Background: Although Etv2 is an essential transcriptional regulator during embryogenesis, the transcriptional regulation of Etv2 gene expression is not well understood.
Results: Multiple signaling cascades were defined as regulators of Etv2 gene expression.
Conclusion: These studies define upstream and downstream networks to regulate Etv2 gene expression.
Significance: This is the first report to comprehensively characterize the transcriptional regulation of Etv2 gene expression.
Keywords: calcineurin, ETS transcription factor family, NFAT transcription factor, transcription regulation, VEGF, transcription activation
Abstract
Etv2 is an essential transcriptional regulator of hematoendothelial lineages during embryogenesis. Although Etv2 downstream targets have been identified, little is known regarding the upstream transcriptional regulation of Etv2 gene expression. In this study, we established a novel methodology that utilizes the differentiating ES cell and embryoid body system to define the modules and enhancers embedded within the Etv2 promoter. Using this system, we defined an autoactivating role for Etv2 that is mediated by two adjacent Ets motifs in the proximal promoter. In addition, we defined the role of VEGF/Flk1-Calcineurin-NFAT signaling cascade in the transcriptional regulation of Etv2. Furthermore, we defined an Etv2-Flt1-Flk1 cascade that serves as a negative feedback mechanism to regulate Etv2 gene expression. To complement and extend these studies, we demonstrated that the Flt1 null embryonic phenotype was partially rescued in the Etv2 conditional knockout background. In summary, these studies define upstream and downstream networks that serve as a transcriptional rheostat to regulate Etv2 gene expression.
Introduction
Recent studies have begun to define regulators and networks that play key roles in the specification of hematoendothelial lineages early during embryogenesis. Flk1 and Flt1 are two important cell surface tyrosine kinase receptors for the Vegf ligand that are required for the development of mesoderm (1–3). The Flk1 mutant embryo is lethal by embryonic day (E)3 8.5 to E9.5 because of a lack of hematopoietic and endothelial lineages in both the embryo proper and the yolk sac, which is similar to the phenotype of the Vegfa null embryo (4–6). In contrast, the targeted deletion of Flt1 results in enhanced production of endothelial lineage progenitors, abnormal “supersized” vasculature, growth arrest, and embryonic lethality by E8.5 (7). However, the deletion of the tyrosine kinase domain of Flt1 does not impact vascular development, which indicates that the kinase activity of Flt1 is not essential for endothelial development (8). Rather, biochemical studies have established that Flt1 has a higher affinity for the Vegf ligand (∼10-fold) than for the Flk1 receptor (9, 10), supporting the notion that Flt1 functions as a Vegf ligand reservoir, thereby inhibiting the Vegf/Flk1 signaling pathway. Flk1 regulates multiple aspects of angiogenesis, including cell migration, cell proliferation, cell survival, and vascular permeability, that are mediated through signaling cascades initiated by tyrosine phosphorylation (1, 2). Flk1 also plays important roles in the transcriptional regulation of gene expression through its downstream effectors, including Creb1 and NFAT (11–13).
NFATs bind to the conserved core sequence (5′-GGAAA-3′) through its Rel homology region domain (14). The NFAT family is composed of five NFAT genes: NFAT1 (NFATc2, NFATp), NFAT2 (NFATc1, NFATc), NFAT3 (NFATc4), NFAT4 (NFATc3, NFATx), and NFAT5 (15, 16). NFAT factors play important roles in a variety of cellular processes, including immune cellular response, fiber-type specification in skeletal muscle, cardiovascular development, and osteoclast differentiation (15–18). Gene disruption technologies have contributed to our understanding of NFAT biology. For example, the Nfat2 null embryo dies because of perturbed cardiac morphogenesis (19, 20). Although Nfat3 knockout mice have no apparent defect, the Nfat3 and Nfat4 double knockout is lethal because of vascular defects (21). The activity of Nfat1–4 factors is regulated by Ca2+/Calcineurin (18, 22), and dephosphorylation of NFAT by Calcineurin leads to its translocation into the nuclear compartment and interaction with other cofactors, such as AP1, Myod, SRF, Rcan1, and CREB-binding protein, to regulate gene expression (14, 23–25). Rcan1 has been identified as a Calcineurin-binding factor and inhibited the activity of Calcineurin (26). Rcan1 has also been identified as a target gene of Vegf/Flk1 signaling through the Calcineurin-NFAT cascade (13, 27).
Recent studies have identified Etv2 as an essential regulator of the hematoendothelial lineages. Etv2 is expressed early during gastrulation (E7.0), up-regulated at E7.5, and then down-regulated from E8.5 during murine embryogenesis (28). Genetic studies in mice and morpholino knockdown strategies in zebrafish and Xenopus have demonstrated that Etv2 plays a critical role in mesodermal lineage specification (29–32). The transition of an Flk1+/Pdgfra+ primitive mesodermal cell population to an Flk1+/Pdgfra− lateral plate mesodermal population is perturbed in the absence of Etv2 during embryogenesis (33). Moreover, conditional knockout studies have revealed that Etv2 also regulates vitelline plexus formation, intra-aortic hematopoiesis, and the remodeling of cranial vascular structures (34). Previous studies have also established that the functional role of Etv2 is mediated, in part, through its interacting factors in a context-dependent fashion. For example, the Etv2-Foxc2 complex recognizes the Ets-Forkhead composite motif in the promoters of 30% of endothelial genes (35). In addition, Gata2 has been shown to serve as an amplifier of Etv2 activity in both endothelial and hematopoietic lineages (36). Other interacting factors, such as Jmjd1a and Ovol2, also influence the function of Etv2 (37, 38). The upstream regulators for Etv2 have been incompletely defined. Recent studies support the notion that Mesp1 and Creb1 are upstream regulators of Etv2 gene expression in the mouse (39–41). In zebrafish, Foxc1 has been reported as an upstream activator of Etv2, whereas Scl and Nkx2.5 repress Etv2 gene expression (42, 43). Interestingly, continued expression of Etv2 in hematoendothelial lineages has been shown to cause overendothelialization, reminiscent of the Flt1 mutant phenotypes (44). Despite the limited studies of Etv2 activation, a comprehensive definition of the positive and negative upstream regulators of the Etv2 gene is lacking, partially because of the lack of a suitable cell line that recapitulates the embryonic context in which Etv2 is expressed. Specifically, Etv2 expression is induced and extinguished rapidly during hematopoietic and endothelial lineage specification from the nascent mesoderm. A system that recapitulates the expression pattern of Etv2 will provide an enhanced understanding of the mechanisms governing Etv2 gene expression.
In this study, we developed a novel strategy, utilizing ES cell lines with stable integration of promoter fragments fused to a reporter, to comprehensively dissect the transcriptional regulatory Etv2 cascade early during embryonic development. Our studies demonstrate that Etv2 autoactivates its own gene expression and that Vegf/Flk1 signaling transactivates Etv2 gene expression through the Vegf/Flk1-Calcineurin-NFAT cascade. We also identified the Flk1-Etv2-Flt1 cascade as the negative feedback mechanism that down-regulates Etv2 gene expression.
Experimental Procedures
Generation of the Etv2 Promoter Mutant (Mut) ES Cells
The 3.9-kb upstream promoter was deleted in V6.5 ES cells using the CRISPR/Cas9 system following protocols described previously (45). The 5′ (AATGCAAGCTTACCCCCAGC) and 3′ (GCCAGAGGTGAGCCACGAAC) guide RNAs (gRNAs) were cloned into the mammalian codon-optimized Cas9 expressing plasmid pX459 (Addgene, plasmid 48139). V6.5 ES cells were transfected with two plasmids expressing the 5′ and 3′ gRNAs targeting the 5′ and 3′ ends of the Etv2 3.9-kb upstream promoter. 24 h after transfection, cells were treated with 2 μg/ml puromycin for 48 h. The cells were then seeded at a clonal density on mouse embryonic fibroblast feeder cells. Individual colonies were selected after 7 days, expanded, and genotyped by PCR (the WT would yield a 4501-bp product, whereas a biallelic deletion of the promoter would yield an ∼454-bp product). The PCR products were cloned into the pCR2.1 TOPO plasmid, and the deletion was verified by sequencing. The clone verified for homozygous deletion of the promoter (Mut) was characterized further by EB differentiation,qRT-PCR, and FACS.
Generation of Etv2 Mut Embryos
For in vitro synthesis of gRNAs, each gRNA was PCR-amplified from the sequence-verified plasmids used for transfections in ES cells with the T7 promoter appended to the 5′ end. The PCR products were used to transcribe the gRNAs in vitro with the MEGAscript T7 transcription kit (Thermo Fisher, catalog no. AM1334). The RNA transcripts were purified using the miRVana miRNA isolation kit (Thermo Fisher, catalog no. AM1561) and stored at −80 °C until use. The Cas9 mRNA was purchased from PNA Bio (catalog no. CR01). For microinjections, sexually immature female C57BL/6J mice (4 weeks old) were superovulated by intraperitoneal injection of 5 IU of pregnant mare serum, followed by 7.5 IU of human chorionic gonadotropin at an interval of 48 h and mated overnight with C57BL/6J male mice that were more than 12 weeks old. Zygotes were collected after 20 h of human chorionic gonadotropin injection by oviductal flushing, and zygotes with pronuclei were placed into M2 medium. Microinjection was performed using a microscope equipped with a microinjector (Eppendorf). Approximately 4 pl of RNA solution containing 40 ng/μl of CAS9 mRNA and 20 ng/μl of each of the 5′ and 3′ gRNA was injected into the cytoplasm of each zygote using continuous pneumatic pressure. Injected zygotes were transferred, on the same day, a into the oviductal ampullae (15 embryos/oviduct) of 7- to 8-week-old female ICR mice mated with vasectomized ICR males. E8.5 embryos were harvested and genotyped using genomic DNA purified from yolk sac biopsy from each embryo. Embryos genotyped for homozygous deletion of the 3.9-kb promoter were sectioned, and immunohistochemistry was performed using anti-endomucin antibody (Abcam, catalog no. ab106100). For qRT-PCR, embryos were stored immediately in TRIzol after collecting the yolk sac sample for genotyping. RNA was extracted from homozygous promoter mutants, and qRT-PCR was performed.
DNA and RNA Manipulation
The 3.9-kb Etv2 promoter-EYFP construct and transgenic mouse model have been reported previously (46). The Etv2 minireporter was constructed by subcloning conserved region (CR) I and CRII into the pGL3TATA vector. The mutation of the designated motifs was constructed in the minireporter using PCR and verified by DNA sequence analysis. The 0.7-kb Flt1 promoter and mutants were fused to pGL3-TATA as the reporter using PCR and confirmed using DNA sequence analysis. The Nfat3 plasmid was purchased from Open Biosystems. RNA extraction and cDNA synthesis were performed as described previously (41). The TaqMan probes utilized for quantitative PCR included Etv2 (Mm00468972_m1), Flk1 (Mm00440085_m1),Flt1 (Mm1210866_m1), Gapdh (4352339E-1208041), Cd31 (Mm01246167_m1), Cdh5 (Mm00486938_m1), and Cd41 (Mm00439741_m1). To detect the endogenous Etv2 gene specifically (not the overexpressed Etv2), the primer pair was designed to include the 5′-UTR region of the Etv2 gene. ZsGreen was measured using custom primers (CTACTTCAAGAACTCCTGCCC and TCGTGGTACATGCAGTTCTC) and Sso Advanced Universal SYBR Green Supermix (Bio-Rad).
ES Cell and Embryoid Body Differentiation and FACS Analysis
ES cells overexpressing HA-tagged Etv2 (iHA-Etv2) have been reported previously (28). HA-Etv2 was induced by doxycycline treatment of iHA-Etv2 EBs on day 4 for the indicated time, and then the gene expression profile was examined using qRT-PCR upon induction of HA-Etv2 (28). To construct the Etv2 promoter-ES cell reporter, the Etv2 promoter was inserted into the pZsGreen1-DR vector, which expresses the destabilized ZsGreen1 protein. The fragment containing the Etv2 promoter-ZsGreen1-DR was then subcloned into the p2Lox vector. The reporter-p2Lox vector was then electroporated into A2Lox-Cre mouse ES cells to generate the ES cell reporter. ES cell maintenance and EB differentiation were performed as described previously (47). EBs were treated with proteasome inhibitor (10 nm MG132) to prevent the degradation of the destabilized ZsGreen1-DR protein for 5 h prior to harvest. FACS analysis was performed to examine the ZsGreen1 protein profile using a BD Biosciences FACSAria II and analyzed with FlowJo software (46).
Calcineurin Inhibition
Cyclosporin A (CSA) was obtained from EMD Chemicals (catalog no. 239835), reconstituted at 25 mg/ml (20.8 mm) in DMSO, and stored at −20 °C. Immediately prior to treatment, the CSA was diluted to 0.32 mm in DMSO and then diluted 1:1000 in mouse EB differentiation medium for a final concentration of 0.32 μm CSA and 0.1% DMSO. FK506 was obtained from Enzo Life Sciences (catalog no. NC9795140) and reconstituted at 1 mg/ml (1.24 mm) in DMSO. Immediately prior to treatment, FK506 was diluted 1:1000 in mouse EB differentiation medium for a final concentration of 1.24 μm FK506 and 0.1% DMSO. ES cells (reporter line 4) were differentiated into EBs as described previously (46). Cells were treated with CSA, FK506, or vehicle (0.1% DMSO) in mouse EB medium from EB D2–4. Cells were harvested on D4. RNA extraction and cDNA synthesis were performed as described above.
ChIP and Transcriptional Assays
Chromatin preparation and ChIP assays were performed using Millipore reagent (36). Antibodies utilized in the ChIP assay included rabbit anti-HA (Y-11) and polyclonal anti-Nfat3 (H-74) sera (Santa Cruz Biotechnology). Quantitative PCR was performed with the immunoprecipitated DNA to examine the enrichment of specific regions and normalized to the Gapdh region. The primer pairs for the Etv2 proximal promoter, the Etv2 distal enhancer, and Gapdh have been reported previously (41). The primers used for the synthesis of the Flt1 proximal promoter included the following: 5′-GCTGAGTAAGCTCGGTGGAG-3′ and 5′-AGGTTCAGGGTCCTTGCTCT-3′; distal promoter, 5′-TTGCTAGGCAAGCAGGAGAT-3′ and 5′-AGACCCTTTCCCCAGAAGTG-3′. 293T cells were maintained in 10% FBS medium and transfected with FuGENE HD reagent (Promega). VEGF treatment was performed as described previously (39). Transcriptional assays were performed using the Dual-Luciferase reporter assay system (Promega) following the directions outlined in the user manual.
Animals and LacZ Staining
The Etv2 conditional knockout has been described previously (41). Flk1-Cre mice were provided by Tom N. Sato (48). Flt1 knockout mice were provided by Guo-Hua Fong (7). LacZ staining was performed as described previously (49). All embryos were stained for 6 h prior to fixation. Histological sections were counterstained with nuclear fast red using standard protocols. All mice were maintained at the University of Minnesota using protocols approved by the Institutional Animal Care and Use Committee and Research Animal Resources.
Statistics
All data represent the mean ± S.D. of at least three replicates. Kruskal-Wallis test, Dunn's test, or Student's t test was performed, and p < 0.05 was deemed to be significant (50).
Results
Characterization of the Etv2 Promoter and Enhancer
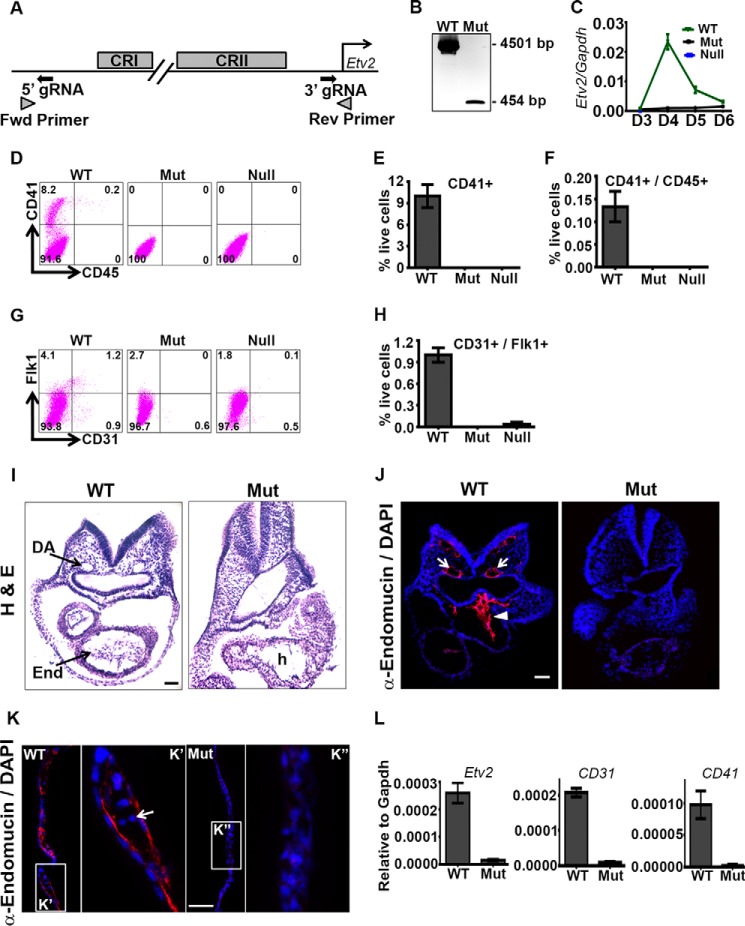
We have previously identified the 3.9-kb Etv2 promoter, which is sufficient to direct the expression of the enhanced yellow fluorescent protein (EYFP) reporter to the endocardial/endothelial and hematopoietic lineages (31, 46). However, whether all the regulatory elements required for Etv2 expression reside in this promoter fragment was not clear. In this study, we used CRISPR/Cas9 technology to delete the 3.9-kb promoter fragment in ES cells (Fig. 1A). The ES cell clone with homozygous deletion of the promoter (Mut) was verified by PCR (Fig. 1B), and the PCR product was sequence-verified for the deletion. We used the ES/EB differentiation system to further characterize the Mut cells. Gene expression analysis using qRT-PCR revealed that the Mut cells have over 95% of reduction of Etv2 compared with the WT cells in D4 EBs (Fig. 1C). We then performed FACS analysis on D6 EBs to determine the differentiation potential for these cells. Mut cells completely lacked the hematopoietic lineage, as determined by CD41/CD45 double-staining and were similar to the Etv2 null (Null) ES cells (Fig. 1, D–F). The endothelial lineage in Etv2 Mut ES cells/EBs was completely absent, as determined by CD31/Flk1 double-staining, whereas single positive cells were reduced severely (Fig. 1, G and H).
FIGURE 1.
Deletion of the 3.9-kb upstream promoter fragment of the Etv2 gene results in lethality and a lack of hematoendothelial lineages. A, schematic of the gRNA and PCR screening primer designs for deleting the Etv2 3.9-kb upstream promoter. Fwd, forward; Rev, reverse. B, PCR genotyping of Mut ES cells demonstrates the detection of only the deleted band. C, gene expression of Etv2 during EB differentiation of WT, Mut, and Null ES cells. D and G, representative FACS profiles of dissociated D6 EBs for expression of hematopoietic and endothelial markers, respectively. E and F, quantitative analysis of the FACS data in D. CD41 single- and CD41/CD45 double-positive cells are completely absent in the Mut as well as the Etv2 Null ES cells. H, quantitative analysis of the FACS data in G. CD31/Flk1 double-positive cells are absent in the Mut. I, histological analyses of the WT and Mut embryo, demonstrating an absence of the endocardial/endothelial lineage (End) and vasculature (dorsal aorta (DA)) in the Mut embryo compared with a WT littermate. h, heart). J, immunohistochemical analysis using endomucin antibody (α-endomucin) in WT and Mut embryos, further demonstrating an absence of the endocardial lineage (arrowhead) and the vasculature (arrows) in the Mut embryo. K, immunohistochemical analysis using anti-endomucin in the WT and Mut yolk sacs showing the presence of endothelium and blood cells (arrow), whereas they are not detectable in the Mut yolk sac. K' and K” are enlarged regions of those indicated by rectangles. L, gene expression of Etv2, CD31, and CD41 in E8.5 WT and Mut embryos. Scale bars = 100 μm (I and J) and 50 μm (K).
To verify the results in vivo, we deleted the 3.9-kb promoter fragment in mouse embryos by injecting zygotes with the Cas9 mRNA and guide RNAs. The Etv2 Mut embryos were nonviable and lacked vasculature and hematopoietic lineages. Histological and immunohistochemical analyses revealed essentially an absence of endothelial/endocardial lineages in the Mut embryos at E8.5 (Fig. 1, I and J). The dorsal aorta and endocardium were not detected in the Mut embryo. We also could not detect endothelium and blood cells in the Mut yolk sac (Fig. 1K). Gene expression analysis revealed an approximately 90% reduction of Etv2 expression in the E8.5 Mut embryo compared with the WT (Fig. 1L). Endothelial marker CD31 and hematopoietic marker CD41 expression was also reduced by more than 90% compared with the WT (Fig. 1L). In summary, these studies indicate that the transcriptional regulatory elements for Etv2 are located within the 3.9-kb promoter region and that deleting this region largely phenocopies the global Etv2 knockout embryos. Importantly, although this Etv2 deletional mutant is nonviable and lacks hematoendothelial lineages, there remains the possibility that other enhancers may transactivate Etv2 gene expression, but at a very low level (i.e. less than 5%). Nevertheless, these gene editing results provided the rationale for examining the 3.9-kb Etv2 promoter upstream region in greater detail.
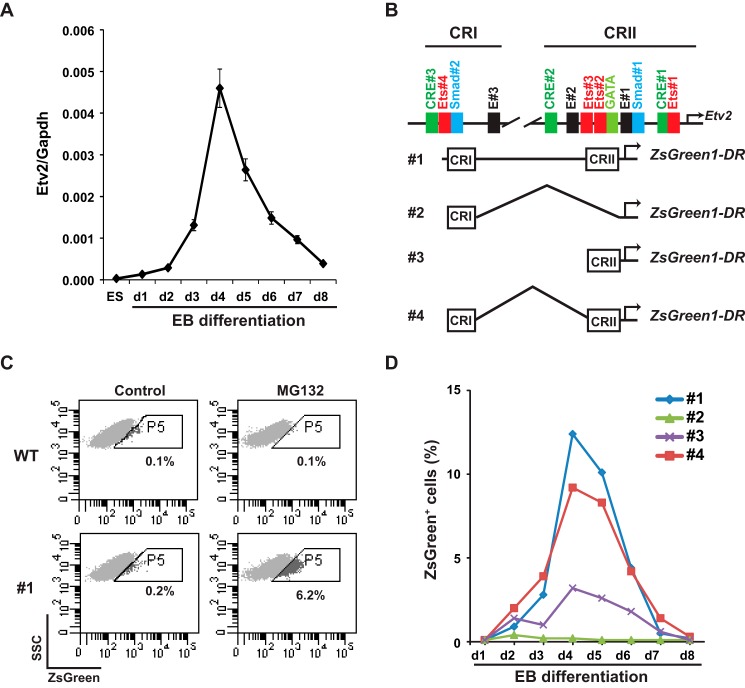
We have reported previously that Etv2 is transactivated by the Flk1-p38-Creb1 signaling cascade and that Mesp1 is an upstream regulator of Etv2 during embryogenesis (39, 41). To further explore the regulation of Etv2 gene expression in a developmental context, we established a system in which promoter activity could be monitored during mouse EB differentiation. We utilized the A2Lox homologous recombination system to establish stably integrated ES cell lines (47). As shown in Fig. 2A, the expression of Etv2 was undetectable in A2Lox ES cells in the undifferentiated state, up-regulated by day 2 of EB differentiation, peaked at day 4, and was then down-regulated. The expression pattern of Etv2 in the A2Lox ES cell/EB system recapitulated the expression pattern observed during mouse embryogenesis, as reported previously (28). We then fused the 3.9-kb Etv2 promoter and its deletional constructs to the ZsGreenDR reporter, which was inserted into the p2Lox vector (Fig. 2B). The promoter-ZSGreenDR constructs were then integrated into A2lox ES cells using homologous recombination. Using these constructs, we initially detected a minimal GFP signal during differentiation of these ES cell lines (Fig. 2C, Control). We hypothesized that the limited signal was due to the rapid degradation of the ZsGreenDR protein. Therefore, we treated the EBs with MG132, a proteasome inhibitor, before analyzing the ES cells/EBs using FACS. We observed no effect of MG132 treatment on wild-type EBs but a robust GFP signal after 5 h of MG132 treatment in EBs from the ES cell line harboring the full-length 3.9-kb construct (#1, Fig. 2C). Therefore, in subsequent experiments, we treated ES cells/EBs with MG132 for 5 h prior to harvesting. As shown in Fig. 2D, ES cell line 1 revealed a similar expression pattern of ZSgreen expression compared with the endogenous Etv2 mRNA. ES cell line 2, which harbored only CRI, had a minimal signal during EB differentiation. ES cell line 3, which harbored only CRII, showed a similar pattern as that of ES cell line 1, although the signal intensity was lower, and the profile of ES cell line 4, which harbored both CRI and CRII but lacked the non-conserved sequence in between, was almost the same as that of ES cell line 1. These results suggested that the critical regulatory motifs in the 3.9-kb upstream region were located within CRI and CRII (construct 4). Therefore, we utilized the CRI-CRII fusion enhancer-promoter as the minireporter of the Etv2 gene in the following experiments (41).
FIGURE 2.
Characterization of the Etv2 cis-regulatory module. A, gene expression of Etv2 during wild-type A2lox ES cell/EB differentiation. Note that Etv2 transcript expression is undetectable in the undifferentiated ES cells. B, schematic of the 3.9-kb Etv2 promoter-reporter constructs. Top panel, two conserved regions were identified in the 3.9-kb upstream fragment and include CRI and CRII. Note that motifs are designated in the CRI and CRII region. Bottom panel, schematic of the ZsGreen1-DR reporters fused with the 3.9-kb Etv2 promoter, CRI, CRII, and CRI+CRII. C, FACS profile of side scatter (SSC) versus ZsGreen in A2lox ES harboring the ZsGreen1-DR reporters during EB differentiation. MG132 treatment has no effect on the WT A2lox ES cells but increases the ZsGreen signal in ES cell #1 upon embryonic differentiation from 0.2% to 6.2%. Controls were treated with DMSO because MG132 was dissolved in DMSO. WT, A2lox ES cells; #1, A2lox ES cells harboring the 3.9-kb fragment linked to the ZsGreen1-DR construct. D, FACS profiles of ES cells harboring constructs #1, #2, #3, and #4 during EB differentiation. Note the similar patterns between the Etv2 gene expression in A and the FACS profile of ES cell line #1. Compared with the profile of ES cell line #1, the ZsGreen+ cells of the ES cell line #3 are reduced. The signal in ES cell line #2 is minimal, whereas that in ES cell line #4 is almost the same as that in ES cell line #1.
Autoactivation of Etv2 Gene Expression
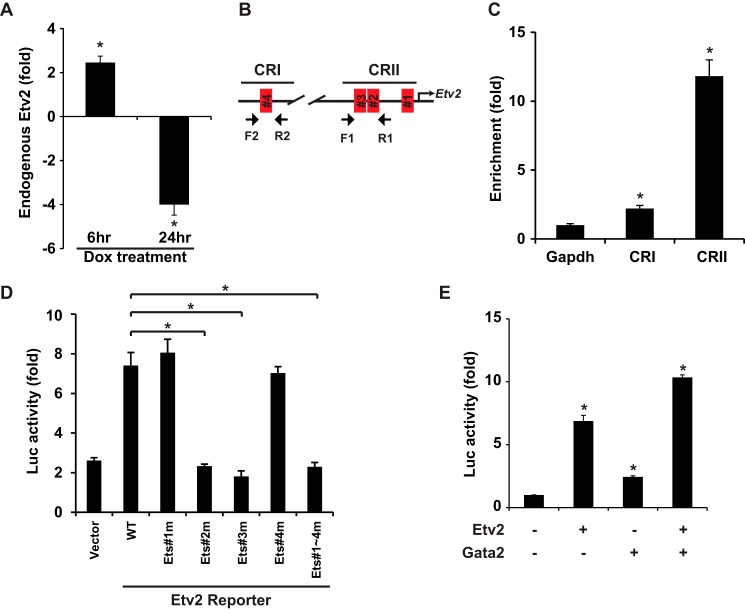
To investigate the autoregulation of Etv2, we examined endogenous Etv2 expression following the induction of Etv2 using iHA-Etv2 ES cells in the ES cell/EB system. As shown in Fig. 3A, early induction of Etv2 resulted in increased endogenous Etv2 expression, whereas later induction of Etv2 repressed endogenous Etv2 expression. Utilizing ChIP assays, we observed that Etv2 interacted with the proximal Etv2 promoter (CRII), whereas interaction with the distal enhancer (CRI) was minimal (Fig. 3, B and C). To examine the contribution of each of the four Ets motifs, we performed transcriptional assays with the reporters harboring the Ets motif mutations. As shown in Fig. 3D, Etv2 transactivated the wild-type reporter ∼8-fold, whereas mutation of the Ets#1 or Ets#4 motifs had no effect. Mutations of the Ets#2 or Ets#3 motifs resulted in abolishment of transactivation to the baseline level (the same level as the control vector), which was comparable with the mutation of all four Ets motifs. These results indicated that the Ets#2 and Ets#3 motifs were essential for the autoactivation of Etv2, whereas the Ets#1 and #4 motifs were dispensable for transactivation by Etv2. We have reported previously that Etv2 and Gata2 synergistically transactivate gene expression in the endothelial and hematopoietic lineages (36). We noted a conserved GATA motif adjacent to the Ets#2 motif in the Etv2 promoter (Fig. 2B). Therefore, we performed transcriptional assays using the Etv2 promoter-reporter construct to examine the effect of Etv2 and Gata2. As shown in Fig. 3E, both Etv2 and Gata2 transactivated the reporter individually, and there was enhanced transactivation of the promoter-reporter construct when Etv2 and Gata2 were coexpressed.
FIGURE 3.
Autoactivation of Etv2 gene expression. A, endogenous Etv2 expression in iHA-Etv2 ES differentiation on day 4 upon doxycycline (Dox) induction for 6 h is increased and inhibited after 24-h treatment. *, p < 0.05. B, schematic of the ChIP strategy. Primer pair F1 and R1 flanks the Ets#2 and Ets#3 motifs, and F2 and R2 flanks the Ets#4 motif. C, ChIP analysis of the interaction between Etv2 and the Etv2 promoter using iHA-Etv2 EBs on day 4. Etv2 preferentially binds to CRII and less so to CRI. *, p < 0.05. D, Etv2 transactivates the Etv2 minireporter ∼8-fold in 293T cells. Mutation of the Ets#1 or Ets#4 motif has no effect on transactivation, whereas mutation of the Ets#2 or Ets#3 motif results in loss of transactivation. Mutation of all four Ets motifs has the same effect as that of the Ets#2 or Ets#3 motif mutants. *, p < 0.05. E, both Etv2 and Gata2 transactivate the Etv2 minireporter in 293T cells ∼6-or 2-fold, respectively. Together, Etv2 and Gata2 transactivate the Etv2 minireporter to ∼10-fold. *, p < 0.05.
Transcriptional Regulation of Etv2 by the Vegf/Flk1-Calcineurin-NFAT Cascade
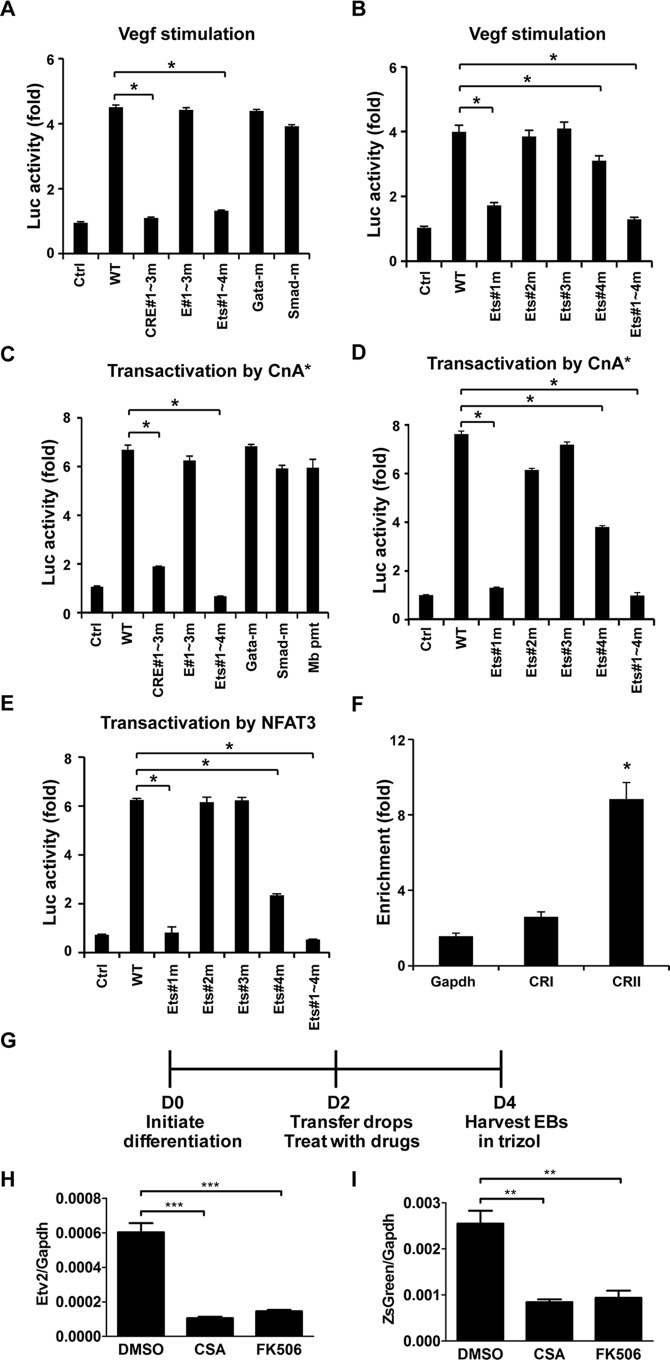
Previous studies in our laboratory have demonstrated that Etv2 is transactivated by the Vegf/Flk1-p38-Creb1 signaling cascade (39). To comprehensively examine the response of Etv2 to Vegf/Flk1 signaling, we performed transcriptional assays using the mutant reporters. As shown in Fig. 4A, mutation of the CREB response element (CRE) motifs (CRE#1–3m) resulted in the loss of transcriptional activation, which was consistent with our previous report (39). We also observed that mutation of the Ets motifs (Ets#1–4m) resulted in a loss of response to Vegf/Flk1 signaling, whereas mutation of the E-box motifs (E#1–3m), Gata motif (Gata-m), or Smad motifs (Smad-m) had minimal effects on the transactivation of the Etv2 promoter-reporter by Vegf/Flk1 signaling (Fig. 4A). Of these four Ets motifs, mutation of Ets#1 had the highest impact on transcriptional activity, which was comparable with the mutation of all four Ets motifs (Fig. 4B). Mutation of Ets#4 also attenuated the activity, but to a lesser extent. However, we did not observe any effect because of the mutation of the Ets#2 or Ets#3 motifs (Fig. 4B). Previous studies have reported that the Calcineurin-NFAT cascade was one of the downstream signaling cascades activated by the Vegf/Flk1 signaling pathway (11). To examine whether Calcineurin was involved in the transactivation of Etv2 gene expression, we utilized the constitutively active Calcineurin (CnA*) construct (51, 52). Myoglobin promoter-reporter construct (Mb pmt) was utilized as a positive control for CnA* (53). As shown in Fig. 4C, CnA* transactivated the wild-type Etv2 promoter reporter 7-fold. Mutation of the CRE motifs impaired the transcriptional activity to 2-fold, and mutation of the Ets motifs resulted in a complete loss of transactivation. Mutation of the E-box motifs, the Gata motif, and the Smad motifs had no effect on transcriptional activity (Fig. 4C). As outlined above, both the Ets#1 and Ets#4 motifs were responsive to Flk1/Vegf stimulation (Fig. 4B). To identify the Ets motif responsive to CnA* stimulation, we performed transcriptional assays using Ets motif mutants. As shown in Fig. 4D, mutation of Ets#1 resulted in minimal transactivation (1.5-fold) and mutation of the Ets#4 motif reduced transactivation to 4-fold, whereas mutation of all of the Ets motifs led to abolishment of transactivation. However, mutation of Ets#2 or #3 motifs has minimal effect. NFAT factors are well characterized downstream effectors of the Calcineurin signaling pathway (26). As shown in Fig. 4E, Nfat3 transactivated the Etv2 wild-type reporter ∼6-fold. Ets#1 mutation resulted in loss of transactivation, whereas mutation of the Ets#4 motif also attenuated the transactivation to 2.5-fold. Neither the Ets#2 nor the Ets#3 motif mutation had any effect on transactivation. To investigate the interaction between Nfat3 and the Etv2 promoter or enhancer, ChIP assays were employed with endogenous Nfat3 in the ES cell/EB system. As shown in Fig. 4F, the CRII region was highly enriched with Nfat3 ChIP (∼10-fold higher than the IgG negative control), whereas CRI region enrichment was minimal. These studies established that Vegf/Flk1 signaling also transactivated the Etv2 promoter through the Calcineurin-NFAT cascade, which was mediated by the Ets#1 and Ets#4 motifs. To assess whether Calcineurin/NFAT signaling was involved in normal development, we utilized the Etv2 minireporter cell line and inhibited this pathway with either CSA or FK506 (Fig. 4G). We measured endogenous Etv2 expression levels and observed a robust decrease in Etv2 expression with treatment of both inhibitors (Fig. 4H). We also measured the levels of ZsGreen transcripts and observed that our reporter was also decreased in response to treatments (Fig. 4I). These results support the hypothesis that Calcineurin signaling is an important regulator of Etv2 and that this regulation occurs within the minireporter. Furthermore, these results validate the idea that our minireporter reflects endogenous expression levels.
FIGURE 4.
Transactivation of Etv2 by the Vegf/Flk1-Calcineurin-NFAT3 cascade. A, the Etv2 minireporter is responsive to Vegf stimulation in 293T cells. Mutation of the E#1–3, Gata, or Smad motifs has no effect on Vegf stimulation. However, mutation of the CRE#1–3 motifs or Ets#1–4 motifs results in loss of response to Vegf stimulation. *, p < 0.05. Ctrl, control; Luc, luciferase. B, mutation of the Ets#2 or Ets#3 motif has no effect on Vegf stimulation in 293T cells. However, mutation of the Ets#1 or #4 motif attenuates the transactivation of Etv2 minireporter stimulation by Vegf. The reduction is further enhanced when all Ets#1–4 motifs were mutated. *, p < 0.05. C, the Etv2 minireporter was transactivated by CnA* ∼7-fold in 293T cells. Mutation of the E#1–3, Gata, or Smad motifs does not affect the transactivation by CnA*. Mutation of the CRE#1–3 motifs or Ets#1–4 motifs results in a reduction of transactivation (∼2-fold) or complete loss of transactivation, respectively. The myoglobin promoter (Mb pmt) reporter serves as a positive control for the transcriptional activation of CnA*. *, p < 0.05. D, mutation of Ets#1 resulted in a severe reduction of transactivation by CnA* (∼1.5-fold), and mutation of Ets#4 impaired the activity (∼4-fold) in 293T cells. However, mutation of the Ets#2 or #3 motif has no significant effect on transactivation by CnA*. Mutation of all Ets #1–#4 motifs resulted in complete loss of transactivation. *, p < 0.05. E, Nfat3 transactivates the Etv2 minireporter ∼6-fold. Mutation of the Ets#1 motif results in complete loss of activity, and mutation of the Ets#4 motif attenuates the activity (2-fold) in 293T cells. Mutation of Ets#2 or Ets#3 has no impact on transactivation. Mutation of all Ets#1–#4 motifs completely abolishes transcription activation. *, p < 0.05. F, ChIP assays using Nfat3 reveal that Nfat3 binds to CRII, whereas the interaction with CRI is minimal in wild-type EBs on day 4. *, p < 0.05. G, experimental schematic in which the cell line harboring the Etv2 minireporter is differentiated and treated with vehicle (DMSO), CSA, or FK506 from EB D2–4. EBs were harvested on D4 to capture peak Etv2 expression. H, Etv2 expression is decreased significantly relative to Gapdh with treatment of either CSA or FK506 compared with control DMSO treatment. ***, p < 0.001. I, ZsGreen reporter expression is decreased significantly relative to Gapdh with treatment of either CSA or FK506 compared with control DMSO treatment. **, p < 0.01.
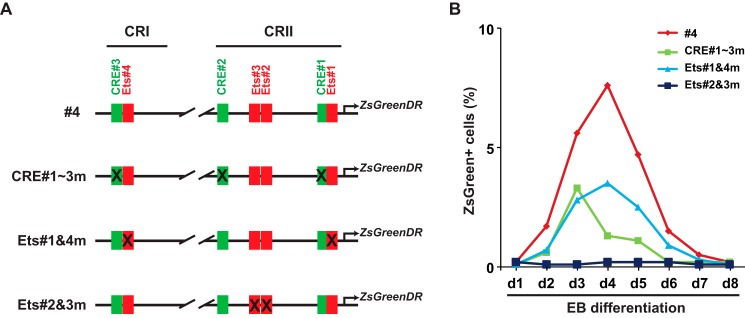
Mutation of Ets Motifs or CRE Motifs
Our results demonstrated that the Ets#2 and #3 motifs were important for the regulation by Etv2, whereas the Ets#1 and #4 motifs were required for Calcineurin-NFAT signaling. Both CRE motifs #1–3 and Ets motifs #1 and #4 were essential for the full activation by the Vegf/Flk1 signaling pathway. To examine the effect of these motifs, we constructed the Etv2 promoter-ZsGreen1-DR reporters with the mutated motifs (Fig. 5A). The #4 reporter is described in Fig. 1B. These constructs were utilized to establish the reporter ES cell lines. FACS profiles of the ZsGreen+ cells were examined after treatment with MG132 for 5 h. As shown in Fig. 4B, mutation of the CRE#1–3 motifs or the Ets#1 and #4 motifs resulted in a decrease in the ZsGreen+ cell population, which may be due to the attenuation of Vegf/Flk1 response. Interestingly, mutation of the Ets#2 and #3 motifs resulted in the loss of reporter activity (Fig. 5B). This profile was similar to that of the CRI reporter (Fig. 2B). Therefore, the Ets#2 and #3 motifs were essential for the function of the promoter.
FIGURE 5.
FACS profile of the reporter ES cell lines harboring the mutated CRI-CRII constructs. A, schematic of the constructs utilized to establish the A2lox ES cell lines (as shown in Fig. 1B). The #4 construct is described in Fig. 1B. Ets#1 and #4 are NFAT-responsive motifs, whereas Ets#2 and #3 are Ets factor-binding motifs. B, FACS profiles of ZsGreen+ cells during ES cell/EB differentiation were performed as described in Fig. 1. Mutation of the CRE motifs (CRE#1–3m) reduces the ZsGreen+ cell population as well as that of the mutation of Ets#1 and #4 (Ets#1&4m). Mutation of the Ets#2 and #3 motifs results in loss of Etv2 reporter expression.
Transactivation of Flt1 by Etv2
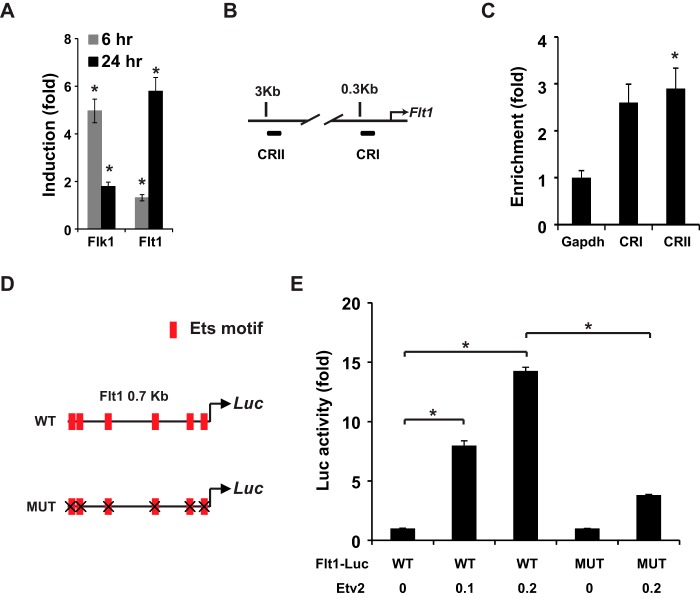
We have previously reported the dysregulation of Flt1 in Etv2 knockout embryos (28). As shown in Fig. 6A, Etv2 induced Flk1 and Flt1 expression, but not Vegfa (data not shown), in inducible ES cells. To examine the interaction between Etv2 and the Flt1 promoter, we performed a ChIP assay with the promoter (CRI) and enhancer (CRII) of the Flt1 upstream regulatory region (Fig. 6B). As shown in Fig. 6C, we observed that Etv2 could bind to both the Flt1 promoter and enhancer. The 0.7-kb proximal promoter of Flt1 plays a critical role in the endothelial expression of Flt1 (54, 55). Therefore, the 0.7-kb proximal CRI region was utilized as the reporter and mutation of all of the Ets motifs as the mutant reporter (Fig. 6D). Etv2 transactivated the wild-type reporter in a dose-dependent fashion (15-fold) (Fig. 6E). However, mutation of the five Ets motifs resulted in an ∼70% decrease in transactivation (4-fold) (Fig. 6, D and E). Therefore, these results supported the notion that Etv2 transactivated Flt1 gene expression directly through the Ets motifs.
FIGURE 6.
Flt1 is a direct downstream target gene of Etv2. A, gene expression profile of Flk1, Flt1, and Vegfa following overexpression of Etv2 using iHA-Etv2 EBs on day 4. Flk1 is up-regulated 5-fold after 6 h of stimulation and 2-fold after 24 h of stimulation. Flt1 is up-regulated ∼2-fold after 6 h of stimulation and 6-fold after 24 h. *, p < 0.05. B, schematic of the ChIP assays using the Flt1 promoter. CRI is located in the 0.3-kb region, whereas CRII is located in the 3-kb region. C, Etv2 interacts with both CRI and CRII of the Flt1 promoter, as revealed using iHA-Etv2 EBs on day 4 and ChIP assays. *, p < 0.05. D, schematic of the 0.7-kb Flt1 promoter in the CRI region, which harbors six Ets motifs. WT, wild-type promoter; MUT, the mutated promoter with all six Ets motifs mutated; Luc, luciferase. E, Etv2 transactivates the 0.7-kb Flt1 reporter in a dose-dependent manner ∼15-fold in 293T cells. The transactivation is reduced to 4-fold when all Ets motifs were mutated. *, p < 0.05.
Partial Rescue of the Phenotype of Flt1 Null in the Etv2 Conditional Knockout
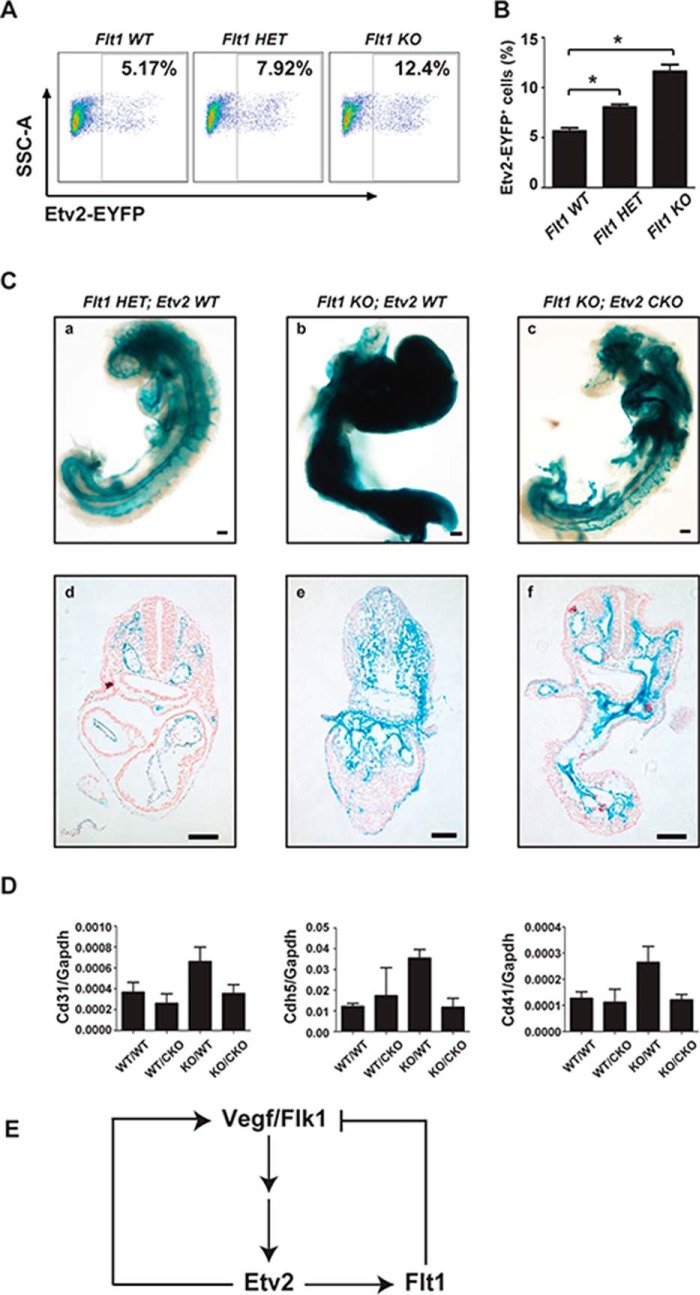
Our studies defined a positive autoregulatory role of Etv2 (Fig. 2). However, the 3.9-kb Etv2-EYFP cell population was increased in the Etv2 null background compared with the wild-type embryonic control. These results supported the notion of an additional negative feedback mechanism by Etv2. Because Flt1 is the negative regulator of Flk1, we hypothesized that the absence of Etv2 in the null embryo resulted in the down-regulation of Flt1. We further hypothesized that decreased Flt1 expression would lead to increased Flk1 activity, thereby up-regulating the 3.9-kb Etv2-EYFP reporter. To test these hypotheses, we crossed the 3.9-kb Etv2-EYFP transgenic mouse model into the Flt1 KO background to examine the activity of the reporter. We observed that, at E8.25, the Etv2-EYFP cell population in the Flt1 WT background (5.7%) was progressively increased in the Flt1 heterozygous (Flt1HET) background (8.0%) and the Flt1 KO background (11.6%) (Fig. 7, A and B).
FIGURE 7.
Partial rescue of the Flt1 null embryonic phenotype in the Etv2 conditional knockout background. A, representative FACS profiles of side scatter area (SSCA) versus EYFP in the 3.9-kb Etv2-EYFP cells in the Flt1 WT, Flt1 HET, and Flt1 KO embryos at E8.25 stage. B, quantitative analysis of the FACS data in A. The percentage of the 3.9-kb Etv2-EYFP cells in the Flt1 WT embryos is ∼5.7%, whereas the percentage is increased to 8.0% in the Flt1 HET embryos and 11.6% in Flt1 KO embryos. *, p < 0.05. C, morphological analysis of representative E8.75 LacZ-stained embryos demonstrating a normal vascular pattern in Flt1 HET (LacZ knockin/knockout) embryos (a) and significant defects, such as lack of vascular patterning and a developmental delay in Flt1 KO embryos (b). Crossing the Flt1 knockout to a Flk1-Cre-mediated Etv2 conditional knockout rescues the Flt1 null embryonic vascular phenotype (c). Further histological analysis of LacZ-stained embryos shows the normal vascular patterning of Flt1 HET (d) and a reduction of lateral plate and paraxial mesoderm in exchange for excessive Flt1-LacZ+ cells in Flt1 KO embryos (e). Flt1 KO; Etv2 CKO embryos featured an intermediate phenotype that includes intermediate levels of non-vascular tissue in the trunk region and formation of multiple vessel structures. Scale bars = 100 μm. D, gene expression in the Flt1 null embryos using qRT-PCR analysis. Expression of CD31, Cdh5, and CD41 was analyzed to validate rescue at the molecular level. Three biologically independent E8.5–9.0 embryos were analyzed in triplicate. WT/WT, Flt1 wild-type/Etv2 wild-type; WT/CKO, Flt1 wild-type/Etv2 conditional knockout; KO/WT, Flt1 knockout/Etv2 wild-type; KO/CKO, Flt1 knockout/Etv2 conditional knockout (n = 3 for each genotype). E, diagram of the negative feedback loop demonstrated by this experiment. Flk1 activates Etv2, whose product, in turn, activates Flt1, along with other target genes. Flt1 competes with the Flk1 signal and results in the reduction of Etv2 expression level. In the absence of Flt1, the Flk1 signal is hyperactivated, resulting in excessive production of the endothelial lineage. Conditionally deleting Etv2 in the vascular lineage reduces this hyperendothelialization phenotype.
Previous studies have suggested that the lethality of the Flt1 null embryo was due to the extended and enhanced Flk1 signal (7). We hypothesized that the induction of Etv2 via the Flk1 signal was partially responsible for the phenotype and that attenuation of Etv2 in the Flt1 null embryo would partially rescue the Flt1 null embryonic phenotype. To test our hypothesis, we bred Flt1 KO mice into the Etv2 conditional knockout background (41). As shown in Fig. 7C, we observed normal vascular patterning in the Flt1 heterozygous embryo at E8.75, as shown by β-galactosidase staining for Flt1-LacZ (Fig. 7C, a and d). In the Flt1 KO embryos, we observed significant defects, including excessive Flt1-LacZ+ cells, supersized vessels, a lack of vascular patterning, and developmental delay (Fig. 7C, b and e). We observed a marked improvement in the phenotype of Flt1 null embryo in the Flk1-Cre-mediated Etv2 conditional knockout (CKO) background compared with the Flt1 KO embryo. The Flt1 KO; Etv2 CKO embryos had an intermediate phenotype that included intermediate levels of non-vascular tissues in the trunk region and the formation of multiple vascular structures (Fig. 7C, c and f). We further examined gene expression in the partially rescued embryos using qRT-PCR analysis. We observed that the expression of Cd31, Cdh5, and Cd41 was affected minimally in Etv2 CKO embryos and increased 2- to 3-fold in Flt1 KO embryos. The expression of the three genes was returned to the normal levels in the Flt1 KO; Etv2 CKO (KO/CKO) embryos (Fig. 7D). Taken together, these data support a partial rescue of vascular and hematopoietic lineage markers in the compound KO/CKO embryos compared with the Flt1 KO; Etv2 WT (KO/WT) embryos.
Discussion
Among the Ets transcription factors, Etv2 has an essential role in the specification of mesodermal derivatives (56). This essential role is further marked by a limited window of expression for Etv2 during mouse embryogenesis. In this study, we deciphered the transcriptional regulation of Etv2 gene expression by defining both positive and negative feedback regulators. One of the positive autoregulatory mechanisms is mediated by the Flk1 signaling cascade. Etv2 is induced by the VEGF signal mediated by the Flk1 receptor and, in turn, transactivates Flk1 gene expression, enhancing the Flk1 signaling cascade (57). We have reported previously that Vegf/Flk1 transactivates Etv2 gene expression through the Flk1-p38-Creb1 cascade through the proximal CRE motif (designated CRE#1 in this study) (39). Our data in this study support the conclusion that the Flk1-calcineurin-NFAT cascade is another important signaling pathway that transactivates Etv2 gene expression upon Vegf/Flk1stimulation. We demonstrate that NFAT activates the Etv2 promoter through the Ets#1 and Ets#4 motifs and that treatment with CSA or FK506 inhibited endogenous Etv2 transcription as well as the reporter gene. In agreement with this observation, it has been shown that Calcineurin mutant embryos have vascular developmental abnormalities, and double knockout embryos that lack Nfat3 and Nfat4 have perturbed vascular structures (21). In addition, studies have demonstrated that the inhibition of Calcineurin activity during a defined E7.5-E8.5 window perturbs vascular development (21). Collectively, these data establish that the Vegf/Flk1 signaling pathway transactivates Etv2 gene expression through two independent pathways: the Flk1-p38-Creb1 and Flk1-Calcineurin-NFAT pathways.
This study also demonstrates another positive feedback-regulatory mechanism because Etv2 interacts and binds to Ets#2 and Ets#3 motifs in its own promoter and transactivates gene expression. In addition, we demonstrate that Gata2 synergistically activates the Etv2 promoter. Therefore, Gata2, in addition to its role in synergizing with Etv2 and activating Etv2 downstream targets (36), may play a role in autoactivation of Etv2. The autoactivation by Etv2 provides an amplification mechanism of gene expression. In this fashion, Etv2 may be able to reach its peak expression level shortly after its initial expression. The autoactivation mechanism is essential for Etv2 gene expression because mutation of the Ets motifs results in loss of promoter activity. There is a possibility that other Ets factors may function as the upstream activators of Etv2. Although we have observed that both overexpressed Ets1 and Ets2 can transactivate the Etv2-luciferase reporter in the transcriptional assays (data not shown), the lack of early hematoendothelial lineage defects in Ets1 mutant mice (58–60) as well as the Ets1 and Ets2 double knockout embryos (61) support the notion that Ets1 and Ets2 are not required for activation of Etv2, at least during the initial expression of Etv2.
Interestingly, after 24 h of forced expression, we observed a negative effect of Etv2 on endogenous Etv2 expression. This suggested an indirect negative feedback mechanism, and we propose a negative feedback loop that is mediated through the Etv2-Flt1-Flk1 cascade (Fig. 7E). Previous studies have shown that Flk1 signaling induces the expression of Flt1, which dampens the Flk1 signal, thereby functioning as a negative feedback mechanism to limit Flk1 activity for the maturation of endothelial cells (62). In this study, we discovered that Etv2 mediates the Flk1 signal to transactivate Flt1 gene expression and is negatively regulated by the feedback mechanism. We demonstrate that the phenotype of the Flt1 null embryo (i.e. superactivation of the Flk1 signal because of the lack of negative feedback) is partially rescued by conditionally deleting Etv2, a mediator of the Flk1 signal, in the endothelial lineage (Fig. 7E).
In summary, we propose that the positive and negative loops that regulate Etv2 gene expression explain the mechanism of tight temporal control of Etv2. Etv2 is initially induced in the mesoderm by the Vegf/Flk1 signaling pathway, which is mediated by Creb, Mesp1, and NFATs (39–41, 64). Etv2 activates its own promoter and Flk1, which initially augment the Etv2 protein level. Because this mechanism relies on the production of Etv2 (as opposed to degradation of a protein), the response is relatively rapid. Flt1, in contrast, is induced at the same time. However, it functions by competing with the pre-existing Flk1 receptors on the cell membrane. Therefore, the response to negative feedback is slower compared with the positive feedback mechanism. This introduces a delay in the negative feedback mechanism and confers rapid induction and extinguishment of the Etv2 gene.
Author Contributions
N. K. N., X. S., T. L. R., S. D., and C. A. W. performed the experiments. N. K. N., X. S., T. L. R., S. D., and D. J. G. analyzed the data. D. J. G. coordinated the study. N. K. N., X. S., and D. J. G. wrote the paper.
Acknowledgments
We thank Rachel M. Gohla for mouse husbandry and Yi Ren for assistance with the FACS analysis. We also thank Tom N. Sato for the Flk1-Cre mice and Guo-Hua Fong for the Flt1 knockout mice.
This work was supported by National Institutes of Health Grants U01 HL100407 and 1R01 HL122576 (to D. J. G.). The authors declare that they have no conflicts of interest with the contents of this article.
- E
- embryonic day
- CRE
- CREB response element
- CREB
- cAMP response element-binding protein
- Mut
- mutant
- gRNA
- guide RNA
- EB
- embryoid body
- mES cells
- mouse embryonic stem cells
- qRT-PCR
- quantitative RT-PCR
- CR
- conserved region
- CSA
- cyclosporin A
- DMSO
- dimethyl sulfoxide
- EYFP
- enhanced yellow fluorescent protein
- CKO
- conditional knockout
- CnA
- Calcineurin
- HET
- heterozygous
- D
- day.
References
- 1.Rossant J., and Howard L. (2002) Signaling pathways in vascular development. Annu. Rev. Cell Dev. Biol. 18, 541–573 [DOI] [PubMed] [Google Scholar]
- 2.Coultas L., Chawengsaksophak K., and Rossant J. (2005) Endothelial cells and VEGF in vascular development. Nature 438, 937–945 [DOI] [PubMed] [Google Scholar]
- 3.Olsson A. K., Dimberg A., Kreuger J., and Claesson-Welsh L. (2006) VEGF receptor signalling: in control of vascular function. Nat. Rev. Mol. Cell Biol. 7, 359–371 [DOI] [PubMed] [Google Scholar]
- 4.Shalaby F., Rossant J., Yamaguchi T. P., Gertsenstein M., Wu X. F., Breitman M. L., and Schuh A. C. (1995) Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature 376, 62–66 [DOI] [PubMed] [Google Scholar]
- 5.Carmeliet P., Ferreira V., Breier G., Pollefeyt S., Kieckens L., Gertsenstein M., Fahrig M., Vandenhoeck A., Harpal K., Eberhardt C., Declercq C., Pawling J., Moons L., Collen D., Risau W., and Nagy A. (1996) Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature 380, 435–439 [DOI] [PubMed] [Google Scholar]
- 6.Ferrara N., Carver-Moore K., Chen H., Dowd M., Lu L., O'Shea K. S., Powell-Braxton L., Hillan K. J., and Moore M. W. (1996) Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature 380, 439–442 [DOI] [PubMed] [Google Scholar]
- 7.Fong G. H., Rossant J., Gertsenstein M., and Breitman M. L. (1995) Role of the Flt-1 receptor tyrosine kinase in regulating the assembly of vascular endothelium. Nature 376, 66–70 [DOI] [PubMed] [Google Scholar]
- 8.Hiratsuka S., Minowa O., Kuno J., Noda T., and Shibuya M. (1998) Flt-1 lacking the tyrosine kinase domain is sufficient for normal development and angiogenesis in mice. Proc. Natl. Acad. Sci. U.S.A. 95, 9349–9354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuh G., Li B., Crowley C., Cunningham B., and Wells J. A. (1998) Requirements for binding and signaling of the kinase domain receptor for vascular endothelial growth factor. J. Biol. Chem. 273, 11197–11204 [DOI] [PubMed] [Google Scholar]
- 10.Shinkai A., Ito M., Anazawa H., Yamaguchi S., Shitara K., and Shibuya M. (1998) Mapping of the sites involved in ligand association and dissociation at the extracellular domain of the kinase insert domain-containing receptor for vascular endothelial growth factor. J. Biol. Chem. 273, 31283–31288 [DOI] [PubMed] [Google Scholar]
- 11.Armesilla A. L., Lorenzo E., Gómez del Arco P., Martínez-Martínez S., Alfranca A., and Redondo J. M. (1999) Vascular endothelial growth factor activates nuclear factor of activated T cells in human endothelial cells: a role for tissue factor gene expression. Mol. Cell. Biol. 19, 2032–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayo L. D., Kessler K. M., Pincheira R., Warren R. S., and Donner D. B. (2001) Vascular endothelial cell growth factor activates CRE-binding protein by signaling through the KDR receptor tyrosine kinase. J. Biol. Chem. 276, 25184–25189 [DOI] [PubMed] [Google Scholar]
- 13.Minami T., Horiuchi K., Miura M., Abid M. R., Takabe W., Noguchi N., Kohro T., Ge X., Aburatani H., Hamakubo T., Kodama T., and Aird W. C. (2004) Vascular endothelial growth factor- and thrombin-induced termination factor, Down syndrome critical region-1, attenuates endothelial cell proliferation and angiogenesis. J. Biol. Chem. 279, 50537–50554 [DOI] [PubMed] [Google Scholar]
- 14.Chen L., Glover J. N., Hogan P. G., Rao A., and Harrison S. C. (1998) Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature 392, 42–48 [DOI] [PubMed] [Google Scholar]
- 15.Rao A., Luo C., and Hogan P. G. (1997) Transcription factors of the NFAT family: regulation and function. Annu. Rev. Immunol. 15, 707–747 [DOI] [PubMed] [Google Scholar]
- 16.Macian F. (2005) NFAT proteins: key regulators of T-cell development and function. Nat. Rev. Immunol. 5, 472–484 [DOI] [PubMed] [Google Scholar]
- 17.Crabtree G. R., and Olson E. N. (2002) NFAT signaling: choreographing the social lives of cells. Cell 109, S67–79 [DOI] [PubMed] [Google Scholar]
- 18.Mancini M., and Toker A. (2009) NFAT proteins: emerging roles in cancer progression. Nat. Rev. Cancer 9, 810–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de la Pompa J. L., Timmerman L. A., Takimoto H., Yoshida H., Elia A. J., Samper E., Potter J., Wakeham A., Marengere L., Langille B. L., Crabtree G. R., and Mak T. W. (1998) Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature 392, 182–186 [DOI] [PubMed] [Google Scholar]
- 20.Ranger A. M., Grusby M. J., Hodge M. R., Gravallese E. M., de la Brousse F. C., Hoey T., Mickanin C., Baldwin H. S., and Glimcher L. H. (1998) The transcription factor NF-ATc is essential for cardiac valve formation. Nature 392, 186–190 [DOI] [PubMed] [Google Scholar]
- 21.Graef I. A., Chen F., and Crabtree G. R. (2001) NFAT signaling in vertebrate development. Curr. Opin. Genet. Dev. 11, 505–512 [DOI] [PubMed] [Google Scholar]
- 22.Hogan P. G., Chen L., Nardone J., and Rao A. (2003) Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 17, 2205–2232 [DOI] [PubMed] [Google Scholar]
- 23.García-Rodríguez C., and Rao A. (1998) Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/CREB-binding protein (CBP). J. Exp. Med. 187, 2031–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez Bosc L. V., Layne J. J., Nelson M. T., and Hill-Eubanks D. C. (2005) Nuclear factor of activated T cells and serum response factor cooperatively regulate the activity of an α-actin intronic enhancer. J. Biol. Chem. 280, 26113–26120 [DOI] [PubMed] [Google Scholar]
- 25.Armand A. S., Bourajjaj M., Martínez-Martínez S., el Azzouzi H., da Costa Martins P. A., Hatzis P., Seidler T., Redondo J. M., and De Windt L. J. (2008) Cooperative synergy between NFAT and MyoD regulates myogenin expression and myogenesis. J. Biol. Chem. 283, 29004–29010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., Rao A., and Hogan P. G. (2011) Interaction of calcineurin with substrates and targeting proteins. Trends Cell Biol. 21, 91–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao Y. G., and Duh E. J. (2004) VEGF selectively induces Down syndrome critical region 1 gene expression in endothelial cells: a mechanism for feedback regulation of angiogenesis? Biochem. Biophys. Res. Commun. 321, 648–656 [DOI] [PubMed] [Google Scholar]
- 28.Koyano-Nakagawa N., Kweon J., Iacovino M., Shi X., Rasmussen T. L., Borges L., Zirbes K. M., Li T., Perlingeiro R. C., Kyba M., and Garry D. J. (2012) Etv2 is expressed in the yolk sac hematopoietic and endothelial progenitors and regulates Lmo2 gene expression. Stem Cells 30, 1611–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sumanas S., and Lin S. (2006) Ets1-related protein is a key regulator of vasculogenesis in zebrafish. PLoS Biol. 4, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee D., Park C., Lee H., Lugus J. J., Kim S. H., Arentson E., Chung Y. S., Gomez G., Kyba M., Lin S., Janknecht R., Lim D. S., and Choi K. (2008) ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell 2, 497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferdous A., Caprioli A., Iacovino M., Martin C. M., Morris J., Richardson J. A., Latif S., Hammer R. E., Harvey R. P., Olson E. N., Kyba M., and Garry D. J. (2009) Nkx2–5 transactivates the Ets-related protein 71 gene and specifies an endothelial/endocardial fate in the developing embryo. Proc. Natl. Acad. Sci. U.S.A. 106, 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neuhaus H., Müller F., and Hollemann T. (2010) Xenopus er71 is involved in vascular development. Dev. Dyn. 239, 3436–3445 [DOI] [PubMed] [Google Scholar]
- 33.Kataoka H., Hayashi M., Nakagawa R., Tanaka Y., Izumi N., Nishikawa S., Jakt M. L., Tarui H., and Nishikawa S. (2011) Etv2/ER71 induces vascular mesoderm from Flk1+PDGFRα+ primitive mesoderm. Blood 118, 6975–6986 [DOI] [PubMed] [Google Scholar]
- 34.Kataoka H., Hayashi M., Kobayashi K., Ding G., Tanaka Y., and Nishikawa S. (2013) Region-specific Etv2 ablation revealed the critical origin of hemogenic capacity from Hox6-positive caudal-lateral primitive mesoderm. Exp. Hematol. 41, 567–581.e9 [DOI] [PubMed] [Google Scholar]
- 35.De Val S., and Black B. L. (2009) Transcriptional control of endothelial cell development. Dev. Cell 16, 180–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi X., Richard J., Zirbes K. M., Gong W., Lin G., Kyba M., Thomson J. A., Koyano-Nakagawa N., and Garry D. J. (2014) Cooperative interaction of Etv2 and Gata2 regulates the development of endothelial and hematopoietic lineages. Dev. Biol. 389, 208–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knebel J., De Haro L., and Janknecht R. (2006) Repression of transcription by TSGA/Jmjd1a, a novel interaction partner of the ETS protein ER71. J. Cell. Biochem. 99, 319–329 [DOI] [PubMed] [Google Scholar]
- 38.Kim J. Y., Lee R. H., Kim T. M., Kim D. W., Jeon Y. J., Huh S. H., Oh S. Y., Kyba M., Kataoka H., Choi K., Ornitz D. M., Chae J. I., and Park C. (2014) OVOL2 is a critical regulator of ER71/ETV2 in generating FLK1+, hematopoietic, and endothelial cells from embryonic stem cells. Blood 124, 2948–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rasmussen T. L., Shi X., Wallis A., Kweon J., Zirbes K. M., Koyano-Nakagawa N., and Garry D. J. (2012) VEGF/Flk1 signaling cascade transactivates Etv2 gene expression. PLoS ONE 7, e50103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamizu K., Matsunaga T., Katayama S., Kataoka H., Takayama N., Eto K., Nishikawa S., and Yamashita J. K. (2012) PKA/CREB signaling triggers initiation of endothelial and hematopoietic cell differentiation via Etv2 induction. Stem Cells 30, 687–696 [DOI] [PubMed] [Google Scholar]
- 41.Shi X., Zirbes K. M., Rasmussen T. L., Ferdous A., Garry M. G., Koyano-Nakagawa N., and Garry D. J. (2015) The transcription factor Mesp1 interacts with cAMP responsive element binding protein 1 (Creb1) and coactivates Ets variant 2 (Etv2) gene expression. J. Biol. Chem. 290, 9614–9625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veldman M. B., Zhao C., Gomez G. A., Lindgren A. G., Huang H., Yang H., Yao S., Martin B. L., Kimelman D., and Lin S. (2013) Transdifferentiation of fast skeletal muscle into functional endothelium in vivo by transcription factor Etv2. PLoS Biol. 11, e1001590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schupp M. O., Waas M., Chun C. Z., and Ramchandran R. (2014) Transcriptional inhibition of etv2 expression is essential for embryonic cardiac development. Dev. Biol. 393, 71–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayashi M., Pluchinotta M., Momiyama A., Tanaka Y., Nishikawa S., and Kataoka H. (2012) Endothelialization and altered hematopoiesis by persistent Etv2 expression in mice. Exp. Hematol. 40, 738–750.e11 [DOI] [PubMed] [Google Scholar]
- 45.Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rasmussen T. L., Kweon J., Diekmann M. A., Belema-Bedada F., Song Q., Bowlin K., Shi X., Ferdous A., Li T., Kyba M., Metzger J. M., Koyano-Nakagawa N., and Garry D. J. (2011) ER71 directs mesodermal fate decisions during embryogenesis. Development 138, 4801–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iacovino M., Bosnakovski D., Fey H., Rux D., Bajwa G., Mahen E., Mitanoska A., Xu Z., and Kyba M. (2011) Inducible cassette exchange: a rapid and efficient system enabling conditional gene expression in embryonic stem and primary cells. Stem Cells 29, 1580–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Motoike T., Markham D. W., Rossant J., and Sato T. N. (2003) Evidence for novel fate of Flk1+ progenitor: contribution to muscle lineage. Genesis 35, 153–159 [DOI] [PubMed] [Google Scholar]
- 49.Bowlin K. M., Embree L. J., Garry M. G., Garry D. J., and Shi X. (2013) Kbtbd5 is regulated by MyoD and restricted to the myogenic lineage. Differentiation 86, 184–191 [DOI] [PubMed] [Google Scholar]
- 50.Caprioli A., Koyano-Nakagawa N., Iacovino M., Shi X., Ferdous A., Harvey R. P., Olson E. N., Kyba M., and Garry D. J. (2011) Nkx2–5 represses Gata1 gene expression and modulates the cellular fate of cardiac progenitors during embryogenesis. Circulation 123, 1633–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Keefe S. J., Tamura J., Kincaid R. L., Tocci M. J., and O'Neill E. A. (1992) FK-506- and CsA-sensitive activation of the interleukin-2 promoter by calcineurin. Nature 357, 692–694 [DOI] [PubMed] [Google Scholar]
- 52.Chin E. R., Olson E. N., Richardson J. A., Yang Q., Humphries C., Shelton J. M., Wu H., Zhu W., Bassel-Duby R., and Williams R. S. (1998) A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 12, 2499–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanatous S. B., and Mammen P. P. (2010) Regulation of myoglobin expression. J. Exp. Biol. 213, 2741–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morishita K., Johnson D. E., and Williams L. T. (1995) A novel promoter for vascular endothelial growth factor receptor (flt-1) that confers endothelial-specific gene expression. J. Biol. Chem. 270, 27948–27953 [DOI] [PubMed] [Google Scholar]
- 55.Jin E., Liu J., Suehiro J., Yuan L., Okada Y., Nikolova-Krstevski V., Yano K., Janes L., Beeler D., Spokes K. C., Li D., Regan E., Shih S. C., Oettgen P., Minami T., and Aird W. C. (2009) Differential roles for ETS, CREB, and EGR binding sites in mediating VEGF receptor 1 expression in vivo. Blood 114, 5557–5566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lammerts van Bueren K., and Black B. L. (2012) Regulation of endothelial and hematopoietic development by the ETS transcription factor Etv2. Curr. Opin. Hematol. 19, 199–205 [DOI] [PubMed] [Google Scholar]
- 57.De Val S., Chi N. C., Meadows S. M., Minovitsky S., Anderson J. P., Harris I. S., Ehlers M. L., Agarwal P., Visel A., Xu S. M., Pennacchio L. A., Dubchak I., Krieg P. A., Stainier D. Y., and Black B. L. (2008) Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell 135, 1053–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bories J. C., Willerford D. M., Grévin D., Davidson L., Camus A., Martin P., Stéhelin D., and Alt F. W. (1995) Increased T-cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature 377, 635–638 [DOI] [PubMed] [Google Scholar]
- 59.Muthusamy N., Barton K., and Leiden J. M. (1995) Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature 377, 639–642 [DOI] [PubMed] [Google Scholar]
- 60.Yamamoto H., Flannery M. L., Kupriyanov S., Pearce J., McKercher S. R., Henkel G. W., Maki R. A., Werb Z., and Oshima R. G. (1998) Defective trophoblast function in mice with a targeted mutation of Ets2. Genes Dev. 12, 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei G., Srinivasan R., Cantemir-Stone C. Z., Sharma S. M., Santhanam R., Weinstein M., Muthusamy N., Man A. K., Oshima R. G., Leone G., and Ostrowski M. C. (2009) Ets1 and Ets2 are required for endothelial cell survival during embryonic angiogenesis. Blood 114, 1123–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sato Y., Kanno S., Oda N., Abe M., Ito M., Shitara K., and Shibuya M. (2000) Properties of two VEGF receptors, Flt-1 and KDR, in signal transduction. Ann. N.Y. Acad. Sci. 902, 201–205; discussion 205–207 [DOI] [PubMed] [Google Scholar]
- 63.Donaldson L. W., Petersen J. M., Graves B. J., and McIntosh L. P. (1996) Solution structure of the ETS domain from murine Ets-1: a winged helix-turn-helix DNA binding motif. EMBO J. 15, 125–134 [PMC free article] [PubMed] [Google Scholar]
- 64.Rasmussen T. L., Martin C. M., Walter C. A., Shi X., Perlingeiro R., Koyano-Nakagawa N., and Garry D. J. (2013) Etv2 rescues Flk1 mutant embryoid bodies. Genesis 51, 471–480 [DOI] [PMC free article] [PubMed] [Google Scholar]