Background: Runx2 is the master regulator of osteoblast differentiation.
Results: JARID1B/KDM5B is a key component of the epigenetic mechanisms that control Runx2 expression during osteoblast and myoblast differentiation. These mechanisms operate at the P1 promoter.
Conclusion: Epigenetic mechanisms regulate Runx2 expression and osteoblast-lineage commitment.
Significance: The work provides new mechanistic insights of Runx2 gene expression control during mesenchymal fate determination.
Keywords: epigenetics, gene expression, histone modification, osteoblast, transcription, Runx2
Abstract
Transcription factor Runx2 controls bone development and osteoblast differentiation by regulating expression of a significant number of bone-related target genes. Here, we report that transcriptional activation and repression of the Runx2 gene via its osteoblast-specific P1 promoter (encoding mRNA for the Runx2/p57 isoform) is accompanied by selective deposition and elimination of histone marks during differentiation of mesenchymal cells to the osteogenic and myoblastic lineages. These epigenetic profiles are mediated by key components of the Trithorax/COMPASS-like and Polycomb group complexes together with histone arginine methylases like PRMT5 and lysine demethylases like JARID1B/KDM5B. Importantly, knockdown of the H3K4me2/3 demethylase JARID1B, but not of the demethylases UTX and NO66, prevents repression of the Runx2 P1 promoter during myogenic differentiation of mesenchymal cells. The epigenetically forced expression of Runx2/p57 and osteocalcin, a classical bone-related target gene, under myoblastic-differentiation is accompanied by enrichment of the H3K4me3 and H3K27ac marks at the Runx2 P1 promoter region. Our results identify JARID1B as a key component of a potent epigenetic switch that controls mesenchymal cell fate into myogenic and osteogenic lineages.
Introduction
Osteoblast lineage commitment is supported and tightly regulated by the coordinated activity of morphogens and their developmentally regulated signaling pathways, including BMPs,3 Wnt-ligands, hormones, growth factors, and cytokines (1). Upon activation in osteoprogenitor cells, these signaling pathways regulate the expression of osteoblast master transcription factors, which in turn control a broad expression program of downstream bone-phenotypic genes, thus establishing the osteoblastic cell component of the skeleton (2).
The Runx2 transcription factor is an essential regulator of bone development as it controls the expression of a number of key target genes in differentiating osteoblastic cells (3). Runx2 null mice exhibit dramatic alterations during skeleton formation as the result of deficient osteoblast differentiation (4, 5). Similarly, hereditary mutations in the human Runx2 gene are linked to skeletal abnormalities as those found in cleidocranial dysplasia (6). Runx2 transcription in osteoblasts is controlled by regulatory elements distributed within the P1 promoter that are recognized by cognate transcription factors during osteogenesis (7–14). Moreover, transcriptional activation of the Runx2 gene in osteoblasts involves a chromatin remodeling event within the proximal 500 bp of the P1 promoter region (15–17). These changes in chromatin structure occur independent of the activities of BRG1- and BRM-containing SWI/SNF complexes (15) and are strongly associated with increased levels of histone H3 and H4 acetylation (15). Nevertheless, the precise epigenetic mechanisms that control this chromatin remodeling event remain undetermined.
Early genetic studies in Drosophila led to the identification of two groups of epigenetic master regulators of gene expression during development: the Trithorax group (TrxG) and the Polycomb Group (PcG) (18, 19). The mammalian counterparts of TrxG function are the Set1 (1A and 1B)- and the mixed lineage leukemia (MLL1, -2, -3, -4, and -5)-containing complexes also denoted as COMPASS (for Complex of Proteins Associated with Set1) and COMPASS-like complexes, respectively (20). TrxG-mediated activation of gene expression involves trimethylation of lysine 4 in histone H3 (H3K4me3), an epigenetic mark strongly associated with gene transcription (21–23). Similarly, COMPASS and COMPASS-like complexes contain the WDR5 (WD Repeat Domain 5) protein subunit, which is required for assembly and stability of these complexes as well as for full methyltransferase activity (24, 25). In addition, MLL3 and MLL4 complexes are particularly enriched in the H3K27 demethylase UTX (KDM6A), a Jumonji C domain-containing enzyme (26, 27). Therefore, recruitment of MLL3/4 COMPASS complexes to target gene sequences can result in transcriptional activation associated with enhanced H3K4me3 and reduced H3K27me3 epigenetic marks. PcG proteins, in contrast, can mediate the formation of repressed chromatin structure and transcriptional silencing. The PcG complex PRC2 (for Polycomb-Repressive Complex 2) is evolutionarily conserved and in mammals mainly consists of the subunits Enhancer of Zeste Homolog 2 (EZH2), Suppressor of Zeste 12 (SUZ12), and Embryonic Ectoderm Development (EED) (28). Among these subunits, EZH2 is the catalytic component of PRC2 as it mediates the H3K27me3 modification (29).
WDR5 is expressed in immortalized marrow stromal cells, osteoblasts, osteocytes, and chondrocytes both in culture and in vivo. Overexpression of WDR5 has been shown to accelerate the osteoblast and chondrocyte differentiation programs both in vivo and in cell culture models (30, 31) in part by activating both the canonical Wnt- and the BMP2-signaling pathways during skeletal development (32, 33). Moreover, WDR5 has been shown to bind to the Runx2 P1 promoter in osteoblastic cells where it can mediate the Runx2 transcriptional up-regulation induced by Wnt signaling. These important early findings on WDR5 need to be integrated with specific mechanisms by which mesenchymal cells epigenetically control activation of Runx2/p57 gene transcription during mesenchymal cell fate determination.
Additionally, EZH2 activity has been recently shown to have a relevant role during commitment of bone marrow-derived mesenchymal stem cells (MSCs) to the osteogenic lineage. Thus, overexpression of EZH2 protein in human MSCs impairs their potential to differentiate to osteoblasts, in part by producing increased levels of H3K27me3 at regulatory regions of bone-related target genes, including Runx2 and osteocalcin/Bglap (34). In agreement with this report, CDK1-dependent phosphorylation of EZH2 at residue Thr-487, which prevents formation of an active PRC2 complex, promotes differentiation of human MSCs to osteoblasts (35). Despite these interesting results, a specific role for EZH2 in the control of Runx2/p57 gene expression remains to be determined.
A potential contribution of enzymes that remove methylation marks at key regulatory histone lysine residues in osteoblasts has only been recently demonstrated. Thus, the histone demethylases JMJD2B (KDM4B) and JMJD3 (KDM6B) appear to be important components in the processes that regulate osteoblast lineage commitment of human MSCs (36). Nevertheless, the presence of osteoblast master regulators among the group of specific direct targets of these enzymes was not confirmed, indicating that the enzymes may be operating through an indirect mechanism or, alternatively, that unidentified demethylases are directly involved. NO66, another JMJC domain-containing demethylase, has been also reported to regulate gene expression during osteoblast differentiation. This enzyme is present in all bone tissues and may function by inhibiting transcription initiation and/or RNA polymerase II elongation by catalyzing H3K4me3 and/or H3K36me3 demethylation, respectively, of Osterix (Osx/Sp7) target genes (37). Osx/Sp7 is a well established osteoblast master transcription factor that is expressed downstream of Runx2 (38). Based on these current findings, the next critical step is to identify histone lysine demethylases that directly control the expression of the bone-specific P1 promoter of the Runx2 gene.
JARID1B (KDM5B) belongs to the Jumonji-Homology Domain demethylase family that functions as a repressor by catalyzing the removal of the H3K4me3 mark (to H3K4me2 and H3K4me1) at target genes (39). JARID1B has been shown to form multienzymatic complexes that co-regulate transcriptional repression by associating with histone deacetylases (40) and lysine methylases (KMTs) like G9a and EZH2 (41). Therefore, JARID1B-containing complexes may mediate gene repression by catalyzing histone deacetylation and H3K4me3 demethylation as well as deposition of H3K9me3 and H3K27me3. However, its contribution during osteoblast differentiation has not been addressed.
In this study we define epigenetic components that modulate transcription of the osteoblast-specific Runx2 P1 promoter. This promoter mediates expression of the bone-related Runx2/p57 isoform during differentiation to the osteogenic lineage in vivo (42). We report that key components of the Trithorax/COMPASS-like and PRC2 complexes together with the arginine methylase PRMT5 and lysine demethylase JARID1B mediate deposition and elimination of histone marks that accompany transcriptional activation and repression of the Runx2 gene during differentiation to the osteoblast and myoblast lineages. Importantly, we determine that erasing the repressive histone marks that are enriched at the Runx2 P1 promoter during muscle differentiation does not suffice to prevent transcriptional silencing of this gene in myoblastic cells. Moreover, we find that maintaining increased levels of the H3K4me3 and H3K27ac marks at the P1 promoter results in expression of Runx2/p57 in cells induced to differentiate to myoblasts.
Experimental Procedures
Cell Culture
C2C12 skeletal muscle progenitor cells (43) were maintained in Dulbecco's modified Eagle's medium with F-12 (DMEM/F-12, Life Technologies) supplemented with 10% fetal bovine serum (Life Technologies) and 1.2 g/liter NaHCO3. To induce osteoblastic differentiation, proliferating C2C12 cells were treated with 300 ng/ml BMP-2 (R&D Biosystems, MN) for up to 72 h (44). To induce myoblastic differentiation, confluent C2C12 cells were cultured in medium supplemented with 10% horse serum for up to 96 h. To detect alkaline phosphatase (AP) activity, the cells were washed with ice-cold phosphate-buffered saline (PBS), fixed with 37% paraformaldehyde, ethanol (1:1), and stained with NBT/BCIP (nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (Roche Applied Science) for 30 min at 37 °C.
Lentivirus Production and Infection of C2C12 Cells
HEK293FT cells (Life Technologies) were grown in 60-mm culture plates to 80–90% confluence; Lipofectamine 2000 reagent (Life Technologies) was used to transfect cells (following the manufacturer's instructions) with the pCMV-VSVg, pCMV-dR8.91, and pLKO.1-shRNA plasmids (at a 1:2:3 ratio, respectively; plasmids donated by Dr. Pedro Zamorano, P. Universidad Catolica de Chile, Santiago, Chile) with a maximum total DNA of 10 μg per plate. pLKO.1 EV (empty vector) was used as a control. After 16–18 h, the culture medium was replaced, and cells were maintained at 32 °C for 48 h. Supernatants containing pseudo-typed particles were collected, filtered through a PVDF filter (0.45-μm pore size). Aliquots of supernatants were immediately stored at −80 °C. C2C12 cells were plated in 6-well culture plates and infected for 48 or 72 h with 300 μl of lentiviral particles containing shRNA-WDR5, shRNA-NO66, shRNA-JARID1B, shRNA-JARID1C, shRNA-EZH2, shRNA-UTX, shRNA-PRMT5, shRNA-P300, or EV plasmids. All short hairpin-containing plasmids were acquired at Open Biosystems (GE Healthcare).
Nuclear Extracts and Protein Expression Analyses
Nuclear extracts were prepared as reported previously (45). The protein levels were quantified using the Bradford assay using bovine serum albumin as a standard. For Western blot analyses, 5–10 μg of total protein was subjected to SDS-polyacrylamide gel electrophoresis and then transferred to nitrocellulose. Immunoblotting was performed with secondary antibodies conjugated to horseradish peroxide and enhanced chemiluminescence solutions (PerkinElmer Life Sciences). Primaryantibodies used were the following: αTFIIB C-18 (sc-225, Santa Cruz Biotechnology), RNA-PolII N-20 (sc-899, Santa Cruz Biotechnology), Runx2 S-19 (sc-12488, Santa Cruz Biotechnology), WDR5 (ab56919, Abcam), NO66 3354c5a (sc-81341, Santa Cruz Biotechnology), JARID1B (ab50958, Abcam), EZH2 (39901, Active Motif), UTX/KDM6A (ab91231, Abcam), PRMT5/JBP1 (611539, BD Biosciences), P300 N-15 (sc-584, Santa Cruz Biotechnology).
Reverse Transcriptase and Quantitative Real-time PCR (qRT-PCR)
Total RNA was extracted with TRIzol (Life Technologies) according to the manufacturer's protocol. An equal amount of each sample (2 μg) was used for reverse transcription. qPCR was performed using Brilliant II SYBR® Green QPCR Master Mix (Agilent Technologies). Data are presented as relative mRNA levels of the gene of interest normalized to GAPDH mRNA levels. Primer used were: RUNX2-II/p57 forward (TCT GGA AAA AAA AGG AGG GAC TAT G) and reverse (GGT GCT CGG ATC CCA AAA GAA); mouse OC forward (CTG AGT CTG ACA AAG CCT TC) and reverse (CTG GTC TGA TAG CTC GTC AC); rat OC forward (CTG AGT CTG ACA AAG CCT TC) and reverse (GTG GTC CGC TAG CTC GTC AC); MYOG forward (TGG AGC TGT ATG AGA CAT CCC) and reverse (TGG ACA ATG CTC AGG GGT CCC); WDR5 (forward (CCA GTC CAA CCT CAT CGT CT) and reverse (CAT CAC GGT TGA AAT GAA CG); NO66 forward (ACT TTG CTC CTG TCG ATG CT) and reverse (AGC ATG TGG ACT TGG GTT TC); JARID1B forward (AGT GGC TTT CCT GTT CGA GA) and reverse (AAG CAC ATG CCC ACA TAC AA); EZH2 forward (CAT TTC ATA CGC TCT TCT GTC GAC) and reverse (CCC TCC AGA TGC TGG TAA CAC T); UTX forward (ACA GTA ATA CGT GGC CTT GCT GGA) and reverse (TTC ATC TGC TGG TTG TAA CAA CTG); PRMT5 forward (GGA ACT CTG AAG CGG CTA TG) and reverse (GTG TGT AGT CGG GGC ATT CT); GAPDH forward (CAT GGC CTT CCG TGT TCC TA) and reverse (CCT GCT TCA CCA CCT TCT TGA T).
Chromatin Immunoprecipitation (ChIP)
ChIP assays were performed in cross-linked chromatin samples as described earlier (46) with the following modifications: C2C12 cell cultures (100-mm diameter plates) were incubated for 10 min with 1% formaldehyde gentle agitation at room temperature. The cells were washed with 10 ml of PBS 3 times. For ChIP against chromatin-modifying enzymes, we used double cross-linking with EGS (ethylene glycol-bis(succinic acid N-hydroxysuccinimide ester), E3257, Sigma) (47); the formaldehyde cross-linked cells were incubated with EGS for 1 h at room temperature with gentle agitation, washed 3 times with cold PBS, resuspended in 1 ml of cell lysis buffer (5 mm Hepes, pH 8.0, 85 mm KCl, Triton X-100, and proteinase inhibitors), and homogenized with a Dounce homogenizer (∼25 times using a tight pestle). The cell extract was collected by centrifugation at 3000 × g for 5 min, resuspended in 0.5 ml of sonication buffer (50 mm Hepes, pH 7.9, 140 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% deoxycholate acid, 0.1% SDS, and a mixture of proteinase inhibitors), and incubated for 10 min on ice. Chromatin was sheared in a water bath sonicator Bioruptor (Diagenode Inc.) to obtain fragments of 500 bp or smaller. Extracts were sonicated at high power for four pulses of 10 min each and centrifuged at 16,000 × g for 15 min at 4 °C. Supernatant was collected, aliquoted, frozen in liquid nitrogen, and stored at −80 °C; one aliquot was used for A260 measurements. Chromatin size was confirmed by electrophoretic analysis. Cross-linked chromatin extracts (2 A260 units) were resuspended in sonication buffer to a final volume of 500 μl; samples were precleared by incubating with 2–4 μg of normal IgG and 50 μl of protein A/G-agarose beads (Santa Cruz Biotechnology) for 1 h at 4 °C with agitation. Chromatin was centrifuged at 4000 × g for 5 min, and the supernatant was collected and immunoprecipitated with specific antibodies (see the list below for antibodies used) for 12–16 h at 4 °C. The immune complexes were recovered with the addition of 50 μl of protein A or G-agarose beads followed by incubation for 1 h at 4 °C with gentle agitation. Immunoprecipitated complexes were washed once with sonication buffer, twice with LiCl buffer (100 mm Tris-HCl, pH 8.0, 500 mm LiCl, 0.1% Nonidet P-40, and 0.1% deoxycholic acid), and once with Tris-EDTA buffer, pH 8.0 (2 mm EDTA and 50 mm Tris-HCl, pH 8.0), each time for 5 min at 4 °C; this was followed by centrifugation at 4000 × g for 5 min. The protein-DNA complexes were eluted by incubation with 100 μl of elution buffer (50 mm NaHCO3 and 1% SDS) for 15 min at 65 °C. Extracts were centrifuged at 10,000 × g for 5 min, and the supernatant was collected and incubated for 12–16 h at 65 °C to reverse the cross-linking. Proteins were digested with 100 μg/ml proteinase K for 2 h at 50 °C, and the DNA was recovered by phenol/chloroform extraction and ethanol precipitation using glycogen (20 μg/ml) as a precipitation carrier. The qPCR primers used to evaluate the Runx2-P1 promoter region were: forward, 5′-GTG GTA GGC AGT CCC ACT TT-3′; reverse, 5′-TGT TTG TGA GGC GAA TGA AG-3′.
The following antibodies were used in ChIP assays: WDR5 (ab56919, Abcam), polyclonal NO66 3354c5a (sc-81341, Santa Cruz Biotechnology), JARID1B (ab50958, Abcam), EZH2 (07–689, Merck Millipore), UTX/KDM6A (ab91231, Abcam), PRMT5/JBP1 (611539, BD Biosciences), H3K27me3 (07–449, Merck Millipore), H3K4me (ab8895, Abcam), H3K4me3 (ab8580, Abcam), H4R3me2s (ab5823, Abcam), H3K27ac (ab4729, Abcam).
Statistical Analyses
For ChIP assays, we used a one-way analysis of variance analysis followed by the Dunnett's post-test to compare significant changes with respect to control. For mRNA expression analysis, we used the Student's t test. In all figures error bars represent the mean ± S.E.; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Results
Expression and Repression of the Runx2/p57 Gene in Mesenchymal Cells Involves Changes in Epigenetic Histone Marks
Transcription of the Runx2/p57 gene, a master regulator of osteoblast differentiation, is controlled by the P1 promoter sequence, in particular by the most proximal 500-bp region, which suffers chromatin remodeling in osteoblastic cells expressing Runx2 (15). This promoter region is highly conserved and has been shown to include regulatory elements that are functional within human, rat, and mouse osteoblastic cells (7). To gain an understanding of the cellular process that controls osteoblast-specific Runx2/p57 expression, we assessed the contribution of epigenetic mechanisms.
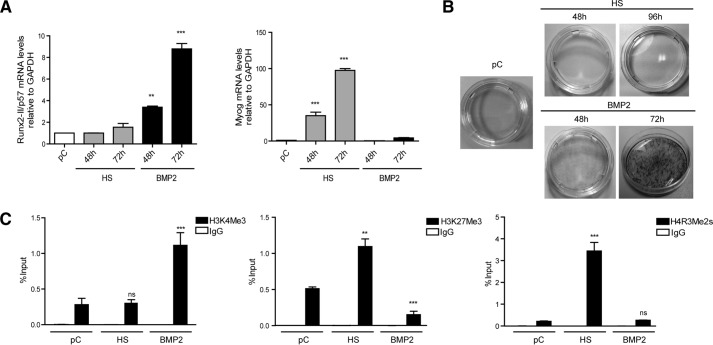
In the presence of BMP2, proliferating mesenchymal multipotent C2C12 cells can differentiate to the osteoblast lineage, as reflected by expression of early and late bone-related markers (44, 48, 49). In contrast, when pre-confluent C2C12 cells are grown in the presence of low serum media (e.g. horse serum), they differentiate toward the myoblastic lineage (43). Therefore, these cells represent a useful and reliable system to study mechanisms that control both activation and repression of either bone- or muscle-related genes. Under our experimental conditions, incubation of proliferating pre-confluent (pC) C2C12 cells with 300 ng/ml BMP2 for 48 or 72 h resulted in increasingly higher Runx2/p57 gene expression as measured by mRNA levels (Fig. 1A, left panel). As expected, this treatment with BMP2 was not accompanied by the expression of myogenin mRNA (Fig. 1A, right panel), a myoblast phenotype gene marker (50) that is strongly expressed in these cells when they are grown in the presence of horse serum for 48 or 72 h (Fig. 1A, right panel). The ability of C2C12 cells to differentiate to osteoblasts in response to BMP2 was also confirmed by the increase in AP activity associated with the extracellular matrix (Fig. 1B, lower panels).
FIGURE 1.
Modulations in Runx2/p57 expression are paralleled by changes in histone post-translational modifications at the P1 promoter region in differentiating mesenchymal cells. C2C12 multipotent cells were differentiated to the myoblast or osteoblast lineage by incubating with 10% HS or 300 ng/ml BMP-2, respectively, for the times indicated on each graph (pC)). A, total RNA was analyzed for Runx2-II/p57 mRNA levels by qRT-PCR using specific primers for this Runx2 isoform. Values were normalized against GAPDH mRNA. Myogenin mRNA levels were determined as controls for myoblast differentiation. B, alkaline phosphatase staining was performed to control for osteoblast extracellular matrix maturation. C, ChIP assays were performed using antibodies against H3K4Me3, H3K27Me3, and H4R3Me2s. Results are expressed as % input ± S.E. using normal IgG as specificity control. Statistical analyses were performed with respect to pC cells. **, p < 0.01; ***, p < 0.001; ns = non-statistically significant differences.
In agreement with the results described above, osteoblastic cells generated from C2C12 cells exhibited at the P1 promoter post-translational modifications at histone H3 (histone H3 marks), which is characteristic of transcriptionally active genes. Thus, we found enrichment of H3K4me3 and reduced levels of H3K27me3 and H4R3me2s at the Runx2 P1 promoter (Fig. 1C). H3K4me3 was found present (albeit at lower levels) in both pre-confluent and in muscle-differentiated (horse serum (HS)-treated) C2C12 cells, indicating that this modification may precede elevated transcriptional activity of the Runx2 P1 promoter in mesenchymal cells and that H3K4me3 is functionally coupled to Runx2 gene up-regulation. In contrast, H3K27me3 (Fig. 1C) was found at the P1 promoter in pre-confluent cells and then was significantly enriched after differentiation of the C2C12 cells toward the myogenic lineage (Fig. 1C). Thus, before full transcriptional activation, sequences spanning the transcriptional start site of the Runx2 gene P1 promoter are organized into chromatin that exhibits histone marks consistent with a bivalent epigenetic pattern (51). This bivalent pattern subsequently evolves into a predominantly H3K4me3- or H3K27me3-enriched condition depending on whether the Runx2 gene is transcriptionally active or silenced, respectively.
On the other hand, H4R3me2s was only enriched at the Runx2 P1 promoter region in C2C12 cells that are driven to differentiate into the myogenic lineage (Fig. 1C). Hence, this epigenetic mark corresponds with silencing of the P1 promoter during commitment to a non-osteoblast lineage.
Runx2 P1 Proximal Promoter Was Recognized by Epigenetic Regulators during Mesenchymal Cell Lineage Commitment
To identify regulatory components mediating the epigenetic changes associated with transcriptional control of the Runx2/p57 gene during osteoblast differentiation, we carried out ChIP analyses against epigenetic regulators. We used specific antibodies against proteins that are expressed in our C2C12 differentiation systems and that mediate the relevant post-translational modifications of histones detected.
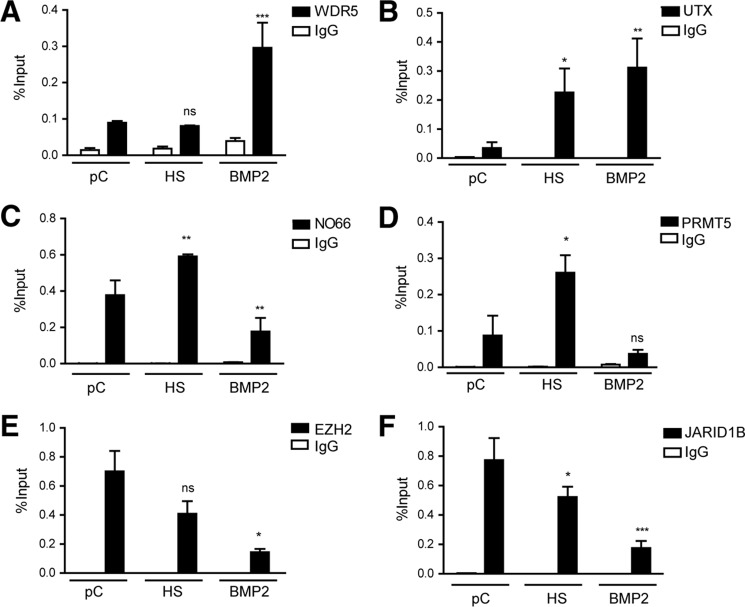
We found that upon induction of osteoblast differentiation WDR5 and UTX, two epigenetic regulators exhibited a significantly higher enrichment at the proximal Runx2 P1 promoter sequence (Fig. 2, A and B, respectively). Both proteins were found weakly associated with the Runx2 P1 promoter in untreated preconfluent C2C12 cells, and UTX was also found at this promoter in cells treated with HS. On the other hand, a reduced interaction of NO66 with the Runx2 P1 promoter region was detected in these BMP2-treated cells for up to 72 h (Fig. 2C). Importantly, this protein was found enriched at the P1 proximal promoter after 72 h of HS-mediated differentiation of C2C12 cells toward the myoblast phenotype (Fig. 2C), concomitant with Runx2 gene silencing.
FIGURE 2.
The Runx2 P1 promoter is recognized by chromatin-modifying enzymes during mesenchymal cell lineage commitment. C2C12 cells were differentiated as described in the previous figure legend, and chromatin samples were collected. ChIP assays were then performed using antibodies against the chromatin-modifying proteins WDR5 (A), UTX (B), NO66 (C), PRMT5 (D), EZH2 (E), and JARID1B (F). Results and statistical analyses are shown as described in the previous figure legend. Normal IgG was used as specificity control. *p < 0.05; **p < 0.01; ***p < 0.001; ns = non-significant differences.
Similarly, we determined that PRMT5 protein exhibited binding to the Runx2 P1 promoter in C2C12 cells engaged in myogenic lineage differentiation for 72 h (Fig. 2D). Moreover, PRMT5 was not detected at the P1 promoter when these C2C12 cells were treated with BMP2 (Fig. 2D). Together, these results indicate that recruitment of PRMT5 to the P1 promoter likely represents a molecular event associated with transcriptional silencing of the Runx2 gene in non-osteoblastic (e.g. myogenic) cells.
We next determined by ChIP analyses that the presence of EZH2 at the P1 promoter is evidenced in both pre-confluent and myogenically differentiated C2C12 cells, whereas EZH2 was not found associated with this promoter in cells treated with BMP2 (Fig. 2E). These results suggested that EZH2 activity may be tightly associated with transcriptional repression of the Runx2 gene and that a decrease in EZH2 binding to the P1 promoter may contributed to transcriptional activation of the Runx2 gene during osteoblast differentiation.
ChIP analyses reveal that in pre-confluent C2C12 cultures expressing reduced levels of Runx2/p57 (Fig. 1A), JARID1B was enriched at the Runx2 P1 proximal promoter region (Fig. 2F). This interaction was significantly decreased in C2C12 cells that were induced to differentiate toward the osteoblastic lineage (BMP2-treated cells) for 72 h but only slightly reduced when the cells differentiated to the myogenic lineage (Fig. 2F). Together, these results indicated that the demethylase JARID1B may participate in the down-regulation of Runx2/p57 expression in non-osteoblastic cells. Additionally, our results indicate that this protein is released from the Runx2 P1 promoter as Runx2/p57 gene transcription during osteoblast lineage commitment is increased.
Epigenetic Regulators Mediate Runx2/p57 Transcriptional Activation and Silencing during Lineage Differentiation of Mesenchymal Cells
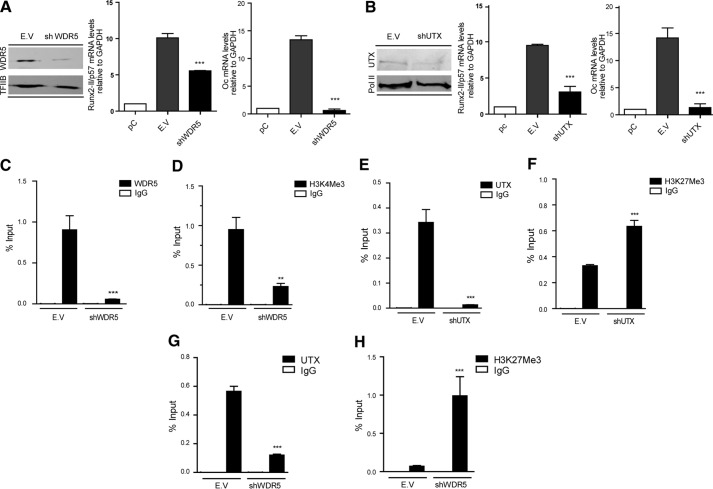
We next addressed whether Runx2/p57 transcription in C2C12 cells forced to engage in osteoblast differentiation is modulated by the epigenetic regulators identified as bound to the Runx2 P1 promoter region. We carried out specific shRNA-mediated knockdowns of epigenetic regulators in C2C12 cells incubated with BMP2. Down-regulation of WDR5 for 72 h (Fig. 3A, left panel) inhibits transcription of Runx2/p57 mRNA (Fig. 3A, central panel). This inhibition prevents further osteoblast differentiation because the expression of osteocalcin mRNA, a well established osteoblast late phenotypic marker that is directly up-regulated by Runx2 (52), is significantly abrogated (Fig. 3A, right panel). Knockdown of WDR5 was also evident from a significant reduction in WDR5 binding to the Runx2 P1 proximal promoter sequence (Fig. 3C) and accompanied by a significant decrease of H3K4me3 enrichment at this same promoter region (Fig. 3D). Similarly, knockdown of UTX in BMP2-treated C2C12 cells (Fig. 3B, left panel) resulted in down-regulation of Runx2/p57 mRNA expression (Fig. 3B, central panel), a decrease that is also accompanied by significant inhibition of osteocalcin gene transcription (Fig. 3B, right panel). Knockdown of UTX was also reflected by a reduced interaction of UTX protein with the Runx2 P1 promoter region (Fig. 3E) and enrichment of the H3K27me3 epigenetic silencing mark (Fig. 3F). WDR5 and UTX proteins have been shown to be components of MLL/COMPASS-like complexes, where WDR5 plays a relevant role maintaining the integrity of the complexes (24). Therefore, we assessed whether knockdown of WDR5 also affects binding of UTX to the Runx2 P1 promoter region. shRNA-mediated reduction of WDR5 in the BMP2-treated cells also resulted in a significant decrease in the association of UTX with the Runx2 promoter together with a drastic increase in the H3K27me3 mark (Fig. 3, G and H, respectively). These results further indicate that binding of WDR5 and UTX proteins is critical during the methylation of H3K4me3 and demethylation of H3K27me3 events that accompany transcriptional activity of the Runx2 gene in osteoblasts.
FIGURE 3.
Binding of WDR5 and UTX is accompanied by H3K4me3 deposition, H3K27me3 demethylation, and transcriptional activation of the Runx2 gene in osteoblasts. C2C12 cells were differentiated to the osteoblast lineage by incubating with 300 ng/ml BMP-2. pC cells were infected with lentiviral particles coding for shRNAs against WDR5 (A, C, D, G, and H) and UTX (B, E, and F). Effective down-regulation of each chromatin modifier was confirmed by Western blot analyses 72 h post infection. TFIIB or RNA-polymerase II protein levels were used as loading controls. Runx2-II/p57 and osteocalcin (Oc/Bglap) mRNA levels were measured by qRT-PCR 72h after infection (and 48 h of differentiation) and normalized against GAPDH mRNA. ChIP assays were performed using antibodies against WDR5 and UTX proteins or against H3K4me3 and H3K27me3 histone marks. Results are expressed as % Input ± S.E., using normal IgG as specificity control. Statistical analyses were performed with respect to the cells infected with virus generated with the pLKO.1 empty vector (E.V). **p < 0.01; ***p < 0.001.
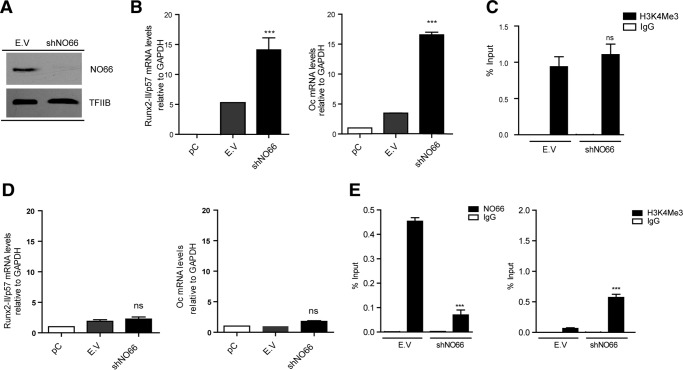
shRNA-mediated down-regulation of NO66 expression (Fig. 4A) resulted in enhanced Runx2/p57 (Fig. 4B, left panel) and osteocalcin (Fig. 4B, right panel) gene transcription. This shNO66-dependent increase in Runx2/p57 expression during C2C12 osteoblast differentiation, however, is not accompanied by further enrichment in H3K4me3 at this promoter region (Fig. 4C).
FIGURE 4.
Knockdown of NO66 demethylase and Runx2/p57 expression during mesenchymal cell lineage commitment. C2C12 cells were differentiated to the osteoblast (B and C) or myoblast (D and E) lineage as described in Fig. 1. pC cells were infected with lentiviral particles coding for shRNAs against NO66. Effective down-regulation was confirmed by Western blot analyses 72 h post infection. TFIIB protein levels were used as loading control (A). Runx2-II/p57 and osteocalcin (Oc/Bglap) mRNA levels were measured by qRT-PCR 72 h after infection (and 48 h of differentiation) and normalized against GAPDH mRNA (B and D). ChIP assays were performed using antibodies against NO66 protein or against the H3K4Me3 histone mark (C and E). Results are expressed as % input ± S.E. using normal IgG as specificity control. Statistical analyses were performed with respect to the cells infected with virus generated with the pLKO.1 empty vector (E.V.). ***, p < 0.001; ns = non-significant differences.
Because of the potential role of NO66 as a negative regulator of Runx2/p57 transcription, we assessed whether a shRNA-mediated knockdown of NO66 interferes with the Runx2/p57 gene repression during C2C12 differentiation to the myogenic lineage. Interestingly, although the absence of NO66 at the Runx2 P1 promoter (Fig. 4E, left panel) was reflected by an increase in H3K4me3 compared with control samples (Fig. 4E, right panel), no significant expression of either Runx2/p57 (Fig. 4D, left panel) or osteocalcin (Fig. 4D, right panel) mRNAs was detected under these conditions. Together, these results suggest that during myogenic differentiation, transcriptional repression of the Runx2/p57 gene did not depend on the binding of NO66 to the P1 promoter.
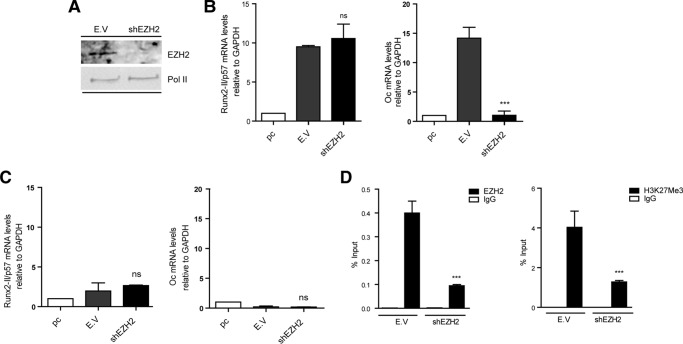
We also determined that knockdown of EZH2 (Fig. 5A) did not affect the expression of Runx2/p57 mRNA in BMP2-treated C2C12 cells (Fig. 5B, left panel). However, the expression of osteocalcin mRNA was found to be drastically reduced when EZH2 were depleted for 72 h (Fig. 5B, right panel). Whether the inhibition of osteocalcin expression reflected de-repression of EZH2-controlled genes whose products prevent transcription of bone phenotypic genes expressed downstream of Runx2 is currently being investigated. Using ChIP analyses, we determined that shRNA-mediated reduction of the EZH2 bound to the Runx2 P1 promoter in C2C12 cells differentiated to the myoblastic lineage (Fig. 5D, left panel) and correlated with a significant decrease in the H3K27me3 mark (Fig. 5D, right panel). Nevertheless, transcription of the Runx2/p57 mRNA (as well as of osteocalcin mRNA) remained strongly inhibited in these cells (Fig. 5C). Together, these results suggest that although EZH2-mediated H3K27 tri-methylation is an epigenetic component of Runx2/p57 gene silencing in non-osteoblastic cells, this modification may not represent the main mechanism to repress the expression of this bone master gene during myoblast differentiation.
FIGURE 5.
Knockdown of EZH2 methyltransferase and Runx2/p57 expression during mesenchymal cell lineage commitment. C2C12 cells were differentiated to the osteoblast (B) or myoblast (C and D) lineage as described in Fig. 1. pC cells were infected with lentiviral particles coding for shRNAs against EZH2. Effective down-regulation was confirmed by Western blot analyses 72 h post infection. RNA polymerase II protein levels were used as loading control (A). Runx2-II/p57 and Osteocalcin (Oc/Bglap) mRNA levels were measured as described in Fig. 4 (B and C). ChIP assays were performed using antibodies against EZH2 protein or against the H3K27Me3 histone mark (D). Results and statistics are expressed as described in Fig. 4. ***, p < 0.001; ns = non-significant differences.
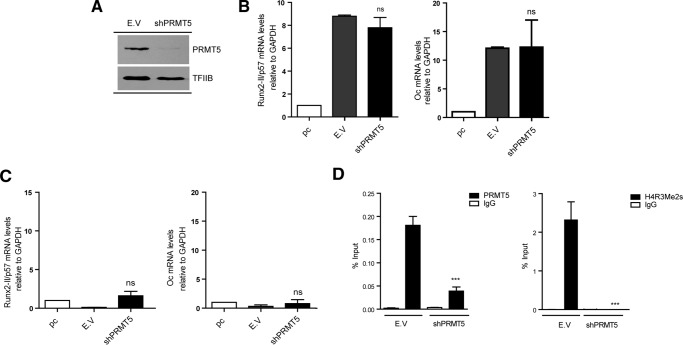
Knockdown of PRMT5 in BMP2-treated C2C12 cells (Fig. 6A) did not affect Runx2/p57 (Fig. 6B, left panel) or osteocalcin (Fig. 6B, right panel) mRNA expression levels. Importantly, PRMT5-depleted C2C12 cells that were differentiated to myoblasts and exhibited a drastic reduction in the enrichment of both PRMT5 protein and H4R3me2s at the Runx2 P1 promoter (Fig. 6D, left and right panels, respectively) remained silent for Runx2/p57 mRNA expression (Fig. 6C). These results indicate that PRMT5-mediated symmetric di-methylation of H4R3 (H4R3me2s) at the Runx2 P1 promoter is not a necessary component of the repressive epigenetic machinery that prevents Runx2/p57 expression in non-osteoblastic cells.
FIGURE 6.
Knockdown of PRMT5 methyltransferase and Runx2/p57 expression during mesenchymal cell lineage commitment. C2C12 cells were differentiated to the osteoblast (B) or myoblast (C and D) lineage as described in Fig. 1. pC cells were infected with lentiviral particles coding for shRNAs against PRMT5. Effective down-regulation was confirmed by Western blot analyses 72 h post infection. TFIIB protein levels were used as loading control (A). Runx2-II/p57 and Osteocalcin (Oc/Bglap) mRNA levels were measured as described in Fig. 4, B and C. ChIP assays were performed using antibodies against PRMT5 protein or against the H4R3Me2-symmetrical histone mark (D). Results and statistics are expressed as described in Fig. 4. ***, p < 0.001; ns = non-significant differences.
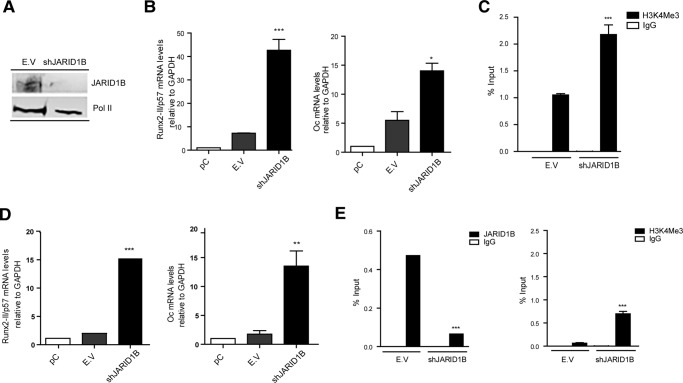
The contribution of the H3K4me3 demethylase JARID1B during transcriptional control of Runx2/p57 was analyzed by knockdown of this enzyme during osteoblast differentiation (Fig. 7A). The results revealed increased expression of Runx2/p57 mRNA (and of osteocalcin; Fig. 7B, right panel) compared with control samples, reaching an 8-fold up-regulation (Fig. 7B, left panel). This enhanced expression of Runx2/p57 mRNA was reflected also by changes in the Runx2 P1 promoter as shown by a 2-fold enrichment of the H3K4me3 mark in BMP2-treated cells when JARID1B expression was abrogated (Fig. 7C). We then analyzed whether the absence of JARID1B in C2C12 cells differentiating to the myogenic lineage affects the ability of these cells to repress Runx2/p57 expression. As expected, shRNA-mediated knockdown of JARID1B resulted in decreased association of this enzyme with the Runx2 P1 promoter (Fig. 7E, left panel) together with an increase in the H3K4me3 mark (Fig. 7E, right panel). Remarkably, JARID1B knockdown prevented Runx2/p57 silencing in HS-treated myogenic C2C12 cells (Fig. 7D, left panel), as Runx2/p57 mRNA concentration reached values that were comparable to those found in C2C12 cells treated with BMP2 (see Fig. 3). The JARID1B-depleted cells also exhibited high osteocalcin mRNA levels (Fig. 7D, right panel) suggesting that Runx2/p57 expression together with the absence of JARID1B promotes the expression of genes associated with the osteoblast differentiation program under conditions where these mesenchymal cells normally are fully engaging muscle differentiation.
FIGURE 7.
Knockdown of JARID1B demethylase prevents Runx2/p57 repression during myoblast differentiation. C2C12 cells were differentiated to the osteoblast (B and C) or myoblast (D and E) lineage as described in Fig. 1. pC cells were infected with lentiviral particles coding for shRNAs against JARID1B. Effective down-regulation was confirmed by Western blot analyses 72 h post infection. RNA Polymerase II protein levels were used as loading control (A). Runx2-II/p57 and osteocalcin (Oc/Bglap) mRNA levels were measured as described in Fig. 4, B and D. ChIP assays were performed using antibodies against JARID1B protein or against the H3K4me3 histone mark (C and E). Results and statistics are expressed as described in Fig. 4. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Runx2/p57 Expression Required the Presence of H3K4me3 and H3K27ac at the P1 Promoter
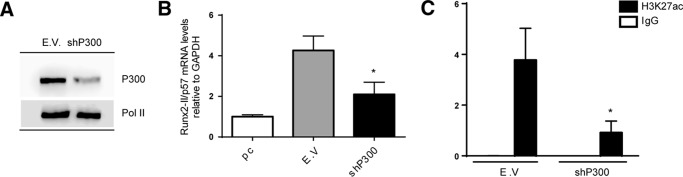
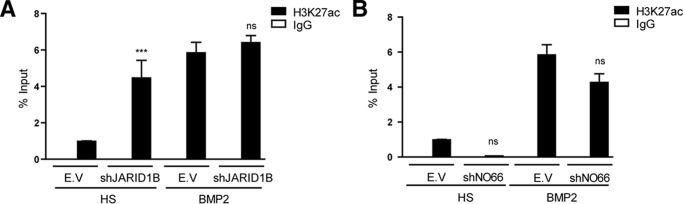
Our NO66 knockdown experiments indicated that an increase in H3K4me3 at the Runx2 P1 promoter in C2C12 cells differentiated to myoblasts is not sufficient to generate Runx2/p57 gene transcription (see Fig. 4, D and E). Therefore, it is necessary to determine whether knockdown of JARID1B (and not of NO66) provides additional epigenetic context at this promoter that facilitates Runx2/p57 transcription. Previous reports indicate that histone H3 acetylation at the Runx2 P1 promoter accompanies Runx2/p57 transcription in osteoblastic cells (15, 17). Interestingly, here we determined that acetylation at the H3K27 residue (H3K27ac), which has been shown to be catalyzed by the co-activator p300 (53, 54), represents a critical epigenetic component associated with Runx2/p57 expression. Thus, shRNA-mediated knockdown of p300 during osteoblast differentiation (Fig. 8A) resulted in a significant reduction of both Runx2/p57 mRNA expression (Fig. 8B) and H3K27ac enrichment at the Runx2 P1 promoter (Fig. 8C). Importantly, depletion of JARID1B (Fig. 9A), but not of NO66 (Fig. 9B), during myogenic differentiation resulted in a significant enrichment of the H3K27ac mark at the Runx2 P1 promoter region. This enrichment is comparable to that obtained in these mesenchymal cells after the induction of osteogenic differentiation. As a control and in agreement with the release of JARID1B from the P1 promoter during osteoblast differentiation (Fig. 2F), shRNA-mediated depletion of this enzyme did not affect the H3K27ac enrichment in cells treated with BMP2 (Fig. 9A). Together, these results indicate that the transcriptional activity of the Runx2/p57 gene necessarily requires elevated H3K27ac at the P1 proximal promoter. Additionally, our data suggest that binding of a JARID1B-containing regulatory complex at this promoter prevents enrichment of the H3K4me3 and H3K27ac marks that accompany transcription of the Runx2/p57 gene.
FIGURE 8.
p300-mediated enrichment of H3K27ac at the Runx2 P1 promoter accompanies Runx2 expression in osteoblasts. C2C12 cells were infected with lentiviral particles coding for shRNAs against p300 or control (E.V.) for 72 h and then differentiated to the osteoblast lineage as described earlier. A, P300 knockdown was confirmed by Western blot using specific antibodies. Detection of RNA Pol II was used to control for equal loading. B, Runx2-II/p57 mRNA levels were measured by qRT-PCR 72h after infection (and 48 h of differentiation) and normalized against GAPDH. C, chromatin samples were analyzed by ChIP using an antibody against H3K27ac. Results are expressed as % input ± S.E., and normal IgG was used as specificity control. Statistical analyses were performed with respect to the cells infected with virus generated with the pLKO.1 empty vector (E.V). *, p < 0.05.
FIGURE 9.
Knockdown of JARID1B results in increased H3K27ac levels at the Runx2 P1 promoter region during myoblast differentiation. C2C12 cells infected with lentiviruses coding for shRNAs against JARID1B (A) or NO66 (B) and differentiated to the myoblast or osteoblast lineages were analyzed by ChIP using antibodies against H3K27Ac. Statistical analyses were performed with respect to the cells infected with virus generated with the pLKO.1 empty vector (E.V). ***, p < 0.001; ns, = non-significant differences.
Discussion
In this work we have addressed epigenetic parameters regulating transcription of the Runx2/p57 bone master gene during commitment of mesenchymal cells toward the osteoblast lineage. We demonstrate the contribution of key components of the Trithorax/COMPASS-like and Polycomb PRC2 complexes, which together with the methylase PRMT5 and demethylase JARID1B mediate deposition and removal of histone marks at the Runx2 P1 proximal promoter region in direct association with expression and silencing of this gene (see Table 1 for a summary).
TABLE 1.
Summary of the effects on Runx2/p57 mRNA expression and enrichment of the H3K4me3, H3K27ac, H3K27me3, and H4R3me2s marks at the Runx2-II/p57 P1 promoter after knockdown of the different chromatin modifying enzymes assessed in this study
ND, not determined during the study.
| Knockdown | Parameter Analyzed | Effect during osteoblast differentiation | Effect during myoblast differentiation |
|---|---|---|---|
| shWDR5 | Runx2 mRNA expression | Inhibited | ND |
| H3K4me3 enrichment | Decreases | ND | |
| shNO66 | Runx2 mRNA expression | Enhanced | Unaffected (remains repressed) |
| H3K4me3 enrichment | Unaffected (remains elevated) | Increases | |
| H3K27ac enrichment | Unaffected (remains elevated) | Unaffected (remains low) | |
| shJARID1B | Runx2 mRNA expression | Enhanced | Enhanced |
| H3K4me3 enrichment | Increases | Increases | |
| H3K27ac enrichment | Unaffected (remains elevated) | Increases | |
| shP300 | Runx2 mRNA expression | Inhibited | ND |
| H3K27ac enrichment | Decreases | ND | |
| shEzh2 | Runx2 mRNA expression | Unaffected (remains elevated) | Unaffected (remains repressed) |
| H3K27me3 enrichment | ND | Decreases | |
| shUTX | Runx2 mRNA expression | Inhibited | ND |
| H3K27me3 enrichment | Increases | ND | |
| shPRMT5 | Runx2 mRNA expression | Unaffected (remains elevated) | Unaffected (remains repressed) |
| H4R3me2s enrichment | ND | Decreases |
We find that there is a critical contribution of JARID1B during the transcriptional repression of the Runx27/p57 gene that accompanies the commitment of mesenchymal multipotent C2C12 cells toward the myoblast lineage. Thus, shRNA-mediated knockdown of this enzyme prevents silencing of the Runx2/p57 gene in cells induced to differentiate to myoblasts. This expression of Runx2/p57 is also accompanied by the expression of late bone phenotypic gene markers like osteocalcin, further indicating that the JARID1B knockdown condition results in an environment in these cells that may facilitate responsiveness to extracellular stimuli leading to osteoblast lineage commitment. Potential biomedical implications of this result are currently under extensive investigation by our group, as targeting JARID1B activity may represent a future therapeutic strategy to promote bone formation in patients suffering from reduced osteoblast differentiation from bone marrow MSCs.
We detected a slight, but statistically significant decrease in the enrichment of JARID1B at the Runx2 P1 promoter in cells differentiated to myoblasts with respect to uncommitted cells. Although any significant conclusion based on this observation will require further experimental analyses, it is tempting to propose that these JARID1B levels are sufficient to maintain the reduced levels of the H3K4me3 mark that accompany Runx2 repression in myoblasts. Nevertheless, it is necessary to also recognize that a reduction in JARID1B binding at the Runx2 promoter may alternatively suggest the contribution of an additional H3K4me3 demethylase (e.g. NO66), which may be operating together with JARID1B to maintain the reduced H3K4me3 enrichment detected at the Runx2 gene in myoblasts.
Similarly, we find that EZH2 enrichment at the Runx2 P1 promoter remains mostly constant (if not slightly reduced) during myoblast differentiation, albeit the levels of the H3K27me3 mark are significantly enhanced. Whether this result implies that PRC2 enzymatic activity (and not necessarily EZH2 binding) is stimulated upon myoblast lineage commitment, will need further clarification. It has been shown that control of the EZH2 activity is a relevant regulatory step during linage commitment of mesenchymal cells. Thus, CDK1-dependent phosphorylation of EZH2 at residue Thr-487 inhibits formation of an active PRC2-EZH2 complex and, therefore, promotes differentiation of human mesenchymal stem cells to osteoblasts (35). Alternatively, an additional contribution to the H3K27me3 enrichment at the Runx2 promoter by a PRC2 complex carrying EZH1 as the catalytic subunit needs to be considered. It has been shown that during myoblast differentiation nuclear PRC2-EZH1 concentrations are drastically increased (55), where this complex is found to mediate important regulatory functions. Remarkably, the increased in H3K27me3 enrichment at the Runx2 P1 promoter during myoblast differentiation occurs in the presence of the H3K27me3 demethylase UTX. This result brings further support to the idea that enhanced PRC2 activity at the Runx2 P1 promoter leads to elevated H3K27me3.
In agreement with previous reports in the literature (17, 34), we find that modifications like H3K4me3, H3K27me3, and H4R3me2s are strongly associated with Runx2/p57 gene expression control. Nevertheless, we additionally determined that, by manipulating their levels at the P1 promoter, each of these marks independently is not sufficient to modulate Runx2/p57 transcription. Thus, although an enrichment of H3K4me3 at the Runx2/p57 P1 proximal promoter region always accompanies Runx2 transcription in osteoblastic cells and that this mark is well recognized as a main epigenetic component of transcriptionally active genes (21, 22), we find that its sole enrichment does not result in transcriptional activation of Runx2/p57 in mesenchymal cells differentiated to myoblasts. Importantly, this inability of H3K4me3 to promote Runx2/p57 mRNA expression is observed even when the P1 promoter exhibits reduced levels of H3K27me3 (and/or reduced H3K9me34). In recent years there has been an exciting discussion (e.g. see Voigt et al. (56)) about the role of epigenetic bivalency (H3K4me3/H3K27me3; Ref. 51) at proximal gene regulatory regions and the potential of these genes to be subsequently expressed. There seems to be strong line of evidence indicating that a bivalent and repressed gene condition (as in the HOX gene cluster) in embryonic stem cells can rapidly turn into a anH3K4me3-rich/K27me3-poor condition that leads to transcriptional activity (27). Our results, in contrast, indicate that the presence of this H3K4me3-rich/K27me3-poor condition in the Runx2 P1 promoter does not produce transcription of this osteoblast master regulator. Rather, our results support a working model (see Fig. 10), where erasing the H3K27me3 mark in the P1 promoter needs to be followed by deposition of H3K27ac, thus generating a chromatin context that facilitates Runx2/p57 transcriptional activity. This model is in agreement with previous findings from our group and from various other investigators, indicating that Runx2/p57 transcription involves chromatin remodeling events at the proximal P1 promoter region that are both independent of SWI/SNF activity (15) and strongly dependent of histone acetylation (15, 17, 57, 58).
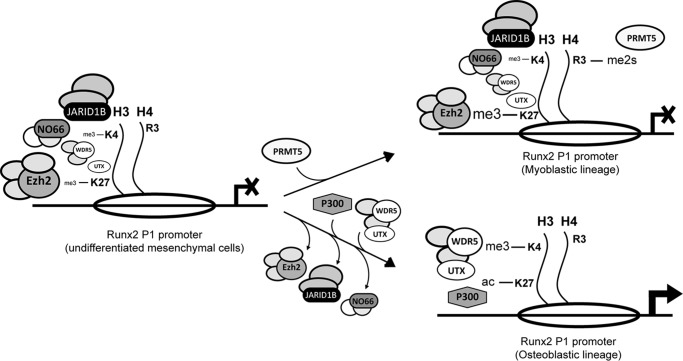
FIGURE 10.
Proposed model describing epigenetic regulation of the Runx2-II/p57 gene in differentiating mesenchymal cells. Changes in histone marks and binding of different chromatin modifiers at the proximal Runx2 P1 promoter during osteoblast or myoblast lineage commitment are indicated. The arrowhead represents the transcription start site and the open ellipses nucleosomes. The size of the figures representing the different complexes bound to the P1 promoter indicates their relative levels of enrichment.
We are currently investigating whether there is a coordinated mechanism that prevents the presence of the H3K4me3/H3K27ac marks when mesenchymal precursor cells differentiate to the myoblastic lineage. Importantly, JARID1B has been shown to form multienzymatic complexes that cooperate to mediate gene repression. These complexes can include, in addition to JARID1B, histone deacetylases (40) as well as EZH2 (41). We propose the existence of a coordinated mechanism (Fig. 10) where recruitment of a complex containing all three JARID1B/histone deacetylase/EZH2 activities to the P1 promoter region results in decreased H3K4me3 and H3ac marks together with an increase in the H3K27me3 mark. Our results indicating that shRNA-mediated knockdown of JARID1B not only results in increased H3K4me3 but also in elevated H3K27ac strongly supports this hypothesis.
Reports from several groups have established that H3K27ac at gene promoters and enhancers is principally mediated by the intrinsic HAT activity of the transcriptional co-activator p300/CBP (53, 59, 60). Our results also support this relevant role of p300 in Runx2 expression, as this protein contributes to maintain H3K27ac levels at the P1 promoter and Runx2/p57 transcription in osteoblastic cells. It remains to be determined whether a coordinated deposition of H3K4me3 and H3K27ac together with elimination of H3K27me3 at the Runx2 P1 promoter is mediated by a complex that includes Trithorax/COMPASS and p300; such a complex would be recruited during mesenchymal cell commitment to the osteoblast lineage (Fig. 10). Previous studies have reported the formation of complexes containing UTX and CBP that bind to specific target genes and promote transcription by increasing H3K27ac (61). Nonetheless, the mechanism mediating the specific recruitment of all of these complexes to the Runx2 P1 promoter remains to be elucidated.
PRMT5-mediated symmetric di-methylation of H4R3 has been involved in transcriptional repression (62–64). Surprisingly, we find that although PRMT5 is bound to, and H4R3me2s is strongly enriched at the Runx2 P1 proximal promoter in cells that do not express Runx2/p57, the contribution of this histone mark is not required during silencing of this gene. The ability of PRMT5 to repress transcription has been mainly attributed to its interaction with DNA methyl transferases like Dnmt3A (65). Moreover, a recent report indicates that PRMT5 has a critical role during suppression of transposable elements in pre-implantation embryos (63). Therefore, one could hypothesize that as the Runx2 P1 promoter sequence is not CpG-methylated during transcriptional repression (66), PRMT5 function as an H4R3 symmetric di-methylase may be disposable during commitment to the myoblastic lineage.
In summary, our results provide an understanding of the epigenetic components that support commitment of mesenchymal cells to the osteoblast lineage. Knowledge of the enzymes that contribute to an epigenetic program of histone modifications that control bone-related genes can potentially lead to new therapeutic targets to treat de-regulations associated with skeletal disorders.
Author Contributions
A. R., B. H., and R. A. designed and performed the experiments and analyzed the data. J. B. L., J. L. S., G. S. S., A. J. v W., and B. v Z. provided technical and conceptual advice and analyzed the results. M. L. A. and M. M. supervised the research and analyzed the data. A. R., R. A., and M. M. wrote the main parts of the manuscript.
This work was supported by grants from Fondo de Finaciamiento de Centros de Investigación en Áreas Prioritarias (15090007; to M. L. A. and M. M.), Fondo Nacional de Ciencia y Tecnologia (FONDECYT 1130706; to M. M.), and NIH Grants R01 AR049069 (to A. J. vW.) and R01 AR039588 (to G. S. S.). The authors declare that they have no conflicts of interest with the contents of this article
R. Aguilar, B. Henriquez, B. van Zundert, and M. Montecino, unpublished results.
- BMP
- bone morphogenetic protein
- TrxG
- Trithorax group
- PcG
- Polycomb Group
- MLL
- mixed lineage leukemia
- MSC
- mesenchymal stem cell
- AP
- alkaline phosphatase
- qRT
- quantitative real-time PCR
- pC
- pre-confluent
- HS
- horse serum
- EV
- empty vector.
References
- 1.Lian J. B., Javed A., Zaidi S. K., Lengner C., Montecino M., van Wijnen A. J., Stein J. L., and Stein G. S. (2004) Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit. reverse. Eukaryot. Gene Expr. 14, 1–41 [PubMed] [Google Scholar]
- 2.Marie P. (2008) Transcription factors controlling osteoblastogenesis. Arch. Biochem. Biophys. 473, 98–105 [DOI] [PubMed] [Google Scholar]
- 3.Stein G. S., Lian J. B., van Wijnen A. J., Stein J. L., Montecino M., Javed A., Zaidi S. K., Young D. W., Choi J. Y., and Pockwinse S. M. (2004) Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene 23, 4315–4329 [DOI] [PubMed] [Google Scholar]
- 4.Komori T., Yagi H., Nomura S., Yamaguchi A., Sasaki K., Deguchi K., Shimizu Y., Bronson R. T., Gao Y. H., Inada M., Sato M., Okamoto R., Kitamura Y., Yoshiki S., and Kishimoto T. (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89, 755–764 [DOI] [PubMed] [Google Scholar]
- 5.Otto F., Thornell A. P., Crompton T., Denzel A., Gilmour K. C., Rosewell I. R., Stamp G. W., Beddington R. S., Mundlos S., Olsen B. R., Selby P. B., and Owen M. J. (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89, 765–771 [DOI] [PubMed] [Google Scholar]
- 6.Otto F., Kanegane H., and Mundlos S. (2002) Mutations in the RUNX2 gene in patients with cleidocranial dysplasia. Hum. Mut. 19, 209–216 [DOI] [PubMed] [Google Scholar]
- 7.Drissi H., Luc Q., Shakoori R., Chuva De Sousa Lopes S., Choi J. Y., Terry A., Hu M., Jones S., Neil J. C., Lian J. B., Stein J. L., Van Wijnen A. J., and Stein G. S. (2000) Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J. Cell. Physiol. 184, 341–350 [DOI] [PubMed] [Google Scholar]
- 8.Zambotti A., Makhluf H., Shen J., and Ducy P. (2002) Characterization of an osteoblast-specific enhancer element in the CBFA1 gene. J. Biol. Chem. 277, 41497–41506 [DOI] [PubMed] [Google Scholar]
- 9.Lengner C. J., Hassan M. Q., Serra R. W., Lepper C., van Wijnen A. J., Stein J. L., Lian J. B., and Stein G. S. (2005) Nkx3.2-mediated repression of Runx2 promotes chondrogenic differentiation. J. Biol. Chem. 280, 15872–15879 [DOI] [PubMed] [Google Scholar]
- 10.Gaur T., Lengner C. J., Hovhannisyan H., Bhat R. A., Bodine P. V., Komm B. S., Javed A., van Wijnen A. J., Stein J. L., Stein G. S., and Lian J. B. (2005) Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J. Biol. Chem. 280, 33132–33140 [DOI] [PubMed] [Google Scholar]
- 11.Lee M. H., Kim Y. J., Yoon W. J., Kim J. I., Kim B. G., Hwang Y. S., Wozney J. M., Chi X. Z., Bae S. C., Choi K. Y., Cho J. Y., Choi J. Y., and Ryoo H. M. (2005) Dlx5 specifically regulates Runx2 type II expression by binding to homeodomain-response elements in the Runx2 distal promoter. J. Biol. Chem. 280, 35579–35587 [DOI] [PubMed] [Google Scholar]
- 12.Hassan M. Q., Tare R. S., Lee S. H., Mandeville M., Morasso M. I., Javed A., van Wijnen A. J., Stein J. L., Stein G. S., and Lian J. B. (2006) BMP2 commitment to the osteogenic lineage involves activation of Runx2 by DLX3 and a homeodomain transcriptional network. J. Biol. Chem. 281, 40515–40526 [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y., Hassan M. Q., Xie R.-L., Hawse J. R., Spelsberg T. C., Montecino M., Stein J. L., Lian J. B., van Wijnen A. J., and Stein G. S. (2009) Co-stimulation of the bone-related Runx2 P1 promoter in mesenchymal cells by SP1 and ETS transcription factors at polymorphic purine-rich DNA sequences (Y-repeats). J. Biol. Chem. 284, 3125–3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henriquez B., Hepp M., Merino P., Sepulveda H., van Wijnen A. J., Lian J. B., Stein G. S., Stein J. L., and Montecino M. (2011) C/EBPβ binds the P1 promoter of the Runx2 gene and up-regulates Runx2 transcription in osteoblastic cells. J. Cell. Physiol. 226, 3043–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cruzat F., Henriquez B., Villagra A., Hepp M., Lian J. B., van Wijnen A. J., Stein J. L., Imbalzano A. N., Stein G. S., and Montecino M. (2009) SWI/SNF-independent nuclease hypersensitivity and an increased level of histone acetylation at the P1 promoter accompany active transcription of the bone master gene Runx2. Biochemistry 48, 7287–7295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hovhannisyan H., Zhang Y., Hassan M. Q., Wu H., Glackin C., Lian J. B., Stein J. L., Montecino M., Stein G. S., and van Wijnen A. J. (2013) Genomic occupancy of HLH, AP1 and Runx2 motifs within a nuclease-sensitive site of the Runx2 gene. J. Cell. Physiol. 228, 313–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tai P. W., Wu H., Gordon J. A., Whitfield T. W., Barutcu A. R., van Wijnen A. J., Lian J. B., Stein G. S., and Stein J. L. (2014) Epigenetic landscape during osteoblastogenesis defines a differentiation-dependent Runx2 promoter region. Gene 550, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., and Cavalli G. (2007) Genome regulation by Polycomb and Trithorax proteins. Cell 128, 735–745 [DOI] [PubMed] [Google Scholar]
- 19.Ringrose L., and Paro R. (2007) Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 134, 223–232 [DOI] [PubMed] [Google Scholar]
- 20.Shilatifard A. (2012) The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. reverse. Biochem. 81, 65–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li B., Carey M., and Workman J. L. (2007) The role of chromatin during transcription. Cell 128, 707–719 [DOI] [PubMed] [Google Scholar]
- 22.Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 23.Nagy P. L., Griesenbeck J., Kornberg R. D., and Cleary M. L. (2002) A Trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc. Natl. Acad. Sci. U.S.A. 99, 90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dou Y., Milne T. A., Ruthenburg A. J., Lee S., Lee J. W., Verdine G. L., Allis C. D., and Roeder R. G. (2006) Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 13, 713–719 [DOI] [PubMed] [Google Scholar]
- 25.Wysocka J., Swigut T., Milne T. A., Dou Y., Zhang X., Burlingame A. L., Roeder R. G., Brivanlou A. H., and Allis C. D. (2005) WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121, 859–872 [DOI] [PubMed] [Google Scholar]
- 26.Lee M. G., Villa R., Trojer P., Norman J., Yan K.-P., Reinberg D., Di Croce L., and Shiekhattar R. (2007) Demethylation of H3K27 regulates Polycomb recruitment and H2A ubiquitination. Science 318, 447–450 [DOI] [PubMed] [Google Scholar]
- 27.Agger K., Cloos P. A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A. E., and Helin K. (2007) UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449, 731–734 [DOI] [PubMed] [Google Scholar]
- 28.Margueron R., and Reinberg D. (2011) The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao R., and Zhang Y. (2004) The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 14, 155–164 [DOI] [PubMed] [Google Scholar]
- 30.Gori F., Divieti P., and Demay M. B. (2001) Cloning and characterization of a novel WD-40 repeat protein that dramatically accelerates osteoblastic differentiation. J. Biol. Chem. 276, 46515–46522 [DOI] [PubMed] [Google Scholar]
- 31.Gori F., and Demay M. B. (2004) BIG-3, a novel WD-40 repeat protein, is expressed in the developing growth plate and accelerates chondrocyte differentiation in vitro. Endocrinology 145, 1050–1054 [DOI] [PubMed] [Google Scholar]
- 32.Gori F., Friedman L. G., and Demay M. B. (2006) Wdr5, a WD-40 protein, regulates osteoblast differentiation during embryonic bone development. Dev. Biol. 295, 498–506 [DOI] [PubMed] [Google Scholar]
- 33.Zhu E. D., Demay M. B., and Gori F. (2008) Wdr5 is essential for osteoblast differentiation. J. Biol. Chem. 283, 7361–7367 [DOI] [PubMed] [Google Scholar]
- 34.Hemming S., Cakouros D., Isenmann S., Cooper L., Menicanin D., Zannettino A., and Gronthos S. (2014) EZH2 and KDM6A act as an epigenetic switch to regulate mesenchymal stem cell lineage specification. Stem Cells 32, 802–815 [DOI] [PubMed] [Google Scholar]
- 35.Wei Y., Chen Y. H., Li L. Y., Lang J., Yeh S. P., Shi B., Yang C. C., Yang J. Y., Lin C. Y., Lai C. C., and Hung M. C. (2011) CDK1-dependent phosphorylation of EZH2 suppresses methylation of H3K27 and promotes osteogenic differentiation of human mesenchymal stem cells. Nat. Cell Biol. 13, 87–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ye L., Fan Z., Yu B., Chang J., Al Hezaimi K., Zhou X., Park N. H., and Wang C. Y. (2012) Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell 11, 50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sinha K. M., Yasuda H., Coombes M. M., Dent S. Y., and de Crombrugghe B. (2010) Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. EMBO J. 29, 68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sinha K. M., and Zhou X. (2013) Genetic and molecular control of osterix in skeletal formation. J. Cell. Biochem. 114, 975–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Secombe J., Li L., Carlos L., and Eisenman R. N. (2007) The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev. 21, 537–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barrett A., Santangelo S., Tan K., Catchpole S., Roberts K., Spencer-Dene B., Hall D., Scibetta A., Burchell J., Verdin E., Freemont P., and Taylor-Papadimitriou J. (2007) Breast cancer associated transcriptional repressor PLU-1/JARID1B interacts directly with histone deacetylases. Int. J. Cancer 121, 265–275 [DOI] [PubMed] [Google Scholar]
- 41.Banck M. S., Li S., Nishio H., Wang C., Beutler A. S., and Walsh M. J. (2009) The ZNF217 oncogene is a candidate organizer of repressive histone modifiers. Epigenetics 4, 100–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao Z. S., Hjelmeland A. B., and Quarles L. D. (2004) Selective deficiency of the “bone-related” Runx2-II unexpectedly preserves osteoblast-mediated skeletogenesis. J. Biol. Chem. 279, 20307–20313 [DOI] [PubMed] [Google Scholar]
- 43.Yaffe D., and Saxel O. (1977) Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature 270, 725–727 [DOI] [PubMed] [Google Scholar]
- 44.Katagiri T., Yamaguchi A., Komaki M., Abe E., Takahashi N., Ikeda T., Rosen V., Wozney J. M., Fujisawa-Sehara A., and Suda T. (1994) Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J. Cell Biol. 127, 1755–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paredes R., Arriagada G., Cruzat F., Villagra A., Olate J., Zaidi K., van Wijnen A., Lian J. B., Stein G. S., Stein J. L., and Montecino M. (2004) Bone-specific transcription factor Runx2 interacts with the 1α,25-dihydroxyvitamin D3 receptor to up-regulate rat osteocalcin gene expression in osteoblastic cells. Mol. Cell. Biol. 24, 8847–8861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soutoglou E., and Talianidis I. (2002) Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295, 1901–1904 [DOI] [PubMed] [Google Scholar]
- 47.Zeng P. Y., Vakoc C. R., Chen Z. C., Blobel G. A., and Berger S. L. (2006) In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. Biotechniques 41, 694. [DOI] [PubMed] [Google Scholar]
- 48.Lee M. H., Javed A., Kim H. J., Shin H. I., Gutierrez S., Choi J. Y., Rosen V., Stein J. L., van Wijnen A. J., Stein G. S., Lian J. B., and Ryoo H. M. (1999) Transient upregulation of CBFA1 in response to bone morphogenetic protein-2 and transforming growth factor β1 in C2C12 myogenic cells coincides with suppression of the myogenic phenotype but is not sufficient for osteoblast differentiation. J. Cell. Biochem. 73, 114–125 [PubMed] [Google Scholar]
- 49.Balint E., Lapointe D., Drissi H., van der Meijden C., Young D. W., van Wijnen A. J., Stein J. L., Stein G. S., and Lian J. B. (2003) Phenotype discovery by gene expression profiling: mapping of biological processes linked to BMP-2-mediated osteoblast differentiation. J. Cell. Biochem. 89, 401–426 [DOI] [PubMed] [Google Scholar]
- 50.Wright W. E., Sassoon D. A., and Lin V. K. (1989) Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell 56, 607–617 [DOI] [PubMed] [Google Scholar]
- 51.Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., and Lander E. S. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 52.Banerjee C., Hiebert S. W., Stein J. L., Lian J. B., and Stein G. S. (1996) An AML-1 consensus sequence binds an osteoblast-specific complex and transcriptionally activates the osteocalcin gene. Proc. Natl. Acad. Sci. U.S.A. 93, 4968–4973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tie F., Banerjee R., Stratton C. A., Prasad-Sinha J., Stepanik V., Zlobin A., Diaz M. O., Scacheri P. C., and Harte P. J. (2009) CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136, 3131–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ogryzko V. V., Schiltz R. L., Russanova V., Howard B. H., and Nakatani Y. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87, 953–959 [DOI] [PubMed] [Google Scholar]
- 55.Stojic L., Jasencakova Z., Prezioso C., Stützer A., Bodega B., Pasini D., Klingberg R., Mozzetta C., Margueron R., Puri P. L., Schwarzer D., Helin K., Fischle W., and Orlando V. (2011) Chromatin regulated interchange between Polycomb repressive complex 2 (PRC2)-Ezh2 and PRC2-Ezh1 complexes controls myogenin activation in skeletal muscle cells. Epigenetics Chromatin 4, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Voigt P., Tee W. W., and Reinberg D. (2013) A double take on bivalent promoters. Genes Dev. 27, 1318–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee H. W., Suh J. H., Kim A. Y., Lee Y. S., Park S. Y., and Kim J. B. (2006) Histone deacetylase 1-mediated histone modification regulates osteoblast differentiation. Mol. Endocrinol. 20, 2432–2443 [DOI] [PubMed] [Google Scholar]
- 58.Bradley E. W., McGee-Lawrence M. E., and Westendorf J. J. (2011) Hdac-mediated control of endochondral and intramembranous ossification. Crit. reverse. Eukaryot. Gene Expr. 21, 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pasini D., Malatesta M., Jung H. R., Walfridsson J., Willer A., Olsson L., Skotte J., Wutz A., Porse B., Jensen O. N., and Helin K. (2010) Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 38, 4958–4969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schwartz Y. B., Kahn T. G., Stenberg P., Ohno K., Bourgon R., and Pirrotta V. (2010) Alternative epigenetic chromatin states of Polycomb target genes. PLoS Genet. 6, e1000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tie F., Banerjee R., Conrad P. A., Scacheri P. C., and Harte P. J. (2012) Histone demethylase UTX and chromatin remodeler BRM bind directly to CBP and modulate acetylation of histone H3 lysine 27. Mol. Cell. Biol. 32, 2323–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fabbrizio E., El Messaoudi S., Polanowska J., Paul C., Cook J. R., Lee J. H., Negre V., Rousset M., Pestka S., Le Cam A., and Sardet C. (2002) Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 3, 641–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim S., Günesdogan U., Zylicz J. J., Hackett J. A., Cougot D., Bao S., Lee C., Dietmann S., Allen G. E., Sengupta R., and Surani M. A. (2014) PRMT5 protects genomic integrity during global DNA demethylation in primordial germ cells and preimplantation embryos. Mol. Cell 56, 564–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pal S., Vishwanath S. N., Erdjument-Bromage H., Tempst P., and Sif S. (2004) Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 24, 9630–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao Q., Rank G., Tan Y. T., Li H., Moritz R. L., Simpson R. J., Cerruti L., Curtis D. J., Patel D. J., Allis C. D., Cunningham J. M., and Jane S. M. (2009) PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat. Struct. Mol. Biol. 16, 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ezura Y., Sekiya I., Koga H., Muneta T., and Noda M. (2009) Methylation status of CpG islands in the promoter regions of signature genes during chondrogenesis of human synovium-derived mesenchymal stem cells. Arthritis Rheum. 60, 1416–1426 [DOI] [PubMed] [Google Scholar]