Abstract
Organoids have tremendous therapeutic potential. They were recently defined as a collection of organ-specific cell types, which self-organize through cell-sorting, develop from stem cells, and perform an organ specific function. The ability to study organoid development and growth in culture and manipulate their genetic makeup makes them particularly suitable for studying development, disease, and drug efficacy. Organoids show great promise in personalized medicine. From a single patient biopsy, investigators can make hundreds of organoids with the genetic landscape of the patient of origin. This genetic similarity makes organoids an ideal system in which to test drug efficacy. While many investigators assume human organoids are the ultimate model system, we believe that the generation of epithelial organoids of comparative model organisms has great potential. Many key transport discoveries were made using marine organisms. In this paper, we describe how deriving organoids from the spiny dogfish shark, zebrafish, and killifish can contribute to the fields of comparative biology and disease modeling with future prospects for personalized medicine.
Keywords: organoids, cystic fibrosis, Squalus acanthias, Danio rerio, Fundulus heteroclitus
Introduction
Organoids are a new research tool with remarkable basic and translational potential [1-6]. Once loosely described as “that which resembles an organ,” organoids recently have been defined as a collection of organ-specific cells types developing from stem cells or induced pluripotent stem cells. These cells self organize to resemble the inner architecture of the organ from which they were derived [1]. Most of our present knowledge about organoid generation and function stems from epithelium focused research. Cells derived from intestines will assemble to form crypt-villus structures with central lumens [7], while cells from the optic cup assemble into organoids with the unique structure of the optic cup [8]. The cells have the engrained memory to reassemble the structures from which they were derived. To be defined as an organoid, the arranged cells also must perform a function specific to their corresponding organ. Intestinal organoids absorb and secrete fluids [9], and liver organoids produce and secrete albumin [10]. These properties make organoids excellent model systems for understanding the development of organs and their diseases. Organoids also can be used therapeutically to test drug efficacy [11] and toxicity [12,13] and have potential for human organ replacement [1,11-14] (Figure 1).
Figure 1.
Key events in the history of various organoid methodologies. From: Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345(6194):1247125. Katharine Sutliff/Science. Reprinted with permission from Science.
Cell biology, toxicity testing, and cancer research all rely on cell culture models [15,16]. Polarized cells, cultured as flat sheets (2-D) on filters, were used to define key transport properties of epithelia through techniques that include short circuit measurements using the Ussing chamber, which allows drugs and agents to be applied from both basolateral (serosal) and apical (mucosal) sides and patch clamping of these polarized cells to determine the properties of ion channels. Cell culture has drawbacks, however [17]. Normal cell lines, such as human mammary epithelial lines (HMEC), cannot be passed infinitely and vary genetically due to telomere attrition senescence [18]. Similarly, hepatocytes in culture have not shown long-term expansion [19,20].
When permanent cell lines are established from kidney epithelia (MDCK cells, LLC-PK1 cells, HEK cells, A6 cells) they have abnormal numbers of chromosomes and physiologic characteristics not present in the tissue of origin. One solution to this problem is using differentiated human embryonic stem (hES) cells and human induced pluripotent stem (hiPS) cells; however, hES and hiPS are genetic and epigenetically unstable [21-24].
Organoids made from epithelial cells hold numerous advantages compared to classic cell culture. A single mouse lung or a biopsy from a patient provides an inexhaustible source of epithelia cells when cultured three dimensionally as organoids. Organoids stemming from a single tissue have been passed over 55 times with genetic stability, making them useful for high throughput studies and personalized medicine, as researchers can conduct tests on organoids in which the genome reflects that of the patient [9,25,26].
The epigenetic and genetic stability of organoids can be attributed to their growth in 3-D culture and their stem cell-like properties [25]. The media in which organoids are cultured imitates the environment auxiliary cells, such as Paneth cells, created for stem cells intestinal crypts [4,7,9]. A cocktail of tissue-specific growth factors is needed to culture organoids of different organs (Table 1). In the presence of growth factors, hepatic organoids can be cultured for over 12 months with weekly passaging. Without these factors, organoids deteriorate after one week [27]. The genetic stability during continuous passaging as well as the closer resemblance to an in vivo environment that organoids provide has attracted many research groups to develop organoid culture protocols. Organoids have been cultured from the thyroid [28], lung [29], pancreas [2,30], liver [27], stomach [31,32], tongue [33], intestine [4,7,9,34], heart [35-37], retina [38], prostate [39], kidney [40-42], and brain [43] of humans.
Table 1. Growth Factors and Culture Medium for Human Organoids.
| Hepatic Organoids [25] | Intestinal Organoids [9] | Brain Organoids [43] |
| DMEM/F12 AdDMEM/F12 (Invitrogen) | DMEM/F12 | low concentration basic fibroblast growth factor (4 ng ml−1) |
| 1% N2 and 1% B27 (GIBCO) | penicillin and streptomycin | 50 μM Rho-associated protein kinase (ROCK) inhibitor49 (Calbiochem) |
| 1.25 mM N-Acetylcysteine (Sigma) | 10 mM HEPES | 100 N2 supplement (Invitrogen) |
| 10 nM gastrin (Sigma) | Glutamax N2, B27 (all from Invitrogen) | Glutamax (Invitrogen) |
| 50 ng/ml EGF (Peprotech) | 1 μM N-acetylcysteine (Sigma) | minimum essential media-nonessential amino acids (MEM-NEAA) |
| 10% RSPO1 conditioned media (homemade) | 50 ng ml−1 mouse epidermal growth factor (mEGF) | 1 μg ml−1 heparin50 (Sigma) |
| 100 ng/ml FGF10 (Peprotech) | 50% Wnt3a-conditioned medium (WCM) | 1:1 mixture of DMEM/F12 |
| 25 ng/ml HGF (Peprotech) | 10% noggin-conditioned medium (NCM) | 1:200 N2 supplement (Invitrogen) |
| 10 mM Nicotinamide (Sigma) | 20% Rspo1-conditioned medium | 1:100 B27 supplement without vitamin A (Invitrogen) |
| 5 uM A83.01 (Tocris) | 10 μM nicotinamide (Sigma) | 3.5 μl l−1 2-mercaptoethanol |
| 10 uM FSK (Tocris) | 10 nM gastrin (Sigma) | 1:4,000 insulin (Sigma) |
| Noggin (Peprotech) | 500 nM A83-01 (Tocris) | 1:100 Glutamax (Invitrogen) |
| 30% Wnt CM (homemade) | 10 μM SB202190 (Sigma) | 1:200 MEM-NEAA |
| 10 uM (Y27632, Sigma Aldrich) | Matrigel | B27 supplement with vitamin A (Invitrogen) |
| Matrigel | Matrigel |
Organoids generated from all of these organs have the potential to help investigators and physicians customize treatments. In the Netherlands, researchers developed large bio-banks of intestinal organoids from the rectal biopsies of cystic fibrosis (CF) patients, replicating the exact genetic defect causing the patient’s CF [9]. CF results from a lack of Cl- and HCO3- transport through an adenosine 3′, 5′-cyclic monophosphate (cAMP)-Protein kinase A (PKA) regulated Cl− channel, named CFTR, expressed in the apical membrane of many epithelial tissues [44]. Organoids made from wild-type patients without CF express CFTR, which secrete anions and fluid into the organoid’s lumen, causing swelling. To enhance this CFTR-dependent swelling, organoids can be stimulated with the adenylyl cyclase activator, forskolin. Organoids with mutated CFTR show much less swelling, and organoids from CFTR knockouts maintain their initial size [4,7,9,45]. Response to drugs among patients with cystic fibrosis varies, even among groups with similar genetic mutations [46]. Applying the forskolin-induced swelling (FIS) assay on CF rectal organoids from biopsies that have been exposed ex vivo to various drugs will allow researchers to identify effective combinations of CFTR correctors for each patient [45]. Colonic organoids also have been used to pre-select optimal drug cocktails to treat patients with colorectal cancer [47]. Organoids generated from humans allow researchers to personalize treatment plans for their patients. While this customizability appears extremely attractive, these organoids are only as effective as the drugs that have already been created. Diseases such as CF do not have a wide variety of therapeutic options or underlying mechanisms that are completely understood.
The Danish physiologist August Krogh, winner of the Nobel Prize in Physiology in 1920, called the scientific communities’ attention to the benefits of model organisms. His principle theorem states, “For a large number of problems there will be some animal of choice, or a few such animals, on which it can be most conveniently studied” [48]. This concept has been central to disciplines such as comparative physiology and functional genomics. While many assume that human organoids are the ultimate model system, particularly for the purpose of modeling epithelial systems in order to understand and treat diseases such as CF, organoids of comparative model organisms have great potential. Many key transport discoveries came from studying marine organisms as model systems. In 1929, E.K. Marshall used the aglomerular goosefish and toadfish to demonstrate tubular epithelial secretion [49]. Homer Smith used the kidney of the aglomerular goosefish to further knowledge of renal physiology [50]. The chemical nature of the glomerular filtrate was first discovered in studies of the frog by A.N. Richards through renal micropuncture experiments [51].
Marine model organisms continue to be of great interest for modeling development and disease. The shark rectal gland (SRG) of the spiny dogfish shark (Squalus acanthias) [52,53], the intestine of the zebrafish (Danio rerio) [54,55], and the operculum of killifish (Fundulus heteroclitus) are currently used in functional studies of the cystic fibrosis transmembrane conductance regulator (CFTR) chloride channel, which is defective in the human genetic chloride channel disease cystic fibrosis (CF). While important findings have come from studies using these model organisms, we propose that future research could be expedited and more finely controlled if done with organoids isolated from comparative model species.
Organoids allow for study of auto-regulation of the epithelium without complex effects of other homeostatic regulators in the body. This enables separation of extrinsic versus intrinsic factors on organ function and pathology in a realistic model. Beyond chicks and mice, limited research has been done on developing organoids from comparative model organisms. To our knowledge, organoids have not yet been generated from marine species. In this review, we outline the advantages of deriving organoids from the spiny dogfish shark rectal gland, zebrafish intestine, and killifish operculum. Organoids from these three model organisms have the potential to be used in functional studies exploiting FIS assay.
SRG of Spiny Dogfish Shark
The shark rectal gland has been used for decades as a model organism for epithelial transport, particularly of chloride [56,57]. SRG has the highest concentration of sodium: potassium: 2 chloride co-transporter (NKCC1), Na-K-ATPase, and CFTR recorded in the literature. These properties have led to many significant discoveries, many of which were made at Mount Desert Island Biological Laboratory [56,58-64]. The lab’s location on the coast makes research on spiny dogfish shark possible during summer months. In order to bring properties of the SRG rectal gland to labs across the world, multiple attempts were made to establish cell lines isolated from the SRG. Despite continuous efforts, a permanent cell line has not yet been established [65], leading to our attempt to expand ex vivo non-transformed SRG epithelial cells by generating organoids and to use them in the FIS assay.
Our attempt to generate these organoids from native rectal gland of the spiny dogfish shark was based on the protocol reported by investigators at the University of Utrecht and University of Rotterdam for establishing organoids from murine and human intestine and lungs [9,66]. Temperature, growth factors, salt concentration, and osmolality were adapted to mimic the cellular environment of the more primitive dogfish shark. Many of these adaptations were done blindly as much less is known about the location and function of stem cells of the SRG than that of the human or murine intestine or lung (Figure 2).
Figure 2.
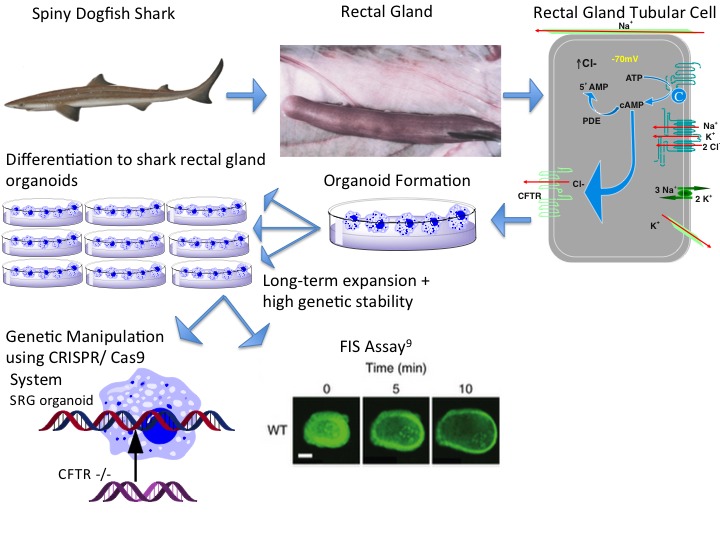
Procedure and Uses of Organoids from Comparative Model Organisms. Sacrifice of a dogfish shark provides a rectal gland. The gland is minced into SRG tubular cells, which are suspended in Matrigel. Organoids of the SRG are generated and passed continuously with high genetic stability. These organoids can be studied using the FIS assay. The CRISPR/Cas9 system can be used to put CFTR-/- or F508del-CFTR 508 into the organoids to examine if the SRG organoids will develop CF or if other amino acids in the shark channel could provide the function of the missing amino acids.
While the development of the SRG organoid is still in its nascent stages, through our attempts, we have realized that the benefit of the organoids, derived from a model organism such as the dogfish shark, extends far beyond becoming independent of a fresh source of the species. They will allow for comparative physiology between different species and genetic manipulation of the donor organisms, creating new disease models for CF and other ion transport diseases.
Intestine of the Zebrafish
Unlike the spiny dogfish shark, the zebrafish has a genome that has been sequenced, making it easier to identify the growth factors necessary to generate organoids [67]. Further, the zebrafish intestine is analogous to the human intestine with segmentation of the small and large intestine. The zebrafish recently has been proposed as a model for inflammatory bowel diseases (IBDs), as it resembles the human intestine in key ways. It displays innate and adaptive immunity, an epithelial barrier, and microbiota [54]. Murine and human intestinal organoids have already been identified as a promising system for IBD research [34]. One important advantage of extending such studies to the zebrafish relies on its potential for in vivo genetic manipulation. Gene expression of zebrafish, unlike that of previously used mouse models, can be easily altered using morpholino antisense technology [68]. In this way, zebrafish could be genetically engineered to down-regulate components of the immune system (IBD-relevant) or to create a CF fish model (using CFTR-directed morpholinos). Alternatively, organoids from wild-type zebrafish could be used to create CFTR mutant organoids or other CF-relevant models using the CRISPR-Cas9 technology for genetic engineering [7]. We hypothesize that organoids derived from those zebrafish would show impaired swelling in the previously described FIS assay. Bagnat et al. demonstrated that zebrafish are a forward genetics model to study CFTR biology, both translatable to mammalian models and useful for the identification of novel regulators of CFTR [69]. We can reasonably argue that organoids isolated from zebrafish would show these traits and provide a more detailed understanding of the CFTR mechanism of action.
Operculum of the Killifish
Another organ of a small marine organism that, if successfully reproduced in organoid cultures, would likely show significant swelling in the FIS assay is the operculum of the killifish. The killifish tolerates fresh and salt water by regulating CFTR chloride secretion by its gill and operculum. Gill epithelial cells create a Na+ gradient across the basolateral membrane using the Na+-K+ -ATPase. This gradient allows cells to take up chloride across the basolateral membrane through the N-K-C-C1 co-transporter. The Cl- is secreted across the apical membrane by the CFTR channel. Chloride secretion results in apical negative transepithelial voltage allowing Na+ secretion by the paracellular pathway. Salinity increases result in a two-pronged response in killifish. The acute response is movement of CFTR from an intracellular pool to the plasma membrane of the operculum [70]. The long-range response is to up regulate expression of the CFTR gene.
These responses combine to allow the killifish to survive in environments of up to 360 percent salinity [71]. We hypothesize that the FIS assay on organoids generated from the killifish operculum at different salinities would show different amounts of swelling, as the amount of Cl- secretion would change. This study would directly measure function of and confirm the cell surface biotinylation and western blot analysis (indirect measures of function) previously done to determine the amount of CFTR in the plasma membrane of the operculum [70] and could be done on a much larger scale.
Another option to create interesting new in vitro models of the killifish operculum is through genetic manipulation of the organoids. A previous limitation of killifish research was the paucity of feasible genetic approaches. For example, researchers studying CFTR killifish recorded an increase in serum and glucocorticoid — inducible kinase (SGK1) mRNA and protein just prior the increased expression of CFTR at the plasma membrane of killifish. However, a definitive correlation could not be attributed to SGK1’s effect on CFTR because no SGK1-inhibitors, siRNA for killifish, or transgenic killifish have been developed. The researchers developed a morpholino to be injected in vivo, which confirmed SGK1’s involvement in the acute trafficking of CFTR from intracellular vesicles to the plasma membrane in gill mitochondrion rich cells of killifish during acclimation to seawater. However, this study involved the synthesis of specific morpholinos as well as western blots on injected killifish to determine CFTR concentration [72,73]. The FIS assay in killifish organoids modified to a CF version using lentivirus-shRNA or CRISPR/Cas9 technology would likely provide a more efficient approach to study the role of CFTR in salt-water adaptation. Moreover, frozen organoids, in contrast to native tissues, could be shared by marine laboratories worldwide and would make research on marine animals less dependent on local facilities and expertise.
Conclusions
Organoid protocols have been developed only for mammalian systems [2,4,7,27,28]. Model marine model organisms have vastly improved the understanding of many principles of transport [44-51,55-63,58,71]. We believe that organoids of model organisms will allow for a fuller understanding of the mechanisms of epithelial systems — allowing us to monitor auto-regulation of these systems and monitor their real time development.
We owe many groundbreaking findings to model organisms, and we do not suggest organoids as a replacement for model organisms. We do, however, want to urge collaboration between the rapidly growing research world of organoids and those researchers using model organisms (Figure 3). The control, convenience, construction, and consistency that organoids provide, combined with the unique properties model epithelial organisms possess, would likely lead to a better understanding of epithelial systems and diseases.
Figure 3.
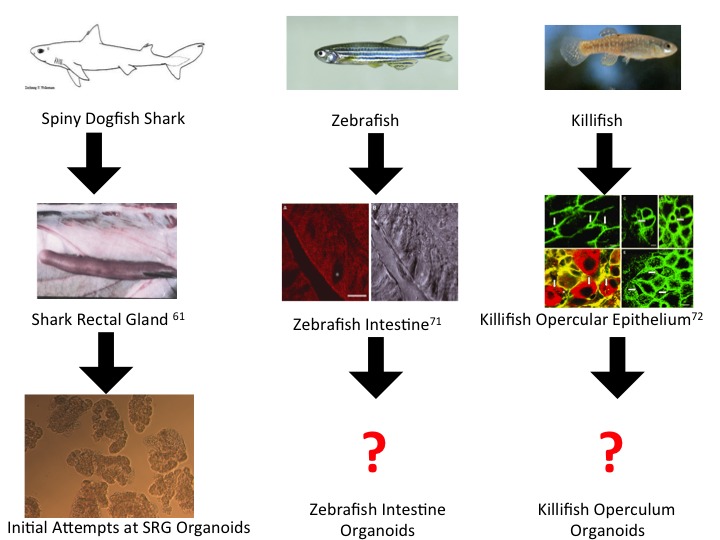
Future Steps of Organoid Development from Comparative Model Organisms.
Abbreviations
- SRG
shark rectal gland
- FIS
forskolin induced swelling
- CFTR
cystic fibrosis transmembrane conductance regulator
- IBD
inflammatory bowel disease
- hES
human embryonic stem cells
- hiPS
human induced pluripotent stem cells
- NKCC1
sodium, potassium 2 chloride transporter 1
- cAMP
adenosine 3’, 5’ –cyclic monophosphate
- PKA
protein kinase A
- CRISPR/Cas9
clustered regularly interspaced short palindromic repeats/crispr associated protein 9
- HMEC
human mammary epithelial cell
- MDCK
Malvin-Darby canine kidney
- LLC-PK1
pig kidney epithelial cells
- HEK
human embryonic kidney
- A6
Xenopus laevis kidney cells
Author contributions
Schwarz: experiments, text, figures, proofreading; De Jonge: experiments, text, proofreading, Forrest: experiments, text, figures, proofreading
Funding
This work was supported by NIH grants DK34208, NIEHS 5 P30 ES03828 (Center for Membrane Toxicology Studies) to JNF; NSF grant DBI-0139190 (REU site at MDIBL), and an MDIBL New Investigator Award to H.RdJ.
References
- Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345(6194):1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- Boj SF, Hwang C, Baker LA, Chio II, Engle DD, Corbo V. et al. Organoid Models of Human and Mouse Ductal Pancreatic Cancer. Cell. 2015;160(1-2):324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pin C, Parker A, Gunning AP, Ohta Y, Johnson IT, Carding SR. et al. An individual based computational model of intestinal crypt fission and its application to predicting unrestrictive growth of the intestinal epithelium. Integrative biology : quantitative biosciences from nano to macro. Integrative Biology. 2015;7(2):213–228. doi: 10.1039/c4ib00236a. [DOI] [PubMed] [Google Scholar]
- Dekkers R, Vijftigschild LA, Vonk AM, Kruisselbrink E, de Winter-de Groot KM, Janssens HM. et al. A bioassay using intestinal organoids to measure CFTR modulators in human plasma. J Cyst Fibros. 2015;14(2):178–181. doi: 10.1016/j.jcf.2014.10.007. [DOI] [PubMed] [Google Scholar]
- Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A. et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159(1):176–187. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ader M, Tanaka EM. Modeling human development in 3D culture. Curr Opin Cell Biol. 2014;31:23–28. doi: 10.1016/j.ceb.2014.06.013. [DOI] [PubMed] [Google Scholar]
- Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T. et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13(6):653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S. et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472(7341):51–56. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- Dekkers JF, Wiegerinck CL, de Jonge HR, Bronsveld I, Janssens HM, de Winter-de Groot KM. et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat Med. 2013;19(7):939–945. doi: 10.1038/nm.3201. [DOI] [PubMed] [Google Scholar]
- Au SH, Chamberlain MC, Mahesh S, Sefton MV, Wheeler AR. Hepatic organoids for microfluidic drug screening. Lab Chip. 2014;14(17):3290–3299. doi: 10.1039/c4lc00531g. [DOI] [PubMed] [Google Scholar]
- Unger C, Kramer N, Walzl A, Scherzer M, Hengstschlager M, Dolznig H. Modeling human carcinomas: Physiologically relevant 3D models to improve anti-cancer drug development. Adv Drug Deliv Rev. 2014;79-80C:50–67. doi: 10.1016/j.addr.2014.10.015. [DOI] [PubMed] [Google Scholar]
- Astashkina A, Grainger DW. Critical analysis of 3-D organoid in vitro cell culture models for high-throughput drug candidate toxicity assessments. Adv Drug Deliv Rev. 2014;69-70:1–18. doi: 10.1016/j.addr.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Astashkina AI, Mann BK, Prestwich GD, Grainger DW. A 3-D organoid kidney culture model engineered for high-throughput nephrotoxicity assays. Biomaterials. 2012;33(18):4700–4711. doi: 10.1016/j.biomaterials.2012.02.063. [DOI] [PubMed] [Google Scholar]
- Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X. et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5(+) stem cell. Nat Med. 2012;18(4):618–623. doi: 10.1038/nm.2695. [DOI] [PubMed] [Google Scholar]
- Stampfer MR, Bartley JC. Human mammary epithelial cells in culture: differentiation and transformation. Cancer Treat Res. 1988;40:1–24. doi: 10.1007/978-1-4613-1733-3_1. [DOI] [PubMed] [Google Scholar]
- Stampfer MR, Yaswen P. Growth, differentiation, and transformation of human mammary epithelial cells in culture. Cancer Treat Res. 1994;71:29–48. doi: 10.1007/978-1-4615-2592-9_2. [DOI] [PubMed] [Google Scholar]
- Ramirez RD, Morales CP, Herbert BS, Rohde JM, Passons C, Shay JW. et al. Putative telomere-independent mechanisms of replicative aging reflect inadequate growth conditions. Genes Dev. 2001;15(4):398–403. doi: 10.1101/gad.859201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe JC, Bhattacharya S, Merchant B, Bassett E, Swisshelm K, Feiler HS. et al. Molecular distinctions between stasis and telomere attrition senescence barriers shown by long-term culture of normal human mammary epithelial cells. Cancer Res. 2009;69(19):7557–7568. doi: 10.1158/0008-5472.CAN-09-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitaka T. The current status of primary hepatocyte culture. Int J Exp Pathol. 1998;79(6):393–409. doi: 10.1046/j.1365-2613.1998.00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan J, Schwartz RE, Ross NT, Logan DJ, Thomas D, Duncan SA. et al. Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat Chem Biol. 2013;9(8):514–520. doi: 10.1038/nchembio.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Zhang Y. Genetic and epigenetic variations in iPSCs: potential causes and implications for application. Cell Stem Cell. 2013;13(2):149–159. doi: 10.1016/j.stem.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund RJ, Narva E, Lahesmaa R. Genetic and epigenetic stability of human pluripotent stem cells. Nat Rev Genet. 2012;13(10):732–744. doi: 10.1038/nrg3271. [DOI] [PubMed] [Google Scholar]
- Pera MF. Stem cells: The dark side of induced pluripotency. Nature. 2011;471(7336):46–47. doi: 10.1038/471046a. [DOI] [PubMed] [Google Scholar]
- Bayart E, Cohen-Haguenauer O. Technological overview of iPS induction from human adult somatic cells. Curr Gene Ther. 2013;13(2):73–92. doi: 10.2174/1566523211313020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM. et al. Long-Term Culture of Genome-Stable Bipotent Stem Cells from Adult Human Liver. Cell. 2015;160(1-2):299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141(5):1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- Huch M, Dorrell C, Boj SF, van Es JH, Li VS, van de Wetering M. et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494(7436):247–250. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonica F, Kasprzyk DF, Opitz R, Iacovino M, Liao XH, Dumitrescu AM. et al. Generation of functional thyroid from embryonic stem cells. Nature. 2012;491(7422):66–71. doi: 10.1038/nature11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Bhang DH, Beede A, Huang TL, Stripp BR, Bloch KD. et al. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell. 2014;156(3):440–455. doi: 10.1016/j.cell.2013.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greggio C, De Franceschi F, Figueiredo-Larsen M, Gobaa S, Ranga A, Semb H. et al. Artificial three-dimensional niches deconstruct pancreas development in vitro. Development. 2013;140(21):4452–4462. doi: 10.1242/dev.096628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stange DE, Koo BK, Huch M, Sibbel G, Basak O, Lyubimova A. et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell. 2013;155(2):357–368. doi: 10.1016/j.cell.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartfeld S, Bayram T, van de Wetering M, Huch M, Begthel H, Kujala P. et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148(1):126–36 e6. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisha H, Tanaka T, Kanno S, Tokuyama Y, Komai Y, Ohe S. et al. Establishment of a novel lingual organoid culture system: generation of organoids having mature keratinized epithelium from adult epithelial stem cells. Sci Rep. 2013;3:3224. doi: 10.1038/srep03224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuratnik A, Giardina C. Intestinal organoids as tissue surrogates for toxicological and pharmacological studies. Biochem Pharmacol. 2013;85(12):1721–1726. doi: 10.1016/j.bcp.2013.04.016. [DOI] [PubMed] [Google Scholar]
- Hirt MN, Hansen A, Eschenhagen T. Cardiac tissue engineering: state of the art. Circ Res. 2014;114(2):354–367. doi: 10.1161/CIRCRESAHA.114.300522. [DOI] [PubMed] [Google Scholar]
- Iyer RK, Odedra D, Chiu LL, Vunjak-Novakovic G, Radisic M. Vascular endothelial growth factor secretion by nonmyocytes modulates Connexin-43 levels in cardiac organoids. Tissue Eng Part A. 2012;18(17-18):1771–1783. doi: 10.1089/ten.tea.2011.0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkumatov A, Baek K, Kong H. Matrix rigidity-modulated cardiovascular organoid formation from embryoid bodies. PLoS One. 2014;9(4):e94764. doi: 10.1371/journal.pone.0094764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Ando S, Takata N, Kawada M, Muguruma K, Sekiguchi K. et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10(6):771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Phillips R. Innovation: Organoids-a better model for prostate cancer. Nat Rev Urol. 2014;11(11):604. doi: 10.1038/nrurol.2014.269. [DOI] [PubMed] [Google Scholar]
- Humphreys BD. Kidney structures differentiated from stem cells. Nat Cell Biol. 2014;16(1):19–21. doi: 10.1038/ncb2904. [DOI] [PubMed] [Google Scholar]
- Taguchi A, Kaku Y, Ohmori T, Sharmin S, Ogawa M, Sasaki H. et al. Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell. 2014;14(1):53–67. doi: 10.1016/j.stem.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Takasato M, Er PX, Becroft M, Vanslambrouck JM, Stanley EG, Elefanty AG. et al. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nat Cell Biol. 2014;16(1):118–126. doi: 10.1038/ncb2894. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME. et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501(7467):373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawczak V, Getsy P, Zaidi A, Sun F, Zaman K, Gaston B. Novel Approaches for Potential Therapy of Cystic Fibrosis. Curr Drug Targets. 2015;16(9):923–936. doi: 10.2174/1389450116666150102113314. [DOI] [PubMed] [Google Scholar]
- Ikpa PT, Bijvelds MJC, de Jonge HR. Cystic fibrosis: Toward personalized therapies. Int J Biochem Cell Biol. 2014;52:192–200. doi: 10.1016/j.biocel.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Wainwright CE, Elborn JS, Ramsey BW, Marigowda G, Huang X, Cipolli M. et al. Lumacaftor-Ivacaftor in Patients with Cystic Fibrosis Homozygous for Phe508del CFTR. N Engl J Med. 2015;373(3):220–231. doi: 10.1056/NEJMoa1409547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161(4):933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A. The progress of physiology. Science. 1929;70:200–204. doi: 10.1126/science.70.1809.200. [DOI] [PubMed] [Google Scholar]
- Marshall EK. The aglomerular kidney of the toadfish (Opsanus tau) Bull Johns Hopkins Hosp. 1929;45:95–101. [Google Scholar]
- Vize PD, Smith HW. A Homeric view of kidney evolution: A reprint of H.W. Smith’s classic essay with a new introduction. Evolution of the kidney. 1943. Anat Rec A Discov Mol Cell Evol Biol. 2004;277(2):344–354. doi: 10.1002/ar.a.20017. [DOI] [PubMed] [Google Scholar]
- Wearn JT, Richards AN. Observations on the composition of glomerular urinem, with particular reference to the problem of reabsorption in the renal tubules. Am J Physiol. 1924;71(1):209–227. [Google Scholar]
- Stahl M, Stahl K, Brubacher MB, Forrest JN Jr.. Divergent CFTR orthologs respond differently to the channel inhibitors CFTRinh-172, glibenclamide, and GlyH-101. Am J Physiol Cell Physiol. 2012;302(1):C67–C76. doi: 10.1152/ajpcell.00225.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jonge HR, Tilly BC, Hogema BM, Pfau DJ, Kelley CA, Kelley MH. et al. cGMP inhibition of type 3 phosphodiesterase is the major mechanism by which C-type natriuretic peptide activates CFTR in the shark rectal gland. Am J Physiol Cell Physiol. 2014;306(4):C343–C353. doi: 10.1152/ajpcell.00326.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Tomkovich S, Jobin C. Could a swimming creature inform us on intestinal diseases? Lessons from zebrafish. Inflamm Bowel Dis. 2014;20(5):956–966. doi: 10.1097/01.MIB.0000442923.85569.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navis A, Marjoram L, Bagnat M. Cftr controls lumen expansion and function of Kupffer's vesicle in zebrafish. Development. 2013;140(8):1703–1712. doi: 10.1242/dev.091819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrich RW, Aller SG, Webster P, Marino CR, Forrest JN Jr.. Vasoactive intestinal peptide, forskolin, and genistein increase apical CFTR trafficking in the rectal gland of the spiny dogfish, Squalus acanthias. Acute regulation of CFTR trafficking in an intact epithelium. J Clin Invest. 1998;101(4):737–745. doi: 10.1172/JCI803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan JR, Forbush B 3rd, Hanrahan JW. The molecular basis of chloride transport in shark rectal gland. J Exp Biol. 1994;196:405–418. doi: 10.1242/jeb.196.1.405. [DOI] [PubMed] [Google Scholar]
- Aller SG, Lombardo ID, Bhanot S, Forrest JN Jr.. Cloning, characterization, and functional expression of a CNP receptor regulating CFTR in the shark rectal gland. Am J Physiol. 1999;276(2 Pt 1):C442–C449. doi: 10.1152/ajpcell.1999.276.2.C442. [DOI] [PubMed] [Google Scholar]
- Forrest JN Jr., Aller SG, Wood SJ, Ratner MA, Forrest JK, Kelley GG. Cadmium disrupts the signal transduction pathway of both inhibitory and stimulatory receptors regulating chloride secretion in the shark rectal gland. Journal of Experimental Zoology. 1997;279(5):530–536. doi: 10.1002/(sici)1097-010x(19971201)279:5<530::aid-jez17>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Ratner MA, Decker SE, Aller SG, Weber G, Forrest JN Jr.. Mercury toxicity in the shark (Squalus acanthias) rectal gland: apical CFTR chloride channels are inhibited by mercuric chloride. J Exp Zool A Comp Exp Biol. 2006;305(3):259–267. doi: 10.1002/jez.a.257. [DOI] [PubMed] [Google Scholar]
- Waldegger S, Fakler B, Bleich M, Barth P, Hopf A, Schulte U. et al. Molecular and functional characterization of s-KCNQ1 potassium channel from rectal gland of Squalus acanthias. Pflugers Arch. 1999;437(2):298–304. doi: 10.1007/s004240050783. [DOI] [PubMed] [Google Scholar]
- Weber GJ, Mehr AP, Sirota JC, Aller SG, Decker SE, Dawson DC. et al. Mercury and zinc differentially inhibit shark and human CFTR orthologues: involvement of shark cysteine 102. Am J Physiol Cell Physiol. 2006;290(3):C793–C801. doi: 10.1152/ajpcell.00203.2005. [DOI] [PubMed] [Google Scholar]
- Yang T, Forrest SJ, Stine N, Endo Y, Pasumarthy A, Castrop H. et al. Cyclooxygenase cloning in dogfish shark, Squalus acanthias, and its role in rectal gland Cl secretion. Am J Physiol Regul Integr Comp Physiol. 2002;283(3):R631–R637. doi: 10.1152/ajpregu.00743.2001. [DOI] [PubMed] [Google Scholar]
- Forrest JN Jr.. Cellular and molecular biology of chloride secretion in the shark rectal gland: regulation by adenosine receptors. Kidney Int. 1996;49(6):1557–1562. doi: 10.1038/ki.1996.224. [DOI] [PubMed] [Google Scholar]
- Pfau DP, Poeschla M, Poeschla EM, Forrest JN Jr.. Strategies to establish a continuous cell line in the shark rectal gland of Squalus acanthias. The Bulletin, MDI Biological Laboratory. 2014;53:3. [Google Scholar]
- Hynds RE, Giangreco A. Concise review: the relevance of human stem cell-derived organoid models for epithelial translational medicine. Stem Cells. 2013;31(3):417–422. doi: 10.1002/stem.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M. et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496(7446):498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart MA, Klee EW. Zebrafish approaches enhance the translational research tackle box. Transl Res. 2014;163(2):65–78. doi: 10.1016/j.trsl.2013.10.007. [DOI] [PubMed] [Google Scholar]
- Bagnat M, Navis A, Herbstreith S, Brand-Arzamendi K, Curado S, Gabriel S. et al. Cse1l is a negative regulator of CFTR-dependent fluid secretion. Curr Biol. 2010;20(20):1840–1845. doi: 10.1016/j.cub.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JR, Sato JD, VanderHeide J, LaCasse T, Stanton CR, Lankowski A. et al. The role of SGK and CFTR in acute adaptation to seawater in Fundulus heteroclitus. Cell Physiol Biochem. 2008;22(1-4):69–78. doi: 10.1159/000149784. [DOI] [PubMed] [Google Scholar]
- Stanton CR, Thibodeau R, Lankowski A, Shaw JR, Hamilton JW, Stanton BA. Arsenic inhibits CFTR-mediated chloride secretion by killifish (Fundulus heteroclitus) opercular membrane. Cell Physiol Biochem. 2006;17(5-6):269–278. doi: 10.1159/000094139. [DOI] [PubMed] [Google Scholar]
- Notch EG, Shaw JR, Coutermarsh BA, Dzioba M, Stanton BA. Morpholino gene knockdown in adult Fundulus heteroclitus: role of SGK1 in seawater acclimation. PLoS One. 2011;6(12):e29462. doi: 10.1371/journal.pone.0029462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notch EG, Chapline C, Flynn E, Lameyer T, Lowell A, Sato D. et al. Mitogen activated protein kinase 14-1 regulates serum glucocorticoid kinase 1 during seawater acclimation in Atlantic killifish, Fundulus heteroclitus. Comp Biochem Physiol A Mol Integr Physiol. 2012;162(4):443–448. doi: 10.1016/j.cbpa.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]



