem R. K., Formstecher E., Coppey-Moisan M., and Galli T. (2012) A molecular network for the transport of the TI-VAMP/VAMP7 vesicles from cell center to periphery. Dev. Cell 23, 166–180 [DOI] [PubMed] [Google Scholar]
- 33.Poullet P., Carpentier S., and Barillot E. (2007) myProMS, a web server for management and validation of mass spectrometry-based proteomic data. Proteomics 7, 2553–2556 [DOI] [PubMed] [Google Scholar]
- 34.Vojtek A. B., and Hollenberg S. M. (1995) Ras-Raf interaction: two-hybrid analysis. Methods Enzymol. 255, 331–342 [DOI] [PubMed] [Google Scholar]
- 35.Bartel P. L. (1993) in Cellular Interactions in Development: A Practical Approach (Hartley D. A., ed) pp. 153–179, Oxford University Press, Oxford, UK [Google Scholar]
- 36.Fromont-Racine M., Rain J. C., and Legrain P. (1997) Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat. Genet. 16, 277–282 [DOI] [PubMed] [Google Scholar]
- 37.de Chaumont F., Dallongeville S., Chenouard N., Hervé N., Pop S., Provoost T., Meas-Yedid V., Pankajakshan P., Lecomte T., Le Montagner Y., Lagache T., Dufour A., and Olivo-Marin J. C. (2012) Icy: an open bioimage informatics platform for extended reproducible research. Nat. Methods 9, 690–696 [DOI] [PubMed] [Google Scholar]
- 38.Chaineau M., Danglot L., Proux-Gillardeaux V., and Galli T. (2008) Role of HRB in clathrin-dependent endocytosis. J. Biol. Chem. 283, 34365–34373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burgo A., Sotirakis E., Simmler M. C., Verraes A., Chamot C., Simpson J. C., Lanzetti L., Proux-Gillardeaux V., and Galli T. (2009) Role of Varp, a Rab21 exchange factor and TI-VAMP/VAMP7 partner, in neurite growth. EMBO Rep. 10, 1117–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pryor P. R., Jackson L., Gray S. R., Edeling M. A., Thompson A., Sanderson C. M., Evans P. R., Owen D. J., and Luzio J. P. (2008) Molecular basis for the sorting of the SNARE VAMP7 into endocytic clathrin-coated vesicles by the ArfGAP Hrb. Cell 134, 817–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kent H. M., Evans P. R., Schäfer I. B., Gray S. R., Sanderson C. M., Luzio J. P., Peden A. A., and Owen D. J. (2012) Structural basis of the intracellular sorting of the SNARE VAMP7 by the AP3 adaptor complex. Dev. Cell 22, 979–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takáts S., and Juhász G. (2013) A genetic model with specifically impaired autophagosome-lysosome fusion. Autophagy 9, 1251–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rossi V., Picco R., Vacca M., D'Esposito M., D'Urso M., Galli T., and Filippini F. (2004) VAMP subfamilies identified by specific R-SNARE motifs. Biol. Cell 96, 251–256 [DOI] [PubMed] [Google Scholar]
- 44.Stynen B., Tournu H., Tavernier J., and Van Dijck P. (2012) Diversity in genetic in vivo methods for protein-protein interaction studies: from the yeast two-hybrid system to the mammalian split-luciferase system. Microbiol. Mol. Biol. Rev. 76, 331–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dascher C., Matteson J., and Balch W. E. (1994) Syntaxin 5 regulates endoplasmic reticulum to Golgi transport. J. Biol. Chem. 269, 29363–29366 [PubMed] [Google Scholar]
- 46.Prekeris R., Klumperman J., Chen Y. A., and Scheller R. H. (1998) Syntaxin 13 mediates cycling of plasma membrane proteins via tubulovesicular recycling endosomes. J. Cell Biol. 143, 957–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown A., Bernier G., Mathieu M., Rossant J., and Kothary R. (1995) The mouse dystonia musculorum gene is a neural isoform of bullous pemphigoid antigen 1. Nat. Genet. 10, 301–306 [DOI] [PubMed] [Google Scholar]
- 48.Leung C. L., Sun D., Zheng M., Knowles D. R., and Liem R. K. (1999) Microtubule actin cross-linking factor (MACF): a hybrid of dystonin and dystrophin that can interact with the actin and microtubule cytoskeletons. J. Cell Biol. 147, 1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kondo A., Hashimoto S., Yano H., Nagayama K., Mazaki Y., and Sabe H. (2000) A new paxillin-binding protein, PAG3/Papα/KIAA0400, bearing an ADP-ribosylation factor GTPase-activating protein activity, is involved in paxillin recruitment to focal adhesions and cell migration. Mol. Biol. Cell 11, 1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morelli E., Ginefra P., Mastrodonato V., Beznoussenko G. V., Rusten T. E., Bilder D., Stenmark H., Mironov A. A., and Vaccari T. (2014) Multiple functions of the SNARE protein Snap29 in autophagy, endocytic, and exocytic trafficking during epithelial formation in Drosophila. Autophagy 10, 2251–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu D., Joglekar A. P., Williams A. L., and Hay J. C. (2000) Subunit structure of a mammalian ER/Golgi SNARE complex. J. Biol. Chem. 275, 39631–39639 [DOI] [PubMed] [Google Scholar]
- 52.Liu Y., and Barlowe C. (2002) Analysis of Sec22p in endoplasmic reticulum/Golgi transport reveals cellular redundancy in SNARE protein function. Mol. Biol. Cell 13, 3314–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thayanidhi N., Helm J. R., Nycz D. C., Bentley M., Liang Y., and Hay J. C. (2010) α-Synuclein delays endoplasmic reticulum (ER)-to-Golgi transport in mammalian cells by antagonizing ER/Golgi SNAREs. Mol. Biol. Cell 21, 1850–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siddiqi S. A., Siddiqi S., Mahan J., Peggs K., Gorelick F. S., and Mansbach C. M. 2nd. (2006) The identification of a novel endoplasmic reticulum to Golgi SNARE complex used by the prechylomicron transport vesicle. J. Biol. Chem. 281, 20974–20982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stauber T., Simpson J. C., Pepperkok R., and Vernos I. (2006) A role for kinesin-2 in COPI-dependent recycling between the ER and the Golgi complex. Curr. Biol. 16, 2245–2251 [DOI] [PubMed] [Google Scholar]
- 56.Kakinuma T., Ichikawa H., Tsukada Y., Nakamura T., and Toh B. H. (2004) Interaction between p230 and MACF1 is associated with transport of a glycosyl phosphatidylinositol-anchored protein from the Golgi to the cell periphery. Exp. Cell Res. 298, 388–398 [DOI] [PubMed] [Google Scholar]
- 57.Ishizaki R., Shin H. W., Mitsuhashi H., and Nakayama K. (2008) Redundant roles of BIG2 and BIG1, guanine-nucleotide exchange factors for ADP-ribosylation factors in membrane traffic between the trans-Golgi network and endosomes. Mol. Biol. Cell 19, 2650–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chia J., Goh G., Racine V., Ng S., Kumar P., and Bard F. (2012) RNAi screening reveals a large signaling network controlling the Golgi apparatus in human cells. Mol. Syst. Biol. 8, 629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suga K., Hattori H., Saito A., and Akagawa K. (2005) RNA interference-mediated silencing of the syntaxin 5 gene induces Golgi fragmentation but capable of transporting vesicles. FEBS Lett. 579, 4226–4234 [DOI] [PubMed] [Google Scholar]
- 60.Miyazaki K., Wakana Y., Noda C., Arasaki K., Furuno A., and Tagaya M. (2012) Contribution of the long form of syntaxin 5 to the organization of the endoplasmic reticulum. J. Cell Sci. 125, 5658–5666 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.