Background: eIF2α-P induced GADD34 and constitutively expressed CReP target PP1c to dephosphorylate eIF2α-P to dictate translation control of the ISR.
Results: Differential expression of GADD34 and CReP is regulated by upstream ORF (uORF)-mediated ribosome reinitiation.
Conclusion: uORFs regulate differential expression of GADD34 and CReP and are important for cell adaptation to stress.
Significance: Regulation of eIF2α-P is central for protein homeostasis and cell viability.
Keywords: endoplasmic reticulum stress (ER stress), eukaryotic initiation factor 2 (eIF2), stress response, translation control, translation initiation, CReP, GADD34, Integrated Stress Response, ribosome reinitiation, uORF
Abstract
In the integrated stress response, phosphorylation of eIF2α (eIF2α-P) reduces protein synthesis to conserve resources and facilitate preferential translation of transcripts that promote stress adaptation. Preferentially translated GADD34 (PPP1R15A) and constitutively expressed CReP (PPP1R15B) function to dephosphorylate eIF2α-P and restore protein synthesis. The 5′-leaders of GADD34 and CReP contain two upstream ORFs (uORFs). Using biochemical and genetic approaches we show that features of these uORFs are central for their differential expression. In the absence of stress, translation of an inhibitory uORF in GADD34 acts as a barrier that prevents reinitiation at the GADD34 coding region. Enhanced eIF2α-P during stress directs ribosome bypass of the uORF, facilitating translation of the GADD34 coding region. CReP expression occurs independent of eIF2α-P via an uORF that allows for translation reinitiation at the CReP coding region independent of stress. Importantly, alterations in the GADD34 uORF affect the status of eIF2α-P, translational control, and cell adaptation to stress. These results show that properties of uORFs that permit ribosome reinitiation are critical for directing gene-specific translational control in the integrated stress response.
Introduction
Protein synthesis is dynamic and is modulated in response to a variety of environmental stresses. An important mechanism regulating translation involves phosphorylation of eukaryotic initiation factor 2 (1). During the initiation phase of translation, eIF2 associates with initiator Met-tRNAiMet, GTP, and ultimately the 40S ribosomal subunit to facilitate start codon selection. Phosphorylation of the α subunit of eIF2 at serine 51 (eIF2α-P) inhibits the exchange of eIF2-GDP for eIF2-GTP, blocking delivery of the initiator tRNA that triggers a global reduction in initiation of protein synthesis (2). Reduced protein synthesis serves to conserve energy and resources and allows cells to reconfigure gene expression to alleviate stress damage. Facilitating the reprogramming of gene expression, eIF2α-P also leads to the preferential translation of specific transcripts that facilitate adaptation to a specific stress condition. Because there are multiple mammalian eIF2 kinases, each directing translational control in response to different stress arrangements, this pathway has been referred to as the integrated stress response (ISR)2 (2).
The 5′-leader of mRNAs containing upstream ORFs (uORFs) that precede the coding sequence (CDS) is critical for translational control in response to eIF2α-P. Among the ISR gene transcripts that are subject to preferential translation are those encoding transcription factors ATF4 (CREB2) and CHOP (DDIT3/GADD153) that serve to direct the transcriptome to address cellular stress, and GADD34 (PPP1R15A) that combines with the catalytic subunit of protein phosphatase 1 (PP1c) to target dephosphorylation of eIF2α-P and restore protein synthesis (3–6). GADD34 functions in combination with a constitutively expressed PP1c-targeting subunit CReP (PPP1R15B) (7), and together their expression, along with activities of eIF2 kinases, dictate the amount of eIF2α-P and degree of translational control. Emphasizing the importance of the GADD34 and CReP in the ISR, pharmacological agents have been reported that inhibit their functions and have significant medical implications (8–10). Preferential translation of ATF4, CHOP, and GADD34 ensure that the levels of these short-lived regulatory proteins in the ISR are tightly linked to the levels of eIF2α-P. Additionally, each of these key ISR regulatory genes are transcriptionally induced in response to stress, ensuring the availability of mRNAs for enhanced translation.
The uORFs in the ISR gene transcripts perform specific roles in preferential translation in response to eIF2α-P. For example, two uORFs in ATF4 convey translational control (3). In the “delayed translation reinitiation” model, the short 5′-proximal uORF1 of the ATF4 mRNA serves as a positive-acting element that during eIF2α-P allows for scanning ribosomes to surpass an inhibitory uORF2 that overlaps out-of-frame with the CDS and instead translate the ATF4 polypeptide. This model has features that are conserved with GCN4 translational control in yeast (11). Preferential translation of CHOP during eIF2α-P occurs via a bypass mechanism, where a single inhibitory uORF is bypassed due in part to a less than optimal start codon context (4). Of interest, translation of the CHOP uORF is suggested to trigger an elongation pause, ensuring no translation reinitiation at the downstream CDS.
Although uORFs are central for preferential translation of the ISR genes, the presence of uORFs alone are not sufficient to ensure enhanced translation in response to eIF2α-P. Approximately 40% of mammalian mRNAs contain uORFs, and genome-wide analyses of changes in translation in response to eIF2α-P suggest that uORFs are equally present among those gene transcripts whose translation are enhanced, repressed, or resistant to eIF2α-P (12). These findings suggest that there are specific properties for each uORF that delineate whether the 5′-leader of the mRNA directs activation or repression of translation in response to eIF2α-P. These uORF properties could include the sequences of the uORF coding and/or flanking regions, the length of the uORF, and the proximity of the uORF to the CDS of the transcript and with other uORFs.
In this study, we addressed the nature of uORFs that facilitate preferential translation in response to eIF2α-P. Translation of GADD34 mRNA is enhanced in response to eIF2α-P and serves a central role for feedback regulation of the ISR (5, 13), whereas the related CReP (PPP1R15B) is suggested to be expressed independently of eIF2α-P and functions to target PP1c for dephosphorylation of eIF2α-P under basal conditions (7, 14). Both GADD34 and CReP mRNAs contain two uORFs, and using biochemical and genetic approaches we define the central regulatory features that direct translational control of the two paralogs. We also show that alteration of these regulatory features in GADD34 alter the dynamics of the induction of the ISR, which has significant effects on cell adaptation to stress. This study not only provides a mechanistic understanding of translational control of key ISR genes during eIF2α-P, but also provides for rules to help predict the effects of uORFs in translational control and for new insight into the utility of therapeutic approaches to modulating levels of eIF2α-P and the ISR.
Experimental Procedures
Cell Culture and Generation of Stable Cell Lines
WT and A/A mouse embryonic fibroblast (MEF) cells, which express a WT version of eIF2α and eIF2α-S51A, were cultured in Dulbecco's modified Eagle's medium (DMEM) as previously described (15). GADD34ΔC/ΔC MEF cells were kindly provided by David Ron (University of Cambridge, UK) and were previously described (16). Stable Flp-In GADD34ΔC/ΔC cells lines were generated by using the Flp-In System (Invitrogen) and full-length GADD34 cDNAs including 1-kb of the GADD34 promoter and mutant versions of the GADD34 5′-leader that were integrated into the genome following the manufacturer's instructions. GADD34ΔC/ΔC FRT, GADD34-WT2, GADD34-OPT2, GADD34-AAA2, and GADD34-Δ2 MEFs were grown in DMEM supplemented with 10% (v/v) fetal bovine serum, 100 units/ml of penicillin, 100 μg/ml of streptomycin, 1× nonessential amino acids, and 55 μm β-mercaptoethanol.
Immunoblot Analyses
MEF cells were treated with 1 μm thapsigargin for up to 6 h, or left untreated. Protein lysates were collected and quantitated, and immunoblot analyses were carried out as previously described (17). Quantification of immunoblots was conducted using ImageJ software. Antibodies used for the immunoblot analyses include: GADD34 (Proteintech catalog number 10449-1-AP), CReP (Proteintech catalog number 14634-1-AP), eIF2α-P (Abcam catalog number ab32157), CHOP (Santa Cruz Biotechnology catalog number sc-7351), and β-actin (Sigma catalog number A5441). Monoclonal antibody measuring total eIF2α was kindly provided by Dr. Scott Kimball (Pennsylvania State University College of Medicine, Hershey, PA).
mRNA Measurement by qRT-PCR
RNA was isolated from MEF cells and polysome fractions using TRIzol reagent (Invitrogen) and single-strand cDNA synthesis was performed using TaqMan reverse transcriptase kit (Applied Biosystems) according to the manufacturer's instructions. Transcript levels were measured by qPCR using SYBR Green (Applied Biosystems) on a Realplex2 Master Cycler (Eppendorf). Primers used for measuring transcripts are listed in Table 1.
TABLE 1.
Description of primers used for qPCR in this study
| Primer name | Primer sequence |
|---|---|
| GADD34: forward primer | 5′-AGGACCCCGAGATTCCTCT-3′ |
| GADD34: reverse primer | 5′-CCTGGAATCAGGGGTAAGGT-3′ |
| CReP: forward primer | 5′-GGCTACAGTGGCCTTCTCTG-3′ |
| CReP: reverse primer | 5′-CATCCATCCCTTGCAAATTC-3′ |
| CHOP: forward primer | 5′-CGGAACCTGAGGAGAGAG-3′ |
| CHOP: reverse primer | 5′-CGTTTCCTGGGGATGAGATA-3′ |
| β-Actin: forward primer | 5′-TGTTACCAACTGGGACGACA-3′ |
| β-Actin: reverse primer | 5′-GGGGTGTTGAAGGTCTCAAA-3′ |
| Firefly luciferase: forward primer | 5′-CCAGGGATTTCAGTCGATGT-3′ |
| Firefly luciferase: reverse primer | 5′-AATCTCACGCAGGCAGTTCT-3′ |
Polysome Profiling and Sucrose Gradient Ultracentrifugation
MEF cells were left untreated or treated with 1 μm thapsigargin for 6 h. Cells were incubated in culture media containing 50 μg/ml of cycloheximide just prior to lysate collection. Lysates were collected, sheared, and layered on top of 10–50% sucrose gradients followed by ultracentrifugation as previously described (12, 18). Sucrose gradients were fractionated and whole cell lysate polysome profiles were collected using a Piston Gradient Fractionator (BioComp) and a 254-nm UV monitor with Data Quest Software.
Following fractionation, 10 ng/ml of firefly luciferase control RNA (Promega) was spiked into each pooled sample to generate polysome shifts for specific transcripts normalized to an exogenous RNA control (12, 18). Samples were mixed with 750 μl of TRIzol, and RNA isolation and cDNA generation was performed as described above. Calculations for % total gene transcript and % transcript shifts are as described previously (12). Whole cell lysate polysome profiles and mRNA polysome shifts are representative of three independent biological experiments.
Plasmid Constructions and Luciferase Assays
A 5′-rapid amplification of cDNA ends (5′-RACE; FirstChoice Ambion) was performed using RNA lysates collected from WT MEF cells treated with 1 μm thapsigargin for 6 h, or left untreated, to determine the transcriptional start sites for GADD34 and CReP. The cDNA segments encoding the 5′-leader of GADD34 and CReP were inserted between HindIII and NcoI between the TK-promoter and firefly luciferase CDS in a derivative of plasmid pGL3 (3). The resulting PTK-GADD34-Luc and PTK-CReP-Luc contain the mouse GADD34 and CReP 5′-leaders and the start codon for each CDS fused to a luciferase reporter. Site-directed mutagenesis and subcloning of synthesized cDNAs were used to generate mutant PTK-GADD34-Luc and PTK-CReP-Luc constructs (Table 2) that were sequenced to verify nucleotide substitutions. PTK-GADD34-Luc and PTK-CReP-Luc constructs were transiently co-transfected with a Renilla reporter plasmid into WT or A/A MEF cells for 24 h followed by a 6-h 0.1 μm thapsigargin treatment. Lysates were collected and firefly and Renilla luciferase activities were measured as described previously (3). At least three independent biological experiments were conducted for each luciferase measurement, and relative values are represented with S.D. indicated.
TABLE 2.
Description of GADD34 and CReP mutations used in this study
| Gene construct | Description of mutation |
|---|---|
| GADD34 stem loop insertion | Insertion of CTGCAGCCACCACGGCCCCCAAGCTTGGGCCG-TGGTGGCTGCAG 3′ of GCTCTGAGT |
| GADD34 uORF1 ΔATG | ATG to AGG |
| GADD34 uORF2 ΔATG | ATG to ATA |
| GADD34 uORF2 strong Kozak Context | GGCGACATGA to GCCACCATGA |
| GADD34 uORF2 extension | TGA to GGA |
| GADD34 uORF2 ΔAA 5–25 | Deletion of CTTCGCGAGCAGTCCGGACCCACGATCGCTTTTGGCAACCAGAACCGGCGCTTCAGCCCCCGGGG |
| GADD34 uORF2 ΔAA 15–25 | Deletion of TTTGGCAACCAGAACCGGCGCTTCAGCCCCCGGGG |
| GADD34 uORF2 frameshift | Deletion of A 3′ of ATGA and insertion of A 5′ of GGTGA |
| GADD34 uORF2 PPG to AAA | Mutation of CCCCCGGGGTGA to GCCGCGGCGTGA |
| GADD34 uORF2 insert AAA | Insertion of GCCGCCGCC 3′ of CCCCCGGGG |
| CReP stem loop insertion | Insertion of CTGCAGCCACCACGGCCCCCAAGCTTGGGCCGTGGTGGCTGCAG 3′ of GCGGCCATT |
| CReP uORF1 ΔATG | ATG to AGG |
| CReP uORF2 ΔATG | ATG to AGG |
| CReP uORF2 extension | TAG to GGA |
| CReP uORF2 replaced with GADD34 uORF2 | Replacement of ATGGACCTAACTGTGTAG with ATGAACCCGCCCCCGGGGTGA |
| CReP 21 nucleotides surrounding uORF2 start codon replaced with GADD34 21-nucleotide surrounding uORF2 start codon | Replacement of CTAGCCGGGATGGACCTAACC with GAGGGCGACATGAACCCGCTG |
| CReP 21 nucleotides surrounding uORF2 stop codon replaced with GADD34 21-nucleotide surrounding uORF2 stop codon | Replacement of GAGACTGTGTAGACCTCGGAT with CCCCCGGGGTGACGTGCAGCC |
| CReP 9 nucleotides 5′ of uORF2 stop codon replaced with GADD34 9 nucleotides 5′ of uORF2 stop codon (encoding PPG) | Replacement of GAGACTGTG with CCCCCGGGG |
| CReP 9 nt 3′ of uORF2 stop codon replaced with GADD34 9 nucleotides 3′ of uORF2 stop codon | Replacement of ACCTCGGAT with CGTGCAGCC |
The T7 promoter of sequence, TAATACGACTCACTATAGGGAGA, and the GADD34 5′-leader containing the start codon for the GADD34 CDS were inserted between HindIII and NcoI in the pGL3 basic luciferase vector (Promega) for generation of PT7-GADD34-Luc constructs for in vitro translation assays. Sequencing was used to verify nucleotide substitutions and in vitro assays were conducted as described below.
In Vitro Transcription and Translation Assays
Capped and polyadenylated RNA was synthesized with T7 RNA polymerase using mMESSAGE mMACHINE T7 Ultra (Ambion) from PT7-GADD34-Luc constructs. Synthesized GADD34-Luc mRNA was added to rabbit reticulocyte lysate (Promega) per the manufacturers' instructions. For luciferase assays, in vitro translation reactions with GADD34-Luc mRNA were carried out for 20 min at 30 °C, and firefly luciferase activity was measured.
For primer extension inhibition (Toeprint) assays, reticulocyte lysates were treated with cycloheximide upon addition of the GADD34-Luc mRNA to measure initiating ribosomes (time 0) or 5 min after addition of the transcript to measure ribosomal localization during steady-state translation (time 5). Toeprint assays were conducted as previously described and using primers: 5′-TGAAGCGCCGGTTCTGGTTG-3′ (Fig. 4D) and ZW4, 5′-TCCAGGAACCAGGGCGTA-3′ (Fig. 4E) (19).
FIGURE 4.
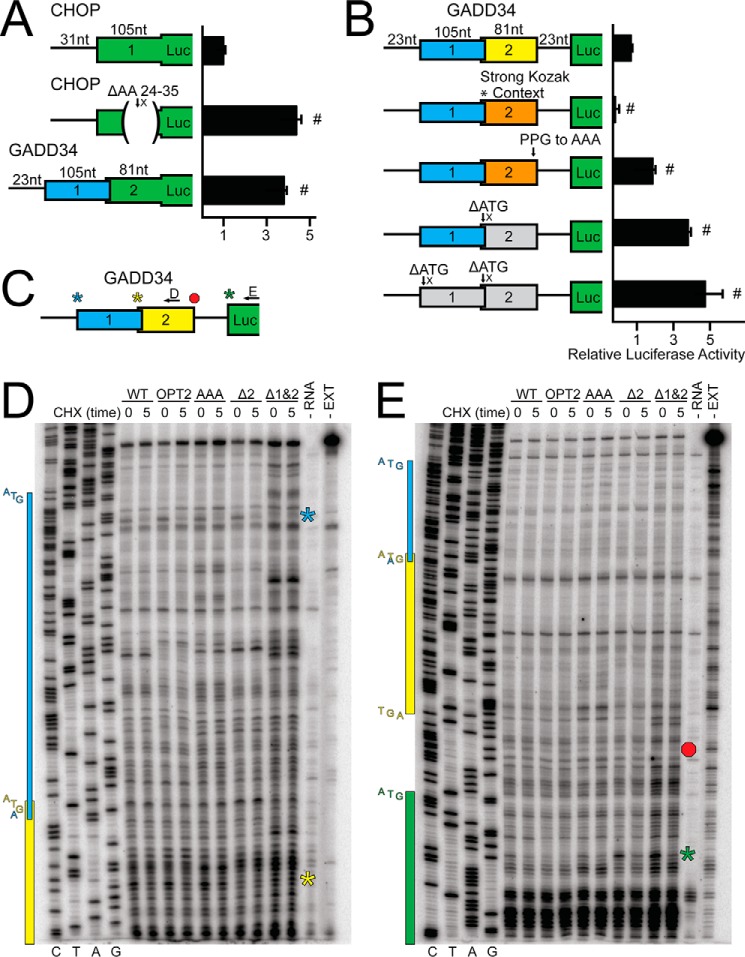
GADD34 translation control involves bypass of an inhibitory uORF. A, in-frame fusions between firefly luciferase reporter with the WT CHOP uORF, CHOP uORF deleted for codons 24–35, or GADD34 uORF2 and firefly luciferase, were transfected into WT MEF cells, treated for 6 h, or left untreated, and measured using a dual luciferase assay. Relative values are represented as histograms for each with the S.D. indicated. B, WT and mutant versions of GADD34-Luc mRNAs were added to in vitro translation reactions and expression was measured using a luciferase assay. Mutant versions of PT7-GADD34-Luc included uORF2 start codon context mutated to the Kozak consensus sequence (*, strong Kozak context), substitution of the codons for Pro-Pro-Gly to codons for Ala-Ala-Ala (PPG to AAA), and deletion of uORF2 or uORF1&2 (ΔATG). Data are representative of three independent biological experiments. C, depiction of the toeprint design using the WT version of GADD34-Luc mRNA. Black arrows represent the location of primers used in panels D and E. Toeprints corresponding to initiation at uORF1, uORF2, or the luciferase CDS are indicated by blue, yellow, or green stars, respectively. Termination at uORF2 is indicated by a red octagon. D and E, reticulocyte lysates were treated with cycloheximide (CHX) upon addition of WT or mutant versions of GADD34-Luc mRNA to measure initiating ribosomes (time 0), or 5 min after addition of the transcript to measure ribosome localization during steady state translation (time 5). Toeprint assays were conducted for each sample and sequencing reactions can be read 5′ to 3′ from top to bottom. The nucleotide residue complementary to the dideoxynucleotide added to each sequencing reaction is listed on the left, below the first four lanes. The products from control primer extension assays in the absence of RNA (−RNA) or absence of rabbit reticulocyte lysate translation mixture (−EXT) are indicated on the right. The green star represents the toeprint corresponding to initiation at the luciferase coding region and the yellow star and red octagon represent the toeprint corresponding to initiation and termination of the GADD34 uORF2. Initiation at uORF1 is indicated by a blue star. The blue, yellow, and green colored boxes on the left span the sequences to corresponding to uORF1, uORF2, and the luciferase CDS, respectively, and are comparable with the GADD34 5′-leader schematic in panel C. The start and stop codons for each ORF are represented to the left of their corresponding colored box. Mutant constructs utilized are the same as listed in panel B and data are representative of three independent biological experiments.
Cell Number and Viability Assays
For cell proliferation assays, GADD34-WT2, GADD34-OPT2, GADD34-AAA2, and GADD34-Δ2 MEFs were seeded at 5,000 cells/well in a 96-well plate. Cells were fixed (3.7% formalin) and stained (10 μg/ml Hoechst) immediately following seeding, or 24 and 48 h after seeding, and fluorescence was measured on a Synergy H1 Microplate reader (BioTek).
MTT assays were carried out by seeding cells at 5,000 cells/well in a 96-well plate. Cells were cultured for 24 h, and MTT activity was measured using CellTiter 96-well Non-radioactive Cell Proliferation Assay (Promega). For measurements of MTT activity after ER stress treatment, cells were seeded, allowed to grow for 24 h, and treated with 0.4 μm thapsigargin with or without 1 μm guanabenz, or left untreated for an additional 24 h.
Statistical Analyses
Values represent the mean ± S.D. and were derived from at least three independent experiments. Statistical significance was calculated using the two-tailed Student's t test. Differences between multiple groups were analyzed using a one-way analysis of variance followed by a post hoc Tukey HSD test. p values less than 0.05 were considered statistically significant and are indicated by “*”, and treatment groups considered statistically significant from WT control are indicated by a “#” sign.
Results
eIF2α-P Is Required for GADD34 Transcription and Translation, but CReP Expression Occurs Independent of eIF2α-P
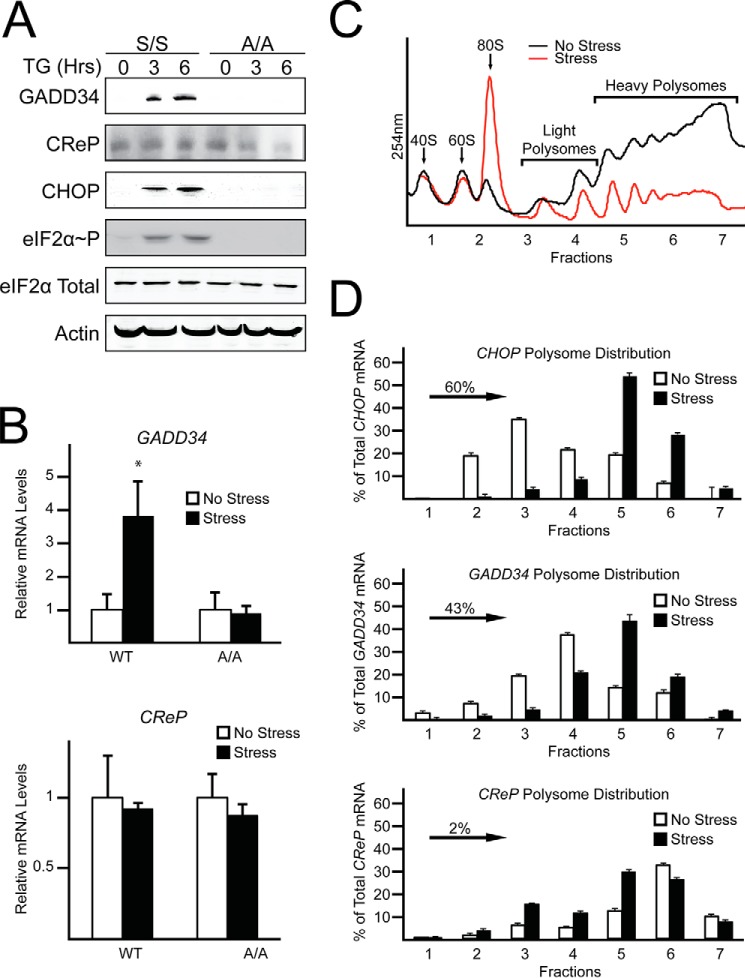
GADD34 expression is enhanced during eIF2α-P, whereas CReP levels are suggested to be independent of eIF2α-P (5, 7). To further explore the role that eIF2α-P and translational control play in the differential expression of GADD34 and CReP, we measured changes in their mRNA and protein levels in wild-type (WT) MEF cells and mutant MEF cells (A/A) expressing eIF2α-S51A that cannot be phosphorylated. eIF2α-P was induced only in WT cells by treatment with thapsigargin, a potent trigger of endoplasmic reticulum (ER) stress (Fig. 1A). Both GADD34 mRNA and protein levels were increased in WT MEF cells, whereas there was no change in GADD34 mRNA and minimal protein expression in A/A cells (Fig. 1, A and B). By contrast, there was no change in the amount of CReP mRNA and protein in WT cells upon ER stress. Of interest, whereas the levels of CReP mRNAs were similar between WT and A/A cells, there was reduced CReP protein in A/A cells during ER stress (Fig. 1, A and B).
FIGURE 1.
eIF2α-P is required for induced GADD34 translation, but CReP expression occurs independent of eIF2α-P. A, WT and A/A MEF cells were treated with thapsigargin, for up to 6 h or left untreated. Lysates were processed and levels of GADD34, CReP, CHOP, eIF2α-P, eIF2α total, and β-actin were measured by immunoblot. B, total RNA was collected from WT and A/A MEFs treated with thapsigargin for 6 h or left untreated and relative levels of GADD34 and CReP mRNA were measured by qRT-PCR. C, WT MEF cells were treated with thapsigargin for 6 h or left untreated. Lysates were collected and layered on top of 10–50% sucrose gradients, followed by ultracentrifugation and analysis of whole lysate polysome profiles at 254 nm. D, total RNA was isolated from sucrose fractions and the percentage of total CHOP, GADD34, and CReP mRNAs was determined by qRT-PCR. Panels C and D are representative of three independent biological experiments.
To explore the role of translational control in the differential expression of GADD34 and CReP, WT MEF cells were subjected to thapsigargin treatment, and lysates were prepared and analyzed by polysome profiling using sucrose density ultracentrifugation. As expected, polysome profiling revealed that ER stress led to reduced global translation initiation as viewed by a decrease in polysomes coincident with increased monosomes (Fig. 1C). GADD34 and CReP mRNAs, along with CHOP mRNA, which is known to be subject to preferential translation, were then measured by qRT-PCR in the polysome fractions. Both GADD34 and CHOP transcripts were predominantly associated with monosomes and disomes in the absence of stress. However, upon thapsigargin treatment and eIF2α-P, there was a significant shift of the GADD34 and CHOP transcripts to heavy polysomes (Fig. 1D). By contrast, CReP mRNA was associated with heavy polysomes in both thapsigargin-treated cells and those not subjected to stress. Together these results suggest that in addition to transcriptional induction, GADD34 is preferentially translated upon stress and eIF2α-P, whereas CReP is largely translated independent of the stress conditions. These results are consistent with earlier reports that indicated that expression of GADD34 is induced upon stress as part of a feedback control of the ISR and CReP is constitutively present (5, 7).
Preferential Translation of GADD34 Features an Inhibitory uORF
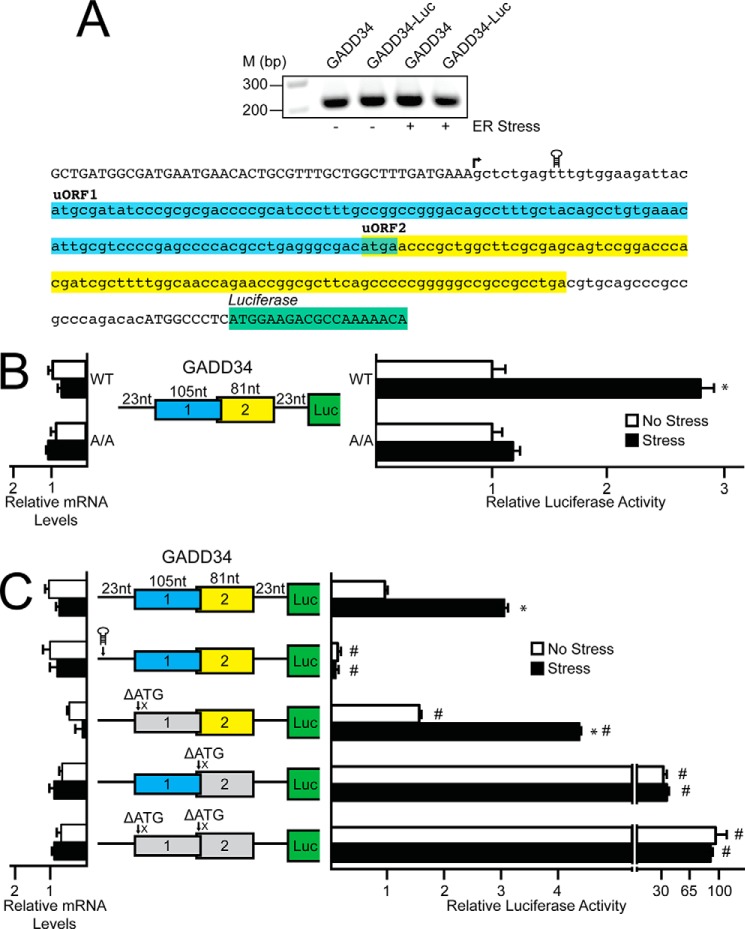
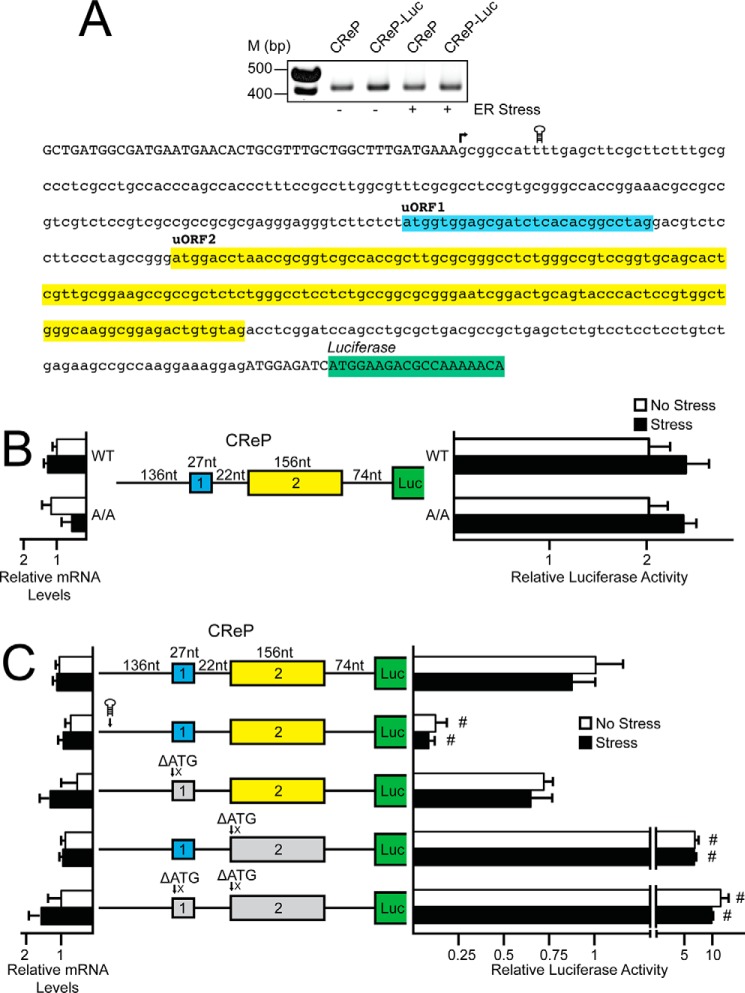
Next we carried out a 5′-RACE to define the transcriptional start site in mouse of the GADD34 gene (Fig. 2A). A cDNA segment encoding the 228-nucleotide sequence of the 5′-leader of GADD34 was then inserted between a minimal TK promoter and the firefly luciferase reporter CDS, generating PTK-GADD34-Luc. This luciferase reporter featured the initiation codon of the GADD34 CDS fused in-frame to the luciferase CDS. Expression of GADD34-Luc was increased 3-fold in WT MEF cells treated with thapsigargin as compared with no change in luciferase activity in A/A cells (Fig. 2B). In these reporter measurements, and subsequent ones discussed below, there was no significant change in the luciferase mRNA. These results indicate that the 5′-leader of GADD34 directs preferential translation in response to eIF2α-P.
FIGURE 2.
Preferential translation of GADD34 features an inhibitory uORF. A, top panel, 5′-RACE was carried out for GADD34 using WT MEFs treated with thapsigargin for 6 h or left untreated; total RNA was prepared and DNA products were separated by gel electrophoresis, with markers of the indicated base pair sizes illustrated on the left. A, bottom panel, Representation of GADD34 5′-leader in lowercase letters, with uppercase letters representing the 5′-linker added during the 5′-RACE procedure and the beginning of the CDS of the GADD34-Luc fusion. Colored boxes represent the GADD34 uORFs and the coding region of the GADD34-Luc fusion. The transcription start site is indicated with an arrow, and location of the stem loop insertion is illustrated. B, the PTK-GADD34-Luc construct and a Renilla luciferase reporter were co-transfected into WT or A/A MEF cells and treated for 6 h with thapsigargin or left untreated. GADD34 translation control was measured via dual luciferase assay and corresponding GADD34-Luc mRNA was measured by qRT-PCR. The PTK-GADD34-Luc construct contains the cDNA sequence corresponding to the GADD34 5-leader fused to the luciferase reporter gene with both GADD34 uORFs and the CDS of the GADD34-Luc fusion indicated with colored boxes. C, WT and mutant versions of PTK-GADD34-Luc were transfected into WT MEFs, treated for 6 h or left untreated, and measured using a dual luciferase assay and qRT-PCR. Mutant versions of PTK-GADD34-Luc include a stem loop insertion and mutation of the initiation codons for uORFs individually or together, as represented by ΔATG. Relative values are represented as histograms for each with the S.D. indicated.
To determine whether enhanced GADD34 translation occurs via ribosome scanning, a palindromic sequence with a predicted free energy of ΔG = −41 kcal/mol was inserted 10 nucleotides downstream of the 5′ cap of the GADD34-Luc mRNA (Fig. 2A). Addition of this stem-loop to the GADD34-Luc transcript significantly decreased luciferase activity independent of stress, indicating that preferential translation mediated by the 5′-leader of GADD34 occurs by ribosome scanning (Fig. 2C). Ribosomes scanning the 5′-leader of GADD34 encounter two uORFs before reaching the start codon for the GADD34 CDS. To determine the contribution of the two uORFs to GADD34 translation regulation, the uORF start codons were mutated from ATG to an AGG or ATA, as indicated by the ΔATG in Fig. 2C. Deletion of uORF1 alone led to a small increase, albeit significant, in the basal luciferase expression, with an induction upon ER stress that was similar to the reporter with the WT version of the GADD34 5′-leader. This result suggests that uORF1 serves to modestly dampen downstream translation regardless of stress. By comparison, deletion of uORF2 led to a 30-fold increase in luciferase activity independent of ER stress, indicating that uORF2 is inhibitory to downstream translation and is the dominant regulatory uORF in the GADD34 5′-leader, a finding consistent with Lee et al. (13). Combined deletion of uORF1 and uORF2 led to an additional increase in luciferase activity, further supporting the roles of uORF1 and uORF2 as repressing elements in GADD34 translational expression (Fig. 2C).
GADD34 Translation Control Involves Bypass of an Inhibitory uORF
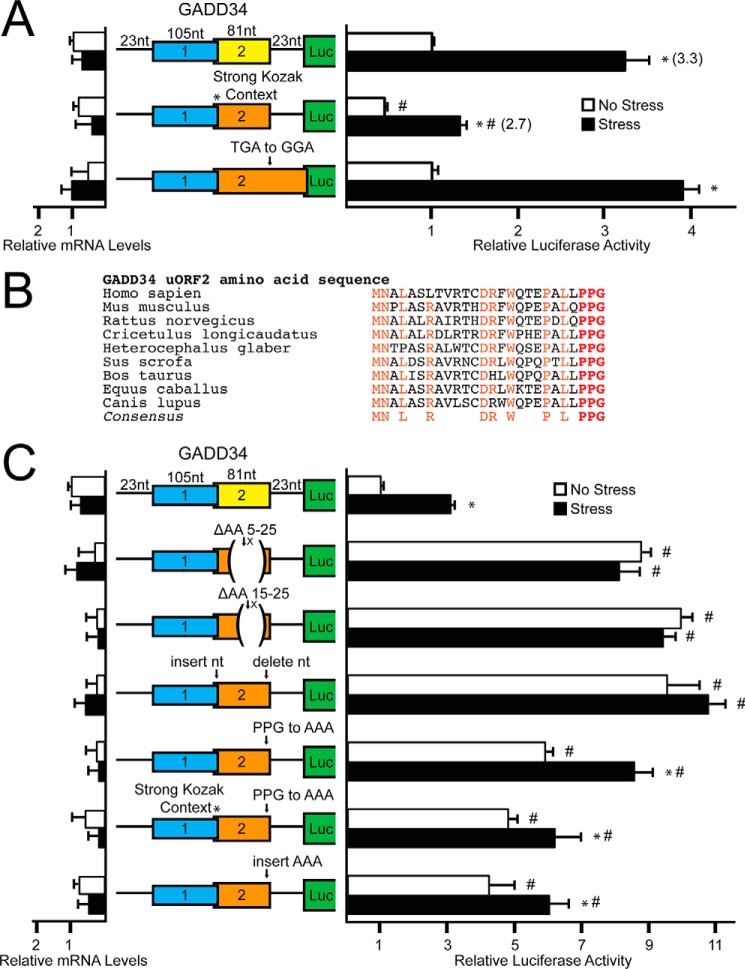
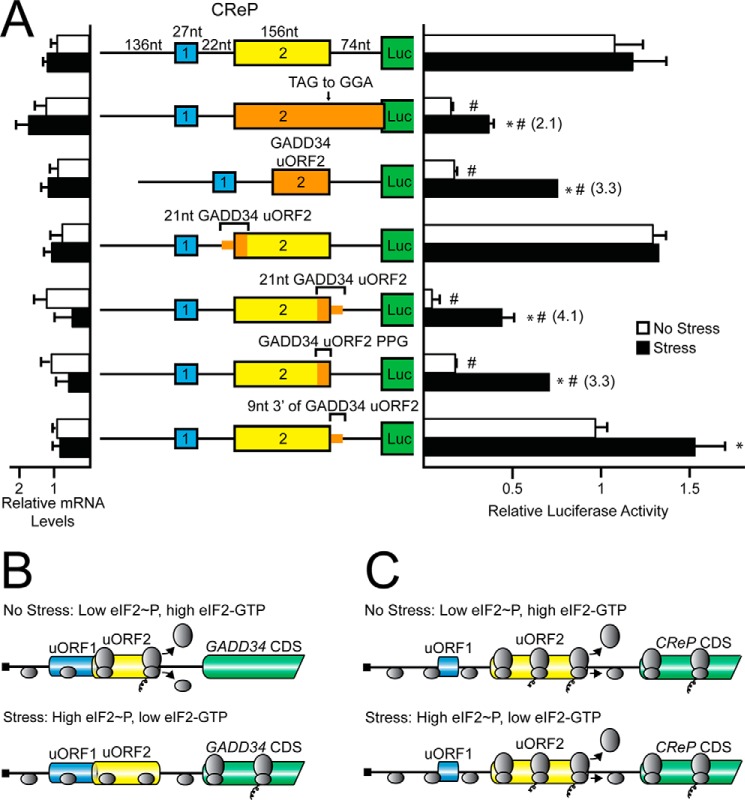
The initiation codon context for the GADD34 uORF2 (GGCGACAUGU) is less than optimal compared with the Kozak consensus sequence (GCC(A/G)CCAUGG), a feature similar to the single uORF present in the CHOP mRNA that is subject to the bypass model of translational control (4). To determine whether context of the start codon plays a role in uORF2-mediated regulation of GADD34 translation, the poor start codon context of uORF2 was mutated to the optimal Kozak consensus. Mutation of uORF2 to the strong Kozak context reduced luciferase expression basally and decreased stress-induced luciferase activity to 2.7-fold as compared with a 3.3-fold induction for the WT GADD34-Luc reporter (Fig. 3A). This finding suggests that uORF2 can be bypassed during eIF2α-P in part due to its poor initiation codon context, thereby enhancing translation of the downstream GADD34 CDS.
FIGURE 3.
GADD34 translation control involves bypass of an inhibitory uORF that relies on a Pro-Pro-Gly juxtaposed to the uORF2 stop codon. A, WT and mutant versions of PTK-GADD34-Luc were transfected into WT MEFs, treated for 6 h or left untreated, and measured using a dual luciferase assay and qRT-PCR. Mutant versions of PTK-GADD34-Luc include mutation of the GADD34 uORF2 poor start codon context to “*, strong Kozak context,” and mutation of the stop codon of uORF2 to generate and overlapping out-of-frame uORF (TGA to GGA). Relative values are represented as histograms for each with the S.D. indicated. B, polypeptide sequence encoded by GADD34 uORF2 from different vertebrates. The uORF2 polypeptide sequences were from cDNAs derived from GADD34 orthologs, including human (GenBankTM accession number NM_014330), mouse (NM_008654), rat (NM_133546), hamster (L28147), naked mole rat (XM_004889808), pig (XM_003127275), cow (NM_001046178), horse (XM_001489532), and dog (XM_533626). Residues conserved between the uORF sequences are listed in the consensus and are highlighted. C, WT and mutant versions of PTK-GADD34-Luc were transfected into WT MEFs, treated for 6 h or left untreated, and measured using a dual luciferase assay and qRT-PCR. Mutant versions of PTK-GADD34-Luc include deletion of codons 5–25 or 15–25 (ΔAA 5–25 or ΔAA 15–25) and a frameshift construct in which a nucleotide was inserted just following the uORF2 ATG start codon and deleted just prior to the uORF2 stop codon. Additional constructs included mutation of the codons encoding Pro-Pro-Gly to codons for Ala-Ala-Ala (PPG to AAA), simultaneous mutation of the uORF2 start codon to Kozak consensus sequence with the Pro-Pro-Gly to Ala-Ala-Ala mutation (* Strong Kozak Context, PPG to AAA), and insertion of codons encoding Ala-Ala-Ala just prior to the uORF2 stop codon (insert AAA). Relative values are represented as histograms for each with the S.D. indicated.
Translation initiation downstream of uORFs can also be dependent on translation reinitiation (2, 11). To determine the contribution of post-uORF2 translation reinitiation in GADD34 translation regulation, the stop codon of uORF2 was mutated from TGA to GGA, resulting in an uORF that overlaps out-of-frame with the luciferase CDS (Fig. 3A). There was no statistically significant difference between the WT PTK-GADD34-Luc and the reporter with the uORF2 overlapping the CDS. This finding argues against significant ribosome reinitiation at the GADD34 CDS following synthesis of the uORF2 polypeptide, and instead supports the idea that preferential translation of GADD34 CDS relies on ribosomal bypass of the inhibitory uORF2.
Inhibitory Function of GADD34 uORF2 Is Reliant on Pro-Pro-Gly Juxtaposed to the uORF2 Stop Codon
Many features of uORFs, including length and coding sequences, can promote the repressing functions of uORFs. To investigate the inhibitory nature of the GADD34 uORF2, in-frame deletions from codons 5–25 and 15–25 were analyzed in the GADD34-Luc reporter. Both deletions in the uORF2 coding sequence increased luciferase expression independent of stress, suggesting that the repressing function of uORF2 lies at least in part within its coding sequence (Fig. 3C).
To address whether the RNA sequence in uORF2 per se contributes to the repressing functions of this uORF, a single nucleotide was deleted just after the ATG start codon in uORF2 and a single nucleotide was inserted just prior to the TGA termination codon. The resulting frameshift thus maintains the uORF2 nucleotide sequence and length, but the uORF now encodes a different polypeptide. Luciferase activity of this frameshift reporter was increased in the presence or absence of stress, consistent with the hypothesis that the encoded uORF2 polypeptide sequence is responsible for the inhibitory function of uORF2 in translational control (Fig. 3C).
A comparison of the uORF2-encoding polypeptide sequences among mammals revealed several conserved segments, including a carboxyl-terminal Pro-Pro-Gly sequence (Fig. 3B). Contiguous prolines and Pro-Pro-Gly sequences have been suggested to be problematic for translation elongation and require eIF5A for efficient protein synthesis (20). Substitution of the uORF2 codons encoding the Pro-Pro-Gly sequence with codons encoding Ala-Ala-Ala resulted in a 5-fold increase in luciferase activity basally, while retaining a modest induction during ER stress (Fig. 3C). Alteration of the uORF2 start codon to an optimal Kozak sequence in the presence of the Pro-Pro-Gly to Ala-Ala-Ala substitution also led to elevated luciferase activity in the absence of stress, along with a modest increase (1.3-fold) upon thapsigargin treatment. These results suggest that bypass of the uORF2 during stress is required for maximal induction of GADD34 translation, because translation of the codons encoding the uORF2 Pro-Pro-Gly residues precludes the ribosome from efficiently reinitiating at the downstream CDS. Insertion of three alanine residues between the Pro-Pro-Gly sequence and the uORF2 termination codon also led to similar increases in luciferase activity in the presence or absence of stress (Fig. 3C), indicating that the ability of the Pro-Pro-Gly sequence in uORF2 to inhibit translation reinitiation involves its juxtaposition to a termination codon.
Translation of the CHOP inhibitory uORF is suggested to trigger an elongation pause that is responsible for lowering downstream translation reinitiation (4). This can be visualized by low expression of an in-frame fusion of the CHOP uORF with the luciferase CDS. Luciferase activity of the CHOP uORF-Luc fusion protein is significantly enhanced upon deletion of its critical inhibitory sequences and alleviation of the elongation pause (Fig. 4A). We therefore asked if the Pro-Pro-Gly sequence in GADD34 uORF2 results in an elongation pause by making a similar in-frame fusion between GADD34 uORF2 and the luciferase CDS. There was elevated expression of the GADD34 uORF-Luc fusion, suggesting that the GADD34 Pro-Pro-Gly sequence does not facilitate a pause in ribosomal elongation (Fig. 4A).
We also analyzed the effects of selected GADD34 uORF2 mutations for translational expression by using T7 RNA polymerase to synthesize GADD34-Luc mRNAs that were introduced into in vitro translation assays using rabbit reticulocyte lysates. Consistent with our analysis of MEF cells, mutations of the initiation codon of uORF2 led to elevated luciferase activity, which was further enhanced by combined loss of uORF1 (Fig. 4B). Substitution of the Pro-Pro-Gly codons to Ala-Ala-Ala in uORF2 also led to significantly enhanced luciferase expression compared with WT. Finally, introduction of an optimized Kozak context for the initiation codon of uORF2 sharply lowered luciferase activity (Fig. 4B). Together these results support the idea that the Pro-Pro-Gly codons are important for the uORF2 inhibition of downstream CDS translation.
As the GADD34 uORF2 fusion construct from Fig. 4A does not take into account the dependence of the Pro-Pro-Gly sequence on juxtaposition to a termination codon for its inhibitory nature, toeprinting experiments using two different primers were used to map the positions of ribosomes along the 5′-leader of GADD34 transcripts during in vitro translation (Fig. 4C). Reticulocyte lysates were treated with cycloheximide upon addition of the GADD34-Luc mRNA to measure where ribosomes first initiate translation (time 0). Alternatively, cycloheximide was added 5 min after addition of the transcript to measure ribosome positions during steady-state translation and active polypeptide synthesis (time 5). At time 0 there were toeprints measuring the ribosomes positioned at the initiation codons of uORF1 (blue star) and uORF2 (yellow star) and the luciferase CDS (green star) (Fig. 4, D and E). Mutation of the initiation codon of uORF2 individually or in combination with uORF1 resulted in lowered toeprint signals at the uORF2, with a similar corresponding increase in initiation at the luciferase CDS (Fig. 4, D and E). Initiation at the luciferase CDS is also observed at time 5 for these mutant transcripts, indicating that translation initiation at the dominant uORF2 precludes ribosome reinitiation at the downstream CDS during steady state translation (Fig. 4E).
Introduction of optimized Kozak context for the initiation codon of uORF2 significantly reduced translation initiation at the CDS at both time 0 and time 5, indicating increased ribosomal preference for the more 5′ optimized start codon of uORF2 largely precludes translation at the downstream CDS (Fig. 4E). A modest toeprint at the uORF2 termination codon is also observed (red hexagon) for both the WT and optimized uORF2 mRNAs at time 5. Substitution of the Pro-Pro-Gly codons to Ala-Ala-Ala resulted in a 32% reduction in the toeprint signal at the uORF2 termination codon as compared with the WT mRNA. Collectively these results indicate that there is not an extended ribosome pause at uORF2, but the Pro-Pro-Gly sequence can promote inefficient ribosome termination (Fig. 4E).
CReP Translation Is Dampened by an Inhibitory uORF in an eIF2α-P Independent Manner
CReP has two uORFs with similar spatial arrangements to the 5′-leader of GADD34 mRNA, yet CReP expression appears to be unchanged in WT cells upon eIF2α-P and stress. We carried out a 5′-RACE to define the transcriptional start site for mouse CReP, and determined that the 5′-leader of the CReP mRNA is 421 nucleotides in length (Fig. 5A). The cDNA encoding the CReP 5′-leader was inserted between the TK promoter and luciferase reporter gene, generating PTK-CReP-Luc. Transfection of PTK-CReP-Luc into WT and A/A MEF cells resulted in similar levels of luciferase activity independent of stress treatment and eIF2α-P (Fig. 5B). Levels of CReP-Luc mRNA in these assays, as well as those described below, were similar, indicating that luciferase activity is a measure of translational expression. These results further support the idea that CReP mRNA is efficiently translated independent of eIF2α-P.
FIGURE 5.
CReP translation is dampened by an inhibitory uORF in an eIF2α-P independent manner. A, top panel, a 5′-RACE was carried out for CReP using WT MEF cells either transfected with PTK-CReP-Luc or left untransfected. MEFs were treated with thapsigargin for 6 h or left untreated and total RNA was prepared. DNA products were separated by gel electrophoresis and a DNA ladder with markers of the indicated base pair sizes is illustrated on the left. A, bottom panel, the sequence of the CReP 5′-leader is represented in lowercase letters with uppercase letters representing the 5′-linker added during the 5′-RACE procedure and the coding region of the CReP-Luc fusion. Colored boxes represent the two CReP uORFs and the coding region of the CReP-Luc fusion. The transcription start site is indicated with an arrow and the location of stem loop insertion is illustrated. B and C, WT and mutant versions of PTK-CReP-Luc and a Renilla luciferase reporter were co-transfected into WT or A/A MEF cells, as indicated, and treated for 6 h with thapsigargin or left untreated. CReP translation control was measured by dual luciferase assay and corresponding CReP-Luc mRNA was measured by qRT-PCR. The PTK-CReP-Luc construct contains the cDNA sequence corresponding to the CReP 5′-leader fused to the luciferase reporter gene with both CReP uORFs and the CDS of the CReP-Luc fusion indicated with colored boxes. Relative values are represented as histograms for each with the S.D. indicated. Mutant versions of PTK-CReP-Luc include a stem loop insertion and deletion of both uORFs individually or together, as represented by ΔATG.
To determine whether CReP translation occurs by ribosome scanning, a stem-loop was inserted 10 nucleotides downstream of the 5′-end of the CReP mRNA (Fig. 5C). Insertion of the stem-loop structure sharply reduced luciferase activity in the presence or absence of ER stress, consistent with the requirement for ribosome scanning for CReP translation. As ribosomes scanning the 5′-leader of CReP would encounter two uORFs, the uORF start codons were changed from ATG to AGG individually or in combination, as indicated by ΔATG in Fig. 5C. Deletion of uORF1 led to a modest reduction in luciferase activity in the presence or absence of stress (Fig. 5C). By contrast, deletion of uORF2 led to a 7-fold increase in luciferase activity independent of stress, suggesting that uORF2 is the dominant repressing uORF in CReP translation, a feature shared with GADD34. Deletion of both uORF1 and uORF2 led to a further increase in luciferase activity.
To address the ability of ribosomes to reinitiate downstream after translation of uORF2, the CReP uORF2 stop codon was mutated from a TGA to a GGA, generating an extended uORF that overlaps out-of-frame with the CReP CDS. The overlapping uORF2 resulted in a 5-fold reduction in basal luciferase activity, which was increased 2-fold upon ER stress (Fig. 6A). These results suggest that although a small portion of the scanning ribosomes can bypass CReP uORF2 and initiate downstream translation, the majority of ribosomes that initiate at the downstream CReP CDS are reinitiating ribosomes that have previously translated uORF2.
FIGURE 6.
Regulatory properties of GADD34 uORF2 are transferable to a heterologous 5′-leader derived from CReP. A, WT and mutant versions of PTK-CReP-Luc were transfected into WT MEFs, treated for 6 h or left untreated, and measured using a dual luciferase assay and qRT-PCR. Mutant versions of PTK-CReP-Luc include mutation of the uORF2 stop codon from TAG to GGA to generate an overlapping out-of-frame uORF (TAG to GGA), insertion of GADD34 uORF2 in place of CReP uORF2 (GADD34 uORF2), insertion of the 21 nucleotides surrounding GADD34 uORF2 start codon in place of the corresponding CReP uORF2 sequence (21nt GADD34 uORF2), insertion of the 21 nucleotides surrounding GADD34 uORF2 stop codon in place of the corresponding CReP uORF2 sequence (21nt GADD34 uORF2), insertion of the codons encoding GADD34 uORF2 Pro-Pro-Gly sequence in place of the corresponding CReP uORF2 sequence, and insertion of the 9 nucleotides 3′ of GADD34 uORF2 in place of the corresponding CReP sequence. Relative values are represented as histograms for each with the S.D. indicated. B, model for GADD34 translational control. In the absence of stress, low eIF2α-P, and high eIF2-GTP, ribosomes scan the 5′-leader of the GADD34 mRNA and initiate translation at GADD34 uORF2. After translation of uORF2, terminating ribosomes are precluded from translation reinitiation downstream and are suggested to dissociate from the mRNA. In the presence of stress, eIF2α-P and low eIF2-GTP levels allow for some scanning ribosomes to bypass the GADD34 uORF2 in part due to poor start codon context, and instead initiate translation at the GADD34 CDS. C, model for CReP translational control. In the presence or absence of stress, ribosomes scan the 5′-leader of the CReP mRNA and initiate translation at the CReP uORF2. After translation of uORF2, a portion of the terminating ribosomes resume scanning and initiate translation downstream at the CReP CDS. It is noted that during stress and high eIF2α-P, a small portion of ribosomes can bypass the uORF2 and initiate translation at CReP CDS. Together these processes are suggested to lead to CReP translation independent of eIF2α-P.
Regulatory Properties of GADD34 uORF2 Are Transferable to a Heterologous 5′-Leader Derived from CreP
We next determined if the regulatory properties of GADD34 uORF2 could be transferred to a heterologous 5′-leader derived from CReP. We began by replacing the CReP uORF2 with the coding sequence of GADD34 uORF2 (Fig. 6A). The proximity of the GADD34 uORF2 in the context of the CReP uORF1 and CDS was also the same as that of WT CReP. Introduction of the GADD34 uORF2 into the CReP-Luc reporter led to a significant reduction in luciferase activity in the absence of stress, which was induced 3.3-fold upon ER stress (Fig. 6A). This result indicates that uORF2 of GADD34 is a transferable element that can direct preferential translation of a heterologous mRNA.
To define critical portions of the GADD34 uORF2 that confer translational control, we introduced smaller portions of the GADD34 uORF2 into the CReP-Luc reporter (Fig. 6A). Exchange of 21 nucleotides centered on the initiation codon of the GADD34 uORF2 for the corresponding sequences in CReP resulted in no significant differences from the WT CReP-Luc reporter (Fig. 6A). We interpret this finding to suggest that the enhanced ability of CReP uORF2 to allow for reinitiation at the downstream CDS diminishes in part the translational control that can be imparted by bypass of the substituted GADD34 start codon.
Exchange of a 21-nucleotide sequence that includes the Pro-Pro-Gly and GADD34 uORF2 stop codon for the corresponding sequences in CReP-Luc resulted in a large decrease in luciferase activity in non-stressed conditions that was enhanced over 4-fold upon ER stress (Fig. 6A). This result indicates that the 3′-portion of the GADD34 uORF2 is sufficient to confer significant preferential translation to CReP upon stress. To delineate further the contribution of this 21-nucleotide sequence for preferential translation, we exchanged only the 9 nucleotides encoding the Pro-Pro-Gly residues from GADD34 uORF2 for the corresponding sequences in the CReP uORF2 in the CReP-Luc reporter (Fig. 6A). Introduction of the nucleotides encoding the Pro-Pro-Gly sequence led to a decrease in basal luciferase activity that was stress-inducible, similar to that observed for exchange of the entire GADD34 uORF2 in CReP. This result suggests that the GADD34 uORF2 Pro-Pro-Gly sequence can serve to block translation reinitiation in the heterologous CReP 5′-leader. The exchange of the 9 nucleotides following the termination codon of the GADD34 uORF2 for the corresponding region of the CReP 5′-leader resulted in luciferase activities similar to WT CReP-Luc, although there was some induction upon ER stress. These findings suggest that the Pro-Pro-Gly sequence encoded in GADD34 uORF2 is the dominant regulator of downstream translation reinitiation and is central to preferential translation, with the 9 nucleotides following the uORF2 of GADD34 playing a modest role in this regulation. Additionally, the proximity of the uORF to the CDS of the transcript does not appear to be a key feature of the regulation imparted by the GADD34 Pro-Pro-Gly sequence due to its ability to regulate expression predictably in the CReP 5′-leader, even though CReP uORF2 is nearly 3 times further from the CDS as is found for GADD34 uORF2. Fig. 6, B and C, illustrate models for GADD34 and CReP, highlighting the differential abilities of the uORF2 from each to allow for translation reinitiation. The translation models for GADD34 and CReP and their broader implications will be further highlighted under “Discussion.”
Alterations in the Regulatory Features of GADD34 uORF2 Affect Cell Viability during ER Stress
GADD34 is central for determining the appropriate levels of eIF2α-P in the ISR during transitions from basal to stress conditions, and vice versa. In turn, the amounts of eIF2α-P can dictate the levels of global and gene-specific translation that determine protein homeostasis and the health of the cell. To determine the role that GADD34 translational control by the uORF2 has on eIF2α-P and cellular adaptation to ER stress, MEF cells were engineered such that they stably expressed GADD34 with WT or selected mutant versions of uORF2. Initially, a Flp recombination target (FRT) site was integrated in a single location in the genome of GADD34 functional knock-out MEF cells (GADD34ΔC/ΔC). Integration of the FRT site was then followed by the insertion and clonal isolation of cells expressing full-length GADD34 cDNAs under the control of 1 kb of the GADD34 promoter, which ensures its proper transcriptional induction in response to ER stress (21).
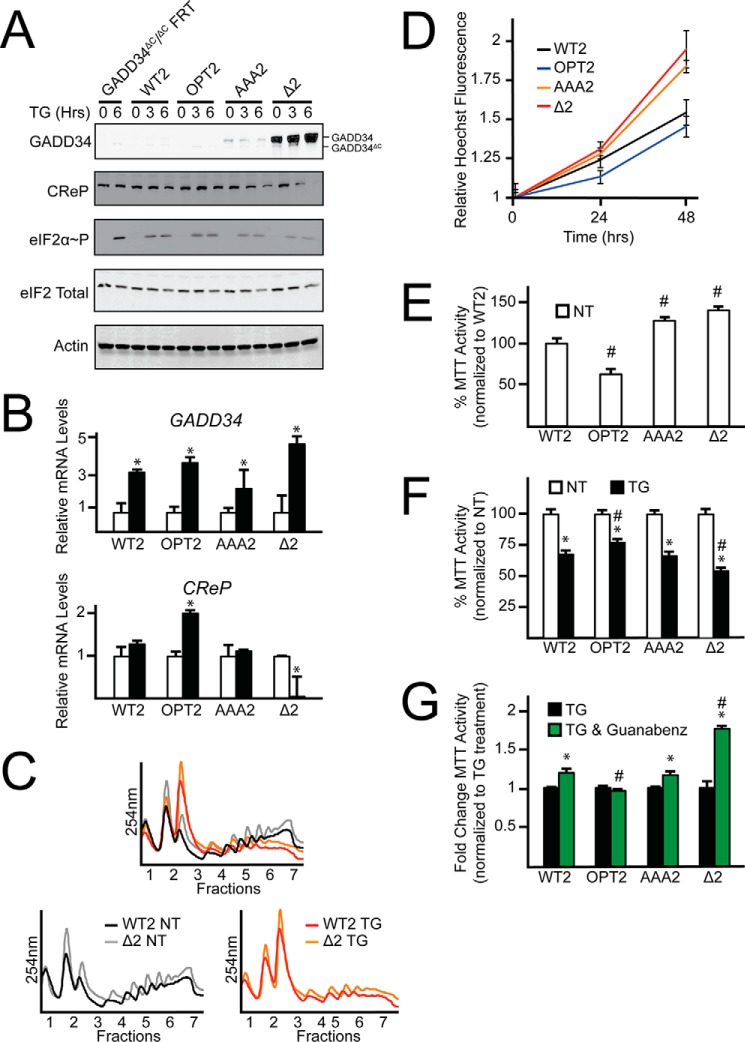
Four different versions of the GADD34 expressing cells were generated using the FRT strategy, including MEF cells expressing GADD34 with a WT uORF2 (WT2), an uORF2 with a mutant initiation codon (Δ2), an uORF2 with an initiation codon with optimal Kozak consensus (OPT2), or an uORF2 with Pro-Pro-Gly substituted to Ala-Ala-Ala (AAA2). This isogenic collection of GADD34-expressing cells was then cultured in the presence or absence of thapsigargin. Measurements of GADD34 protein revealed the predicted pattern of expression based on our analyses of endogenous GADD34 and GADD34-Luc reporters (Fig. 7A). For each, there was a significant increase in GADD34 mRNA upon ER stress, indicating that transcription induction was retained for each version of GADD34 (Fig. 7B). Cells expressing GADD34-Δ2 displayed sharply elevated GADD34 protein in the absence of stress, which was induced 57-fold upon thapsigargin treatment as compared with GADD34-WT2 (Fig. 7A). The GADD34-AAA2 cells presented with GADD34 protein that was expressed independent of stress, which were much lower than that measured in GADD34-Δ2 cells, but greater than that expressed in cells with GADD34-WT2. Finally, GADD34-OPT2 displayed minimal detectable GADD34 protein even during ER stress.
FIGURE 7.
Alterations in the regulatory features of GADD34 uORF2 affect cell viability during ER stress. A, MEF cells functionally deleted for GADD34 via deletion of the GADD34 C-terminal PP1c interacting domain were stably selected to express WT GADD34 (WT2), GADD34 with an optimized uORF2 (OPT2), GADD34 with uORF2 codons encoding Pro-Pro-Gly mutated to Ala-Ala-Ala (AAA2), and GADD34 with uORF2 deleted (Δ2) and treated with ER stress agent, thapsigargin, for up to 6 h or left untreated. Full-length GADD34 is labeled as GADD34. Truncated GADD34 lacking the C-terminal PP1c interacting domain is labeled as GADD34ΔC. Lysates were processed and levels of GADD34, CReP, eIF2α-P, eIF2α total, and β-actin were measured by immunoblot. B, total RNA was collected from WT2, OPT2, AAA2, and Δ2 MEF cells cultured in the presence or absence of thapsigargin and relative levels of GADD34 and CReP mRNA were measured by qRT-PCR. C, WT2 and Δ2 MEF cells were treated with thapsigargin for 6 h or left untreated. Lysates were collected and layered on top of 10–50% sucrose gradients, followed by centrifugation and analysis of whole lysate polysome profiles at 254 nm. D, equal numbers of WT2, OPT2, AAA2, and Δ2 MEF cells were seeded in 96-well plates, cultured for 0, 24, or 48 h, and then fixed using 3.7% formalin with Hoechst stain. Relative values for Hoechst fluorescence are represented with the S.D. indicated. E, equal numbers of WT2, OPT2, AAA2, and Δ2 MEF cells were seeded in 96-well plates, cultured and allowed to grow for 24 h, and then MTT activity was measured. Relative values are represented as histograms for each with the S.D. indicated. F, the WT and mutant GADD34 cells were seeded in 96-well plates, cultured for 24 h, followed by treatment with or without thapsigargin for an addition 24 h. MTT activity was measured by the conversion of tetrazolium to formazan. G, the collection of GADD34 MEF cells were seeded in 96-well plates, cultured for 24 h, followed by treatment with thapsigargin with or without guanabenz for an additional 24 h. MTT activity was measured by the conversion of tetrazolium to formazan.
Expression of these GADD34 uORF variants led to significant changes in the levels of eIF2α-P during stress treatment. Of note, the sharply elevated GADD34 expression in GADD34-Δ2 cells led to a decrease in eIF2α-P in response to ER stress as compared with cells containing the GADD34-WT2 (Fig. 7A). GADD34-AAA2 cells presented with a partial lowering of induced eIF2α-P. Polysome analyses of cells expressing GADD34-WT2 or GADD34-Δ2 supported the translational control effects predicted from the patterns of induced eIF2α-P (Fig. 7C). In both non-stressed and ER stress conditions, the GADD34-Δ2 cells displayed increased polysome levels compared with GADD34-WT2. These results suggest that GADD34 expression is tightly regulated through uORF2-mediated translational control to allow for the optimal amounts of eIF2α-P during stress. It is also noted that whereas CReP mRNA and protein levels are considered to be constitutively expressed independent of ER stress, that there were significant differences in CReP expression among the cells containing the selected uORF2 versions of GADD34. The most dramatic changes were found in cells expressing GADD34-Δ2, where coincident with increased GADD34 protein levels there was a sharp reduction in CReP mRNA and protein levels upon ER stress (Fig. 7, A and B). Furthermore, in GADD34-OPT2 cells there was a 2-fold increase in CReP mRNA upon thapsigargin treatment, although the CReP protein levels appeared to be unchanged. These results suggest that CReP mRNA levels can be modulated depending on the nature of GADD34 translational expression.
To determine how the status of GADD34 translational control by uORF2 affects cell homeostasis, cell nuclei were stained with Hoechst fluorescent dye, and MTT activity was measured to assess cell number and vitality. Both measures were significantly increased in GADD34-Δ2 and GADD34-AAA2 expressing cells compared with GADD34-WT2 (Fig. 7, D and E). GADD34-OPT2 cells were significantly reduced for MTT activity, and trended lower for Hoechst staining, although without significance. These results suggest that the status of GADD34 translational expression can affect cell homeostasis.
Next the collection of GADD34-expressing cells were treated with thapsigargin to determine how the status of the uORF2 and GADD34 translational expression can affect their ability to adapt to ER stress. Although each of the cells showed reduced MTT activity upon ER stress, the GADD34-Δ2 that expressed the highest levels of GADD34 fared the most poorly, whereas the GADD34-OPT2 with the lowest levels of GADD34 protein expression showed the most resistance (Fig. 7F). These results suggest that enhanced GADD34 expression can render cells more sensitive to stress. Supporting this idea, the addition of guanabenz, a potent inhibitor of GADD34 targeting of PP1c dephosphorylation of eIF2α-P (8) to the GADD34-Δ2 cells substantially alleviated its sensitivity to thapsigargin treatment (Fig. 7G).
Discussion
In this study, we address the nature of uORFs that facilitate preferential translation in response to eIF2α-P and the roles that these regulatory elements play in cell adaptation to stress. Levels of GADD34 and CReP expression are critical for determining the amounts of eIF2α-P and expression of the two paralogs has previously been shown to be differentially regulated in response to ER stress (5, 7, 12). The 5′-leaders of GADD34 and CReP mRNAs contain two uORFs, with uORF2 in each serving as the dominant inhibitory element that is suggested to contribute to translational control (13, 14). We define here the central regulatory features by which each of the uORF2 sequences direct translational control of GADD34 and CReP. As illustrated in a model presented in Fig. 6B, GADD34 uORF2 serves as an efficient barrier to downstream CDS translation in basal conditions. Central to this low level of downstream translation reinitiation is an inhibitory Pro-Pro-Gly sequence juxtaposed to the termination codon in GADD34 uORF2. However, during ER stress, eIF2α-P facilitates a bypass of GADD34 uORF2 due, in part, to a poor start codon context, allowing for ribosome initiation at the GADD34 CDS (Fig. 6B). It is important to note that only a small portion of ribosomes bypass the GADD34 uORF2 during ER stress, as deletion of the uORF2 led to over 10 times more luciferase activity as compared with the WT during thapsigargin treatment (Fig. 2C). This level of bypass ensures that there is appropriate expression of GADD34 protein during feedback control of the ISR, which protects against premature restoration of translation during periods of ER stress.
Whereas the uORFs in CReP have some physical and functional similarities with GADD34, there are also several significant differences. Regarding similarities, both GADD34 and CReP have two uORFs of comparable spatial arrangements, with uORF2 having a major repressing function on downstream CDS translation and uORF1 displaying a modest dampening role (Fig. 5D). Furthermore, ribosomes are suggested to bypass uORF2 in both CReP and GADD34, although the bypass occurs to a greater degree in GADD34 (Fig. 6, B and C). The critical difference between GADD34 and CReP lies in the ability of CReP uORF2 to facilitate more ribosome reinitiation at the downstream CDS. By comparing expression of CReP-Luc between WT and ΔuORF2 constructs in the absence of stress (Fig. 4C), we estimate that upwards of 12% of the ribosomes that translate uORF2 reinitiate at the CReP CDS. Using a similar comparison for GADD34-Luc (Fig. 2C), it is estimated that less than 3% of ribosomes translating uORF2 reinitiate at the GADD34 CDS. Together the modest bypass of uORF2 during ER stress and efficient ribosome reinitiation allow for constitutive ribosome translation of the CReP CDS. It is also of note that efficient reinitiation at the CReP CDS occurs with an uORF2 of longer length, 52 codons, which appears to differ with the suggested models whereby uORFs only a few codons in length are necessary for appreciable ribosome reinitiation at a downstream CDS (22).
Roles of uORFs in Regulating the ISR and Cellular Resistance to Stress
Both GADD34 and CReP are responsible for directing PP1c to dephosphorylate eIF2α-P. As the amount of eIF2α-P can dictate the levels of global and gene-specific translation, regulation of GADD34 and CReP expression is central for maintaining protein homeostasis and health of the cell. We showed that alteration of the regulatory features in GADD34 uORF2 results in significant changes in protein synthesis and cell vitality both basally and during ER stress (Fig. 7). Of note, deletion of GADD34 uORF2 resulted in a dramatic increase in GADD34 expression, which then lowered levels of both eIF2α-P and translational control that coincided with increased sensitivity of the cells to ER stress. These results suggest that aberrant regulation of GADD34 expression alters the dynamics of the ISR, which would not allow sufficient time for stressed cells to induce ISR-target genes to alleviate stress damage before resumption of global translation. Paradoxically, functional deletion of GADD34 and chronically low levels of global translation have been previously shown to also result in increased sensitivity of cells to ER stress (23), which further emphasizes the importance of the mechanisms regulating GADD34 and CReP expression in the timing and magnitude of ISR induction. Interestingly, mice deleted for GADD34 are resistant to renal toxicity upon ER stress treatment, suggesting that in tissues there are further complexities to the dynamics of the ISR (24). The mechanisms underlying differential regulation of GADD34 and CReP translation also have implications for the utility of emerging drugs to modulate the ISR and its control of cell adaptation to intracellular and extracellular stresses (8–10).
uORFs Have Different Functions in Translational Control
Although uORFs are central for the preferential translation of key ISR genes, including ATF4, CHOP, and GADD34, the mere presence of an uORF is not sufficient to confer preferential translation in response to eIF2α-P. In fact, uORFs are suggested to be prevalent among gene transcripts whose translation is enhanced, repressed, or resistant to eIF2α-P. These findings suggest that specific features of each uORF delineate its ability to activate or repress downstream translation. This study suggests that the repressing function of GADD34 uORF2 is dependent on low levels of ribosome reinitiation due to an inhibitory Pro-Pro-Gly sequence juxtaposed to a termination codon. There are different strategies for thwarting reinitiation, with the ATF4 uORF2 overlapping out-of-frame with the downstream CDS and translation of the CHOP uORF leading to an elongation pause (3, 4). Interestingly, the sole GADD34/CReP paralog in Drosophila melanogaster has a single uORF that overlaps out-of-frame with the CDS (25), which suggests that this model organism has established an inhibitory uORF in a bypass mechanism through an alternative strategy from its GADD34 mammalian counterpart. The rules for regulation of ribosome reinitiation examined here for GADD34 and CReP provide new insight into uORF-mediated differences in expression in response to eIF2α-P. This has exciting implications for genome-wide assessments of translation, where the features of uORFs can be used to predict the translation control properties for a given mRNA during basal conditions and those inducing eIF2α-P.
Author Contributions
S. K. Y. conceived the study, designed, performed, and analyzed experiments, and wrote the manuscript. J. A. W. designed, performed, and analyzed experiments shown in Fig. 7. C. W. designed, performed, and analyzed experiments shown in Fig. 4, C–E. M. S. S. conceived, designed, and analyzed experiments shown in Fig. 4, C–E, and contributed to the preparation of the manuscript. R. C. W. conceived and coordinated the study, designed and analyzed experiments, and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Dr. Eric Jan and members of the Wek laboratory for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant GM049164 (to R. C. W.) and the Ralph W. and Grace M. Showalter Research Trust Fund.
- ISR
- integrated stress response
- uORF
- upstream ORF
- CDS
- coding sequence
- MEF
- mouse embryonic fibroblast
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- ER
- endoplasmic reticulum
- TK
- thymidine kinase
- FRT
- Flp recombination target
- qPCR
- quantitative PCR.
References
- 1.Hinnebusch A. G. (2014) The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 83, 779–812 [DOI] [PubMed] [Google Scholar]
- 2.Baird T. D., and Wek R. C. (2012) Eukaryotic initiation factor 2 phosphorylation and translational control in metabolism. Adv. Nutr. 3, 307–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vattem K. M., and Wek R. C. (2004) Reinitiation involving upstream open reading frames regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 101, 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palam L. R., Baird T. D., and Wek R. C. (2011) Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem. 286, 10939–10949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Novoa I., Zeng H., Harding H. P., and Ron D. (2001) Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J. Cell Biol. 153, 1011–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walter P., and Ron D. (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 [DOI] [PubMed] [Google Scholar]
- 7.Jousse C., Oyadomari S., Novoa I., Lu P., Zhang Y., Harding H. P., and Ron D. (2003) Inhibition of a constitutive translation initiation factor 2a phosphatase, CReP, promotes survival of stressed cells. J. Cell Biol. 163, 767–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsaytler P., Harding H. P., Ron D., and Bertolotti A. (2011) Selective inhibition of a regulatory subunit of protein phosphatase 1 restores proteostasis. Science 332, 91–94 [DOI] [PubMed] [Google Scholar]
- 9.Boyce M., Bryant K. F., Jousse C., Long K., Harding H. P., Scheuner D., Kaufman R. J., Ma D., Coen D. M., Ron D., and Yuan J. (2005) A selective inhibitor of eIF2α dephosphorylation protects cells from ER stress. Science 307, 935–939 [DOI] [PubMed] [Google Scholar]
- 10.Das I., Krzyzosiak A., Schneider K., Wrabetz L., D'Antonio M., Barry N., Sigurdardottir A., and Bertolotti A. (2015) Preventing proteostasis diseases by selective inhibition of a phosphatase regulatory subunit. Science 348, 239–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinnebusch A. G. (2005) Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 59, 407–450 [DOI] [PubMed] [Google Scholar]
- 12.Baird T. D., Palam L. R., Fusakio M. E., Willy J. A., Davis C. M., McClintick J. N., Anthony T. G., and Wek R. C. (2014) Selective mRNA translation during eIF2 phosphorylation induces expression of IBTKα. Mol. Biol. Cell 25, 1686–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee Y. Y., Cevallos R. C., and Jan E. (2009) An upstream open reading frame regulates translation of GADD34 during cellular stresses that induce eIF2α phosphorylation. J. Biol. Chem. 284, 6661–6673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andreev D. E., O'Connor P. B., Fahey C., Kenny E. M., Terenin I. M., Dmitriev S. E., Cormican P., Morris D. W., Shatsky I. N., and Baranov P. V. (2015) Translation of 5′ leaders is pervasive in genes resistant to eIF2 repression. eLife 4, e03971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang H. Y., Wek S. A., McGrath B. C., Lu D., Hai T., Harding H. P., Wang X., Ron D., Cavener D. R., and Wek R. C. (2004) Activating transcription factor 3 (ATF3) is integral to the eIF2 kinase stress response. Mol. Cell. Biol. 24, 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harding H. P., Zhang Y., Scheuner D., Chen J. J., Kaufman R. J., and Ron D. (2009) Ppp1r15 gene knockout reveals an essential role for translation initiation factor 2α (eIF2α) dephosphorylation in mammalian development. Proc. Natl. Acad. Sci. U.S.A. 106, 1832–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teske B. F., Fusakio M. E., Zhou D., Shan J., McClintick J. N., Kilberg M. S., and Wek R. C. (2013) CHOP induces activating transcription factor 5 (ATF5) to trigger apoptosis in response to perturbations in protein homeostasis. Mol. Biol. Cell 24, 2477–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teske B. F., Baird T. D., and Wek R. C. (2011) Methods for analyzing eIF2 kinases and translational control in the unfolded protein response. Methods Enzymol. 490, 333–356 [DOI] [PubMed] [Google Scholar]
- 19.Wang Z., and Sachs M. S. (1997) Ribosome stalling is responsible for arginine-specific translation attenuation in Neurospora crassa. Mol. Cell. Biol. 17, 4904–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutierrez E., Shin B. S., Woolstenhulme C. J., Kim J. R., Saini P., Buskirk A. R., and Dever T. E. (2013) eIF5A promotes translation of polyproline motifs. Mol. Cell 51, 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma Y., and Hendershot L. M. (2003) Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J. Biol. Chem. 278, 34864–34873 [DOI] [PubMed] [Google Scholar]
- 22.Jackson R. J., Hellen C. U., and Pestova T. V. (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novoa I., Zhang Y., Zeng H., Jungreis R., Harding H. P., and Ron D. (2003) Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 22, 1180–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marciniak S. J., Yun C. Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H. P., and Ron D. (2004) CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18, 3066–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malzer E., Szajewska-Skuta M., Dalton L. E., Thomas S. E., Hu N., Skaer H., Lomas D. A., Crowther D. C., and Marciniak S. J. (2013) Coordinate regulation of eIF2α phosphorylation by PPP1R15 and GCN2 is required during Drosophila development. J. Cell Sci. 126, 1406–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]