Abstract
Essential tremor (ET) is the most prevalent movement disorder, affecting millions of people in the USA. Although a positive family history is one of the most important risk factors for ET, the genetic causes of ET remain unknown. In an attempt to identify genetic causes for ET, we performed whole-exome sequencing analyses in a large Spanish family with ET, in which two patients also developed epilepsy. To further assess pathogenicity, site-directed mutagenesis, mouse and human brain expression analyses, and patch clamp techniques were performed. A disease-segregating mutation (p.Gly1537Ser) in the SCN4A gene was identified. Posterior functional analyses demonstrated that more rapid kinetics at near-threshold potentials altered ion selectivity and facilitated the conductance of both potassium and ammonium ions, which could contribute to tremor and increase susceptibility to epilepsy, respectively. In this report, for the first time, we associated the genetic variability of SCN4A with the development of essential tremor, which adds ET to the growing list of neurological channelopathies.
Introduction
Essential tremor (ET) is one of the most common neurological disorders and the most frequent adult-onset movement disorder, with an increased age-dependent prevalence of up to 5% in people 65 years old and over (1). Although its core motor symptom is a bilateral postural and kinetic tremor of the hands and arms, some patients also develop other motor and non-motor manifestations, including parkinsonism, myoclonus, dystonia, cerebellar dysfunction, sensory abnormalities, sleep disorders and cognitive and psychiatric features (2). The neuropathology of ET is not well understood and whether there is an underlying neurodegenerative process of the cerebellum in ET remains controversial (3). There is growing evidence that ET may not be a single disease but rather a family of diseases (4).
Although many concerns arise from methodological issues, there is a consensus that a positive family history is common in patients with ET (5). The genetic basis of ET remains elusive, however, partly due to the lack of stringent diagnostic criteria, the lack of biomarkers, a high phenocopy rate and high locus heterogeneity in presumably monogenic ET (5). In addition, misdiagnosis is a common feature in ET, with 37–50% of ET patients reportedly misdiagnosed (6). Although these associations have not been consistently replicated, genetic variants within the LINGO1 (MIM #609791), DRD3 (MIM #126451), and HS1-BP3 (MIM #609359) genes have been reported to render susceptibility to ET (5,7). More recently, polymorphisms at the SLC1A2 (MIM #600300) locus have also been shown to lend susceptibility to ET (8), and the FUS (MIM #137070), HTRA2 (MIM #606441) and SORT1 (MIM #602458) genes have been reported as causative ET genes using whole-exome sequencing (WES) analyses (9–11). Whereas the disease-associated mutation in the FUS gene was only present in 54% of individuals classified as having ‘possible’ ET, it fully segregated with disease in individuals with definite and probable ET (9).
In this study, we clinically examined a Spanish family with ET to identify the causative gene mutation. We utilized WES, which has been successfully demonstrated to identify the underlying genetic defect in clinically and genetically heterogeneous diseases in which traditional genetic approaches have failed (12,13).
Results
Clinical examination
Here, we report on a large family featuring ET in which two affected individuals also manifested epilepsy. The inheritance pattern was autosomal dominant. There were seven affected individuals over four different generations, and full clinical evaluation was conducted in five patients (Patients 1–5) and three unaffected family members (Fig. 1A). Two individuals were reported as having ET but were not examined by us, and DNA testing was not possible (Fig. 1A).
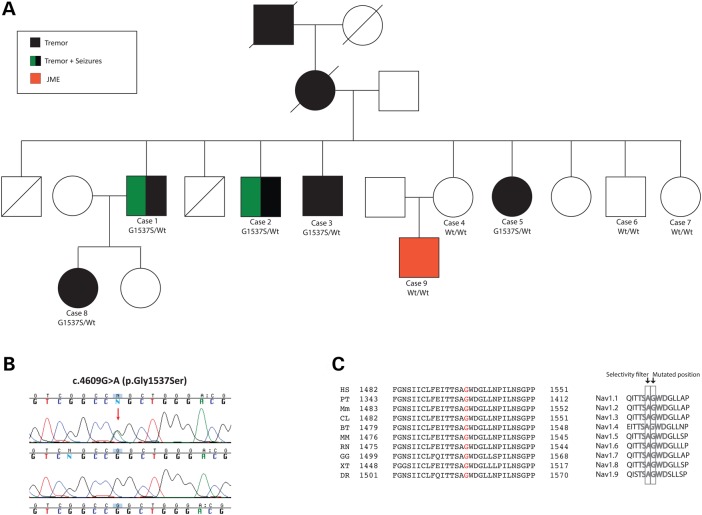
Figure 1.
Autosomal dominant ET due to a SCN4A change-of-function mutation. (A) Pedigree structure. Heterozygous mutation carriers are represented as G1537S/Wt and non-carriers as Wt/Wt (A). (B) Sanger chromatogram sequences. Wild-type sequences are shown at the bottom and mutant sequences at the top. (C) Alignment of the SCN4A p.G1537S mutation. Left: alignment with different species; right: alignment with the whole sodium channel protein family each encoded by a different gene. HS: H. sapiens; PT: P. troglodytes; Mm: M. musculus; CL: C. lupus; BT: B. taurus; MM: M. mulatta; RN: R. norvegicus; GG: G. gallus; XT: X. tropicalis; DR: D. rerio.
Case 1
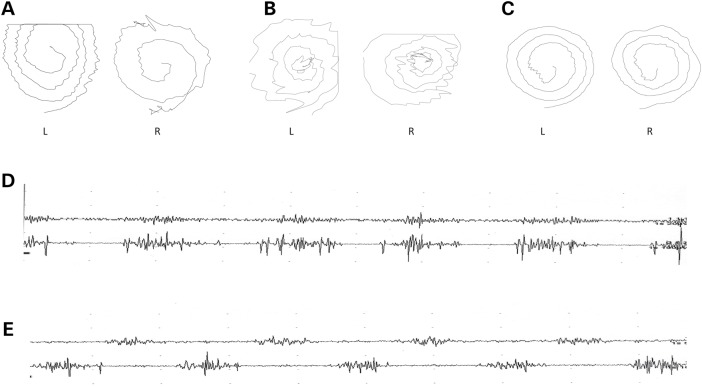
Patient 1, who is now 73 years old, began suffering from generalized seizures precipitated by sleep deprivation at the age of 10 years. At the age of 30, he developed tremor of both hands. On physical examination, he showed postural tremor of both hands, head tremor, mild resting tremor and a mild ataxic gait without any other cerebellar, rigid–akinetic or dystonic signs. His tremor incapacitates him from writing and drinking. Electroencephalography (EEG) showed no pathological changes at rest without a photoparoxysmal response. Somatosensory evoked potential (SEP) exhibited normal amplitude, and C-waves were not obtained. EMG recorded in the arms showed a tremor 8 Hz in frequency and 48–385 µV in amplitude with the co-contraction of agonist and antagonist muscles (Fig. 2D and E). He scored 27 on the MoCA test and 36 on the Fahn-Tolosa-Marin tremor rating scale (TRS).
Figure 2.
Archimedes spiral tests and EMG recordings of ET patients described in this study. (A–C) Archimedes spirals. Archimedes spirals were drawn on a digitizing tablet. L represents the non-dominant left hand, and R represents the dominant right hand. Left- and right-hand spirals of patients 1(A), 2 (B) and 4 (C) are shown. (D and E) Surface EMG (SEMG) recordings. SEMG recordings of Patient 1 at the postural position at rest and with the arms outstretched are illustrated. The upper register of each figure corresponds to the biceps and the lower to the triceps. SEMGs show a tremor with the synchronous pattern at rest (D) and an alternating pattern with the arms outstretched (E).
Case 2
Patient 2, who is 71 years old, began suffering from rare, generalized seizures precipitated by alcohol intake at the age of 20 years. He was treated with antiepileptic drugs until ∼20 years ago, and since then, no attack has been experienced. He had an isolated myoclonus, and since 2005, he has suffered from tremor of both hands that improves with alcohol intake. He recently developed head tremor. On examination, he had postural tremor of both hands and head tremor, but no other cerebellar, rigid–akinetic or dystonic symptoms. Although his hand tremor incapacitates him from writing and drinking, he takes no medication. EMG recorded in the arms showed a tremor 3.5–4.5 Hz in frequency and 66–503 µV in amplitude with the co-contraction of agonist and antagonist muscles. He scored 25 on the MoCA test and 40 on the TRS.
Case 3
Patient 3, who is 69 years old, has suffered from a symmetrical tremor of both hands since the age of 62. He initially had both postural and action tremor and later developed voice tremor. EMG recorded in the arms showed a tremor 6.6 Hz in frequency and 298 µV in amplitude with the co-contraction of agonist and antagonist muscles. He scored 26 on the MoCA test and 29 on the TRS.
Case 4
Unaffected member 1 is now 68 years old and has a history of chronic anxiety without evidence of epileptic seizures or tremors. She showed no tremor.
Case 5
Patient 4, who is now 66 years old, suffers from a slightly asymmetric tremor of both hands that is more intense in the right hand, which has rendered her unable to write since the age of 54. On physical examination, she showed a mild head tremor but no other cerebellar, rigid–akinetic or dystonic signs. EEG showed no pathological changes at rest without a photoparoxysmal response. EMG recorded in the arms showed a tremor 6.6 Hz in frequency and 15.8 µV in amplitude with the co-contraction of agonist and antagonist muscles. She scored 25 on the MoCA test and 16 on the TRS.
Case 6
Unaffected member 2 has a history of diabetes, ischaemic heart disease, depression, anxiety and alcohol abuse. He has no evidence of epileptic seizures or tremors and showed no tremor. He scored 15 on the MoCA test and is now 61 years old.
Case 7
Unaffected member 3 showed no tremor on examination and has no history of epileptic seizures or tremors. She scored 25 on the MoCA test and is now 59 years old.
Case 8
Patient 5 has suffered from head tremor since she started smoking tobacco at the age of 23. On recent examination, she showed a mild head tremor without tremor of the voice or limbs and no evidence of epileptic seizures. She is now 48 years old.
Case 9
Patient 6, who is 48 years old, began suffering from myoclonic jerks at the age of 15. In 2003, he also suffered from a generalized seizure after sleep deprivation, and an EEG recorded at the time showed generalized spike-wave discharges. His seizures showed a beneficial response to treatment with valproate, but he experienced a mild distal hand tremor with increasing amplitude at the end of the movement of the finger-nose test during treatment. EMG recorded in the arms showed an alternating tremor 10 Hz in frequency, probably secondary to treatment with valproate, as he showed typical characteristics of valproate-induced tremor, such as high frequency, low amplitude, short burst duration and a synchronous burst pattern (14). He was accordingly diagnosed with juvenile myoclonic epilepsy (JME).
Identification of SCN4A as the disease-causing gene
WES approaches were performed on four family members (Cases 2, 4, 8, 9), including two cases affected with ET (Cases 2 and 8), a case diagnosed with JME (Case 9) and an unaffected case (Case 4; Fig. 1A). Between 92.09% (Case 8) and 89.12% (Case 2) of the target exome at 20-fold coverage or higher was captured for all sequenced samples. Including only novel missense and nonsense coding variations, 972 single nucleotide variations (SNVs) for Case 9, 969 SNVs for Case 2, 942 SNVs for Case 4 and 849 SNVs for Case 8 were identified. Because her mother did not present any clinical symptoms, we considered that the individual with JME may also carry pathogenic mutations in the same disease gene, which may have reduced penetrance; thus, we searched for common variations between all four family members sequenced but failed to identify any genetic variation of this type. We then searched for common genetic variation between both ET patients subjected to WES approaches (Patients 2 and 5; Fig. 1A) and identified seven novel SNVs common to both of them. After validation through Sanger sequencing and the examination of these novel genetic variants in the remaining family members, only one was found to segregate with disease status (Fig. 1B). This disease-segregating mutation, which resulted in a G-to-A transversion at nucleotide 4609 and created a p.Gly1537Ser mutation, was localized in the SCN4A gene that encodes for a voltage-gated sodium channel (Nav1.4) protein. The Nav1.4 p.Gly1537Ser mutation was found to not only be conserved across other species but also across the α subunits of human voltage-gated sodium channel (SCN) genes (Fig. 1C). It was later proved to be absent in the normal population, including public databases (n > 10 000) and ethnicity-matched controls (n > 188 chromosomes), and was predicted to be pathogenic by Alamut software and two other computational methods (MutPred score = 0.999; SNPs&Go score = Disease). While writing this article, we observed that this variant is currently listed in dbSNP (rs571210585) but without validation or frequency data and that it is now reported with very low frequency (6.63 E–05; 8/120 668) in the Exome Aggregation Consortium (ExAC) data, which contains exome data for a variety of rare diseases, including rare neuromuscular diseases (15). This frequency is consistent with the disease, as the prevalence of ET in the general population is ∼4.6% in individuals aged 65 years and older (1).
Although mutations in SCN4A (MIM #603967) are known to cause normo- (NormoKPP), hyper- (HyperKPP2) and hypokalemic periodic paralysis type 2 (HypoKPP2), congenital paramyotonia, myasthenic syndrome and severe neonatal episodic laryngospasm, the genetic variability of SCN4A is not associated with the development of tremor (16–21). On the other hand, pathogenic mutations in other genes of the SCN family have been reported in epileptic syndromes (SCN1A, SCN2A, SCN3A, SCN5A, SCN8A) and cardiac conduction defects (SCN1A, SCN3A, SCN5A) (18). Indeed, the p.Gly1537 amino acid is mutated in both SCN1A (p.Gly1725Cys) and SCN5A (p.Gly1712Ser) proteins, causing Dravet syndrome (MIM #601144) and Brugada syndrome (MIM #607208), respectively (22). Taken together, it is very likely that the SCN4A p.Gly1537Ser mutation is the disease-associated mutation responsible for the tremor phenotype described in our family. The SCN4A p.Gly1537Ser mutation was then examined in 76 sporadic and 25 familial Spanish patients with ET but was not detected. The entire coding region of the SCN4A gene was also examined in 22 additional cases with familial ET, but no other pathogenic mutation was identified.
SCN4A is expressed in both the mouse and human brain
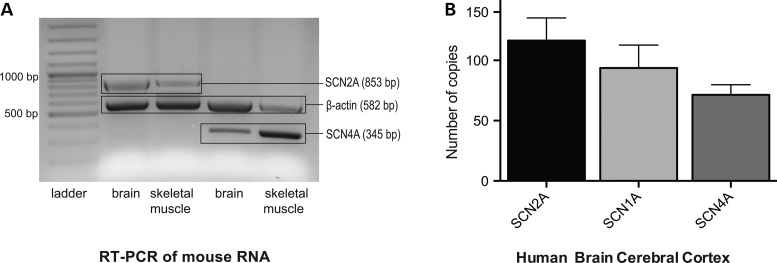
Because SCN4A is generally considered to be expressed specifically in skeletal muscle (23), we first tested its expression in the mouse brain in comparison with skeletal muscle. The SCN2A gene that encodes a classical neuronal sodium channel was used as a control, and all reaction mixtures contained ACTB primers encoding β-actin to verify the reactivity of the reagents. Both SCN2A and SCN4A were amplifiable from both the muscle and brain (Fig. 3A). The expression of SCN4A was then confirmed in the human cerebral cortex (Fig. 3B), which strongly suggested the relevant expression of Nav1.4 in neuronal tissues.
Figure 3.
SCN4A expression in the mouse and human brain. (A) Mouse brain expression. Column 1: 100 bp ladder; Column 2: multiplex RT-PCR products of SCN2A and ACTB from brain RNA; Column 3: multiplex RT-PCR products of SCN2A and ACTB from muscle RNA; Column 4: multiplex RT-PCR products of SCN4A and ACTB from brain RNA; Column 5: multiplex RT-PCR products of SCN4A and ACTB from muscle RNA (Column 5). Note that SCN2A and SCN4A are expressed in both skeletal muscle and brain. The ubiquitously expressed ACTB encoding β-actin served as a control for the reactivity of the mix only. (B) Human brain expression. The expressions of SCN4A and other SCN genes known to be expressed in the human brain were evaluated by absolute quantification using the standard curve method. The y-axis represents the mRNA expression (in number of copies), and the x-axis represents the SCN genes tested in the cerebral cortex of the human brain.
The SCN4A p.G1537S mutation did not affect kinetics and voltage-dependent gating but altered ion selectivity
The SCN4A mutation identified in our family (p.G1537S) is situated in the portion of the IVS5-S6 loop that dips into the membrane and forms the lining of the pore, which is important for ion selectivity (24,25). To test for the possible functional significance of p.G1537S in the context of the ET phenotype, we performed whole-cell patch-clamp studies in a heterologous expression system. Increasing channel activation by 12.5 ms depolarization from a holding potential of −140 mV yielded an inward sodium current of overall similar density and voltage dependence as the wild type (Fig. 4A and B). From the current maxima, steady-state activation parameters were determined by fitting the current–voltage relationship with the equation I = Gmax*(V − Vrev)/(1 + exp((V0.5 − V)/k)), whereby Vrev and Gmax are the reversal potential and maximum conductance of channels, respectively; V0.5 is the potential for half-maximal current; V is the test potential and k is the slope factor. Although there was a tendency for mutant channels to have a lower threshold, no significant differences in any of the activation parameters were found between mutant and wild type (Fig. 4C and Table 1).
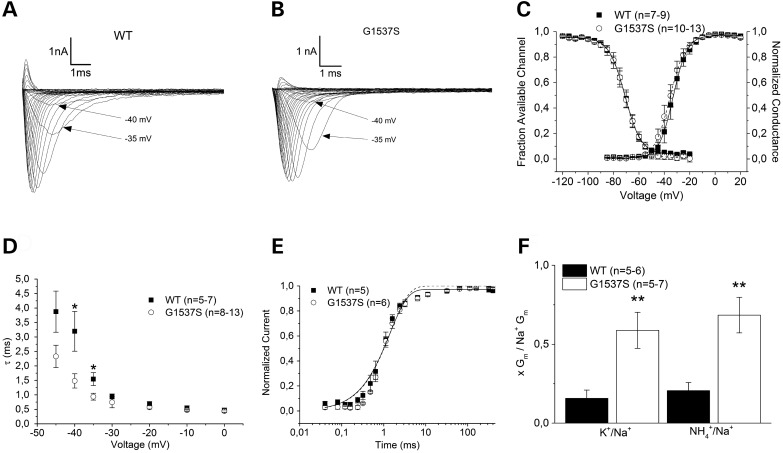
Figure 4.
Functional study. (A and B) Sodium currents from heterologously expressed wild-type Nav1.4 (A) and G1537S (B) channels are shown. Note the more rapid kinetics of the mutant at potentials near the threshold, as highlighted by the arrows. (C) Conductance–voltage relationships from Boltzmann fits of steady-state activation and inactivation, which are similar for the WT and mutant. (D) Kinetics of inactivation determined by monoexponential fit. Note the significant acceleration of the mutant inactivation at −40 and −35 mV. (E) Recovery from fast inactivation as determined by a single exponential fit, which was similar for the WT and mutant. (F) Relative maximal conductance from solutions containing K+ or NH4+ normalized to the maximal Na+ conductance as calculated from Boltzmann fits. Note the 3.2- to 3.9-fold increased relative conductance of the mutant. All error bars represent the means ± SEM. The P values are *<0.05 and **<0.01.
Table 1.
Fit parameters of the functional study
| Parameters | WT | ±SEM | G1537S | ±SEM | |
|---|---|---|---|---|---|
| Activation | Gmax (pA/mV) | 42.48 | 5.26 | 43.58 | 5.14 |
| n = 7–9 | Vrev (mV) | 61.93 | 3.77 | 59.98 | 4.17 |
| V0.5 (mV) | −33.07 | 2.15 | −35.24 | 2.05 | |
| k (mV) | 4.32 | 0.64 | 4.01 | 0.55 | |
| Time to peak | at −45 mV (ms) | 2.67 | 0.5 | 2.12 | 0.29 |
| Inactivation | A | 0.92 | 0.01 | 0.97 | 0.02 |
| n = 8 | V0.5 (mV) | −71.54 | 2.03 | −70.47 | 1.61 |
| k (mV) | −5.45 | 0.20 | −5.93 | 0.24 | |
| C | 0.04 | 0.01 | 0.01 | 0.02 | |
| Recovery | A | 0.97 | 0.02 | 0.99 | 0.00 |
| n = 5–6 | τ (ms) | 1.40 | 0.21 | 1.55 | 0.13 |
From the same data set, the time constants of fast inactivation (τh) were obtained by fitting a single exponential function to the current decay, I = exp(−t/τh) + C, whereby t is the time and C is the asymptote. For the majority of the voltage ranges, there were no significant differences in τh; however, near the threshold at −40 and −35 mV, the mutant channels inactivated significantly more rapidly than the wild type, i.e. 1.48 ± 0.25 ms versus 3.2 ± 0.69 ms (−40 mV) and 0.94 ± 0.12 ms versus 1.54 ± 0.22 ms (−35 mV) with P < 5% (Fig. 4D). Additionally, we found a tendency towards the faster activation of mutant channels, i.e. reduced time-to-peak (Table 1). This faster gating in the near-threshold potential range (arrows in Fig. 4A and B) may contribute to the gradual amplitude decrease of repetitive action potential series during the neuronal oscillations that are typical for tremor. Steady-state inactivation was examined by a family of 50 ms test pulses from −120 mV to +20 mV following an inactivating 100 ms prepulse to −10 mV. Boltzmann functions were fitted to the data by the equation I/Imax = A/(1 + exp((V0.5 − V)/k)) + C, whereby V represents the pre-pulse potential, V0.5 represents the potential at which half of channels are inactivated, C represents the fraction of non-inactivated channels and k represents the slope factor. None of these parameters differed significantly between mutant and wild type (Fig. 4C and Table 1).
Recovery from the fast inactivation state was determined using a double pulse protocol with varying intermediate repolarization times to −140 mV with an inactivating 100 ms followed by a 50-ms test pulse, both to the same potential of −10 mV. The recovery from inactivation was analysed by fitting the data with a single exponential function, I/Imax = A*(1 − exp(−t/τ)), whereby τ denotes a time constant, A the fractional amplitude and C the level of non-inactivating sodium current. No significant difference was found for any of these parameters (Fig. 4E and Table 1).
Because of the close proximity of the p.G1537S mutation to the Nav1.4 selectivity filter of Domain IV at position A1536, we performed monovalent ion selectivity experiments by exchanging the permeable bath ion (sodium) for potassium and ammonium and eliminating all permeable ions from the pipette. Using the same activation protocol and Boltzmann fits as described above, we calculated the quotients of the maximal conductance (Gmax_K/Gmax_Na and Gmax_NH4/Gmax_Na) for the mutant and wild type (Fig. 4F). In both cases, the relative conductivities were significantly larger for the mutant, namely by a factor of 3.2–3.9, for ammonium (0.68 ± 0.11 versus 0.21 ± 0.5) and for potassium (0.59 ± 0.11 versus 0.15 ± 0.5), both with P < 1% and n = 5–7. These results suggest that SCN4A p.G1537S behaves as a change-of-function sodium channel mutation.
Discussion
Here, we described the phenotypic characteristics of a family featuring ET and epilepsy (two members) in which the age at the onset of symptoms ranged from 10 to 62 years. The five patients reported were clinically heterogeneous. Two patients featured generalized seizures and tremor occurring in the hands and head, two patients showed tremor of both hands, which progressed to head tremor in one patient and voice tremor in the other, and one patient (the youngest) showed only head tremor. Although these phenotypic heterogeneities are common in patients with familial cortical myoclonic tremor and epilepsy (FCMTE), in whom the onset of tremor and epileptic seizures could be simultaneous or occur within an interval that could extend up to several decades and in whom the age at onset may vary from 11 to 70 years (26), the electrophysiological studies performed did not support this diagnosis and showed the typical findings of ET. One family member was diagnosed with JME. We believe that the epilepsy in this subject was incidental, as it is not consistent with a dominant pattern of inheritance; however, we cannot rule out that the epilepsy in this case may also be related to the seizure activity seen in the other two patients, whereby the Nav1.4 mutation may act as an exacerbating factor.
A disease-segregating mutation (p.Gly1537Ser) was identified in the SCN4A gene. Until now, primarily myogenic disorders have been shown to be caused by SCN4A mutations, such as dyskalemic periodic paralyses, myasthenic syndromes and different forms of myotonia and paramyotonia (27). Therefore, Nav1.4 was thought to be a muscle-specific channel; however, our RT-PCR results confirm abundant levels of mRNA in neuronal tissue and suggest functional significance. A contribution of a change in Nav1.4 to a tremor phenotype may not be surprising considering that other sodium channels of the Nav family have been shown to produce tremor; for example, mice carrying SCN8A mutations not only present with paralysis, dystonia and muscle weakness but also with tremor-ataxia phenotypes (28). Although some channelopathies produce relatively uniform phenotypes, SCN4A phenotypic variability is known to be associated with different presenting phenotypes, even in patients carrying the same pathogenic mutations, which clearly suggests that the SCN4A-associated phenotypic expression may be influenced by other genetic or epigenetic factors (18,29). This may explain why two of our SCN4A mutation carriers also developed epileptic seizures. It is also worth mentioning that two novel SCN4A mutations (p.Glu974Asp, p.Lys1126Ile) have recently been reported in a patient featuring NormoKPP with involuntary movements and generalized epilepsy (30). Our functional analyses of the p.Gly1537Ser mutation in whole-cell patch-clamp studies demonstrated a tendency towards faster activation and a significantly faster inactivation in near-threshold potentials of mutated channels, which may gradually decrease the amplitudes in repetitive action potential firing and facilitate the thalamic oscillations previously described for tremor (31). In this regard, it has been shown experimentally that increased sodium channel kinetics enhance oscillation after adding a rapidly gating Nav1.7 sodium channel (similarly rapid to Nav1.4) to the several times slower Nav1.8 sodium channel (involved in tremor in mice (28,32)), which resulted in larger amplitude subthreshold oscillation and increased repetitive firing (33). Such an oscillatory mechanism can be further enhanced by voltage-sensitive potassium conductance (34). Therefore, it is conceivable that the decreased ion selectivity along with the increased potassium conductance of the mutant enhances oscillation.
Our results also show significantly higher conductivity for ammonium ions in the mutated protein versus its wild-type counterpart, suggesting change-of-function behaviour. This is not unexpected, as our previous functional study showed that a cysteine mutation of the rat homologue of G1537 (rat-G1530C) exhibited an increased relative conductance of cations, such as lithium, potassium and ammonium (35,36). Such changes in ion selectivity are compatible with its location next to the known selectivity filter residue, A1536 (24). In fact, intoxication with ammonia (NH4+), which is released with glutamate during neuronal firing, or potassium has been shown to depolarize GABAergic neurons, impairing cortical inhibition and consequently causing neurological dysfunction and seizure activity (37,38), which could explain the presence of epileptic seizures in two of our reported SCN4A mutation carriers. The involvement of the inhibitory neurotransmitter GABA in the genesis of tremor is further supported by the GABAergic action of some drugs with tremorlytic properties, the reduced GABA levels in the cerebrospinal fluid (CSF) of ET patients (39), the abnormally increased GABAA-receptor binding in the cerebellothalamic pathways of ET patients (40,41) and the fact that deficiency of GABAA-receptor α1 subunits produces an ET-like phenotype in mice (42).
The association of sodium channel genes with multiple phenotypes is not surprising because they are vital for the functioning of excitable tissue across various organ systems, including the nervous system, heart and muscle, and are responsible for generating and orchestrating the electrical signals passing through the brain, heart and muscle (43). In this study, we report the first autosomal dominant pedigree featuring ET due to a pathogenic change-of-function SCN4A mutation. Although we cannot prove that the reported SCN4A mutation is responsible for the seizure activity seen in two mutation carriers, given the SCN4A-associated phenotypic heterogeneity and the known role of the SCN family in epileptic syndromes, we conclude that SCN4A genetic variability may lead to increased seizure susceptibility. In conclusion, the pathogenicity of the p.Gly1537Ser mutation is supported by its disease segregation status, its absence in ethnicity-matched neurologically normal individuals, its prediction as pathogenic by several computational methods, its location in relevant protein functions, and its effects on ion conductivity and selectivity. This finding associates the SCN4A mutation with the development of tremor and incorporates ET into the growing list of neurological channelopathies.
Materials and Methods
Subjects
A detailed family pedigree with ET was constructed by collecting clinical histories on all putatively affected and unaffected relatives. Written informed consent, fully approved by the local ethics committee of the Hospital Universitario Donostia, was obtained from all participants. All family members' DNA samples were isolated from whole blood using standard procedures.
Ninety-four DNA samples belonging to ethnicity-matched neurologically normal individuals (49 females and 45 males) without a family history of any movement disorders were also available. The age at sample collection of the control individuals ranged from 60 to 93 years with an average of 69.1 years. DNA samples from 25 familial ET patients and 76 sporadic patients were also examined for the disease-causing mutation identified in this study, and the entire coding region of the SCN4A gene was additionally examined in 22 familial ET cases.
Clinical evaluation
All participants in this study underwent a series of structured questionnaires and a comprehensive neurological and neuropsychological assessment undertaken at the Movement Disorders Unit by three experienced movement disorder specialists (A.B., J.R.M. and J.F.M.M.). Each patient received a diagnosis of ET after the first evaluation from a neurologist specializing in movement disorders. This diagnosis was subsequently confirmed by consensus with the rest of the team based on a review of the clinical data and electrophysiological records from the second evaluation using the formerly described diagnostic criteria. The Archimedes spiral tests used for evaluation are shown in Figure 2 and Supplementary Material, Figure S1.
Physical examination
The following standardized protocol was used: (1) demographic variables; (2) personal and family history and general medical health [Cumulative Illness Rating Scale score (range: 0–42), total number of prescription medications]; (3) neurological assessment: a subjective motor complaints questionnaire, the Activities-Specific Balance Confidence (ABC) scale, a motor examination including UPDRS part III and a general examination in order to detect dystonia, myoclonus, ataxia and polyneuropathy, as well as a specific tremor exploration that includes one test for postural tremor and five tests for kinetic tremor; (4) the use of medication (yes versus. no); (5) additional variables of interest (e.g. age of symptom onset); (6) the SCOPA-AUT assessment of autonomic dysfunction (44) and (7) the Pittsburgh Sleep Quality Index (PSQI) (45). In an effort to minimize diagnostic pitfalls, the clinical criteria were comprehensively reviewed by the Consensus Statement of the Movement Disorder Society on Tremor (46) and the Washington Heights–Inwood Genetic Study of Essential Tremor criteria (47,48).
Neuropsychological and electrophysiological examination
In addition to the standard clinical exploration, the evaluation of subjects was carried out by recording the drawing of an ‘Archimedes spiral’ and using the Fahn-Tolosa-Marin TRS (49,50). The Montreal Cognitive Assessment (MoCA), which assesses different cognitive domains (http://www.mocatest.org/), was used to determine possible cognitive dysfunction (normal value >25), and DSM-IV criteria were used for the diagnosis of depression and anxiety disorders (51). EEGs were obtained after the application of electrodes and conducting jelly using the international 10–20 system of electrode placement. Standard techniques for nerve conduction studies were used. Peroneal and tibial motor responses and sural sensory responses were recorded. For SEPs, the median nerve at the wrist was stimulated, and upper limb SEPs were recorded at the contralateral scalp (C3′ and C4′; 2 cm posterior to C3 and C4 on the international 10–20 system). The stimulation rate was 3 Hz, with a duration of 0.2 ms. Digital averaging was performed using 200 samples; the filters were set at a high cut of 500 Hz and a low cut of 10 Hz. The latencies of the N20 and P25 peaks and the interpeak amplitudes of N20–P25 were analysed. Averaging was typically performed three times to ensure reproducibility. Surface electromyographies (EMGs) were recorded from wrist extensor and flexor muscles using surface electrodes placed over the muscle bellies 3 cm apart. The filters were set with a band pass of 10 Hz–1 kHz. A triaxial accelerometer was placed over the first dorsal interosseous muscle of the hand. C-reflexes were recorded from the abductor pollicis brevis muscle after delivering a supramaximal stimulus over the median nerve in the wrist.
WES analyses
Four DNA samples were subject to WES analyses: two cases featuring ET (Cases 2, 8), one case featuring JME (Case 9) and an unaffected case (Case 4) (Fig. 1A). The SureSelect Human All exon 50 Mb exon-capture kit was used for library enrichment (Agilent Technologies Inc., Santa Clara, CA), and captured libraries were sequenced on the HiSeq2000 according to the manufacturer's instructions for paired-end 100-bp reads (Illumina Inc., San Diego, CA), using a single flow cell lane per sample. Sequencing data were put through a computational pipeline for WES data processing and analysis following the general workflow adopted by the 1000 Genomes Project (52). Briefly, raw sequence reads were aligned to the human reference genome sequence (NCBI GRCh37.p13) using a fast lightweight Burrows–Wheeler Alignment Tool (BWA) (53), followed by a base-quality recalibration and local realignment through the Genome Analysis Toolkit (GATK v1.5-16-g58245bf); the GATK Unified Genotyper tool was used to call single-nucleotide substitutions (SNP/SNV) and short insertions/deletions (INDELs). Calls were then filtered based on mapping quality (q30 or higher) and depth of coverage (d10 or higher), and resulting calls were lastly annotated with an exhaustive genome annotation toolkit (AnnTools) (54). Each exome's statistics were conducted in PICARD (http://picard.sourceforge.net/).
Potential mutations observed as common variations (frequency >3%) in the latest dbSNP137 build, 1000 Genomes Project Phase 1, the Exome Variant Server of the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (http://evs.gs.washington.edu/EVS/) (55) or exomes generated in-house (13,56) were removed from further analyses. Genetic variants mapping to intra-genic, intronic and non-coding exonic regions, with the exception of those variants mapping close to splice sites, were also removed because they were unlikely to be causative.
Alamut Visual 2.4.2 software (http://www.interactive-biosoftware.com/alamut-visual/) was used to perform in silico analyses of the novel, disease-segregating SCN4A mutation identified in this study. Computational methods for the prediction of pathogenicity that were not included in the Alamut software, such as MutPred (http://mutpred.mutdb.org/) and SNPs&Go (http://snps-and-go.biocomp.unibo.it/snps-and-go/), were also employed. The professional Human Gene Mutation Database (HGMD) (https://portal.biobase-international.com/hgmd/pro/start.php) and the NCBI ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/) were used to determine whether the SCN4A p.Gly1537Ser mutation was already known to be associated with a disease-related phenotype.
Candidate gene screening
Genomic primers for PCR amplifications of the RIMS1 exon 21, KMT2C exon 30, UNC13B exon 8, AGAP5 exon 8, NFATC4 exon 2, ANKFN1 exon 16 and all SCN4A exons were designed using a public primer design website (http://ihg.gsf.de/ihg/ExonPrimer.html) (primer sequences available upon request). All purified PCR products were sequenced in both the forward and reverse directions with Applied Biosystems BigDye terminator v3.1 sequencing chemistry as per the manufacturer's instructions. The resulting sequencing reactions were resolved on an ABI3730 genetic analyser (Applied Biosystems, Foster City, CA) and were analysed using Sequencher 5.2 software (Gene Codes Corporation, Ann Arbor, MI).
Gene expression assays
Reverse transcription polymerase chain reaction (RT-PCR) of brain (2-month-old mouse brain, Zyagen) and skeletal muscle mouse RNA (BioChain) was performed with the RT-PCR Kit from Qiagen and the primers are described in Supplementary Material, Table S1. The PCR products were run on a 1.5% agarose gel and stained with Red Safe (iNtRON Biotechnology) and sequenced for confirmation as previously described.
Total RNA from the human cerebral cortex was acquired from Clontech (Clontech, Mountain View, CA) and reverse transcribed into cDNA using the Omniscript reverse transcription kit according to the manufacturer's instructions (Qiagen, Hilde, Germany). Absolute quantification using a standard curve generated with seven serial dilutions of cDNA (0.4–425 ng) was used to quantify the expression levels of the SCN1A, SCN2A and SCN4A genes. The SCN1A and SCN2A genes were used as control genes. All qPCR assays were performed on an Eco Real-Time PCR System (Illumina, San Diego, CA) using SYBR green PCR master mix (Applied Biosystems, Foster City, CA) and the oligonucleotide sequences are described in Supplementary Material, Table S1. The samples were run in quadruplicate, and non-template controls were included in each run. The data were automatically analysed with Eco software.
Functional analyses
Site-directed mutagenesis was performed by PCR of an SCN4A-containing pCRC-CMV-EGFP plasmid. TsA201 cells were transfected with wild-type or mutant cDNA using the jetPEI kit. Standard whole-cell recordings were performed with a –p/4 protocol to subtract the leak current. Data acquired with pCLAMP 10 (Molecular Devices, Sunnyvale, CA) were filtered at 5 kHz and analysed by a combination of pCLAMP and ORIGIN (MicroCal) (Malvern Instruments, Westborough, MA). For the activation, inactivation and recovery experiments, the patch electrodes contained the standard solution (in mM): CsF 105, NaCl 35, EGTA 10 and HEPES 10. The bath contained the standard solution (in mM): NaCl 150, KCl 2, CaCl2 1.5, MgCl2 1 and HEPES 10. The patch electrodes for the ion selectivity experiments contained the following (in mM): NMDG 140, EGTA 10 and HEPES 10. The bath contained the following (in mM): NaCl 152, CaCl2 1.5, MgCl2 1 and HEPES 10. Additionally, the monovalent cations K+ or NH4+ replaced Na+ as chloride salts for further permeability tests. Solutions were adjusted to pH 7.4, and experiments were performed at room temperature (20–25°C). All data are presented as the means ± SEM. Statistical significance was assessed by Welch's t-test for normally distributed data with criteria of P < 0.05 and P < 0.01.
Supplementary Material
Funding
This work is supported in part by the ‘Instituto de Salud Carlos III’ (FIS PI10/02714; J.F.M.M.), the non-profit Hertie foundation (K.J.R.), the IonNeurONet of the German Federal Research Ministry of Research (K.J.R.) and the National Institute of Neurological Disorders and Stroke of the National Institutes of Health (R21NS082881, R01NS079388; C.P.R.).
Supplementary Material
Acknowledgements
We thank the patients, relatives and other participants for their cooperation in this study.
Conflict of Interest statement. The authors declare no conflicts of interest.
References
- 1.Louis E.D., Ferreira J.J. (2010) How common is the most common adult movement disorder? Update on the worldwide prevalence of essential tremor. Mov. Disord., 25, 534–541. [DOI] [PubMed] [Google Scholar]
- 2.Louis E.D. (2010) Essential tremor: evolving clinicopathological concepts in an era of intensive post-mortem enquiry. Lancet Neurol., 9, 613–622. [DOI] [PubMed] [Google Scholar]
- 3.Jellinger K.A. (2014) Is there cerebellar pathology in essential tremor? Mov. Disord., 29, 435–436. [DOI] [PubMed] [Google Scholar]
- 4.Benito-Leon J. (2014) Essential tremor: a neurodegenerative disease? Tremor Other Hyperkinet. Mov., 4, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhlenbaumer G., Hopfner F., Deuschl G. (2014) Genetics of essential tremor: meta-analysis and review. Neurology, 82, 1000–1007. [DOI] [PubMed] [Google Scholar]
- 6.Schrag A., Munchau A., Bhatia K.P., Quinn N.P., Marsden C.D. (2000) Essential tremor: an overdiagnosed condition? J. Neurol., 247, 955–959. [DOI] [PubMed] [Google Scholar]
- 7.Zimprich A. (2011) Genetics of Parkinson's disease and essential tremor. Curr. Opin. Neurol., 24, 318–323. [DOI] [PubMed] [Google Scholar]
- 8.Thier S., Lorenz D., Nothnagel M., Poremba C., Papengut F., Appenzeller S., Paschen S., Hofschulte F., Hussl A.C., Hering S. et al. (2012) Polymorphisms in the glial glutamate transporter SLC1A2 are associated with essential tremor. Neurology, 79, 243–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merner N.D., Girard S.L., Catoire H., Bourassa C.V., Belzil V.V., Riviere J.B., Hince P., Levert A., Dionne-Laporte A., Spiegelman D. et al. (2012) Exome sequencing identifies FUS mutations as a cause of essential tremor. Am. J. Hum. Genet., 91, 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Unal Gulsuner H., Gulsuner S., Mercan F.N., Onat O.E., Walsh T., Shahin H., Lee M.K., Dogu O., Kansu T., Topaloglu H. et al. (2014) Mitochondrial serine protease HTRA2 p.G399S in a kindred with essential tremor and Parkinson disease. Proc. Natl. Acad. Sci. USA, 111, 18285–18290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sánchez E., Bergareche A., Krebs C.E., Gorostidi A., Makarov V., Ruiz-Martinez J., Chorny A., Lopez de Munain A., Martí-Massó J.F., Paisán-Ruiz C. (2015) SORT1 mutation resulting in sortilin deficiency and p75NTR upregulation in a family with essential tremor. ASN Neuro, 7, pii: 1759091415598290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krebs C.E., Paisan-Ruiz C. (2012) The use of next-generation sequencing in movement disorders. Front. Genet., 3, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marti-Masso J.F., Bergareche A., Makarov V., Ruiz-Martinez J., Gorostidi A., Lopez de Munain A., Poza J.J., Striano P., Buxbaum J.D., Paisan-Ruiz C. (2013) The ACMSD gene, involved in tryptophan metabolism, is mutated in a family with cortical myoclonus, epilepsy, and parkinsonism. J. Mol. Med. (Berl.), 91, 1399–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehndiratta M.M., Satyawani M., Gupta S., Khwaja G.A. (2005) Clinical and surface EMG characteristics of valproate induced tremors. Electromyogr. Clin. Neurophysiol., 45, 177–182. [PubMed] [Google Scholar]
- 15.Exome Aggregation Consortium (ExAC). (2015) Cambridge, MA. Accessed at http://exac.broadinstitute.org (07/2015).
- 16.Venance S.L., Jurkat-Rott K., Lehmann-Horn F., Tawil R. (2004) SCN4A-associated hypokalemic periodic paralysis merits a trial of acetazolamide. Neurology, 63, 1977. [DOI] [PubMed] [Google Scholar]
- 17.Fontaine B., Khurana T.S., Hoffman E.P., Bruns G.A., Haines J.L., Trofatter J.A., Hanson M.P., Rich J., McFarlane H., Yasek D.M. et al. (1990) Hyperkalemic periodic paralysis and the adult muscle sodium channel alpha-subunit gene. Science, 250, 1000–1002. [DOI] [PubMed] [Google Scholar]
- 18.Brunklaus A., Ellis R., Reavey E., Semsarian C., Zuberi S.M. (2014) Genotype phenotype associations across the voltage-gated sodium channel family. J. Med. Genet., 51, 650–658. [DOI] [PubMed] [Google Scholar]
- 19.Lion-Francois L., Mignot C., Vicart S., Manel V., Sternberg D., Landrieu P., Lesca G., Broussolle E., Billette de Villemeur T., Napuri S. et al. (2010) Severe neonatal episodic laryngospasm due to de novo SCN4A mutations: a new treatable disorder. Neurology, 75, 641–645. [DOI] [PubMed] [Google Scholar]
- 20.Ptacek L.J., George A.L. Jr, Barchi R.L., Griggs R.C., Riggs J.E., Robertson M., Leppert M.F. (1992) Mutations in an S4 segment of the adult skeletal muscle sodium channel cause paramyotonia congenita. Neuron, 8, 891–897. [DOI] [PubMed] [Google Scholar]
- 21.Tsujino A., Maertens C., Ohno K., Shen X.M., Fukuda T., Harper C.M., Cannon S.C., Engel A.G. (2003) Myasthenic syndrome caused by mutation of the SCN4A sodium channel. Proc. Natl. Acad. Sci. USA, 100, 7377–7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh R., Peters N.S., Cook S.A., Ware J.S. (2014) Paralogue annotation identifies novel pathogenic variants in patients with Brugada syndrome and catecholaminergic polymorphic ventricular tachycardia. J. Med. Genet., 51, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Catterall W.A., Goldin A.L., Waxman S.G. (2005) International Union of Pharmacology. XLVII. Nomenclature and structure–function relationships of voltage-gated sodium channels. Pharmacol. Rev., 57, 397–409. [DOI] [PubMed] [Google Scholar]
- 24.Heinemann S.H., Terlau H., Stuhmer W., Imoto K., Numa S. (1992) Calcium channel characteristics conferred on the sodium channel by single mutations. Nature, 356, 441–443. [DOI] [PubMed] [Google Scholar]
- 25.Terlau H., Heinemann S.H., Stuhmer W., Pusch M., Conti F., Imoto K., Numa S. (1991) Mapping the site of block by tetrodotoxin and saxitoxin of sodium channel II. FEBS Lett., 293, 93–96. [DOI] [PubMed] [Google Scholar]
- 26.van Rootselaar A.F., van Schaik I.N., van den Maagdenberg A.M., Koelman J.H., Callenbach P.M., Tijssen M.A. (2005) Familial cortical myoclonic tremor with epilepsy: a single syndromic classification for a group of pedigrees bearing common features. Mov. Disord., 20, 665–673. [DOI] [PubMed] [Google Scholar]
- 27.Nicole S., Fontaine B. (2015) Skeletal muscle sodium channelopathies. Curr. Opin. Neurol., 28, 508–514. [DOI] [PubMed] [Google Scholar]
- 28.Meisler M.H., Kearney J., Escayg A., MacDonald B.T., Sprunger L.K. (2001) Sodium channels and neurological disease: insights from Scn8a mutations in the mouse. Neuroscientist, 7, 136–145. [DOI] [PubMed] [Google Scholar]
- 29.Lee S.C., Kim H.S., Park Y.E., Choi Y.C., Park K.H., Kim D.S. (2009) Clinical diversity of SCN4A-mutation-associated skeletal muscle sodium channelopathy. J. Clin. Neurol., 5, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao L., Li X., Hong D. (2014) Normokalemic periodic paralysis with involuntary movements and generalized epilepsy associated with two novel mutations in SCN4A gene. Seizure, 24, 134–136. [DOI] [PubMed] [Google Scholar]
- 31.Shaikh A.G., Miura K., Optican L.M., Ramat S., Tripp R.M., Zee D.S. (2008) Hypothetical membrane mechanisms in essential tremor. J. Transl. Med., 6, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu X., Guo F., Lv X., Feng R., Min D., Ma L., Liu Y., Zhao J., Wang L., Chen T. et al. (2013) Abnormal changes in voltage-gated sodium channels Na(V)1.1, Na(V)1.2, Na(V)1.3, Na(V)1.6 and in calmodulin/calmodulin-dependent protein kinase II, within the brains of spontaneously epileptic rats and tremor rats. Brain Res. Bull., 96, 1–9. [DOI] [PubMed] [Google Scholar]
- 33.Choi J.S., Waxman S.G. (2011) Physiological interactions between Na(v)1.7 and Na(v)1.8 sodium channels: a computer simulation study. J. Neurophysiol., 106, 3173–3184. [DOI] [PubMed] [Google Scholar]
- 34.Amir R., Liu C.N., Kocsis J.D., Devor M. (2002) Oscillatory mechanism in primary sensory neurones. Brain, 125, 421–435. [DOI] [PubMed] [Google Scholar]
- 35.Chiamvimonvat N., Perez-Garcia M.T., Ranjan R., Marban E., Tomaselli G.F. (1996) Depth asymmetries of the pore-lining segments of the Na+ channel revealed by cysteine mutagenesis. Neuron, 16, 1037–1047. [DOI] [PubMed] [Google Scholar]
- 36.Struyk A.F., Cannon S.C. (2002) Slow inactivation does not block the aqueous accessibility to the outer pore of voltage-gated Na channels. J. Gen. Physiol., 120, 509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye X., Robinson M.B., Pabin C., Quinn T., Jawad A., Wilson J.M., Batshaw M.L. (1997) Adenovirus-mediated in vivo gene transfer rapidly protects ornithine transcarbamylase-deficient mice from an ammonium challenge. Pediatr. Res., 41, 527–534. [DOI] [PubMed] [Google Scholar]
- 38.Rangroo Thrane V., Thrane A.S., Wang F., Cotrina M.L., Smith N.A., Chen M., Xu Q., Kang N., Fujita T., Nagelhus E.A. et al. (2013) Ammonia triggers neuronal disinhibition and seizures by impairing astrocyte potassium buffering. Nat. Med., 19, 1643–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mally J., Baranyi M., Vizi E.S. (1996) Change in the concentrations of amino acids in CSF and serum of patients with essential tremor. J. Neural. Transm., 103, 555–560. [DOI] [PubMed] [Google Scholar]
- 40.Boecker H., Weindl A., Brooks D.J., Ceballos-Baumann A.O., Liedtke C., Miederer M., Sprenger T., Wagner K.J., Miederer I. (2010) GABAergic dysfunction in essential tremor: an 11C-flumazenil PET study. J. Nucl. Med., 51, 1030–1035. [DOI] [PubMed] [Google Scholar]
- 41.Gironell A., Figueiras F.P., Pagonabarraga J., Herance J.R., Pascual-Sedano B., Trampal C., Gispert J.D. (2012) Gaba and serotonin molecular neuroimaging in essential tremor: a clinical correlation study. Parkinsonism Relat. Disord., 18, 876–880. [DOI] [PubMed] [Google Scholar]
- 42.Kralic J.E., Criswell H.E., Osterman J.L., O'Buckley T.K., Wilkie M.E., Matthews D.B., Hamre K., Breese G.R., Homanics G.E., Morrow A.L. (2005) Genetic essential tremor in gamma-aminobutyric acidA receptor alpha1 subunit knockout mice. J. Clin. Invest., 115, 774–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ackerman M.J., Clapham D.E. (1997) Ion channels—basic science and clinical disease. N. Engl. J. Med., 336, 1575–1586. [DOI] [PubMed] [Google Scholar]
- 44.Visser M., Marinus J., Stiggelbout A.M., Van Hilten J.J. (2004) Assessment of autonomic dysfunction in Parkinson's disease: the SCOPA-AUT. Mov. Disord., 19, 1306–1312. [DOI] [PubMed] [Google Scholar]
- 45.Buysse D.J., Reynolds C.F. III, Monk T.H., Berman S.R., Kupfer D.J. (1989) The Pittsburgh Sleep Quality Index: a new instrument for psychiatric practice and research. Psychiatry Res., 28, 193–213. [DOI] [PubMed] [Google Scholar]
- 46.Deuschl G., Bain P., Brin M. (1998) Consensus statement of the Movement Disorder Society on Tremor. Ad Hoc Scientific Committee. Mov. Disord., 13 (Suppl 3), 2–23. [DOI] [PubMed] [Google Scholar]
- 47.Benito-Leon J., Louis E.D. (2006) Essential tremor: emerging views of a common disorder. Nat. Clin. Pract. Neurol., 2, 666–678; quiz 662p following 691. [DOI] [PubMed] [Google Scholar]
- 48.Louis E.D., Ottman R., Ford B., Pullman S., Martinez M., Fahn S., Hauser W.A. (1997) The Washington Heights-Inwood Genetic Study of essential tremor: methodologic issues in essential-tremor research. Neuroepidemiology, 16, 124–133. [DOI] [PubMed] [Google Scholar]
- 49.Elble R.J., Sinha R., Higgins C. (1990) Quantification of tremor with a digitizing tablet. J. Neurosci. Methods, 32, 193–198. [DOI] [PubMed] [Google Scholar]
- 50.Fahn S., Tolosa E., Marin C. (2003) Clinical Rating Scale for Tremor. Williams and Wilkins, Baltimore, USA. [Google Scholar]
- 51.American Psychiatric Association. (1994) Diagnostic and Statistical Manual of Mental Disorders: DSM-IV. American Psychiatric Association, USA. [Google Scholar]
- 52.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet., 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H., Durbin R. (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Makarov V., O'Grady T., Cai G., Lihm J., Buxbaum J.D., Yoon S. (2011) AnnTools: a comprehensive and versatile annotation toolkit for genomic variants. Bioinformatics, 28, 724–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Exome Variant Server. (2015) NHLBI Exome Sequencing Project (ESP), Seattle, WA. Accessed at http://evs.gs.washington.edu/EVS/ (07/2015).
- 56.Krebs C.E., Karkheiran S., Powell J.C., Cao M., Makarov V., Darvish H., Di Paolo G., Walker R.H., Shahidi G.A., Buxbaum J.D. et al. (2013) The Sac1 domain of SYNJ1 identified mutated in a family with early-onset progressive Parkinsonism with generalized seizures. Hum. Mutat., 34, 1200–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.