Background: The Gun4 protein stimulates activity of magnesium chelatase, and it is important for chlorophyll biosynthesis.
Results: Mechanism of porphyrin binding by Gun4 was proposed, and a Gun4 mutant was characterized in detail.
Conclusion: Gun4 controls substrate channeling into chlorophyll biosynthesis.
Significance: In silico, in vitro, and in vivo data were integrated to explain the function of Gun4 protein.
Keywords: chlorophyll, cyanobacteria, docking, molecular modeling, porphyrin, Gun4, Mgprotoporphyrin IX, Synechocystis 6803
Abstract
In oxygenic phototrophs, chlorophylls, hemes, and bilins are synthesized by a common branched pathway. Given the phototoxic nature of tetrapyrroles, this pathway must be tightly regulated, and an important regulatory role is attributed to magnesium chelatase enzyme at the branching between the heme and chlorophyll pathway. Gun4 is a porphyrin-binding protein known to stimulate in vitro the magnesium chelatase activity, but how the Gun4-porphyrin complex acts in the cell was unknown. To address this issue, we first performed simulations to determine the porphyrin-docking mechanism to the cyanobacterial Gun4 structure. After correcting crystallographic loop contacts, we determined the binding site for magnesium protoporphyrin IX. Molecular modeling revealed that the orientation of α6/α7 loop is critical for the binding, and the magnesium ion held within the porphyrin is coordinated by Asn-211 residue. We also identified the basis for stronger binding in the Gun4-1 variant and for weaker binding in the W192A mutant. The W192A-Gun4 was further characterized in magnesium chelatase assay showing that tight porphyrin binding in Gun4 facilitates its interaction with the magnesium chelatase ChlH subunit. Finally, we introduced the W192A mutation into cells and show that the Gun4-porphyrin complex is important for the accumulation of ChlH and for channeling metabolites into the chlorophyll biosynthetic pathway.
Introduction
Tetrapyrrole biosynthesis ranks among the most fundamental pathways in living systems, leading to production of heme, siroheme, and vitamin B12. Synthesis of heme is conserved among all three domains of life, starting with synthesis of δ-aminolevulinic acid (ALA),4 which is converted through several enzymatic steps into protoporphyrin IX (PIX), a metal-free porphyrin ring. PIX is a substrate for ferrochelatase, which catalyzes the insertion of iron to produce heme.
The tetrapyrrole biosynthetic pathway is more complex in photosynthetic organisms because, apart from heme, they usually require large quantities of other tetrapyrrole end products. (Bacterio)chlorophylls are critical cofactors of the photosynthetic apparatus, and cyanobacteria and red algae also produce large amounts of linear phycobilins serving for light capture in phycobilisomes. The synthesis of (bacterio)chlorophylls, hemes, and bilins shares the same pathway up until PIX, at which point the ferrochelatase competes for the same substrate with magnesium chelatase (MgCh). Insertion of Mg2+ by MgCh leads to magnesium protoporphyrin IX (MgP), the first intermediate on the (bacterio)chlorophyll branch. This step is followed by methylation of MgP by MgP methyltransferase enzyme (ChlM).
Tetrapyrroles are phototoxic, so their biosynthesis must be tightly regulated to prevent accumulation of intermediates. However, there are special features of this pathway in organisms performing oxygenic photosynthesis (cyanobacteria, algae, and plants) that cause its regulation to be particularly important and complex. These organisms produce chlorophyll (Chl) and heme under light and in the presence of oxygen, which is a dangerous environment for dealing with high concentration of tetrapyrroles (1, 2). Moreover, the relative levels of heme and Chl are quite different, and the rate of their formation depends on actual growth conditions (3, 4). Photosynthetic cells must control the total flow through the tetrapyrrole pathway while precisely distributing PIX into heme and Chl branches. There is likely a complex regulatory network securing the synthesis of tetrapyrrole end products in required amounts while keeping the intermediate concentration extremely low (reviewed in Refs. 5 and 6).
It is accepted that both ferrochelatase and MgCh play an important regulatory role in allocation of metabolites into the heme/Chl branches (6–9). The mechanism by which this occurs, however, is far from clear. Ferrochelatase is a single-subunit enzyme but MgCh requires three subunits (ChlH, ChlI, and ChlD) for activity. In bacteriochlorophyll-producing bacteria, these three subunits are sufficient for a highly active MgCh enzyme when assayed in vitro (10). In contrast, MgCh from cyanobacteria, algae, and plants requires a protein called Gun4 that strongly enhances MgCh activity in vitro (11–13) and very likely also in vivo (14, 15).
Gun4 was shown to bind various porphyrins, exhibiting the highest affinity to MgP, the product of MgCh enzyme, and a weaker affinity to PIX (13, 16). There is evidence that Gun4 physically interacts with ChlH (11, 14, 17), which is the porphyrin-binding MgCh subunit that presumably also contains the active site for chelation (18). Available in vitro kinetic data imply that Gun4 stimulates MgCh by substantially reducing the PIX and Mg2+ concentrations required for catalysis (12). Inactivation of the gun4 gene perturbs Chl biosynthesis and accumulation of Chl-binding proteins in cyanobacteria (14), green algae (15), and plants (11, 19). However, phenotypes of available Gun4 mutants are difficult to explain simply by a defect in MgCh activity, and the in vivo activities of Gun4 appear to be complex (15, 19, 20).
Although it is widely accepted that the role of Gun4 depends on tight porphyrin binding (20, 21), there is no conclusive model of how the Gun4-porphyrin complex is implicated in the control of tetrapyrrole biosynthesis. Here, we employed in silico modeling, which allows us to identify the MgP-binding pocket and provides an atomic level description of the MgP-binding mechanism for cyanobacterial Gun4. Having a detailed view on interactions between Gun4 and MgP, we replaced the Trp-192 residue to obtain a Gun4 variant (W192A) with significantly weakened affinity for porphyrins. The W192A mutation was explored in vitro in a MgCh assay and in vivo in the cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis). These experiments revealed that the weaker affinity of W192A-Gun4 to porphyrins compromised the formation of Gun4-ChlH complex. Because the Synechocystis gun4-W192A strain was deficient in Chl precursors and exhibits a defect in accumulation of ChlH and ChlM, the porphyrin-driven interaction between Gun4-ChlH appears to be crucial for the formation and maintenance of an active MgCh enzyme.
Experimental Procedures
Molecular Dynamics
Simulations were performed with the AMBER11 molecular modeling suite (22). All systems were first prepared at pH 7.7 using the Protein Preparation Wizard from Schrödinger (23). The parm99 force field was used to define the parameters of the proteins in combination with a truncated octahedron water box containing ∼24,000 TIP3P water molecules. Na+ and Cl− ions were added to neutralize and reach an ionic strength of 0.15 m. After standard equilibration, we performed 200 ns of molecular dynamics at constant pressure and temperature (NPT ensemble) using the Berendsen barostat and thermostat. Root mean square fluctuation analysis was performed using the Python PRODY library, and cross-correlation maps were computed using the ptraj tool from the Ambertools 13 package.
Loop Prediction
The Prime loop prediction tool (Prime, version 3.8; Schrödinger) was used to model conformations of the α2/α3 and α6/α7 loops in the Gun4 core domain. Because of the size of the loops, we used the extended loop prediction options. Side chains with less than 7.5 Å of separation were also refined, and the 10 lowest energy structures were minimized. The crystal symmetry option was used to predict the effect of crystal contacts. Dielectric constants were 1 and 80 for the (solvent-exposed) internal and external environments, respectively.
Simulation of the Porphyrin Binding to Gun4 by Protein Energy Landscape Exploration (PELE)
PELE, a Monte Carlo algorithm developed specifically to explore protein ligand interactions (24), was used to model porphyrin binding. In PELE, each Monte Carlo step comprises two main parts: perturbation and relaxation. Perturbation consists of a random translation and rotation of the ligand plus an anisotropic network model procedure to model backbone fluctuations. Relaxation involves optimization of the side chains local to the ligand and a global minimization. Then the step is rejected or accepted using a metropolis criterion to generate a realistic distribution of the configurations generated.
Entrance simulations initially placed the ligand in the bulk solvent, outside of the binding site. Using the spawning algorithms in PELE, the ligand was then directed to enter into the active site. This algorithm constrains the ligand random search within a sphere around 18 Å from a given (chosen) point. If the ligand escapes this sphere, it will be placed in the closest conformation to the reference given point found by any explorer (trajectory). The Alpha carbon in Phe-160 was used as the spawning center (representative of an active site position).
PELE was able to find binding poses in ∼24 h using 48 trajectories (with 1 trajectory per computing core), using random translations and rotations in the 1–7 Å and 0–90° ranges, respectively. Once the ligand reached the active site, local refinement explorations used a smaller spawning sphere, 10 Å, and a smaller translation of 0.5 Å to better explore the binding energy profile in the bound region. A total of ∼50,000 accepted Monte Carlo steps were produced in this refinement process, requiring ∼240 trajectories, each running 24 h.
Construction of the Synechocystis Mutant Strains and the Growth Conditions
To prepare Synechocystis Δgun4, we made a linear PCR construct by the method of (25) containing the Zeocin resistance cassette fused with gun4 flanking sequences (∼300 bp) to allow homologous recombination. After transformation into WT cells and segregation, the Zeocin cassette replaced almost the entire gun4 gene (bp 119–694).
For the construction of the gun4-W192A strain, we cloned the chromosomal fragment containing the gun4 gene and 300-bp DNA upstream and downstream flanking regions into pUC18 vector (Life Technologies) between EcoRI and BamHI restriction sites. The W192A mutation was inserted into the cloned gun4 gene using the QuikChange II site-directed mutagenesis kit (Agilent Technologies). The pUC18-gun4-W192A plasmid was transformed into the above-described Synechocystis Δgun4 strain, and photoautotrophic transformants were selected on BG11 agar plates at 28 °C and 30 μmol photons m−2 s−1. Synechocystis cells were grown in liquid culture in 250-ml Erlenmeyer flasks on a rotary shaker at 28 °C and bubbled with air.
Absorption Spectra, Determination of Chl Content, and Quantification of Chl Precursors
Absorption spectra of whole cells were measured at room temperature using a Shimadzu UV-3000 spectrophotometer (Kyoto, Japan). Chl was extracted from cell pellets (1 ml, OD750 = ∼0.4) with 100% (v/v) methanol, and its concentration was measured spectrophotometrically according to Ref. 26. For the quantitative determination of Chl precursors in Synechocystis cells, 2 ml of culture at OD750 ∼ 0.4 were quickly harvested, and the extracted pigments were analyzed essentially as described in Ref. 27.
Electrophoresis and Immunoblotting
To analyze protein levels, Synechocystis cells in exponential growth phase (OD750 = ∼0.4) were harvested by centrifugation; washed; resuspended in the buffer containing 25 mm HEPES/NaOH, pH 7.4, 5 mm CaCl2, 10 mm MgCl2, and 20% (w/v) glycerol; and broken using glass beads. Cell extracts were further denatured in 2% SDS and 1% dithiothreitol for 30 min at room temperature and separated in a 10% polyacrylamide gel using a mini Bio-Rad system. For immunoblotting, proteins were transferred onto a PVDF membrane (Sigma), probed with specific primary antibodies followed by the secondary antibody conjugated to horseradish peroxidase (Sigma). The primary antibodies raised against recombinant Synechocystis Gun4 were kindly provided by Prof. Annegret Wilde (Universität Freiburg, Freiburg, Germany).
Circular Dichroism Spectra, Tryptophan Quenching, and in Vitro MgCh Enzyme Assays
To obtain recombinant Gun4 and W192A proteins, the Synechocystis gun4 gene was cloned into the pET14b plasmid and purified as for the Thermosynechococcus elongatus protein as reported previously (28). The W192A mutation was introduced into the pET14(b):gun4 plasmid by the same procedure used for construction of the Synechocystis gun4-W192A strain (see above). Recombinant subunits of Synechocystis MgCh were expressed and purified as described in Ref. 29.
CD spectra were recorded using a JASCO-810 spectrometer (JASCO, Great Dunmow, UK). Protein was dissolved in water to a concentration of 1 mg ml−1 and analyzed in a cuvette with a 0.2-cm path length. Spectra were recorded stepwise, from 250 to 190 nm in 1-nm increments, 4 s/nm.
Porphyrin-binding studies were carried out on a Jobin Yvon Fluoromax 3 spectofluorometer fitted with a temperature-controlled cuvette holder. The samples consisted of 1 μm protein in 50 mm MOPS/KOH, 11 mm KCl, 1 mm DTT, 0.3 m glycerol, pH 7.7, at 34 °C. Titrations were performed by the addition of porphyrin and incubated for 2 min before recording fluorescence (λex = 280 ± 4.5 nm, λem = 350 ± 18 nm). Changes in volume were <5% and were taken into account in calculating dissociation constants by using the titration mode in DynaFit 4 (Biokin Ltd.).
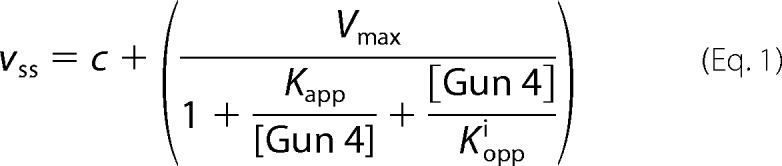
Functional MgCh enzyme assays were performed essentially as described previously (17). Calibration curves using known standard concentrations of magnesium deuteroporphyrin IX (MgD) were created for each concentration of Gun4 analyzed. Plots of MgCh steady state activity with respect to Gun4 concentration were fitted using Igor Pro (Wavemetrics Inc) to provide apparent constants for the formation of the MgCh-Gun4 complex (Kapp). At high (8 μm) deuteroporphyrin IX (DIX) concentrations, the activity of MgCh is inhibited when Gun4 reaches high concentrations. These data were fitted to Equation 1, which allows for this observed inhibition by calculating an apparent inhibition constant (Kiapp). At low concentrations of DIX (0.8 μm), no inhibition is found in the Gun4 concentration range observed; therefore Equation 2 was used to obtain estimates of complex formation. In Equations 1 and 2, νss is the observed rate of MgCh, c is the rate of MgCh in absence of Gun4, and Vmax is the theoretical maximum rate.
 |
 |
Results
Comparison of Available Crystal Structures of Gun4 Proteins and the Prediction of Loops
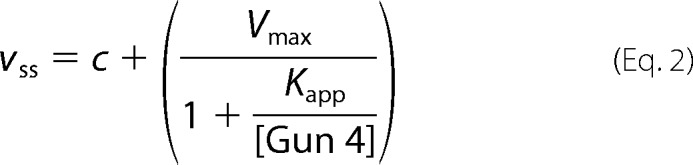
To study the role of porphyrin binding by the Gun4 protein, we performed in silico modeling, where recent developments (mainly motivated by the pharmacological industry) have allowed an accurate all-atom description of the ligand binding mechanism for complex systems such as Gun4 (30–32). First, we inspected the three available Gun4 crystal structures, all in the porphyrin-free form, deposited in the Protein Data Bank when this project commenced: the Synechocystis Gun4 protein (13), the Gun4 from T. elongatus, and so called Gun4-1 mutant (L105F) also from T. elongatus (12). We note that two more structures have been reported since (33, 34). As previously reported (12), T. elongatus Gun4 and L105F structures are practically identical except for the L105F change. Synechocystis and T. elongatus WT structures, however, show large differences in orientation of α2/α3 and α6/α7 loops (Fig. 1), part of the highly conserved Gun4 core domain (13). Based on an extensive analysis of site-directed Synechocystis Gun4 mutant proteins and NMR chemical shift measurements, this core domain was proposed as the porphyrin binding pocket (Fig. 1 and Ref. 13). However, it should be noted that docking of MgP into the binding pocket as proposed in Ref. 13 is not possible for Synechocystis Gun4. The tight packing derived from the above loop orientations introduces severe steric clashes that return no bound poses from standard docking approaches.
FIGURE 1.
An overview of original Gun4 structures and the prediction of α2/α3 and α6/α7 loops. A, comparison of two cyanobacterial Gun4 crystal structures: T. elongatus (blue) and Synechocystis (gold) corresponding to the Protein Data Bank codes 1Z3X and 1Y6I, respectively (12, 13). Loops involved in opening of the binding site (see main text) are highlighted. Ovals indicate sites of crystal contacts in the symmetric units. The inset shows the position of Leu-105 (T. elongatus) buried among α1, α2, and α4 helixes. B, alignment of the original T. elongatus structure (beige) and the Synechocystis (gold) and T. elongatus (blue) structures after loop predictions by Prime. Note the similar orientation of Trp-192 and Tyr-197 residues. C, the original loop orientation and the position of Trp-192 residue in the Synechocystis Gun4 structure (green) compared with the lowest energy predictions by Prime without crystal mates (red). The α2/α3 loop corresponds to Pro-122–Phe-132 residues, and the highlighted part of the α6/α7 loop corresponds to Ser-185–Gly-195.
Interestingly, although the α2/α3 loop has high atomic crystal β-factors, the α6/α7 one presents quite low scores, indicating its rigidity (not common in such a large loop) or constrained nature. Indeed, inspection of the x-ray symmetric unit indicated critical crystallographic contacts capable of defining (constraining) the α6/α7 loop position. The number of interactions in this region is larger in Synechocystis Gun4 and their nature (salt bridges) is also stronger. In particular, in Synechocystis Gun4 we find Trp-192–Glu-7, Arg-191–Glu-49 and Thr-190–Glu-49 contacts, whereas in the T. elongatus structure, we only observe the Lys-192–His-69 one. The conformational mobility of the α6/α7 loop appears to drive a change in the mobile α2/α3 adjacent loop, which further restricts the porphyrin access into the putative binding pocket (Fig. 1).
To check whether these contacts produce crystallographic artifacts in the loops, we performed loop prediction in the presence and absence of crystal mates. In solution, where no crystal mates are included, the predicted α6/α7 loop for T. elongatus Gun4 is more closed but still similar to the original structure with an overall open conformation. The predicted Synechocystis Gun4 structure, however, drastically changes from the crystal structure (Fig. 1B), with an overall alpha carbon RMSD of 6.1 Å and an open structure similar to the one obtained from T. elongatus. Additionally, the α2/α3 loop also significantly moved in the predicted Synechocystis Gun4, further opening the cavity (Fig. 1C). We also noted that the conserved aromatic Trp-192 residue, located at the α6/α7 loop, markedly turned and adopted a similar position to the homologous Tyr-197 residue in T. elongatus Gun4 (Fig. 1B). The Trp-192/Tyr-197 residue has been reported to be important for porphyrin binding (12), and therefore we selected a Trp-192 mutant Gun4 for in silico, in vitro, and in vivo analysis.
The Prime loop prediction software allows running simulations that take into account the crystallographic symmetry by adding neighboring units into the model. Assuming neighboring side chains help to constrain loop structures, one would expect to reproduce the crystal structure. This is actually what happens in Gun4, where simulations with crystal mates produce loops with a ∼1–2 Å alpha carbon RMSD with respect to the crystallographically determined structures (hardly distinguishable from the experimental structures). Taken together, these results on both systems point to a strong bias in the loop conformation mediated by crystallographic contacts; thus, the conformation of functionally important loops in Gun4 cannot be assessed purely on the basis of crystallographic data, and study/correction with in silico methods (now a well established practice; Ref. 35) is required. Importantly, this bias, originating from the structural data for crystallographic Gun4, produces a conformation with the loop restricting the access to the expected binding pocket. Thus, we adopted the corrected semi-open loop conformation, as modeled in solution by Prime, for the next round of simulations.
Molecular Simulations of Porphyrin Binding into WT and Mutant Synechocystis Gun4 Proteins
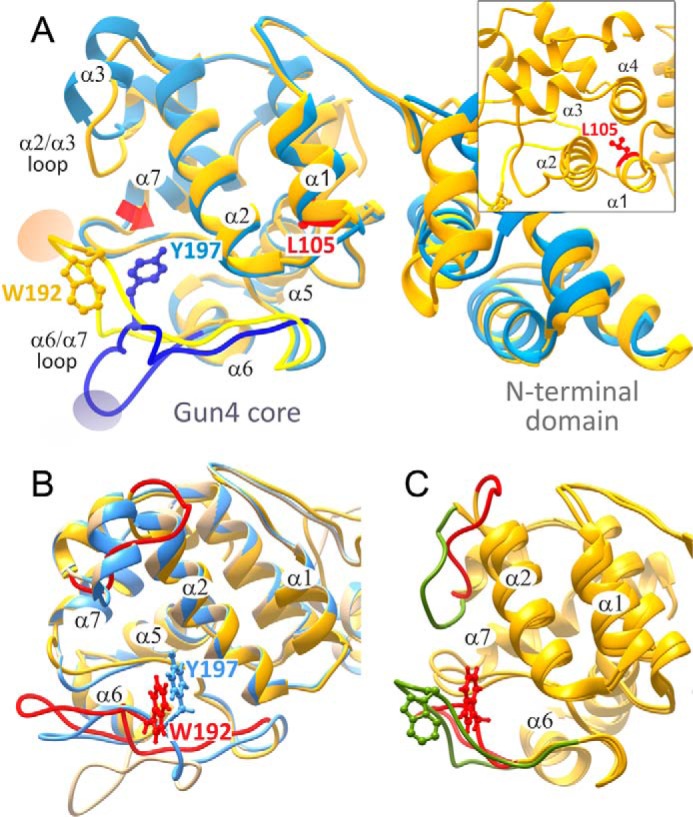
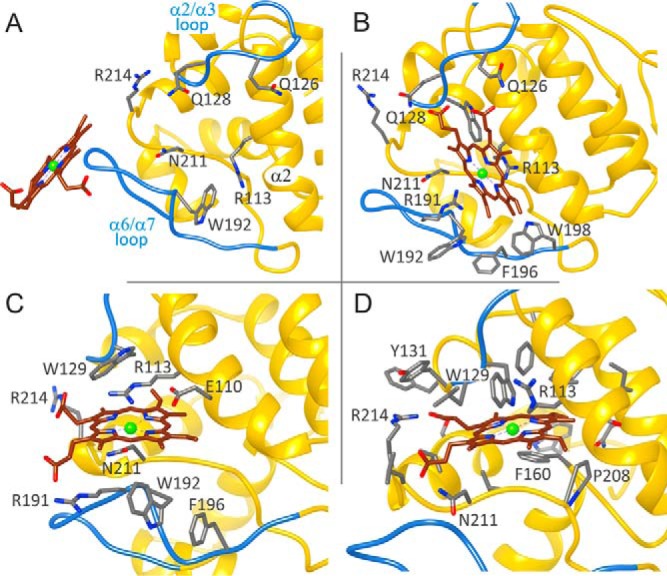
To map the porphyrin binding mechanism, we performed simulations in which the ligand, starting in the bulk solvent, is asked to enter the binding site. We focused on the Synechocystis Gun4 structure because a detailed analysis of site-directed mutants has been already performed together with NMR measurements (13). In addition, Synechocystis Gun4 mutants can be explored using both in vitro and in vivo systems. Fig. 2 shows four different snapshots underlining the porphyrin docking mechanism into Synechocystis Gun4 as observed in the PELE simulation (the entire migration movie can be seen in supplemental Movie S1). In Fig. 2A, we show the initial structure where we placed the porphyrin ligand in the bulk solvent outside the Gun4 binding pocket. Fig. 2B shows a recurrent predocking pose for the protein surface observed for both metal and nonmetal porphyrins. In this pose, the porphyrin stacks onto the Arg-113 side chain and forms hydrogen bonds with two glutamines (Gln-126 and Gln-128) present in the α2/α3 loop. From this predocking site, the porphyrin moves into the binding pocket forming an interesting axial coordination-like motif with Asn-211 and Arg-113; we named this site binding pocket A (Figs. 2C and 3A). As shown below, this structure represents a steady state in protein-ligand interaction energy. From this pose, however, the porphyrin can migrate deeper into the binding pocket with less ligand exposure to solvent and with porphyrin propionate groups anchored by interactions with Arg-214 and Asn-211; we named this site binding pocket B (Figs. 2D and 3B).
FIGURE 2.
Four snapshots along the porphyrin migration pathway and binding site entrance in Synechocystis Gun4. A, initial structure with a bulk solvent-exposed ligand. B, predocking pose with stacking onto Arg-113 and hydrogen bonds with Gln-126 and Gln-128 in the α2/α3 loop. C, pocket A bound structure with an iron axial coordination-like motif with Asn-211 and Arg-113. D, pocket B bound deeper structure.
FIGURE 3.
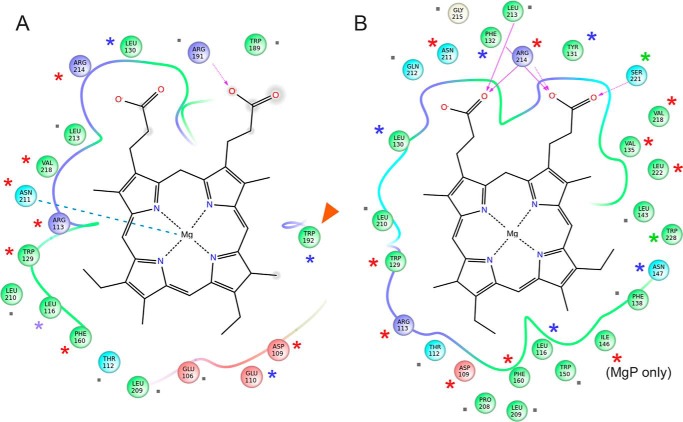
Interaction between MgP and Gun4 residues in two putative binding pockets identified by the PELE simulation. The diagrams in A and B correspond to the position of MgP depicted in C and D of Fig. 2, respectively. Line discontinuities in the vicinity of the propionate groups indicate larger exposure to solvent. Arrows represent hydrogen bonds, the dashed line in A highlights coordination of MgP by the Asn-211 residue, and the red arrowhead shows the Trp-192 residue selected for mutagenesis. Residues marked by asterisks were analyzed by site-directed mutagenesis in previous reports (12, 13) and in this report for Trp-192; green asterisks indicate minimal effect of the replacement of given residue on porphyrin binding (an increase of Kd < 2 for MgP or PIX), blue asterisks mean a moderate effect (2–5), and red asterisks mark residues, which mutation increased Kd more than five times. Gray dots mark residues for which no data are available.
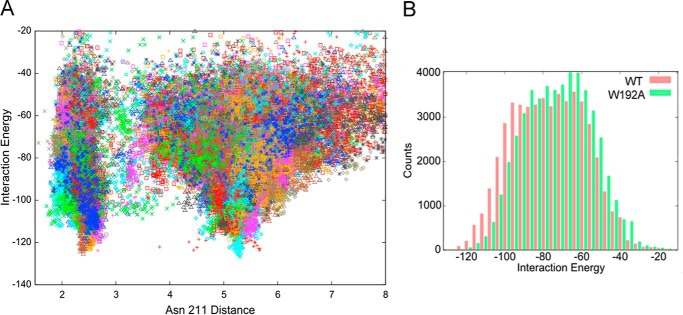
To better characterize the porphyrin binding mode, we performed a refinement search where MgP, starting at pocket A, is allowed to explore a distance of up to 10 Å (measured as the ligand center of mass displacement to the Asn-211 side chain oxygen). Fig. 4A shows the protein-MgP interaction energies for the 50,000 configurations sampled in this refinement procedure. We clearly observe the existence of two minima, which correspond to binding pockets A and B, with similar (almost degenerate) interaction energies. We note that these interaction energies are derived from a classical force field not including any metal coordination component (beyond electrostatic attraction).
FIGURE 4.
Analyses of the protein-MgP interaction energies by PELE. A, protein-ligand interaction energy against the distance to Asn-211 (side chain oxygen atom) along the refinement process for the Synechocystis Gun4. B, the result of a 4 kcal/mol binning of the interaction energies for the Gun4 (red) and the W192A (green) mutant protein.
To distinguish which of these two minima might better represent the biological system, we compared interacting residues with previously reported studies of side-directed Gun4 mutants (12, 13). Indeed, a number of residues that affect binding are involved in both positions (Fig. 3). However, a mutation in Asn-211, which is placed in a coordination position with MgP in the less deep pocket A (Fig. 3A), has been shown to exert a significant effect on the affinity of the MgP analog MgD than for DIX (12). Additionally, the residue homologous to Trp-192 in T. elongatus (Tyr-197), which participates directly in binding only in pocket A (Fig. 3A), is required for high affinity porphyrin binding (Ref. 12; also see below). On the other hand, replacement of Ser-221 has almost no effect on binding (13), which is not in agreement with the situation in pocket B, where Ser-221 forms a hydrogen bond to MgP (Fig. 3B). These are strong indications that the less deep docking position A is closer to the real binding pocket.
To verify our model further, we studied the W192A mutant. This residue and Tyr-197 are oriented differently in the original Synechocystis and T. elongatus structures (Fig. 1) but, after loop prediction and docking, Trp-192 and Tyr-197 are both exposed into the proposed binding pocket A (Figs. 1B and 3A). First, we compared recombinant Synechocystis WT and Gun4-W192A proteins. Circular dichroism spectra showed that the WT and W192A Gun4 have very similar secondary and tertiary structures (Fig. 5). However, a difference in the 200–210-nm region indicates a slightly more compact fold for the W192A protein. Indeed, the affinities of the mutated Gun4 for DIX and MgD were two and three times weaker than the WT, respectively (Table 1). Then we repeated the PELE local refinement search for W192A, observing that the mutant has slightly higher interaction energies and lower density of points at the bottom of the minima. To better quantify this, we binned all points in groups of 4 kcal/mol (Fig. 4B). By doing so, we can now appreciate more clearly the weaker (shift in) interaction energies for the W192A mutant. The direct role of Trp-192 on porphyrin binding is evident from our detailed atomic simulations: inspecting the WT structures along the refinement trajectories, we observe the direct interaction between the Trp-192 side chain and the porphyrin group (Fig. 6 and supplemental Movie S2).
FIGURE 5.
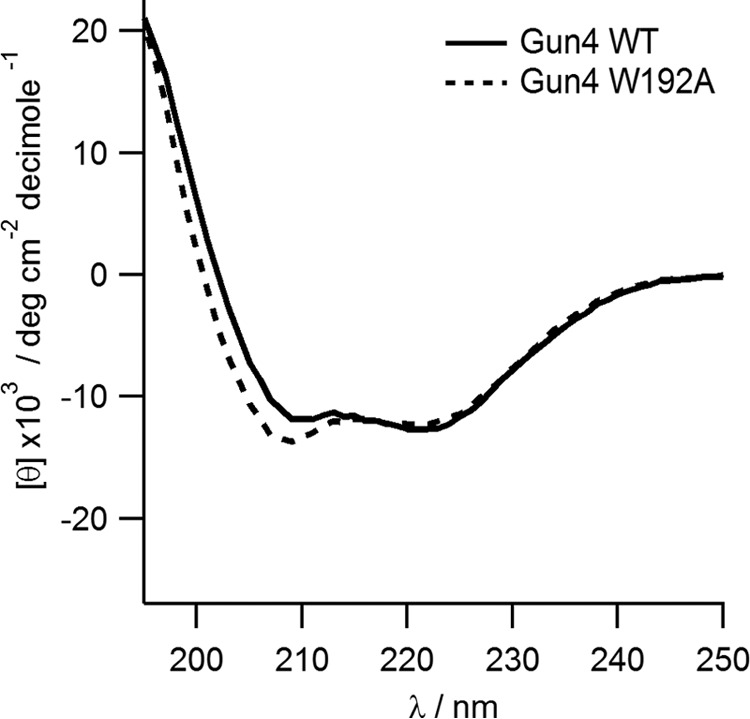
Circular dichroism spectra of the recombinant Synechocystis Gun4 protein and its mutated W192A variant.
TABLE 1.
Dissociation constants (Kd) for the Synechocystis Gun4 protein and W192A mutant
Dissociation constants were calculated for DIX and MgD, monitoring tryptophan fluorescence quenching (see “Experimental Procedures” for details).
| Protein |
Kd |
|
|---|---|---|
| DIX | MgD | |
| μm | ||
| Synechocystis Gun4 | 3.83 ± 0.39 | 2.5 ± 0.09 |
| Synechocystis W192A | 7.32 ± 0.61 | 7.22 ± 0.82 |
FIGURE 6.
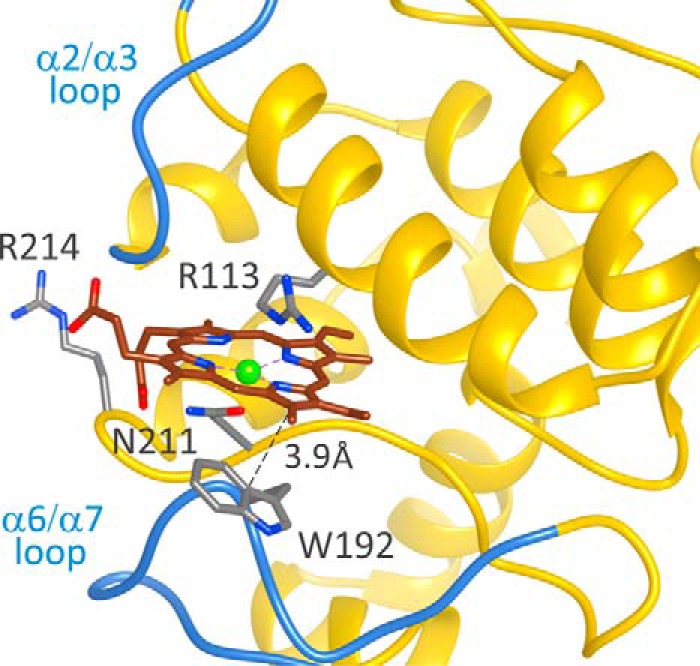
A representative snapshot for the refinement of MgP docking in the Synechocystis Gun4 protein. The direct van der Waals interaction between Trp-192 and the porphyrin ligand is indicated.
Dynamic Simulations of T. elongatus WT and L105F Gun4 Proteins: Cross-correlation Maps
The Gun4-1 mutation was originally identified in a Chl-deficient Arabidopsis mutant with lesions of signaling between the chloroplast and nucleus (11, 37). The basis for the effect of the Leu → Phe (L105F in T. elongatus Gun4) mis-sense mutation has not been elucidated. Our model shows that although the Synechocystis Trp-192 residue is in direct contact with the bound porphyrin model, Leu-105 in T. elongatus Gun4 is located far away from the porphyrin binding site (Fig. 1). Moreover, WT and Gun4-1 T. elongatus crystal structures do not show significant differences (12). Therefore, the mechanism by which the enigmatic Gun4-1 mutation confers a much tighter porphyrin binding (Kd for DIX and MgD decreased 15- and 11-fold, respectively) (12) poses a real challenge. To address this issue, and seeking possible dynamic effects, we employed molecular dynamics simulations.
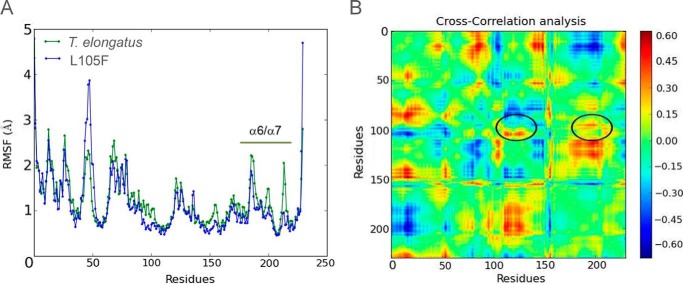
We performed 200-ns molecular dynamics simulations of T. elongatus Gun4 and Gun4-L105F and analyzed the dynamics with root mean square fluctuation and cross-correlation maps. As seen in Fig. 7A, replacement of Leu-105 by Phe produces a significant change in the root mean square fluctuation of several regions, by up to 1Å. The most important change concerning porphyrin binding is the clear loss of mobility in the α6/α7 loop (residues 175–215) for the L105F mutant. The (residue movement) difference cross-correlation map, shown in Fig. 7B, allowed us to establish the mechanism for such a reduction in mobility. If we follow the position around Leu-105, we clearly see two main correlated (red) groups (marked in Fig. 7B). As expected, several residues of helix 2 (residues 110–125) in contact with Leu-105, are correlated in their motion. More importantly, residues of the α6/α7 loop exhibit marked correlation with Leu-105.
FIGURE 7.
Molecular dynamics analysis of the T. elongatus WT Gun4 and L105F mutant. A, root mean square fluctuation (RMSF) for both species. B, cross-correlation differences between T. elongatus WT and the L105F mutant. The black circles highlight the region corresponding to helix 2 and the α6/α7 loop.
Effect of the W192A Mutation on MgCh Activity and Chl Biosynthesis
To understand the physiological role of Gun4 in the cell, we extended the mechanistic description arising from our binding model and performed kinetic and in vivo analyses on the W192A and WT Gun4 proteins. The ability of the W192A mutant to stimulate MgCh activity was assessed by titrating either Synechocystis Gun4 WT or W192A recombinant protein into MgCh assays and monitoring the steady state rate; the more water-soluble porphyrin DIX was used as a substrate instead of PIX. Both Gun4 and W192A stimulated activity ∼3-fold compared with MgCh activity alone. However, at high concentrations of DIX substrate (8 μm), the Kapp (the apparent binding constant for MgCh-Gun4 complex formation) suggests that the Gun4 WT has almost 3-fold tighter binding with MgCh compared with the W192A mutant. At low concentrations (0.8 μm DIX), this effect was even more pronounced with a 4-fold difference in Kapp between WT and W192A and a 50% decrease in Vmax between WT and W192A (Table 2). At 8 μm DIX, there is evidence of inhibition at high concentrations of Gun4 (Fig. 8, closed symbols). Together, these data imply that the lowered affinity of Gun4 to porphyrin leads to a weaker interaction between the mutated Gun4 and the core MgCh complex, and this defect is emphasized at low substrate concentrations.
TABLE 2.
Apparent binding constants of Synechocystis WT and W192A-Gun4 to the core MgCh complex
| Protein | DIX concentration | Vmax | Kapp | Ki app |
|---|---|---|---|---|
| μm | μm min−1 | μm | μm | |
| Gun4 | 8a | 0.444 ± 0.14 | 0.094 ± 0.059 | 0.540 ± 0.356 |
| 0.8b | 0.061 ± 0.005 | 0.043 ± 0.013 | ||
| W192A | 8a | 0.529 ± 0.168 | 0.261 ± 0.133 | 0.523 ± 0.295 |
| 0.8b | 0.0328 ± 0.003 | 0.164 ± 0.063 |
a Fitted to Equation 1 (see “Experimental Procedures” for details).
b Fitted to Equation 2 (see “Experimental Procedures” for details).
FIGURE 8.
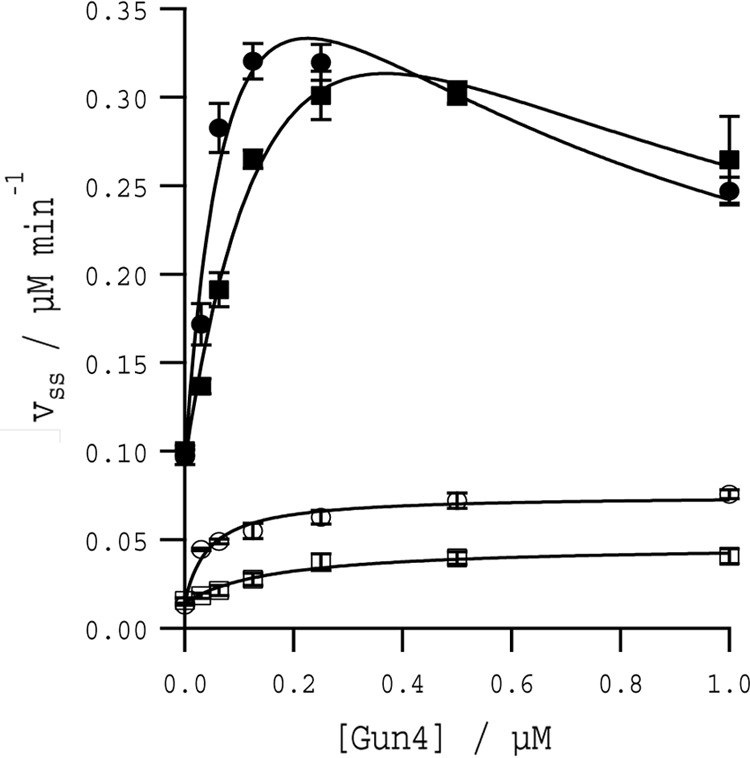
In vitro stimulation of the MgCh activity by Synechocystis Gun4 and W192A proteins. Assembly titrations of MgCh and WT Gun4 (circles) or W192A (squares). Assays contained 0.1 μm ChlD, 0.2 μm ChlI, and 0.4 μm ChlH in 50 mm MOPS/KOH, 0.3 m glycerol, 1 mm DTT, 1 mm free Mg2+, I = 0.1 with KCl, pH 7.7, at 34 °C with 8 μm DIX (closed symbols) and 0.8 μm DIX (open symbols). The curves at 8 μm DIX can be described by Equation 1, and those at 0.8 μm DIX can be described by Equation 2, with characterizing parameters in Table 2.
To monitor possible effects of the W192A mutation in vivo, we constructed a Synechocystis gun4-W192A strain possessing the gun4-W192A allele instead of the original gun4 gene. Mutated cells exhibited no difference in phycobilisome content but contained 20% less Chl under moderate light conditions (30 μE of photons) and 35% less Chl under saturating light (300 μE of photons; Fig. 9A). Under these conditions, the cellular levels of mutated Gun4, ChlH, and ChlM were comparable with WT levels (Fig. 9B), although the mutant contained significantly more ChlI associated with membranes (Fig. 9B). To identify how the W192A mutation affects Chl biosynthesis, we analyzed pools of all intermediates in the tetrapyrrole pathway from coproporphyrin(ogen) III (CoPP) to chlorophyllide. Detection was based on a separation by an HPLC system connected with two fluorescence detectors (Fig. 9C). Notably, even under moderate light intensity, the mutant exhibited significantly decreased levels of MgP, MgP methyl ester, and both the monovinyl and divinyl forms of protochlorophyllide. However, the mutant did not accumulate PIX; in fact, levels of CoPP and PIX were rather lower in the mutant (Fig. 9C).
FIGURE 9.
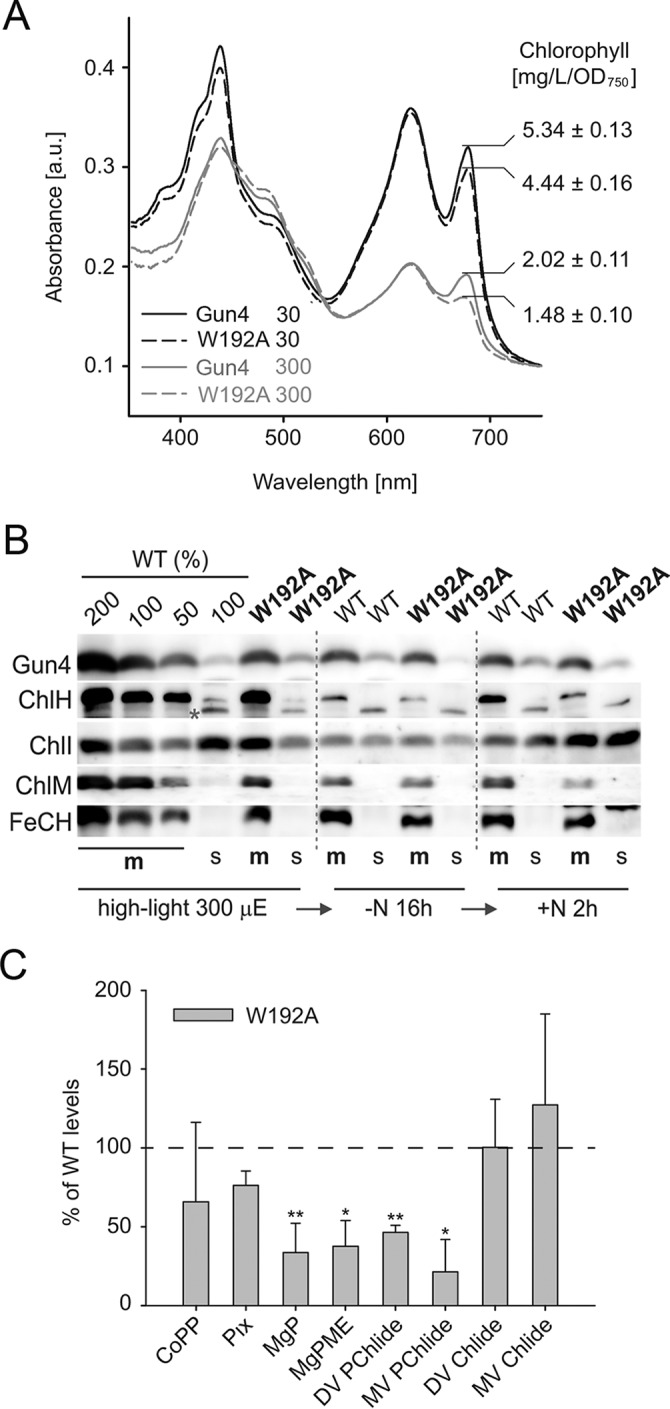
Chl biosynthesis in the Synechocystis gun4-W192A strain. A, whole cell absorbance spectra of the Synechocystis WT and gun4-W192A strain (W192A) grown at 30 and 300 μmol of photons m−2 s−1. Chl is represented by 680-nm peak and phycobilinoproteins by the 625-nm peak. Spectra were normalized to light scattering at 750 nm. The total Chl content per optical density is indicated. B, immunodetection of Gun4, MgCh ChlH and ChlI subunits, and ChlM and ferrochelatase (FeCH) in the WT and gun4-W192A strains grown under saturating light and during nitrogen depletion and regreening (see “Results” and Fig. 10 for details). The membrane (m) and soluble (s) protein fractions were separated by SDS-electrophoresis, blotted, and probed using indicated antibodies. Purple asterisk marks an unspecific cross-reaction or, potentially, a low abundant variant of ChlH. C, the relative levels of Chl precursors in the gun4-W192A strain in comparison to the WT. The values shown represent means ± S.D. from five independent measurements. The asterisks indicate statistically significant differences in precursor levels as tested using a paired Student's t test. *, p = 0.05; **, p = 0.01. Pix, protoporphyrin(ogen) IX; MgPME, MgP methyl ester; DV PChlide, divinyl-protochlorophyllide; MV PChlide, monovinyl-protochlorophyllide; DV Chlide, divinyl-chlorophyllide; MV Chlide, monovinyl-chlorophyllide.
The result of the kinetic study of the MgCh enzyme in vitro (Table 2) indicated that the effect of the W192A mutation in vivo should be more pronounced when the level of MgCh substrate PIX decreases. To mimic such a situation, we depleted pools of Chl precursors by incubating cells for 16 h in a growth medium without nitrogen (Fig. 10, top two panels). After this treatment, the only detectable Chl precursor is monovinyl-chlorophyllide, which, however, originates from dephytolation of existing Chl and not from de novo synthesis (36). Chl biosynthesis was restarted by adding of NaNO3, and after 2 h, a divinyl-protochlorophyllide peak became visible in WT chromatograms (Fig. 10, bottom two panels). In the gun4-W192A strain, Chl biosynthesis was much delayed; there was no detectable protochlorophyllide pool, and the amount of MgP methyl ester is ∼20% of that in the WT. Levels of CoPP and PIX were, however, similar (Fig. 10, bottom two panels) in both strains, suggesting that either the flux through the whole pathway is down-regulated in the mutant and/or any excess of PIX is metabolized by ferrochelatase. Levels of WT and mutated Gun4 changed just slightly during nitrogen starvation, but the ChlI subunit and the ChlM enzyme were significantly reduced, and the ChlH subunit almost completely disappeared in both strains (Fig. 9B). Interestingly, after adding nitrogen, both the ChlH and ChlM started to accumulate, and this accumulation was apparently delayed in the mutant (Fig. 9B). We can therefore conclude that the Gun4-porphyrin complex is important for quick reactivation of Chl biosynthesis after a period of nutrient stress.
FIGURE 10.
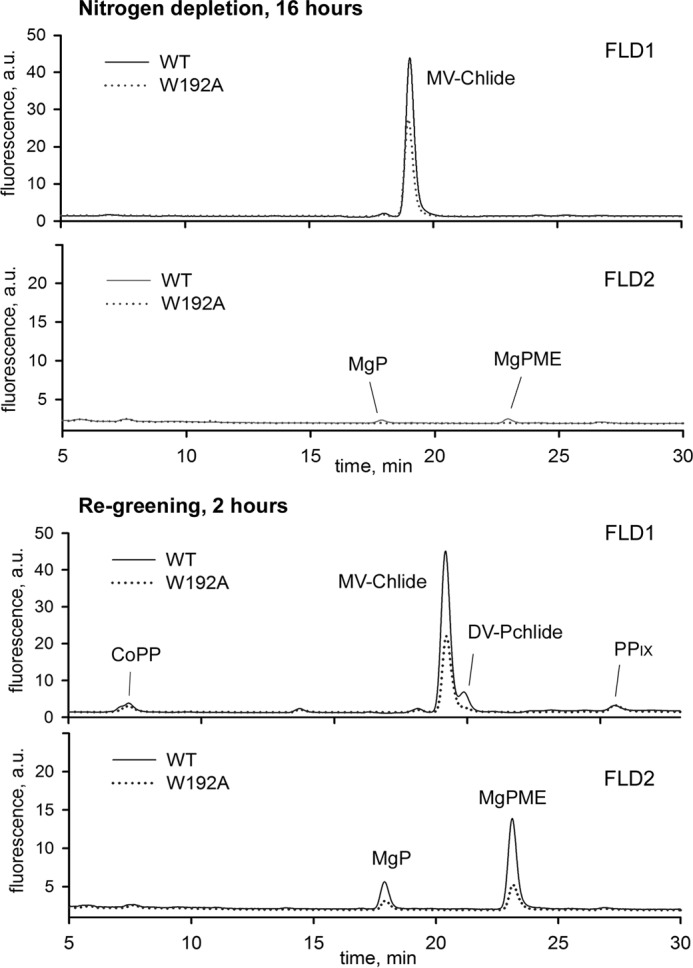
Analysis of Chl precursors in the Synechocystis WT and gun4-W192A strains during recovery from nitrogen starvation. Top two panels, levels of Chl precursors in cells grown under saturating light without nitrogen for 16 h; bottom two panels, 2 h after adding of 10 mm NaNO3. Extracted pigments were analyzed by HPLC equipped with a pair of fluorescence detectors (FLD1 and FLD2) set for different wavelengths to cover all Chl precursors from coproporphyrinogen III (oxidized spontaneously to coproporphyrin III) to monovinyl chlorophyllide. The pigments are labeled as in Fig. 9.
Discussion
Mechanism of Porphyrin Binding to the Gun4 Protein
Gun4 is required for the synthesis of sufficient amounts of Chl in all groups of oxygenic phototrophs. It also appears that Gun4 proteins from virtually all sources stimulate MgCh activity via an interaction with the ChlH subunit and bind porphyrins with the highest affinity to MgP (14, 16, 17). The essential role of porphyrin binding for Gun4 function can be suggested from amino acid alignments (13) because residues in the proposed MgP binding site (α6/α7 loop residues, Arg-214, and Asn-211) are conserved through evolution from cyanobacteria to plants.
NMR data, crystal structures, and site-directed mutagenesis suggested that the porphyrin binding region lies within the pocket formed at the intersection of the hydrophobic α6/α7 loop, the α2/α3 loop and by helices α2 and α7 (Fig. 1). However, the mechanism of the Gun4-porphyrin interaction and how Gun4 discriminates between empty and metal-bound porphyrins remained unknown.
Our data imply that the orientation of the α6/α7 loop is critical for porphyrin binding. After removing the crystallographic artifacts, arising from protein-protein interactions in the crystal, the α6/α7 loop in both Synechocystis and T. elongatus Gun4 adopted similar configurations, indicating that the shape of the loop is conserved (Fig. 1, B and C). The observed flexibility of the α6/α7 loop and its possible conformational changes after porphyrin binding are consistent with changes in NMR-determined chemical shifts upon addition of DIX (13).
An important finding of our simulation is the ability of Asn-211 to coordinate MgP, which offers an explanation of the stronger affinity for MgP than PIX, demonstrated experimentally (Table 1). As described earlier, we expect that the binding pocket A, which includes coordination of MgP by Asn-211 (Fig. 3A), is closer to the real pocket. Interestingly, the replacement of Trp-192 by alanine had a stronger effect on the binding of MgD than to DIX, yielding a Gun4 with almost equal affinity for both these porphyrins (Table 1). It is possible that the Trp-192 is critical for the positioning of MgP in the proximity to Asn-211. It would be interesting to employ PELE to compare binding energies for MgP and PIX in WT and W192A mutant. However, the lack of accuracy when describing metal interactions by classical force fields does not allow a quantitative comparison between these two porphyrins.
While this paper was under review a Synechocystis Gun4 structure with bound MgD has been deposited to the Protein Data Bank (33). In this structure, the loop orientation is more open than predicted by our modeling work, and therefore the bound MgD is in a more planar orientation with respect to the α6/α7 loop, and it lies closer to helix 7. However, the main conclusions we derived from the simulations are valid: the α6/α7 loop residues are crucial for the formation of a relatively shallow binding pocket, MgP is coordinated by Asn-211, and also the Trp-192 residue interacts with MgD essentially as we proposed.
Asn-211 is highly conserved, and we speculate that the coordination is a universal feature of Gun4 proteins. On the other hand, there might be some variability in orientation of porphyrin propionates groups between cyanobacterial and plant Gun4 proteins. According to Ref. 16, the homologue Arg-214 residue in Arabidopsis Gun4 shows a defect in MgP binding, although it is not essential, unlike in the Synechocystis counterpart.
The Gun4-1 mutation was originally identified in Arabidopsis as the one of several genome uncoupled mutations causing modified plastid to nucleus signaling (37). The level of Gun4-1 protein in Arabidopsis is very low (11); however, this mutation was characterized later using recombinant T. elongatus and Synechocystis Gun4-1 proteins that both exhibit an ∼10-fold higher affinity for porphyrins than the WT Gun4 (12). Our results based on molecular dynamic simulations show a clear steric pathway connecting Leu-105 with Helix 2 and the α6/α7 loop (Fig. 7), where the loss of mobility is effectively transmitted. Indeed, restricted flexibility of the Helix 2 and the α6/α7 loop can significantly affect the porphyrin binding, supporting the critical role of these segments in our docking model (Fig. 2).
The Role of Gun4 in Regulation of Tetrapyrrole Biosynthesis
Although Gun4 proteins bind a broad spectrum of porphyrins including heme b or pheophorbide (16), MgP might be the crucial ligand in vivo, because there is evidence that only the binding of MgP (the product of MgCh) but not PIX (MgCh substrate) enhances MgCh activity. This assumption is based on an in vitro analysis of Synechocystis Gun4 proteins mutated at residues critical for binding DIX but not for binding MgD and vice versa. Mutants of Gun4 where DIX binding is abolished are still able to stimulate MgCh activity, whereas Gun4 mutants incapable of binding MgD completely failed to stimulate MgCh (13). These results suggest that Gun4 is implicated in release of the product (MgP) from ChlH. The fact that MgP is coordinated by the highly conserved Asn-211 indicates that Gun4 has evolved to bind MgP (or to bind it better then PIX) and connects the function of Gun4 with binding of this porphyrin. In the in vitro assay at limiting concentrations of DIX, W192A-Gun4 is less effective in stimulating the MgCh enzyme because of the compromised interaction between Gun4 and the MgCh complex (Table 2). It is not known when Gun4 binds to ChlH during the catalytic cycle of MgCh, but it appears that the high affinity of Gun4 to the MgP product facilitates a transient assembly of a Gun4-MgCh complex, stimulating MgCh activity. We expect that a similar defect also occurs in vivo in Synechocystis gun4-W192A cells (see below).
The in vivo results obtained for various Gun4 mutants show complex changes in tetrapyrrole metabolism, and it could be misleading to describe Gun4 simply as an enhancer of the MgCh enzyme. Overexpression of Gun4 in plants causes an aberrant accumulation of Chl, accompanied by elevated steady state levels of PIX and MgP porphyrins (19). In the green alga Chlamydomonas reinhardtii, overexpression of Gun4 also increases Chl content per cell (38), whereas a C. reinhardtii strain possessing a low Gun4 content has reduced both Chl and PIX levels (15, 20). Only after the complete elimination of Gun4 do cyanobacteria, algae, and plants accumulate aberrant concentrations of PIX (14, 20, 39).
To discuss the role of Gun4 in controlling Chl biosynthesis, one has to be careful in comparing data obtained from different organisms and under different growth conditions. However, the Gun4 mutations are generally more pronounced with increasing light intensity (14, 19, 38). The sensitivity of Gun4-less mutants to light and changes in ALA synthesizing capacity led to a hypothesis that Gun4 binds excessive porphyrins, which serves as a signal to down-regulate ALA formation (19, 20). Although this model can explain reduced ALA synthesis in Gun4 or MgCh mutants (40), it is not consistent with the massive accumulation of PIX in ferrochelatase mutants (8, 41).
We propose an alternative model, taking into account that Gun4 has been detected as a part of large membrane complexes (11). Under conditions when Synechocystis cells reach maximal growth rate (saturating light, intensively air-bubbled culture), Gun4 and ChlH are localized almost exclusively in the membrane fraction (Fig. 9B). Thus, the synthesis of MgP is likely located in a membrane-associated complex where Gun4 interacts with ChlH (21) and presumably also with the following enzyme ChlM (42). Although it has been reported that in plants the association of Gun4 with the membrane requires porphyrins (21), we detected Gun4 in the membrane fraction even in cells lacking virtually all Chl intermediates except chlorophyllide (Fig. 9B). Weaker affinity for porphyrins (W192A-Gun4) or a very low content of ChlH (nitrogen depletion) does not prevent Gun4 from associating with membranes and does not significantly alter the Gun4 level (Fig. 9B). This robustness contrasts with the disappearance of ChlH during nitrogen depletion, and its resynthesis once nitrogen becomes available. Because the affinity of Gun4 for porphyrins correlates with affinity of Gun4 for the MgCh enzyme complex during catalysis (Table 2), we propose that in the presence of PIX or MgP the interaction of Gun4 with ChlH helps to stabilize a membrane-associated MgCh/ChlM complex. A Gun4-porphyrin complex can modulate disassembly and reassembly of this enzymatic unit and thereby expose individual components to a post-translational control. It should be noted that in both cyanobacteria and plants, MgCh subunits are present even in the absence of Gun4 (14, 19). Gun4 is not essential for the synthesis/stability of these proteins, but it might be a critical component allowing cells to maintain optimal channeling of PIX into the Chl metabolic branch. This role would be particularly important under fluctuating conditions requiring well tuned deactivation and reactivation of MgCh and ChlM enzymes.
Consistent with this model, the Arabidopsis Gun4-less mutant is much more sensitive to a fluctuating light regime. Under continuous low light, the mutant can be partly complemented by supplementing with ALΑ (24). Interestingly, in Arabidopsis lacking Gun4, the level of ChlM is drastically reduced under fluctuating conditions, but it is restored by increasing the PIX concentration by exogenous ALA (19).
According to the available data, it appears that there is cross-talk between the activity of the MgCh-Gun4 complex and regulation of metabolite flux through the whole tetrapyrrole pathway, keeping the PIX pool small (19, 20, 40, 43). As already discussed, other enzymes in the tetrapyrrole pathway, including ferrochelatase, could be a part of the identical membrane domain (5), so it is not difficult to imagine that the deactivation of MgCh allows ferrochelatase to attenuate ALA formation, e.g. via a heme feedback loop (6). We observed that ferrochelatase is up-regulated during nitrogen starvation (Fig. 9B). The presence of Gun4 in such a complex might be structurally important, which would explain why its complete elimination causes accumulation of PIX. As already noted, a low level of Gun4 is sufficient for balancing the PIX pool (15).
In this work, we demonstrated that replacement of Gun4 by a variant with lower affinity for porphyrins reduced the pools of intermediates in the Chl branch, but CoPP and PIX levels are less affected. As expected from in vitro data, the effect of this mutation is more pronounced once the mutated cells have to deal with fluctuations of metabolites in the tetrapyrrole pathway. The docking of MgP in Gun4 and the model of Gun4-based regulation described here can serve as a framework for future studies addressing Gun4 function.
Author Contributions
J. K. constructed the Synechocystis gun4-W192A strain and together with R. S. performed all in vivo experiments, and I. C. d. V. and V. G. performed molecular simulations and porphyrin docking. P. A. D. prepared plasmid for expression of the recombinant W192A Gun4, N. B. P. A. and A. A. B. designed and performed in vitro kinetic analyses, and N. B. P. A. also determined dissociation constants. R. S., V. G., N. B. P. A., and C. N. H. jointly wrote the article, and R. S. had major responsibility for the project.
Supplementary Material
This work was supported by Project P501/12/G055 of the Czech Science Foundation, by National Programme of Sustainability I Grant LO1416, by ERC Grant 2009-Adg25027-PELE (to V. G.). This work was also supported by Project Algain Grant EE2.3.30.0059 (to J. K.), Awards BB/G021546/1 and BB/M000265/1 from the Biotechnology and Biological Sciences Research Council (to N. B. P. A., P. A. D., A. A. B., and C. N. H.), and Advanced Award 338895 from the European Research Council (to C. N. H.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Movies S1 and S2.
- ALA
- δ-aminolevulinic acid
- PIX
- protoporphyrin IX
- MgCh
- magnesium chelatase
- MgP
- magnesium protoporphyrin IX
- ChlM
- MgP methyltransferase enzyme
- Chl
- chlorophyll
- PELE
- Protein Energy Landscape Exploration
- MgD
- magnesium deuteroporphyrin IX
- CoPP
- coproporphyrin(ogen) III
- DIX
- deuteroporphyrin IX.
References
- 1.Meskauskiene R., Nater M., Goslings D., Kessler F., op den Camp R., and Apel K. (2001) FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 98, 12826–12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apel K., and Hirt H. (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399 [DOI] [PubMed] [Google Scholar]
- 3.Papenbrock J., Mock H. P., Kruse E., and Grimm B. (1999) Expression studies in tetrapyrrole biosynthesis: inverse maxima of magnesium chelatase and ferrochelatase activity during cyclic photoperiods. Planta 208, 264–273 [Google Scholar]
- 4.Kopečná J., Komenda J., Bučinská L., and Sobotka R. (2012) Long-term acclimation of the cyanobacterium Synechocystis sp PCC 6803 to high light is accompanied by an enhanced production of chlorophyll that is preferentially channeled to trimeric Photosystem I. Plant Physiol. 160, 2239–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sobotka R. (2014) Making proteins green; biosynthesis of chlorophyll-binding proteins in cyanobacteria. Photosynth. Res. 119, 223–232 [DOI] [PubMed] [Google Scholar]
- 6.Czarnecki O., and Grimm B. (2012) Post-translational control of tetrapyrrole biosynthesis in plants, algae, and cyanobacteria. J. Exp. Bot. 63, 1675–1687 [DOI] [PubMed] [Google Scholar]
- 7.Sobotka R., Komenda J., Bumba L., and Tichy M. (2005) Photosystem II assembly in CP47 mutant of Synechocystis sp PCC 6803 is dependent on the level of chlorophyll precursors regulated by ferrochelatase. J. Biol. Chem. 280, 31595–31602 [DOI] [PubMed] [Google Scholar]
- 8.Sobotka R., McLean S., Zuberova M., Hunter C. N., and Tichy M. (2008) The C-terminal extension of ferrochelatase is critical for enzyme activity and for functioning of the tetrapyrrole pathway in Synechocystis strain PCC 6803. J. Bacteriol. 190, 2086–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo T., Fan T., Liu Y., Rothbart M., Yu J., Zhou S., Grimm B., and Luo M. (2012) Thioredoxin redox regulates ATPase activity of magnesium chelatase CHLI subunit and modulates redox-mediated signaling in tetrapyrrole biosynthesis and homeostasis of reactive oxygen species in pea plants. Plant Physiol. 159, 118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson L. C., Jensen P. E., and Hunter C. N. (1999) Magnesium chelatase from Rhodobacter sphaeroides: initial characterization of the enzyme using purified subunits and evidence for a BchI-BchD complex. Biochem. J. 337, 243–251 [PMC free article] [PubMed] [Google Scholar]
- 11.Larkin R. M., Alonso J. M., Ecker J. R., and Chory J. (2003) GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299, 902–906 [DOI] [PubMed] [Google Scholar]
- 12.Davison P. A., Schubert H. L., Reid J. D., Iorg C. D., Heroux A., Hill C. P., and Hunter C. N. (2005) Structural and biochemical characterization of Gun4 suggests a mechanism for its role in chlorophyll biosynthesis. Biochemistry 44, 7603–7612 [DOI] [PubMed] [Google Scholar]
- 13.Verdecia M. A., Larkin R. M., Ferrer J. L., Riek R., Chory J., and Noel J. P. (2005) Structure of the Mg-chelatase cofactor GUN4 reveals a novel hand-shaped fold for porphyrin binding. PLos Biol. 3, e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sobotka R., Dühring U., Komenda J., Peter E., Gardian Z., Tichy M., Grimm B., and Wilde A. (2008) Importance of the cyanobacterial GUN4 protein for chlorophyll metabolism and assembly of photosynthetic complexes. J. Biol. Chem. 283, 25794–25802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Formighieri C., Ceol M., Bonente G., Rochaix J. D., and Bassi R. (2012) Retrograde signaling and photoprotection in a gun4 mutant of Chlamydomonas reinhardtii. Mol. Plant 5, 1242–1262 [DOI] [PubMed] [Google Scholar]
- 16.Adhikari N. D., Orler R., Chory J., Froehlich J. E., and Larkin R. M. (2009) Porphyrins promote the association of GENOMES UNCOUPLED 4 and a Mg-chelatase subunit with chloroplast membranes. J. Biol. Chem. 284, 24783–24796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams N. B., Marklew C. J., Qian P., Brindley A. A., Davison P. A., Bullough P. A., and Hunter C. N. (2014) Structural and functional consequences of removing the N-terminal domain from the magnesium chelatase ChIH subunit of Thermosynechococcus elongatus. Biochem. J. 464, 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karger G. A., Reid J. D., and Hunter C. N. (2001) Characterization of the binding of deuteroporphyrin IX to the magnesium chelatase H subunit and spectroscopic properties of the complex. Biochemistry 40, 9291–9299 [DOI] [PubMed] [Google Scholar]
- 19.Peter E., and Grimm B. (2009) GUN4 is required for posttranslational control of plant tetrapyrrole biosynthesis. Mol. Plant 2, 1198–1210 [DOI] [PubMed] [Google Scholar]
- 20.Brzezowski P., Schlicke H., Richter A., Dent R. M., Niyogi K. K., and Grimm B. (2014) The GUN4 protein plays a regulatory role in tetrapyrrole biosynthesis and chloroplast-to-nucleus signalling in Chlamydomonas reinhardtii. Plant J. 79, 285–298 [DOI] [PubMed] [Google Scholar]
- 21.Adhikari N. D., Froehlich J. E., Strand D. D., Buck S. M., Kramer D. M., and Larkin R. M. (2011) GUN4-porphyrin complexes bind the ChlH/GUN5 subunit of Mg-chelatase and promote chlorophyll biosynthesis in Arabidopsis. Plant Cell 23, 1449–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Case D. A., Darden T. E., Cheatham I., Simmerling C. L., Wang J., Duke R. E., Luo R., Walker R. C., Zhang W., Merz K. M., Roberts B., Wang B., Hayik S., Roitberg A., Seabra G., LKolossváry I., Wong K. F., Paesani F., Vanicek J., Liu J., Wu X., Brozell S. R., Steinbrecher T., Gohlke H., Cai Q., Ye X., Wang J., Hsieh M.-J., Cui G., Roe D. R., Mathews D. H., Seetin M. G., Sagui C., Babin V., Luchko T., Gusarov S., Kovalenko A., and Kollman P. A. (2010) Amber 11, University of California, San Francisco [Google Scholar]
- 23.Sastry G. M., Adzhigirey M., Day T., Annabhimoju R., and Sherman W. (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 27, 221–234 [DOI] [PubMed] [Google Scholar]
- 24.Borrelli K. W., Vitalis A., Alcantara R., and Guallar V. (2005) PELE: Protein energy landscape exploration: a novel Monte Carlo based technique. J. Chem. Theory Comput. 1, 1304–1311 [DOI] [PubMed] [Google Scholar]
- 25.Lee J., Lee H. J., Shin M. K., and Ryu W. S. (2004) Versatile PCR-mediated insertion or deletion mutagenesis. BioTechniques 36, 398–400 [DOI] [PubMed] [Google Scholar]
- 26.Porra R. J., Thompson W. A., and Kriedemann P. E. (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 975, 384–394 [Google Scholar]
- 27.Pilný J., Kopečná J., Noda J., and Sobotka R. (2015) Detection and quantification of heme and chlorophyll precursors using a high performance liquid chromatography (HPLC) system equipped with two fluorescence detectors. Bio-protocol 5, e1390 [Google Scholar]
- 28.Adams N. B., Marklew C. J., Brindley A. A., Hunter C. N., and Reid J. D. (2014) Characterization of the magnesium chelatase from Thermosynechococcus elongatus. Biochem. J. 457, 163–170 [DOI] [PubMed] [Google Scholar]
- 29.Jensen P. E., Gibson L. C., Henningsen K. W., and Hunter C. N. (1996) Expression of the chlI, chlD, and chlH genes from the cyanobacterium Synechocystis PCC6803 in Escherichia coli and demonstration that the three cognate proteins are required for magnesium-protoporphyrin chelatase activity. J. Biol. Chem. 271, 16662–16667 [DOI] [PubMed] [Google Scholar]
- 30.Kotev M., Lecina D., Tarragó T., Giralt E., and Guallar V. (2015) Unveiling prolyl oligopeptidase ligand migration by comprehensive computational techniques. Biophys. J. 108, 116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madadkar-Sobhani A., and Guallar V. (2013) PELE web server: atomistic study of biomolecular systems at your fingertips. Nucleic Acids Res. 41, W322–W328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hernández-Ortega A., Borrelli K., Ferreira P., Medina M., Martínez A. T., and Guallar V. (2011) Substrate diffusion and oxidation in GMC oxidoreductases: an experimental and computational study on fungal aryl-alcohol oxidase. Biochem. J. 436, 341–350 [DOI] [PubMed] [Google Scholar]
- 33.Chen X., Pu H., Wang X., Long W., Lin R., and Liu L. (2015) Crystal structures of GUN4 in complex with porphyrins. Mol. Plant 8, 1125–1127 [DOI] [PubMed] [Google Scholar]
- 34.Tarahi Tabrizi S., Langley D. B., Harrop S. J., Duff A. P., and Willows R. D. (2015) Structure of GUN4 from Chlamydomonas reinhardtii. Acta Crystallogr. F Struct. Biol. Commun. 71, 1094–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guallar V., Jacobson M., McDermott A., and Friesner R. A. (2004) Computational modeling of the catalytic reaction in triosephosphate isomerase. J. Mol. Biol. 337, 227–239 [DOI] [PubMed] [Google Scholar]
- 36.Kopečná J., Pilný J., Krynická V., Tomčala A., Kis M., Gombos Z., Komenda J., and Sobotka R. (2015) Lack of phosphatidylglycerol inhibits chlorophyll biosynthesis at multiple sites and limits chlorophyllide reutilization in the cyanobacterium Synechocystis 6803. Plant Physiol. 169, 1307–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Susek R. E., Ausubel F. M., and Chory J. (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74, 787–799 [DOI] [PubMed] [Google Scholar]
- 38.Grovenstein P. B., Wilson D. A., Lankford K. D., Gaston K. A., Perera S., and Mitra M. (2013) Identification and molecular characterization of the second Chlamydomonas gun4 mutant, gun4-II. F1000Res 2, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mochizuki N., Tanaka R., Tanaka A., Masuda T., and Nagatani A. (2008) The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 105, 15184–15189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papenbrock J., Mock H. P., Tanaka R., Kruse E., and Grimm B. (2000) Role of magnesium chelatase activity in the early steps of the tetrapyrrole biosynthetic pathway. Plant Physiol. 122, 1161–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Papenbrock J., Mishra S., Mock H. P., Kruse E., Schmidt E. K., Petersmann A., Braun H. P., and Grimm B. (2001) Impaired expression of the plastidic ferrochelatase by antisense RNA synthesis leads to a necrotic phenotype of transformed tobacco plants. Plant J. 28, 41–50 [DOI] [PubMed] [Google Scholar]
- 42.Shepherd M., McLean S., and Hunter C. N. (2005) Kinetic basis for linking the first two enzymes of chlorophyll biosynthesis. FEBS J. 272, 4532–4539 [DOI] [PubMed] [Google Scholar]
- 43.Papenbrock J., Pfündel E., Mock H. P., and Grimm B. (2000) Decreased and increased expression of the subunit CHL I diminishes Mg chelatase activity and reduces chlorophyll synthesis in transgenic tobacco plants. Plant J. 22, 155–164 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.