Background: ADA3, a conserved component of several HAT complexes, regulates mitosis.
Results: ADA3 associates with the centromere through CENP-B and regulates chromosome segregation by directing centromeric loading of CENP-B.
Conclusion: ADA3 regulates mitosis by its association with the CENP-B/centromere and regulates segregation of chromosomes.
Significance: This study provides the mechanism of how ADA3 regulates mitosis to maintain genomic stability.
Keywords: cell cycle, centromere, chromatin immunoprecipitation (ChIP), mitosis, transcriptional coactivator, ADA3, ATAC, CENP-B, SAGA, chromosome segregation
Abstract
ADA3 (alteration/deficiency in activation 3) is a conserved component of several transcriptional co-activator and histone acetyltransferase (HAT) complexes. Recently, we generated Ada3 knock-out mice and demonstrated that deletion of Ada3 leads to early embryonic lethality. The use of Ada3FL/FL mouse embryonic fibroblasts with deletion of Ada3 using adenovirus Cre showed a critical role of ADA3 in cell cycle progression through mitosis. Here, we demonstrate an association of ADA3 with the higher order repeat region of the α-satellite region on human X chromosome centromeres that is consistent with its role in mitosis. Given the role of centromere proteins (CENPs) in mitosis, we next analyzed whether ADA3 associates with the centromere through CENPs. Both an in vivo proximity ligation assay and immunofluorescence studies confirmed the association of ADA3 with CENP-B protein, a highly conserved centromeric protein that binds to the 17-bp DNA sequences on α-satellite DNA. Deletional analysis showed that ADA3 directly associates with CENP-B through its N terminus, and a CENP-B binding-deficient mutant of ADA3 was incompetent in cell proliferation rescue. Notably, knockdown of ADA3 decreased binding of CENP-B onto the centromeres, suggesting that ADA3 is required for the loading of CENP-B onto the centromeres. Finally, we show that deletion of Ada3 from Ada3FL/FL mouse embryonic fibroblasts exhibited various chromosome segregation defects. Taken together, we demonstrate a novel ADA3 interaction with CENP-B-centromere that may account for its previously known function in mitosis. This study, together with its known function in maintaining genomic stability and its mislocalization in cancers, suggests an important role of ADA3 in mitosis.
Introduction
The centromere is a chromatin region that is essential for driving chromosome segregation in cell division and is responsible for accurate inheritance of eukaryotic chromosomes during this process (1–3). It serves as the site of kinetochore assembly to which microtubule attachment occurs (4). The centromere-kinetochore is a complex network of proteins that work in concert for the faithful segregation of chromosomes (5, 6). A major class of this network of proteins is centromere proteins (CENPs),4 which includes CENP-A, -B, -C, -E, -F, -H, -I, and others (2, 7).
In the CENP group of proteins, CENP-B is highly conserved in several mammalian species (8). CENP-B specifically binds to a 17-bp sequence, known as the CENP-B box through its N-terminal region and dimerizes through its C-terminal region (9, 10). The CENP-B box is conserved in centromeric human α-satellite and the mouse minor satellite region (11). CENP-B is required for de novo assembly of centromere and kinetochore nucleation (12, 13). Yeast CENP-B homolog acts as a site-specific nucleation factor for the formation of centromeric heterochromatin by heterochromatin-specific modifications of histone tails (14). The centromere function mainly entails CENP-A, -B, and -C, in which CENP-B plays a crucial role by recruiting CENP-A and stabilizing CENP-C at centromeres (15–17).
Recent studies from our laboratory and others have shown a critical role of ADA3 (alteration/deficiency in activation 3) in cell cycle regulation (18, 19). ADA3 is an essential component of several transcriptional adaptor and histone acetyltransferase (HAT) complexes conserved among eukaryotes (20). HATs and histone deacetylases are required to maintain steady-state levels of acetylation (21–24). A number of HAT enzymes, including GCN5 (general control non-repressed 5), p300, PCAF (p300/CBP-associated factor), and CBP (CREB-binding protein) have been demonstrated as part of large complexes, such as SAGA (Spt/Ada/Gcn5 acetyltransferase), TFTC (TBP-free TAF complex), and ATAC (Ada2a-containing) complexes in humans (21–24).
Our laboratory previously reported that germ line deletion of Ada3 in mice is embryonic lethal, and lack of ADA3 in mouse embryonic fibroblasts (MEFs) results in a severe proliferation defect, dramatic changes in global histone acetylation, delay in G1 to S phase transition, mitotic defects, and delay in G2/M to G1 transition (18). Furthermore, we have shown a novel role for ADA3 in maintaining DNA repair process and genomic stability by controlling DNA repair checkpoints (25). Consistently, we observed that ADA3 is overexpressed/mislocalized in breast cancers, and its overexpression predicts poor survival and poor prognosis in breast cancer patients, underscoring the critical function of ADA3 in physiology and pathology (26).
To better understand how ADA3 is involved in multiple biological processes, we recently performed chromatin immunoprecipitation followed by sequencing (ChIP-seq) and found that ADA3 was significantly associated with human centromere regions across most chromosomes.5 Interestingly, in yeast, GCN5 has been shown to play an important role in mitosis, by binding to centromeres (27). Given that GCN5 and ADA3 form an integral part of various HAT complexes and based on a clear role of ADA3 protein in mitosis, we explored whether ADA3 associates with centromeres. In this study, using a series of PCR primers corresponding to the centromere region of human X chromosome, we demonstrate that ADA3 specifically binds to the higher order repeat (HOR) region of centromere, which is the site of kinetochore attachment.
Given the known role of CENPs in centromere regulation, we examined whether ADA3 associates with centromeric proteins, such as CENP-A and CENP-B. We observed that ADA3 is associated with CENP-B, and this interaction of ADA3 with CENP-B was noticed throughout all phases of the cell cycle. Significantly, centromere binding of CENP-B was decreased with knockdown of ADA3. More importantly, in contrast to wild type ADA3, an ADA3 mutant that lacks the ability to bind to CENP-B failed to rescue cell proliferation defects caused by the deletion of endogenous Ada3. Finally, we demonstrate that ablation of Ada3 leads to defective chromosomal segregation with increase in anaphase bridges and lagging chromosomes. Taken together, these results provide a novel connection for the role of ADA3 in mitosis.
Experimental Procedures
Constructs
To generate N-terminally GST-tagged FLAG-ADA3 bacterial expression vector, full-length FLAG-ADA3 was PCR-amplified from the pMSCV puro FLAG-ADA3 construct (18). Subsequently, BglII-SalI-digested FLAG-ADA3 PCR amplicon was cloned into BamHI-SalI sites of pGEX6P-1 vector (GE Healthcare). Similarly, GST-tagged ADA3 C-terminal deletion (residues 1–369, 1–214, and 1–110) and N-terminal deletion (residues 111–432) bacterial expression vectors were constructed by cloning the respective BglII-SalI-digested PCR amplicons into BamHI-SalI sites of pGEX6P-1 vector. Generation of the retroviral pMSCV puro FLAG-ADA3 construct has been described previously (18). A retroviral construct for FLAG-ADA3(111–432) was generated by cloning BglII-SalI-digested FLAG-ADA3(111–432) PCR amplicon into the BglII-XhoI site of the pMSCV puro vector (Clontech).
Cell Culture, Transfections, and Viral Infections
76NTERT cells were cultured in DFCI medium as described before (28). Ada3FL/FL MEFs were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Ada3FL/FL MEFs stably expressing full-length FLAG-ADA3 or FLAG-ADA3(111–432) were generated as described previously (18). Adenoviruses expressing EGFP-Cre or EGFP alone were purchased from University of Iowa (gene transfer vector core). Cre-mediated deletion of Ada3 was performed as described previously (18). For ADA3 knockdown experiments in 76NTERT, cells were transfected with 50 nm control (sc-37007, Santa Cruz Biotechnology, Inc.) or ADA3 siRNA (sc-78466, Santa Cruz Biotechnology), using DharmaFECT 1 transfection reagent (T-2001–03, Dharmacon).
Chromatin Immunoprecipitation (ChIP)
The ChIP assay was performed using the ChIP-IT Express kit from Active Motif according to the manufacturer's protocol with slight modifications in fixation and sonication conditions. 76NTERT cells were washed twice with 1× PBS and fixed in ethylene glycol bis(succinimidyl succinate) and formaldehyde at room temperature. In particular, cells were incubated in 1.5 mm ethylene glycol bis(succinimidyl succinate) in 1× PBS on a shaking platform for 15 min. To this formaldehyde (1% working concentration) was added for another 15 min. The fixation reaction was then stopped by 1× glycine at room temperature for 5 min. Chromatin from 76NTERT cells was isolated and sonicated for 12 min to obtain a fragment size of 200–1000 bp. The remaining steps for ChIP were followed exactly as per the manufacturer's protocol. Antibodies used for ChIP assay were anti-ADA3 monoclonal antibody (18), anti-CENP-A (ab13939, Abcam), and anti-CENP-B antibody (07-735, EMD Millipore). PCR amplification was performed using primers as described in Table 1.
TABLE 1.
PCR primer sets used for ChIP-PCR
PCR primer sequences were aligned with human genome (hg19). Sequences of PCR primer sets of γ-ALR jxn, Xp mono-HOR jxn, HOR satellite (D′), Xq mono-HOR jxn, and Xq sat jxn were from Mravinac et al. (32). NA, not applicable.
| Genomic site | Description | Position (hg19) |
|---|---|---|
| A: γ-ALR jxn | Forward: 5′-agcccgaggaaaatactggtgagg-3′ | chX: 58319092–58319315 |
| Reverse: 5′-gctgtctttctagtttttgtcgtgggttat-3′ | ||
| B: Xp mono sat | Forward: 5′-tgcagagggatatttgtaagcat-3′ | chX: 58514700–58514948 |
| Reverse: 5′-tgcttctgtctaattttcgtgtg-3′ | ||
| C: Xp mono-HOR jxn | Forward: 5′-aacgctgcgctatcaaagggaaagt-3′ | chX: 58560457–58560769 |
| Reverse: 5′-ggacatgtggagcgctttgtgc-3′ | ||
| D: HOR satellite | Forward: 5′-aaagggtgtttcgaacctga-3′ | chX: 61718416–61718622 |
| Reverse: 5′-tgaacatgccttttgatgga-3′ | ||
| D′: HOR satellite | Forward: 5′-ataatttcccataactaaacaca-3′ | chX: 58605896–58606430 |
| Reverse: 5′-tgtgaagataaaggaaaaggctt-3′ | ||
| E: Xq mono-HOR jxn | Forward: 5′-gacctcaaagcactctaaatacac-3′ | chX: 61725908–61726403 |
| Reverse: 5′-cttcacataaaaactagacagacag-3′ | ||
| F: Xq mono sat | Forward: 5′-aaaattgaggtttcaaaactgct-3′ | chX: 61745729–61745968 |
| Reverse: 5′-ttccttttcatagcgcactt-3′ | ||
| G: Xq sat jxn | Forward: 5′-cctgctgaatcaaaacaatggt-3′ | chX: 62045588–62045976 |
| Reverse: 5′-caaagaaggctgggtgagaag-3′ | ||
| Universal α satellite | Forward: 5′-cattctcagaaacttctttgtg-3′ | NA |
| Reverse: 5′-cttctgtctagtttttatgtga-3′ |
Immunofluorescence
For immunofluorescence, 76NTERT cells were grown to 50% confluence on glass coverslips in 12-well plates. After knocking down ADA3 in 76NTERT cells by siRNA, the coverslips were fixed in 4% paraformaldehyde for 20 min. Staining was performed as described earlier (25). The primary antibodies used were FITC-labeled human anti-centromere antibody (ACA) (15-235-F, Antibodies Inc.), anti-CENP-B antibody (ab25734 (Abcam) or 07-735 (EMD Millipore), and anti-ADA3 antibody (18). Secondary antibodies used were Alexa Fluor 488, Alexa Fluor 594, and Alexa Fluor 647 from Life Technologies, Inc. Nuclei were counterstained with DAPI. The coverslips were then placed on slides using the mounting medium. Fluorescent images were captured using an LSM 510 META confocal fluorescence microscope (Zeiss).
DuoLink in Situ Proximity Ligation Assay
Anti-mouse proximity ligation assay (PLA) probe plus, anti-rabbit PLA probe minus, and detection kit Red 563 were purchased from OLink Bioscience. 4% formaldehyde-fixed cells were blocked with PBS containing 10% goat serum and 0.001% Triton X-100 for 1 h and incubated with primary antibodies for ADA3 and CENP-B (07-735 (EMD Millipore) or ab25734 (Abcam)) or p53 (sc-6243, Santa Cruz Biotechnology) overnight at 4 °C. PLA probes were diluted 1:8 in blocking solution. Detection of the PLA signals was carried out with an LSM 510 META Confocal fluorescence microscope (Zeiss).
In Vitro Binding Assays
GST-FLAG-ADA3 full-length or various GST-ADA3 truncated mutants were purified from bacterial lysates based on the protocol by Frangioni and Neel (29). 1 μg of GST, GST-FLAG ADA3 full-length, or GST-ADA3 deletion mutants non-covalently bound to glutathione beads was incubated with 300 ng of purified His6-tagged CENP-B (ab73636, Abcam) in NETN buffer (20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.5% Nonidet P-40, 0.1 mm Na4VO3, 1 mm NaF, and protease inhibitor mixture) for 2 h at 4 °C and washed five times with NETN buffer. The bound proteins were resolved by SDS-PAGE, transferred to PVDF membrane, and immunoblotted using anti-CENP-B antibody (07-735 (EMD Millipore) or ab25734 (Abcam)).
CellTiter-Glo® Luminescent Cell Viability Assay
The assay was performed three times independently. 2 × 105 Ada3FL/FL/Vector, Ada3FL/FL/FLAG-ADA3(Full-Length), or Ada3FL/FL/FLAG-ADA3(111–432) MEFs were plated in p100 dishes. After overnight attachment, cells were infected either with Adeno-EGFP or Adeno-EGFP-Cre as described previously (18). 24 h after infection (day 1), each plate was divided into 6-well plates and 96-well plates (one plate for each day). In particular, 1 × 104 cells were plated in one well of a 6-well plate (for Western blotting), whereas 150 cells/well were plated in six replicates of a 96-well plate (for luminescence). Cells were cultured with change of medium every alternate day. At days 1, 3, 5, 7, and 9, cell viability was measured by a CellTiter-Glo® luminescent cell viability assay (Promega) following the manufacturer's protocol. To confirm the deletion of endogenous Ada3 and ectopic expression of ADA3 full-length and ADA3(111–432), cells were also harvested for Western blotting at the aforementioned days and immunoblotted with the indicated antibodies.
Colony Formation Assay
Cells were infected with either Adeno-EGFP or Adeno-EGFP-Cre as described above. 24 h after infection (day 1), 10,000 cells/well were plated in a 6-well plate and cultured until day 9 with a change of medium every alternate day. At day 9, cells were fixed and stained with crystal violet solution (0.25% crystal violet in 25% methanol) and imaged as described previously (18).
Cell Fractionation and Immunoblotting
72 h after infecting Ada3FL/FL MEFs with control or Cre adenovirus, cells were trypsinized, collected, and washed once with PBS. Cell fractionation was performed according to previously published protocols with modifications (30, 31). A fraction of the harvested cells was used to make whole cell extracts. The remaining cell pellet was suspended in lysis buffer (10 mm HEPES, pH 7.4, 10 mm KCl, 0.05% Nonidet P-40, 0.1 mm Na4VO3, 1 mm NaF, 10 mm nicotinamide, 2 μm trichostatin A, and protease inhibitor mixture), incubated on ice for 30 min, and vortexed twice at high speed, followed by centrifugation at 14,000 rpm for 10 min at 4 °C. The supernatant obtained was kept as the cytoplasmic fraction, and the pellet containing nuclei was washed once with lysis buffer. Nuclei were then resuspended in low salt buffer (10 mm Tris-HCl, pH 7.4, 0.2 mm MgCl2, 1% Triton X-100, 0.1 mm Na4VO3, 1 mm NaF, 10 mm nicotinamide, 2 μm trichostatin A, and protease inhibitor mixture) and incubated on ice for 15 min, followed by centrifugation at 14,000 rpm for 10 min at 4 °C. The supernatant was stored as the nucleoplasmic fraction, and the pellet was resuspended in 0.2 n HCl and incubated on ice for 20 min. The soluble fraction was neutralized with 1 m Tris-HCl, pH 8, and used as the chromatin fraction. The cell fractions were quantitated using the BCA protein assay reagent (Pierce). The proteins were resolved by SDS-PAGE and transferred onto the PVDF membrane. Immunoblotting was performed with primary antibodies against ADA3 (mouse monoclonal antibody (18) or rabbit polyclonal antibody (HPA042250, Sigma)), CENP-B (ab25734, Abcam), FLAG (A8592, Sigma), HSC70 (sc-7298, Santa Cruz Biotechnology), GAPDH (MAB374, EMD Millipore), and histone H3 (06-755, EMD Millipore).
Chromosome Mis-segregation Analyses
For analyzing chromosome mis-segregations, Ada3FL/FL MEFs were infected with control or Cre adenovirus. 24 h after infection, cells were trypsinized and plated on 18-mm coverslips in 12-well plates. The following day (48 h after infection), cells were synchronized in S phase by double thymidine block (18 h first block with 2 mm thymidine and then release for 9 h in complete medium and then second block with 2 mm thymidine for another 18 h). After the second block, the cells were released in complete medium for 6 h (approximately the time for cells to go into mitosis), followed by fixing cells in 4% paraformaldehyde and mounting the coverslips in DAPI-containing mounting medium (Vectashield). The images were captured at ×63 using an LSM 510 META confocal fluorescence microscope (Zeiss).
Statistical Analyses
The cell viability assay was performed three times independently. For each independent experiment, the luminescence from six replicates was recorded and averaged. The S.E. values were calculated using luminescence from three independent experiments, and p values were computed by Student's t test (two-tailed, unpaired) using Microsoft Excel 2010, and p ≤ 0.05 was considered as statistically significant. For the effect of ADA3 depletion on CENP-B and ACA co-localization, a total of 120 cells were counted in control or ADA3 siRNA without bias and cells with >5 CENP-B and ACA co-localization foci were considered positive for co-localization. For chromosomal abnormalities, at least 50 anaphase chromosomes were counted in control or Ada3-deleted cells without bias and examined for segregation defects. The χ2 test was performed using SAS 9.3 (SAS Institute, Cary, NC), and p ≤ 0.05 was considered as statistically significant.
Results
ADA3 Associates with HOR Region of X Chromosome Centromere
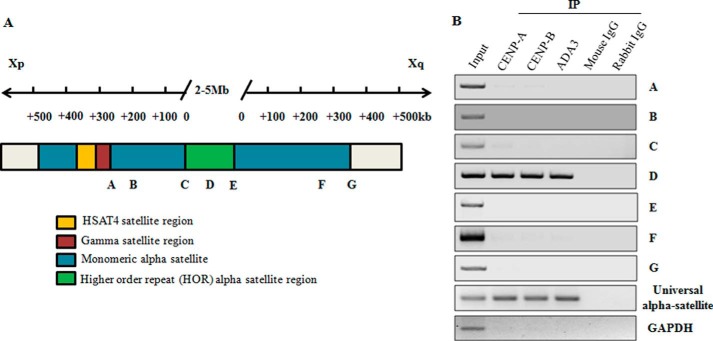
As mentioned above, we observed association of ADA3 with centromeric regions of most human chromosomes by performing ChIP-seq.5 Because the centromere is an essential chromosomal domain that is required for chromosome segregation and ensures the faithful inheritance of the chromosome during cell division (1–4), we assessed ADA3 interaction with the centromere. We first confirmed the binding of ADA3 with the centromere by performing ADA3 ChIP-PCR using a series of PCR primers on the human X chromosome centromere region in an immortal human mammary epithelial cell line, 76NTERT (32) (Table 1). To test the specificity of the ChIP primers as well as the binding of ADA3 to centromeres, we performed ChIP using anti-CENP-A or CENP-B antibodies because both CENP-A and CENP-B are known to bind to HORs in the centromeres (33). As expected, we observed binding of CENP-A and CENP-B only to HOR regions of the centromere as reported previously. Interestingly, ADA3 also associated with the α-satellite region in HORs, which is the site of kinetochore assembly (Fig. 1, A and B) (34). The universal primer set that recognizes the α-satellite region on the human genome was also amplified in ADA3 ChIP-PCR (Fig. 1, A and B), suggesting that ADA3 may maintain genomic stability by regulating chromosome separation through association with the centromere.
FIGURE 1.
Association of ADA3 with HOR region of human X chromosome centromeric α satellite region by ChIP-PCR. A, structure of the centromere of human X chromosome along with putative CENP-A, CENP-B, and ADA3 binding region shown in green on the centromere of X chromosome as obtained from B. Note that the diagram is not drawn to scale. Xp, short arm; Xq, long arm. B, ADA3 associates with the HOR region of human X chromosome centromere: ADA3, CENP-A, or CENP-B protein was immunoprecipitated from cross-linked chromatin-protein complex prepared from 76NTERT cells. Associated chromatin was then eluted and amplified by PCR using primers against centromere regions depicted in A (also see Table 1). Mouse and rabbit IgGs were used as negative controls for immunoprecipitation. Universal α satellite primers were used as positive control whereas GAPDH primers were used as negative control in PCR. IP, immunoprecipitation.
ADA3 Associates with CENP-B Protein at the Centromere
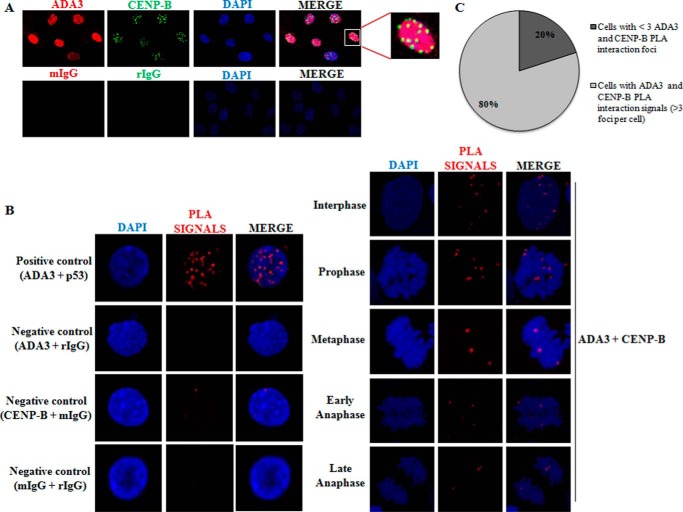
Several studies have identified a number of protein components that associate with the centromere, including CENPs (2). Given the fact that ADA3 is a transcriptional co-activator and itself does not contain DNA binding domain, we hypothesized that some other centromeric protein may mediate ADA3 interaction with the centromere. Among the well known centromere-binding proteins, CENP-B is highly conserved in several mammalian species, specifically binds to a 17-bp sequence (CENP-B box) that is conserved in the centromeric human α-satellite region, and associates with centromeric heterochromatin (10, 11). Because ADA3 associated with the HOR region in the α-satellite region on the centromere where CENP-B also binds, we examined whether ADA3 co-localizes with centromere marker CENP-B. A co-immunofluorescence assay using anti-ADA3 and anti-CENP-B antibodies clearly demonstrated that CENP-B and ADA3 are co-localized in the nucleus (Fig. 2A). Although the specificity of ADA3 antibody used in this assay has been extensively determined in our previous publications (18, 25, 26), we used ADA3 siRNA knockdown cells in this experiment to ensure specificity of the ADA3 antibody in the immunofluorescence assay (supplemental Fig. S1). To further confirm ADA3 association with centromeres, we analyzed ADA3 interaction with CENP-B by DuoLink in situ PLA. In addition to CENP-B, we assessed whether ADA3 also interacts with another centromeric protein, CENP-A. To determine the specificity of interactions in PLA, we used p53 protein, known to directly interact with ADA3 (35, 36), and rabbit or mouse IgG as negative control (Fig. 2B). In this assay, primary antibodies raised in different species are used against two interacting proteins, and when species-specific secondary antibodies linked with complementary DNA probes come in close proximity (30–40 nm), the linked DNA can be amplified and visualized with a fluorescent probe as distinct foci. ADA3 and CENP-A/CENP-B were immunostained with anti-mouse and anti-rabbit secondary antibodies, respectively, that were linked to complementary oligonucleotides. PLA exhibited only CENP-B and not CENP-A (data not shown) interaction with ADA3, particularly in interphase of cell cycle (Fig. 2B). Of the total cells quantified in various cell cycle phases, ∼80% of cells showed more than three ADA3-CENP-B interaction foci (Fig. 2C), and the interaction signals persisted when cells entered into prophase, but the signals were reduced when cells entered into metaphase and anaphase (Fig. 2B). Taken together, these results demonstrate that ADA3 associates with CENP-B during interphase and prophase, but its interaction with CENP-B is reduced in metaphase and anaphase, suggesting an important role of ADA3 in the early phase of mitosis.
FIGURE 2.
ADA3 associates with CENP-B protein at the centromeres. A, co-localization of ADA3 and CENP-B in 76NTERT cells. Cells were cultured on coverslips, fixed with 4% formaldehyde, and co-immunostained for CENP-B and ADA3, followed by fluorescence microscopy using appropriate filters (red, ADA3; green, CENP-B; blue, DAPI). Mouse and rabbit IgG (mIgG or rIgG) served as negative controls. B, interaction of ADA3 with CENP-B during the cell cycle using the PLA. 76NTERT cells were treated for 16.5 h with 100 ng/ml nocodazole, released in culture medium, and harvested at different time points. Fixed cells were incubated with antibodies against ADA3 and CENP-B followed by DuoLink in situ PLA and fluorescence microscopy using appropriate filters (blue, DAPI; red, PLA signals). Interaction of ADA3 with p53 is shown as a positive control, whereas mouse or rabbit IgG served as negative controls for PLA. C, quantification of cells with ADA3-CENP-B PLA interaction signals from B. More than 100 cells in different cell cycle phases were quantified for PLA interaction foci present in the nucleus. Cells with greater than three PLA interaction signals were considered positive for ADA3-CENP-B interaction.
ADA3 Directly Interacts with CENP-B through Its N Terminus
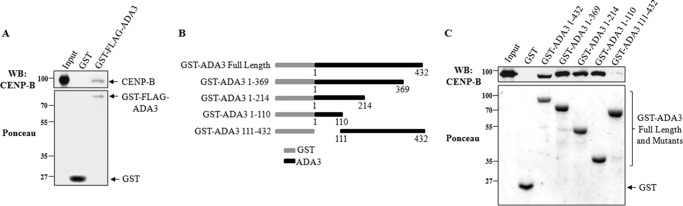
Next, to assess whether ADA3 and CENP-B directly interact, we performed a GST pull-down assay using purified GST-tagged human FLAG-ADA3 and His6-CENP-B protein. As shown in Fig. 3A, CENP-B is detected in the GST pull-down lysates after incubation of GST-FLAG-tagged ADA3 with CENP-B protein, demonstrating that ADA3 directly interacts with CENP-B in vitro. Next, to map the region of ADA3 essential for binding to CENP-B, we generated a series of GST-ADA3 constructs (GST-ADA3(1–369), GST-ADA3(1–214), and GST-ADA3(1–110)), in which a coding region from the C terminus was sequentially removed along with the one, GST-ADA3(111–432), in which codons that code for the first 110 amino acids from the N terminus were deleted (Fig. 3B). Recombinant proteins from these constructs were used as baits, and purified CENP-B was used as prey in our GST pull-down assays. Immunoblotting with anti-CENP-B antibody showed that all ADA3 fragments were able to efficiently pull down CENP-B except for the 111–432 fragment of ADA3, suggesting that the N terminus of ADA3 is critical for its interaction with CENP-B (Fig. 3C).
FIGURE 3.
ADA3 directly interacts with CENP-B through its N terminus. A, in vitro binding assays were performed to determine whether ADA3 directly interacts with CENP-B. 1 μg of GST or GST-FLAG ADA3 bound to glutathione beads was incubated with 300 ng of purified CENP-B protein. After washes, the beads were loaded onto SDS-PAGE, transferred to PVDF membrane, and immunoblotted using anti-CENP-B antibody. Input is 100%. B, schematic representation of GST-ADA3 constructs used to determine the region in ADA3 required for its interaction with CENP-B. C, in vitro GST pull-down assays were performed as in A using GST-ADA3 constructs shown in B. Input is 10%. WB, Western blot.
CENP-B Binding-defective ADA3 Mutant Fails to Rescue Cell Proliferation Arrest Caused by the Deletion of Endogenous Ada3
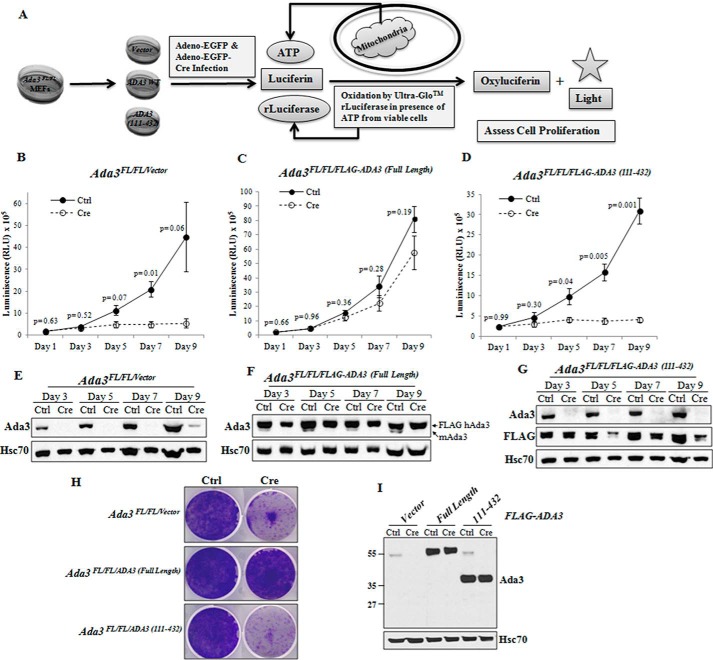
We have previously demonstrated that conditional deletion of endogenous Ada3 from Ada3FL/FL MEFs causes cell proliferation arrest (18). However, Ada3FL/FL/ADA3 MEFs in which human ADA3 is ectopically expressed rescue the cell proliferation defects. Therefore, we tested the ability of the 111–432 fragment of ADA3, which lacks the binding ability with CENP-B, to rescue cell cycle arrest. For this purpose, FLAG-tagged ADA3 full-length or 111–432 fragment was stably expressed in Ada3FL/FL MEFs, followed by the deletion of endogenous Ada3 by adenovirus expressing Cre recombinase, and cell proliferation was assessed at regular intervals up to 9 days by the CellTiter-Glo® luminescent cell viability assay, which determines the number of viable cells in culture based on quantitation of ATP (Fig. 4A). Deletion of endogenous Ada3 and expression of ectopically expressed ADA3 was confirmed by immunoblotting (Fig. 4, E–G). Consistent with our previous report (18), we observed that in contrast to vector alone, wild type full-length ADA3 was able to restore the cell proliferation arrest caused by the deletion of endogenous Ada3 (Fig. 4, B and C). Interestingly, we observed that the CENP-B binding-defective 111–432 mutant failed to rescue the cell cycle defect (Fig. 4D). To further confirm that 111–432 ADA3 mutant is defective in cell proliferation rescue, a colony formation assay was also performed at day 9. Similar results were obtained in this assay (Fig. 4, H and I), indicating that CENP-B and ADA3 interaction may be required for the ability of ADA3 to regulate cell proliferation.
FIGURE 4.
CENP-B binding-defective ADA3 mutant fails to rescue cell proliferation arrest caused by the deletion of endogenous Ada3. A, strategy used to perform ADA3 rescue cell proliferation assays. B–D, cell viabilities of Ada3FL/FL/Vector (B), Ada3FL/FL/FLAG-ADA3(Full-Length) (C), and Ada3FL/FL/FLAG-ADA3(111–432) (D) MEFs after control adenovirus (Ctrl) or Cre adenovirus (Cre) infection obtained using a CellTiter-GLO luminescent cell viability assay as described under “Experimental Procedures.” Data shown here are mean ± S.E. (error bars) from three independent experiments performed in six replicates, and p values were computed using Student's t test. E–G, ADA3 protein levels at different time points after Cre adenovirus infection of the indicated cell lines. Note that full-length or mutant ADA3 reconstituted control cells express both mouse ADA3 and human FLAG-ADA3 proteins, whereas only human FLAG-ADA3 is seen in Cre adenovirus-infected cells. H, colony formation assay. Crystal violet staining of the indicated cells infected with control virus or Cre adenovirus grown for 9 days is shown. I, Western blotting of lysates from H showing exogenous and endogenous ADA3.
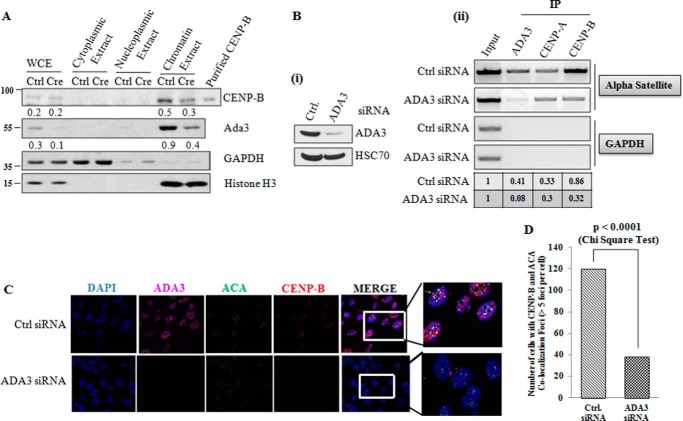
Depletion of ADA3 Abrogates CENP-B Recruitment to Centromeres
Given the critical roles of CENPs in centromere regulation, we examined the role of ADA3 in CENP-B association to centromeres. For this purpose, we deleted Ada3 from Ada3FL/FL MEFs by Cre-mediated deletion, fractionated cell compartments, and assessed the levels of CENP-B in whole cells and cytoplasmic, nucleoplasmic, or chromatin fractions. As seen in Fig. 5A, CENP-B protein was only observed in whole cell extract (WCE) and chromatin fraction. Notably, the levels of CENP-B were not altered in the whole cell extracts, consistent with our recent report where mRNA levels of CENP-B do not change upon deletion of Ada3 in a microarray (18); however, a significant decrease in CENP-B levels was seen in the chromatin fraction (Fig. 5A). To confirm the effect of ADA3 on the ability of CENP-B to bind to centromere, we performed a ChIP assay using PCR primers specific to the HOR region on the centromere of human X chromosome (primer D′ in Table 1) and observed that binding of CENP-B to the centromere of the X chromosome is dramatically decreased upon ADA3 knockdown. Notably, 80% knockdown of ADA3 led to 63% reduction in CENP-B recruitment to the X-chromosome HOR region (Fig. 5B). However, no significant change in the recruitment of CENP-A onto the HOR region upon ADA3 knockdown was noticed. In a different strategy, we knocked down ADA3 in 76NTERT cells using siRNA and then examined the binding of CENP-B to centromeres by co-immunofluorescence using anti-CENP-B and ACAs. As seen in Fig. 5, C and D, we observed a significant reduction (χ2 test p value of <0.0001) in co-localization of CENP-B with ACA after knockdown of ADA3. Taken together, our results demonstrate that ADA3 is required for association of CENP-B with centromeres.
FIGURE 5.
Depletion of ADA3 abrogates CENP-B recruitment to centromeres. A, Ada3FL/FL MEFs were infected with control (Ctrl) or Cre adenovirus. 72 h after infection, cells were trypsinized and harvested. Cell compartments were fractionated as described under “Experimental Procedures.” The fractions were run on SDS-PAGE, transferred to PVDF membrane, and immunoblotted with the indicated antibodies. The values indicate intensities measured using ImageJ. The intensities were normalized against GAPDH (for whole cell extract (WCE) and cytoplasmic extract) or histone H3 (for chromatin extract). B, 76NTERT cells were transfected with control or ADA3 siRNA. i, Western blot showing ADA3 depletion by ADA3 siRNA. ii, ADA3, CENP-A, or CENP-B protein was immunoprecipitated from cross-linked chromatin-protein complex, and ChIP-PCR was performed using PCR primer D′, corresponding to the higher order repeat α satellite region on the X chromosome (Table 1). GAPDH was used as a negative control. The table shows the band intensities estimated by ImageJ software, normalized against inputs of control and ADA3 siRNA independently. C, representative immunofluorescence images of the co-localization of CENP-B and ACA performed in 76NTERT cells after transfecting with either control siRNA or siRNA against ADA3. Cells were fixed on 18-mm coverslips with 4% paraformaldehyde after 48 h of transfection. The indicated antibodies were used for immunofluorescence. ACA staining was used to identify centromeres. D, quantification of cells exhibiting CENP-B and ACA co-localization from C. Control or ADA3 siRNA-infected cells co-stained with ACA and CENP-B were quantified based on ACA-CENP-B co-localization foci. Cells with more than five CENP-B-ACA interaction foci were considered positive (n = 120). IP, immunoprecipitation.
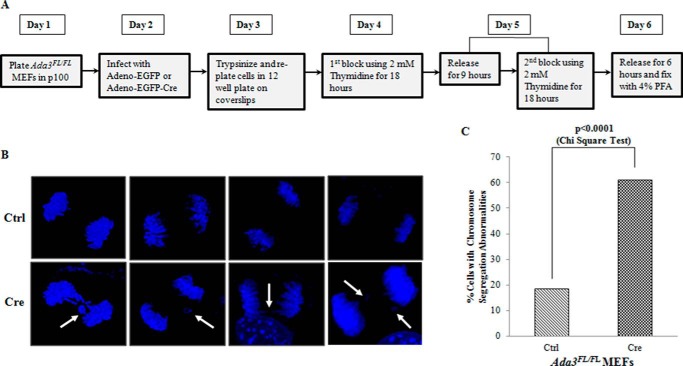
Deletion of Ada3 Causes Defects in Chromosome Segregation
Previous studies have demonstrated that CENP-B protein is important in ensuring faithful chromosome segregation during mitosis and thus assuring the highest fidelity of centromere function (16, 17) because deletion of CENP-B causes significant elevation in chromosome mis-segregation (17). Because ADA3 knockdown/deletion in cells causes significant reduction in CENP-B recruitment onto centromeres, we speculated that deletion of Ada3 might cause chromosome mis-segregation in cells as seen upon depletion of CENP-B. To test this, we deleted Ada3 from Ada3FL/FL MEFs and measured defects in chromosome segregation. To enrich cells in mitosis, we synchronized control and Cre-infected cells by double thymidine block in S phase and then fixed cells after release with complete medium for 6 h (Fig. 6A). Analyses of DAPI-stained anaphase chromosomes in both control and Cre-infected cells by confocal microscopy revealed a significantly higher percentage of anaphase bridges and lagging chromosomes in Ada3-deleted cells compared with control cells (χ2 test p value of <0.0001) (Fig. 6, B and C). These results reveal a novel role of ADA3 in the process of chromosome segregation through its binding to centromeres. Thus, ADA3 is required for maintaining the fidelity of chromosome segregation in mitosis (Fig. 7).
FIGURE 6.
Deletion of Ada3 causes defects in chromosome segregation. A, strategy used to assess chromosome mis-segregation in Ada3-deleted cells. B, representative confocal images of DAPI-stained anaphase chromosomes from control or Cre-infected Ada3FL/FL MEFs. Note that Ada3-deleted cells show various chromosomal segregation abnormalities, such as lagging chromosomes and anaphase bridges (indicated by white arrows). C, quantification of chromosomal segregation defects in control or Cre-infected Ada3FL/FL MEFs from B. Note that at least 50 anaphase cells each from control (Ctrl) and Cre-infected Ada3FL/FL MEFs were analyzed for quantification.
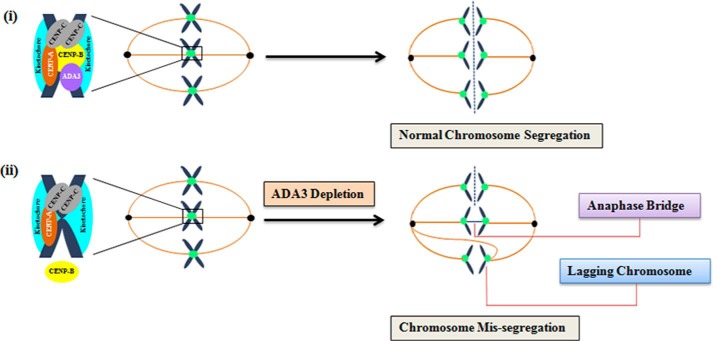
FIGURE 7.
Model displaying the role of ADA3 in chromosome segregation. i, ADA3 associates with the centromere by directly binding to CENP-B that interacts with centromeric proteins CENP-A and CENP-C to form functional centromere, which is required for proper chromosome segregation. ii, in conditions where ADA3 protein is depleted, the recruitment of CENP-B at the centromere is diminished, leading to chromosomal segregation abnormalities, viz. anaphase bridges and lagging chromosomes. Note that for the simplicity of the model, the association of ADA3 and centromeric proteins with the centromere is enlarged at only one chromosome and not shown for each chromosome.
Discussion
Human centromeres are multimegabase regions of highly ordered arrays of α satellite DNA that are separated from chromosome arms by unordered α satellite monomers and other repetitive sequences. The centromere is an essential chromosomal domain that is required for chromosome segregation and ensures the faithful inheritance of the chromosome during cell division (1–4).
Our recent study where conditional deletion of Ada3 from Ada3FL/FL MEFs using the adenovirus Cre system demonstrated a critical role of ADA3 in cell cycle progression with defect in mitosis and another study where knockdown of Ada3 in NIH3T3 cells demonstrated the role of ATAC (ADA3-associated complex) in mitosis provided the rationale to examine the mechanism of ADA3 regulation of mitosis (18, 19). We performed ChIP-seq followed by a ChIP assay using anti-ADA3 antibody and demonstrated that ADA3 associates with the higher order repeat region in α satellite of X chromosome centromere, which is the site of kinetochore assembly. Our results, together with a recent study that showed direct binding of GCN5 to the centromere (27), suggest a role of the ATAC/SAGA complex in mitosis through regulation of centromere function.
Several CENPs, including CENP-B, are known to bind to the centromere and regulate centromere function and thus chromosome segregation (33, 37). Among centromeric proteins, CENP-B is the only protein that has DNA sequence-specific binding ability that is conferred by a 17-bp sequence, known as the CENP-B box, in the α satellite region of the human and minor satellite region of the mouse centromere (8, 38), suggesting a role of CENP-B in centromere identity. Whereas studies of human artificial chromosome formation have revealed an essential role of CENP-B in de novo centromerization (12, 13), the less severe mitotic defects in Cenpb knock-out mice (39–41) and lack of CENP-B boxes in Y chromosome and neocentromeres (42, 43) have led to the idea that CENP-B might be dispensable in centromere function. However, the two recent ne plus ultra studies (16, 17) by Cleveland and colleagues have provided several lines of evidence that uncover the role of CENP-B in centromere function. They observed an increased rate of chromosome mis-segregation in Cenpb null MEFs and CENP-B-devoid Y and neocentromeres, a new concept according to which CENP-B ensures the highest fidelity of chromosome segregation.
Our results using PLA and GST pull-down assays showed direct interaction of ADA3 with CENP-B. Further, to determine the region of ADA3 required for its interaction with CENP-B, we generated various GST-tagged truncated mutants of ADA3 and demonstrated that the N-terminal 110 amino acids of ADA3 are essential for its interaction with CENP-B. Accordingly, previous studies from our laboratory and others have demonstrated that the N-terminal half of ADA3 facilitates its interaction with transcription factors such as p53, AATF, estrogen receptor, ANCO-1, and other non-HAT-complex components, whereas the C terminus of ADA3 is essential for its incorporation into HAT complexes through its direct interaction with the ADA2 subunit and is also required for its association with p300 HAT (36, 44–49). Thus, our finding that the N terminus of ADA3 directly interacts with CENP-B defines a new interactor of ADA3 N terminus. More importantly, we showed that the ADA3 truncated mutant that does not interact with CENP-B failed to rescue the cell proliferation defects caused upon deletion of endogenous Ada3 in Ada3FL/FL MEFs. These findings imply that ADA3-CENP-B interaction is important for cell proliferation.
Furthermore, subcellular protein fractionation, ChIP assay, and direct immunofluorescence demonstrated the role of ADA3 in CENP-B recruitment to centromeres. These results along with published findings implicate the role of ADA3 in regulation of the centromere. Nevertheless, how ADA3 controls the localization of CENP-B on chromatin still remains to be answered. We speculated that ADA3 might regulate the localization of CENP-B by mediating its acetylation by various KATs; however, in our assays, neither p300 nor GCN5 was able to acetylate CENP-B protein (data not shown). One possibility is that direct interaction of ADA3 with CENP-B might enhance its DNA binding ability without the acetylation of CENP-B, as we have demonstrated in the case of estrogen receptor (46, 50). The second possibility is that ADA3 might regulate the DNA binding ability of CENP-B indirectly. A recent report suggests that α-N-trimethylation of CENP-B protein enhances its binding ability to the CENP-B box (51), and the role of ADA3 in this context is the subject of future studies that might reveal a novel function of ADA3.
Additionally, based on recent studies that show a vital role of CENP-B in maintaining the fidelity of chromosome segregation, we examined whether ADA3 could regulate the chromosome segregation process through its interaction with CENP-B/centromere. Analyses of DAPI-stained anaphase chromosomes in Ada3-deleted cells revealed a dramatic increase in the chromosome mis-segregation events compared with control cells, suggesting an essential role of ADA3 in maintaining the faithful segregation of chromosomes during mitosis (Fig. 7). These findings also provide a rationale for our earlier study that demonstrated an important role of ADA3 in maintaining genomic stability (25). In this study, we showed that deletion of Ada3 caused an increase in chromosomal aberrations, such as chromosome breaks, fragments, deletions, and translocations. Because a defect in the process of chromosome segregation is an important element leading to chromosomal aberrations in cells (3), the genomic instability observed in Ada3-deleted cells could be attributed to the critical role of ADA3 in regulating the process of chromosome segregation through its centromere binding ability.
Although we demonstrated ADA3 association with the centromere via CENP-B in the present study, given the role of ADA3 as a mediator protein of global acetylation of histones, it is possible that ADA3 also regulates histone acetylation at the centromeric region. Indeed, histone H3 Lys-9 and histone H4 acetylation have been shown to regulate kinetochore assembly (52, 53). However, in our ChIP experiments, we were unable to detect any significant changes in the levels of histone H3 Lys-9 acetylation or histone H3 Lys-9 trimethylation at the X-chromosome HOR region upon ADA3 knockdown (data not shown). However, we cannot rule out the possibility that ADA3 knockdown may change histone acetylation at centromeres of other chromosomes. Besides regulating the acetylation of histone proteins, ADA3 also promotes the acetylation of non-histone proteins (35), and we cannot rule out the possibility that ADA3 might play a role in acetylation of other centromeric proteins.
This study, together with our previous studies where we demonstrated a halt in the cell cycle and genomic instability upon deletion of Ada3 (18, 25), clearly demonstrates an important role for ADA3 in the regulation of mitosis. Given that ADA3 level/localization alter in breast cancers and predict poor prognosis and poor survival in patients (26), this study underscores the important role of ADA3 in abnormal proliferation that leads to oncogenesis.
Author Contributions
S. Mohibi conducted and analyzed in vitro binding assays, chromatin fractionation, and chromosome segregation defects and generated stable MEF cell lines. S. S. carried out the ChIP analysis for ADA3, CENP-A, and CENP-B binding to X chromosome centromere in Figs. 1 and 5B. J. W.-F. made the initial observations that ADA3 associates with centromeres and performed the PLA and co-immunofluorescence assays in Fig. 2. S. Mirza performed and quantified the CENP-B and ACA co-localization after ADA3 knockdown (Fig. 5, C and D) and also quantified ADA3-CENP-B PLA interaction experiments (Fig. 2C). S. Mohibi, S. S., S. Mirza, and X. Z. performed the rescue proliferation assays. S. Mohibi, S. S., J. W. F., H. B., and V. B. wrote the paper. H. B. and V. B. directed the overall study. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Janice A. Taylor and James R. Talaska (Advanced Microscopy Core Facility, University of Nebraska Medical Center) for providing assistance with confocal microscopy.
Work in our laboratories was supported by National Institutes of Health Grants CA96844 and CA144027 (to V. B.) and CA87986 and CA99163 (to H. B.); Department of Defense Grants W81XWH-07-1-0351 and W81XWH-11-1-0171 (to V. B.); and Fred and Pamela Buffett Cancer Center Support Grant P30CA036727. The Confocal Core Facility is supported by the Nebraska Research Initiative and Eppley Cancer Center Grant P30CA036727. The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Fig. S1.
J. Wang-France, H. Band, and V. Band, unpublished observation.
- CENP
- centromere protein
- HAT
- histone acetyltransferase
- MEF
- mouse embryonic fibroblast
- ChIP-seq
- chromatin immunoprecipitation followed by sequencing
- CREB
- cAMP-response element-binding protein
- HOR
- higher order repeat
- ACA
- anti-centromere antibody
- PLA
- proximity ligation assay
- TBP
- TATA-binding protein
- TAF
- TBP-associated factor.
References
- 1.Cleveland D. W., Mao Y., and Sullivan K. F. (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112, 407–421 [DOI] [PubMed] [Google Scholar]
- 2.Verdaasdonk J. S., and Bloom K. (2011) Centromeres: unique chromatin structures that drive chromosome segregation. Nat. Rev. Mol. Cell Biol. 12, 320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganem N. J., and Pellman D. (2012) Linking abnormal mitosis to the acquisition of DNA damage. J. Cell Biol. 199, 871–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheeseman I. M., and Desai A. (2008) Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9, 33–46 [DOI] [PubMed] [Google Scholar]
- 5.Maiato H., DeLuca J., Salmon E. D., and Earnshaw W. C. (2004) The dynamic kinetochore-microtubule interface. J. Cell Sci. 117, 5461–5477 [DOI] [PubMed] [Google Scholar]
- 6.Chan G. K., Liu S. T., and Yen T. J. (2005) Kinetochore structure and function. Trends Cell Biol. 15, 589–598 [DOI] [PubMed] [Google Scholar]
- 7.Earnshaw W. C. (2015) Discovering centromere proteins: from cold white hands to the A, B, C of CENPs. Nat. Rev. Mol. Cell Biol. 16, 443–449 [DOI] [PubMed] [Google Scholar]
- 8.Earnshaw W. C., Sullivan K. F., Machlin P. S., Cooke C. A., Kaiser D. A., Pollard T. D., Rothfield N. F., and Cleveland D. W. (1987) Molecular cloning of cDNA for CENP-B, the major human centromere autoantigen. J. Cell Biol. 104, 817–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoda K., Kitagawa K., Masumoto H., Muro Y., and Okazaki T. (1992) A human centromere protein, CENP-B, has a DNA binding domain containing four potential α helices at the NH2 terminus, which is separable from dimerizing activity. J. Cell Biol. 119, 1413–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muro Y., Masumoto H., Yoda K., Nozaki N., Ohashi M., and Okazaki T. (1992) Centromere protein B assembles human centromeric α-satellite DNA at the 17-bp sequence, CENP-B box. J. Cell Biol. 116, 585–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sullivan K. F., and Glass C. A. (1991) CENP-B is a highly conserved mammalian centromere protein with homology to the helix-loop-helix family of proteins. Chromosoma 100, 360–370 [DOI] [PubMed] [Google Scholar]
- 12.Ohzeki J., Nakano M., Okada T., and Masumoto H. (2002) CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 159, 765–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okada T., Ohzeki J., Nakano M., Yoda K., Brinkley W. R., Larionov V., and Masumoto H. (2007) CENP-B controls centromere formation depending on the chromatin context. Cell 131, 1287–1300 [DOI] [PubMed] [Google Scholar]
- 14.Nakagawa H., Lee J. K., Hurwitz J., Allshire R. C., Nakayama J., Grewal S. I., Tanaka K., and Murakami Y. (2002) Fission yeast CENP-B homologs nucleate centromeric heterochromatin by promoting heterochromatin-specific histone tail modifications. Genes Dev. 16, 1766–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ando S., Yang H., Nozaki N., Okazaki T., and Yoda K. (2002) CENP-A, -B, and -C chromatin complex that contains the I-type α-satellite array constitutes the prekinetochore in HeLa cells. Mol. Cell Biol. 22, 2229–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fachinetti D., Folco H. D., Nechemia-Arbely Y., Valente L. P., Nguyen K., Wong A. J., Zhu Q., Holland A. J., Desai A., Jansen L. E., and Cleveland D. W. (2013) A two-step mechanism for epigenetic specification of centromere identity and function. Nat. Cell Biol. 15, 1056–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fachinetti D., Han J. S., McMahon M. A., Ly P., Abdullah A., Wong A. J., and Cleveland D. W. (2015) DNA sequence-specific binding of CENP-B enhances the fidelity of human centromere function. Dev. Cell 33, 314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohibi S., Gurumurthy C. B., Nag A., Wang J., Mirza S., Mian Y., Quinn M., Katafiasz B., Eudy J., Pandey S., Guda C., Naramura M., Band H., and Band V. (2012) Mammalian alteration/deficiency in activation 3 (Ada3) is essential for embryonic development and cell cycle progression. J. Biol. Chem. 287, 29442–29456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orpinell M., Fournier M., Riss A., Nagy Z., Krebs A. R., Frontini M., and Tora L. (2010) The ATAC acetyl transferase complex controls mitotic progression by targeting non-histone substrates. EMBO J. 29, 2381–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohibi S., Srivastava S., Band H., and Band V. (2014) in Nuclear Signaling Pathways and Targeting Transcription in Cancer, pp. 33–55, Springer, New York [Google Scholar]
- 21.Nagy Z., and Tora L. (2007) Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 26, 5341–5357 [DOI] [PubMed] [Google Scholar]
- 22.Carrozza M. J., Utley R. T., Workman J. L., and Côté J. (2003) The diverse functions of histone acetyltransferase complexes. Trends Genet. 19, 321–329 [DOI] [PubMed] [Google Scholar]
- 23.Roth S. Y., Denu J. M., and Allis C. D. (2001) Histone acetyltransferases. Annu. Rev. Biochem. 70, 81–120 [DOI] [PubMed] [Google Scholar]
- 24.Lee K. K., and Workman J. L. (2007) Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell Biol. 8, 284–295 [DOI] [PubMed] [Google Scholar]
- 25.Mirza S., Katafiasz B. J., Kumar R., Wang J., Mohibi S., Jain S., Gurumurthy C. B., Pandita T. K., Dave B. J., Band H., and Band V. (2012) Alteration/deficiency in activation-3 (Ada3) plays a critical role in maintaining genomic stability. Cell Cycle 11, 4266–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirza S., Rakha E. A., Alshareeda A., Mohibi S., Zhao X., Katafiasz B. J., Wang J., Gurumurthy C. B., Bele A., Ellis I. O., Green A. R., Band H., and Band V. (2013) Cytoplasmic localization of alteration/deficiency in activation 3 (ADA3) predicts poor clinical outcome in breast cancer patients. Breast Cancer Res. Treat. 137, 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vernarecci S., Ornaghi P., Bâgu A., Cundari E., Ballario P., and Filetici P. (2008) Gcn5p plays an important role in centromere kinetochore function in budding yeast. Mol. Cell Biol. 28, 988–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Band V., Zajchowski D., Kulesa V., and Sager R. (1990) Human papilloma virus DNAs immortalize normal human mammary epithelial cells and reduce their growth factor requirements. Proc. Natl. Acad. Sci. U.S.A. 87, 463–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frangioni J. V., and Neel B. G. (1993) Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 210, 179–187 [DOI] [PubMed] [Google Scholar]
- 30.Zhong L., Martinez-Pastor B., Silberman D. M., Sebastian C., and Mostoslavsky R. (2013) Assaying chromatin sirtuins. Methods Mol. Biol. 1077, 149–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang J., Huen M. S., Kim H., Leung C. C., Glover J. N., Yu X., and Chen J. (2009) RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat. Cell Biol. 11, 592–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mravinac B., Sullivan L. L., Reeves J. W., Yan C. M., Kopf K. S., Farr C. J., Schueler M. G., and Sullivan B. A. (2009) Histone modifications within the human X centromere region. PLoS One 4, e6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perpelescu M., and Fukagawa T. (2011) The ABCs of CENPs. Chromosoma 120, 425–446 [DOI] [PubMed] [Google Scholar]
- 34.Schueler M. G., Higgins A. W., Rudd M. K., Gustashaw K., and Willard H. F. (2001) Genomic and genetic definition of a functional human centromere. Science 294, 109–115 [DOI] [PubMed] [Google Scholar]
- 35.Nag A., Germaniuk-Kurowska A., Dimri M., Sassack M. A., Gurumurthy C. B., Gao Q., Dimri G., Band H., and Band V. (2007) An essential role of human Ada3 in p53 acetylation. J. Biol. Chem. 282, 8812–8820 [DOI] [PubMed] [Google Scholar]
- 36.Wang T., Kobayashi T., Takimoto R., Denes A. E., Snyder E. L., el-Deiry W. S., and Brachmann R. K. (2001) hADA3 is required for p53 activity. EMBO J. 20, 6404–6413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choo K. H. (2000) Centromerization. Trends Cell Biol. 10, 182–188 [DOI] [PubMed] [Google Scholar]
- 38.Masumoto H., Masukata H., Muro Y., Nozaki N., and Okazaki T. (1989) A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J. Cell Biol. 109, 1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hudson D. F., Fowler K. J., Earle E., Saffery R., Kalitsis P., Trowell H., Hill J., Wreford N. G., de Kretser D. M., Cancilla M. R., Howman E., Hii L., Cutts S. M., Irvine D. V., and Choo K. H. (1998) Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. J. Cell Biol. 141, 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kapoor M., Montes de Oca Luna R., Liu G., Lozano G., Cummings C., Mancini M., Ouspenski I., Brinkley B. R., and May G. S. (1998) The cenpB gene is not essential in mice. Chromosoma 107, 570–576 [DOI] [PubMed] [Google Scholar]
- 41.Perez-Castro A. V., Shamanski F. L., Meneses J. J., Lovato T. L., Vogel K. G., Moyzis R. K., and Pedersen R. (1998) Centromeric protein B null mice are viable with no apparent abnormalities. Dev. Biol. 201, 135–143 [DOI] [PubMed] [Google Scholar]
- 42.Miga K. H., Newton Y., Jain M., Altemose N., Willard H. F., and Kent W. J. (2014) Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res. 24, 697–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choo K. H. (1997) Centromere DNA dynamics: latent centromeres and neocentromere formation. Am. J. Hum. Genet. 61, 1225–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horiuchi J., Silverman N., Marcus G. A., and Guarente L. (1995) ADA3, a putative transcriptional adaptor, consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex. Mol. Cell Biol. 15, 1203–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Candau R., and Berger S. L. (1996) Structural and functional analysis of yeast putative adaptors. Evidence for an adaptor complex in vivo. J. Biol. Chem. 271, 5237–5245 [DOI] [PubMed] [Google Scholar]
- 46.Meng G., Zhao Y., Nag A., Zeng M., Dimri G., Gao Q., Wazer D. E., Kumar R., Band H., and Band V. (2004) Human ADA3 binds to estrogen receptor (ER) and functions as a coactivator for ER-mediated transactivation. J. Biol. Chem. 279, 54230–54240 [DOI] [PubMed] [Google Scholar]
- 47.Li C. W., Dinh G. K., Zhang A., and Chen J. D. (2008) Ankyrin repeats-containing cofactors interact with ADA3 and modulate its co-activator function. Biochem. J. 413, 349–357 [DOI] [PubMed] [Google Scholar]
- 48.Gamper A. M., Kim J., and Roeder R. G. (2009) The STAGA subunit ADA2b is an important regulator of human GCN5 catalysis. Mol. Cell Biol. 29, 266–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zencir S., Sike A., Dobson M. J., Ayaydin F., Boros I., and Topcu Z. (2013) Identification of transcriptional and phosphatase regulators as interaction partners of human ADA3, a component of histone acetyltransferase complexes. Biochem. J. 450, 311–320 [DOI] [PubMed] [Google Scholar]
- 50.Germaniuk-Kurowska A., Nag A., Zhao X., Dimri M., Band H., and Band V. (2007) Ada3 requirement for HAT recruitment to estrogen receptors and estrogen-dependent breast cancer cell proliferation. Cancer Res. 67, 11789–11797 [DOI] [PubMed] [Google Scholar]
- 51.Dai X., Otake K., You C., Cai Q., Wang Z., Masumoto H., and Wang Y. (2013) Identification of novel α-n-methylation of CENP-B that regulates its binding to the centromeric DNA. J. Proteome Res. 12, 4167–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bergmann J. H., Jakubsche J. N., Martins N. M., Kagansky A., Nakano M., Kimura H., Kelly D. A., Turner B. M., Masumoto H., Larionov V., and Earnshaw W. C. (2012) Epigenetic engineering: histone H3K9 acetylation is compatible with kinetochore structure and function. J. Cell Sci. 125, 411–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minoda A., Saitoh S., Takahashi K., and Toda T. (2005) BAF53/Arp4 homolog Alp5 in fission yeast is required for histone H4 acetylation, kinetochore-spindle attachment, and gene silencing at centromere. Mol. Biol. Cell 16, 316–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.