Summary
The torpedo model of transcription termination asserts that the exonuclease Xrn2 attacks the 5′PO4-end exposed by nascent RNA cleavage and chases down the RNA polymerase. We tested this mechanism using a dominant-negative human Xrn2 mutant and found that it delayed termination genome-wide. Xrn2 nuclease inactivation caused strong termination defects downstream of most poly(A) sites and modest delays at some histone and U snRNA genes suggesting that the torpedo mechanism is not limited to poly(A) site-dependent termination. A central untested feature of the torpedo model is that there is kinetic competition between the exonuclease and the pol II elongation complex. Using pol II rate mutants, we found that slow transcription robustly shifts termination upstream, and fast elongation extends the zone of termination further downstream. These results suggest that kinetic competition between elongating pol II and the Xrn2 exonuclease is integral to termination of transcription on most human genes.
Introduction
Transcription termination is an essential step integral to the universal cycle of initiation, elongation and termination of RNA chains. It is required to recycle polymerase and to prevent interference between adjacent transcription units. Termination requires dismantling of the stable ternary complex of DNA template, polymerase and RNA transcript. At most genes termination of transcription by RNA polymerase II (pol II) is triggered by recognition of the poly(A) site (PAS) thereby ensuring that it occurs after the transcript is complete (Birse et al., 1998; Connelly and Manley, 1988; Logan et al., 1987; Whitelaw and Proudfoot, 1986). Pol II transcription termination occurs at diffuse sites at variable distances as much as 10 kb downstream of genes and is generally preceded by a peak of pol II density that is also usually quite diffuse (Lian et al., 2008). We refer to this region of 3′ flanking sequences as the “termination zone”. The peak of pol II occupancy in the termination zone has been interpreted as a pause that may facilitate co-transcriptional 3′ end processing and coupled polymerase release from the template (Glover-Cutter et al., 2008; Gromak et al., 2006) but other interpretations are also possible (Anamika et al., 2014; Grosso et al., 2012). The length of the termination zone in the 3′ flanking region varies between genes and is especially short for histone genes (Anamika et al., 2012) that make non-adenylated transcripts by a mechanism that uses that same CPSF73 endonuclease that cuts poly(A) sites (Dominski et al., 2005; Kolev and Steitz, 2005). What determines the length of the termination zone is not known. Nor is it known for PAS-containing genes whether the length of the termination zone is relatively constant and gene-specific or something that varies between cell types. Recent evidence shows that in some cases termination downstream of poly(A) sites can be greatly delayed in response to stress stimuli (Vilborg et al., 2015).
The mechanism of poly(A) site dependent termination has been a topic of intense interest for almost three decades. The debate is centered on two predominant models that are not mutually exclusive, the allosteric model and the torpedo model (Kuehner et al., 2011; Luo et al., 2006; Richard and Manley, 2009). The allosteric model (Logan et al., 1987) posits a conformational change in the polymerase after encountering the poly(A) site that weakens the ternary complex and favors termination. The allosteric model is supported by in vitro experiments in the presence of an anti-pol II antibody that induces termination, but its relevance in vivo remains to be established (Zhang et al., 2015; Zhang et al., 2004). The torpedo model (Connelly and Manley, 1988), on the other hand, proposes that the 5′ phosphate end exposed by poly(A) site cleavage provides an entry point for an RNA 5′-3′ exonuclease that tracks down the nascent transcript and when it encounters the polymerase, helps to dislodge it from the DNA template. The requirement for a nuclear 5′-3′ RNA exonuclease predicted by the torpedo model has been compellingly validated in budding yeast where inactivation of the Rat1 exonuclease substantially delays termination downstream of genes with poly(A) sites. Furthermore exonucleolytic activity of Rat1 is required for transcription termination in vivo as demonstrated by mutation of the active site (Kim et al., 2004) whereas in vitro there are conflicting reports about whether enzymatic activity is required for termination (Park et al., 2015; Pearson and Moore, 2013). In fission yeast, the 3′-5′ exonucleases in the exosome function have been proposed to act as backup termination factors at protein coding genes but the importance of a 5′-3′ “torpedo” in this organism is not known (Lemay et al., 2014). Whether the 5′-3′ torpedo mechanism is widely used for poly(A) site dependent termination of transcription in metazoan cells remains unresolved. Unexpectedly, shRNA knock-down of mammalian Xrn2 exonuclease, the homologue of Rat1, failed to inhibit termination downstream of a β-actin reporter gene (Banerjee et al., 2009). On the other hand Xrn2 knockdown did inhibit termination at a β-globin reporter gene where termination is thought to be initiated by self cleavage of the transcript at a CoTC element downstream of the poly(A) site (West et al., 2004). At this exceptional class of CoTC containing genes (Nojima et al., 2013), cleavage and polyadenylation are proposed to occur post-transcriptionally and uncoupled from termination (West et al., 2008). Surprisingly, Xrn2 knockdown does not affect termination downstream of PAS-containing chromosomal genes (Brannan et al., 2012; Nojima et al., 2015), although we and others found evidence for an effect on premature termination within genes (Brannan et al., 2012; Wagschal et al., 2012). Based on these investigations it has been suggested that “Xrn2 plays a unique role” in premature termination near the transcription start site, but not in termination at 3′ ends (Nojima et al., 2015). Alternative candidate termination factors that might act downstream of poly(A) sites include the nuclear exosome and the helicase senataxin (Sen1) that both function as terminators in yeast by distinct mechanisms (Fox et al., 2015; Lemay et al., 2014; Porrua and Libri, 2013; Schaughency et al., 2014; Steinmetz et al., 2006).
Pol II termination downstream of genes that make non-adenylated transcripts is also poorly understood in metazoans. Histone mRNA and U snRNA 3′ ends are made by endonucleolytic cleavage that that is uncoupled from polyadenylation (Dominski, 2007). In addition miRNA precursors can be co-transcriptionally cleaved by microprocessor, Drosha/DCGR8 (Morlando et al., 2008) which is required for transcription termination of a few long non-coding RNAs (lnc RNAs) that host miRNAs (Dhir et al., 2015). The 3′ ends of the MALAT1 and MENβ lnc RNAs are created by the tRNA processing endonuclease RNAseP (Wilusz and Spector, 2010). In principle, endonucleolytic cleavage at any these sites of 3′ end formation could provide entry points for an exonuclease torpedo that terminates transcription, but this idea has yet to be confirmed (Baillat et al., 2005; Dominski et al., 2005; O’Reilly et al., 2014; Skaar et al., 2015; Weiner, 2005). It has been reported in a few cases however, that transcripts extending downstream of a co-transcriptional microprocessor cleavage site are stabilized by Xrn2 knockdown (Ballarino et al., 2009; Wagschal et al., 2012). Whether transcription termination by a torpedo mechanism commonly occurs downstream of microprocessor cleavage sites within miRNA precursor genes is not known.
An integral component of the torpedo model is the implied kinetic competition between two moving parts, the transcription elongation complex and the 5′-3′ RNA exonuclease that is pursuing it. This aspect of the model makes a clear prediction that has remained untested, namely that fast elongation will delay termination and slow elongation will advance it. Kinetic competition of this type was first defined in E. coli where the relative rates of RNA polymerase elongation and rho helicase translocation determine where transcription termination occurs (Jin et al., 1992). There is also strong evidence for kinetic competition between RNA pol II elongation and the terminator helicase Sen1 at the short non-coding snoRNA genes in yeast (Hazelbaker et al., 2013). Slow and fast mutants in the trigger loop of the pol II large subunit produced shortened or lengthened pre-snoRNA transcripts respectively consistent with kinetic competition between pol II and Sen1 that tracks along the nascent transcript (Hazelbaker et al., 2013; Porrua and Libri, 2013). Pol II elongation rate differs substantially between genes (Danko et al., 2013; Fuchs et al., 2014; Larson et al., 2011; Veloso et al., 2014), but whether this variation affects where termination occurs in 3′ flanking regions is not known.
Here we investigate the universality of the torpedo model of pol II termination in human cells and its prediction that termination and transcription elongation are engaged in a kinetic competition with one another. We tested the importance of the 5′-3′ exonuclease Xrn2 for pol II termination genome-wide by expressing an inducible dominant-negative mutant in the exonuclease active site. This mutant in combination with shRNA mediated knockdown of endogenous Xrn2 caused a general inhibition of pol II termination that shifted the termination zone further downstream of genes demonstrating that the torpedo mechanism operates widely. To test for kinetic competition, we monitored termination genome-wide in cells expressing slow and fast mutants of human pol II. The results showed that at most genes with poly(A) sites the site of termination is highly dependent on elongation rate. Slow and fast elongation evoked early/upstream and late/downstream termination respectively, as predicted by the torpedo model.
Results
Xrn2 nuclease is required for poly(A) site-dependent pol II termination genome-wide
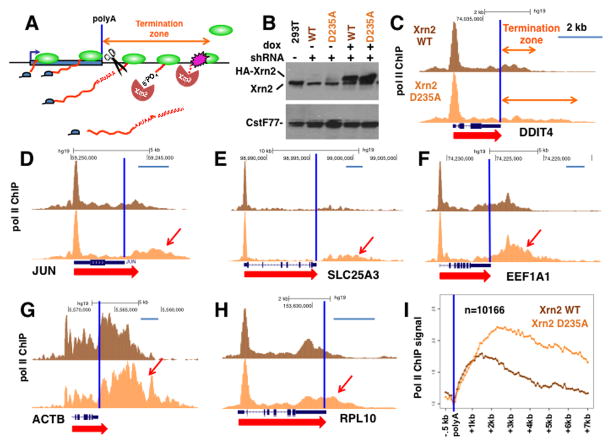
To determine whether the Xrn2 exonuclease plays a widespread role in termination in human cells, we wanted to inhibit its activity and determine whether profiles of pol II in 3′ flanking regions were altered. Because we and others (Banerjee et al., 2009; Brannan et al., 2012; Nojima et al., 2015) failed to detect a defect in termination when Xrn2 expression was knocked down, we turned to a dominant-negative approach guided by a mutation of the Xrn1 active site that inactivates the enzyme while preserving the ability to form a stable complex with RNA through interactions with a conserved 5′ PO4 binding pocket (Jinek et al., 2011). We mutated the homologous position in the active site of human Xrn2 (D235A) and expressed the HA-tagged mutant protein under doxycycline control after site-specific integration into HEK293 Flp-in cells (see Experimental Procedures). By analogy with the Xrn1 mutant, we expect Xrn2(D235A) to form stable complexes with RNA 5′ PO4 ends in vivo and to delay degradation by sterically interfering with endogenous WT Xrn2. As a control we made isogenic cells expressing HA-tagged wild type Xrn2. Both lines induced expression of HA-tagged protein at approximately equal levels that were several fold above endogenous Xrn2 (Fig. 1B). Pol II ChIP-seq analysis of these cells after doxycycline induction showed a very small delay in termination in the Xrn2(D235A) mutant expressing cells (Fig. S1A). Over-expression of the WT Xrn2 did not strongly affect termination relative to cells in which it is not over-expressed (Fig. S1A). To enhance the potential dominant-negative effect, we stably knocked down endogenous Xrn2 by over 75% (Fig. 1B) with a lentiviral shRNA targeted to the 3′ UTR in cells with HA-Xrn2 WT and HA-Xrn2 (D235A) transgenes. These cells were used for all other experiments. To confirm that the over-expressed WT and D235A Xrn2 are recruited to endogenous genes we performed anti-HA ChIP of the tagged proteins. The results in Figure S1B show that the WT and D235A Xrn2 are recruited to the termination region of the GAPDH gene and the TSS of the RPL32 gene at approximately equivalent levels. We conclude that the D235A mutation does not inadvertently inhibit the ability of Xrn2 to associate with transcription complexes.
Figure 1. Exonuclease dead Xrn2 causes widespread delays in poly(A) site dependent termination.
A. The torpedo model of transcription termination by Xrn2 in kinetic competition with elongating pol II in the “termination zone” downstream of the poly(A) site.
B. Induction of HA-tagged WT and exonuclease dead (D235A) Xrn2 with doxycycline (24hr) in HEK293 Flp-in TREX cells. Western blot signals with anti-Xrn2 antibody were normalized to the CstF77 loading control using the Bio-Rad ChemiDoc MP imaging system. Xrn2 shRNA knockdown was >75% (lanes 2,3 vs lane 1) and over-expression of WT and D235A HA-tagged Xrn2 over endogenous was approximately 2.5 fold (lanes 4, 5).
C–H. Anti-Pol II ChIP-seq shows delayed termination downstream of most genes with poly(A) sites in cells expressing D235A dominant negative mutant Xrn2 relative to WT. Endogenous Xrn2 is stably knocked down with an shRNA targeted to the 3′ UTR. UCSC genome browser screen shots are shown with arrows indicating 3′ extension of the termination zone caused by Xrn2 (D235A). Poly(A) sites are marked by vertical blue lines. Scale bars represent 2kb. A biological replicate ChIP-seq is shown in Figs. S1D–I.
I. Metaplot of mean pol II ChIP signals normalized to the poly(A) site for over 10,000 well separated genes (>5kb apart) in cells expressing WT and dominant negative Xrn2 (D235A). Note the downstream shift in average pol II density caused by expression of mutant Xrn2. A biological replicate is shown in Figure S1C.
Notably we found that expression of Xrn2 (D235A) in the context of endogenous Xrn2 knockdown specifically extended regions of pol II occupancy beyond the poly(A) site by up to several kilobases relative to the WT Xrn2 control (Fig. 1C–H) and this result was reproduced in a biological replicate (Fig. S1D–I). The effect of inactivating the Xrn2 nuclease on transcription is likely dose-dependent because the combination of endogenous Xrn2 knockdown with expression of mutant Xrn2 had a markedly greater impact than either treatment alone. Furthermore the effect of Xrn2 enzymatic inactivation is widespread as shown by the shift in pol II density further into 3′ flanking sequences in a metaplot of several thousand well-separated genes (Figs. 1I, S1C). We interpret this result to mean that poly (A) site dependent termination is generally delayed but not entirely prevented by interfering with Xrn2 access to 5′ PO4 ends. Pol II density normally increases in flanking sequences relative to the 3′ end of the gene consistent with a transcriptional slow-down or pause prior to termination. Xrn2 nuclease inactivation extended the length of termination zone without reducing the density of pol II in that region. Thus while inactivating Xrn2 nuclease activity delays termination, it does not diminish the apparent pause that precedes termination. In summary, Xrn2 exonuclease is required for timely pol II termination downstream of poly(A) sites genome-wide in human cells as predicted by the torpedo model.
The position of the termination zone is a gene-specific property
To address whether the length of the termination zone is a specific property of individual genes, we analyzed pol II density profiles in the 3′ flanking regions of highly expressed, well-separated non-histone genes. Genes were initially clustered into 2 groups based on relative pol II occupancy in their 3′ flanking regions in HEK293 cells (Fig. 2A). Gene groups I and II (Table S1) have peaks of relative pol II density approximately 1kb and 3kb downstream of the most 3′ poly(A) site. We compared pol II ChIP-seq profiles in 3′ flanking regions for HEK293, Hela, 21NT breast epithelial and HAP1 myeloid leukemia lines and found a remarkable conservation of the termination groups between these lines (Fig. 2B, C). This observation suggests that the location of the termination zone is a relatively constant intrinsic gene-specific feature. Groups I and II did not differ markedly in the strength of their poly(A) sites but group I had slightly higher frequencies of AATAAA and ATTAAA consensus sequences. We next asked how Xrn2 affects termination at group I and group II genes, and found that Xrn2 (D235A) caused a robust termination delay in both gene groups (Fig 2D, E). These results therefore suggest that Xrn2 plays an important role in termination at both proximal and distal positions downstream of poly(A) sites.
Figure 2. The termination zone is a reproducible gene-specific feature influenced by Xrn2 activity.
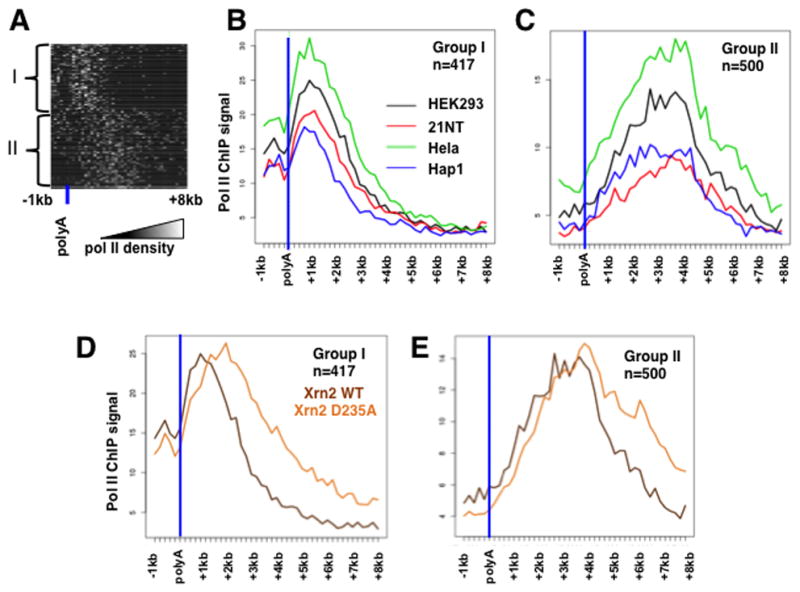
A. Two groups of genes (I, II, Table S1) that terminate at proximal, and distal positions relative to the poly(A) site. Pol II ChIP seq profiles for well separated (>8kb) non-histone genes in HEK293 cells expressing WT HA-Xrn2 were clustered by k-means clustering (kmeans function in R) after converting a distribution into binary features below and above 75 percentile.
B. C. Proximal and distal termination groups I and II are conserved between cell types. Mean Pol II ChIP seq signals for HEK293 cells as in A, Hela, HAP1 myeloid leukemia and 21NT breast epithelial cells are shown. Note that termination distance is an intrinsic property of genes conserved between cell types.
D, E. Xrn2(D235A) impairs termination at proximal and distal positions relative to the poly(A) site. Mean Pol II ChIP-seq signals are plotted from the experiment in Fig. 1.
Xrn2 facilitates termination at some genes with polyA-transcripts
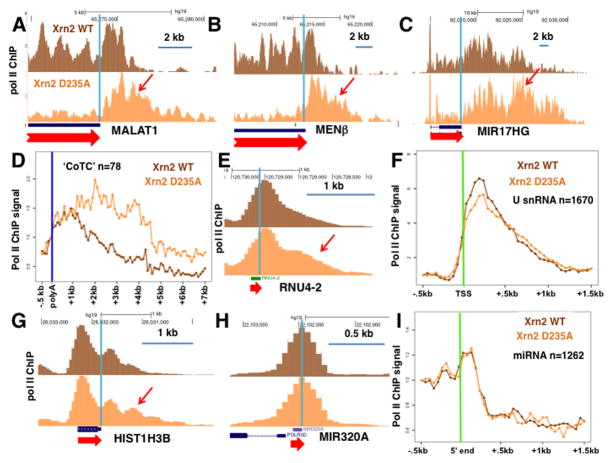
We examined whether Xrn2 functions in pol II termination at genes whose RNA 3′ ends are formed by cleavage mechanisms that are not coupled to polyadenylation. Mutation of Xrn2 robustly delayed termination of the non-adenylated miRNA host gene MIR17HG processed by the microprocessor (Dhir et al., 2015) and the MALAT and MENβ lncRNAs whose 3′ ends are formed by RNAseP cleavage (Wilusz and Spector, 2010) (Fig. 3A–C, Fig. S2A–C). Consistent with a direct Xrn2 function in termination at these genes, ChIP experiments show that it is recruited to their 3′ ends (Fig. 4A–C). Xrn2 (D235A) also markedly delayed termination at a cohort of over 70 genes that are candidates for termination mediated by self-cleaving CoTC elements (Nojima et al., 2013) (Fig. 3D). Together these results strongly suggest that the Xrn2 exonuclease torpedo can mediate termination at genes where 3′ ends are made by cleavage mechanisms other than canonical cleavage coupled to polyadenylation.
Figure 3. Mutant Xrn2 inhibits termination at a subset of genes with alternative modes of 3′ end formation.
A–C. Xrn2(D235A) impairs termination at lnc RNA genes with RNAseP- (A, B) and microprocessor-mediated (C) 3′ end cleavage. UCSC genome browser screen shots of pol II ChIP-seq results as in Fig. 1C. Arrows indicate 3′ extension of the termination zone caused by Xrn2 nuclease inactivation. 3′ ends are marked by vertical pale blue lines. These results are reproduced in a biological replicate ChIP-seq experiment shown in Figure S2A–C.
D. Xrn2 D235A delays termination at candidate CoTC containing genes (Nojima et al., 2013). Metaplot of mean pol II ChIP signals normalized to poly(A) site.
E. Xrn2 D235A delays termination at a U4 snRNA gene. (See also Figure S2D).
F. Xrn2 D235A delays termination at U snRNA genes. Metaplot of mean pol II ChIP signals normalized to −500 bp relative to the TSS. Note a modest but detectable average delay in termination caused by the Xrn2 (D235A) mutant.
G. Xrn2 D235A delays termination at a replication-coupled histone H3 gene. (See also Figure S2E).
H. No detectable effect of Xrn2 mutation on termination at the non-canonical miR320A gene whose transcript is not cleaved by microprocessor (Xie et al., 2013).
I. No general effect of Xrn2 mutation on putative termination downstream of canonical miRNA genes that are cleaved by microprocessor. Metaplots of mean pol II ChIP signals normalized to −500 bp relative to the 5′ ends of miRNA genes. Note that pol II density drops downstream of miRNA genes consistent with termination, but the density profile is not noticeably affected by Xrn2 (D235A).
Figure 4. Xrn2 localizes to 3′ ends of genes with non-adenylated transcripts.
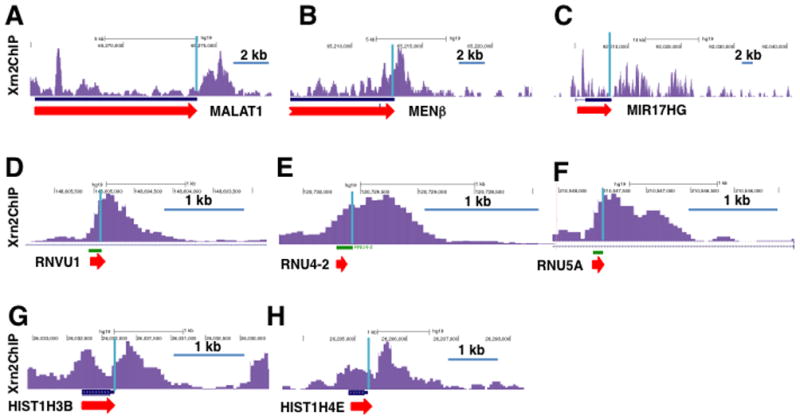
Xrn2 ChIP-seq of doxycycline induced HEK293 cells expressing WT Xrn2 as in Fig. 1. A–C. lnc RNA genes with RNAseP- (A, B) and microprocessor-mediated (C) 3′ end cleavage. D–F. U snRNA genes. G, H. replication coupled histone genes. UCSC genome browser screen shots of pol II ChIP-seq results as in Fig. 1C. Blue lines mark 3′ ends.
Pol II ChIP-seq showed that the Xrn2 (D235A) dominant negative mutant does cause a modest shift in pol II occupancy into the 3′ flanking regions of some histone and U snRNA genes consistent with delayed termination (Fig. 3E, G, Fig. S2D, E). Metaplots of the average pol II profile for several hundred U snRNA genes showed a small but detectable delay in termination in the Xrn2 D235A mutant (Fig. 3F) that is consistent with the presence of Xrn2 on these genes as detected by ChIP (Fig. 4D–F). On the other hand, average pol II density over about 70 well-transcribed histone genes did not suggest a widespread delay in termination when Xrn2 was inhibited (not shown), despite the fact that Xrn2 is recruited to histone genes (Fig. 4G, H). These results are consistent with a redundant termination mechanism(s) operating side-by-side with Xrn2 at histone and U snRNA genes or that these genes are less sensitive to reduced Xrn2 activity.
We mapped pol II density across over 1200 intragenic and intergenic miRNA genes and found a drop in pol II density at their 3′ ends consistent with transcription termination, or possibly an acceleration of pol II elongation rate (Fig. 3I). However, the Xrn2 mutant had little or no overall effect on the putative termination 3′ of miRNA genes (Fig. 3I). Similarly termination at the non-canonical miRNA320A gene that is not a substrate of microprocessor (Xie et al., 2013) is unaffected by Xrn2 nuclease inactivation suggesting that an independent termination mechanism is involved (Fig. 3H). In summary the results in Figures 1–4 indicate that Xrn2 makes different contributions to termination at different classes of pol II transcribed genes. It has a major influence of termination at most human genes with poly(A) sites, and also has a detectable positive effect on termination at some genes with alternative forms of RNA 3′ end formation (Fig. 3) where it may act redundantly with other termination factors (Rondon et al., 2009).
Elongation rate has widespread effects on termination downstream of poly(A) sites
To investigate whether kinetic competition affects the timing of transcription termination downstream of mRNA coding genes, we mapped pol II by ChIP in previously described inducible cell lines expressing α-amanitin resistant mutants of the human large subunit that accelerate or decelerate elongation. The R749H substitution in the funnel domain slows elongation whereas the E1126G trigger loop mutation accelerates transcription. Average in vivo elongation rates in these cell lines were 1.7 kb/min for the WT Amr pol II, 0.5 kb/min for R749H and 1.9 kb/min for E1126G (Fong et al., 2014). Pol II ChIP was conducted in cells treated with doxycycline to induce the WT or mutant Amr large subunits, and then with α-amanitin for 42 hr. to inhibit and degrade endogenous pol II large subunit. Notably, pol II density profiles in the 3′ flanking regions of several protein coding genes (Fig. 5A–F) show that the slow R749H mutant provokes proximal termination and the fast E1126G mutant provokes distal termination relative to WT Amr pol II. The shifting of termination sites in the pol II mutants is substantial, often increasing or decreasing the length of the transcribed 3′ flanking regions by several kilobases (e.g. Figs. 5C–E). Furthermore the effects of slow and fast elongation mutants on where termination occurs are general, as evidenced by average pol II densities in a group of over 9000 well-separated genes (Fig. 5G). The results of this meta-analysis show that the slow R749H mutant shifted the average pol II profile in 3′ flanking regions upstream relative to WT by 1–2kb. The fast E1126G mutant on the other hand caused an overall flattening of the pol II profile in the termination zone that replaced the clear drop-off in pol II density that occurs with WT pol II. This flattened profile is consistent with reduced pausing, delayed termination and consequent broadening of the “termination zone”. We examined the effects of elongation rate at Group I and II genes that terminate at different positions relative to the poly(A) site (Fig. 2A–C) and found that slow and fast mutants shift sites of termination proximally and distally respectively in both groups (Fig. 5H, I).
Figure 5. Pol II elongation rate has widespread effects on poly(A) site dependent termination.
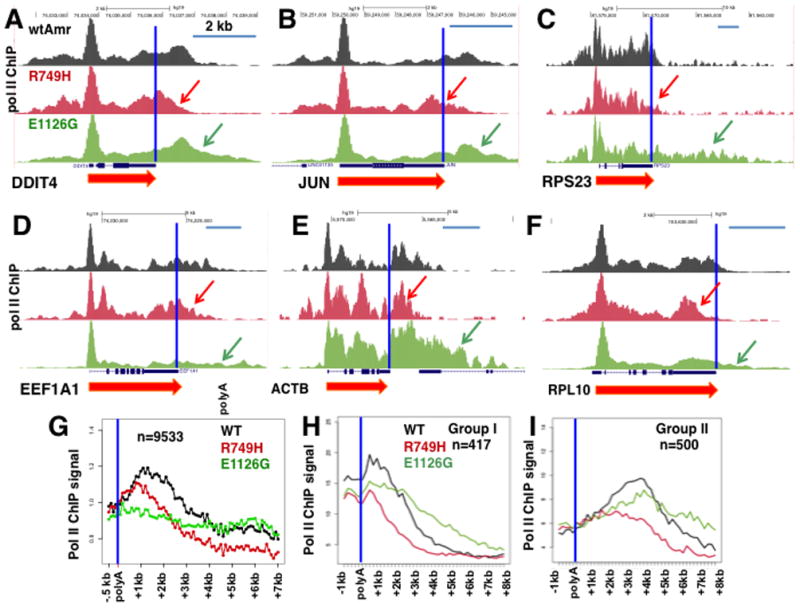
A–F. Anti-Pol II ChIP-seq in HEK 293 Flp-in TREX cells expressing WT, slow (R749H, red) and fast (E1126G, green) α-amanitin resistant (Amr) pol II large subunits. Cells were treated with doxycycline to induce expression of Amr large subunits, then with α-amanitin for 42 hrs to inhibit and degrade endogenous pol II prior to ChIP with anti-pol II antibody. UCSC genome browser screen shots are shown with red and green arrows indicating shortening and extension of the termination zone caused by slow and fast mutants. Poly(A) sites are marked by vertical blue lines. Scale bars represent 2kb. These results are reproduced in biological replicate ChIP-seq experiments shown in Figure S3.
G. Metaplots of mean pol II ChIP signals normalized to the poly(A) site as in Fig. 1I for well-separated genes (>5kb apart) in cells expressing WT (black) slow (R749H, red) and fast (E1126G, green) α-amanitin resistant (Amr) pol II large subunits. Note more proximal termination in the slow mutant relative to WT and delayed termination in the fast mutant and a less prominent peak of pol II density 3′ of the PAS.
H–I. Metaplots of mean pol II ChIP signals for proximal and distal termination groups I and II as in Figure 2. Note effects of elongation rate on both proximal and distal termination.
In summary, the ways in which slow and fast pol II mutants shifted termination at thousands of genes, are most simply accounted for by kinetic competition between elongating RNA polymerase and a “torpedo” pursuing it. These results therefore provide a new independent line of evidence that validates a previously untested prediction of the original “torpedo” model of poly(A) site dependent termination (Connelly and Manley, 1988).
Elongation rate effects on termination at genes with polyA-transcripts
We asked whether transcription elongation rate also affected termination at genes with modes of RNA 3′ end formation other than cleavage/polyadenylation. The results in Figure 6 show that termination at histone and U snRNA genes can be affected by elongation rate in a similar way to genes with poly(A) sites. The fast mutant pol II extended further past the 3′ ends of these genes than the slow mutant, though the distances travelled into 3′ flanking sequences are much less than at most poly (A) site-containing genes, consistent with previous work (Anamika et al., 2012). In summary these results suggest that kinetic competition with transcription elongation is a functional determinant of where termination occurs at least at some genes with non-canonical modes of 3′ end formation. This conclusion is supported by the effects of the dominant negative Xrn2 (Fig. 3) and by the presence of Xrn2 at histone and U snRNA genes as determined by ChIP (Fig. 4).
Figure 6. Pol II elongation rate effects on termination at histone and U snRNA genes.
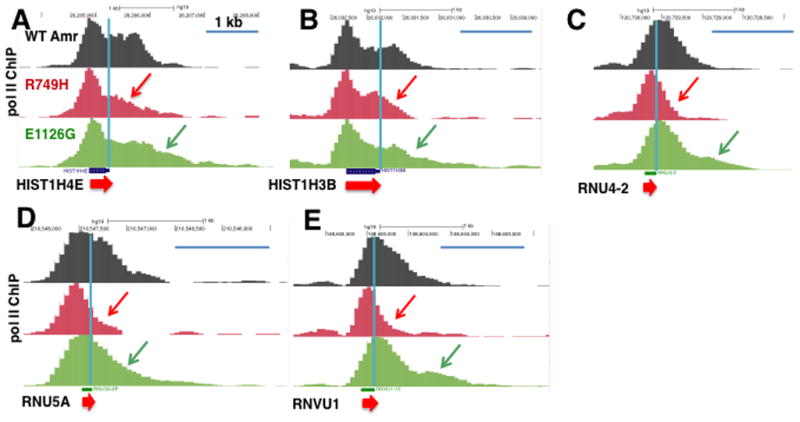
A–E. UCSC genome browser screen shots of pol II ChIP-seq results for selected histone (A, B) and U snRNA genes (C–F) are shown with red and green arrows indicating shortening and extension of the termination zone caused by slow and fast mutants. Vertical blue lines mark RNA 3′ ends. Blue scale bars represent 1kb. These results are reproduced in biological replicate ChIP-seq experiments shown in Figure S4.
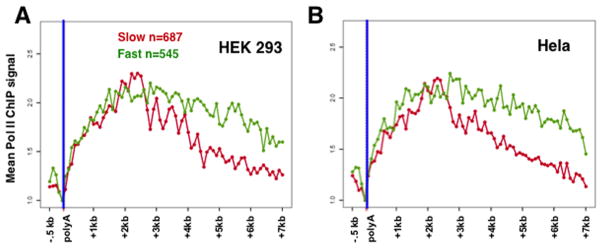
Natural variation in elongation rate correlates with location of the termination zone
The pol II mutants show that changing the elongation rate of pol II mutants can influence where transcription terminates downstream of most genes. This observation begs the question of whether the variation in elongation rate between genes influences where transcription normally terminates. To test this idea we compared termination on genes with fast and slow average elongation rates as determined by in vivo pulse labeling of nascent RNA after release of a DRB transcription block (Fuchs et al., 2014). Remarkably we found that the fastest genes identified in that study (>4kb/min) had markedly different termination profiles than the slowest genes (<3.5kb/min) as determined by pol II ChIP seq in their 3′ flanking regions. The analysis in Figure 7 showed that the genes with slow elongation rates have more proximal termination than those with fast elongation both in Hela and HEK293 cells. These results therefore argue that one important determinant of the length of the termination zone at a particular gene is the natural rate of transcription elongation. Notably the effects of natural variation in elongation rate, and manipulated changes in rate due to mutation of Rpb1, are identical. In both cases fast elongation is associated with delayed termination relative to slow elongation.
Figure 7. Elongation rate correlates with the extent of the termination zone.
A, B. Metaplots of mean pol II ChIP-seq signal normalized to the poly(A) site for genes with fast (>4kb/min) or slow (<3.5 kb/min) elongation rate as defined in Hela cells by Fuchs et al. (Fuchs et al., 2014). Note that slow elongation correlates with more proximal and fast elongation with more distal termination and this is conserved between Hela and HEK293 cells. HEK293 pol II ChIP is from cells expressing WT HA tagged Xrn2 (Fig. 1). Hela pol II ChIPseq data (GSE30895)is from (Kim et al., 2011).
Discussion
In this report we tested two central predictions of the torpedo model for RNA pol II transcription termination (Connelly and Manley, 1988) in human cells: a) that termination requires the nuclear 5′-3′ exonuclease Xrn2 and b) that there is kinetic competition between transcript elongation by pol II and transcript degradation by the exonuclease. In budding yeast the Rat1 homologue of Xrn2 has an established role in termination downstream of poly(A) sites but it has not been clear if this is a general paradigm. In fission yeast the nuclear exosome, a 3′-5′ exonuclease, functions as a terminator and the importance of Rat1 in this process is not known (Lemay et al., 2014). Unexpectedly, knockdown of Xrn2 has little or no effect on termination downstream of poly(A) sites on natural chromosomal genes (Brannan et al., 2012) leading to the suggestion that it may function exclusively in premature termination near 5′ ends (Nojima et al., 2015). We re-examined the role of Xrn2 in termination downstream of genes using a combination of shRNA knockdown and expression of a dominant negative active site mutant, D235A that is predicted to lack exonuclease activity and obstruct access of WT Xrn2 to substrate 5′ PO4 ends (Jinek et al., 2011). Termination was analyzed genome-wide by anti-pol II ChIP-seq which permits mapping of pol II occupancy in the “termination zone” downstream of the 3′ ends of genes before it is released from the template (Fig. 1A). The salient result of these experiments is that Xrn2 is indeed required genome-wide for proper termination downstream of thousands of poly(A) site containing genes (Figs. 1, 2). Termination was not prevented, but was delayed by as much as 5kb or more on some genes when Xrn2 exonuclease activity was inhibited (e.g. Fig. 1E). This observation is in agreement with results in in budding yeast, showing that inactivation of Rat1 nuclease delays, but does not eliminate termination both in vivo (Kim et al., 2004) (B.E. and D.B. unpublished) and in vitro (Park et al., 2015). We conclude that the 5′-3′ RNA exonuclease, Xrn2, is a major effector of termination downstream of poly(A) sites throughout the human genome. Dominant negative Xrn2 also delayed termination at some genes that make poly(A)-transcripts including U snRNAs that are cleaved by the Integrator complex and the long non-coding RNAs MALAT1 and MENβ that are cleaved by RNAseP (Fig. 3). These results suggest that the Xrn2 torpedo can load onto 5′ PO4 ends exposed not only by CPSF mediated cleavage coupled to polyadenylation, but also by other endonucleolytic mechanisms.
An integral feature of the torpedo model is that after the exonuclease engages the cleaved nascent transcript, it must pursue the polymerase and catch up to it before it can promote destabilization of the transcription elongation complex. This scheme implies an obligate kinetic competition between polymerase translocation by transcript elongation and torpedo translocation by 5′-3′ exonucleolytic digestion. We tested for kinetic competition by using fast and slow pol II mutants to determine whether elongation rate affects where termination occurs. These experiments demonstrated that slow elongation causes proximal termination and conversely fast elongation causes distal termination at thousands of genes with poly(A) sites (Fig. 5, Fig. S3). Termination at some histone and U snRNA genes that make non-adenylated transcripts was similarly affected by the pol II mutants (Fig. 6, Fig. S4). Importantly, these effects of transcription rate on termination are most easily accounted for by kinetic competition between elongating pol II and a 5′-3′ translocating torpedo that follows it. Previously kinetic competition between RNA polymerase and 5′-3′ RNA helicases was reported for Rho in E. coli (Jin et al., 1992) and Sen1 at yeast snoRNA genes (Hazelbaker et al., 2013). We can not absolutely eliminate the possibility that the slow and fast pol II mutants affect termination by an unknown mechanism unrelated to their effects on elongation rate. However two lines of evidence argue against this possibility: 1) If the mutants worked independently of elongation rate, then it is not clear why the slow and fast mutants would have opposite effects on termination (Fig. 5); 2) Wild-type pol II terminates later on genes with fast elongation than on genes with slow elongation (Fig. 7).
The size of the termination zone downstream of a poly(A) site varies between genes (Anamika et al., 2012) and is relatively constant between cell types (Fig. 2B, C) suggesting that it is an intrinsic gene-specific property. We found that genes with fast elongation have more distal sites of termination than genes with slow elongation (Fig. 7) consistent with the effects of fast and slow pol II mutants. Natural variation in elongation rate (Danko et al., 2013; Fuchs et al., 2014; Veloso et al., 2014) is therefore a likely determinant of where termination occurs downstream of genes. It remains to be tested whether Group I genes with proximal termination sites (Fig. 2) have slower average elongation rates than Group II genes with distal termination sites. Control of the extent of the zone of termination downstream of genes by elongation rate could be of functional importance in cases where it affects interference with adjacent sense or antisense transcription units, or collisions with replication forks (Alzu et al., 2012). It will be of interest in future to investigate whether modulation of “torpedo” function has a regulatory role in determining where transcription termination occurs.
Our results demonstrate that Xrn2 exonuclease dependent termination operates widely among pol II transcribed genes in human cells, but they do not eliminate the possibility that other modes of termination also at work on these genes. Redundant termination mechanisms might be required for example if there were 3′ flanking sequences that block Xrn2 translocation. Such redundancy is consistent with the fact that inhibition of Xrn2 did not eliminate termination, but only delayed it. Additional candidate terminators include the DNA helicase TTF2, the RNA helicase senataxin and the RNA binding protein p54nrb that interact with Xrn2 (Brannan et al., 2012; Kaneko et al., 2007; Skourti-Stathaki et al., 2011), as well as the nuclear exosome (Lemay et al., 2014) and the pol II CTD-binding protein, Pcf11 (Zhang and Gilmour, 2006). Our results are not consistent with widespread in vivo use of the poly(A) site-cleavage independent termination mechanism reported in vitro (Zhang et al., 2015), because such a mechanism is predicted not to be affected by Xrn2 activity. The effects of fast and slow elongation on termination are consistent with any exonuclease or helicase “torpedo” that translocates 5′ to 3′ along the nascent transcript and ultimately catches up with the polymerase to dislodge it (Connelly and Manley, 1988). However, as originally predicted (Connelly and Manley, 1988), the termination defects caused by an Xrn2 active site mutant at most pol II transcribed genes strongly implicate a 5′-3 RNA exonuclease torpedo mechanism together with kinetic competition that determines where termination occurs.
Experimental Procedures
Plasmids
The human Xrn2 ORF sequence was inserted into the pcDNA5/FRT/TO (Invitrogen) with a 6XHA C-terminal epitope tag. The D235A mutation was introduced by mismatch overlap extension PCR.
Cell lines
Site-specific integration of pcDNA5 Xrn2 expression plasmids into HEK293-Flp-in T-REX cells (Invitrogen) was mediated by Flp recombinase. The cells were maintained in DMEM media supplemented with 10% FBS, 200μg/ml hygromycin B, 6.5μg/ml blasticidin, and pen/strep. Expression was induced with doxycycline (2.0 μg/ml) for 24 hr. before harvesting for ChIP. These cells were infected with lentivirus pLK0.1 expressing shRNA targeting the Xrn2 3′ UTR (TRCN0000293640 target sequence GTGTATTCTAGATCATCTAAG) and were stably selected in puromycin (2.0 μg/ml) as described(Brannan et al., 2012). HEK293 cells expressing WT, slow (R749H) and fast (E1126G) α-amanitin resistant mutants of Rpb1 have been described (Fong et al., 2014). ChIP extracts were prepared from these cells after induction with doxycycline (2.0 μg/ml) for 12–16 hr. and treatment with α-amanitin (2.5 μg/ml) for a further 42hr. 21NT human breast cells (Band et al., 1990) were grown in DMEM, 10mM Hepes, 1mM sodium pyruvate, 1X non-essential amino acids, 2mM L-glutamine, 1% pen/strep, 10% fetal bovine serum, 1ng/ml insulin, 1ug/ml hydrocortisone and 12.5ng/ml human epidermal growth factor. HAP1 myeloid leukemia cells (Carette et al., 2012) were grown in IMDM 10% FBS, 4mM L-glutamine, and 1%pen/strep.
Antibodies
Rabbit anti-pan pol II CTD and anti-Xrn2 were described previously (Brannan et al., 2012; Schroeder et al., 2000). Anti-HA rabbit polyclonal was affinity purified..
ChIP-seq
ChIP has been described (Kim et al., 2011). Enzymatic steps and size fractionation of libraries were done on AMPure XP SPRI beads (Beckman Coulter Genomics) (Fisher et al., 2011) and sequenced on the Illumina Genome Analyzer IIx or Hi-Seq platforms. Reads were mapped to the hg19 UCSC human genome (Feb. 2009) with Bowtie version 0.12.5 (Langmead et al., 2009) (Table S2). We generated bed and wig profiles using 50bp bins and 200bp windows assuming a 180bp fragment size shifting effect. Results were viewed with the UCSC genome browser and meta plots were generated using R. Metaplots include all genes in common between the relevant data sets for which a minimum ChIP signal was obtained in all bins. The heatmap was made using the 543_heatmap function in HMTools https://github.com/actor0/hmtools.
Supplementary Material
Acknowledgments
This work was supported by NIH grant GM063873 to D.B. K.B. and R.S. were supported by NIHT32-GM08730. We thank UC Denver Nextgen sequencing facility, B. Gao, T. Shade and K. Diener for sequencing and T. Blumenthal, R. Davis, J. Hesselberth, and members of our lab for helpful discussions and comments on the manuscript.
Footnotes
Accession Numbers ChIP-seq data sets are deposited at GEO accession GSE72800.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alzu A, Bermejo R, Begnis M, Lucca C, Piccini D, Carotenuto W, Saponaro M, Brambati A, Cocito A, Foiani M, et al. Senataxin Associates with Replication Forks to Protect Fork Integrity across RNA-Polymerase-II-Transcribed Genes. Cell. 2012;151:835–846. doi: 10.1016/j.cell.2012.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anamika K, Gyenis À, Poidevin L, Poch O, Tora L. RNA polymerase II pausing downstream of core histone genes is different from genes producing polyadenylated transcripts. PLoS ONE. 2012;7:e38769. doi: 10.1371/journal.pone.0038769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anamika K, Gyenis À, Tora L. How to stop. Transcription. 2014;4:7–12. doi: 10.4161/trns.22300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Ballarino M, Pagano F, Girardi E, Morlando M, Cacchiarelli D, Marchioni M, Proudfoot NJ, Bozzoni I. Coupled RNA processing and transcription of intergenic primary microRNAs. Mol Cell Biol. 2009;29:5632–5638. doi: 10.1128/MCB.00664-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band V, Zajchowski D, Swisshelm K, Trask D, Kulesa V, Cohen C, Connolly J, Sager R. Tumor progression in four mammary epithelial cell lines derived from the same patient. Cancer Res. 1990;50:7351–7357. [PubMed] [Google Scholar]
- Banerjee A, Sammarco MC, Ditch S, Wang J, Grabczyk E. A novel tandem reporter quantifies RNA polymerase II termination in mammalian cells. PLoS ONE. 2009;4:e6193. doi: 10.1371/journal.pone.0006193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse CE, Minvielle-Sebastia L, Lee BA, Keller W, Proudfoot NJ. Coupling termination of transcription to messenger RNA maturation in yeast. Science. 1998;280:298–301. doi: 10.1126/science.280.5361.298. [DOI] [PubMed] [Google Scholar]
- Brannan K, Kim H, Erickson B, Glover-Cutter K, Kim S, Fong N, Kiemele L, Hansen K, Davis R, Lykke-Andersen J, et al. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol Cell. 2012;46:311–324. doi: 10.1016/j.molcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2012;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly S, Manley JL. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 1988;2:440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- Danko CG, Hah N, Luo X, Martins AL, Core L, Lis JT, Siepel A, Kraus WL. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol Cell. 2013;50:212–222. doi: 10.1016/j.molcel.2013.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhir A, Dhir S, Proudfoot NJ, Jopling CL. Microprocessor mediates transcriptional termination of long noncoding RNA transcripts hosting microRNAs. Nat Struct Mol Biol. 2015;22:319–327. doi: 10.1038/nsmb.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z. Nucleases of the metallo-beta-lactamase family and their role in DNA and RNA metabolism. Crit Rev Biochem Mol Biol. 2007;42:67–93. doi: 10.1080/10409230701279118. [DOI] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Marzluff WF. The Polyadenylation Factor CPSF-73 Is Involved in Histone-Pre-mRNA Processing. Cell. 2005;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, Young G, Fennell TJ, Allen A, Ambrogio L, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 2011;12:R1. doi: 10.1186/gb-2011-12-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Kim H, Zhou Y, Ji X, Qiu J, Saldi T, Diener K, Jones K, Fu XD, Bentley DL. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev. 2014;28:2663–2676. doi: 10.1101/gad.252106.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MJ, Gao H, Smith-Kinnaman WR, Liu Y, Mosley AL. The Exosome Component Rrp6 Is Required for RNA Polymerase II Termination at Specific Targets of the Nrd1-Nab3 Pathway. PLoS Genetics. 2015;10:e1004999. doi: 10.1371/journal.pgen.1004999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Voichek Y, Benjamin S, Gilad S, Amit I, Oren M. 4sUDRB-seq: measuring genomewide transcriptional elongation rates and initiation frequencies within cells. Genome Biol. 2014;15:R69. doi: 10.1186/gb-2014-15-5-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromak N, West S, Proudfoot NJ. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol Cell Biol. 2006;26:3986–3996. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosso AR, de Almeida SF, Braga J, Carmo-Fonseca M. Dynamic transitions in RNA polymerase II density profiles during transcription termination. Genome Res. 2012;22:1447–1456. doi: 10.1101/gr.138057.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbaker DZ, Marquardt S, Wlotzka W, Buratowski S. Kinetic competition between RNA Polymerase II and Sen1-dependent transcription termination. Mol Cell. 2013;49:55–66. doi: 10.1016/j.molcel.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin DJ, Burgess RR, Richardson JP, Gross CA. Termination efficiency at rho-dependent terminators depends on kinetic coupling between RNA polymerase and rho. Proc Natl Acad Sci U S A. 1992;89:1453–1457. doi: 10.1073/pnas.89.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Coyle SM, Doudna JA. Coupled 5′ nucleotide recognition and processivity in Xrn1-mediated mRNA decay. Mol Cell. 2011;41:600–608. doi: 10.1016/j.molcel.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev. 2007;21:1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Kim S, Kim H, Fong N, Erickson B, Bentley DL. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc Natl Acad Sci U S A. 2011;108:13564–13569. doi: 10.1073/pnas.1109475108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Steitz JA. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehner JN, Pearson EL, Moore C. Unravelling the means to an end: RNA polymerase II transcription termination. Nature Rev Mol Cell Biol. 2011;12:283–294. doi: 10.1038/nrm3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011;332:475–478. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemay JF, Larochelle M, Marguerat S, Atkinson S, Bahler J, Bachand F. The RNA exosome promotes transcription termination of backtracked RNA polymerase II. Nat Struct Mol Biol. 2014;21:919–926. doi: 10.1038/nsmb.2893. [DOI] [PubMed] [Google Scholar]
- Lian Z, Karpikov A, Lian J, Mahajan MC, Hartman S, Gerstein M, Snyder M, Weissman SM. A genomic analysis of RNA polymerase II modification and chromatin architecture related to 3′ end RNA polyadenylation. Genome Res. 2008;18:1224–1237. doi: 10.1101/gr.075804.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan J, Falck PE, Darnell JEJ, Shenk T. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse beta maj-globin gene. Proc Natl Acad Sci U S A. 1987;84:8306–8310. doi: 10.1073/pnas.84.23.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Johnson AW, Bentley DL. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: implications for a unified allosteric-torpedo model. Genes Dev. 2006;20:954–965. doi: 10.1101/gad.1409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Dienstbier M, Murphy S, Proudfoot NJ, Dye MJ. Definition of RNA polymerase II CoTC terminator elements in the human genome. Cell Rep. 2013;3:1080–1092. doi: 10.1016/j.celrep.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Gomes T, Grosso ARF, Kimura H, Dye MJ, Dhir S, Carmo-Fonseca M, Proudfoot NJ. Mammalian NET-Seq Reveals Genome-wide Nascent Transcription Coupled to RNA Processing. Cell. 2015;161:526–540. doi: 10.1016/j.cell.2015.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly D, Kuznetsova OV, Laitem C, Zaborowska J, Dienstbier M, Murphy S. Human snRNA genes use polyadenylation factors to promote efficient transcription termination. Nucleic Acids Res. 2014;42:264–275. doi: 10.1093/nar/gkt892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Kang M, Kim M. Unraveling the mechanistic features of RNA polymerase II termination by the 5'-3' exoribonuclease Rat1. Nucleic Acids Res. 2015;43:2625–2637. doi: 10.1093/nar/gkv133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson EL, Moore CL. Dismantling promoter-driven RNA polymerase II transcription complexes in vitro by the termination factor Rat1. J Biol Chem. 2013;288:19750–19759. doi: 10.1074/jbc.M112.434985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrua O, Libri D. A bacterial-like mechanism for transcription termination by the Sen1p helicase in budding yeast. Nat Struct Mol Biol. 2013;20:884–891. doi: 10.1038/nsmb.2592. [DOI] [PubMed] [Google Scholar]
- Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondon AG, Mischo HE, Kawauchi J, Proudfoot NJ. Fail-safe transcriptional termination for protein-coding genes in S. cerevisiae. Mol Cell. 2009;36:88–98. doi: 10.1016/j.molcel.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaughency P, Merran J, Corden JL. Genome-Wide Mapping of Yeast RNA Polymerase II Termination. PLoS Genetics. 2014;10:e1004632. doi: 10.1371/journal.pgen.1004632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder SC, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar JR, Ferris AL, Wu X, Saraf A, Khanna KK, Florens L, Washburn MP, Hughes SH, Pagano M. The Integrator complex controls the termination of transcription at diverse classes of gene targets. Cell Res. 2015;25:288–305. doi: 10.1038/cr.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skourti-Stathaki K, Proudfoot Nicholas J, Gromak N. Human Senataxin Resolves RNA/DNA Hybrids Formed at Transcriptional Pause Sites to Promote Xrn2-Dependent Termination. Mol Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-Wide Distribution of Yeast RNA Polymerase II and Its Control by Sen1 Helicase. Mol Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Veloso A, Kirkconnell KS, Magnuson B, Biewen B, Paulsen MT, Wilson TE, Ljungman M. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res. 2014;24:896–905. doi: 10.1101/gr.171405.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilborg A, Passarelli MC, Yario TA, Tycowski KT, Steitz JA. Widespread Inducible Transcription Downstream of Human Genes. Mol Cell. 2015;59:449–461. doi: 10.1016/j.molcel.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagschal A, Rousset E, Basavarajaiah P, Contreras X, Harwig A, Laurent-Chabalier S, Nakamura M, Chen X, Zhang K, Meziane O, et al. Microprocessor, Setx, Xrn2, and Rrp6 Co-operate to Induce Premature Termination of Transcription by RNAPII. Cell. 2012;150:1147–1157. doi: 10.1016/j.cell.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AM. E Pluribus Unum: 3′ end formation of polyadenylated mRNAs, histone mRNAs, and U snRNAs. Mol Cell. 2005;20:168–170. doi: 10.1016/j.molcel.2005.10.009. [DOI] [PubMed] [Google Scholar]
- West S, Gromak N, Proudfoot NJ. Human 5′ --> 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- West S, Proudfoot NJ, Dye MJ. Molecular dissection of mammalian RNA polymerase II transcriptional termination. Mol Cell. 2008;29:600–610. doi: 10.1016/j.molcel.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw E, Proudfoot N. Alpha-thalassaemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3′ end processing in the human alpha 2 globin gene. EMBO J. 1986;5:2915–2922. doi: 10.1002/j.1460-2075.1986.tb04587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Spector DL. An unexpected ending: noncanonical 3′ end processing mechanisms. RNA. 2010;16:259–266. doi: 10.1261/rna.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Li M, Vilborg A, Lee N, Shu MD, Yartseva V, Sestan N, Steitz JA. Mammalian 5′-capped microRNA precursors that generate a single microRNA. Cell. 2013;155:1568–1580. doi: 10.1016/j.cell.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Rigo F, Martinson HG. Poly(A) Signal-Dependent Transcription Termination Occurs through a Conformational Change Mechanism that Does Not Require Cleavage at the Poly(A) Site. Mol Cell. 2015;59:437–448. doi: 10.1016/j.molcel.2015.06.008. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Gilmour DS. Pcf11 is a termination factor in Drosophila that dismantles the elongation complex by bridging the CTD of RNA polymerase II to the nascent transcript. Mol Cell. 2006;21:65–74. doi: 10.1016/j.molcel.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wu CH, Gilmour DS. Analysis of polymerase II elongation complexes by native gel electrophoresis. Evidence for a novel carboxyl-terminal domain-mediated termination mechanism. J Biol Chem. 2004;279:23223–23228. doi: 10.1074/jbc.M402956200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.