Background: APOBEC3B-catalyzed DNA cytosine deamination causes mutations in cancer.
Results: We present the first APOBEC3B catalytic domain crystal structures including a dCMP-bound form.
Conclusion: A closed active site conformation distinguishes APOBEC3B from related enzymes and suggests that conformational changes are central to the overall single-stranded DNA binding mechanism.
Significance: These high resolution structures provide a foundation for inhibitor development.
Keywords: cancer, crystal structure, DNA binding protein, DNA repair, enzyme mutation, APOBEC3B, DNA deaminase, DNA editing, Mutation, cancer mutagenesis
Abstract
Functional and deep sequencing studies have combined to demonstrate the involvement of APOBEC3B in cancer mutagenesis. APOBEC3B is a single-stranded DNA cytosine deaminase that functions normally as a nuclear-localized restriction factor of DNA-based pathogens. However, it is overexpressed in cancer cells and elicits an intrinsic preference for 5′-TC motifs in single-stranded DNA, which is the most frequently mutated dinucleotide in breast, head/neck, lung, bladder, cervical, and several other tumor types. In many cases, APOBEC3B mutagenesis accounts for the majority of both dispersed and clustered (kataegis) cytosine mutations. Here, we report the first structures of the APOBEC3B catalytic domain in multiple crystal forms. These structures reveal a tightly closed active site conformation and suggest that substrate accessibility is regulated by adjacent flexible loops. Residues important for catalysis are identified by mutation analyses, and the results provide insights into the mechanism of target site selection. We also report a nucleotide (dCMP)-bound crystal structure that informs a multistep model for binding single-stranded DNA. Overall, these high resolution crystal structures provide a framework for further mechanistic studies and the development of novel anti-cancer drugs to inhibit this enzyme, dampen tumor evolution, and minimize adverse outcomes such as drug resistance and metastasis.
Introduction
APOBEC3B (A3B)3 is a member of a larger family of zinc-dependent DNA deaminases that convert cytosine bases into uracils in single-stranded DNA and RNA (reviewed in Refs. 1 and 2). Seven APOBEC3 (A3) enzymes, A3A/B/C/D/F/G/H, encoded by a tandem gene array on human chromosome 22 provide overlapping protection against a variety of DNA-based parasites including viruses and transposable elements (reviewed in Refs. 3 and 4). A3B is the only family member that is predominantly nuclear through the duration of the cell cycle, except in the M phase, in which it is excluded from metaphase chromosomes, and after cell division and formation of daughter cell nuclei, it is then actively reimported to the nuclear compartment (5, 6). Two related enzymes shuttle transiently into the nuclear compartment to reach biologically relevant nucleic acid substrates. Activation-induced deaminase (AID) is transported into the nucleus to deaminate immunoglobulin gene DNA and drive the antigen-driven processes of class switch recombination and somatic hypermutation (reviewed in Refs. 7 and 8). APOBEC1 shuttles into the nucleus to deaminate a variety of RNA substrates including cytosines in the coding region of mRNAs (such as the APOB mRNA), as well as a wide variety of noncoding regions (reviewed in Refs. 9 and 10). A3B is thought to have retained the nuclear import mechanism of an ancestral AID/APOBEC1 enzyme but lost the capacity to be exported actively into the cytoplasm from the nuclear compartment (11–13).
In contrast to its beneficial function in innate immunity, a variety of studies have combined to demonstrate a role for A3B in cancer mutagenesis (reviewed in Refs. 14–16). First, A3B is up-regulated in breast, head/neck, lung, bladder, cervical, and several other cancers, and up-regulation correlates with higher frequencies of cytosine mutation (17–21). Second, the patterns of both the dispersed and clustered (kataegis) cytosine mutations that dominate the mutational landscapes in these cancers in vivo closely resemble the intrinsic biochemical preferences of A3B-catalyzed DNA cytosine deamination in vitro (17–19, 21–23). Specifically, the most frequently mutated cytosines in these cancers are in 5′-TCA, 5′-TCG, and 5′-TCT trinucleotide motifs, which resemble the actual biochemical preferences of recombinant A3B (5′-TCA = 5′-TCG > 5′-TCT) (17, 21). Third, high A3B mRNA expression levels correlate with poor outcomes for estrogen receptor-positive breast cancer (24, 25), renal cancer (26), and multiple myeloma (27). Fourth, the spectrum of activating mutations in PIK3CA is biased toward a potential A3B deamination hot spot in A3B-high, HPV-infected head/neck tumors, in comparison to A3B-low, virus-negative tumors (20). Taken together, these and other studies demonstrate that A3B is a significant endogenous source of genomic mutation in cancer, which promotes tumor evolution, heterogeneity, and poor clinical outcomes and is induced by some types of viral infection.
Given this role in cancer mutagenesis, A3B has become a promising target for anti-cancer drug development. The attractiveness of A3B is further enhanced by the fact that it is a nonessential enzyme (28). However, progress toward developing methods to inhibit A3B mutagenesis will require high resolution crystal structures, and to date, only homology models are possible because this enzyme has resisted structural determination (i.e. only structures of related human DNA deaminases have been reported (29–40)). Here we present multiple crystal structures of the A3B catalytic domain, including one bound by dCMP, which represents the first nucleotide-bound structure of any polynucleotide cytosine deaminase. These structures reveal an active site pocket occluded by loop regions, which highlights the importance of conformational flexibility in the ssDNA deamination mechanism.
Experimental Procedures
Expression Constructs and Sequence Alignments
The bacterial expression plasmids used in this study were generated by inserting a codon-optimized gene for the full-length human A3B (residues 1–382) or A3B C-terminal domain (A3Bctd, residues 187–378) between the NdeI/XhoI sites of the pET24a vector (Novagen) for C-terminally His6-tagged proteins or between the BsaI/XbaI sites of the pE-SUMO vector (Lifesensors) for the N-terminally SUMO-tagged proteins. The A3B protein sequence used in this study matches UniProt ID Q9UH17. Derivatives encoding amino acid substitution mutants such as the quadruple mutant (QM) of A3Bctd were constructed by standard site-directed mutagenesis procedures and verified by DNA sequencing (University of Minnesota Genomics Center).
Protein Purification
A3Bctd-QM (F200S/W228S/L230K/F308K), A3Bctd-QMΔloop3, and its variant A3Bctd-QMΔloop3-A3Aloop1 were expressed with a noncleavable C-terminal His6 tag (LEHHHHHH) using the pET24a-based expression vector in Escherichia coli strain C43(DE3)pLysS. A3Bctd-DM (L230K/F308K) was expressed as a SUMO fusion. Protein expression was induced by adding isopropyl β-d-1-thiogalactopyranoside at a final concentration of 0.5 mm into the E. coli culture in LB medium during the mid-log growth phase, supplemented by 100 μm of zinc chloride at the time of induction. E. coli were collected after ∼16 h of incubation at 18 °C and lysed by sonication, and soluble protein was purified by nickel affinity and Superdex 200 size exclusion chromatography. The purified proteins suspended in 20 mm Tris-HCl (pH 7.4), 0.5 m NaCl, and 5 mm β-mercaptoethanol produced single peak in size exclusion chromatography corresponding to a monomer (see “Results”). Additional zinc was not included in buffers during purification, but the purified protein contained a stoichiometric amount of zinc as confirmed by a colorimetric zinc quantification assay (Abcam). For A3Bctd-DM, the SUMO tag was removed by Ulp1 treatment prior to the size exclusion chromatography step. C-terminally myc-tagged APOBEC3A was expressed in 293T cells and purified as described (39, 41, 42).
Crystallization and Structure Determination
A3Bctd-QMΔloop3 was concentrated to ∼20 mg/ml by ultrafiltration and crystallized by the hanging drop vapor diffusion method, using a reservoir solution including 20% polyethylene glycol 3,350 and 0.1 m MES-NaOH buffer (pH 6.5). Crystals grew as clusters of fine needles. Both the orthorhombic and monoclinic space group crystals were obtained in essentially the same condition, and they had similar crystal morphologies. Isolated crystals were cryoprotected by transfer into a reservoir solution supplemented by 25% glycerol or ethylene glycol and flash-cooled in liquid nitrogen. X-ray diffraction data were collected at the Advanced Photon Source NE-CAT Beamlines 24-ID-C and 24-ID-E and processed using HKL2000 (43) or XDS (44). The structures were solved by molecular replacement phasing using a previously reported A3Gctd crystal structure (39) (PDB code 3V4K) as the search model, with the program PHASER (45). Model building and refinement was done using the program COOT (46) and PHENIX (47), respectively. The nucleotide (dCMP)-bound structure of A3Bctd-QMΔloop3 (5CQH) was obtained by soaking the crystal in the orthorhombic crystal form in 200 mm dCMP during cryoprotection with ethylene glycol. The glycerol-bound structure (PDB code 5CQI) was obtained by soaking the crystal with a small fragment of ssDNA (5-mer) during cryoprotection with glycerol, although electron density was not observed for the ssDNA. The glycerol/PEG-bound structure (PDB code 5CQK) was obtained by cryoprotection with glycerol, without any nucleotide ligands. The dCMP and PEG bound to overlapping sites on the protein surface. Summaries of x-ray diffraction data and model refinement statistics are listed in Table 1. Active site volumes were estimated using Sitemap (48).
TABLE 1.
X-ray data collection and refinement statistics
Statistics for the highest resolution shells are shown in parentheses.
| A3B-dCMP (ethylene glycol) | A3B (glycerol) | A3B (glycerol + PEG) | A3B (monoclinic) | |
|---|---|---|---|---|
| Protein Data Bank code | 5CQH | 5CQI | 5CQK | 5CQD |
| Diffraction data | ||||
| Wavelength (Å) | 0.979 | 0.979 | 0.979 | 0.979 |
| Resolution range (Å) | 40.7–1.73 | 40.2–1.68 | 40.3–1.88 | 40.9–2.08 |
| (1.79–1.73) | (1.74–1.68) | (1.95–1.88) | (2.16–2.08) | |
| Space group | P 21 21 21 | P 21 21 21 | P 21 21 21 | P 21 |
| Unit cell | ||||
| a, b, c (Å) | 50.84, 52.45, 68.04 | 51.02, 54.29, 65.41 | 50.63, 54.04, 66.42 | 53.44, 50.84, 70.15 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 101.83, 90 |
| Total reflections | 91,583 (7314) | 180,705 (6315) | 106,363 (7997) | 98,439 (3214) |
| Unique reflections | 19,558 (1922) | 20,099 (1451) | 15,315 (1520) | 21,151 (1644) |
| Multiplicity | 4.7 (3.8) | 9.0 (4.4) | 6.9 (5.3) | 4.7 (2.0) |
| Completeness (%) | 100.0 (100.0) | 94.0 (69.0) | 100.0 (99.0) | 94.0 (73.0) |
| Mean I/σI | 17.3 (2.30) | 14.2 (1.77) | 7.73 (1.00) | 9.81 (1.47) |
| Rmerge (%) | 9.0 (56.7) | 15.0 (94.9) | 20.7 (168.0) | 7.6 (44.6) |
| CC1/2 | 0.995 (0.661) | 0.993 (0.426) | 0.992 (0.314) | 0.995 (0.659) |
| Refinement | ||||
| Reflections | 19,545 (1922) | 20,099 (1449) | 15,315 (1519) | 21,151 (1640) |
| Test set reflections | 998 (90) | 1012 (67) | 771 (74) | 1057 (82) |
| Rwork (%) | 15.2 (23.9) | 16.0 (29.2) | 17.4 (31.5) | 18.8 (32.8) |
| Rfree (%) | 18.6 (29.9) | 19.8 (33.3) | 20.3 (35.2) | 22.7 (34.2) |
| Number of atoms | 1760 | 1765 | 1633 | 3183 |
| Macromolecules | 1557 | 1529 | 1506 | 3075 |
| Ligands | 18 | 19 | 34 | 32 |
| Protein residues | 183 | 182 | 181 | 371 |
| Root mean square deviation | ||||
| Bond lengths (Å) | 0.018 | 0.005 | 0.009 | 0.003 |
| Bond angles (°) | 1.65 | 0.82 | 0.97 | 0.57 |
| Ramachandran | ||||
| Favored (%) | 98.3 | 97.8 | 98.3 | 97.2 |
| Allowed (%) | 1.7 | 2.2 | 1.7 | 2.8 |
| Outliers (%) | 0 | 0 | 0 | 0 |
| Average B-factor (Å2) | 23.69 | 19.66 | 27.85 | 53.02 |
| Macromolecules | 22.09 | 17.96 | 27.34 | 53.07 |
| Ligands | 35.16 | 28.82 | 39.69 | 50.80 |
| Solvents | 36.04 | 30.78 | 31.78 | 51.66 |
Rifampicin Resistance Mutation Assay
E. coli strain C43(DE3) was transformed with each plasmid and plated on LB plates containing either ampicillin (100 μg/ml) or kanamycin (50 μg/ml). The rpoB forward mutation assay was performed as described (49, 50). Briefly, single colonies were picked and inoculated into 1 ml of LB with appropriate antibiotic. Cultures were grown to an A600 of 0.4–0.6 followed by plating 0.1–0.5 ml of neat culture onto LB rifampicin (100 μg/ml) or appropriate dilutions onto LB plates. After an overnight incubation, colonies were counted, and mutation frequency was calculated by dividing the number of rifampicin-resistant colonies per ml by the total number of cells per ml. The sequence preference of rpoB mutagenesis was determined by colony PCR using 5′-TTGGCGAAATGGCGGAAAACC and 5′-CACCGACGGATACCACCTGCTG, treatment of the PCR products with shrimp alkaline phosphatase and Exonuclease I (New England Biolabs, Ipswich, MA), and sequencing using the forward rpoB primer. Mutations were analyzed using Sequencher version 5.0.1 (GeneCodes, Ann Arbor, MI). Most constructs tested in the rpoB assay were Sumo-fused (pE-SUMO vector) because they elicited slightly higher mutation frequencies than the corresponding His6-tagged constructs (pET24a vector), except for A3AL1 for which the Sumo fusion construct could not be generated because of toxicity in E. coli.
In Vitro DNA Deamination Assay
800 nm oligonucleotide 5′-ATTATTATTATTCAAATGGATTTATTTATTTATTTATTTATTT-fluorescein was treated with purified proteins for 1 h at 37 °C, followed by treatment with 0.06 units uracil-DNA glycosylase (New England Biolabs) for 10 min at 37 °C, and treatment with 100 mm NaOH for 10 min at 95 °C (39, 42). Reaction products were separated by 15% denaturing PAGE and scanned on a Typhoon FLA 7000 imager (GE Healthcare Life Sciences).
Results
Solubility and Activity of the A3B Catalytic Domain
The full-length human A3B enzyme consists of two tandem zinc-coordinating domains: the N-terminal domain with a pseudo-active site and the C-terminal domain with catalytic activity (51). Initial purification attempts with full-length A3B did not succeed because it is expressed at low levels and is poorly soluble. We therefore created a deletion construct that retained residues 187–378 to focus on characterizing the catalytic domain (A3Bctd) (5, 11, 12, 52). These deletion boundaries were informed by comparisons with the related enzymes A3G and A3F, where analogous approaches led to successful catalytic domain structures (29–31, 33, 37–39).
The E. coli-based rifampicin resistance mutation assay was used to assess DNA cytosine deaminase activity (49). The median number of rifampicin-resistant mutants for each condition provides a quantitative comparison of the mutation frequency relative to control plasmid expressing cells. Single colonies were grown overnight, and then aliquots of saturated cultures were plated on rifampicin-containing plates to select for rpoB mutants and additional aliquots were diluted and plated on rich growth medium to assess viability. The A3Bctd construct elicited a slightly higher mutation frequency than full-length A3B (residues 1–382), and E. coli cells expressing either of these constructs showed mutation frequencies over 100-fold higher than cells with the vector control or expressing a catalytic mutant (E255A) (Fig. 1A). These data demonstrated that the C-terminal domain alone is sufficient for DNA editing.
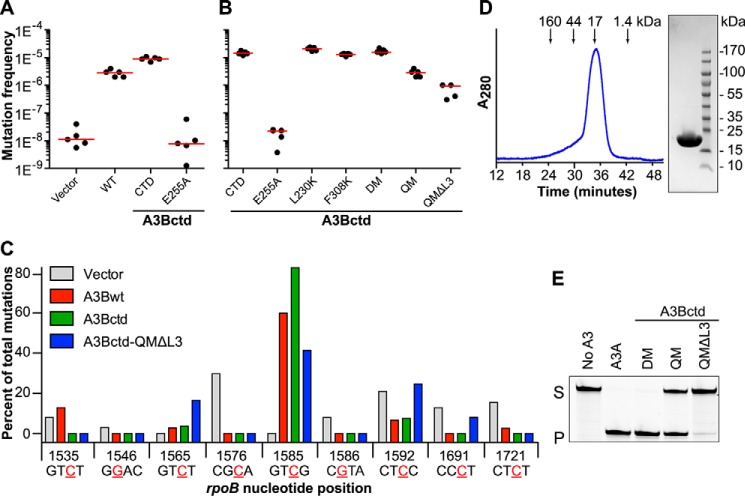
FIGURE 1.
Generation of an active and soluble A3B catalytic domain construct. A and B, rifampicin resistance (rpoB) mutation frequency of E. coli expressing the indicated Sumo-A3B constructs. Each dot represents data from an individual culture (n = 5 per condition), and the horizontal line reports the median value for each condition. Vector, pE-Sumo-Amp; WT, full-length wild-type A3B; CTD, A3Bctd residues 187–378. Amino acid substitution mutants are derivatives of A3Bctd. C, rifampicin resistance (rpoB) mutation spectrum of E. coli expressing WT, A3Bctd, and A3Bctd-QMΔloop3 in comparison to the vector control. A histogram summarizing the percentage of C to T transitions occurring at mutable positions in rpoB (shown in tetranucleotide context with mutated base colored red, underlined, and numbered according to position in the rpoB gene). D, size exclusion chromatography profile of purified A3Bctd-QMΔloop3 and Coomassie Brilliant Blue-stained SDS-PAGE analysis of purified A3Bctd-QMΔloop3. The protein concentration was adjusted to 1.5 mg ml−1 prior to injection into a Superdex 200 10/300 GL column. The molecular mass standards used were as follows; bovine γ-globulin (160 kDa), chicken ovalbumin (44 kDa), horse myoglobin (17 kDa), and vitamin B12 (1.4 kDa). E, comparison of the in vitro DNA deaminase activities of recombinant A3A purified from 293 cells and 10 μm A3Bctd-DM, A3Bctd-QM, and A3Bctd-QMΔloop3 purified from E. coli. S, substrate; P, product.
We next focused on improving A3Bctd solubility in E. coli by introducing four amino acid substitutions: F200S, W228S, L230K, and F308K (QM). Three of these substitutions (F200S, W228S, and L230K) changed A3Bctd residues to the corresponding sequence of the homologous A3A protein, which recently yielded to structural studies (34, 40). F308K mimics the F302K and F310K substitutions used in structural studies of A3Fctd and A3Gctd, respectively (29–31, 33). In addition, to reduce conformational entropy and facilitate crystallization, we replaced the region spanning Ala-242 to Tyr-250, which corresponds to the flexible loop 3 of A3A and A3G (53), with a single serine residue (designated Ser-250 in the structural coordinates and from here on). The resulting construct, A3Bctd-QMΔloop3, was confirmed to be active by eliciting an rpoB mutation frequency nearly 100-fold higher than background and only modestly reduced compared with the parental construct (Fig. 1B).
Next, individual rifampicin-resistant mutants were subjected to high fidelity colony PCR and DNA sequencing. Importantly, the distribution of cytosine mutations within the rpoB gene was similar for full-length A3B (wild type), A3Bctd, and A3Bctd-QMΔloop3 demonstrating again that the C-terminal domain governs catalytic activity and intrinsic local deamination preferences and that the amino acid changes required for solubility did not compromise these important properties (Fig. 1C). The major transition mutation hot spot for A3B activity in rpoB is a 5′-GTCG (C1585) consistent with the previously reported in vitro preferences (17, 21). In addition, purified A3Bctd-QMΔloop3 showed a monodisperse distribution by size exclusion chromatography, single band by SDS-PAGE, and ssDNA deaminase activity in vitro (Fig. 1, D and E).
A3B Catalytic Domain Crystal Structures
A3Bctd-QMΔloop3 crystallized in two different crystal forms, and molecular replacement was used to determine the structures (Fig. 2 and Table 1). The three structures determined in the orthorhombic space group (P212121) were different only in terms of bound ligands. The structure refined at 1.73 Å resolution was bound by a single dCMP nucleotide and four ethylene glycol molecules. The 1.68 Å resolution structure had ordered glycerol molecules bound inside and near the active site pocket. The 1.88 Å resolution structure has glycerol in the active site, as well as an ordered partial PEG chain on the protein surface. The structure refined in the monoclinic space group (P21) also showed an ordered glycerol molecule occupying the active site pocket of both of the protein molecules in the asymmetric unit.
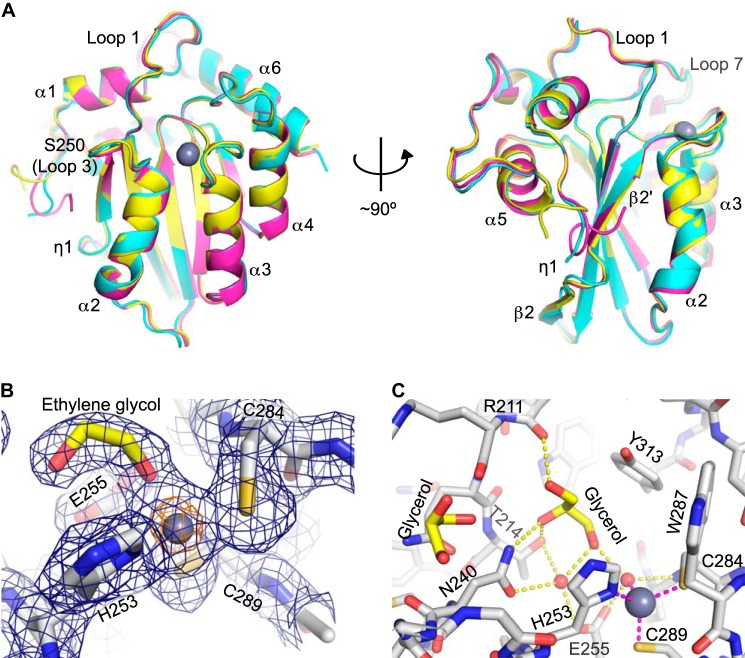
FIGURE 2.
A3B catalytic domain structures. A, superposition of three independent A3Bctd-QMΔloop3 structures. The structure refined in the orthorhombic space group (P212121) is in cyan, and the two crystallographically independent molecules in the monoclinic space group (P21) are shown in magenta and yellow. A gray sphere represents each zinc ion. B, A3B active site residues involved in zinc coordination. The fourth ligand of zinc coordination in this case is ethylene glycol (yellow and red). The final model is overlaid with the simulated annealing, composite omit 2Fo − Fc map contoured at 1.0 σ (blue mesh) or 10.0 σ (orange mesh). C, the network of interactions in the glycerol-occupied active site pocket. Hydrogen bonds are shown in yellow, and the zinc-coordinating interactions are shown as magenta dotted lines. The fourth ligand of zinc coordination in this case is a water molecule (hydroxyl ion).
The refined atomic models of A3Bctd-QMΔloop3 are very similar between the two crystal forms and between the two crystallographically independent molecules in the monoclinic space group (Fig. 2A). The backbone root mean square deviations for all pairwise comparisons are smaller than 0.5 Å. The overall architecture of A3Bctd-QMΔloop3 closely resembles those of other A3 members and consists of a single-layered, five-stranded β-sheet surrounded by six α-helices and connecting loops (Fig. 2A). As observed for the closely related Z1-type deaminase domains A3A and A3Gctd, β2 on the exposed edge of the β-sheet is interrupted by a short 310 helical segment (helix η1 in Fig. 2A; A3A (34, 40); A3Gctd (30, 31, 33, 37, 39)). Correspondingly, the C-terminal end of α2, which interacts with this helical segment, is bent toward the discontinuous β2 (Fig. 2A). Although functional dimerization has been proposed for closely related A3 proteins A3A (40) and A3Gctd (33), the A3Bctd crystal structures showed no evidence for oligomerization through homologous motifs. For instance, the interfaces appear mostly distinct between the two crystal forms, with the only recurring crystallographic dimer interface observed in both crystal forms facilitated by Ser-250 hydrogen bonding (the residue that had been introduced to replace loop 3). Moreover, at high protein concentrations, A3Bctd-QMΔloop3 behaved as a monomer in size exclusion chromatography (Fig. 1D). Our data combine to indicate that the A3Bctd is active as a monomer, although we have not addressed the possibility that it might oligomerize upon substrate engagement, and it is notable that full-length protein is capable of forming higher order oligomers in a cellular context, most likely through its insoluble N-terminal domain (54).
Closed Conformation of the A3B Active Site
In the structure of A3Bctd-QMΔloop3, a single zinc ion is bound between the N-terminal ends of α2 and α3, coordinated by two cysteines (Cys-284 and Cys-289) and a histidine (His-253; Fig. 2, A–C). The mode of zinc coordination resembles those of other A3 members and zinc-dependent nucleoside/mononucleotide deaminase enzymes. The single zinc ion in A3Bctd-QMΔloop3 is partially accessible to solvent through a deep active site pocket (Fig. 3, A–E). The exposed face of the zinc ion interacts with a water molecule serving as the fourth ligand of zinc coordination in the glycerol-bound active site (described below) or, alternatively, is bound by an ethylene glycol molecule when it was used as the cryoprotectant (Figs. 2, B and C, and 3, A–D). This water or ethylene glycol molecule is also hydrogen-bonded to Glu-255 in the bottom of the pocket; the observed geometry supports a catalytic mechanism in which Glu-255 activates the water molecule for nucleophilic attack of the C4 position of cytosine, and the zinc ion directly coordinates and stabilizes the transition state of the deamination intermediate. In the active site pocket, several additional well ordered water molecules were also observed (Fig. 3, A and B) or, depending on the cryoprotectant used, a well ordered glycerol molecule (Figs. 2C and 3, C and D). The glycerol molecule bound in the pocket makes either direct or water-mediated hydrogen bonds with the polar side chains (Thr-214 and Asn-240) or main chain groups (Arg-211 carbonyl and Cys-284 amide) lining the active site pocket, which may mimic interactions made by the cytosine substrate.
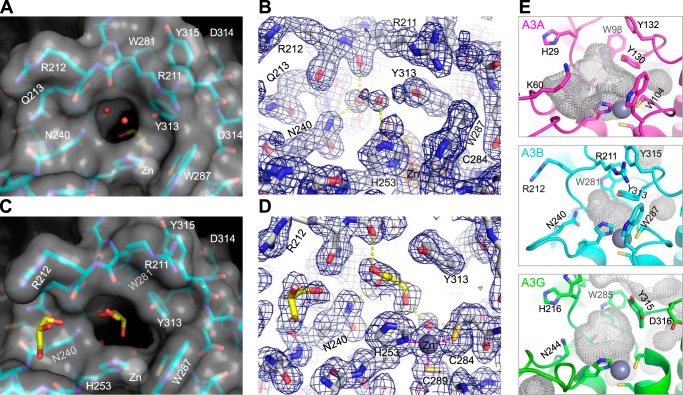
FIGURE 3.
The closed conformation of the A3B active site. A, molecular surface around the active site of A3Bctd-QMΔloop3 showing a small portal into the catalytic pocket. The active site pocket contains ordered water molecules represented by red spheres and an ethylene glycol molecule in yellow as shown in Fig. 2B. B, electron density for the region shown in A. The final model is overlaid with the simulated annealing, composite omit 2Fo − Fc map as in Fig. 2B. C, a view of the active site pocket with glycerol molecules shown in yellow and red sticks. D, electron density for the region shown in C. The simulated annealing composite omit 2Fo − Fc map is contoured at 1.0 σ (blue mesh). E, comparison of active site conformations for A3A (PDB code 4XXO), A3Bctd-QMΔloop3 (this study), and A3Gctd (PDB code 3V4K). The gray wireframes show pockets/cavities in each protein.
Previous structural and functional studies of A3 proteins have demonstrated that loop 1 (between α1 and β1) and loop 7 (between β4 and α4) surround the active site and suggested that these flexible regions play important roles in binding single-stranded DNA substrates (30, 53, 55–58). The structures of A3Bctd-QMΔloop3 in both crystal forms show a tightly closed conformation of the active site compared with those observed for other A3 proteins (Fig. 3, A and C). In particular, Arg-211 from loop 1 and Tyr-315 from loop 7 stack on each other to stabilize a collapsed conformation of these loops. The Arg-211 and Tyr-315 side chains in turn stack on Tyr-313 that is positioned over the active site pocket. In addition, the short bulge connecting the two zinc-coordinating cysteines (Cys-284 and Cys-289) that precedes α3 is positioned proximal to the active site pocket, where Ser-286 and Trp-287 form a hydrogen bond and van der Waals contacts, respectively, with Asp-316 and Arg-211 to further stabilize the closed conformation of loops 1 and 7. The tightly packed side chains around the active site leave a limited opening for single-stranded DNA cytosines to enter the active site pocket, and this does not appear large enough for a nucleotide substrate to pass through (Fig. 3, A and C). This closed conformation results in an active site pocket of ∼104 Å3 in comparison with the larger active site volumes of 272 Å3 for A3A and 184 Å3 for A3Gctd (Fig. 3E). Thus, substrate binding into the A3B active site is likely to require considerable rearrangement of the surrounding residues from the closed conformation observed in our A3Bctd-QMΔloop3 crystal structures into a more open and catalytically active conformation.
Roles of A3B Active Site Residues
To probe roles of the amino acid residues around the active site mentioned above, we tested the deaminase activities of mutant A3B proteins using the E. coli-based mutation assay. The amino acid substitutions were introduced into the A3Bctd-L230K/F308K background, because it had already proven to be one of the most active constructs (Fig. 1B). Of all amino acid substitutions tested, Y313A showed the strongest effect, with a complete loss of activity (Fig. 4A). On the other hand, Y313F caused no change of mutation frequency, suggesting that the aromatic moiety of Tyr-313, but not the hydroxyl group of the tyrosine side chain, is critical for function. Tyr-313 from loop 7 is conserved across most A3 proteins, and aromatic residues at the equivalent position in various mononucleotide/nucleoside cytosine deaminases, provided in trans (intermolecularly) in some cases, have been observed to make direct contact with the pyrimidine moiety occupying the active site pocket (59, 60). Substitution of Arg-211 for alanine in loop 1, another well conserved residue among A3 members, caused a strong but incomplete reduction of mutation frequency (Fig. 4A). W287A did not affect the mutation frequency, suggesting that Trp-287 is not an essential residue for the catalytic activity of A3B. This result, however, is not too surprising because Trp-287 and the adjacent Gly-288 are particular to A3B and the related subfamily member A3A, suggesting that these amino acid residues may have a function beyond substrate binding and catalysis.
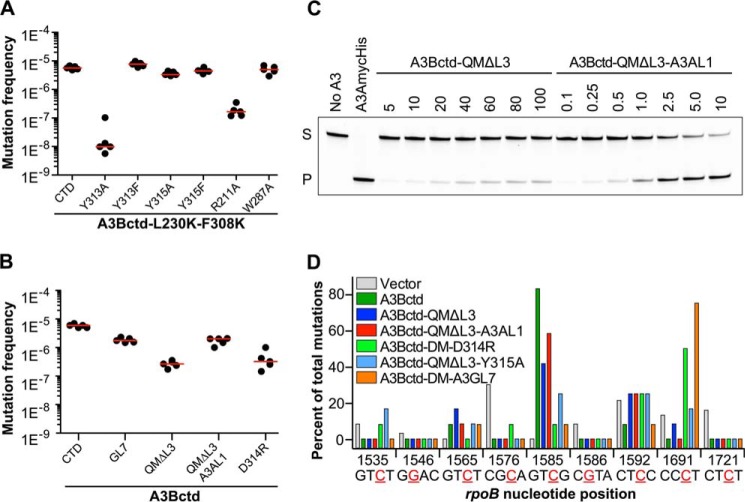
FIGURE 4.
Enzymatic activity and local preferences in E. coli. A, rifampicin resistance (rpoB) mutation frequencies of E. coli expressing the indicated Sumo-A3Bctd constructs (active site single amino acid substitution mutants). Performed and labeled as described for experiments in Fig. 1. B, rifampicin resistance (rpoB) mutation frequency of E. coli expressing the indicated chimeric A3B constructs. Each dot represents data from an individual culture (n = 5 per condition), and the horizontal line reports the median value for each condition. C, comparison of the in vitro DNA deaminase activities of purified A3Bctd-QMΔloop3 and A3Bctd-QMΔloop3-A3Aloop1 over the indicated ranges of protein concentrations (in μm). S, substrate; P, product. D, rifampicin resistance (rpoB) mutation spectra for select constructs from C. The histogram summarizes the percentage of C to T transitions occurring at mutable positions in the rpoB gene shown in tetranucleotide context with mutated base colored red, underlined, and numbered according to position in the coding sequence. To facilitate comparisons, the vector, A3Bctd, and A3Bctd-QMΔL3 data are reproduced from Fig. 1.
We also examined the mutagenic activity of chimeric A3Bctd proteins (Fig. 4B). Replacing loop 7 residues of A3Bctd with the corresponding sequence from A3G (GL7) only slightly reduced mutation frequency, indicating that the precise identities of loop 7 residues are not essential for catalytic activity. Although A3B and A3G residues around the active site are generally well conserved, their loop 1 and 7 sequences differ significantly, likely reflecting the different ssDNA substrate sequence preferences of these enzymes (elaborated further below). A3A and A3B have essentially identical sets of active site residues, except that their loop 1 sequences differ. Replacing A3B loop 1 with the corresponding residues from A3A led to enhancement of mutation frequency in the rpoB assay and to significantly higher in vitro deaminase activity of the purified proteins (Fig. 4, B and C). This improved activity relative to the A3Bctd-QMΔloop3 parental construct is consistent with the more open active site conformation reported recently for A3A (34, 40) (Fig. 3E).
A3B Sequence Preferences
The deamination target sequence profile of various A3B constructs were examined to gain insights into the determinants of sequence selectivity. Wild-type A3B and A3Bctd preferentially deaminated a single 5′-GTCG-3′ hot spot motif in the rpoB gene (Figs. 1C and 4D), consistent with previous reports for recombinant A3Bctd in vitro (17, 21). Thus, the N-terminal domain of A3B is not important for activity or intrinsic target selectivity. The crystallized protein A3Bctd-QMΔloop3 lacking the flexible loop 3 showed the same target preference as the wild-type protein, suggesting that the loop 3 is not essential for either catalytic activity or local target sequence selectivity of A3B (Figs. 1C and 4D). The more active protein, A3Bctd-QMΔloop3-A3AL1, which has loop 1 from A3A, also showed a similar 5′-TC-3′ sequence preference (Fig. 4D).
Previous studies have demonstrated the importance of loop 7 residues in sequence selectivity of related A3 family members, AID, A3A, A3F, and A3G (53, 55–58). Consistent with these reports, replacing loop 7 of A3B with that from A3G (GL7) converted the target sequence preference to 5′-CC-3′ preferred by A3G (cytosine 1691 in rpoB; Fig. 4D). Interestingly, single amino acid residue change, D314R, which is a conserved residue in loop 7 between A3A (Asp-131), A3B (Asp-314), and A3G (Asp-316), was sufficient to cause the change of preference at the −1 position from T to C (Fig. 4D). Although the precise mechanism of this change in selectivity is unknown, the observation underscores the importance of A3B loop 7 in target sequence selectivity. Another amino acid substitution within loop 7, Y315A, did not cause a clear conversion of target sequence preference but appeared to relax it somewhat (Fig. 4D).
Nucleotide Binding Site and an A3B-ssDNA Model
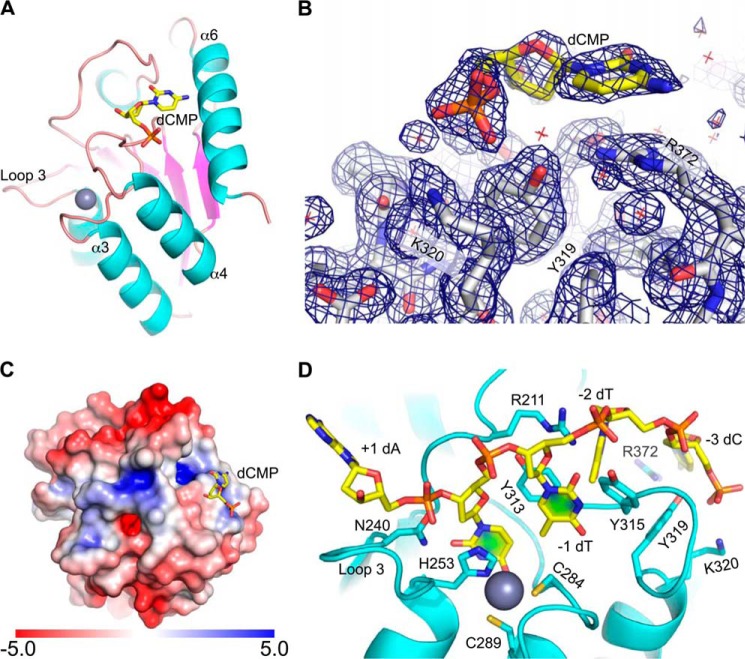
Soaking the A3Bctd-QMΔloop3 crystal in the orthorhombic space group with dCMP as an attempt to capture the nucleotide in the active site yielded no additional electron density in the active site pocket. Unexpectedly, however, we observed clear electron density at 1.7 Å resolution for the nucleotide bound at a positively charged surface close to but outside of the active site (Fig. 5, A–C). The dCMP molecule lies flat on the protein surface; it is bound at this position through hydrogen bonding of the 5′-phosphate group with Tyr-319 and Lys-320 side chains from α4 and cation π stacking between the pyrimidine ring and Arg-372 side chain from α6. The guanidinium group of Arg-372 forms a salt bridge with Asp-314. In addition to these interactions, the 3′-OH group of dCMP is hydrogen-bonded to a side chain from a symmetry-related protein molecule as part of the crystallographic lattice. The observed interactions do not appear sequence (cytosine)-specific.
FIGURE 5.
A dCMP-bound structure and a model for A3Bctd-ssDNA complex formation. A, a ribbon schematic of A3Bctd-QMΔloop3 with dCMP bound on the protein surface shown as sticks. B, a close-up view of the bound dCMP molecule (yellow) and the interacting protein residues (gray) as sticks, overlaid with the simulated annealing, composite omit 2Fo − Fc map contoured at 1.0 σ (blue mesh). C, A3Bctd-QMΔloop3 surface colored according to the electrostatic potential (−5.0 to +5.0 kT/e), with the bound dCMP as sticks. A3Bctd-QMΔloop3 is oriented to position the active site pocket approximately in the middle of the view. D, a hypothetical model of A3Bctd bound to a 5-mer ssDNA, based on the crystallographically observed nucleotide. See main text for details.
We reasoned that the bound dCMP molecule may represent a nucleotide 5′ to the target cytosine within a ssDNA. Based on the orientation of the bound dCMP and its distance from the likely position of the deamination target cytosine and the likelihood that loop 7 residues located between the dCMP and the active site dictate the identity of the −1 base, we further reasoned that this dCMP represents the −3 base in a typical single-stranded DNA substrate (i.e. −3 relative to a 0 position target cytosine). Thus, we built a hypothetical model of A3B-ssDNA complex by connecting the crystallographically observed nucleotide, which occupies the −3 position, and a nucleotide in the active site (0 position). The nucleotide in the active site pocket was modeled based on superposition of a 2′-deoxycytidylate deaminase crystal structure (PDB code 1VQ2 (59)) with ligand bound in the active site onto the A3Bctd-QMΔloop3 crystal structure. As described above, the A3Bctd-QMΔloop3 crystal structures show a closed conformation not compatible with substrate binding into the active site pocket. Thus, to accommodate the ssDNA substrate including the intervening nucleotides (at the −1 and −2 positions), the conformation of loop 7 was modified to mimic the more open state observed in the A3Gctd crystal structure (Fig. 3E) based on superpositioning of the two structures, followed by energy minimization to regularize geometry.
In the resulting ssDNA-A3B model (Fig. 5D), the cytosine base in the active site pocket stacks on the zinc-coordinating residue His-253, whereas the C2 carbonyl group of the pyrimidine base potentially interacts with Asn-240. Tyr-313 stacks on the thymine base at the −1 position, which fits tightly within a limited space surrounded by aromatic side chains that would only accommodate a pyrimidine base. This thymine (−1) base is modeled so that its C5 methyl group makes a van der Waals contact with the cytosine base in the active site, whereas its C4 carbonyl group is positioned close to Tyr-315. When A3G with the 5′-CC-3′ target preference binds its cognate sequence, the C4 amino group of the cytosine base at this (−1) position could hydrogen bond with the aspartate side chain at the corresponding position (Asp-317), which has been proposed to serve as a helix-capping residue to help define the cytosine-binding pocket (53). Arg-211 interacts with a backbone phosphate of the DNA. The ssDNA shown in Fig. 5D has thymine at −2 and adenine at +1 positions, but the model would accommodate other sequences at these positions as well. The possible significance of this hypothetical ssDNA binding mechanism is discussed below.
Discussion
We have generated a highly soluble A3B catalytic domain construct (A3Bctd-QMΔloop3) and determined its high resolution crystal structures in two crystal forms with distinct molecular packing environments. Importantly, A3Bctd-QMΔloop3 is catalytically active and has the same target DNA sequence preference as the wild-type enzyme, ensuring functional relevance of the observed structural features. Our crystal structures reveal a closed active site conformation that is likely to prevent substrate entry into the active site pocket. The high similarity between the structures of three crystallographically independent molecules, across two crystal forms, suggests that the observed closed conformation is the most thermodynamically stable state of the enzyme (Fig. 2). The closed conformation is stabilized by a stacking interaction between Arg-211 and Tyr-315 from loops 1 and 7, respectively. Previous studies together with the mutation analyses presented here demonstrate that these loop regions are important for catalytic activity and dinucleotide target sequence selectivity, respectively, strongly implicating direct interactions with ssDNA substrates (30, 53, 55–58). Based on these observations, we hypothesize that the binding of a ssDNA substrate carrying the preferred 5′-TCG or 5′-TCA target sequence triggers conformational changes of structural elements including loops 1 and 7, which in turn stabilize the open conformation of the active site and allow entry of the target cytosine base into the active site pocket. Together with the fact that related enzymes, A3A and A3Gctd, have been crystallized in more open states (Fig. 3E) (33, 39, 40), the closed conformation of our A3Bctd structures suggests that the transition between the two states may be regulated in a manner that maximizes innate antiviral activity and minimizes damage to genomic DNA.
Currently, no nucleotide- or polynucleotide-bound structures exist for any member of the APOBEC family from humans or any other species (i.e. AID, APOBEC1, APOBEC2,APOBEC3A-H, and APOBEC4). Thus, the high resolution, dCMP-bound structure of A3BctdQMΔ3 is of interest as a starting point and model to guide future studies (Fig. 5). The single dCMP molecule binds the A3Bctd surface and makes several contacts with the enzyme, but it is also assisted by crystal lattice contacts involving symmetry-related molecules (see PDB entry 5CQH), and thus the significance of this result should be interpreted conservatively. Nonetheless, the general location and orientation of the bound nucleotide appear consistent with previous models of A3 proteins binding ssDNA substrates (30, 34, 37, 38, 61, 62). A3Gctd and A3A NMR chemical shift perturbation experiments, as well as mutational analyses of A3Fctd and A3Gctd, although differing in details, all suggest ssDNA binding paths spanning the catalytic domain surface and extending from the active site toward the α-helix 6, which overlaps with the crystallographically observed dCMP interacting with Arg-372 from the α-helix 6 and Tyr-319/Lys-320 from the N terminus of α-helix 4 (A3Bctd data in Fig. 5; prior studies with other family members (30, 34, 37, 38, 61, 62)). Curiously, this nucleotide-binding site on the A3Bctd surface is accessible even when the active site is in the closed conformation. Therefore, this site could serve as an initial contact point to recruit ssDNA substrates, which in turn may allow downstream nucleotides to form more sequence-specific interactions with ligands in loops 1 and 7 around the active site. The nucleotide bound at this position may remain engaged during catalysis (as in Fig. 5D) or, alternatively, may transition to a more stable binding mode concomitant with conformational changes of the active site as discussed above. In either scenario, we propose a model in which ssDNA binding by A3Bctd entails at least two distinct steps: initial nonspecific binding that involves the exposed binding site followed by a more sequence-specific, stable interactions that require conformational changes of the loops adjacent to the active site pocket.
Comprehensive future experiments will be required to test this model rigorously and rule out potential alternatives ranging from the nucleotide binding site being a co-factor binding site, a protein-protein interaction motif, or a region of low functional relevance. Many scenarios are possible given precedents from functionally relevant ligand-bound structures of related enzymes including those of the human ADAR2/IP6 complex (PDB code 1ZY7 (63)), where the ligand serves as an essential co-factor to maintain the enzyme fold, the Staphylococcus aureus TadA/tRNA complex (PDB code 2B3 (64)), where an RNA polynucleotide substrate was captured in complex with the enzyme, and the E. coli cytidine deaminase/nucleoside complex (PDB code 1CTU (60)), the Bacillus subtilis cytidine deaminase/nucleoside complex (PDB code 1JTK (65)), and the bacteriophage T4 2′-deoxycytidylate deaminase/nucleotide complex (PDB code 1VQ2 (59)), where transition state analogs were captured in the active sites. By contrast, our observed nucleotide-binding site could be part of a larger ssDNA binding path, and accordingly, analogous future experiments may be even more challenging. In any event, high resolution crystal structures of the enzyme in complex with ssDNA substrates will be needed to unambiguously resolve the substrate binding mechanism.
A3B is a significant source of somatic mutations in multiple human cancers (reviewed in Refs. 14–16). Together with recently published clinical outcome data (24–27), it is likely that A3B causes a chronic mutator phenotype that drives tumor evolution and facilitates mutation dependent hallmarks of cancer such as drug resistance and metastasis. Our high resolution crystallographic studies provide a framework for further mechanistic studies on this enzyme. Our structures are also likely to facilitate the development of inhibitors of this enzyme, which may be useful as mutation-suppressing adjuvants in combination with targeted cancer treatments. An A3B inhibitor could target the active site pocket to compete with substrate binding or allosterically block substrate entry by stabilizing the closed conformation of the active site.
Author Contributions
H. A. and R. S. H. designed the studies and wrote the manuscript. K. S. purified and crystallized A3B and determined its x-ray structures. M. A. C. performed E. coli mutation and in vitro editing experiments. K. K. assisted with mutant construction, protein purification, and crystallization. All authors analyzed the results and approved the final version of the manuscript.
Acknowledgments
We thank Aihara and Harris lab members, as well as Drs. Harki, Amaro, and Demir for critical discussions.
This work was supported by National Institutes of Health Grants GM095558 and GM109770 from the NIGMS (to the Aihara lab), AI064046 from the NIAID and GM091743 from the NIGMS (to the Harris lab), GM110734 from the NIGMS (to M. A. C.), and P41 GM103403 from the NIGMS (to Advanced Photon Source NE-CAT beamlines). Advanced Photon Source is a U.S. Department of Energy Office of Science User Facility operated by Argonne National Laboratory under contract DE-AC02–06CH11357. R. S. H. is a co-founder of ApoGen Biotechnologies Inc. The other authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (codes 5CQH, 5CQI, 5CQK, and 5CQD) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- A3B
- APOBEC3B
- A3
- APOBEC3
- AID
- activation-induced deaminase
- A3Bctd
- A3B C-terminal domain
- QM
- quadruple mutant
- PDB
- Protein Data Bank.
References
- 1.Conticello S. G. (2008) The AID/APOBEC family of nucleic acid mutators. Genome Biol. 9, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Refsland E. W., and Harris R. S. (2013) The APOBEC3 family of retroelement restriction factors. Curr. Top. Microbiol. Immunol. 371, 1–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris R. S., and Dudley J. P. (2015) APOBECs and virus restriction. Virology 479–480, 131–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desimmie B. A., Delviks-Frankenberrry K. A., Burdick R. C., Qi D., Izumi T., and Pathak V. K. (2014) Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J. Mol. Biol. 426, 1220–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogerd H. P., Wiegand H. L., Hulme A. E., Garcia-Perez J. L., O'Shea K. S., Moran J. V., and Cullen B. R. (2006) Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. U.S.A. 103, 8780–8785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lackey L., Law E. K., Brown W. L., and Harris R. S. (2013) Subcellular localization of the APOBEC3 proteins during mitosis and implications for genomic DNA deamination. Cell Cycle 12, 762–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Noia J. M., and Neuberger M. S. (2007) Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76, 1–22 [DOI] [PubMed] [Google Scholar]
- 8.Robbiani D. F., and Nussenzweig M. C. (2013) Chromosome translocation, B cell lymphoma, and activation-induced cytidine deaminase. Annu. Rev. Pathol. 8, 79–103 [DOI] [PubMed] [Google Scholar]
- 9.Fossat N., and Tam P. P. (2014) Re-editing the paradigm of cytidine (C) to uridine (U) RNA editing. RNA Biol. 11, 1233–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith H. C., Bennett R. P., Kizilyer A., McDougall W. M., and Prohaska K. M. (2012) Functions and regulation of the APOBEC family of proteins. Semin. Cell Dev. Biol. 23, 258–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stenglein M. D., Matsuo H., and Harris R. S. (2008) Two regions within the amino-terminal half of APOBEC3G cooperate to determine cytoplasmic localization. J. Virol. 82, 9591–9599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pak V., Heidecker G., Pathak V. K., and Derse D. (2011) The role of amino-terminal sequences in cellular localization and antiviral activity of APOBEC3B. J. Virol. 85, 8538–8547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lackey L., Demorest Z. L., Land A. M., Hultquist J. F., Brown W. L., and Harris R. S. (2012) APOBEC3B and AID have similar nuclear import mechanisms. J. Mol. Biol. 419, 301–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swanton C., McGranahan N., Starrett G. J., and Harris R. S. (2015) APOBEC enzymes: mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 5, 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts S. A., and Gordenin D. A. (2014) Hypermutation in human cancer genomes: footprints and mechanisms. Nat. Rev. Cancer 14, 786–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson S., and Fenton T. (2015) APOBEC3 genes: retroviral restriction factors to cancer drivers. Trends Mol. Med. 21, 274–284 [DOI] [PubMed] [Google Scholar]
- 17.Burns M. B., Lackey L., Carpenter M. A., Rathore A., Land A. M., Leonard B., Refsland E. W., Kotandeniya D., Tretyakova N., Nikas J. B., Yee D., Temiz N. A., Donohue D. E., McDougle R. M., Brown W. L., Law E. K., and Harris R. S. (2013) APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 494, 366–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burns M. B., Temiz N. A., and Harris R. S. (2013) Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 45, 977–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts S. A., Lawrence M. S., Klimczak L. J., Grimm S. A., Fargo D., Stojanov P., Kiezun A., Kryukov G. V., Carter S. L., Saksena G., Harris S., Shah R. R., Resnick M. A., Getz G., and Gordenin D. A. (2013) An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 45, 970–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson S., Chakravarthy A., Su X., Boshoff C., and Fenton T. R. (2014) APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep. 7, 1833–1841 [DOI] [PubMed] [Google Scholar]
- 21.Leonard B., Hart S. N., Burns M. B., Carpenter M. A., Temiz N. A., Rathore A., Vogel R. I., Nikas J. B., Law E. K., Brown W. L., Li Y., Zhang Y., Maurer M. J., Oberg A. L., Cunningham J. M., Shridhar V., Bell D. A., April C., Bentley D., Bibikova M., Cheetham R. K., Fan J. B., Grocock R., Humphray S., Kingsbury Z., Peden J., Chien J., Swisher E. M., Hartmann L. C., Kalli K. R., Goode E. L., Sicotte H., Kaufmann S. H., and Harris R. S. (2013) APOBEC3B upregulation and genomic mutation patterns in serous ovarian carcinoma. Cancer Res. 73, 7222–7231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexandrov L. B., Nik-Zainal S., Wedge D. C., Aparicio S. A., Behjati S., Biankin A. V., Bignell G. R., Bolli N., Borg A., Børresen-Dale A. L., Boyault S., Burkhardt B., Butler A. P., Caldas C., Davies H. R., Desmedt C., Eils R., Eyfjörd J. E., Foekens J. A., Greaves M., Hosoda F., Hutter B., Ilicic T., Imbeaud S., Imielinski M., Jäger N., Jones D. T., Jones D., Knappskog S., Kool M., Lakhani S. R., López-Otín C., Martin S., Munshi N. C., Nakamura H., Northcott P. A., Pajic M., Papaemmanuil E., Paradiso A., Pearson J. V., Puente X. S., Raine K., Ramakrishna M., Richardson A. L., Richter J., Rosenstiel P., Schlesner M., Schumacher T. N., Span P. N., Teague J. W., Totoki Y., Tutt A. N., Valdes-Mas R., van Buuren M. M., van 't Veer L., Vincent-Salomon A., Waddell N., Yates L. R., Zucman-Rossi J., Futreal P. A., McDermott U., Lichter P., Meyerson M., Grimmond S. M., Siebert R., Campo E., Shibata T., Pfister S. M., Campbell P. J., and Stratton M. R. (2013) Signatures of mutational processes in human cancer. Nature 500, 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Bruin E. C., McGranahan N., Mitter R., Salm M., Wedge D. C., Yates L., Jamal-Hanjani M., Shafi S., Murugaesu N., Rowan A. J., Grönroos E., Muhammad M. A., Horswell S., Gerlinger M., Varela I., Jones D., Marshall J., Voet T., Van Loo P., Rassl D. M., Rintoul R. C., Janes S. M., Lee S. M., Forster M., Ahmad T., Lawrence D., Falzon M., Capitanio A., Harkins T. T., Lee C. C., Tom W., Teefe E., Chen S. C., Begum S., Rabinowitz A., Phillimore B., Spencer-Dene B., Stamp G., Szallasi Z., Matthews N., Stewart A., Campbell P., and Swanton C. (2014) Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 346, 251–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sieuwerts A. M., Willis S., Burns M. B., Look M. P., Meijer-Van Gelder M. E., Schlicker A., Heideman M. R., Jacobs H., Wessels L., Leyland-Jones B., Gray K. P., Foekens J. A., Harris R. S., and Martens J. W. (2014) Elevated APOBEC3B correlates with poor outcomes for estrogen-receptor-positive breast cancers. Hormones Cancer 5, 405–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cescon D. W., Haibe-Kains B., and Mak T. W. (2015) APOBEC3B expression in breast cancer reflects cellular proliferation, while a deletion polymorphism is associated with immune activation. Proc. Natl. Acad. Sci. U.S.A. 112, 2841–2846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu L., Chang Y., An H., Zhu Y., Yang Y., and Xu J. (2015) High APOBEC3B expression is a predictor of recurrence in patients with low-risk clear cell renal cell carcinoma. Urol. Oncol. 33, e1–e8 [DOI] [PubMed] [Google Scholar]
- 27.Walker B. A., Wardell C. P., Murison A., Boyle E. M., Begum D. B., Dahir N. M., Proszek P. Z., Melchor L., Pawlyn C., Kaiser M. F., Johnson D. C., Qiang Y. W., Jones J. R., Cairns D. A., Gregory W. M., Owen R. G., Cook G., Drayson M. T., Jackson G. H., Davies F. E., and Morgan G. J. (2015) APOBEC family mutational signatures are associated with poor prognosis translocations in multiple myeloma. Nat. Commun. 6, 6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kidd J. M., Newman T. L., Tuzun E., Kaul R., and Eichler E. E. (2007) Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 3, e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bohn M. F., Shandilya S. M., Albin J. S., Kouno T., Anderson B. D., McDougle R. M., Carpenter M. A., Rathore A., Evans L., Davis A. N., Zhang J., Lu Y., Somasundaran M., Matsuo H., Harris R. S., and Schiffer C. A. (2013) Crystal structure of the DNA cytosine deaminase APOBEC3F: the catalytically active and HIV-1 Vif-binding domain. Structure 21, 1042–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen K. M., Harjes E., Gross P. J., Fahmy A., Lu Y., Shindo K., Harris R. S., and Matsuo H. (2008) Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature 452, 116–119 [DOI] [PubMed] [Google Scholar]
- 31.Harjes E., Gross P. J., Chen K. M., Lu Y., Shindo K., Nowarski R., Gross J. D., Kotler M., Harris R. S., and Matsuo H. (2009) An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model. J. Mol. Biol. 389, 819–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kouno T., Luengas E. M., Shigematsu M., Shandilya S. M., Zhang J., Chen L., Hara M., Schiffer C. A., Harris R. S., and Matsuo H. (2015) Structure of the Vif-binding domain of the antiviral enzyme APOBEC3G. Nat. Struct. Mol. Biol. 22, 485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shandilya S. M., Nalam M. N., Nalivaika E. A., Gross P. J., Valesano J. C., Shindo K., Li M., Munson M., Royer W. E., Harjes E., Kono T., Matsuo H., Harris R. S., Somasundaran M., and Schiffer C. A. (2010) Crystal structure of the APOBEC3G catalytic domain reveals potential oligomerization interfaces. Structure 18, 28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Byeon I. J., Ahn J., Mitra M., Byeon C. H., Hercík K., Hritz J., Charlton L. M., Levin J. G., and Gronenborn A. M. (2013) NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nat. Commun. 4, 1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitamura S., Ode H., Nakashima M., Imahashi M., Naganawa Y., Kurosawa T., Yokomaku Y., Yamane T., Watanabe N., Suzuki A., Sugiura W., and Iwatani Y. (2012) The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat. Struct. Mol. Biol. 19, 1005–1010 [DOI] [PubMed] [Google Scholar]
- 36.Krzysiak T. C., Jung J., Thompson J., Baker D., and Gronenborn A. M. (2012) APOBEC2 is a monomer in solution: implications for APOBEC3G models. Biochemistry 51, 2008–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holden L. G., Prochnow C., Chang Y. P., Bransteitter R., Chelico L., Sen U., Stevens R. C., Goodman M. F., and Chen X. S. (2008) Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature 456, 121–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siu K. K., Sultana A., Azimi F. C., and Lee J. E. (2013) Structural determinants of HIV-1 Vif susceptibility and DNA binding in APOBEC3F. Nat. Commun. 4, 2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li M., Shandilya S. M., Carpenter M. A., Rathore A., Brown W. L., Perkins A. L., Harki D. A., Solberg J., Hook D. J., Pandey K. K., Parniak M. A., Johnson J. R., Krogan N. J., Somasundaran M., Ali A., Schiffer C. A., and Harris R. S. (2012) First-in-class small molecule inhibitors of the single-strand DNA cytosine deaminase APOBEC3G. ACS Chem. Biol. 7, 506–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bohn M. F., Shandilya S. M., Silvas T. V., Nalivaika E. A., Kouno T., Kelch B. A., Ryder S. P., Kurt-Yilmaz N., Somasundaran M., and Schiffer C. A. (2015) The ssDNA mutator APOBEC3A is regulated by cooperative dimerization. Structure 23, 903–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stenglein M. D., Burns M. B., Li M., Lengyel J., and Harris R. S. (2010) APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat. Struct. Mol. Biol. 17, 222–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carpenter M. A., Li M., Rathore A., Lackey L., Law E. K., Land A. M., Leonard B., Shandilya S. M., Bohn M. F., Schiffer C. A., Brown W. L., and Harris R. S. (2012) Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J. Biol. Chem. 287, 34801–34808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 44.Kabsch W. (2010) XDS. Acta Crystallogr. D. Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 47.Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., and Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halgren T. A. (2009) Identifying and characterizing binding sites and assessing druggability. J. Chem. Inf. Model. 49, 377–389 [DOI] [PubMed] [Google Scholar]
- 49.Harris R. S., Petersen-Mahrt S. K., and Neuberger M. S. (2002) RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell 10, 1247–1253 [DOI] [PubMed] [Google Scholar]
- 50.Haché G., Liddament M. T., and Harris R. S. (2005) The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J. Biol. Chem. 280, 10920–10924 [DOI] [PubMed] [Google Scholar]
- 51.Bogerd H. P., Wiegand H. L., Doehle B. P., and Cullen B. R. (2007) The intrinsic antiretroviral factor APOBEC3B contains two enzymatically active cytidine deaminase domains. Virology 364, 486–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McDougle R. M., Hultquist J. F., Stabell A. C., Sawyer S. L., and Harris R. S. (2013) D316 is critical for the enzymatic activity and HIV-1 restriction potential of human and rhesus APOBEC3B. Virology 441, 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rathore A., Carpenter M. A., Demir Ö., Ikeda T., Li M., Shaban N. M., Law E. K., Anokhin D., Brown W. L., Amaro R. E., and Harris R. S. (2013) The local dinucleotide preference of APOBEC3G can be altered from 5′-CC to 5′-TC by a single amino acid substitution. J. Mol. Biol. 425, 4442–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li J., Chen Y., Li M., Carpenter M. A., McDougle R. M., Luengas E. M., Macdonald P. J., Harris R. S., and Mueller J. D. (2014) APOBEC3 multimerization correlates with HIV-1 packaging and restriction activity in living cells. J. Mol. Biol. 426, 1296–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carpenter M. A., Rajagurubandara E., Wijesinghe P., and Bhagwat A. S. (2010) Determinants of sequence-specificity within human AID and APOBEC3G. DNA Repair (Amst.) 9, 579–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang M., Rada C., and Neuberger M. S. (2010) Altering the spectrum of immunoglobulin V gene somatic hypermutation by modifying the active site of AID. J. Exp. Med. 207, 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kohli R. M., Abrams S. R., Gajula K. S., Maul R. W., Gearhart P. J., and Stivers J. T. (2009) A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase. J. Biol. Chem. 284, 22898–22904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kohli R. M., Maul R. W., Guminski A. F., McClure R. L., Gajula K. S., Saribasak H., McMahon M. A., Siliciano R. F., Gearhart P. J., and Stivers J. T. (2010) Local sequence targeting in the AID/APOBEC family differentially impacts retroviral restriction and antibody diversification. J. Biol. Chem. 285, 40956–40964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Almog R., Maley F., Maley G. F., Maccoll R., and Van Roey P. (2004) Three-dimensional structure of the R115E mutant of T4-bacteriophage 2′-deoxycytidylate deaminase. Biochemistry 43, 13715–13723 [DOI] [PubMed] [Google Scholar]
- 60.Betts L., Xiang S., Short S. A., Wolfenden R., and Carter C. W. Jr. (1994) Cytidine deaminase. The 2.3 angstrom crystal structure of an enzyme: transition-state analog complex. J. Mol. Biol. 235, 635–656 [DOI] [PubMed] [Google Scholar]
- 61.Furukawa A., Nagata T., Matsugami A., Habu Y., Sugiyama R., Hayashi F., Kobayashi N., Yokoyama S., Takaku H., and Katahira M. (2009) Structure, interaction and real-time monitoring of the enzymatic reaction of wild-type APOBEC3G. EMBO J. 28, 440–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shindo K., Li M., Gross P. J., Brown W. L., Harjes E., Lu Y., Matsuo H., and Harris R. S. (2012) A Comparison of two single-stranded DNA binding models by mutational analysis of APOBEC3G. Biology 1, 260–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Macbeth M. R., Schubert H. L., Vandemark A. P., Lingam A. T., Hill C. P., and Bass B. L. (2005) Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309, 1534–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Losey H. C., Ruthenburg A. J., and Verdine G. L. (2006) Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat. Struct. Mol. Biol. 13, 153–159 [DOI] [PubMed] [Google Scholar]
- 65.Johansson E., Mejlhede N., Neuhard J., and Larsen S. (2002) Crystal structure of the tetrameric cytidine deaminase from Bacillus subtilis at 2.0 angstrom resolution. Biochemistry 41, 2563–2570 [DOI] [PubMed] [Google Scholar]