In a recent report, Roehle et al (2008) described the global microRNA (miRNA) expression signature of B-cell lymphomas. In that study, the authors were also able to investigate the impact of miRNAs expression on the survival of diffuse large B-cell lymphomas (DLBCL). However, although several miRNAs segregated with outcome in their series, expression of the oncogenic MIRN155 played no role on the overall or event-free survival of DLBCL patients.
MiRNAs are abundant non-protein coding RNAs that attenuate gene expression by pairing to the 3′UTR of target transcripts. MiRNAs are critical to a variety of physiologic processes and their dysfunction, at a structural or regulatory level, is frequently found in cancer. MIRN155 is transcribed from the BIC locus, originally cloned at a viral integration site in an avian model of B-cell lymphoma. The development of high-grade lymphoma in Eμ-MIRN155 transgenic mice (Costinean et al, 2006) and the identification of a MIRN155 orthologue in the Kaposi sarcoma-associated herpesvirus (KSHV)(Gottwein et al, 2007), which is linked to B-cell tumours, further reinforced the putative lymphomagenic nature of this miRNA. Recently, we and others reported that MIRN155 is overexpressed in the aggressive activated B-cell-like (ABC) subtype of DLBCL (Rai et al, 2008). In mechanistic support to these findings, the nuclear factor (NF)-κB and Janus kinase JNK pathways, which are constitutively active in ABC-DLBCL, have been linked to the transcriptional control of MIRN155 (Rai et al, 2008; Yin et al, 2008). Therefore, considering the known association of ABC-DLBCL with poor outcome, it was of interest to validate the findings of Roehle et al (2008) in a larger collection of DLBCL and test if MIRN155 expression per se influences survival in this disease. From a biological standpoint, the characterization of this putative association is relevant because it may offer insight in the mechanism of transformation associated with MIRN155 overexpression and, because every miRNA is predicted to regulate the expression of hundreds of genes, it may present multiple opportunities to the development of targeted therapeutic strategies. Also, as the expression of a single or small group of miRNAs has been show to be of prognostic relevance in cancer, including the overexpression of MIRN155 in lung adenocarcinomas (Yanaihara et al, 2006), these data could improve prognostic models for DLBCL.
Herein, we analysed the impact of MIRN155 on the outcome of a large cohort of 129 DLBCLs; the expression of MIRN155 and its association with ABC-DLBCL in this tumour collection was recently reported (Rai et al, 2008). Kaplan–Meier estimates of survival were generated according to MIRN155 expression levels (high versus low dichotomized at the median) and survival distributions were compared using the log-rank test; the Z-score was used to test the significance of difference of Kaplan–Meier survival probability estimates at 60 months. Association between outcome (cured or fatal/refractory) and MIRN155 expression was defined with Fisher’s exact test.
In agreement with the data of Roehle et al (2008), we found that MIRN155 expression did not correlate with DLBCL outcome (cured versus fatal/refractory, P > 0.4 Fisher’s exact test) or overall survival distributions (P > 0.6, Log-Rank test, Fig 1, top panel). However, as we had demonstrated that MIRN155 expression was significantly higher in ABC-DLBCL (Rai et al, 2008) we decided to evaluate the impact of MIRN155 expression exclusively within the ABC molecular subgroup (n = 24). Surprisingly, we found a marked trend towards better survival rates for those patients expressing higher levels of this miRNA (Fig 1, bottom panel), with a survival probability at 60 months of 15% and 53% for patients with low and high MIRN155 levels, respectively (P = 0.04, Z-score test). Importantly, the distribution of International Prognostic Index (IPI) scores in these two groups was nearly identical (P > 0.5, Fisher’s exact test) indicating that intrinsic differences in the patient population did not account for these findings.
Fig 1.
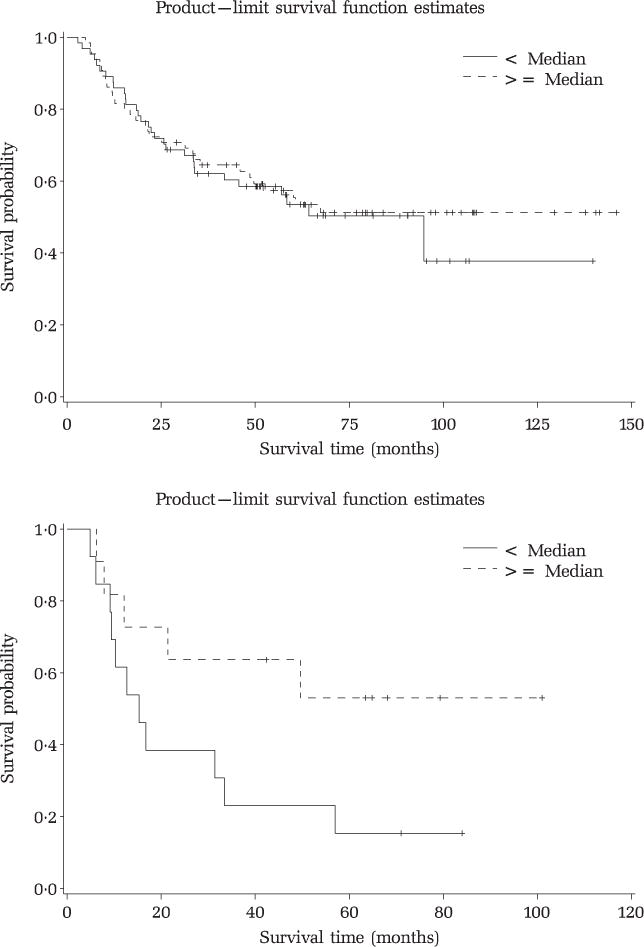
Kaplan–Meier survival curves according to MIRN155 expression for the entire DLBCL population (top) or ABC tumours only (bottom).
The reasons for the association between MIRN155 overexpression and a better outcome in ABC-DLBCL are not immediately clear. However, it should be expected that the concerted action of genes targeted by this miRNA contribute to this finding. Indeed, within the limited number of bona-fide MIRN155 targets described thus far, some, including IKBKE, PIK3CA and AICDA, may be related to these effects (Desiderio, 2008; Gottwein et al, 2007). The first two are positive regulators of major oncogenic pathways, implying that their downregulation in MIRN155 overexpressing ABC-DLBCL could play a role in the less aggressive behaviour of these tumours. The latter, AICDA, a key component of the somatic hypermutation and class switch recombination processes, is known to induce lymphomagenic chromosomal translocations and to be required for germinal centre – derived tumorigenesis (Pasqualucci et al, 2008). Thus, although AICDA is primarily linked to B-cell tumour initiation and does not associate with discrete DLBCL subtypes (Pasqualucci et al, 2004), it may also play a role in tumour progression suggesting that its downregulation by MIRN155 could favourably impact on DLBCL outcome. Finally, it should be noted that heterogeneity within the ABC-DLBCL molecular subtype exists and genetic markers, such as loss of chromosome 9p21, clearly define those with a particularly ominous outcome (Tagawa et al, 2005).
In summary, our data validate and expand on the information provided by Roehle et al (2008) by demonstrating that MIRN155 expression does not globally influence the outcome in DLBCL but, rather, it may define a unique sub-group of ABC-DLBCL with a less aggressive behaviour. Evidently, these findings need to be reconciled with the described oncogenic properties of MIRN155 but they imply that additional studies, including the characterisation of MIRN155 expression in a large number of ABC-DLBCL, are needed to clarify the underlying basis for this association and to firmly establish the role of this miRNA in lymphomagenesis.
Acknowledgments
Supported by a grant from the Elsa U. Pardee Foundation.
References
- Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiderio S. Along came a spider: AID escapes a microRNA web. Immunity. 2008;28:596–598. doi: 10.1016/j.immuni.2008.04.012. [DOI] [PubMed] [Google Scholar]
- Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, Cullen BR. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Guglielmino R, Houldsworth J, Mohr J, Aoufouchi S, Polakiewicz R, Chaganti RS, Dalla-Favera R. Expression of the AID protein in normal and neoplastic B cells. Blood. 2004;104:3318–3325. doi: 10.1182/blood-2004-04-1558. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Bhagat G, Jankovic M, Compagno M, Smith P, Muramatsu M, Honjo T, Morse HC, III, Nussenzweig MC, Dalla-Favera R. AID is required for germinal center-derived lymphomagenesis. Nature Genetics. 2008;40:108–112. doi: 10.1038/ng.2007.35. [DOI] [PubMed] [Google Scholar]
- Rai D, Karanti S, Jung I, Dahia PL, Aguiar RC. Coordinated expression of microRNA-155 and predicted target genes in diffuse large B-cell lymphoma. Cancer Genetics and Cytogenetics. 2008;181:8–15. doi: 10.1016/j.cancergencyto.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roehle A, Hoefig KP, Repsilber D, Thorns C, Ziepert M, Wesche KO, Thiere M, Loeffler M, Klapper W, Pfreundschuh M, Matolcsy A, Bernd HW, Reiniger L, Merz H, Feller AC. MicroRNA signatures characterize diffuse large B-cell lymphomas and follicular lymphomas. British Journal of Haematology. 2008;142:732–744. doi: 10.1111/j.1365-2141.2008.07237.x. [DOI] [PubMed] [Google Scholar]
- Tagawa H, Suguro M, Tsuzuki S, Matsuo K, Karnan S, Ohshima K, Okamoto M, Morishima Y, Nakamura S, Seto M. Comparison of genome profiles for identification of distinct subgroups of diffuse large B-cell lymphoma. Blood. 2005;106:1770–1777. doi: 10.1182/blood-2005-02-0542. [DOI] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique micro-RNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Yin Q, Wang X, McBride J, Fewell C, Flemington E. B-cell receptor activation induces BIC/miR-155 expression through a conserved AP-1 element. Journal of Biological Chemistry. 2008;283:2654–2662. doi: 10.1074/jbc.M708218200. [DOI] [PMC free article] [PubMed] [Google Scholar]


