Abstract
Background. Human cytomegalovirus is a leading cause of congenital infection, and there are limited data on prognosis markers in disease development. We aimed to study 3 virology targets (glycoprotein [g]B, gN, and UL144) to assess their correlation with congenital infection and various organ system involvement.
Methods. Forty-eight congenital cases and 58 postnatally infected children were included (2003–2014). Genotyping for the 3 targets and distribution among the cohorts were investigated, and the relationship between the gB, gN, and UL144 types with clinical manifestations in congenital infection was also studied.
Results. All of the genotypes were similarly represented among cohorts, and the most prevalent were the UL144B, gB1, and gN1 genotypes. The gB2 genotype was associated with abnormal image findings by ultrasound and/or magnetic resonance in congenital infection (odds ratio [OR], 6.2; 95% confidence interval [CI], 1.1–34.3; P = .036); the gN1 genotype was associated with an elevated risk of developing neurological disorders (OR, 7.0; 95% CI, 1.1–45.9; P = .043). Both gN1 and gB2 were independent factors for symptomatic infection. Statistical analyses showed no association between any UL144 genotype and disease severity.
Conclusions. All of the genotypes can be involved in congenital infection, although the gB2 and gN1 genotypes might be associated with a more serious illness.
Keywords: congenital infection, cytomegalovirus, gB, genotypes, gN, UL144
Human cytomegalovirus (CMV) is a leading cause of congenital infection worldwide. In industrialized countries, it affects an average 0.6%–0.7% of newborns, and approximately 10%–15% of these present with symptoms at birth [1, 2]. Disease severity in symptomatic children is variable, ranging from mild symptoms to severe sensorineural hearing loss (SNHL) and long-term neurological sequelae [3–5].
Primary infections during pregnancy could lead to intrauterine transmission of CMV, although a transmission in previously seropositive women due to a reactivation or reinfection is the major route [6–8]. It is not typically possible to recognize CMV seroimmune status or to establish the moment of acquisition for documented primary infection because universal screening during pregnancy does not exist in most countries. In addition, there is no clear disease severity correlation between newborns born from women who are primarily or not primarily infected [4, 5].
Some viral factors have been postulated to be CMV pathogenicity markers [9–12]. Envelope glycoproteins are implicated in the recognition and input to the host cell, playing a potential role in the virulence of the various clinical isolates. Glycoprotein (g)B and gN are involved in the initial attachment of the virion to the cell surface, and the polymorphisms in these genes (UL55 and UL73, respectively) have been widely studied. Previous reports have associated particular gB or gN genoypes with severe congenital CMV (cCMV) infection, but information is scarce and is contradictory between studies [13–20]. There is also emerging interest in the study of other viral regions involving the host immune response. This interest is applied to UL144, a truncated tumor necrosis factor-α-like receptor gene, which contributes to the ability of CMV to escape immune clearance [9]. The association between the UL144 CMV genotype and the outcome in congenital infection has been investigated, but the heterogeneity between studies and the small number of congenital cases included have had inconsistent results [21–25].
We aimed to study the viral UL55, UL73, and UL144 genes to assess their correlation with cCMV infection and the involvement of various organ systems in CMV disease. The circulating strains of viral CMV in our geographic area were also studied, and a group of children with postnatal primary infection was included for this purpose.
MATERIAL AND METHODS
Study Population and Clinical Isolates
A retrospective study was conducted at Hospital Universitario 12 de Octubre, a tertiary care facility in Madrid, Spain, whose virology laboratory is a reference laboratory for the Southern area of Madrid, and which routinely receives samples from other hospitals (Hospital Universitario Severo Ochoa and Hospital Universitario de Getafe). The subjects involved in the study were identified from the virology laboratory database. We reviewed all of the positive CMV results from urine cultures and positive polymerase chain reaction (PCR) detections (amniotic fluid, fetal blood, and dried blood spots) from 2003 to 2014. Patients were included based upon virological results, detailed clinical records, and a specimen available at the moment of the study (kept frozen at −80°C). Approval from our hospital's ethics committee was obtained, and informed consent was given by the source hospitals.
A total of 48 congenital cases were included in the study, consisting of 36 newborns and 12 infected fetuses. Demographical and clinical data from all congenitally infected patients were recorded, including sex, gestational age at birth, presence of abnormal image findings by ultrasound (US) and/or magnetic resonance (MR) compatible with cCMV infection, clinical signs, neurologic abnormalities, SNHL, chorioretinitis, and laboratory findings at birth.
Included in the study were maternal age, human immunodeficiency virus (HIV) infection and other immunodeficiencies, treatment with CMV-specific hyperimmune globulin (HIG) during pregnancy, and primary maternal CMV infection, as defined by seroconversion and/or positive immunoglobulin (Ig)M and IgG with low CMV-specific IgG avidity.
Congenital CMV infection was confirmed by virus isolation from the patient's urine or saliva within the first 3 weeks of life (conventional cell culture and shell-vial culture [Vircell CMV-MAb, Granada, Spain] in MRC-5 cell lines) and/or by real-time PCR detection in amniotic fluid, fetal blood, or dried blood spots [26]. Viral load quantification in the blood and amniotic fluid was performed using the CMV R-gene test (Argene, Verhailes, France).
A group of 58 postnatally infected children from the same period of time was also included in the study. Laboratory and clinical records were reviewed. Cytomegalovirus infection was diagnosed based on consistent clinical symptoms (fever, mononucleoside syndrome) and the presence of specific CMV IgM in the patient's serum, as well as the isolation of the virus from the patient's urine. Among the 58 patients, 12 were younger than 4 months of age, and congenital infection was discharged in these cases because a previously negative urine CMV culture was available.
Clinical Definitions
Abnormal image findings in cranial US or cranial MR at birth or during pregnancy were defined as the presence of hydrocephalus, ventriculomegaly, periventricular cysts, white matter abnormalities, cerebral or cerebellar hypoplasia, hippocampal dysplasia, neuronal migration abnormalities, calcifications, ischemic lesions, lissencephaly, and microcephaly. All of the children infected with cCMV underwent a fundoscopy and cranial US at birth. All of the children with clinical symptoms and/or abnormal cranial US underwent cranial MR.
Neurologic abnormalities were defined as the presence of seizures, hypotonia, paresis, or spasticity.
Abnormal physical findings at birth were defined as the presence of microcephaly (head circumference <2 standard deviations below the mean for age and birth weight [World Health Organization]), hepatosplenomegaly, petechiae, and/or purpura.
Hearing function was assessed by the brainstem auditory evoked response (BAER) test, and SNHL was defined as a unilateral or bilateral hearing threshold of >20 decibels in the test.
The abnormal laboratory findings were defined as a hemoglobin value <9.5 g/dL (<8 g/dL in preterm infants), a neutrophil count <1000 cells/mm3, a platelet count <100 000 cells/mm3, alanine aminotransferase levels of >80 UI/L, or direct bilirubin levels of >2 mg/dL.
Symptomatic congenital CMV disease at birth was defined as the presence of at least 1 of the following: (1) abnormal physical exam, (2) abnormal image findings in US or MR, (3) SNHL, (4) neurologic abnormalities, (5) chorioretinitis, or (6) laboratory abnormalities. The presence of isolated intrauterine growth retardation, small for gestational age at birth, preterm delivery, or lenticulostriate vasculopathy in cranial US were not considered to be symptomatic criteria.
Characterization of Glycoprotein B, Glycoprotein N, and UL144 Genomic Variants
Genomic viral DNA was extracted from 200 µL of the specimens, which were stored at −80°C, using the NucliSENS easyMAG instrument (Biomèrieux Diagnostics, Marcy l'Etoile, France). Three nested PCR targeting the UL55 (gB), UL73 (gN), and UL144 genes were performed according to published protocols [27–29]. The amplicons were sequenced by using Big Dye 3.1 sequencing technology (3130 Genetic Analyzer; Applied Biosystems, Austin, TX). Genotypes for each target were determined by comparing the divergence of each amplicon with the reference sequences previously submitted to GenBank via the neighbor-joining tree-building method.
The sequences obtained were deposited at GenBank under the following accession numbers: KR992723-KR992835 for UL144 sequences; KR992836-KR992947 for gB sequences; and KR992948-KR993060 for gN sequences.
Statistical Analysis
The data were collected and analyzed using SPSS software version 15.0 (SPSS, Chicago, IL). Genotype distribution among congenitally and postnatally infected patients, the relationship between the gB, gN, and UL144 genotype, and the outcome of CMV congenital infections were analyzed using a 2-tailed χ2 test or Fisher's exact test for categorical variables and the Mann–Whitney U test for continuous variables. Logistic regression analysis was used to assess the associated risk between particular genotypes and the variables of the study. An unadjusted and adjusted (for treatment with HIG during pregnancy) odds ratio (OR) and associated 95% confidence interval (CI) were obtained. A P value < .05 was considered statistically significant.
RESULTS
Clinical Data on Congenitally Infected Patients
The clinical records of the 36 congenitally infected newborns are summarized in Table 1. We found a 50% proportion of symptomatic neonates. Sensorineural hearing loss, neuroimaging findings, and neurologic abnormalities were found in 83.3%, 83.3%, and 66.7% of the symptomatic patients, respectively. The most common pathological image findings found were white matter disease (66.7%), intracranial calcifications (38.9%), periventricular cysts (27.8%), ventricular adhesions (16.7%), and hydrocephalus (16.7%). Of symptomatic newborns in the study, 72.2% were treated with antiviral therapy (ganciclovir or valganciclovir).
Table 1.
Clinical and Laboratory Findings at Birth and Pathologic Obstetric Evaluations From Newborns Congenitally Infected With Cytomegalovirus
| Variable | Total (N = 36) |
|---|---|
| Male sex (%) | 18 (50.0) |
| Gestational age (wks)a | 38.0 (IQR, 37.0–41.0) |
| Weight (g)a | 2755.0 (IQR, 2250.0–3295.0) |
| Length (cm)a | 48.0 (IQR, 45.0–50.0) |
| Head circumference (cm)a | 34.0 (IQR, 31.75–35.0) |
| Neuroimaging findings (%)b | 15 (41.7) |
| Sensorineural hearing loss (%) | 15 (41.7) |
| Chorioretinitis (%) | 1 (2.8) |
| Microcephaly (%) | 3 (8.3) |
| Small for gestational age (%) | 5 (13.9) |
| Cardiomegaly (%) | 4 (11.1) |
| Hyperechogenic bowel lesions (%) | 1 (2.8) |
| Amniotic fluid levels altered (%) | 2 (5.6) |
| Petechiae (%) | 6 (16.7) |
| Hepatomegaly (%) | 2 (5.6) |
| Splenomegaly (%) | 2 (5.6) |
| Neurologic abnormalities (%)c | 12 (33.3) |
| Laboratory findings (%)d | 3 (8.3) |
Abbreviations: IQR, interquartile range; MR, magnetic resonance; US, ultrasound.
a Data expressed as median (IQR).
b Neuroimaging findings in cranial US or cranial MR were defined as the presence of hydrocephalus, ventriculomegaly, periventricular cysts, white matter abnormalities, cerebral or cerebellar hypoplasia, hippocampal dysplasia, neuronal migration abnormalities, calcifications, ischemic lesions, lissencephaly, and microcephaly.
c Neurologic abnormalities were defined as the presence of seizures, hypotonia, paresis, or spasticity.
d Laboratory findings were defined as a hemoglobin value <9.5 g/dL (<8 g/dL in preterm infants), a neutrophil count <1000 cells/mm3, a platelet count <100 000 cells/mm3, alanine aminotransferase levels of >80 UI/L, or direct bilirubin levels of >2 mg/dL.
Regarding the 12 cases of pregnancies that were terminated, 1 was a spontaneous abortion, and the rest were interrupted on the basis of pathologic ultrasound findings (N = 10) and/or alterations in the cord blood analytical parameters (thrombopenia and elevated β2 microglobulin) (N = 8). The primary image abnormalities found in the fetus were hepatomegaly (66.7%), cardiomegaly (41.7%), ascitis (41.7%), splenomegaly (33.3%), hydrops (33.3%), periventricular cysts (33.3%), central nervous system (CNS) parenchymal lesions (16.7%), and cerebral ischemic damage (16.7%).
Pregnancy Information
Primary CMV infection was confirmed in 45.8% of the pregnant women, whereas it was not possible to determine the moment of acquisition in the remaining 54.2% of the cases. The women had a median age of 32.5 years (interquartile range [IQR]), 27.0–35.5), 88.9% were born in Spain, and 11.1% were from South America. Four mothers (8.3%) were infected with HIV, but no transmission to the fetus occurred in those cases. Nine (19.6%) mothers with a positive CMV result in amniotic fluid decided to be treated with HIG to prevent the development of symptomatic congenital CMV infection.
Cytomegalovirus Genotyping
Genotyping was performed on 106 viral isolates (48 from congenital cases and 58 from postnatal cases) that were obtained from urine (N = 86), fetal blood (N = 9), amniotic fluid (N = 6), dried blood spots (N = 4), and saliva (N = 1). The median viral load in the fetal blood, amniotic fluid, and dried blood spots were as follows: 145 781 IU/mL (IQR, 86 000–1 984 550), 23 617 933.5 IU/mL (IQR, 16 671 083.5–27 235 908), and 8619 IU/mL (IQR, 1864–13 780), respectively.
All of the UL144 (A, B, C, A/C, A/B), gB (1–5), and gN (1, 2, 3a, 3b, 4a, 4b, 4c) genotypes were detected within the study population. The UL144 genotype B (36.8%), gB genotype 1 (41.6%), and gN genotype 1 (21.7%) were the most prevalent genomic variants. In contrast, the recombinant UL144 genotype A/B, gB genotype 5, and gN genotype 4b were found at the lowest frequencies (1.9%, 0.9%, and 8.5%, respectively). The genotype distribution for the 3 targets was similar between the congenital and postnatal groups, although gB genotype 4 was more frequent among the congenitally infected patients (12.5% vs 1.7%; P = .045), and gN genotype 4b was more frequently reported among the postnatally infected children (13.8% vs 2.1%; P = .038). However, gB genotype 5 and UL144 genotype A/B were not detected in congenital cases (Table 2).
Table 2.
Frequencies of Glycoprotein (g)B, gN, and UL144 Genotypes Among Congenitally and Postnatally Infected Patients
| All Isolates (N = 106) | Congenital (N = 48) | Postnatal (N = 58) | P Value | |
|---|---|---|---|---|
| gB genotype (%) | ||||
| 1 | 44 (41.6) | 17 (35.4) | 27 (46.6) | .322 |
| 2 | 28 (26.4) | 13 (27.1) | 15 (25.9) | 1.00 |
| 3 | 26 (24.5) | 12 (25.0) | 14 (24.1) | 1.00 |
| 4 | 7 (6.6) | 6 (12.5) | 1 (1.7) | .045 |
| 5 | 1 (0.9) | 0 | 1 (1.7) | 1.00 |
| gN genotype (%) | ||||
| 1 | 23 (21.7) | 12 (25.0) | 11 (19.0) | .486 |
| 2 | 16 (15.1) | 9 (18.75) | 7 (12.1) | .418 |
| 3a | 15 (14.1) | 6 (12.5) | 9 (15.5) | .782 |
| 3b | 13 (12.3) | 5 (10.4) | 8 (13.8) | .768 |
| 4a | 15 (14.1) | 9 (18.75) | 6 (10.3) | .268 |
| 4b | 9 (8.5) | 1 (2.1) | 8 (13.8) | .038 |
| 4c | 15 (14.1) | 6 (12.5) | 9 (15.5) | .782 |
| UL144 genotype (%) | ||||
| A | 23 (21.7) | 9 (18.75) | 14 (24.1) | .637 |
| B | 39 (36.8) | 16 (33.3) | 23 (39.7) | .548 |
| C | 24 (22.6) | 14 (29.2) | 10 (17.2) | .167 |
| A/C | 18 (17.0) | 9 (18.75) | 9 (15.5) | .796 |
| A/B | 2 (1.9) | 0 | 2 (3.5) | .500 |
Distribution of Viral Isolates Over Time (2003–2014)
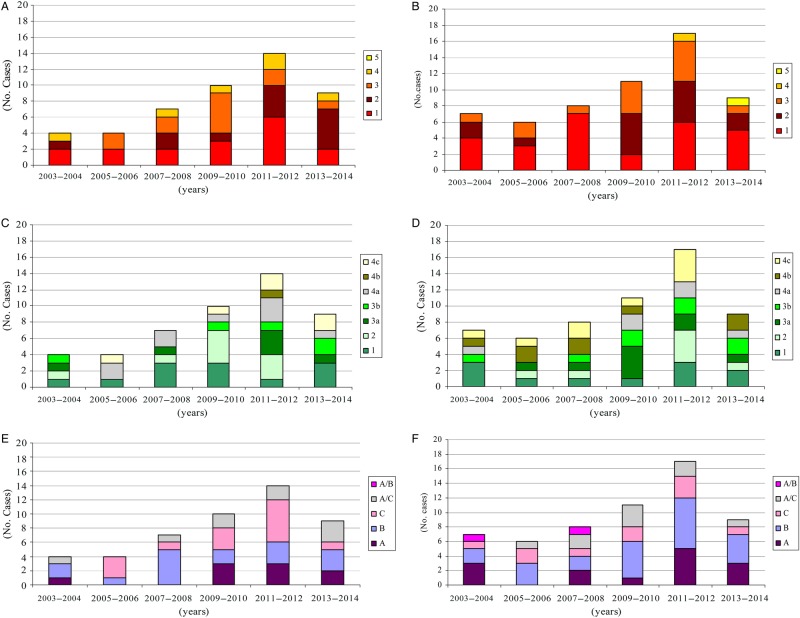
We studied genotype distribution patterns in periods of 2 years, observing a similar distribution of genotypes between cohorts. We did not find any combination of particular gB/gN, gB/UL144, or gN/UL144 genotypes among clinical isolates. We also reported the displacement of circulating viral strains over years. An increasing trend in the number of congenital cases occurred until 2011 (Figure 1).
Figure 1.
Distribution of viral glycoprotein ([g]B and gN) and UL144 genotypes over a 12-year study period (2003–2014). (A) The gB genotype distribution in congenital cases. (B) The gB genotype distribution in postnatal group. (C) The gN genotype distribution in congenital cases. (D) The gN genotype distribution in postnatal group. (E) UL144 genotype distribution in congenital cases. (F) UL144 genotype distribution in postnatal group.
Genotype Association With Congenital Infection
We aimed to study the involvement of particular CMV genotypes in the development of symptomatic disease at birth, as well as its association with image findings, SNHL, neurological disorders, and analytical parameters. The statistical analysis showed no association between any UL144 genotype and specific clinical manifestations. However, regarding the UL55 gene, the gB2 genotype was associated with the presence of abnormal findings in image studies at birth (P = .046). After including in the analysis the fetuses from interrupted pregnancies with CNS abnormalities on fetal US, significant differences remained (P = .030). In the logistic regression analysis, the OR (adjusted by HIG treatment during pregnancy) for gB2 and abnormal image findings were 6.2 (95% CI, 1.1–34.3; P = .036) (Table 3).
Table 3.
Association of Viral Genotypes and Outcome in Congenital Infection
| Clinical Manifestation | Genotype | OR (95% CI) | P Value | Adjusted OR (95% CI)c | Adjusted P Valuec |
|---|---|---|---|---|---|
| Abnormal image findingsa | gB2 | 6.3 (1.2–33.4) | .030 | 6.2 (1.1–34.3) | .036 |
| gB4 | –d | .015 | –d | –d | |
| gN1 | 4.5 (.8–24.0) | .081 | 3.9 (.7–22.3) | .123 | |
| Neurological disordersb | gB2 | 5.0 (.9–26.5) | .059 | 4.5 (.8–24.6) | .082 |
| gB4 | 0.5 (.4–4.6) | .504 | 0.4 (.04–4.7) | .510 | |
| gN1 | 7.9 (1.2–49.8) | .029 | 7.0 (1.1–45.9) | .043 | |
| SNHLb | gB2 | 3.0 (.6–15.3) | .186 | 2.4 (.4–13.9) | .314 |
| gB4 | –e | .062 | –e | –e | |
| gN1 | 4.7 (.8–29.0) | .092 | 3.7 (.5–25.2) | .179 |
Abbreviations: CI, confidence interval; CMV, cytomegalovirus; MR, magnetic resonance; OR, odds ratio; SNHL, sensorineural hearing loss; US, ultrasound.
a Data from 46 patients (36 congenitally infected newborns and 10 fetuses from pregnancies interrupted by pathological image findings).
b Data from 36 congenitally infected newborns.
c Data adjusted for treatment with CMV-specific hyperimmune globulin during pregnancy.
d It was not possible to calculate OR (95% CI) because none of the patients with the gB4 genotype presented abnormal image findings in US/MR.
e It was not possible to calculate OR (95% CI) because none of the patients with the gB4 genotype presented SNHL at birth.
On the other hand, gB4 was linked with a better prognosis, and no children or fetuses with this genotype presented abnormal image findings (P = .015). It was not possible to calculate the OR and 95% CI for this association (Table 3). Patients with the gB4 genotype presented higher platelet counts at birth (324 200 cells/L vs 187 600 cells/L; P = .067).
The study of the UL73 gene showed an association between the gN1 genotype and neurologic findings at birth (P = .029); in the logistic regression analysis, adjusted OR was 7.0 (95% CI, 1.1–45.9; P = .043) (Table 3). The gN1 genotype was also associated with a lower platelet count at birth compared with other gN variants (120 400 cells/L vs 238 550 cells/L; P = .037). The gN1 genotype was not associated with abnormal image findings (P = .081) (Table 3).
Finally, we did not find any association between a particular gB or gN genotype and SNHL.
DISCUSSION
Although cCMV infection is a major cause of long-term sequelae in children worldwide, in most countries there is no current strategy for general screening during pregnancy or in newborns. Moreover, most cases are asymptomatic at birth; however, even in these cases, the rate of long-term sequelae is important [30, 31]. In this scenario, it is of paramount importance to establish sensitive and accurate prognostic markers to guide the management of congenitally infected patients.
As with other viral infections, organ system involvement caused by CMV is a balance between the pathogenicity of the strain and the host's immune ability to neutralize the infection [32]. The viral load in blood or urine has been reported to be a prognostic marker of CMV infection. The levels of viral load in urine have been observed to be higher in congenital than in acquired infection [15], and 1 study has recently reported that CMV DNAemia could predict CMV sequelae in asymptomatic congenitally infected newborns [33]. Further studies with larger and homogeneous populations are required to establish a viral load cutoff, because there is evidence that DNAemia and urine excretion could be high and prolonged during CMV infection [34]. In our case, data on the CMV DNA viral load in urine were not available because we used viral cultures and did not quantify samples. For amniotic fluids, fetal blood, and dried blood spots, the viral load values obtained varied among specimens, ranging from 103 logarithm values in fetal blood to 107 logarithm values in amniotic fluid. The number of determinations was low, and we could not estimate the association with CMV disease.
The linkage between specific envelope glycoproteins and CMV disease has been previously investigated, and data on genotype distribution among congenitally infected patients are variable between studies [13, 15, 17–19]. As we report, the gB1 type has been found to be the most prevalent in congenital infection [14, 16, 17]. We also identified this genotype as the predominant variant among postnatally infected children, which indicates it is the primary genotype circulating in our geographic area. We found that genotype distribution among cohorts was similar, and that all gB genotypes can be involved in congenital infection, as other authors have also reported [13–15, 17]. Genotype 5 is a much less common circulating genotype, and we only determined 1 patient to be postnatally infected by this variant, so we cannot conclude that there is an association of this genotype with congenital infection. Yan et al [16] reported greater frequencies of the gB3 genotype in congenital than in acquired infection, and they found this variant to be associated with SNHL among congenital cases. Gandhoke et al [20] reported that the gB2 genotype was associated with CNS disease and SNHL among CMV-infected babies, although the study design and the criteria to define the study population (congenital and noncongenital cases) were not clearly established, thus the results should be carefully considered.
Our research revealed that the gB2 genotype was associated with abnormal image findings in the fetus and newborns with congenital infection, whereas gB4 might be associated with a lower risk of abnormal image findings. However, the number of infants with the gB4 genotype is small and the study is not sufficiently powered to assess this association. Abnormal cranial image findings in fetuses and newborns with cCMV infection are well known prognostic factors of long-term sequelae, including SNHL [35–37], and, to our knowledge, this is the first report that finds a relationship with a particular gB genotype.
Regarding gN types, we found a similar distribution of genotypes among cohorts, with gN1 being the most prevalent in congenital infection and gN3a the most prevalent in the postnatal group. Other authors found the gN4c or gN3a genotypes the most representative in congenital infection [17, 19]. Although the frequencies are similar to ours, the differences could reflect geographical distribution and the time of the studies. We found all genotypes both in congenital and acquired infection, although genotype 4b showed a trend toward a higher prevalence among noncongenital patients (P = .038), suggesting a reduced ability of this type to be transmitted in utero.
Some authors have documented an association between particular gN genotypes and disease severity in congenital infection. Pignatelli et al [19] found the gN1 and gN3a genotypes more frequently represented in asymptomatic patients with a favorable long-term outcome, and the gN4 genotypes more frequently represented in patients with symptoms at birth and more sequelae. Paradowska et al [18] also reported the detection of the gN2 or the gN4 genotypes in the children who were most seriously affected. Our results do not confirm prior observations, and, in contrast, we found the gN1 genotype more frequently in newborns with neurological disorders and lower platelet counts at birth.
We also investigated the UL144 gene due to its role in the virus's ability to escape the immune response. Previous studies have reported an association between UL144 genotypes and congenital infection [21, 23, 24], and other authors have reported no linkage between the congenital picture and disease severity [16, 22, 25, 29]. We did not find any association between a particular UL144 genotype and symptomatic disease. The distribution of UL144 genotypes in our population is consistent with previously reported data: the B genotype is the primary genotype represented both in congenital and acquired infection. However, we found higher frequencies of A, C, and A/C genotypes than other studies reported [16, 22, 23].
Genomic variation among CMV strains and intrahost viral diversity have recently been characterized through next-generation sequencing [38, 39], and the extensive variability found could be comparable to many RNA viruses. Mixed populations of the virus have been documented in congenitally infected fetuses and newborns as well as in pregnant women [14, 17, 39–41]. Our study characterizes the primary circulating genotypes in the affected patients, and these genotypes are hypothetically the most important viral populations that could ultimately lead to disease severity.
CONCLUSIONS
Although the data presented here are compelling, this study has some limitations. First, this is a retrospective research, and due to the patient's inclusion criteria and the lack of a current strategy to identify congenital cases, we obtained a high rate of symptomatic infections among live-born infants (50%), which does not represent the prevalence in Madrid (Spain). We have probably detected the most serious infections. Second, the information about the moment of the infection during pregnancy was not available in all cases, so we cannot know whether women were primarily or not-primarily infected. In spite of this, we present a 12-year study that provides a good depiction of the main circulating strains of CMV among congenitally and postnatally infected children in Madrid, Spain. We included patients based on very strict criteria and clinical data collected is complete and thoroughly detailed, which allows us to establish associations between genotypes and outcome in congenital infection. Of the 3 viral targets that we have investigated, gB seems to be the most appropriate because we have found some evidence about certain genotypes and the abnormal image findings present in the infected patients. Further studies are warranted to analyze the evolution of patients in time, both symptomatic and asymptomatic, and the implication of genotypes during the follow up.
Acknowledgments
We thank Luis Manuel Prieto from Hospital Universitario de Getafe, María Isabel González-Tomé and Pablo Rojo on behalf of the study Group of congenital cytomegalovirus from Hospital Universitario 12 de Octubre for clinical assistance, and David Lora from the Instituto de Investigación Biomédica “i+12” for statistical analysis support.
Financial support. This work was supported by a grant from the Instituto de Salud Carlos III, FEDER, Ministerio de Economía y Competitividad, Spain (FIS13/02147).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1.Kenneson A, Cannon MJ. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev Med Virol 2007; 17:253–76. [DOI] [PubMed] [Google Scholar]
- 2.Dollard SC, Grosse SD, Ross DS. New estimates of the prevalence of neurological and sensory sequelae and mortality associated with congenital cytomegalovirus infection. Rev Med Virol 2007; 17:355–63. [DOI] [PubMed] [Google Scholar]
- 3.Dreher AM, Arora N, Fowler KB et al. . Spectrum of disease and outcome in children with symptomatic congenital cytomegalovirus infection. J Pediatr 2014; 164:855–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manicklal S, Emery VC, Lazzarotto T et al. . The “silent” global burden of congenital cytomegalovirus. Clin Microbiol Rev 2013; 26:86–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boppana SB, Ross SA, Fowler KB. Congenital cytomegalovirus infection: clinical outcome. Clin Infect Dis 2013; 57:S178–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boppanna SB, Rivera LB, Fowler KB et al. . Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N Engl J Med 2001; 344:1366–71. [DOI] [PubMed] [Google Scholar]
- 7.Mussi-Pinhata MM, Yamamoto AY, Moura Brito RM et al. . Birth prevalence and natural history of congenital cytomegalovirus infection in a highly seroimmune population. Clin Infect Dis 2009; 49:522–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Vries JJ, van Zwet EW, Dekker FW et al. . The apparent paradox of maternal seropositivity as a risk factor for congenital cytomegalovirus infection: a population-based prediction model. Rev Med Virol 2013; 23:241–9. [DOI] [PubMed] [Google Scholar]
- 9.Benedict CA, Butrovich KD, Lurain NS et al. . Cutting edge: a novel viral TNF receptor superfamily member in virulent strains of human cytomegalovirus. J Immunol 1999; 162:6967–70. [PubMed] [Google Scholar]
- 10.Miller WE, Zagorski WA, Brenneman A et al. . US28 is a potent activator of phospholipase C during HCMV infection of clinically relevant target cells. PLoS One 2012; 7:e50524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freed DC, Tang Q, Tang A et al. . Pentameric complex of viral glycoprotein H is the primary target for potent neutralization by a human cytomegalovirus vaccine. Proc Natl Acad Sci U S A 2013; 110:E4997–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pignatelli S, Dal Monte P, Rossini G, Landini MP. Genetic polymorphisms among human cytomegalovirus (HCMV) wild-type strains. Rev Med Virol 2004; 14:383–410. [DOI] [PubMed] [Google Scholar]
- 13.Picone O, Costa JM, Lereuz-Ville M et al. . Cytomegalovirus (CMV) DNA load in the amniotic fluid of infected fetus. Prenat Diagn 2004; 24:1001–6. [DOI] [PubMed] [Google Scholar]
- 14.Pati SK, Pinninti S, Novak Z et al. . Genotypic diversity and mixed infection in newborn disease and hearing loss in congenital cytomegalovirus infection. Pediatr Infect Dis J 2013; 32:1050–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nijman J, Mandemaker FS, Verboon-Maciolek MA et al. . Genotype distribution, viral load and clinical characteristics of infants with postnatal or congenital cytomegalovirus infection. PLoS One 2014; 9:e108018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan H, Koyano S, Inami Y et al. . Genetic variations in the gB, UL144 and UL149 genes of human cytomegalovirus strains collected from congenitally and postnatally infected Japanese children. Arch Virol 2008; 153:667–74. [DOI] [PubMed] [Google Scholar]
- 17.Ross SA, Novak Z, Pati S et al. . Mixed infection and strain diversity in congenital cytomegalovirus infection. J Infect Dis 2011; 204:1003–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paradowska E, Jabłońska A, Studzińska M et al. . Distribution of cytomegalovirus gN variants and associated clinical sequelae in infants. J Clin Virol 2013; 58:271–5. [DOI] [PubMed] [Google Scholar]
- 19.Pignatelli S, Lazzarotto T, Gatto MR et al. . Cytomegalovirus gN genotypes distribution among congenitally infected newborns and their relationship with symptoms at birth and sequelae. Clin Infect Dis 2010; 51:33–41. [DOI] [PubMed] [Google Scholar]
- 20.Gandhoke I, Hussain SA, Pasha ST et al. . Glycoprotein B genotyping in congenital/perinatal cytomegalovirus infection in symptomatic infants. Indian Pediatr 2013; 50:663–7. [DOI] [PubMed] [Google Scholar]
- 21.Arav-Boger R, Willoughby RE, Pass RF et al. . Polymorphisms of the cytomegalovirus (CMV)-encoded tumor necrosis-α and β-chemokine receptors in congenital CMV disease. J Infect Dis 2002; 186:1057–64. [DOI] [PubMed] [Google Scholar]
- 22.Picone O, Costa JM, Chaix ML et al. . Human cytomegalovirus UL144 gene polymorphisms in congenital infections. J Clin Microbiol 2005; 43:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waters A, Hassan J, deGascun C et al. . Human cytomegalovirus UL144 is associated with viremia and infant development sequelae in congenital infection. J Clin Microbiol 2010; 48:3956–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arav-Boger R, Battaglia CA, Lazzarotto T et al. . Cytomegalovirus (CMV)-encoded UL144 (truncated tumor necrosis factor receptor) and outcome of congenital CMV infection. J Infect Dis 2006; 194:464–73. [DOI] [PubMed] [Google Scholar]
- 25.Bale JF Jr, Petheram SJ, Robertson M et al. . Human cytomegalovirus a sequence and UL144 variability in strains from infected children. J Med Virol 2001; 65:90–6. [PubMed] [Google Scholar]
- 26.Watzinger F, Suda M, Preuner S et al. . Real-time quantitative PCR assays for detection and monitoring of pathogenic human viruses in immunosuppressed pediatric patients. J Clin Microbiol 2004; 42:5189–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stranska R, Schuurman R, Toet M et al. . Application of UL144 molecular typing to determine epidemiology of cytomegalovirus infections in preterm infants. J Clin Microbiol 2006; 44:1108–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pignatelli S, Dal Monte P, Rossini G et al. . Latency-associated human cytomegalovirus glycoprotein N genotypes in monocytes from healthy blood donors. Transfusion 2006; 46:1754–62. [DOI] [PubMed] [Google Scholar]
- 29.Lurain NS, Kapell KS, Huang DD et al. . Human cytomegalovirus UL144 open reading frame: sequence hypervariability in low-passage clinical isolates. J Virol 1999; 73:10040–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fowler KB, Boppana SB. Congenital cytomegalovirus (CMV) infection and hearing deficit. J Clin Virol 2005; 35:226–31. [DOI] [PubMed] [Google Scholar]
- 31.Williamson WD, Demmler GJ, Percy AK, Catlin FI. Progressive hearing loss in infants with asymptomatic congenital cytomegalovirus infection. Pediatrics 1992; 90:862–6. [PubMed] [Google Scholar]
- 32.Hanley PJ, Bollard CM. Controlling cytomegalovirus: helping the immune system take the lead. Viruses 2014; 6:2242–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forner G, Abate D, Mengoli C et al. . High cytomegalovirus (CMV) DNAemia predicts CMV sequelae in asymptomatic congenitally infected newborns born to women with primary infection during pregnancy. J Infect Dis 2015; 212:67–71. [DOI] [PubMed] [Google Scholar]
- 34.Cannon MJ, Hyde TB, Schmid DS. Review of cytomegalovirus shedding in bodily fluids and relevance to congenital cytomegalovirus infection. Rev Med Virol 2011; 21:240–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guerra B, Simonazzi G, Puccetti C et al. . Ultrasound prediction of symptomatic congenital cytomegalovirus infection. Am J Obstet Gynecol 2008; 198:380.e1–7. [DOI] [PubMed] [Google Scholar]
- 36.Ancora G, Lanari M, Lazzarotto T et al. . Cranial ultrasound scanning and prediction of outcome in newborns with congenital cytomegalovirus infection. J Pediatr 2007; 150:157–61. [DOI] [PubMed] [Google Scholar]
- 37.Capretti MG, Lanari M, Tani G et al. . Role of cerebral ultrasound and magnetic resonance imaging in newborns with congenital cytomegalovirus infection. Brain Dev 2014; 36:203–11. [DOI] [PubMed] [Google Scholar]
- 38.Sijmoms S, Van Ranst M, Maes P. Genomic and functional characteristics of human cytomegalovirus revealed by next-generation sequencing. Viruses 2014; 6:1049–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renzette N, Bhattacharjee B, Jensen JD et al. . Extensive genome-wide variability of human cytomegalovirus in congenitally infected infants. PLoS Pathog 2011; 7:e1001344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamoto AY, Mussi-Pinhata MM, de Deus Wagatsuma VM et al. . Human cytomegalovirus glycoprotein B genotypes in Brazilian mothers and their congenitally infected infants. J Med Virol 2007; 79: 1164–8. [DOI] [PubMed] [Google Scholar]
- 41.Rycel M, Wujcicka W, Zawilinska B et al. . Mixed infections with distinct cytomegalovirus glycoprotein B genotypes in Polish pregnant women, fetuses and newborns. Eur J Clin Microbiol Infect Dis 2015; 34:585–91. [DOI] [PMC free article] [PubMed] [Google Scholar]