Abstract
Pharmacogenomic testing in mental health has not yet reached its full potential. An important reason for this involves differentiating individual gene testing (IGT) from a combinatorial pharmacogenomic (CPGx) approach. With IGT, any given gene reveals specific information that may, in turn, pertain to a smaller number of medications. CPGx approaches attempt to encompass more complete genomic information by combining moderate risk alleles and synergistically viewing the results from the perspective of the medication. This manuscript will discuss IGT and CPGx approaches to psychiatric pharmacogenomics and review the clinical validity, clinical utility, and economic parameters of both.
Keywords: combinatorial, pharmacogenomics, mental health, clinical validity, clinical utility, economic outcomes
Introduction
The tantalizing promise of pharmacogenomics to utilize individual genetic markers to improve efficacy and decrease side effects has been suggested by researchers [1-3] and the media [4] for years. Precision medicine offers the possibility of moving away from “blockbuster” drugs to a personalized modality of treatment that attempts to utilize individual genetic markers to improve efficacy and decrease side effects for psychiatric patients. Iterations of this phenomenon include individual and panel P450 testing for pharmacokinetic (PK) indicators as well as pharmacodynamic (PD) markers relating to psychiatric pharmacology.
Individual gene testing (IGT) is best conceptualized as testing pharmacogenomic markers ad hoc and revealing each gene’s results to a clinician in an attempt to positively intervene in the medication management of a patient. Combinatorial gene testing (CPGx) is the approach of combining genetic markers into sophisticated risk categories as an attempt to collect more relevant information to improve the clinical utility of pharmacogenomic testing. This manuscript will discuss IGT and its clinical validity, clinical utility, and economic outcomes related to mental health. Additionally, we will outline some approaches to CPGx in mental health and share some of the literature on its clinical validity, clinical utility, and economic outcomes.
Individual Gene Testing
Individual clinical pharmacogenomic testing began in earnest in 2004 with Food and Drug Administration (FDA) approval of Roche’s amplichip testing for CYP2D6 and CYP2C19 [5]. While this amplichip gives information about two genes, it is most accurately conceptualized as an IGT because each gene’s resultant phenotypes are reported individually and not synergized to provide clinically actionable information from the perspective of the medication. Another important aspect to this microarray was that newly found alleles could not easily be added to the technology given that it was an FDA-approved test. It became frozen in time, an anachronism when further clinically important alleles were discovered for CYP2D6 [6]. Since that time, several laboratories have offered individual gene testing either a la carte [7] or in the form of a panel of genes [8]. Most have updated their testing for current clinically significant alleles; however, both a la carte and panels of genes share the limitations commensurate with IGT.
Attempting to create clinical utility from large amounts of disparate information is part of the art and science of medicine. However, psychiatric pharmacogenomic testing has a classic “big data” problem. There are few people who would suggest that sharing raw genomic data — from a full exome sequence or a million SNP microarray — would engender clinically meaningful change to practice. IGT represents a truncated version of giving clinicians raw genomic data. In these panels, individual genes are selected for their potential to illuminate clinically relevant information. The clinician is expected to put this information in an appropriate statistical and clinical context. Increasing amounts of information necessitate the ability to parse out clinically important information to create utility. Given an explosion in our understanding of genes and alleles, it is not shocking that the transition of these steps toward clinical utility has come into question.
In IGT panels, each enzyme and resultant phenotypes provide information about a multitude of medications. For example, CYP2D6 is involved in the metabolism of many antidepressants and antipsychotics [9]. When a clinician orders a CYP2D6 test for a patient, the genotype, phenotype (UM, EM, IM, PM), and a list of medications that are partially or mostly metabolized by CYP2D6 are typically provided. As multiple enzymes are tested for, the burden falls on the clinician to “reverse engineer” the effects of several genes on medications.
Multiple genes on a pharmacogenomic panel create more complexity and increase the requirement for analysis on the part of the clinician who might have very little experience with pharmacogenomics [10]. Predictably, with IGT, the clinical relevance has been only validated in medications that are metabolized by one — or maybe two — enzymes (e.g., tricyclic antidepressants, paroxetine, aripiprazole etc.). The Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines reinforce this narrowed scope of testing for individual pharmacokinetic genes [11].
Combinatorial Pharmacogenomic Testing
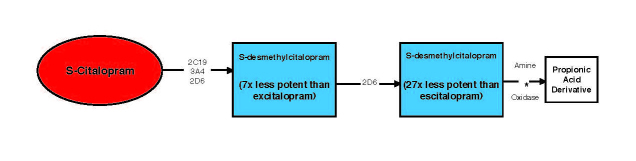
Neuropsychiatric medications are metabolized by multiple enzymes and interact with multiple neuropathways in order to attain clinical response. The genes that inform this response often do so in piecemeal fashion. As an example, a physician might order pharmacogenomic testing to inform their choice and dosing of escitalopram. Figure 1 shows that escitalopram is significantly metabolized by three different P450 enzymes — CYP2D6, CYP2C19, and CYP3A4 [12]. In a hypothetical scenario, a patient might be a CYP2D6 ultrarapid (UM) metabolizer, CYP2C19 poor metabolizer (PM), and a CYP3A4 extensive metabolizer (EM). The clinical information provided to the physician would be conveyed from the perspective of each of the genes tested. For CYP2D6, the clinical inference might be to increase the dose due to the patient’s ultrarapid metabolic capacity. However, for CYP2C19, the suggestion might be to lower the dose due to the patient’s poor metabolizing status. The clinical path forward is unclear. These opposing clinical suggestions may lead to conflicting treatments and likely contribute to the dilution of the clinical effect of pharmacogenomics in psychiatry.
Figure 1.
Metabolism flowchart for escitalopram including relevant P450 enzymes.
The clinician is wholly interested in the fate of the medication in the body, not the enzyme. Examining pharmacogenomic information in light of each medication’s unique metabolic profile — as opposed to each enzyme’s profile — reveals the most useful clinical information. CPGx testing is the process of simultaneously assessing the combined effects of multiple pharmacokinetic (PK) and pharmacodynamic (PD) genes for a given medication so that the information from each relevant gene is conveyed in a way that gives integrated information about the pharmacology of the medication for an individual.
The Process of Creating a Combinatorial Pharmacogenomic Test
The groundwork for creating a combinatorial pharmacogenomic test is laid down by research on the effects of each gene individually. For PK genes, this information is gathered by in vitro and in vivo studies looking at the effect of each enzyme on the metabolism of a medication. In vivo studies confirm that blood levels of a given medication increase or decrease based on relevant CYP alleles. These studies are now routinely done for potential new medications, and requisite warnings are labeled by the FDA based on the functionality of each CYP enzyme for a given medication [13]. Once individual genes show clinical validity, the study of combinatorial effects are the next logical step.
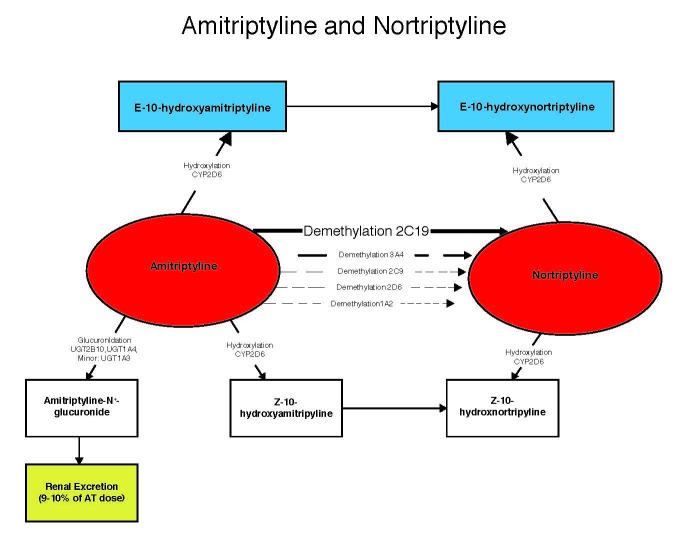
The important question is whether combinations of these genes give a more complete clinical picture compared to each gene reported individually. Steimer et al. exemplified this by using a limited combinatorial approach combining CYP2C19 and CYP2D6 for depressed patients taking amitriptyline [14]. They categorized the patients into low, medium low, medium high, and high risk groups for side effects based on the number of functional CYP2C19 and CYP2D6 alleles. The interesting caveat in this study was that nortriptyline levels were highly correlated with side effects, but not amitriptyline levels. This increased risk for nortriptyline-induced side effects has previously been suggested by other investigators [15,16]. The conversion of amitriptyline to nortriptyline by several CYP enzymes in the liver [16] introduces an important clinical scenario in which the variance in key PK enzymes involved in this process will have differing effects on nortriptyline levels. Because nortriptyline levels — and not amitriptyline levels — correlate directly with side effects, the combination of metabolizing phenotypes into categories (low, medium low, medium high, and high) based on resultant nortriptyline levels yield the most clinically actionable information about side effects from amitriptyline.
Figure 2 shows the metabolic pathway of amitriptyline, which is converted to nortriptyline by CYP2C19. Nortriptyline is converted, in turn, to its inactive metabolite by CYP2D6. Given that nortriptyline levels are associated with toxicity, decreased CYP2D6 activity in conjunction with increased CYP2C19 activity would be associated with more side effects, whereas increased CYP2D6 activity and decreased CYP2C19 activity would be associated with lower side effects. This is exactly what Steimer et al. observed in their population. The lowest risk group had two active CYP2D6 alleles and at least one inactive CYP2C19 allele, and the highest risk group had at least one inactive CYP2D6 allele and two active CYP2C19 alleles. Thus, the combinatorial approach accurately categorized the patients into higher and lower risk groups based on the simultaneous assimilation of pharmacogenomic information.
Figure 2.
Metabolism flowchart for amitriptyline and nortriptyline including relevant P450 enzymes.
Approaching the problem from the perspective of one gene-one enzyme-one medication (i.e., IGT) does not account for the multiple pathways and creates a simplified approach that at best obscures the information and at worst gives information that is antithetical to the true clinical message. With an IGT approach, most clinicians would ostensibly surmise that CYP2C19 PMs would have more side effects and change medications accordingly. However, the combinatorial categorical approach showed that side effects were most accurately predicted when both enzymes were taken into account. The highest side effect burden was realized when someone had deficiencies in CYP2D6 but over-activity in CYP2C19. The combinatorial risk categories of low, medium low, medium high, and high had more predictive ability for side effect burden than either gene in isolation.
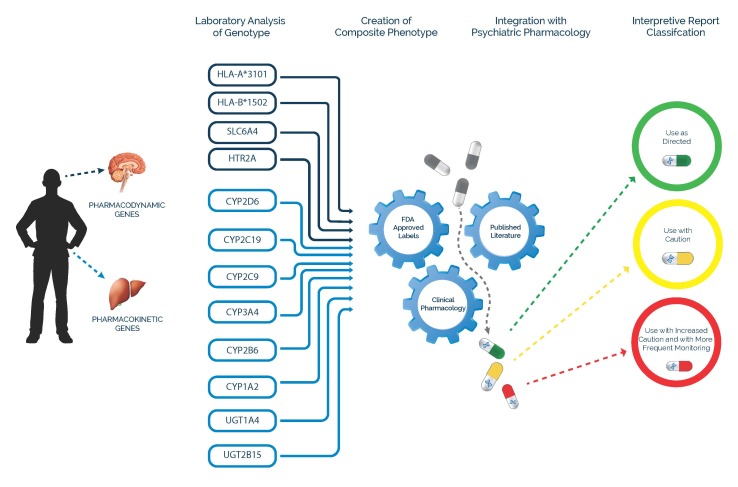
An extrapolation of this algorithmic process was conceived at the Mayo Clinic and Cincinnati Children’s Hospital Medical Center and refined over several years to include the most commonly prescribed psychotropic medications. The GeneSight Psychotropic test has been developed to provide practicing clinicians with a suite of preselected psychiatric genes yielding a composite phenotype that integrates the known pharmacology of 38 medications compromising the large majority of psychotropic medication prescriptions in the United States (Figure 3).
Figure 3.
GeneSight combinatorial process.
The GeneSight test includes pharmacokinetic and pharmacodynamic genes, which address both the safety and efficacy of psychiatric medications. This combinatorial stratification process categorizes antidepressant and antipsychotic medications into three color-coded categories for each genotyped individual: little or no gene-drug interaction (green “use as directed”), moderate gene-drug interaction (yellow “use with caution”), and severe gene-drug interaction (red “use with caution and with more frequent monitoring”). This combinatorial approach accounts for both single and multiple metabolic pathways for each medication synergistically combined with genetic changes to brain pathways affecting the mechanism of drug action. Additional information is supplied to the clinician through the use of drug specific footnotes for those medications in the yellow and red advisory categories, which supply the details of the gene-drug interactions. Genotype and phenotype results with interpretive comments for each of the genes provide a further level of detail. This layering process allows for complex genomic and pharmacologic data to be presented in discrete, increasingly informative layers that provide useful clinical insight (example results for one individual patient are shown in Figure 4).
Figure 4.
Example GeneSight Psychotropic results for antidepressants for an individual patient.
Clinical Validity and Utility of Individual Psychiatric Pharmacogenomics
Clinical validity is the accuracy of a genetic test to predict a stated clinical outcome such as relative blood levels or the likelihood of response to treatment. Single gene pharmacogenomic testing has been significantly studied — particularly for PK genes [17] and a select number of PD genes [18]. The clinical validity for the prediction of blood levels has been fairly well characterized [19]. Clinical utility represents the improvement in outcomes in PGx-tested individuals compared to standard of care. Individual psychiatric gene studies have historically produced mixed results regarding clinical outcomes [20], though recent guidelines have brought attention to a few clinically important single gene-drug interactions [11,21].
Recognizing the limitations of IGT, the Centers for Medicare & Medicaid Services (CMS) has engaged in a comprehensive review of the literature to address the monetary and clinical value of individual gene testing for specific indications. After their exhaustive review, they have concluded that individual gene testing has clinical validity with only a limited number of antidepressant and antipsychotic medications [22]. Not surprisingly, all of these medications are significantly metabolized by one enzyme. One viable reason for the lack of overwhelming evidence for IGT is that the combined (i.e., combinatorial) effects of multiple genes give the most accurate pharmacologic picture for a given medication. Without this full combinatorial picture, the clinical effect may be muted creating mixed outcomes.
Economic Utility of Individual Gene Testing
The economic viability of IGT for mental illness has been assessed with several studies. The first study looked at the economic impact of CYP2D6 testing in a severely mentally ill population [23]. They reported that the treatment costs in patients who had an “extreme phenotype” (either PM or UM) for CYP2D6 were $4,000 to $6,000 more than EM or IM phenotypes. Additionally, Herbild investigated a population specifically enriched for extreme phenotypes for both CYP2D6 and CYP2C19. They were separated into a treatment as usual (TAU) and pharmacogenomic-informed treatment group. The individual pharmacogenomic-tested group comparatively reduced costs for the extreme metabolizers [24]. Ruaño et al. (2013) retrospectively found that individuals with reduced CYP2D6 function averaged hospitalization length of stay that was 2 days longer than those with normal or increased CYP2D6 function [25]. Fagerness et al. also looked at the economic utility of a panel of individual genes in a psychiatric setting compared to matched controls [26]. Their results suggested the IGT was associated an increase in pharmacy costs with a concomitant decrease in health care utilization. Finally, Olgiati and Seretti have modeled the use of a PD gene, SLC6A4, and suggested that testing would be cost effective for antidepressant treatment [27,28].
Clinical Validity of Combinatorial Psychiatric Pharmacogenomics
Several case studies have suggested the clinical validity of combinatorial pharmacogenomic deficiencies relating to PK side effects in patients on various psychiatric medications [29,31], including two fatalities due to extremely high blood levels [32,33]. Other studies have prospectively assessed the clinical validity of combinatorial pharmacogenomics as they relate to side effects and suggested a role for combined PK genetic testing [14,34-36].
Data from three prospective clinical trials (two open-label trials and one placebo-controlled double-blind trial) have demonstrated the clinical validity of treatment guided by GeneSight combinatorial pharmacogenomics in Major Depressive Disorder [37-39]. The outcomes of the standard of care groups were analyzed to assess the clinical validity and predictive capability of the pharmacogenomic report. In the standard of care groups, the patients and the clinicians were blinded to their genetic information so that the treatment was not guided by CPGx testing. However, after completion of the study, the CPGx information was revealed to determine the clinical validity (predictive ability of response and intolerable treatment) of the categorical approach. In all three prospective clinical trials, patients on red category medications, indicating the most severe gene-drug interaction, experienced the worst outcomes based on the 17-item Hamilton Rating Scale for Depression (HAM-D17) (Figure 5). In a pooled analysis of the standard of care arm in the three studies, combinatorial pharmacogenomics showed this ability to predict poor outcomes, whereas single genes were not predictive [40]. The clinical validity of the GeneSight CPGx information is shown by its predictive capability to determine that patients who are placed on red category medications have increased likelihood of poorer outcomes.
Figure 5.
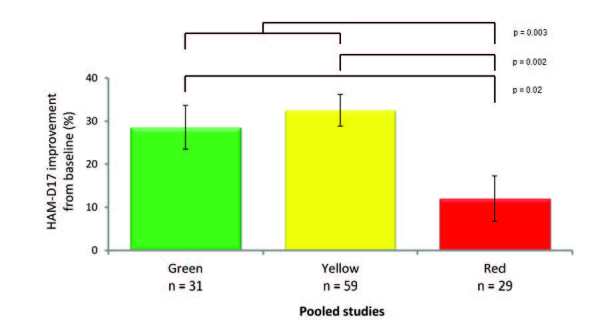
Clinical outcomes of blinded subjects treated without pharmacogenomic testing by GeneSight CPGx advisory category (pooled data from three trials).
Clinical Utility, Algorithms, and Ease of Use
The framework from which complex pharmacogenomic technology is communicated has an extensive effect on its clinical utility. Various methods have been employed to bring combinatorial methods from the bench to the bedside. In the non-psychiatric literature, warfarin blood levels have long been known to be susceptible to combined effects of variations in both VKORC1 and CYP2C9 (i.e., clinical validity) [41]. However, this information proved cumbersome for practicing physicians. Understanding the need to bridge this known combinatorial gap, the FDA created tables to help with clinical translation. Subsequently, online genetics-based algorithms were developed to expand and improve upon the utility of warfarin pharmacogenomic testing [42].
Current CPIC guidelines specifically address this education/translation problem and recommend an algorithmic approach stating that, “Dosing algorithms using genetics outperform non-genetic clinical algorithms and fixed-dose approaches in dose prediction ... genetics-based algorithms also better predict warfarin dose than the FDA-approved warfarin label table. Therefore, the use of pharmacogenetic algorithm-based dosing is recommended when possible” [2]. Not only does CPGx testing help aggregate the moderate clinical effects from genetics, but this suggests it also helps to simplify the pertinent information into discrete categories improving clinical utility by simplifying complex information for physicians while concomitantly making the end product more amenable to comparative outcomes research.
Clinical Utility of Combinatorial Psychiatric Pharmacogenomics
While the clinical validity of a few previously mentioned limited combinatorial algorithms for mental illness have been reported, GeneSight testing is the only combinatorial pharmacogenomic information prospectively tested in “real world” clinical settings known to authors, and, therefore, comprises all of the literature on the clinical utility of combinatorial pharmacogenomics. Three clinical trials have studied the clinical utility of GeneSight combinatorial testing compared to standard of care. The first prospective, open-label trial identified a significant reduction in the GeneSight-guided group compared to the standard of care group based on the HAM-D17 as well as the 16-item Clinician Rated Quick Inventory of Depressive Symptomatology (QIDS-C16) [37]. This was replicated by a much larger study, which resulted in a significantly improved response on the QIDS-C16 and HAM-D17, as well as the patient reported nine-item Patient Health Questionnaire (PHQ-9), in the GeneSight guided group compared to standard of care [38]. Finally, the smaller placebo-controlled, double-blind study trended toward similar clinical significance, showing improvement in the GeneSight group compared to standard of care with double the likelihood of response [39].
Economic Utility of Combinatorial Psychiatric Pharmacogenomics
In a retrospective chart review using the GeneSight Psychotropic report, patients who were on medications predicted to yield the greatest gene-drug complications (i.e., “red category” medications) presented with significantly increased total health care visits, medical absence days, and disability claims compared to patients taking green or yellow category medications, resulting in nearly $5,200 greater health care expenditures than those on genetically appropriate medications [43].
The largest economic study to date on pharmacogenomics for mental illness was done with GeneSight CPGx testing (n = 2,166 patients with genetic testing and n = 10,880 standard of care controls). This study analyzed total medication expenditures with the GeneSight test compared to standard of care in a prospective design. In this study, the pharmacy costs in the GeneSight group were $1,035.60 lower per patient per year compared to the propensity matched standard of care group. Given the large population, this difference was highly significant (p = 0.0007). These costs improved even more for non-psychiatrists, patients whose physicians followed the genetic report, and patients with anxiety disorders [44].
Conclusions and Outlook
The ultimate purpose of psychiatric pharmacogenomic testing is to improve patients’ lives by accurately predicting response to treatment (clinical validity) and then significantly improving outcomes with this prediction (clinical utility). An enticing third characteristic is that CPGx testing might clinically improve patient lives while simultaneously saving health care dollars. The promise of this appealing triumvirate has been presaged since the publication of the full sequenced human genome.
Recognizing the ongoing accumulation of clinical data in the psychiatric pharmacogenomic field and the enticing potential of lower costs and improved outcomes for its patient population, the Centers for Medicare & Medicaid Services extensively evaluated the clinical validity, clinical utility, and economic data for both IGT and CPGx testing presented here. After this comprehensive process, CMS released a specific coverage decision for the CPGx GeneSight Psychotropic test [45] while simultaneously narrowing the reimbursed indications for IGT for antidepressants and antipsychotics [22]. Multiple private insurance companies and the U.S. Department of Veterans Affairs have also made decisions to cover the GeneSight combinatorial test.
The distinction between IGT panel testing and CPGx testing is an important one. With IGT, any given gene reveals specific information that may, in turn, pertain to a small number of medications. Additionally, it requires a sophisticated level of knowledge to accurately weigh the information so that it is not over- or under-valued in clinical decision making. Given this limited scope, IGT has demonstrated a narrowed clinical impact for psychiatric medications. Up to 40 percent of the inter-individual differences in antidepressant response may be explained by common genetic variations [46]. However, this is juxtaposed with the fact that the clinical utility of individual genes has not been robust. Combinations of genes — each with a small to moderate effect — likely represent the problem and solution to this conundrum. Additionally, the way in which the clinician interfaces with complex genomic information likely has an important impact on clinical utility as well.
The importance of the curation and formulation of relevant genes and biomarkers comprising a CPGx test is underscored by the expense of proving its clinical utility. While individual genetic information is becoming less expensive, establishing true clinical utility is a much more involved endeavor. With terabytes of data at our disposal, we are now beyond an era where it is feasible to “dump” information on a clinician in the hopes that it will yield positive results. Future biomarkers, genetic information, and “big data” necessitate combining a precision approach with the appropriate translation of this data to help guide treatment. This translational combinatorial approach must then demonstrate efficacy through prospective clinical trials. Future iterations of the CPGx process could incorporate drug-drug interactions, other biomarkers, and electronic medical record-derived clinical data overlaid on the combinatorial pharmacogenomics platform. We believe that a true phase shift forward in psychiatric practice rests in CPGx testing and precision algorithms that demonstrate clinical utility through replicated outcome studies.
Abbreviations
- IGT
individual gene testing
- CPGx
combinatorial pharmacogenomics
- PK
pharmacokinetic
- PD
pharmacodynamics
- CPIC
Clinical Pharmacogenetics Implementation Consortium
- UM
ultrarapid
- PM
poor metabolizer
- EM
extensive metabolizer
- CMS
Centers for Medicare & Medicaid Services
- TAU
treatment and usual
- HAM-D17
17-item Hamilton Rating Scale for Depression
- QIDS-C16
16-item Clinician Rated Quick Inventory of Depressive Symptomatology
- FDA
Food and Drug Administration
Authors' note
Drs. Winner and Dechairo work for Assurex Health Inc. Dr. Winner is the Medical Director; Dr. Dechairo is Senior Vice President of Medical Affairs and Clinical Development.
References
- Evans WE, Johnson JA. Pharmacogenomics: the inherited basis for interindividual differences in drug response. Annu Rev Genomics Hum Genet. 2001;2:9–39. doi: 10.1146/annurev.genom.2.1.9. [DOI] [PubMed] [Google Scholar]
- Johnson JA, Gong L, Whirl-Carrillo M, Gage BF, Scott SA, Stein CM. et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 Genotypes and Warfarin Dosing. Clin Pharmacol Ther. 2011;90(4):625–629. doi: 10.1038/clpt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebert DW, Bingham E. Pharmacogenomics: Out of the lab and into the community. Trends Biotechnol. 2001;19(12):519–523. doi: 10.1016/s0167-7799(01)01805-4. [DOI] [PubMed] [Google Scholar]
- Friedman RA. On the Horizon, Personalized Depression Drugs. The New York Times. 2007 June 19;
- Phillips K, Van Bebber SL. Measuring the value of pharmacogenomics. Nat Rev Drug Discov. 2005;4(6):500–509. doi: 10.1038/nrd1749. [DOI] [PubMed] [Google Scholar]
- Raimundo SI, Toscano C, Klein K, Fischer J, Griese EU, Eichelbaum M. et al. A novel intronic mutation, 2988g>A, with high predictivity for impaired function of cytochrome P450 2D6 in white subjects. Clin Pharmacol Ther. 2004;76(2):128–138. doi: 10.1016/j.clpt.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Pharmacogenetics. LapCorp [Internet] [2015 Sept. 11]. Available from: https://www.labcorp.com/wps/portal/!ut/p/c1/04_SB8K8xLLM9MSSzPy8xBz9CP0os_hQV5NgQ09LYwMDS38nAyMv8zAjC6cgI_cAA6B8pFl8oIWZX6CRn5GBha-Rm4GRsZmTgamzq6GBgQEB3X4e-bmp-gW5EeUAMTMnVQ!!/dl2/d1/L0lDU0lKSWdrbUEhIS9JRFJBQUlpQ2dBek15cXchL1lCSkoxTkExTkk1MC01RncvN19VRTRTMUk5MzAwRjcyMDJKTkRWRUZFMjAwNy9MX19fXzQ!/?WCM_PORTLET=PC_7_UE4S1I9300F7202JNDVEFE2007_WCM&WCM_GLOBAL_CONTEXT=/wps/wcm/connect/labcorp+content/LabCorp/Provider/Resources/Services/Pharmacogenetics .
- Tests. Genelex [Internet] [2015 Sept. 11]. Available from: http://genelex.com/pharmacogenetic-tests/ .
- Flockhart DA. Drug Interactions: Cytochrome P450 Drug Interaction Table. Indiana University Department of Medicine [Internet] 2007. Available from: http://medicine.iupui.edu/clinpharm/DDIs/ .
- Winner JG, Goebert D, Matsu C, Mrazek DA. Training in psychiatric genomics during residency: a new challenge. Acad Psychiatry. 2010;34(2):115–118. doi: 10.1176/appi.ap.34.2.115. [DOI] [PubMed] [Google Scholar]
- Hicks J, Bishop J, Sangkuhl K, Müller D, Ji Y, Leckband S. et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin Pharmacol Ther. 2015;98(2):127–134. doi: 10.1002/cpt.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M-H, Lin K-M, Hsiao M-C, Shen WW, Lu M-L, Tang H-S. et al. Genetic polymorphisms of cytochrome P450 enzymes influence metabolism of the antidepressant escitalopram and treatment response. Pharmacogenomics. 2010;11(4):537–546. doi: 10.2217/pgs.09.168. [DOI] [PubMed] [Google Scholar]
- Guidance for industry. Drug interaction studies study design, data analysis, implications for dosing, and labeling recommendations. Food and Drug Administration [Internet] 2012. Available from: http://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM292362.pdf .
- Steimer W, Zöpf K, Von Amelunxen S, Pfeiffer H, Bachofer J, Popp J. et al. Amitriptyline or not, that is the question: Pharmacogenetic testing of CYP2D6 and CYP2C19 identifies patients with low or high risk for side effects in amitriptyline therapy. Clin Chem. 2005;51(2):376–385. doi: 10.1373/clinchem.2004.041327. [DOI] [PubMed] [Google Scholar]
- Jornil J, Jensen KG, Larsen F, Linnet K. Risk assessment of accidental nortriptyline poisoning: The importance of cytochrome P450 for nortriptyline elimination investigated using a population-based pharmacokinetic simulator. Eur J Pharm Sci. 2011;44(3):265–272. doi: 10.1016/j.ejps.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Haufroid V, Hantson P. CYP2D6 genetic polymorphisms and their relevance for poisoning due to amfetamines, opioid analgesics and antidepressants. Clin Toxicol (Phila) 2015;53(6):501–510. doi: 10.3109/15563650.2015.1049355. [DOI] [PubMed] [Google Scholar]
- The Human Cytochrome P450 (CYP) Allele Nomenclature Committee. The Human Cytochrome P450 (CYP) Allele Nomenclature Database [Internet] [cited 2015 Sept 11]. Available from: http://www.cypalleles.ki.se/ .
- Mrazek DA. Psychiatric Pharmacogenomics. New York: Oxford University Press; 2010. [Google Scholar]
- Müller DJ, Kekin I, Kao ACC, Brandl EJ. Towards the implementation of CYP2D6 and CYP2C19 genotypes in clinical practice: update and report from a pharmacogenetic service clinic. Int Rev Psychiatry. 2013;25(5):554–571. doi: 10.3109/09540261.2013.838944. [DOI] [PubMed] [Google Scholar]
- Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. Recommendations from the EGAPP Working Group: testing for cytochrome P450 polymorphisms in adults with nonpsychotic depression treated with selective serotonin reuptake inhibitors. Genet Med. 2007;9(12):819–825. doi: 10.1097/gim.0b013e31815bf9a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks JK, Swen JJ, Thorn CF, Sangkuhl K, Kharasch ED, Ellingrod VL. et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Tricyclic Antidepressants. Clin Pharmacol Ther. 2013;93(5):402–408. doi: 10.1038/clpt.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Human Cytochrome P450 (CYP) Allele Nomenclature CommitteeLocal Coverage Determination (LCD): CYP2C19, CYP2D6, CYP2C9, and VKORC1 Genetic Testing (L35327) Centers for Medicare & Medicaid Services [Internet] [cited 2015 Sept 11]. Available from: http://www.mediquant.com/policy/L35327_20150828.pdf .
- Chou WH, Yan FX, de Leon J, Barnhill J, Rogers T, Cronin M. et al. Extension of a pilot study: impact from the cytochrome P450 2D6 polymorphism on outcome and costs associated with severe mental illness. J Clin Psychopharmacol. 2000;20(2):246–251. doi: 10.1097/00004714-200004000-00019. [DOI] [PubMed] [Google Scholar]
- Herbild L, Andersen SE, Werge T, Rasmussen HB, Jürgens G. Does Pharmacogenetic Testing for CYP450 2D6 and 2C19 Among Patients with Diagnoses within the Schizophrenic Spectrum Reduce Treatment Costs? Basic Clin Pharmacol Toxicol. 2013;113(4):266–272. doi: 10.1111/bcpt.12093. [DOI] [PubMed] [Google Scholar]
- Ruano G, Szarek BL, Villagra D, Gorowski K, Kocherla M, Seip RL. et al. Length of psychiatric hospitalization is correlated with CYP2D6 functional status in inpatients with major depressive disorder. Biomark Med. 2013;7(3):429–439. doi: 10.2217/bmm.13.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerness J, Fonesca E, Hess GP. et al. Pharmacogenetic-guided psychiatric intervention associated with increased adherence and cost savings. Am J Manag Care. 2014;20(5):e146–e156. [PubMed] [Google Scholar]
- Serretti A, Olgiati P, Bajo E, Bigelli M, De Ronchi D. A model to incorporate genetic testing (5-HTTLPR) in pharmacological treatment of major depressive disorders. World J Biol Psychiatry. 2011;12(7):501–515. doi: 10.3109/15622975.2011.572998. [DOI] [PubMed] [Google Scholar]
- Olgiati P, Bajo E, Bigelli M, De Ronchi D, Serretti A. Should pharmacogenetics be incorporated in major depression treatment? Economic evaluation in high- and middle-income European countries. Prog Neuropsychopharmacol Biol Psychiatry. 2012;36(1):147–154. doi: 10.1016/j.pnpbp.2011.08.013. [DOI] [PubMed] [Google Scholar]
- Chua EW, Foulds J, Miller AL, Kennedy MA. Novel CYP2D6 and CYP2C19 variants identified in a patient with adverse reactions towards venlafaxine monotherapy and dual therapy with nortriptyline and fluoxetine. Pharmacogenet Genomics. 2013;23(9):494–497. doi: 10.1097/FPC.0b013e328363688d. [DOI] [PubMed] [Google Scholar]
- Stephan P, Sirot E, Mueller B, Eap C, Baumann P. Adverse Drug Reactions Following Nonresponse in a Depressed Patient with CYP2D6 Deficiency and Low CYP 3A4/5 Activity. Pharmacopsychiatry. 2006;39(4):150–152. doi: 10.1055/s-2006-946705. [DOI] [PubMed] [Google Scholar]
- Harris E, Eng HY, Kowatch R, Delgado SV, Saldaña SN. Disinhibition as a Side Effect of Treatment with Fluvoxamine in Pediatric Patients with Obsessive-Compulsive Disorder. J Child Adolesc Psychopharmacol. 2010;20(4):347–353. doi: 10.1089/cap.2009.0126. [DOI] [PubMed] [Google Scholar]
- Jornil J, Nielsen TS, Rosendal I, Ahlner J, Zackrisson AL, Boel LWT. et al. A poor metabolizer of both CYP2C19 and CYP2D6 identified by mechanistic pharmacokinetic simulation in a fatal drug poisoning case involving venlafaxine. Forensic Sci Int. 2013;226(1-3):e26–e31. doi: 10.1016/j.forsciint.2012.12.020. [DOI] [PubMed] [Google Scholar]
- Neukamm MA, Vogt S, Hermanns-Clausen M, Naue J, Thierauf A, Auwärter V. Fatal doxepin intoxication – Suicide or slow gradual intoxication? Forensic Sci Int. 2013;227(1-3):82–84. doi: 10.1016/j.forsciint.2012.08.050. [DOI] [PubMed] [Google Scholar]
- Emoto C, Fukuda T, Venkatasubramanian R, Vinks AA. The impact of CYP3A5*3 polymorphism on sirolimus pharmacokinetics: insights from predictions with a physiologically-based pharmacokinetics model. Br J Clin Pharmacol. 2015 doi: 10.1111/bcp.12743. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlpine DE, Biernacka JM, Mrazek DA, OʼKane DJ, Stevens SR, Langman LJ. Effect of Cytochrome P450 Enzyme Polymorphisms on Pharmacokinetics of Venlafaxine. Ther Drug Monit. 2011;33(1):14–20. doi: 10.1097/FTD.0b013e3181fcf94d. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Sawamura K, Someya T. Polymorphisms in the 5-hydroxytryptamine 2A receptor and cytochrome P4502D6 genes synergistically predict fluvoxamine-induced side effects in Japanese depressed patients. Neuropsychopharmacology. 2006;31(4):825–831. doi: 10.1038/sj.npp.1300919. [DOI] [PubMed] [Google Scholar]
- Hall-Flavin DK, Winner JG, Allen JD. et al. Using a pharmacogenomic algorithm to guide the treatment of depression. Transl Psychiatry. 2012;2:e172. doi: 10.1038/tp.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Flavin DK, Winner JG, Allen JD, Carhart JM, Proctor B, Snyder KA. et al. Utility of integrated pharmacogenomic testing to support the treatment of major depressive disorder in a psychiatric outpatient setting. Pharmacogenet Genomics. 2012;23(10):535–548. doi: 10.1097/FPC.0b013e3283649b9a. [DOI] [PubMed] [Google Scholar]
- Winner JG, Carhart JM, Altar CA, Allen JD, Dechairo BM. A prospective, randomized, double-blind study assessing the clinical impact of integrated pharmacogenomic testing for major depressive disorder. Discov Med. 2013;16(89):219–227. [PubMed] [Google Scholar]
- Altar CA, Carhart JM, Allen JD, Hall-Flavin DK, Dechairo BM, Winner JG. Clinical validity: Combinatorial pharmacogenomics predicts antidepressant responses and healthcare utilizations better than single gene phenotypes. Pharmacogenomics J. 2015;15(5):443–451. doi: 10.1038/tpj.2014.85. [DOI] [PubMed] [Google Scholar]
- FDA Clears Genetic Lab Test for Warfarin Sensitivity. US Food and Drug Administration [Internet] 2007. Available from: http://www.fda.gov/newsevents/newsroom/pressannouncements/2007/ucm108984.htm .
- WarfarinDosing.org [Internet] St. Louis: Washington University; [2015 Sept 14]. Available from: http://warfarindosing.org/Source/Home.aspx . [Google Scholar]
- Winner J, Allen JD, Altar CA, Spahic-Mihajlovic A. Psychiatric pharmacogenomics predicts health resource utilization of outpatients with anxiety and depression. Transl Psychiatry. 2013;3:e242. doi: 10.1038/tp.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winner JG, Carhart JM, Altar CA, Goldfarb S, Allen JD, Lavezzari G. et al. Combinatorial pharmacogenomic guidance for psychiatric medications reduces overall pharmacy costs in a one year prospective evaluation. Curr Med Res Opin. 2015;18:1–30. doi: 10.1185/03007995.2015.1063483. [DOI] [PubMed] [Google Scholar]
- CGS Administrators. MoPath: GeneSight Assay for Refractory Depression (L35437) MediQuant.com [Internet] Available from: http://www.mediquant.com/policy/L35437_20141024.pdf .
- Tansey KE, Guipponi M, Hu X, Domenici E, Lewis G, Malafosse A. et al. Contribution of Common Genetic Variants to Antidepressant Response. Biol Psychiatry. 2013;73:7–679. doi: 10.1016/j.biopsych.2012.10.030. [DOI] [PubMed] [Google Scholar]