Background: PLOD2 expression and subsequent collagen pyridinoline cross-links are induced in fibrotic tissues by TGFβ1.
Results: TGFβ1 induces changes to histone modifications. SP1 and SMAD3 but not P300/CBP are responsible for PLOD2 induction.
Conclusion: SMAD3 regulates TGFβ1-induced PLOD2 expression via histone-modifying enzymes other than the currently known downstream effectors.
Significance: Unraveling PLOD2 transcriptional regulation would provide new ways to interfere in fibrotic processes.
Keywords: E1A binding protein p300 (P300), epigenetics, fibrosis, histone deacetylase 1 (HDAC1), histone modification, SMAD transcription factor, transcription coactivator, transforming growth factor beta (TGF-B)
Abstract
PLOD2 (procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2) hydroxylates lysine residues in collagen telopeptides and is essential for collagen pyridinoline cross-link formation. PLOD2 expression and subsequent pyridinoline cross-links are increased in fibrotic pathologies by transforming growth factor β-1 (TGFβ1). In this report we examined the molecular processes underlying TGFβ1-induced PLOD2 expression. We found that binding of the TGFβ1 pathway related transcription factors SMAD3 and SP1-mediated TGFβ1 enhanced PLOD2 expression and could be correlated to an increase of acetylated histone H3 and H4 at the PLOD2 promoter. Interestingly, the classical co-activators of SMAD3 complexes, p300 and CBP, were not responsible for the enhanced H3 and H4 acetylation. Depletion of SMAD3 reduced PLOD2 acetylated H3 and H4, indicating that another as of yet unidentified histone acetyltransferase binds to SMAD3 at PLOD2. Assessing histone methylation marks at the PLOD2 promoter depicted an increase of the active histone mark H3K79me2, a decrease of the repressive H4K20me3 mark, but no role for the generally strong transcription-related modifications: H3K4me3, H3K9me3 and H3K27me3. Collectively, our findings reveal that TGFβ1 induces a SP1- and SMAD3-dependent recruitment of histone modifying enzymes to the PLOD2 promoter other than the currently known TGFβ1 downstream co-activators and epigenetic modifications. This also suggests that additional activation strategies are used downstream of the TGFβ1 pathway, and hence their unraveling could be of great importance to fully understand TGFβ1 activation of genes.
Introduction
Resolving tissue injury is a multifactorial process that often ends with scarring, a process also known as fibrosis. A common denominator of fibrosis is the presence of elevated levels of fibrillar collagen type I, which is deposited by fibroblasts that are activated by profibrotic cytokines such as transforming growth factor β-1 (TGFβ1)3 (1–3). Examination of collagen derived from tissues and fibroblasts isolated from patient material show enhanced levels of pyridinoline cross-links that function to stabilize collagen fibrils (4–7). Formation of these cross-links is initiated by procollagen-lysine 2-oxoglutarate 5-dioxygenase 2 (PLOD2), which specifically hydroxylates lysine residues of the collagen telopeptide area (4, 5). Lysyl hydroxylase activities of the other family members, PLOD1 and PLOD3, are restricted to helical region (8). The presence of helical hydroxylysines are not required for pyridinoline cross-link formation, whereas the telopeptide hydroxylysines are a prerequisite (9). After collagen deposition in the extracellular space, lysyl oxidases catalyze the formation of pyridinoline cross-links by converting the telopeptide hydroxylysyl into hydroxyallysyl (10, 11). Collagen containing pyridinoline cross-links are difficult to degrade by collagenases and thereby are able to accumulate at the fibrotic lesion (12). This enables a possible feedback loop whereby the enhanced collagen deposition increases tissue stiffness, and in turn stimulates myofibroblast differentiation and additional collagen production (13–15). Reducing the levels of pyridinoline cross-links in scarring pathologies may have the potential to circumvent this fibrotic feedback loop and enable the formation of a non-fibrotic matrix. Therefore, understanding transcriptional regulation of PLOD2 is essential for paving the way to therapeutically prevent pyridinoline cross-links in scarred tissues.
Epigenetic modifications are generally described as chemical changes to chromatin that are able to affect the transcriptional state of a gene. Recent evidence indicates that in addition to transcription factors, changes in epigenetic modifications of chromatin (DNA and histones) have a fundamental role in the initiation and progression of fibrotic pathologies (16–21). DNA methylation is catalyzed by DNA methyltransferases DNMT1, 3A and 3B in a CpG context and is implicated to have determining roles during embryonic development, and is linked to various diseases such as cancer (22, 23). Hypermethylated CpG islands represented in the promoter or first exon of protein-coding genes can have strong repressive outcomes on its transcriptional activity (24). Many types of posttranslational histone modifications have been recognized, and each type of modification and their specific location on the histone can have various outcomes on transcriptional activity of genes (25, 26). Histone lysine methylation and acetylation are among the most extensively studied histone modifications, and are widely found on the histone tails extruding from the nucleosome. Both types of chemical modifications are able to affect the compaction of chromatin either directly via charge differences of the histone tails that are connected to the DNA, or indirect by attracting chromatin remodeling complexes. These changes in chromatin compaction are thought to interfere with transcriptional activity by affecting the accessibility of transcription factors and transcription initiation complexes or even elongation of RNA polymerase II complexes (27–30). The acetylation status of histones are balanced by histone acetyltransferases (HATs) that add acetyl groups and are related to gene activation, whereas histone deacetylases (HDACs) that cleave acetyl groups (31, 32) are related to gene repression. Histone methylation is unique compared with most other types of modifications since they can occur in a mono-, di-, or tri-methyl form. For instance, tri-methylated lysine 4 of histone 3 (H3K4me3) and H3K79me2 present in a promoter of a gene are correlated to gene expression, whereas H3K9me3, H3K27me3, and H4K20me3 are correlated to gene repression. The large repertoire of methylated lysine possibilities are normally balance by numerous histone lysine methyltransferases (HKMTs) and lysine demethylases (KDMs) that, respectively, add or remove methyl groups from a histone lysine (27, 33–35).
PLOD2 expression and subsequent pyridinoline cross-linking by fibroblasts is strongly enhanced by members of the TGFβ family, most notably by TGFβ1 (36–38). However, it is not clear how PLOD2 is transcriptionally regulated in activated fibroblasts in relation to scarring processes, e.g. which transcription factors and epigenetic factors play a role. In this study, we report that TGFβ1-induced PLOD2 expression is accompanied by a dynamic change of several histone modifications, and that binding of the transcription factors SMAD3 and SP1 to PLOD2 is essential to these processes.
Experimental Procedures
Cell Culture
Human (adult) skin fibroblasts (ATCC: CCD-1093 SK), were cultured up to 12 passages in EMEM (Lonza, Basel, Switzerland) supplemented with penicillin/streptomycin (Life Technologies, Carlsbad, CA), 2 mm l-glutamine (Lonza), and 10% heat-inactivated fetal calf serum (FCS) (Thermo Scientific, Waltham, MA) and incubated at 37 °C with 5% CO2. Recombinant human TGFβ1 (PeproTech, Rocky Hill, NJ) was reconstituted in 10 mm citric acid pH 3.0 and diluted 20-fold by PBS supplemented with 0.1% bovine serum albumin to reach a concentration of 5 μg ml−1. Cells were serum-starved with medium containing 0.5% FCS 18 h prior to TGFβ1 stimulations. Medium containing TGFβ1 (10 ng ml−1) or equal amount of vehicle control was refreshed daily.
Western Blotting
Total protein was extracted with RIPA buffer (Thermo Scientific) and separated by SDS-PAGE and transferred to nitrocellulose membranes. The blots were incubated for 2 h at room temperature with primary antibodies against PLOD2 (R&D systems, Minneapolis, MN), SMAD3 (Abcam, Cambridge, UK), ASH1L (Santa Cruz Biotechnology, Dallas, TX), and YWHAZ (Abcam) as a loading control. Next, the blots were incubated for 1 h with secondary antibodies goat-anti-rabbit-HRP or rabbit-anti-mouse-HRP (DAKO, Glostrup, Denmark), after which chemiluminescence was detected with ECL (Thermo Scientific).
Cloning, Electroporation, and Transient Transfection
PLOD2 promoter fragments (kind gift of Dr. R. Stoop, TNO, Leiden, The Netherlands) were subcloned into the CpG-free reporter plasmid pCpGL (obtained from Dr. M. Rehli, Regensburg, Germany) with restriction enzymes SacI and HindIII and were transformed into PIR1 competent bacteria. Sequence validated plasmids were electroporated with the NHDF Nucleofection kit (Lonza). In total 3 μg plasmids, considering vector sizes and adjusted with pFASTBAC (Life Technologies), as well as 10% of total plasmid concentration of pCMV-LacZ (Clontech, Mountain View, CA) was added as a transfection control. All plasmids were mixed with 100 μl transfection reagent per 6 × 105 fibroblasts and electroporated with a Nucleofector device. Afterward, cell were seeded in fresh medium and allowed to recover for 24 h. For overexpression experiments, plasmids containing cDNA of SP1 (SC101137; OriGene Technologies, Rockville, MD) and SMAD3 (SC321871; OriGene Technologies) were transfected into fibroblasts with Lipofectamine Plus LTX (Life Technologies). For knockdown experiments, esiRNA against SMAD3, ASH1L, CBP, P300, or eGFP-control (Sigma Aldrich) was transfected into fibroblasts with RNAiMAX Lipofectamine (Life Technologies).
Luciferase/LacZ Reporter Assay
After 48 h of TGFβ1 stimulation, electroporated fibroblasts were washed with PBS and lysed with passive lysis buffer (Promega, Madison, WI) followed by two freeze thaw cycles. Firefly luciferase activity was determined with LARII substrate (Promega), and measured in a luminescence reader. β-Galactosidase activity, used as control for transfection efficiency, was detected by adding equal amounts of lysate and assay buffer (Promega) to a 96-well plate and incubated at 37 °C until visible change of color. The absorbance was measured at 420 nm. Relative luciferase activity was calculated by dividing absolute values from firefly luciferase by LacZ.
Molecular Inhibition Assays
SB431542, C646, Suberoylanilide hydroxamic acid (SAHA), SIS3, and Mithramycin A (all from Sigma Aldrich) were dissolved in DMSO. Fibroblasts were seeded at 15 × 103/cm2. For SAHA treatment, the day after seeding 0.5% FCS containing medium supplemented with SAHA was added to the cells for 72 h, and refreshed daily. For chemical inhibition of SP1, SMAD3, P300, and ALK5, cells were pre-incubated 1 h with the inhibitors SIS3, Mithramycin A, C646, or SB-431542, respectively, after which medium containing the inhibitor and TGFβ1 or vehicle control was added for additional 24 h. In all cases equal volumes of DMSO were added as a control.
Quantitative Real-time PCR
RNA was isolated with the RNeasy Plus mini kit (Qiagen, Venlo, The Netherlands) and cDNA was synthesized by reverse transcription with the RevertAid kit (Thermo Scientific). Standardized amounts of cDNA were combined with a mix of Sybr Green (Bio-Rad) and the following primers: PLOD1, 5′-GAAGCTCTACCCCGGCTACT-3′ (forward) and 5′-CTTGTAGCGGACGACAAAGG-3′(reverse); PLOD2, 5′-GGGAGTTCATTGCACCAGTT-3′ (forward) and 5′-GAGGACGAAGAGAACGCTGT-3′ (reverse); PLOD3, 5′-CACTACGGCCAGTGGTCAG-3′ (forward)and 5′-.GTGGGCACATTCTCGTAGC-3′ (reverse); CBP, 5′-GGAGAGGAAAAAGGAAGAGAGC-3′ (forward) and 5′-CTTGCTGTCGCCCTGACT-3′ (reverse); P300, 5′-GATCTGTGTCCTTCACCATGAG (forward) and 5′-AAACAGCCATCACAGACGAA-3′ (reverse). GAPDH, 5′-AGCCACATCGCTCAGACAC-3′ (forward) and 5′-GCCCAATACGACCAAATCC-3′ (reverse); Quantitative real time PCR analysis was performed with the ViiA7 platform (Life Technologies). Expression values were normalized to GAPDH values using the ΔΔCt method.
Chromatin Immunoprecipitation
Cells treated with TGFβ1 or vehicle were harvested and counted, after which they were fixed in 1% formaldehyde for 8 min at room temperature and quenched with 125 mm glycine. The cells were lysed on ice for 20 min with SDS lysis buffer (1% SDS, 50 mm Tris-HCl pH 8.0, 10 mm EDTA) supplemented with proteinase inhibitor mixture (Sigma Aldrich), PMSF and sodium butyrate and sonicated with a Bioruptor® device (Diagenode, Liège, Belgium) to fragment the chromatin. After clearing the chromatin by centrifugation, the supernatant was diluted 10 fold with RIPA buffer (0.1% SDS, 0.1% Na-deoxycholate, 1% Triton-X100, 1 mm EDTA, 10 mm Tris-HCl pH 7.5, 140 mm NaCl, 0.5 mm EGTA) supplemented with proteinase and phosphatase inhibitor mixture, PMSF and sodium butyrate. Next, 40 μl Protein-G or -A Dynabeads® (Life Technologies) were coated with 5 μg antibodies per ChIP reaction; H3ac (Merck Millipore, Billerica, MA), H4ac (Merck Millipore), H3K4me3 (Merck Millipore), H3K9me3 (Abcam), H3K27me3 (Merk Millipore), H3K79me2 (Abcam), H4K20me3 (Abcam), CBP (Abcam), P300 (Merck Millipore), HDAC1 (Santa Cruz Biotechnology), HDAC2 (Santa Cruz Biotechnology), SP1 (Abcam), SMAD3 (Abcam), RNA pol II CTD repeat (phospho S5) (Abcam) and normal rIgG (Abcam) as control. The sheared chromatin was incubated with antibody-bound beads overnight at 4 °C. Chromatin of 0.250 × 106 fibroblasts per ChIP was used to detect histone modifications, whereas for transcription factors and other histone modifying enzymes 1 × 106 cells were used. Next day, the beads were washed three times after which DNA-protein complexes were eluted from the beads. Cross-links of eluted complexes were reversed with NaCl and treatment with RNase for 4 h at 62 °C (Roche, Basel, Switzerland) and subsequently treated with Proteinase K (Roche) for 1 h at 62 °C. ChIP DNA fragments were isolated by spin column purification (Qiagen) and quantified by qRT-PCR, with ROX mix (Abgene, Surrey, UK) and the following primers and probe: PLOD2 −73/+50, 5′-GAGCAAATTCTCACCCTTCG-3′ (forward), 5′-TTTGGGAGAGGGAGGAGGA-3′ (reverse) and 5′-[FAM]AGACGGGAACACCGCCCTCC[TAMRA] (probe); PLOD2 −792/−657, 5′-ACAAAACGTGATCATAATGGAA-3′ (forward), 5′-TTCTGGAATCTCCTGCCTAAAT-3′ (reverse) and 5′-FAM]TCAAAGGCCCAGAGTTATAACGGGTG[TAMRA]-3′ (probe). qChIP data were calculated as percent of input.
Bisulfite Sequencing
Genomic DNA, isolated by chloroform-ethanol with subsequent Proteinase K and RNase treatment, was bisulfite converted with the Methyl Gold kit (Zymo Research). Amplified fragments of PLOD2 with TA overhang were obtained by a PCR reaction with primers against region I: 5′-TTAAAGTTAAGTGTAGGTTTTT-3′ (forward) and 5′-AAAACAACAACTAAAACTTC-3′ (reverse); region II, 5′-GATTTGATGAGTTAAAAGATATATTAGGT-3′ (forward) and 5′-ACATTTCTACTTAAATATTCCATTAACA-3′ (reverse). The fragments were ligated in TOPO vector pCR2.1 of the TA Cloning kit (Life Technologies) and transformed into TOP10 competent bacteria. Individual clones confirmed by blue/white screening were Sanger sequenced (BaseClear B.V., the Netherlands) with standard M13 primers.
Results
TGFβ1-induced PLOD2 Expression Depends on ALK5 and Proximal Promoter Activation
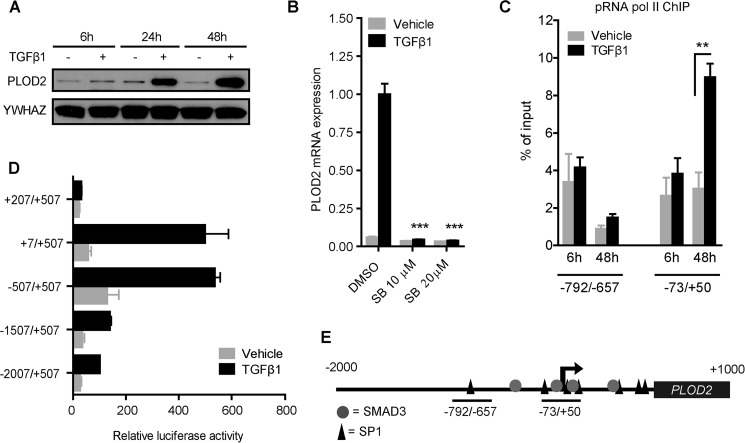
To confirm that PLOD2 expression is enhanced by TGFβ1 in our skin fibroblasts, we performed Western blot staining. We observed a strong increase of PLOD2 protein levels over time when TGFβ1 was added to the cells (Fig. 1A). Next, we wanted to identify whether this TGFβ1-induced PLOD2 expression occurs via the canonical or non-canonical pathway. Therefore, the activin receptor-like kinase 5 (ALK5) was inhibited by SB431542 while co-treating with TGFβ1. PLOD2 mRNA expression was completely reduced when either 10 μm or 20 μm inhibitor was added (Fig. 1B), confirming the canonical ALK5 pathway as the main signaling pathway.
FIGURE 1.
TGFβ1-induced PLOD2 expression depends on ALK5 signaling and proximal promoter activation. A, PLOD2 Western blot from fibroblasts treated with TFGβ1 or vehicle for 6, 24, and 48 h. B, qRT-PCR of fibroblasts treated with SB431542 or DMSO control, and co-treated with TGFβ1 or vehicle. C, ChIP on phosphorylated RNA polymerase II of fibroblasts treated with TGFβ1 or vehicle for 6 and 48 h, quantified by qPCR of two regions in the PLOD2 gene (depicted by horizontal bars, Fig. 1E). D, reporter assays of PLOD2 promoter fragments, measured from the TSS (+1 bp) to translation start site (+507 bps), and treated with TGFβ1 or vehicle. E, schematic overview of in silico detected SP1 and SMAD3 binding sites in PLOD2 from −2000 bps to +1000 bps of the TSS. Data are mean ± S.E. (n = 3), **, p < 0.01; ***, p < 0.001, unpaired Student's t test.
To explore whether the mechanism of TGFβ1-enhanced PLOD2 expression is indeed related to enhanced transcriptional initiation and not via indirect translational processes, we performed chromatin immunoprecipitation (ChIP) on phosphorylated RNA Polymerase II at two time points and two locations on the promoter. After 6 h of TGFβ1 stimulation there was no difference in enrichment of phosphorylated RNA polymerase II, whereas a significant enrichment was detected at the transcription start site (TSS) (73/+50 bp) after 48 h TGFβ1 stimulation (Fig. 1C). These results support our findings that the increased PLOD2 protein expression over time is related to increased transcription. Furthermore, RNA polymerase enrichment was higher at the TSS than in the region more upstream (−792/−657 bp) which is in accordance to current genome wide understanding of transcriptional regulation (39). To pinpoint regulatory elements encoded in the PLOD2 promoter, we performed reporter assays with deletion fragments of the PLOD2 promoter. After transfection of the constructs in fibroblasts and stimulation with TGFβ1, luciferase activity was highest with the proximal promoter regions of +7/+505 and −507/+507 fragments (Fig. 1D). The −1507/+507 and −2007/+507 fragments induced a considerable lower luciferase activity than the size-controlled +7/+507 and −507/+507 fragments, suggesting the presence of repressive elements in the distal promoter. In silico analysis of putative transcription factor (TF) binding sites known to act downstream of the ALK5 pathway revealed several binding motifs for the downstream transcription factors SP1 and SMAD3 at PLOD2. Most notably in the area around the transcription start site (Fig. 1E) that correlated with the PLOD2 promoter fragments that induced the highest reporter activity.
Transcription Factors SP1 and SMAD3 Drive Enhanced PLOD2 Expression
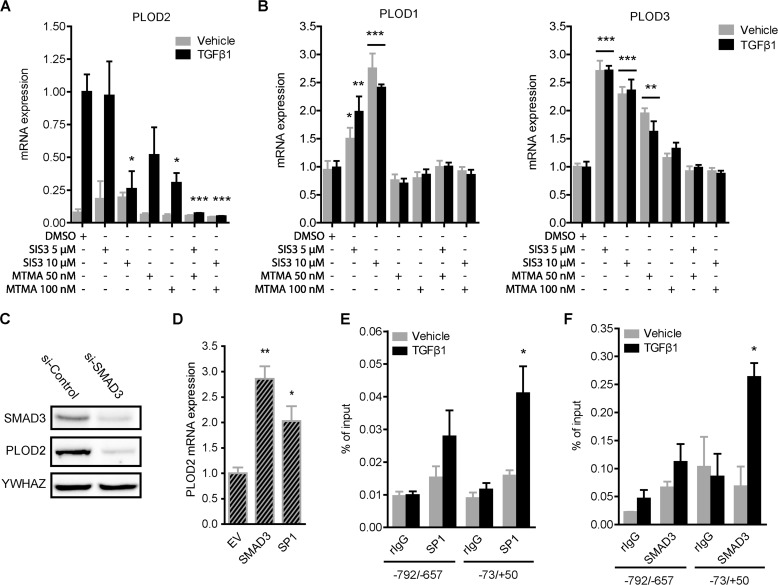
We wanted to assess in more depth the individual contribution of SP1 and SMAD3 to TGFβ1-induced PLOD2 expression. To prevent the actions of both TFs, we treated fibroblasts with increasing dosage of SIS3 to inhibit SMAD3 phosphorylation and its subsequent translocation to the nucleus, and with Mithramycin A to inhibit the DNA-binding ability of SP1. Both compounds were successful in attenuating TGFβ1-induced PLOD2 expression significantly up to, respectively, 74 and 70% (Fig. 2A). Also, to address possible synergistic effects of both inhibitors on PLOD2 expression, fibroblasts were treated with combinations of SIS3 and Mithramycin A. We observed that the co-treatment with the two inhibitors reduced TGFβ1-induced PLOD2 expression toward unstimulated levels. The results clearly indicate that the transcriptional effects of both inhibitors were TGFβ1 pathway related, since no significant effects were observed on the basal PLOD2 expression in unstimulated fibroblasts. The other PLOD family members, PLOD1 and PLOD3, are not responsive to TGFβ1 stimulation (38). To prove that their transcriptional activation is independent on SP1 and SMAD3, we analyzed mRNA expression of PLOD1 and PLOD3 after treatment with SIS3 and Mithramycin A. Indeed, in contrast to PLOD2, we found that both inhibitors did not negatively affect expression of both genes in both stimulated or unstimulated conditions (Fig. 2B). Instead, both TGFβ1-stimulated and unstimulated fibroblasts showed a significant increased PLOD1 and PLOD3 expression when treated with SIS3. Furthermore, PLOD3 expression increased when fibroblasts were treated with 50 nm Mithramycin A. In both cases, transcriptional effects revealed not to be TGFβ1 specific. To confirm our PLOD2 observations regarding both inhibitors, we transfected fibroblasts with esiRNA against SMAD3 or SP1, and checked for protein expression by Western blotting. PLOD2 protein expression was strongly reduced when SMAD3 was depleted (Fig. 2C). Unfortunately, efforts to deplete SP1 protein with esiRNA or shRNA were unsuccessful (data not shown). Next, we overexpressed SMAD3 and SP1 in unstimulated fibroblasts. Both constructs were able to significantly increase PLOD2 mRNA expression (resp. 2.8 and 2.1-fold) compared with empty vector control (Fig. 2D). In order to decipher if the observed transcriptional effects of both TFs on PLOD2 were indirect or due to binding the PLOD2 promoter, ChIP was performed. We observed that SP1 (Fig. 2E) and SMAD3 (Fig. 2F) bind PLOD2 at the transcription start site in TGFβ1-stimulated fibroblasts, while they were not detected (similar to rIgG values) in unstimulated fibroblasts. The region upstream of the TSS (−792/−657) had strong reduced presence of both TFs and indicates that binding is restricted to the TSS area.
FIGURE 2.
TGFβ1-enhanced PLOD2 expression is regulated via SP1 and Smad3. qRT-PCR analysis of (A) PLOD2 or (B) PLOD1 and PLOD3 mRNA expression from vehicle or TGFβ1-stimulated fibroblasts co-treated with SMAD3 phosphorylation inhibitor (SIS3) or SP1 inhibitor Mitramycin A (MTMA). C, representative Western blot of PLOD2 and SMAD3 from fibroblasts transfected with esiRNA against SMAD3 or control and treated with TGFβ1 for 48 h. D, PLOD2 mRNA expression of fibroblasts transfected with plasmids to overexpress SMAD3, SP1 or empty vector (EV) control. qChIP performed on fibroblasts treated with TGFβ1 or control for 48 h with antibodies against: (E) SP1 and (F) SMAD3. Normal rIgG was used as control, and enrichment for PLOD2 regions −73/+50 and −792/−657 was checked by qPCR and normalized to input. Data are mean ± S.E. (n = 3); *, p < 0.05; **, p < 0.01, unpaired Student's t test. Horizontal bars indicate both sets are significant to their individual control.
Histone Acetylation Associates with PLOD2 Expression
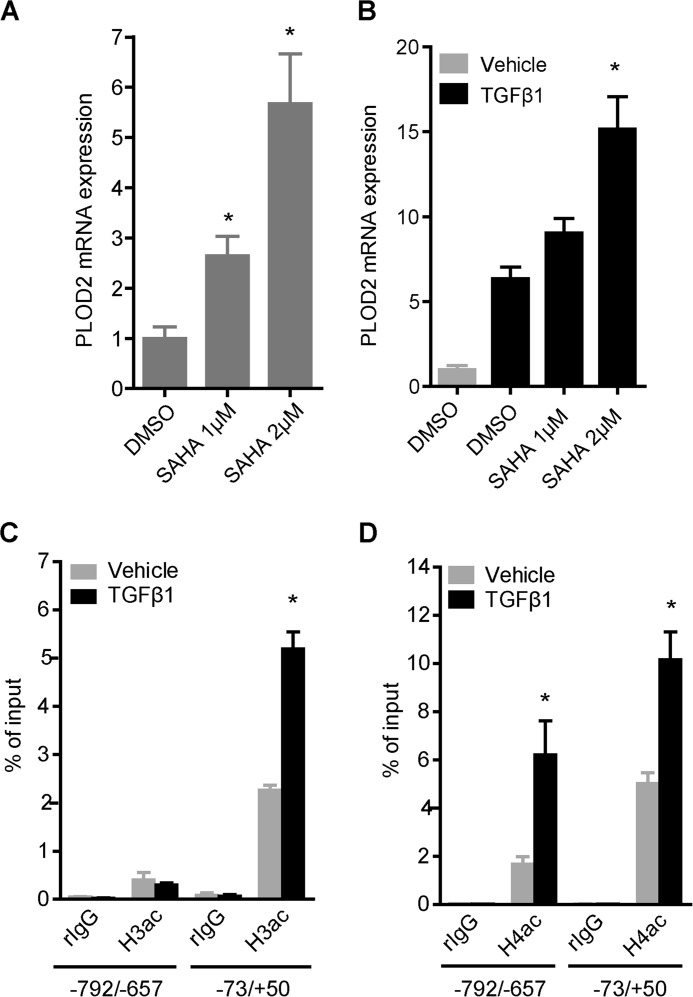
Next, we set out to explore whether TGFβ1-induced PLOD2 expression is related to changes in histone acetylation levels and if so, which enzymes can be accounted for this. We started by assessing PLOD2 expression after inhibiting histone deacetylases (HDACs) with the HDAC inhibitor SAHA. Unstimulated fibroblasts were treated for 3 days with increasing concentrations of SAHA, after which mRNA expression was detected by qRT-PCR. PLOD2 mRNA expression was significantly increased (up to 5.6-fold) after SAHA treatment (Fig. 3A). Interestingly, co-treatment of SAHA with TGFβ1 further enhanced PLOD2 expression (Fig. 3B). Since we could not exclude these transcriptional effects to be related to off-target effects caused by SAHA, we performed ChIP to detect actual histone acetylation levels at the PLOD2 promoter. Total acetylation of histone H3 (H3ac) and H4 (H4ac) were significantly more enriched at the TSS when fibroblasts were stimulated with TGFβ1 (Fig. 3, C and D), whereas histone acetylation levels were reduced for the more upstream regions. These results suggest that changes in PLOD2 expression are indeed related to histone acetylation.
FIGURE 3.
Histone acetylation levels correlate with PLOD2 expression. PLOD2 mRNA expression of fibroblasts treated with increasing dosage of SAHA for 72 h in unstimulated conditions (A) or TGFβ1 co-treated for 48 h (B). C, qChIP on fibroblasts treated with TGFβ1 or vehicle for 48 h, with antibodies against total acetylated histone H3 and (D) total acetylated histone H4. Normal rIgG was used as control, and enrichment for PLOD2 regions −73/+50 and −792/−657 was checked by qPCR, and normalized to input. Data are represented as mean ± S.E. (n = 3); *, p < 0.05; **, p < 0.01 unpaired Student's t test.
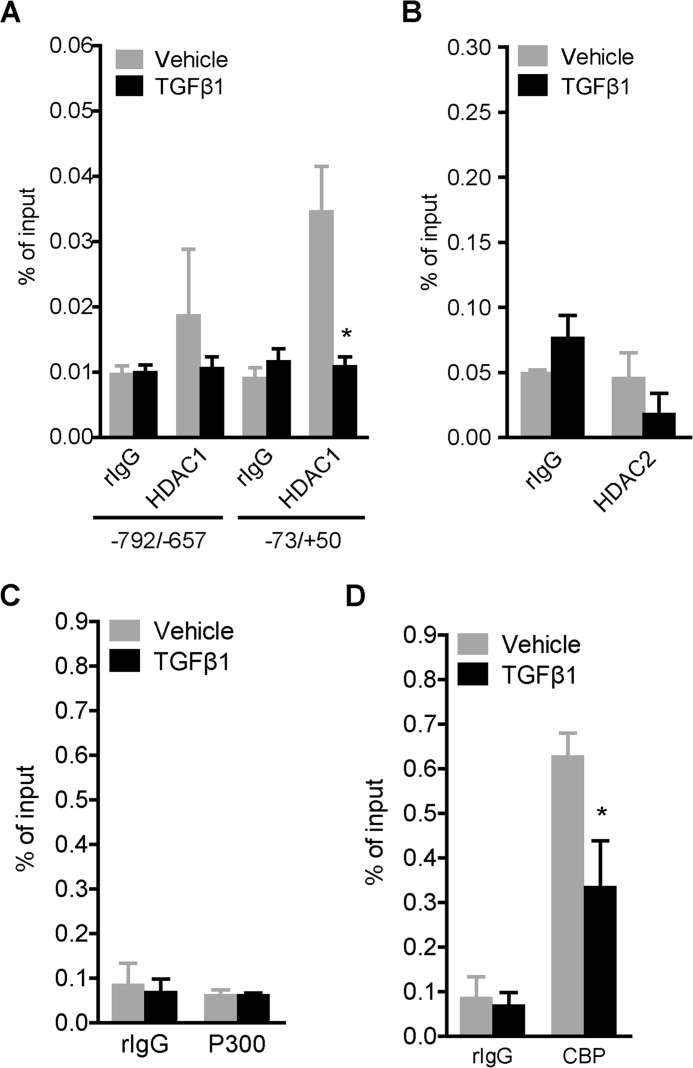
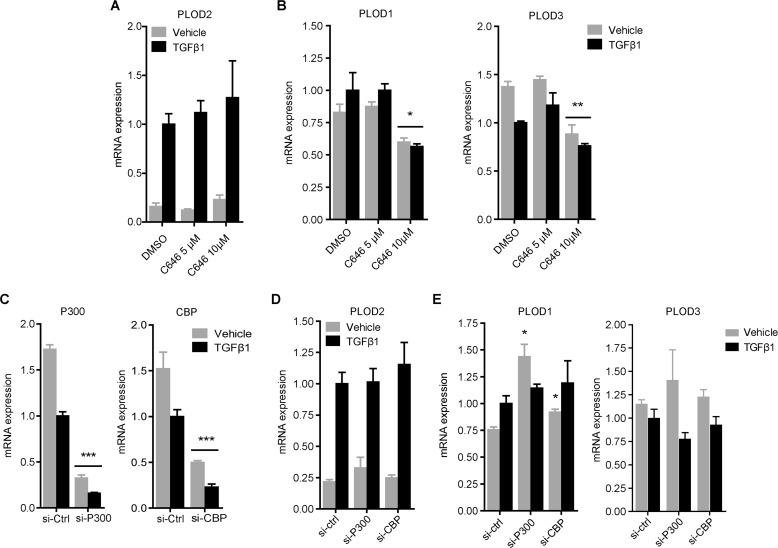
Since SAHA inhibition of HDACs resulted in enhanced PLOD2 expression, we wanted to find out if this could be related to PLOD2 binding of HDACs previously described to influence various fibrotic processes (40–42); namely HDAC1 and HDAC2. ChIP experiments revealed that HDAC2 was not present at the PLOD2 TSS (Fig. 4B), whereas HDAC1 binding was detected solely at the TSS in unstimulated fibroblasts and significantly reduced in TGFβ1-stimulated fibroblasts (Fig. 4A). Our data indicate that in TGFβ1-stimulated conditions, histones at PLOD2 are actively acetylated. Therefore we explored the involvement of histone acetyltransferases (HATs) P300 and CBP, which are known co-activators of the TGFβ1 pathway. We performed ChIP on both HATs and found that P300 was not detected above background levels and therefore concluded it does not bind to PLOD2 in both conditions (Fig. 4C). In contrast, CBP did bind to PLOD2 in unstimulated cells, but was significantly reduced by TGFβ1 stimulation (Fig. 4D). To further confirm that TGFβ1-induced PLOD2 expression was independent of P300 and CBP activity, we treated fibroblasts with the P300/CBP inhibitor C646 and assessed mRNA expression by qRT-PCR. We observed no effects on PLOD2 expression toward increasing dosage of C646 in both TGFβ1 and vehicle-treated cells (Fig. 5A). In contrast, both PLOD1 and PLOD3 mRNA expression were significantly reduced after treatment with 10 μm C646 (Fig. 5B). To confirm these results, we depleted CBP and P300 with esiRNA (Fig. 5C) and checked for mRNA expression of the PLOD family members. PLOD2 mRNA expression was not affected by depletion of either CBP or P300 in vehicle or TGFβ1 stimulated fibroblasts (Fig. 5D). PLOD1 mRNA expression was significantly increased in unstimulated fibroblasts when either P300 or CBP were depleted, whereas no effects were seen for PLOD3 mRNA expression in both conditions (Fig. 5E).
FIGURE 4.
Assessing PLOD2 binding of HDACs and HATS. qChIP on fibroblasts treated with TGFβ1 or vehicle for 48 h, with antibodies against (A) HDAC1, (B) HDAC2, (C) P300, and (D) CBP. Normal rIgG was used as control, and enrichment for PLOD2 region −73/+50, or both −73/+50 and −792/−657 for HDAC1, was checked by qPCR and normalized to input. Data are represented as mean ± S.E. (n = 3); *, p < 0.05; **, p < 0.01 unpaired Student's t test.
FIGURE 5.
CBP and P300 inhibition and depletion assert differential effects on expression of PLOD family members. A, PLOD2 and B, PLOD1 and PLOD3 mRNA expression of fibroblasts pre-treated for 1 h with 5 μm or 10 μm P300 inhibitor C646 or DMSO control, and co-treated with TGFβ1 or vehicle for additional 24 h. C, P300 and CBP mRNA expression of fibroblasts transfected with esiRNA against P300, CBP, or control. D, PLOD2 and E, PLOD1 and PLOD3 mRNA expression of fibroblasts transfected with CBP, P300, or control esiRNA and subsequently treated with TGFβ1 or vehicle for 48 h. Data are represented as mean ± S.E. (n = 3); *, p < 0.05; **, p < 0.01; ***, p < 0.001 unpaired Student's t test. Horizontal bars indicate both sets are significant to their individual control.
Histone Methylation but Not DNA Methylation Correlates with Enhanced PLOD2 Expression
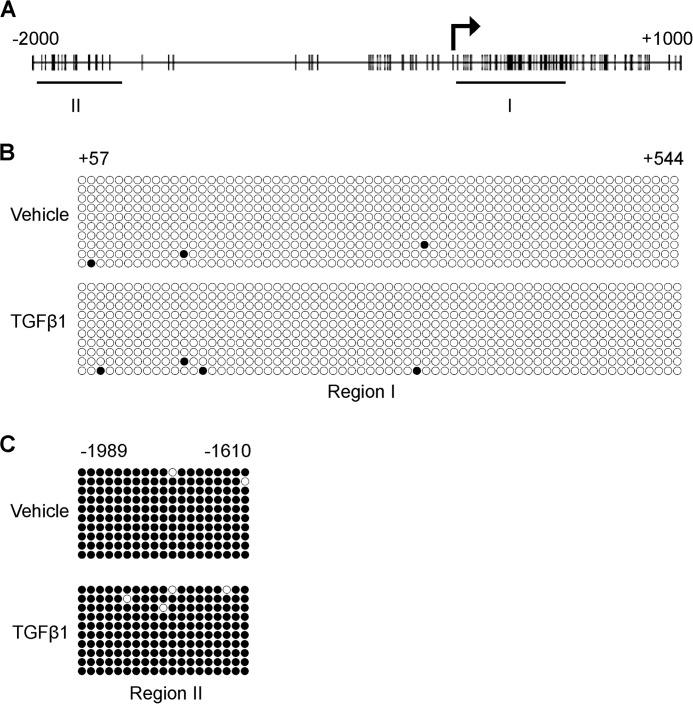
DNA methylation is strongly associated with gene repression, and since PLOD2 harbors many CpG sites in its promoter (Fig. 6A), we investigated whether differential methylation of these CpG's could contribute to changes in PLOD2 transcriptional activity. We isolated genomic DNA from fibroblasts and performed bisulfite sequencing. We observed that PLOD2 was hypomethylated in the region stretching from TSS into the first exon (Fig. 6B) under both TGFβ1 and control conditions. Interestingly, the distal promoter was hypermethylated, independent from TGFβ1 stimulation (Fig. 6C). However, since there was no difference between TGFβ1 and the untreated condition this indicates that there was no correlation between the DNA methylation state and PLOD2 expression.
FIGURE 6.
DNA methylation pattern of PLOD2 supports gene expression. A, schematic overview of CpG sites (vertical bars) on the PLOD2 gene ranging from −2000 to +1000 of the TSS. The horizontal bars depict the two sequencing areas (Regions I and II). Bisulfite sequencing results of fibroblasts treated with TGFβ1 or vehicle for 48 h and checked for PLOD2 region I (+57/+544 bp) (B) or region II (−1989/−1610 bp) (C). Open circles represent unmethylated CpGs and closed circles methylated CpGs from 10 individual clones each.
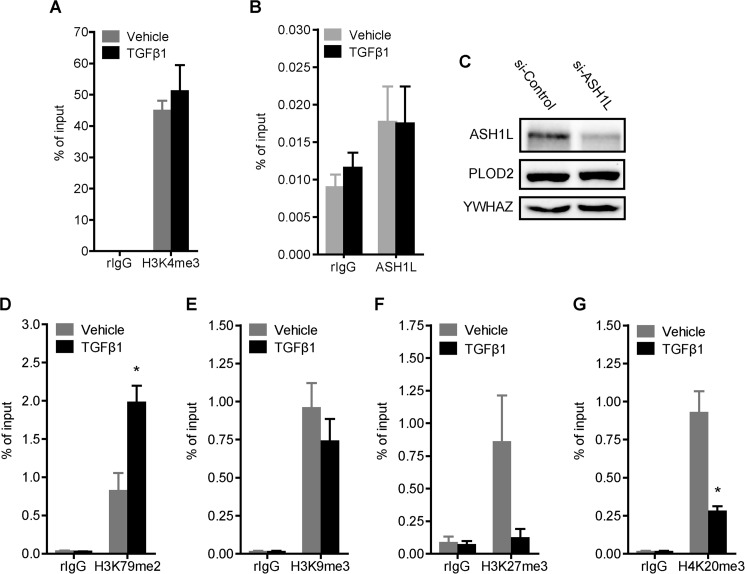
Methylation of various histone lysine residues can have a profound effect on transcriptional activity of genes. Therefore, we wanted to identify whether certain lysyl methylation marks have a function in PLOD2 expression. The presence of tri-methylated histone 3 lysine 4 (H3K4me3) is strongly correlated to transcriptional activation. In this respect, ASH1L, a H3K4me3 HKMT, has been linked to increased levels of H3K4me3 at fibrosis-related genes in liver fibrosis and as such is crucial for their transcriptional activation (16). To dissect whether ASHL1 and H3K4me3 are also associated with TGFβ1-induced PLOD2 expression we performed ChIP experiments on both. Against our expectations, H3K4me3 was present on the PLOD2 promoter at high levels in unstimulated fibroblast, and no further increase was observed when fibroblasts were stimulated with TGFβ1 (Fig. 7A). We did detect ASH1L at the PLOD2 promoter although at low levels (Fig. 7B). However, esiRNA knockdown of ASH1L had no effects on PLOD2 protein expression (Fig. 7C). Next we checked for other methylated lysine residues. H3K79me2, generally associated with promoters of activated genes, was significantly increased after TGFβ1 stimulation (Fig. 7D). With respect to the three gene repression-related histone modifications; H3K9me3, H3K27me3, and H4K20me3 (resp. Fig. 7, E, F, and G), only the presence of H4K20me3 was significantly reduced upon TGFβ1 stimulation.
FIGURE 7.
PLOD2 promoter histone methylation is affected by TGFβ1 stimulation. A, qChIP against H3K4me3 from fibroblasts treated with TGFβ1 or vehicle for 48 h (B) qChIP on fibroblast treated with TGFβ1 or vehicle for 48 h, with antibodies against ASH1L. C, Western blot of fibroblasts transfected with esiRNA against ASH1L or control and treated with TGFβ1 or vehicle for 48 h. Blots were stained with antibodies against ASH1L, PLOD2 and YWAHZ as loading control. qChIP on fibroblasts treated with TGFβ1 or vehicle for 48 h, with antibodies against gene activation related (D) H3K79me2, and gene repression related modifications: (E) H3K9me3, (F) H3K27me3, and (G) H3K20me3. In all ChIP experiments normal rIgG was used as control and enrichment for PLOD2 −73/+50 was checked by qPCR, and normalized to input. Data are represented as mean ± S.E. (n = 3), *, p < 0.05; **, p < 0.01, unpaired Student's t test.
SMAD3 Recruits a Histone Acetyltransferase to the PLOD2 Promoter
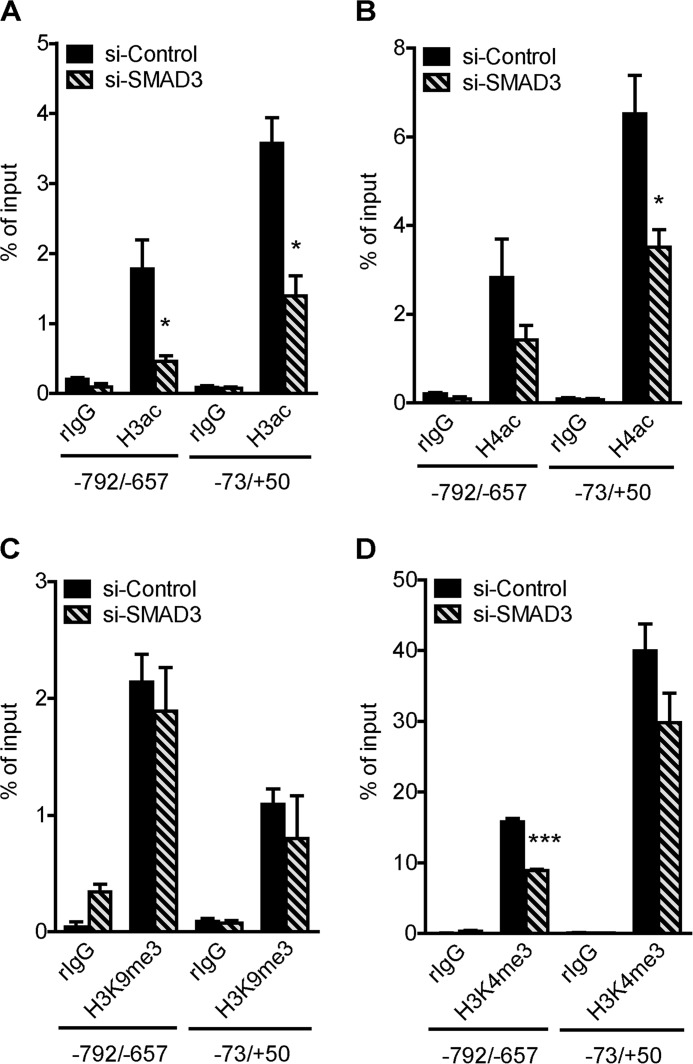
Histone modifying enzymes can be recruited to specific sites in the genome by transcription factors. Since inhibition and knockdown of SMAD3 attenuated TGFβ1-induced PLOD2 expression, we hypothesized that SMAD3 recruits histone modifying enzymes to the PLOD2 promoter for introducing activating histone marks. To explore this, we performed ChIP on various histone modifications in TGFβ1-stimulated fibroblasts that were transfected with esiRNA to deplete SMAD3. Compared with esiRNA controls, we observed significant reductions in H3ac and H4ac throughout the analyzed regions (Fig. 8, A and B), whereas H3K4me3 and H3K9me3 did not change at the TSS (Fig. 8, C and D). Upstream of the TSS SMAD3 KD did result in reduced H3K4me3 levels, possibly due to a cross-talk effect induced by the strong reduced H3 and H4 acetylation levels. These findings suggest that SMAD3 targets one or several histone acetyltransferases to PLOD2 that actively acetylate histones at the PLOD2 promoter. As expected, SMAD3 did not target enzymes to the TSS responsible for H3K4me3 and H3K9me3 to PLOD2, since these marks were not responsive to TGFβ1 and thereby SMAD3 binding.
FIGURE 8.
Depletion of Smad3 reduces PLOD2 promoter histone acetylation. Fibroblasts transfected with esiRNA against SMAD3 or control and treated with TGFβ1 for 48 h and used for qChIP with antibodies against: (A) H3ac, (B) H4ac, (C) H3K4me3, and (D) H3K9me3. Normal rIgG was used as control and enrichment for PLOD2 regions −73/+50 and −792/−657 was checked by qPCR, and normalized to input. Data are represented as mean ± S.E. (n = 3), *, p < 0.05, unpaired Student's t test.
Discussion
PLOD2 is considered a fibrotic marker since its expression and the resulting pyridinoline cross-links are increased in various scarred tissues and in vitro in fibroblast by profibrotic cytokines such as TGFβ1 (4, 5, 38, 43, 44). In addition, PLOD2 expression is enhanced in various types of cancers (e.g. glioma, bone, cervical, and liver) either or not directly caused by cytokines such as TGFβ1, and is strongly linked to the progression of these diseases likely by affecting cross-links of collagen that surrounds these tumors (45–49). Therefore, dissecting the downstream pathway of TGFβ1-mediated PLOD2 regulation is of fundamental interest to develop strategies that can tackle a wide range of scarring-related pathologies. Downstream of the canonical ALK5 pathway, the transcription factors SP1 and SMAD2/3, together with histone acetyltransferases P300 and CBP, are considered as the core regulators mediating expression of TGFβ1-responsive genes (50–52). We aimed to identify whether these or other factors play a role in TGFβ1-induced PLOD2 expression in skin fibroblasts. TGFβ1-induced PLOD2 expression in synovial fibroblasts was recently linked to phosphorylated SMAD2/3 (36). However, direct evidence of SMAD2/3 binding and information concerning recruitment of additional transcriptional modifiers to PLOD2 was so far lacking. In our study with skin fibroblasts, we found that both SP1 and SMAD3 are at the center of regulating PLOD2 expression, since inhibition of SP1, SMAD3, and knockdown of SMAD3 strongly attenuated TGFβ1-induced PLOD2 expression. More importantly, we show that both TFs bind directly at the PLOD2 TSS. By itself, SMAD3 has a weak DNA binding affinity, and it depends on complex formation with additional transcription factors for more effective DNA binding (53–55). It is therefore likely that, since SP1 and SMAD3 are known to form complexes together (51, 52), SP1 recruits SMAD3 and as a complex they bind to the PLOD2 promoter. This was essentially confirmed by our observation of the strong synergistic effects after co-treatment with SIS3 and Mithramycin A. PLOD2 is the only one of its family members that is TGFβ1-responsive. In contrast to PLOD2, PLOD1, and PLOD3 expression was increased in both unstimulated and TGFβ1 stimulated fibroblasts when phosphorylation of SMAD3 was inhibited. Furthermore, SP1 inhibition resulted in enhanced PLOD3 expression. Since SMAD3 is able to target both co-activators and co-repressors to their target genes, we speculate that TGFβ1 independent activation and translocation of SMAD3 targets a co-repressor complex to PLOD1 and PLOD3. Even though SP1 inhibition increased PLOD3 expression, we observed no synergistic effects of both inhibitors. Therefore we do not expect SP1 and SMAD3 to act as a complex on both genes.
Histone acetyltransferase CBP and P300 are co-activators that are widely found in complexes that contain SMAD3 or SP1. However, our ChIP, molecular inhibition and knockdown results indicate that P300 and CPB are not responsible for the increased histone acetylation at the PLOD2 promoter after TGFβ1 stimulation. This is in sharp contrast to other TGFβ1-responsive genes (56–58). Furthermore, knockdown of SMAD3 achieved reduced PLOD2 expression and PLOD2 promoter histone acetylation. The resulting effects of SMAD3 depletion therefore point to the inability to target a co-activator to PLOD2. Since P300 and CBP do not affect TGFβ1induced PLOD2 expression, we conclude that another -as of yet unidentified- HAT functions as the preferred SMAD3 co-activator that gets targeted to the PLOD2 promoter in TGFβ1 stimulated fibroblasts. To the best of our knowledge there are no reports stating that HATs other than p300 and CBP bind to SMAD3. Interestingly, both PLOD1 and PLOD3 expression were downregulated when treated with the CBP/P300 inhibitor, but PLOD1 expression was increased in unstimulated fibroblast when P300 and CBP was depleted by esiRNA. In some circumstances C646 can enhance P300-induced acetylation, and also its specificity toward P300/CBP is under debate (59). Therefore, the discrepancy between inhibitor and knockdown effects on PLOD1 and PLOD3 might be related to such issues.
CBP and HDAC1 were both detected at the PLOD2 promoter in unstimulated fibroblast, whereas both enzymes were reduced or depleted at the same location in TGFβ1-stimulated fibroblasts. We therefore hypothesize that both HDAC1 and CBP are present at the PLOD2 promoter in unstimulated fibroblasts to maintain a balance of histone acetylation to allow basal expression of PLOD2. Inhibition of class I and II HDACs by SAHA indeed increased PLOD2 expression in unstimulated fibroblasts and was even further enhanced when supplemented together with TGFβ1. These effects could directly be related to an imbalance in PLOD2 histone acetylation. However, since HDACs are known to also affect acetylation of non-histone proteins, such as SP1 transcription factor, we cannot exclude such indirect effects.
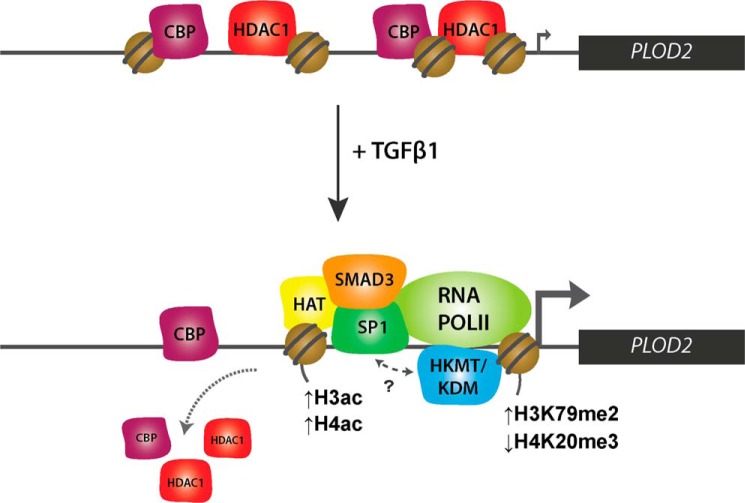
H3K4me3 is present in promoters of transcriptionally active genes and strongly correlates to gene expression (60, 61). Although H3K4me3 levels have previously been shown to increase after TGFβ1 stimulation for several other TGFβ1 responsive genes (62), this was not the case for PLOD2. This suggests that modifications on top of H3K4me3 are likely important in supporting PLOD2 transcriptional activation. Of interest is the decrease in H4K20me3, which is involved in heterochromatin formation (63, 64), and the increase of H3K79me2, which is related to gene expression and transcription elongation (29, 65). Although the precise roles of H3K79me2 and H4K20me3 in transcriptional regulation are still unclear, these modifications appear to be more associated with TGFβ1-induced PLOD2 expression than the classical marks H3K4me3, H3K9me3, and H3K27me3, which were not significantly affected by TGFβ1. As H4K20me3 and H3K79me2 have never before been linked to be responsive to TGFβ1 signaling, this finding requires further investigation. In cancer cells, presence of H3K79me2 frequently coincides with H3K4me3 at unmethylated promoters of tumor suppressor genes and is lost upon de novo DNA methylation (66). The increase of H3K79me2 after TGFβ1 treatment therefore might suggest additional transcriptional activating roles of this modification unrelated to changes in H3K4me3. Based on our observations we propose a model of PLOD2 transcriptional regulation (Fig. 9) whereby HDAC1, CBP, and a H4K20me3 HKMT ensure a chromatin state to maintain a basal PLOD2 expression level in unstimulated fibroblasts. Upon TGFβ1 stimulation, binding of HDAC1 and CBP is reduced, while SMAD3 and SP1 target HAT(s) and potentially multiple HKMTs/HDMs to PLOD2 to remodel the local chromatin in order to support or drive enhanced transcription. It is evident from our data that TGFβ1 stimulation does not affect the DNA methylation status of PLOD2, thereby confirming our view that the combined action of TFs and certain histone modifications are determining PLOD2 transcription.
FIGURE 9.
PLOD2 transcriptional regulation pathway. Schematic overview of how PLOD2 is regulated by transcription factors SP1 and SMAD3 in combination with histone-modifying enzymes during TGFβ1 stimulation.
To conclude, the work presented here identified novel regulators of PLOD2 expression in human fibroblasts that could be of potential interest for designing therapies that enable specific attenuation of TGFβ1-induced PLOD2 expression. Also, our data depict a regulatory mechanism of PLOD2 upon TGFβ1 stimulation that is independent on classical TGFβ1 transcriptional activation-related modifications (H3K4me3 and H3K27me3) and co-activators (P300 and CBP). These insights suggest that alternative TGFβ1 downstream regulatory mechanisms do exist, and underscores the need to obtain further insights to fully understand this pathway in various pathologies.
Author Contributions
R. A. F. G. conducted most of the experiments. S. D. R. conducted the HDACi experiments. R. A. F. G., M. G. R., and R. A. B. conceived the idea for the project, analyzed the results, and wrote the paper.
Acknowledgments
We thank Dr. R. Stoop (TNO, Leiden, The Netherlands) for providing the PLOD2 promoter fragments and Dr. M. Rehli (Universitätsklinikum Regensburg, Germany) for providing the pCpGL plasmid.
This work was supported by Netherlands Institute for Regenerative Medicine (NIRM) Grant FES0908 (to R. A. B.) and NWO/VIDI 91786373 (to M. G. R.). The authors declare that they have no conflicts of interest with the contents of this article.
- TGF
- transforming growth factor
- PLOD
- procollagen-lysine, 2-oxoglutarate 5-dioxygenase
- HAT
- histone acetyltransferase
- HDAC
- histone deacetylase.
References
- 1.Branton M. H., and Kopp J. B. (1999) TGF-beta and fibrosis. Microbes Infect. 1, 1349–1365 [DOI] [PubMed] [Google Scholar]
- 2.Namazi M. R., Fallahzadeh M. K., and Schwartz R. A. (2011) Strategies for prevention of scars: what can we learn from fetal skin? Int. J. Dermatol. 50, 85–93 [DOI] [PubMed] [Google Scholar]
- 3.Wynn T. A. (2008) Cellular and molecular mechanisms of fibrosis. J. Pathol. 214, 199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Slot A. J., Zuurmond A. M., Bardoel A. F., Wijmenga C., Pruijs H. E., Sillence D. O., Brinckmann J., Abraham D. J., Black C. M., Verzijl N., DeGroot J., Hanemaaijer R., TeKoppele J. M., Huizinga T. W., and Bank R. A. (2003) Identification of PLOD2 as telopeptide lysyl hydroxylase, an important enzyme in fibrosis. J. Biol. Chem. 278, 40967–40972 [DOI] [PubMed] [Google Scholar]
- 5.van der Slot A. J., Zuurmond A. M., van den Bogaerdt A. J., Ulrich M. M., Middelkoop E., Boers W., Karel Ronday H., DeGroot J., Huizinga T. W., and Bank R. A. (2004) Increased formation of pyridinoline cross-links due to higher telopeptide lysyl hydroxylase levels is a general fibrotic phenomenon. Matrix Biol. 23, 251–257 [DOI] [PubMed] [Google Scholar]
- 6.Ricard-Blum S., Esterre P., and Grimaud J. A. (1993) Collagen cross-linking by pyridinoline occurs in non-reversible skin fibrosis. Cell Mol. Biol. 39, 723–727 [PubMed] [Google Scholar]
- 7.Moriguchi T., and Fujimoto D. (1979) Crosslink of collagen in hypertrophic scar. J. Invest. Dermatol. 72, 143–145 [DOI] [PubMed] [Google Scholar]
- 8.Takaluoma K., Lantto J., and Myllyharju J. (2007) Lysyl hydroxylase 2 is a specific telopeptide hydroxylase, while all three isoenzymes hydroxylate collagenous sequences. Matrix Biol. 26, 396–403 [DOI] [PubMed] [Google Scholar]
- 9.Yamauchi M., and Sricholpech M. (2012) Lysine post-translational modifications of collagen. Essays Biochem. 52, 113–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith-Mungo L. I., and Kagan H. M. (1998) Lysyl oxidase: properties, regulation and multiple functions in biology. Matrix Biol. 16, 387–398 [DOI] [PubMed] [Google Scholar]
- 11.Reiser K., McCormick R. J., and Rucker R. B. (1992) Enzymatic and nonenzymatic cross-linking of collagen and elastin. FASEB J. 6, 2439–2449 [DOI] [PubMed] [Google Scholar]
- 12.van der Slot-Verhoeven A. J., van Dura E. A., Attema J., Blauw B., Degroot J., Huizinga T. W., Zuurmond A. M., and Bank R. A. (2005) The type of collagen cross-link determines the reversibility of experimental skin fibrosis. Biochim. Biophys. Acta. 1740, 60–67 [DOI] [PubMed] [Google Scholar]
- 13.Wiegand C., and White R. (2013) Microdeformation in wound healing. Wound Repair Regen. 21, 793–799 [DOI] [PubMed] [Google Scholar]
- 14.Balestrini J. L., Chaudhry S., Sarrazy V., Koehler A., and Hinz B. (2012) The mechanical memory of lung myofibroblasts. Integr. Biol. 4, 410–421 [DOI] [PubMed] [Google Scholar]
- 15.Georges P. C., Hui J. J., Gombos Z., McCormick M. E., Wang A. Y., Uemura M., Mick R., Janmey P. A., Furth E. E., and Wells R. G. (2007) Increased stiffness of the rat liver precedes matrix deposition: implications for fibrosis. Am. J. Physiol. Gastrointest. Liver Physiol. 293, G1147–G1154 [DOI] [PubMed] [Google Scholar]
- 16.Perugorria M. J., Wilson C. L., Zeybel M., Walsh M., Amin S., Robinson S., White S. A., Burt A. D., Oakley F., Tsukamoto H., Mann D. A., and Mann J. (2012) Histone methyltransferase ASH1 orchestrates fibrogenic gene transcription during myofibroblast transdifferentiation. Hepatology 56, 1129–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko Y. A., Mohtat D., Suzuki M., Park A. S., Izquierdo M. C., Han S. Y., Kang H. M., Si H., Hostetter T., Pullman J. M., Fazzari M., Verma A., Zheng D., Greally J. M., and Susztak K. (2013) Cytosine methylation changes in enhancer regions of core pro-fibrotic genes characterize kidney fibrosis development. Genome Biol. 14, R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheen-Chen S. M., Lin C. R., Chen K. H., Yang C. H., Lee C. T., Huang H. W., and Huang C. Y. (2013) Epigenetic histone methylation regulates transforming growth factor β-1 expression following bile duct ligation in rats. J. Gastroenterol. 49, 1285–1297 [DOI] [PubMed] [Google Scholar]
- 19.Watson C. J., Collier P., Tea I., Neary R., Watson J. A., Robinson C., Phelan D., Ledwidge M. T., McDonald K. M., McCann A., Sharaf O., and Baugh J. A. (2014) Hypoxia-induced epigenetic modifications are associated with cardiac tissue fibrosis and the development of a myofibroblast-like phenotype. Hum. Mol. Genet. 23, 2176–2188 [DOI] [PubMed] [Google Scholar]
- 20.Coward W. R., Feghali-Bostwick C. A., Jenkins G., Knox A. J., and Pang L. (2014) A central role for G9a and EZH2 in the epigenetic silencing of cyclooxygenase-2 in idiopathic pulmonary fibrosis. FASEB J. 28, 3183–3196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tampe B., Tampe D., Muller C. A., Sugimoto H., Lebleu V., Xu X., Muller G. A., Zeisberg E. M., Kalluri R., and Zeisberg M. (2014) Tet3-Mediated Hydroxymethylation of Epigenetically Silenced Genes Contributes to Bone Morphogenic Protein 7-Induced Reversal of Kidney Fibrosis. J. Am. Soc. Nephrol. 25, 905–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reik W. (2007) Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447, 425–432 [DOI] [PubMed] [Google Scholar]
- 23.Goding C. R., Pei D., and Lu X. (2014) Cancer: pathological nuclear reprogramming? Nat. Rev. Cancer 14, 568–573 [DOI] [PubMed] [Google Scholar]
- 24.Brenet F., Moh M., Funk P., Feierstein E., Viale A. J., Socci N. D., and Scandura J. M. (2011) DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS ONE 6, e14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bannister A. J., and Kouzarides T. (2011) Regulation of chromatin by histone modifications. Cell Research 21, 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campos E. I., and Reinberg D. (2009) Histones: annotating chromatin. Annu. Rev. Genet. 43, 559–599 [DOI] [PubMed] [Google Scholar]
- 27.Kooistra S. M., and Helin K. (2012) Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 13, 297–311 [DOI] [PubMed] [Google Scholar]
- 28.Tee W. W., and Reinberg D. (2014) Chromatin features and the epigenetic regulation of pluripotency states in ESCs. Development 141, 2376–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veloso A., Kirkconnell K. S., Magnuson B., Biewen B., Paulsen M. T., Wilson T. E., and Ljungman M. (2014) Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res. 24, 896–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger S. L. (2007) The complex language of chromatin regulation during transcription. Nature 447, 407–412 [DOI] [PubMed] [Google Scholar]
- 31.Ito T. (2007) Role of histone modification in chromatin dynamics. J. Biochem. 141, 609–614 [DOI] [PubMed] [Google Scholar]
- 32.Choi J. K., and Howe L. J. (2009) Histone acetylation: truth of consequences? Biochem. Cell Biol. 87, 139–150 [DOI] [PubMed] [Google Scholar]
- 33.Mosammaparast N., and Shi Y. (2010) Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 79, 155–179 [DOI] [PubMed] [Google Scholar]
- 34.Klose R. J., Kallin E. M., and Zhang Y. (2006) JmjC-domain-containing proteins and histone demethylation. Nature Reviews. Genetics 7, 715–727 [DOI] [PubMed] [Google Scholar]
- 35.Helin K., and Dhanak D. (2013) Chromatin proteins and modifications as drug targets. Nature 502, 480–488 [DOI] [PubMed] [Google Scholar]
- 36.Remst D. F., Blaney Davidson E. N., Vitters E. L., Bank R. A., van den Berg W. B., and van der Kraan P. M. (2014) TGF-ss induces Lysyl hydroxylase 2b in human synovial osteoarthritic fibroblasts through ALK5 signaling. Cell Tissue Res. 355, 163–171 [DOI] [PubMed] [Google Scholar]
- 37.Mia M. M., Boersema M., and Bank R. A. (2014) Interleukin-1beta attenuates myofibroblast formation and extracellular matrix production in dermal and lung fibroblasts exposed to transforming growth factor-beta1. PLoS ONE 9, e91559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Slot A. J., van Dura E. A., de Wit E. C., De Groot J., Huizinga T. W., Bank R. A., and Zuurmond A. M. (2005) Elevated formation of pyridinoline cross-links by profibrotic cytokines is associated with enhanced lysyl hydroxylase 2b levels. Biochim. Biophys. Acta 1741, 95–102 [DOI] [PubMed] [Google Scholar]
- 39.Consortium E. P. (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu C., Chen X., Yang L., Kisseleva T., Brenner D. A., and Seki E. (2014) Transcriptional repression of the transforming growth factor β (TGF-beta) Pseudoreceptor BMP and activin membrane-bound inhibitor (BAMBI) by Nuclear Factor κB (NF-κB) p50 enhances TGF-β signaling in hepatic stellate cells. J. Biol. Chem. 289, 7082–7091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryu J. K., Kim W. J., Choi M. J., Park J. M., Song K. M., Kwon M. H., Das N. D., Kwon K. D., Batbold D., Yin G. N., and Suh J. K. (2013) Inhibition of histone deacetylase 2 mitigates profibrotic TGF-beta1 responses in fibroblasts derived from Peyronie's plaque. Asian J. Androl. 15, 640–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fitzgerald O'Connor E. J., Badshah II, Addae L. Y., Kundasamy P., Thanabalasingam S., Abioye D., Soldin M., and Shaw T. J. (2012) Histone deacetylase 2 is upregulated in normal and keloid scars. J. Invest. Dermatol. 132, 1293–1296 [DOI] [PubMed] [Google Scholar]
- 43.Brinckmann J., Notbohm H., Tronnier M., Açil Y., Fietzek P. P., Schmeller W., Müller P. K., and Bätge B. (1999) Overhydroxylation of lysyl residues is the initial step for altered collagen cross-links and fibril architecture in fibrotic skin. J. Invest. Dermatol. 113, 617–621 [DOI] [PubMed] [Google Scholar]
- 44.Remst D. F., Blaney Davidson E. N., Vitters E. L., Blom A. B., Stoop R., Snabel J. M., Bank R. A., van den Berg W. B., and van der Kraan P. M. (2013) Osteoarthritis-related fibrosis is associated with both elevated pyridinoline cross-link formation and lysyl hydroxylase 2b expression. Osteoarthritis Cartilage 21, 157–164 [DOI] [PubMed] [Google Scholar]
- 45.Bozóky B., Savchenko A., Csermely P., Korcsmáros T., Dúl Z., Pontén F., Székely L., and Klein G. (2013) Novel signatures of cancer-associated fibroblasts. Int. J. Cancer 133, 286–293 [DOI] [PubMed] [Google Scholar]
- 46.Rajkumar T., Sabitha K., Vijayalakshmi N., Shirley S., Bose M. V., Gopal G., and Selvaluxmy G. (2011) Identification and validation of genes involved in cervical tumourigenesis. BMC Cancer 11, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong S., Nutt C. L., Betensky R. A., Stemmer-Rachamimov A. O., Denko N. C., Ligon K. L., Rowitch D. H., and Louis D. N. (2005) Histology-based expression profiling yields novel prognostic markers in human glioblastoma. J. Neuropathol. Exp. Neurol. 64, 948–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blanco M. A., LeRoy G., Khan Z., Alečković M., Zee B. M., Garcia B. A., and Kang Y. (2012) Global secretome analysis identifies novel mediators of bone metastasis. Cell Res. 22, 1339–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noda T., Yamamoto H., Takemasa I., Yamada D., Uemura M., Wada H., Kobayashi S., Marubashi S., Eguchi H., Tanemura M., Umeshita K., Doki Y., Mori M., and Nagano H. (2012) PLOD2 induced under hypoxia is a novel prognostic factor for hepatocellular carcinoma after curative resection. Liver Int. 32, 110–118 [DOI] [PubMed] [Google Scholar]
- 50.Ghosh A. K., and Varga J. (2007) The transcriptional coactivator and acetyltransferase p300 in fibroblast biology and fibrosis. J. Cell. Physiol. 213, 663–671 [DOI] [PubMed] [Google Scholar]
- 51.Shi Y., and Massagué J. (2003) Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 52.Zhang W., Ou J., Inagaki Y., Greenwel P., and Ramirez F. (2000) Synergistic cooperation between Sp1 and Smad3/Smad4 mediates transforming growth factor beta1 stimulation of α2(I)-collagen (COL1A2) transcription. J. Biol. Chem. 275, 39237–39245 [DOI] [PubMed] [Google Scholar]
- 53.Qin H., Chan M. W., Liyanarachchi S., Balch C., Potter D., Souriraj I. J., Cheng A. S., Agosto-Perez F. J., Nikonova E. V., Yan P. S., Lin H. J., Nephew K. P., Saltz J. H., Showe L. C., Huang T. H., and Davuluri R. V. (2009) An integrative ChIP-chip and gene expression profiling to model SMAD regulatory modules. BMC Syst. Biol. 3, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S., and Gauthier J. M. (1998) Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17, 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seoane J., Le H. V., Shen L., Anderson S. A., and Massagué J. (2004) Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117, 211–223 [DOI] [PubMed] [Google Scholar]
- 56.Qiu P., Ritchie R. P., Gong X. Q., Hamamori Y., and Li L. (2006) Dynamic changes in chromatin acetylation and the expression of histone acetyltransferases and histone deacetylases regulate the SM22alpha transcription in response to Smad3-mediated TGFβ1 signaling. Biochem. Biophys. Res. Commun. 348, 351–358 [DOI] [PubMed] [Google Scholar]
- 57.Yuan H., Reddy M. A., Sun G., Lanting L., Wang M., Kato M., and Natarajan R. (2013) Involvement of p300/CBP and epigenetic histone acetylation in TGF-1-mediated gene transcription in mesangial cells. Am. J. Physiol. Renal. Physiol. 304, F601–F613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ihn H., Yamane K., Asano Y., Jinnin M., and Tamaki K. (2006) Constitutively phosphorylated Smad3 interacts with Sp1 and p300 in scleroderma fibroblasts. Rheumatology 45, 157–165 [DOI] [PubMed] [Google Scholar]
- 59.Henry R. A., Kuo Y.-M. M., Bhattacharjee V., Yen T. J., and Andrews A. J. (2015) Changing the selectivity of p300 by acetyl-CoA modulation of histone acetylation. ACS Chem. Biol. 10, 146–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Voigt P., Tee W. W., and Reinberg D. (2013) A double take on bivalent promoters. Genes Dev. 27, 1318–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., and Lander E. S. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 62.Sun G., Reddy M. A., Yuan H., Lanting L., Kato M., and Natarajan R. (2010) Epigenetic histone methylation modulates fibrotic gene expression. J. Am. Soc. Nephrol. 21, 2069–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jørgensen S., Schotta G., and Sørensen C. S. (2013) Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 41, 2797–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Evertts A. G., Manning A. L., Wang X., Dyson N. J., Garcia B. A., and Coller H. A. (2013) H4K20 methylation regulates quiescence and chromatin compaction. Mol. Biol. Cell 24, 3025–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steger D. J., Lefterova M. I., Ying L., Stonestrom A. J., Schupp M., Zhuo D., Vakoc A. L., Kim J. E., Chen J., Lazar M. A., Blobel G. A., and Vakoc C. R. (2008) DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol. Cell. Biol. 28, 2825–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jacinto F. V., Ballestar E., and Esteller M. (2009) Impaired recruitment of the histone methyltransferase DOT1L contributes to the incomplete reactivation of tumor suppressor genes upon DNA demethylation. Oncogene 28, 4212–4224 [DOI] [PubMed] [Google Scholar]