Abstract
Tripartite motif (TRIM) protein TRIM5 of the primate species restricts replication of HIV and other retroviruses. Whereas primates have a single TRIM5 gene, the corresponding locus in the mouse has expanded during evolution, now containing more than eight related genes. Owing to the complexity of the genomic organization, a mouse homolog of TRIM5 has not been fully studied thus far. In the present study, we report that Trim12c (formerly Trim12-2) encodes a TRIM5-like protein with a ubiquitin ligase activity. Similar to the primate TRIM5, TRIM12c is expressed in the cytoplasm as a punctate structure and induced upon IFN and pathogen stimulation in macrophages and dendritic cells. We show that TRIM12c interacts with TRAF6, a key protein in the pathogen recognition receptor signaling, and reciprocally enhances their ubiquitination, leading to cooperative activation of IFN and NF-κB pathways. This study identifies TRIM12c as a mouse TRIM5 equivalent, critical for host innate immunity.
Introduction
Proteins of the tripartite motif (TRIM) superfamily are conserved throughout metazoans and play diverse biological roles (1–3). Many TRIM proteins have characteristic four-domain composition with the RING, B box, coiled-coil, and PRY/SPRY (B30.2) domains. The N-terminal RING domain is attributed to a putative ubiquitin ligase activity, and the C-terminal PRY/SPRY domain is associated with retroviral restriction.
A number of TRIM genes are tandemly arrayed in a genomic region designated the TRIM cluster on chromosome 7 in mice and chromosome 11 in humans. Of the TRIM cluster members, TRIM5α (hereafter TRIM5) has been intensely studied, partly because rehesus monkey TRIM5 was shown to potently restrict HIV life cycle (4–6). TRIM5 in humans, cows, and rabbits also carries antiretroviral activities, which is accounted for by the SPRY domain (7–13). Although humanTRIM5 has a weak activity to restrict HIV replication, a recent report showed that it interacts with TAK1 kinases to activate NF-κB pathway and confers a generalized antiviral state upon the host (14). Similar to many other TRIM proteins, TRIM5 of primates and other species is induced by IFNs, resulting in enhanced antiretroviral activity (15, 16). In addition to TRIM5, other proteins in the TRIM cluster have been shown to inhibit early and late stages of retroviral life cycle (17, 18). Besides retroviral restriction, a number of TRIM proteins, including those in the TRIM cluster, play pivotal roles in host defense through pathogen recognition receptor (PRR) and RIG-I/MDA signaling to regulate NF-κB and IFN pathways (19–23). Some of these TRIM proteins interact with cytoplasmic adaptors or kinases such as TAK1 or IKKs to regulate activity of NF-κB and IFN regulatory factor (IRF) 3/7 (24–28).
The TRIM5 locus of the mouse genome poses interesting, unsolved issues on the evolution of antipathogen activity associated with the TRIM family. Unlike humans and primates that have a single TRIM5 gene, this locus has expanded in the mouse genome, now harboring more than eight Trim5-like genes within a ∼250-kb DNA stretch, called the Trim5 group, that is located between Trim6 and Trim66 of the Trim cluster (29, 30) (see Fig. 1A). This expansion is thought to be a result of gene duplication and positive selection for enhanced antiviral activities (29, 30). Partly owing to the complexity of genomic organization and the presence of multiple Trim genes in this region, a functional mouse homolog of TRIM5 has not been clearly identified so far. For similar reasons, only a few Trim5 group genes, such as Trim30a, have been examined for function (20, 31).
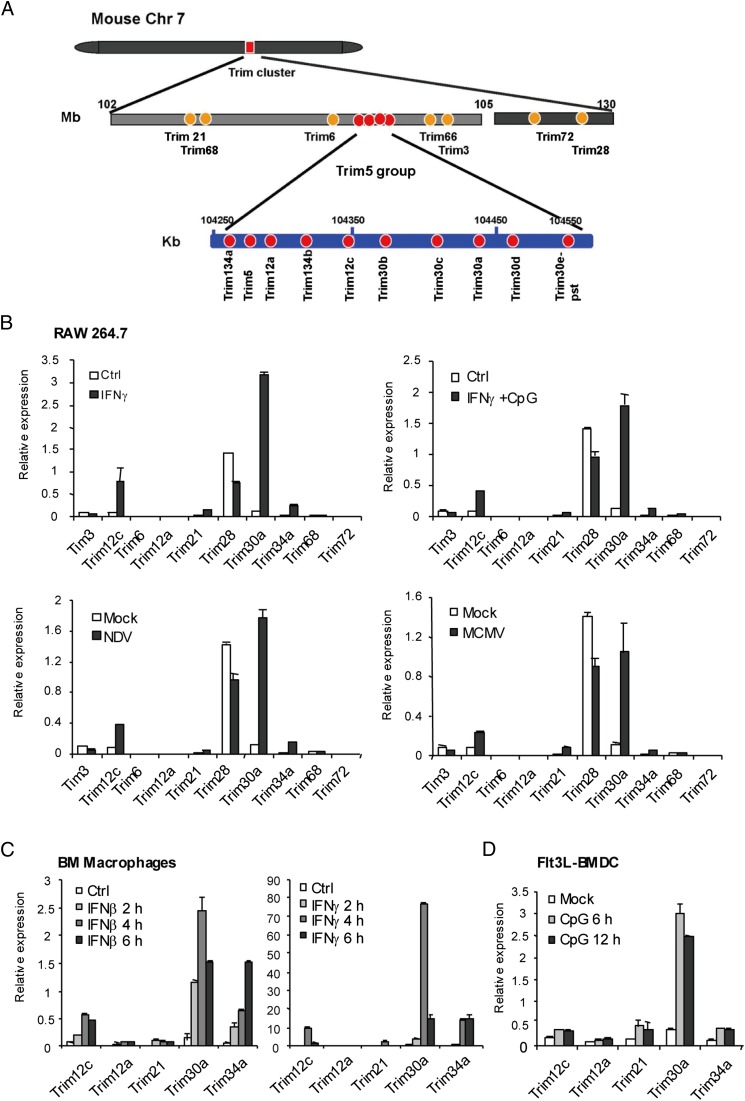
FIGURE 1.
Induction of Trim cluster genes in macrophages and DCs. (A) Schematic diagram of the Trim cluster genes and the Trim5 group region in the mouse genome. The Trim cluster encompasses ∼17 Mb in chromosome 7 (gray bar). The Trim5 region is expanded in the mouse genome to form the Trim5 group, which contains 11 Trim genes within the ∼250-kb space (blue bars with each gene in red). In this study, Trim12c has been analyzed in detail. Trim12a encodes a truncated protein identical to TRIM12c but lacking the SPRY domain. The predicted amino acid sequence of TRIM5 is virtually identical to that of TRIM12c. (B) RAW 264.7 cells stimulated with IFN-γ (100 U/ml) for 24 h, NDV at 640 HA units, or MCMV at a multiplicity of infection of 5 for 24 h or IFN-γ (overnight) plus CpG (6 h, 1 μg/ml), and mRNA levels of indicated Trim genes were measured by qRT-PCR. (C) BM-derived macrophages were stimulated by IFN-β or IFN-γ for indicated times, and mRNA levels of indicated Trim genes were tested as above. (D) Flt3L-derived BMDCs were stimulated with CpG (1 μg/ml) for indicated times, and Trim gene expression was detected as above. mRNA levels of Trim genes were normalized by the hypoxanthine–guanine phosphoribosyltransferase mRNA. Values in this figure represent the average of three independent experiments ± SD.
In this study, we cloned a full-length Trim12c cDNA, among other genes mapped to the Trim5 group. TRIM12c protein has a typical four-domain composition and is structurally very similar to TRIM5 of primates and other species. Trim12c is induced by IFNs and viral infection in macrophages and dendritic cells (DCs), and the protein displays characteristic cytoplasmic localization similar to TRIM5. We show that TRIM12c is a ubiquitin ligase that can interact with TNFR-associated factor (TRAF) 6, a central regulator of NF-κB and IFN pathways. This interaction enhanced ubiquitination of both proteins, leading to increased activation of type I IFN and NF-κB pathways. Collectively, this study identifies TRIM12c as a mouse homolog of TRIM5 with E3 ubiquitin ligase activity through which it activates proinflammatory and IFN responses.
Materials and Methods
Cells and reagents
RAW 264.7 cells (ATCC TIB-71), 293T cells (ATCC CRL-1660) and NIH3T3 cells (ATCC CRL-1658) were grown in DMEM supplemented with 10% FBS. Bone marrow (BM)-derived macrophages and BM-derived DCs (BMDCs) from C57BL/6 mice were grown in the presence of M-CSF or Flt3L as described (32). All mouse procedures were performed according to the protocol approved by the Animal Care and Use Committee of the National Institute of Child Health and Human Development (11-044). Polyinosinic-polycytidylic acid [poly(I:C)], CpG oligodeoxynucleotide 1826 (hereafter CpG), and Escherichia coli–derived bacterial LPS were obtained from InvivoGen and Sigma-Aldrich. Anti-mouse TRIM5/TRIM12c was purchased from R&D Systems (catalog no. AF3550). Abs against Flag-M2, hemagglutinin (HA), and V5 were from Sigma-Aldrich, Roche, and Invitrogen, respectively. Anti-TRAF6 Ab (sc-8409) and anti-IRF3 (sc9086) were from Santa Cruz Biotechnology. Anti-IRF3 pS386 was from Epitomics (catalog no. 2562-1). For transfection, Lipofectamine 2000 from Invitrogen or Dotap from Roche was used. Newcastle disease virus (NDV) and mouse CMV (MCMV) infection were described previously (33, 34).
Cloning of TRIM cDNAs
Full-length cDNAs for Trim5 group genes were cloned from RAW 264.7 cells or BMDCs (both 1 × 106 cells) stimulated with IFN-γ (100 U/ml) for 24 h. First-strand cDNAs were synthesized from 5 μg total RNA prepared by the RNeasy kit (Qiagen) or TRIzol (Invitrogen) and used to amplify full-length cDNA for Trim genes by conventional PCR and cloned into the pcDNA3.1-V5 vector or pmCherry-C1 vector (Clontech) using primers listed in Supplemental Table I. pcDNA3.1 vectors (Invitrogen) containing Flag-tagged mouse IRF3 or IRF7 were described (33, 35). TRAF6 expression vector in p3xFlag-CMV-7.1 (Sigma-Aldrich) was constructed from pUNO-mTRAF6 expression plasmid (InvivoGen). The point mutants of TRIM12c, C15S, and TRIM12c deletions lacking individual domains were constructed in pCDNA3.1-V5 vector. The point mutants of TRAF6 and C70S were constructed in p3xFlag-CMV-7.1 vectors and pUNO TRAF6 vector by site-directed mutagenesis. The stable short hairpin RNA (shRNA) vectors for TRIM12c were constructed in the pSUPER-retro vector (Oligoengine) by inserting TRIM12c-specific or control sequences. The pcDNA3.1-V5 TRIM12c knockdown-resistant (KDR) and TRIM12c C15S KDR constructs were generated from pcDNA3.1-V5 TRIM12c and TRIM12c C15S vectors by single primer amplification–based site-directed mutagenesis within the shRNA target site by using Pfx50 DNA polymerase (Invitrogen). The PCR products were digested by DpnI to remove original vectors and then transformed in XL1-Blue competent cells (Agilent Technologies). The cloned KDR vectors were validated by sequencing. Primers used for cloning, construction of shRNA, or quantitative RT-PCR (qRT-PCR) are listed in Supplemental Table I. The retroviruses for TRIM12c shRNA were produced from 293T cells cotransfected with the above vector along with the packaging vector, pCL10A1 (Imgenex). DCs, macrophages, or NIH3T3 cells were transduced with the retrovirus by spinoculation twice during 2 consecutive days and selected by puromycin (2 μg/ml) for 5 d.
Luciferase reporter analysis and qRT-PCR
293T cells (1 × 105) were transfected with pGL luciferase reporters driven by the IFN-β or NF-κB promoters along with pRL-TK used as an internal control for 24 h (28, 33). In certain experiments the above reporters were cotransfected with expression vectors for TRIMs, IRF3, IRF7, IRF8, or TRAF6 for 24 h. The luciferase activities were detected by Dual-Luciferase assays (Promega) and normalized by activity of Renilla. qRT-PCR was performed as described (33); the primers are listed in Supplemental Table I.
Coimmunoprecipitation, immunoblot, and ubiquitin conjugation assay
Whole-cell extracts were prepared from 106 cells in 400 μl lysis buffer (1% Nonidet P-40, 50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 2.7 mM KCl). Extracts (20 μg) were resolved on 4–12% NuPAGE (Invitrogen) or 10% SDS-PAGE for immunoblotting. For ubiquitination assays, 293T cells were transfected with the expression vector for HA-ubiquitin (35) and TRIM12c or TRAF6 for 30 h. Extract preparations and immunoblotting used for ubiquitination assays were described previously (32, 36). Briefly, extracts were prepared from cells incubated with 20 μM MG132 in the last 4 h in 400 μl SDS buffer (2% SDS, 50 mM Tris-HCl [pH 7.5], 20 mM N-ethylmaleimide plus complete protease inhibitor mixture [Roche]). After sonication, extracts were immunoprecipitated with Ab bound to protein A/G beads (Santa Cruz Biotechnology) or anti-Flag M2–bound beads (Sigma-Aldrich) in renaturation buffer (0.5% Nonidet P-40, 50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5 mM 2-ME plus complete protease inhibitor mixture). The immunoprecipitates were eluted by sample buffer (2% SDS, 50 mM Tris-HCl [pH 6.7], 150 mM NaCl, 20 mM 2-ME, 10% glycerol) and separated in 4–12% NuPAGE (Invitrogen) or 10% SDS-PAGE for immunoblotting with indicated Abs.
Immunostaining and microscopy
NIH3T3 cells (1× 105) were transfected with 1 μg expression plasmid containing TRIM gene cDNA fused to mCherry fluorescence protein for 24 h and then fixed with 4% paraformaldehyde. The red fluorescence of mCherry (excitation, 581 nm; emission, 610 nm) was visualized by fluorescence microscopy with ×400 magnification (Zeiss, Observer A1).
Assay for antiretroviral activity
293T cells were transfected with pMSCV-GFP (Clontech) and pCL-10A1 with and without V5-TRIM12c and TRIM12c C15S, and GFP signals were detected by fluorescence microscopy and flow cytometry analysis. 293T supernatants containing pMSCV-GFP viral particles were used to infect NIH3T3 cells, and GFP signals were detected by fluorescence microscopy and flow cytometry analysis with FACSCalibur.
Results
Mouse Trim12c encodes a TRIM5 homolog that is expressed in macrophages and DCs
The mouse genome contains more than eight Trim5 group genes mapped within the TRIM cluster in chromosome 7 (Fig. 1A) (29, 30). In the updated National Center for Biotechnology Information genome sequence database, this region contains Trim5 (formerly Trim12-1), Trim12a (formerly Trim12), Trim12c (formerly Trim12-2), Trim34a, and Trim30b. For the unusual genomic organization of this region, Trim5 group genes have not been fully studied to date. We isolated a full-length Trim12c cDNA from Flt3L-derived BMDCs, based on the sequence information of the 9230105E10Rik clone, which was available at the beginning of this study. The Trim12c clone we isolated encodes a predicted protein of 497 aa and is identical to the TRIM12c in the National Center for Biotechnology Information database (gene ID 319236, updated MGI:4821183; updated on May 4, 2015) (see Supplemental Fig. 1A and 1B for orthology and sequence similarity among TRIM5 proteins of various species). The nucleotide sequence of Trim12c is 96% identical to another gene in the group Trim5, with changes present mostly in the third codon of the last 600 nucleotides (National Center for Biotechnology Information database, gene ID 667823, MGI:3646853). The TRIM12c amino acid sequence is virtually identical to that of the mouse TRIM5, with three possible changes in the SPRY domain, which may possibly be attributed to sequencing variability. We also isolated full-length TRIM12a, TRIM21, TRIM30a, and TRIM34a from Flt3L-derived BMDCs. However, our cloning effort did not yield a clone that precisely matched Trim5/Trim12-1. This may be due to poor expression of Trim5/Trim12-1 reported previously (29). Interestingly, the amino acid sequence of TRIM12a, a truncated protein without the SPRY domain, is 100% identical to that of TRIM12c. Because Trim30a has been studied for function and is highly induced in macrophages similar to Trim12c (see below), we compared the amino acid sequences of the two proteins (20). Analysis by the T-Coffee software (http://tcoffee.vital-it.ch/apps/tcoffee/index.html) indicated that the two proteins share 56% amino acid identity, significantly less than the similarity between Trim5 and Trim12a (Supplemental Fig. 1C). The difference was more prominent in the C-terminal B30.2/SPRY domain than other domains. These results may suggest rapid divergence of the Trim5 group genes within Mus musculus evolution.
To assess the activity of TRIM cluster genes, we tested expression of 10 genes in macrophages and DCs (Fig. 1B). In the macrophage cell line RAW 264.7, only Trim28 was constitutively expressed, with all other genes at the background level. However, Trim12c and Trim30a, Trim21, and Trim34a were induced at high levels after stimulation with IFN-γ, IFN-γ plus CpG, and infection by NDV or MCMV. Other Trim members, including Trim3, Trim6, Trim12a, Trim68, or Trim 72, were only modestly induced by these stimulations. In agreement with these results, Trim12c, Trim21, Trim30a, and Trim34a transcript expression was induced in primary BM-derived macrophages after IFN-β and IFN-γ stimulation (Fig. 1B). Likewise, these Trim genes were induced in primary BMDCs following stimulation by a TLR ligand, CpG (Fig. 1C). Other TLR ligands, LPS and poly(I:C), also induced Trim12c expression in BMDCs (Supplemental Fig. 2A). Thus, TRIM12c is expressed in macrophages and DCs in response to IFN-γ and virus infection. Immunoblot analysis with a commercially available Ab against the predicted mouse TRIM5/TRIM12c showed induced protein expression in BMDCs after viral infection, indicating that transcript induction results in protein expression (Supplemental Fig. 2B).
TRIM12c protein exhibits cytoplasmic body–like structures
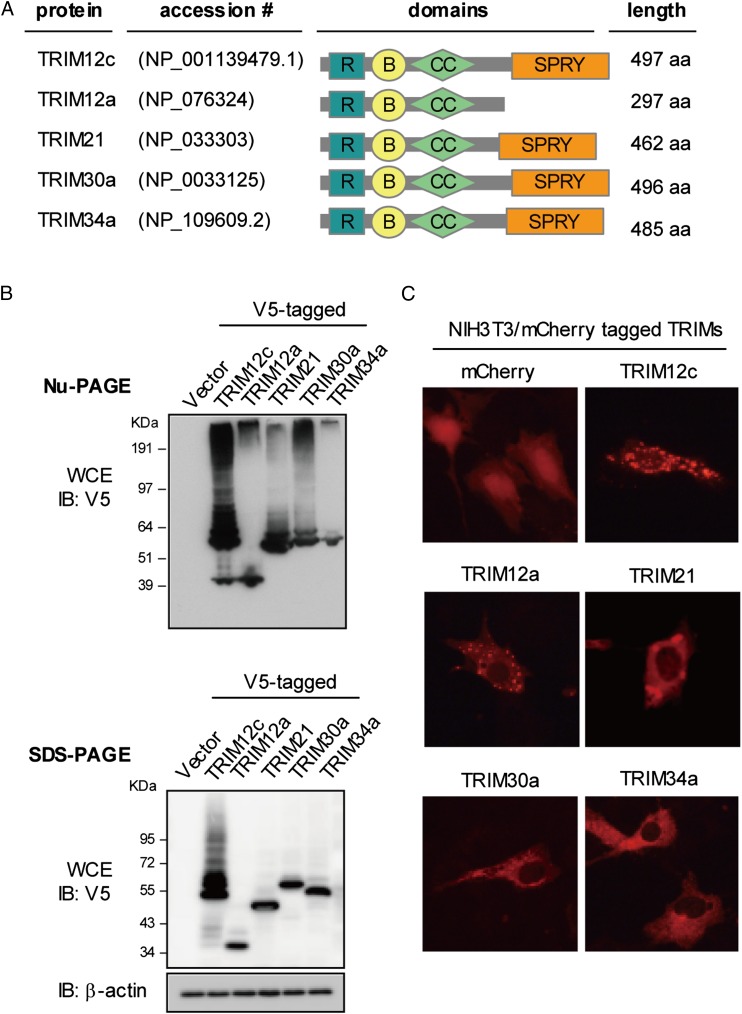
To study the protein expression, the above Trim genes were cloned in expression vectors tagged with V5 or mCherry fluorescent protein and transfected in 293T cells (Fig. 2A). As shown in Fig. 2B of NuPAGE and SDS-PAGE analyses, all TRIM clones produced proteins of expected size, including TRIM12c, which migrated to a position corresponding to ∼53 kDa. The upward trailing may indicate the formation of multimers and/or posttranslational modifications (see below). As expected, other TRIM proteins migrated to a similar position. TRIM12a, however, migrated faster to ∼40 kDa, consistent with the truncation of the SPRY domain (see diagram of domains in Fig. 2A). The mobility of TRIM21, TRIM30a, and TRIM34a was slightly different in the two gel systems, the basis of which is not clear at present. We then examined intracellular localization of these TRIM proteins (Fig. 2C). All mCherry-tagged TRIM proteins localized predominantly to the cytoplasm, whereas free mCherry distributed to all regions in the cells. Interestingly, TRIM12c exhibited prominent punctuate structures throughout the cytoplasm. These structures resemble the cytoplasmic bodies reported for TRIM5 of the human and rhesus macaque, suggesting a similarity with TRIM5 (37, 38). Interestingly, TRIM12a and truncated TRIM12c also showed cytoplasmic body–like structures, whereas other TRIM proteins were diffusely distributed in the cytoplasm. These data indicate that the formation of cytoplasmic body–like structures is dependent on the N-terminal regions of TRIM12c, but not the SPRY domain.
FIGURE 2.
Subcellular distribution of cloned TRIM5 group proteins. (A) Domain composition of TRIM5 group proteins. The protein accession numbers in GenBank are indicated. (B) 293T cells (1 × 106) were transfected with the plasmids containing V5-tagged TRIM cDNAs (2 μg) for 30 h, and whole-cell extracts (WCE, 50 μg) were resolved on on 4–12% NuPAGE (upper panel) or 10% SDS-PAGE (lower panel) for immunoblotting assay with anti-V5 Ab. (C) NIH3T3 cells were transfected with indicated mCherry florescence protein–tagged TRIM genes cloned in pmCherry-C1 vector (1 μg) for 24 h and visualized by fluorescence microscopy at ×400 magnification. The representative images for each TRIM protein are shown. Of >200 cells inspected, >90% of TRIM12c-transfected cells exhibited cytoplasmic body–like structures.
TRIM12c activates type I IFN and NF-κB pathways
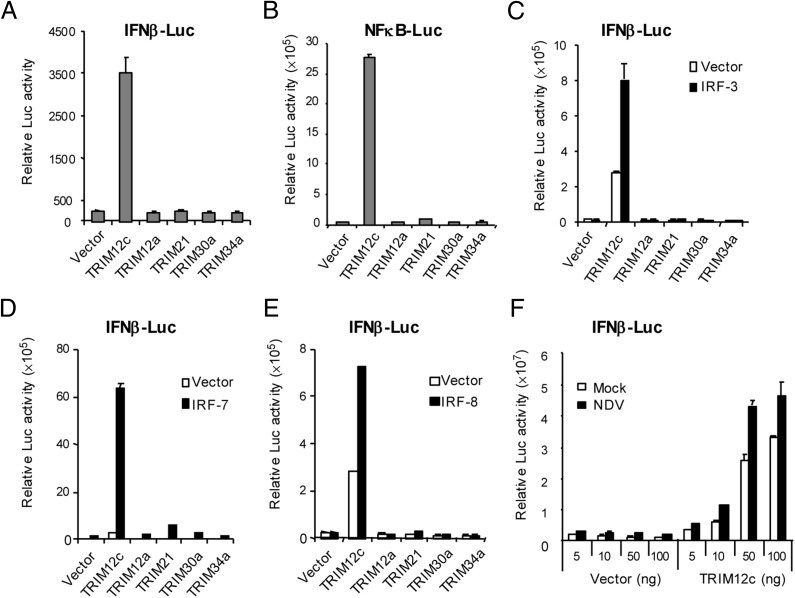
Some TRIM proteins regulate transcription of type I IFN– and NF-κB–dependent genes by acting within the TLR signaling pathway, whereas others regulate NF-κB–dependent transcription through TAK1 (14, 19, 20, 22). We tested whether TRIM12c regulates either the type I IFN or NF-κB pathway or both. The two pathways were tested, because TLRs and other PRRs activate IFN and NF-κB pathways through shared as well as distinct routes (24, 25). Cells transfected with expression vectors for TRIM12c and other TRIMs were tested for activity of luciferase reporters driven by type I IFN-β or NF-κB promoter. As seen in Fig. 3A and 3B, TRIM12c activated both IFN-β and NF-κB reporter activity by >10-fold. In contrast, other TRIMs tested (TRIM12a, TRIM21, TRIM30a, TRIM34a) did not activate either reporter to a measurable level. Immunoblot data in Supplemental Fig. 2C verified that TRIM12c and other TRIM members were expressed at similar levels. As shown in Fig. 3C–E, cotransfection of TRIM12c along with IRF3, IRF7, and IRF8 led to greater reporter activities relative to TRIM12c alone. None of the other TRIM members significantly enhanced promoter activity in the presence of the IRFs (note equivalent expression of IRFs and TRIM members in Supplemental Fig. 2C). In Fig. 3F, IFN-β reporter activity was tested after NDV infection with or without TRIM12c. Reporter activity was significantly increased when cells were cotransfected with TRIM12c in a dose-dependent manner. These results indicate that TRIM12c activates both IFN and NF-κB pathways of transcription in response to PRR signaling.
FIGURE 3.
TRIM12c activates IFN-β and NF-κB promoter activity. 293T (1 × 105) cells were transfected with indicated TRIM expression vectors (pcDNA3.1 V5-tagged, 200 ng for TRIM12c and 400 ng for other TRIM vectors), along with 400 ng IFN-β–Luc (A and C–F) or NF-κB–Luc reporter (B) and pRL-TK (40 ng). In (C)–(E), cells were transfected with above TRIM constructs along with 10 ng pcDNA3.1 Flag-IRF3 or IRF7 plasmids. See the levels of TRIM and IRF proteins expressed in transfected cells in Supplemental Fig. 2C. (F) Cells were transfected with increasing amounts of TRIM12c and vector (5, 10, 50, and 100 ng), IFN-β–Luc (400 ng), and pRL-TK (40 ng) for 24 h, and infected with NDV (640 HA units) for 24 h. Values represent the average of three independent experiments ± SD.
TRIM12c stimulates expression of endogenous type I IFN and inflammatory cytokine genes
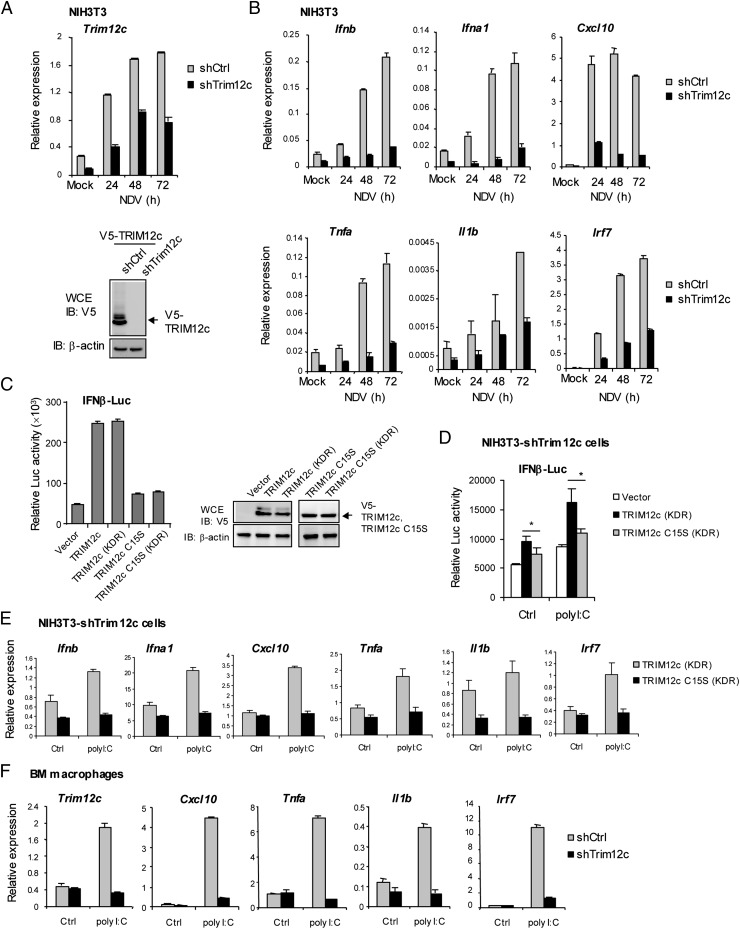
To further validate the role of TRIM12c in stimulating type I IFN and NF-κB pathways, we stably knocked down TRIM12c expression in NIH3T3 cells by shRNA and tested for expression of various genes stimulated by NDV. Trim12c mRNA was expressed in NIH3T3 cells as reported, and it strongly induced after NDV stimulation (29) (Fig. 4A). Trim12c shRNA, but not control shRNA, reduced Trim12c mRNA expression >50% both in untreated and NDV-stimulated cells (Fig. 4A, left panel). Immunoblot analysis of cells expressing V5-tagged TRIM12c showed that Trim12c shRNA, but not control shRNA, inhibited TRIM12c protein expression (Fig. 4A, right panel). As shown in Fig. 4B, NDV infection led to robust induction of type I IFN (Ifnb, ifna1) and proinflammatory genes (Cxcl10, Tnfa, Il1b) as well as Irf7 in cells expressing control shRNA. However, induction of these genes was markedly lower in cells expressing TRIM12c shRNA, indicating that TRIM12c increases expression of endogenous cytokine genes. To rule out possible shRNA off-target effects, we performed rescue experiments using KDR constructs for TRIM12c as well as TRIM12c C15S, a RING domain point mutant that lacks ubiquitin ligase activity (see Materials and Methods). As expected, TRIM12c and TRIM12c KDR both enhanced IFN-β luciferase activity, whereas TRIM12c C15S and TRIM12c C15S KDR did not, confirming functionality of the KDR constructs (Fig. 4C, left panel). Further, immunoblotting data showed similar protein expression levels in the original and KDR constructs (Fig. 4C, right panel). Rescue experiments were performed with NIH3T3 cells expressing shTrim12c where transfection of TRIM12c KDR, but not TRIM12c C15S KDR, substantially enhanced IFN-β reporter activity (Fig. 4D). Moreover, expression of type I IFN and inflammatory cytokine genes was also rescued in NIH3T3 cells expressing Trim12c shRNA upon transfection of TRIM12c KDR, but not TRIM12c C15S KDR (Fig. 4E). These data discounted nonspecific effects of Trim12c shRNA, and they supported the possibility that TRIM12c with intact ubiquitin ligase activity plays a key role in stimulating type I IFN and cytokine responses. We also examined the effect of Trim12c shRNA in BM-derived macrophages (Fig. 4F). We found that induction of Cxcl10, Tnfa, Il1b, and Irf7 mRNA by poly(I:C) was greatly reduced in cells expressing Trim12c shRNA relative to cells expressing control shRNA. Additionally, Trim12c shRNA attenuated CpG induction of endogenous type I IFN and inflammatory cytokine genes in DCs (Supplemental Fig. 3A). Further supporting the role of TRIM12c in stimulating type I IFN transcription, Trim12c shRNA inhibited induction of IFN-β promoter activity stimulated by NDV (Supplemental Fig. 3B). Interestingly, Trim12c shRNA also inhibited IFN-β promoter activity stimulated by the mitochondrial antiviral signaling protein (MAVS), which may support the view that Trim12c acts through PRR signaling (39, 40) (Supplemental Fig. 3C). Taken together, these results establish that TRIM12c is a positive regulator of type I IFN– and NF-κB–dependent transcription.
FIGURE 4.
TRIM12c knockdown attenuates type I IFN and proinflammatory cytokine induction. (A) NIH3T3 cells (1 × 104) were transduced with shRNA retrovirus vector for TRIM12c or control vector, followed by puromycin (1 μg/ml) selection. The transduced cells were infected with NDV at 640 HA units for 24 h. Trim12c knockdown efficiency was determined by qRT-PCR (left panel). In the right panel, cells transduced with the above shRNA vectors were transfected with pcDNA3.1-V5-TRIM12c (2 μg) for 30 h, and the whole-cell extracts were analyzed for immunoblotting with anti-V5 Ab. β-Actin was tested as a loading control. (B) NIH3T3 cells (1 × 105) transduced with TRIM12c or control (Ctrl) shRNA were infected with NDV at 640 HA units for indicated times and mRNA levels of the indicated endogenous cytokine genes were measured by qRT-PCR. Values represent the means of three independent experiments ± SD. (C) Left, 293T (1 × 105) cells were transfected with TRIM12c, TRIM12c C15S, and their respective KDR vectors (pcDNA3.1 V5-tagged, 1 μg), along with 600 ng IFN-β–Luc and 60 ng pRL-TK for Dual-Luciferase assay. Right, Immunoblotting analysis of the TRIM protein expression. (D) NIH3T3 cells (1 × 104) transduced with shTRIM12c vector were cotransfected with TRIM12c KDR or TRIM12c C15S KDR expression vectors (1 μg), 600 ng IFN-β–Luc and 60 ng pRL-TK, and followed by poly(I:C) (100 μg/ml) for 6 h, and luciferase activity was measured as above. Values represent the mean of three independent experiments ± SD with two-tailed Student t test. *p < 0.05. (E) NIH3T3 cells transduced with shTRIM12c and cotransfected with TRIM12c KDR or TRIM12c C15S KDR (1 μg) were stimulated by poly(I:C) (100 μg/ml) for 6 h, and mRNA expression of indicated genes was measured by qRT-PCR. Values represent the mean of three independent experiments ± SD. (F) BM-derived macrophages transduced with Trim12c shRNA or control (Ctrl) shRNA were stimulated by poly(I:C) (100 μg/ml) for 8 h and mRNA levels of the indicated genes was measured by qRT-PCR. Values represent the mean of three determinations ± SD.
TRIM12c and TRAF6 interact with each other and cooperatively stimulate type I IFN and NF-κB transcription
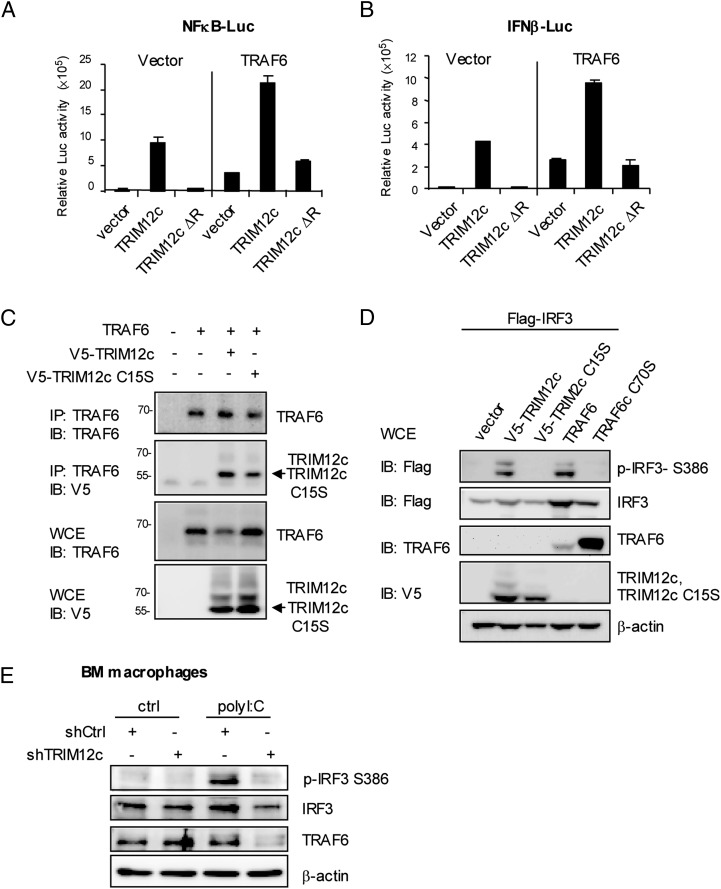
In light of the results that TRIM12c stimulates both arms of transcription, we surmised that TRIM12c acts on an upstream step within the PRR signaling that is common in both type I IFN and NF-κB pathways. PRR signaling, upon ligand binding, first activates adaptors and other upstream molecules and then TRAF6 (and TRAF3 in some cases) (24, 25, 41). TRAF6 is central to PRR signal transduction, as it activates IKK kinases to initiate both NF-κB and IFN pathway transcription (24, 25, 41). To address whether TRIM12c stimulates these pathways by working with TRAF6, we cotransfected TRIM12c and TRAF6 and performed reporter assays for NF-κB and IFNβ cells. As shown in Fig. 5A and 5B, cotransfection of TRIM12c and TRAF6 augmented activity of both reporters more than each factor alone, indicating that the two factors functionally cooperate with each other. This cooperation was, however, not observed when TRIM12c ΔR, a deletion lacking the putative ubiquitin ligase domain, was cotransfected. Supporting functional cooperation, Trim12c shRNA reduced reporter activation by TRAF6 alone by nearly 70% (Supplemental Fig. 3D). These data led us to test the possibility that the two proteins interact with each other. Coimmunoprecipitation assays in Fig. 5C showed that TRAF6 coprecipitated V5-tagged wild-type TRIM12c and the TRIM12c mutant, TRIM12c C15S. Immunoblot analysis of whole-cell extracts verified that TRAF6 and TRIM12c were expressed as expected, indicating that TRAF6 binds to TRIM12c irrespective of its E3 ligase activity.
FIGURE 5.
TRIM12c and TRAF6 interact with each other and cooperatively stimulate type I IFN and NF-κB transcription. (A and B) 293T cells (1 × 106) were transfected with wild-type TRIM12c or TRIM12c with the RING domain deletion (TRIM12c ΔR, 2 μg) with or without TRAF6 vector (2 μg) for 30 h, and reporter activity was measured as in Fig. 3. The values represent the average of three independent experiments ± SD. (C) 293T cells (1 × 106) were cotransfected with the expression vectors pUNO-TRAF6 (2 μg), pcDNA3.1 V5-TRIM12c (2 μg), and TRIM12c C15S (2 μg) for 30 h. Whole-cell extracts (WCE) were imunoprecipitated with anti-TRAF 6 Ab and immunoblotted for TRAF6 or V5-TRIM12c. In lower panels, WCE were immunoblotted for TRAF6 or V5-TRIM12. The position of TRAF6 and TRIM12c is indicated in the right. (D) 293T cells (2 × 106) were cotransfected with pcDNA3.1-Flag-IRF3 (4 μg), pcDNA3.1 V5-TRIM12c (4 μg), TRIM12c C15S (4 μg), pUNO-TRAF6 (4 μg), and pUNO-TRAF6 C70S (4 μg) for 36 h, and WCE were immunoblotted with indicated Ab. (E) BM-derived macrophages (2 × 106) expressing Trim12c or control (Ctrl) shRNA were stimulated by poly(I:C) (100 μg/ml) for 8 h. WCE (50 μg) were immunoblotted with indicated Ab.
To assess the significance of Trim12c–TRAF6 interaction, we next examined phosphorylation of IRF3, an event downstream of PRR signaling that is critical for type I IFN induction. Immunoblot analysis in Fig. 5D (top) showed that Flag-IRF3 was phosphorylated when coexpressed with wild-type TRIM12c and TRAF6. However, IRF3 was not phosphorylated when coexpressed with the E3 ligase mutants TRIM12 C15S and TRAF6 C70S. Consistent with the role of TRIM12c in IRF3 activation, Trim12c shRNA greatly diminished phosphorylation of IRF3 in BM-derived macrophages observed after poly(I:C) stimulation (Fig. 5E). These data reveal that TRIM12c and TRAF6 cooperatively stimulate NF-κB and IFN promoter activity by interacting with each other.
TRIM12c is a ubiquitin ligase that stimulates TRAF6 ubiquitination and vice versa
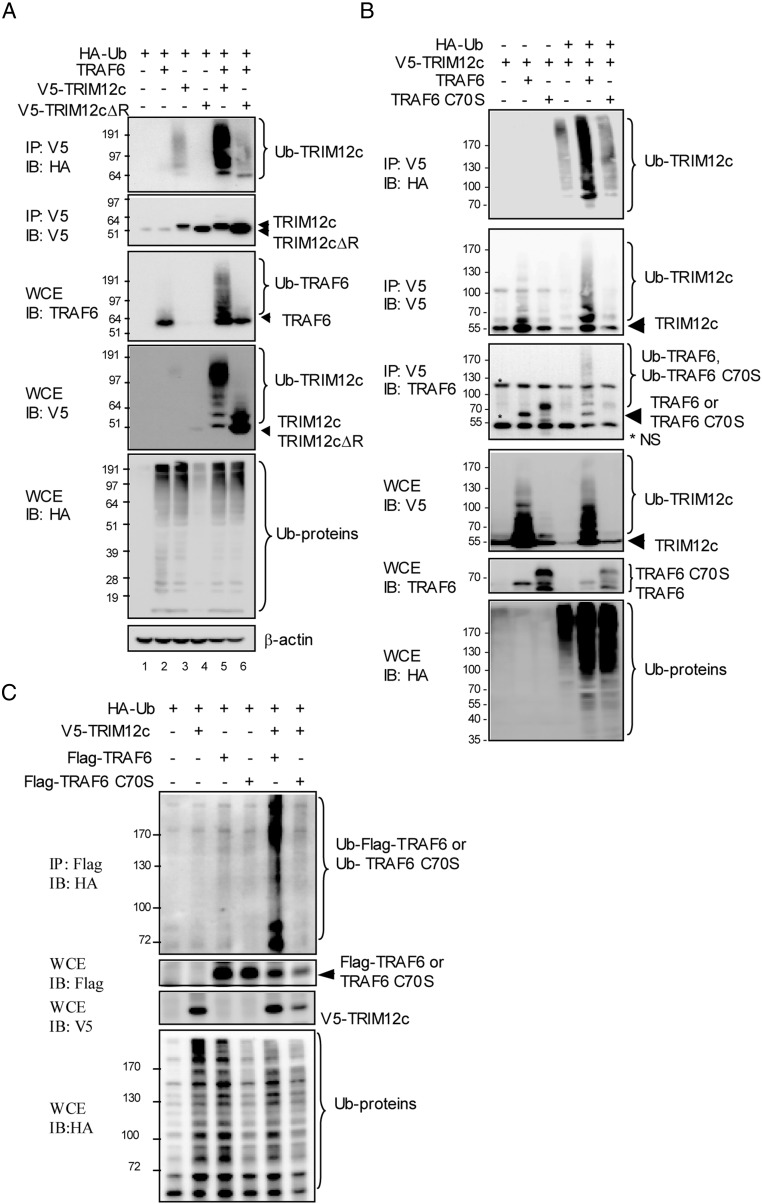
By analogy with TRIM5, TRIM12c may be expected to have an E3 ubiquitin ligase activity (14, 42, 43). TRAF6 is also an E3 ubiquitin ligase, important for PRR signaling (44). We performed in vivo ubiquitin conjugation assays to test ubiquitin ligase activity of the two proteins. Cells transfected with HA-tagged ubiquitin (HA-Ub), TRAF6, V5-tagged TRIM12c, or V5-TRIM12c ΔR were immunoprecipitated with anti-V5 Ab and analyzed by immunoblot for HA-Ub. Data in Fig. 6A (top panel) showed that wild-type TRIM12c, but not TRIM12c ΔR lacking the putative ligase domain, extensively conjugated HA-Ub, indicating TRIM12c autoubiquitination. Importantly, ubiquitination of TRIM12c was markedly increased in the presence of TRAF6. Analysis of whole-cell extracts (Fig. 6A, third panel) showed that TRAF6 was not ubiquitinated significantly when expressed alone under these conditions. However, TRAF6 was copiously ubiquitinated in the presence of wild-type TRIM12c, but not TRIM12c ΔR. Immunoblot of total extracts also showed broad ubiquitination of cellular proteins by TRIM12c and TRAF6 (Fig. 6A, bottom panel). These results indicate that TRIM12c ubiquitinates itself, TRAF6, and other cellular proteins, and that TRIM12c and TRAF6 mutually enhance their ubiquitination.
FIGURE 6.
TRIM12c is a ubiquitin ligase that stimulates TRAF6 ubiquitination and vice versa. (A) 293T cells (1 × 106) were transfected with HA-Ub (0.2 μg), pUNO-TRAF6, V5-TRIM12c, and V5-TRIM12c ΔR (2 μg) for 30 h. Upper two panels, Whole-cell extracts (WCE) (20 μg) were immunoblotted with indicated Ab. Lower two panels, WCE were immunoprecipitated with anti-V5 Ab and immunoblotted with anti-HA or anti-V5 Ab. (B) Cells were transfected with HA-Ub (0.2 μg), pUNO-TRAF6, pcDNA3.1-V5-TRIM12c, and V5-TRIM12c C15S for 30 h followed by MG132 (20 μM) treatment for 4 h. WCE were immunoprecipitated with anti-V5 Ab and immunoblotted with anti-HA or anti-V5 Ab. In the lower panels, WCE were immunoblotted with indicated Abs. (C) Cells were transfected with HA-Ub (0.2 μg), p3XFlag-TRAF6 or TRAF6 C70S (2 μg), and pcDNA3.1-V5-TRIM12c or V5-TRIM12c C15S (2 μg) for 30 h, and the cells were treated by MG132 (20 μM) for 4 h. In the top panel, WCE were immunoprecipitated with anti-Flag Ab and immunoblotted with anti-HA Ab. In the lower panels, WCE were immunoblotted with indicated Ab.
To further analyze the interaction of TRIM12c and TRAF6 and mutual ubiquitination, we performed similar assays using cells expressing wild-type TRAF6 and the catalytic mutant TRAF6 C70S along with V5-tagged TRIM12c and HA-Ub (Fig. 6B). As shown in the top panel, TRIM12c ubiquitin conjugation was markedly increased when coexpressed with wild-type TRAF6, but not with the TRAF6 C70S mutant. Moreover, TRIM12c coprecipitated both wild-type TRAF6 and the catalytic mutant (TRAF6 C70S). These data are in agreement with coimmunoprecipitation data in Fig. 5C and verify that the two proteins interact with each other, irrespective of TRAF6 catalytic activity. We noted that the interaction was clearer in the absence of HA-Ub compared with its presence (bottom second panel, lanes 2 and 3 versus lane 5 and 6). This is likely due to lower TRAF6 expression in the cells expressing HA-Ub. Analysis of whole-cell extracts confirmed that additional cellular proteins were ubiquitinated by TRIM12c and TRAF6 (Fig. 6B, bottom panel).
To corroborate mutual ubiquitination, we then tested Flag-tagged TRAF6 and the mutant for ubiquitin conjugation assay in the presence of TRIM12c. As shown in Fig. 6C (top panel), wild-type TRAF6 was modestly ubiquitinated without TRIM12c. However, with TRIM12c, ubiquitination of wild-type TRAF6 was strongly increased. However, ubiquitination was not increased for the TRAF6 catalytic mutant. Similar experiments performed with the TRIM12c mutant confirmed that cooperative ubiquitination requires the intact catalytic domain of both proteins (Supplemental Fig. 3E). These data substantiate that TRAF6 and TRIM12c boost each other’s ubiquitination.
Ubiquitin ligase activity of TRIM12c and TRAF6 is required for cooperative transcription of IFN and NF-κB pathway genes
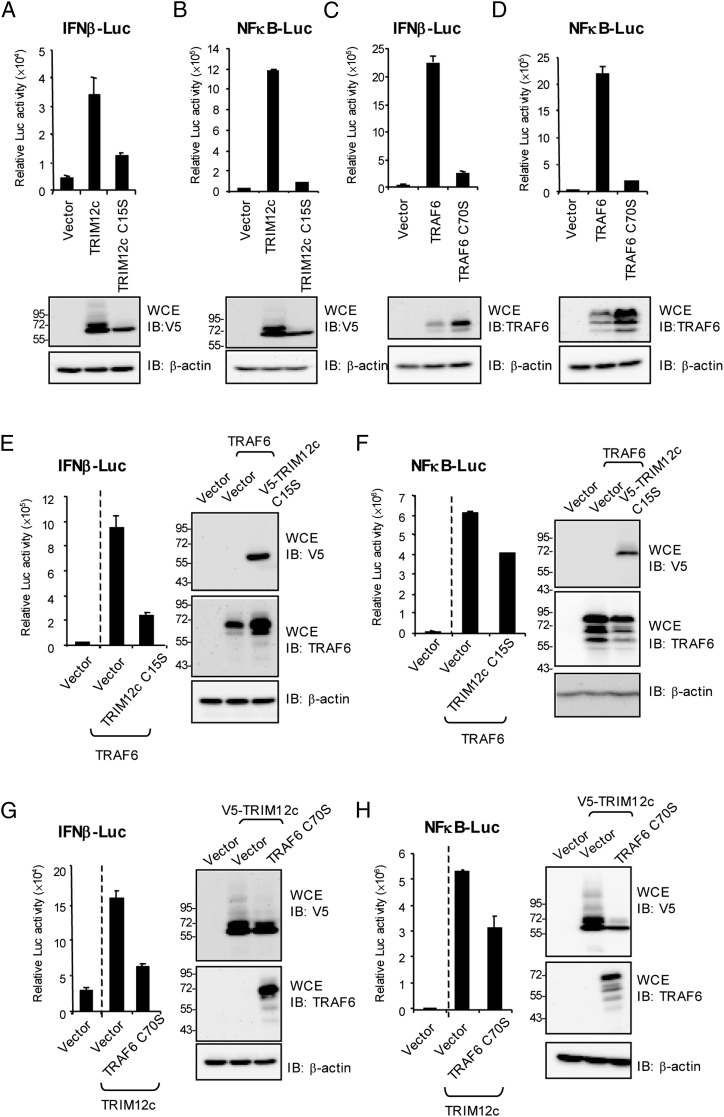
Reporter analysis was performed to ascertain whether ubiquitin ligase activity is required for TRIM12c and TRAF6 to activate type I IFN and NF-κB transcription. As shown in Fig. 7A and 7B (upper panels), wild-type TRIM12c strongly activated both IFN-β and NF-κB reporter activity. However, the catalytic mutant was much less efficient in activating these reporters. Immunoblot analysis, run in parallel (lower panels), showed that wild-type TRIM12c, but not the mutant, produced ubiquitin-conjugated molecules of slower mobility. Note that polyubiquitinated proteins were not observed in these experiments, because cells were not treated with MG132. Similarly, the wild-type TRAF6, but not the C70S mutant, enhanced activity of both reporters (Fig. 7C, 7D, upper panels). Suggesting deficiency in ubiquitin-mediated degradation, the amounts of the C70S mutant were higher than those of wild-type TRAF6 (see immunoblot data in Fig. 7C, 7D). We also found that NF-κB and IFN reporter activation by wild-type TRAF6 was inhibited when the TRIM12 mutant or TRAF6 mutant was cotransfected (Fig. 7E, 7F). Likewise, reporter activation by TRIM12c was inhibited by cotransfection of the TRAF6 mutant or TRIM12c mutant, indicating that the catalytic mutants interfered with the cooperation of endogenous TRIM12c and TRAF6 (Fig. 7G, 7H). These results further support a model where TRAF6 and TRIM12c cooperatively enhance IFN and NF-κB transcription through reciprocal ubiquitination.
FIGURE 7.
Ubiquitin ligase activity of TRIM12c and TRAF6 is required for cooperative enhancement of IFN and NF-κB transcription. (A and B) Upper panel, Cells were cotransfected with TRIM12c or TRIM12c C15S (100 ng), IFN-β–Luc reporter or NF-κB–Luc reporter (200 ng), and pRL-TK (20 ng) for 24 h, and reporter activity was measured as above. Lower panel, Whole-cell extracts (WCE) from above cells were immunoblotted with indicated Abs to confirm expression of the transfected vector for (B)–(G). (C and D) Cells were transfected with TRAF6 and TRAF6 C70S (100 ng), IFN-β–Luc or NF-κB–Luc (200 ng), and pRL-TK (20 ng) for 24 h and reporter activity was measured as above. (E and F) Cells transfected with TRAF6 (50 ng) alone or TARF6 plus TRIM12cC15S along with IFN-β–Luc or NF-κB–Luc reporter (200 ng) and pRL-TK (20 ng), or vector alone (100 ng) for 24 h and reporter activity was measured as above. (G and H) Cells were cotransfected with IFN-β–Luc or NF-κB–Luc reporter (200 ng), pRL-TK (20 ng), TRIM12c alone (100 ng), or with TRIM12c plus TRAF6 or TRAF6 C70S (50 ng) for 24 h and reporter activity was measured. Values represent the average of three independent experiments ± SD.
A number of TRIM proteins have been shown to restrict retroviral growth (2, 17, 18). Antiretroviral activity appears in some cases to be independent of its activity in PRR signaling. In particular, TRIM5 of primate species and its orthologs in cows and rabbits exhibit antiretroviral activity against HIV (4–9). It was of interest to test whether TRIM12c, a likely mouse counterpart of TRIM5, also has antiretroviral activity. We tested this question using a simple model system, the murine stem cell virus encoding GFP (pMSCV-GFP). This virus is used widely as a vector for gene expression in mammalian cells. 293T cells, when transfected with pMSCV-GFP along with the packaging vectors, showed extensive GFP signals, representing MSCV-GFP virions However, GFP signals were substantially reduced when these cells were cotransfected with TRIM12c, but not TRIM12c C15S (Supplemental Fig. 3F). Consistent with these results, immunoblotting analysis showed reduced GFP levels in TRIM12c expressing cells (Supplemental Fig. 3G). Additionally, when NIH3T3 cells were infected with the MSCV-GFP virus harvested from 293 supernatants above, the results of GFP signal expression level in NIH3T3 cells implicated that MSCV-GFP virion production was attenuated in 293T cells expressing TRIM12c, but not TRIM12c C15S (Supplemental Fig. 3H, 3I). Moreover, the infectivity of MSCV-GFP virus was again noticeably lower in NIH3T3 cells expressing TRIM12c, but not TRIM12c S15S, or vector control (Supplemental Fig. 3J, 3K). These data suggest that murine TRIM12c with ubiquitination activity possesses an antiretroviral activity, supporting evolutionary conservation of antiretroviral function associated with TRIM5.
Discussion
We show that murine TRIM12c is encoded by one of the Trim5 group genes in chromosome 7. It is structurally very similar to TRIM5 of other species, including primates, cattle, and rodents. TRIM12c is virtually identical, in the predicted amino acid sequence, to the mouse TRIM5 (listed in the National Center for Biotechnology Information/Mouse Genome Informatics database, formerly TRIM12-1). However, whereas TRIM12c is expressed at significant levels in fibroblasts, macrophages, and DCs, TRIM5 does not seem to be expressed in fibroblasts and macrophages (Ref. 29 and our observation). Thus, it may be assumed that TRIM12c is a major mouse TRIM5 homolog expressed in cells of innate immunity. Nevertheless, it is possible that TRIM5 is also active and functional in cells or stages of development not investigated in the present study. Irrespective of this issue, we found that TRIM12c is inducible by IFNs and PRR stimulation and forms cytoplasmic body–like structures, a characteristic feature of the TRIM5 protein (4, 15, 37, 45, 46). This feature is interesting, because TRIM5 cytoplasmic bodies are suggested to be relevant to the antiretroviral activity (38). Another interesting finding made in this work is that Trim12a encodes a truncated protein that is identical to TRIM12c in the N-terminal domains, but lacking the C-terminal SPRY domain. Intriguingly, whereas TRIM12a and TRIM12c are encoded by separate genes in the mouse, a similar truncated TRIM5 occurs in primates, presumably due to RNA or protein processing, which acts as a silencer of TRIM5 (47). This raises the possibility that TRIM12a also induced by IFNs and TLR signaling may act as a negative regulator of TRIM12c.
We show that TRIM12c is a strong activator of both type I IFN and NF-κB pathway transcription. Among other TRIM cluster genes tested, only TRIM12c activated both IFN and NF-κB reporters, an activity further reinforced by downregulation of endogenous type I IFN and inflammatory genes by Trim12c shRNA. This indicates that TRIM12c can broadly boost innate immunity, activating not only NF-κB, but also IFN signaling. This result is interesting, given that some TRIM proteins are reported to preferentially activate the NF-κB pathway, but possibly not the IFN pathway (20, 28, 48).
Our effort to study underlying mechanisms led us to show that TRIM12c is a ubiquitin ligase that facilitates ubiquitination of TRAF6, itself a ubiquitin ligase belonging to another E3 ligase family. Consequently, TRIM12c was found to be a substrate of TRAF6 and ubiquitinated upon interaction with TRAF6. TRAF6 is a focal point of PRR signaling, acting at the stage where PRR pathways bifurcate into separate avenues of transcription, IFN and NF-κB. TRAF6, through its ubiquitin ligase activity, activates IKKα/β critical for activation of NF-κB transcription, as well as IKKε/TBK1 important for IFN transcription by IRF3 and IRF7 (24, 25, 49–53). In addition to these activities, TRAF6 promotes the formation of unanchored polyubiquitin chains that also activate TAK1/IKK complex (54). Our observation that TRIM12c and TRAF6 can interact with each other provides a plausible physical basis of the reciprocal ubiquitination.
We show that this mutual ubiquitination is critical for activation of IFN and NF-κB transcription, observed not only by cooperative reporter activation, but by reduced activation by the ligase-dead mutants of TRIM12c and TRAF6. Taken together, our results indicate that TRIM12c forms a positive ubiquitination loop with TRAF6 and acts at the central point of PRR signaling to stimulate IFN and NF-κB pathways of transcription. In this context, Zhao et al. (48) reported that TRIM38 binds to TRAF6 through the SPRY domain and augments polyubiquitination of TRAF6, leading to downregulation of NF-κB–dependent proinflammatory responses. There are other TRIM proteins shown to act within the PRR pathways, including TRIM25, which interacts with the adaptor for the RIG-I pathway to activate both type I IFN and NF-κB transcription (19, 20). Furthermore, human TRIM5 is shown to interact with TabA/B to activate TAK1, required for activation of IKKα/IKKβ and the NF-κB pathway. Considering that TRAF6 targets TAK1/IKKα/IKKβ for activation, it is possible that TRIM5 in other species also influences TRAF6 activities. This possibility may be supported by the ability of the human TRIM5 to enhance IRF3-dependent IFN reporter activity (14). Whereas both TRIM12c and TRIM5 activate type I and NF-κB transcription, TRIM30a, another TRIM5 group protein in the mouse, is a negative regulator of the NF-κB activity that also acts on the TabA/B–TAK1 complex (20). More recent studies show that TRIM30a negatively regulates inflammasome activation in monocytes/macrophages and affects proliferation and NF-κB activation in CD4 T cells (31, 55). The mechanism by which TRIM5 and TRIM30a cause opposite functional outcomes has not been resolved at present. This dichotomy may be due to limited sequence identity between the two proteins, leading to functional diversification of Trim 5 group genes.
In conclusion, the mouse TRIM12c, structurally very similar toTRIM5 of other mammalian species, is a ubiquitin ligase that intersects with TRAF6 to activate type I IFN and NF-κB transcription pathways, leading to enhanced innate immune responses.
Supplementary Material
Acknowledgments
We thank You-Sheng Lin and Chih-Chi Cheng (Department of Medical Education and Research, Kaohsiung Veterans General Hospital) for technical assistance.
This work was supported in part by Kaohsiung Veterans General Hospital Grant VGHKS102-004 and by Ministry of Science and Technology, Taiwan Grants NSC 99-2320-B-075B-005 and MOST 104-2320-B-075B-005.
The online version of this article contains supplemental material.
- BM
- bone marrow
- BMDC
- bone marrow–derived DC
- CpG
- CpG oligodeoxynucleotide 1826
- DC
- dendritic cell
- HA
- hemagglutinin
- HA-Ub
- HA-tagged ubiquitin
- IRF
- IFN regulatory factor
- KDR
- knockdown-resistant
- MCMV
- mouse CMV
- NDV
- Newcastle disease virus
- poly(I:C)
- polyinosinic-polycytidylic acid
- PRR
- pattern recognition receptor
- qRT-PCR
- quantitative RT-PCR
- shRNA
- short hairpin RNA
- TRAF
- TNFR-associated factor
- TRIM
- tripartite motif.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Nisole S., Stoye J. P., Saïb A. 2005. TRIM family proteins: retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 3: 799–808. [DOI] [PubMed] [Google Scholar]
- 2.Ozato K., Shin D. M., Chang T. H., Morse H. C., III 2008. TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 8: 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meroni G. 2012. Genomics and evolution of the TRIM gene family. Adv. Exp. Med. Biol. 770: 1–9. [DOI] [PubMed] [Google Scholar]
- 4.Stremlau M., Owens C. M., Perron M. J., Kiessling M., Autissier P., Sodroski J. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427: 848–853. [DOI] [PubMed] [Google Scholar]
- 5.Maillard P. V., Ecco G., Ortiz M., Trono D. 2010. The specificity of TRIM5α-mediated restriction is influenced by its coiled-coil domain. J. Virol. 84: 5790–5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakuma R., Noser J. A., Ohmine S., Ikeda Y. 2007. Rhesus monkey TRIM5α restricts HIV-1 production through rapid degradation of viral Gag polyproteins. Nat. Med. 13: 631–635. [DOI] [PubMed] [Google Scholar]
- 7.Kaiser S. M., Malik H. S., Emerman M. 2007. Restriction of an extinct retrovirus by the human TRIM5α antiviral protein. Science 316: 1756–1758. [DOI] [PubMed] [Google Scholar]
- 8.Yap M. W., Nisole S., Stoye J. P. 2005. A single amino acid change in the SPRY domain of human Trim5α leads to HIV-1 restriction. Curr. Biol. 15: 73–78. [DOI] [PubMed] [Google Scholar]
- 9.Schaller T., Hué S., Towers G. J. 2007. An active TRIM5 protein in rabbits indicates a common antiviral ancestor for mammalian TRIM5 proteins. J. Virol. 81: 11713–11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Si Z., Vandegraaff N., O’huigin C., Song B., Yuan W., Xu C., Perron M., Li X., Marasco W. A., Engelman A., et al. 2006. Evolution of a cytoplasmic tripartite motif (TRIM) protein in cows that restricts retroviral infection. Proc. Natl. Acad. Sci. USA 103: 7454–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biris N., Yang Y., Taylor A. B., Tomashevski A., Guo M., Hart P. J., Diaz-Griffero F., Ivanov D. N. 2012. Structure of the rhesus monkey TRIM5α PRYSPRY domain, the HIV capsid recognition module. Proc. Natl. Acad. Sci. USA 109: 13278–13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diaz-Griffero F., Qin X. R., Hayashi F., Kigawa T., Finzi A., Sarnak Z., Lienlaf M., Yokoyama S., Sodroski J. 2009. A B-box 2 surface patch important for TRIM5α self-association, capsid binding avidity, and retrovirus restriction. J. Virol. 83: 10737–10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sebastian S., Grütter C., Strambio de Castillia C., Pertel T., Olivari S., Grütter M. G., Luban J. 2009. An invariant surface patch on the TRIM5α PRYSPRY domain is required for retroviral restriction but dispensable for capsid binding. J. Virol. 83: 3365–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pertel T., Hausmann S., Morger D., Züger S., Guerra J., Lascano J., Reinhard C., Santoni F. A., Uchil P. D., Chatel L., et al. 2011. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472: 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakuma R., Mael A. A., Ikeda Y. 2007. Alpha interferon enhances TRIM5α-mediated antiviral activities in human and rhesus monkey cells. J. Virol. 81: 10201–10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carthagena L., Parise M. C., Ringeard M., Chelbi-Alix M. K., Hazan U., Nisole S. 2008. Implication of TRIMα and TRIMCyp in interferon-induced anti-retroviral restriction activities. Retrovirology 5: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mische C. C., Javanbakht H., Song B., Diaz-Griffero F., Stremlau M., Strack B., Si Z., Sodroski J. 2005. Retroviral restriction factor TRIM5α is a trimer. J. Virol. 79: 14446–14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uchil P. D., Quinlan B. D., Chan W. T., Luna J. M., Mothes W. 2008. TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog. 4: e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gack M. U., Shin Y. C., Joo C. H., Urano T., Liang C., Sun L., Takeuchi O., Akira S., Chen Z., Inoue S., Jung J. U. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446: 916–920. [DOI] [PubMed] [Google Scholar]
- 20.Shi M., Deng W., Bi E., Mao K., Ji Y., Lin G., Wu X., Tao Z., Li Z., Cai X., et al. 2008. TRIM30α negatively regulates TLR-mediated NF-κB activation by targeting TAB2 and TAB3 for degradation. Nat. Immunol. 9: 369–377. [DOI] [PubMed] [Google Scholar]
- 21.Kamitani S., Ohbayashi N., Ikeda O., Togi S., Muromoto R., Sekine Y., Ohta K., Ishiyama H., Matsuda T. 2008. KAP1 regulates type I interferon/STAT1-mediated IRF-1 gene expression. Biochem. Biophys. Res. Commun. 370: 366–370. [DOI] [PubMed] [Google Scholar]
- 22.Liang Q., Deng H., Li X., Wu X., Tang Q., Chang T. H., Peng H., Rauscher F. J., III, Ozato K., Zhu F. 2011. Tripartite motif-containing protein 28 is a small ubiquitin-related modifier E3 ligase and negative regulator of IFN regulatory factor 7. J. Immunol. 187: 4754–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uchil P. D., Hinz A., Siegel S., Coenen-Stass A., Pertel T., Luban J., Mothes W. 2013. TRIM protein-mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. J. Virol. 87: 257–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawai T., Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11: 373–384. [DOI] [PubMed] [Google Scholar]
- 25.Kawai T., Akira S. 2011. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity 34: 637–650. [DOI] [PubMed] [Google Scholar]
- 26.Mallery D. L., McEwan W. A., Bidgood S. R., Towers G. J., Johnson C. M., James L. C. 2010. Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21). Proc. Natl. Acad. Sci. USA 107: 19985–19990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Espinosa A., Dardalhon V., Brauner S., Ambrosi A., Higgs R., Quintana F. J., Sjöstrand M., Eloranta M. L., Ní Gabhann J., Winqvist O., et al. 2009. Loss of the lupus autoantigen Ro52/Trim21 induces tissue inflammation and systemic autoimmunity by disregulating the IL-23-Th17 pathway. J. Exp. Med. 206: 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshimi R., Chang T. H., Wang H., Atsumi T., Morse H. C., III, Ozato K. 2009. Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-κB-dependent cytokine expression in fibroblasts. J. Immunol. 182: 7527–7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tareen S. U., Sawyer S. L., Malik H. S., Emerman M. 2009. An expanded clade of rodent Trim5 genes. Virology 385: 473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson W. E., Sawyer S. L. 2009. Molecular evolution of the antiretroviral TRIM5 gene. Immunogenetics 61: 163–176. [DOI] [PubMed] [Google Scholar]
- 31.Hu Y., Mao K., Zeng Y., Chen S., Tao Z., Yang C., Sun S., Wu X., Meng G., Sun B. 2010. Tripartite-motif protein 30 negatively regulates NLRP3 inflammasome activation by modulating reactive oxygen species production. J. Immunol. 185: 7699–7705. [DOI] [PubMed] [Google Scholar]
- 32.Chang T. H., Xu S., Tailor P., Kanno T., Ozato K. 2012. The small ubiquitin-like modifier-deconjugating enzyme sentrin-specific peptidase 1 switches IFN regulatory factor 8 from a repressor to an activator during macrophage activation. J. Immunol. 189: 3548–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang T. H., Kubota T., Matsuoka M., Jones S., Bradfute S. B., Bray M., Ozato K. 2009. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog. 5: e1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tailor P., Tamura T., Kong H. J., Kubota T., Kubota M., Borghi P., Gabriele L., Ozato K. 2007. The feedback phase of type I interferon induction in dendritic cells requires interferon regulatory factor 8. Immunity 27: 228–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong H. J., Anderson D. E., Lee C. H., Jang M. K., Tamura T., Tailor P., Cho H. K., Cheong J., Xiong H., Morse H. C., III, Ozato K. 2007. Cutting edge: autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF-8 and enhances cytokine expression in macrophages. J. Immunol. 179: 26–30. [DOI] [PubMed] [Google Scholar]
- 36.Jaffray E. G., Hay R. T. 2006. Detection of modification by ubiquitin-like proteins. Methods 38: 35–38. [DOI] [PubMed] [Google Scholar]
- 37.Campbell E. M., Dodding M. P., Yap M. W., Wu X., Gallois-Montbrun S., Malim M. H., Stoye J. P., Hope T. J. 2007. TRIM5α cytoplasmic bodies are highly dynamic structures. Mol. Biol. Cell 18: 2102–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Connor C., Pertel T., Gray S., Robia S. L., Bakowska J. C., Luban J., Campbell E. M. 2010. p62/sequestosome-1 associates with and sustains the expression of retroviral restriction factor TRIM5α. J. Virol. 84: 5997–6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437: 1167–1172. [DOI] [PubMed] [Google Scholar]
- 40.Seth R. B., Sun L., Ea C. K., Chen Z. J. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell 122: 669–682. [DOI] [PubMed] [Google Scholar]
- 41.Ben-Neriah Y., Karin M. 2011. Inflammation meets cancer, with NF-κB as the matchmaker. Nat. Immunol. 12: 715–723. [DOI] [PubMed] [Google Scholar]
- 42.Kim J. Y., Ozato K. 2009. The sequestosome 1/p62 attenuates cytokine gene expression in activated macrophages by inhibiting IFN regulatory factor 8 and TNF receptor-associated factor 6/NF-κB activity. J. Immunol. 182: 2131–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Z. J. 2005. Ubiquitin signalling in the NF-κB pathway. Nat. Cell Biol. 7: 758–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhoj V. G., Chen Z. J. 2009. Ubiquitylation in innate and adaptive immunity. Nature 458: 430–437. [DOI] [PubMed] [Google Scholar]
- 45.Campbell E. M., Perez O., Anderson J. L., Hope T. J. 2008. Visualization of a proteasome-independent intermediate during restriction of HIV-1 by rhesus TRIM5α. J. Cell Biol. 180: 549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hwang C. Y., Holl J., Rajan D., Lee Y., Kim S., Um M., Kwon K. S., Song B. 2010. Hsp70 interacts with the retroviral restriction factor TRIM5α and assists the folding of TRIM5α. J. Biol. Chem. 285: 7827–7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maegawa H., Nakayama E. E., Kuroishi A., Shioda T. 2008. Silencing of tripartite motif protein (TRIM) 5α mediated anti-HIV-1 activity by truncated mutant of TRIM5α. J. Virol. Methods 151: 249–256. [DOI] [PubMed] [Google Scholar]
- 48.Zhao W., Wang L., Zhang M., Yuan C., Gao C. 2012. E3 ubiquitin ligase tripartite motif 38 negatively regulates TLR-mediated immune responses by proteasomal degradation of TNF receptor-associated factor 6 in macrophages. J. Immunol. 188: 2567–2574. [DOI] [PubMed] [Google Scholar]
- 49.Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., Chen Z. J. 2000. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103: 351–361. [DOI] [PubMed] [Google Scholar]
- 50.Häcker H., Redecke V., Blagoev B., Kratchmarova I., Hsu L. C., Wang G. G., Kamps M. P., Raz E., Wagner H., Häcker G., et al. 2006. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature 439: 204–207. [DOI] [PubMed] [Google Scholar]
- 51.Kawai T., Sato S., Ishii K. J., Coban C., Hemmi H., Yamamoto M., Terai K., Matsuda M., Inoue J., Uematsu S., et al. 2004. Interferon-α induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 5: 1061–1068. [DOI] [PubMed] [Google Scholar]
- 52.Song Y. J., Izumi K. M., Shinners N. P., Gewurz B. E., Kieff E. 2008. IRF7 activation by Epstein-Barr virus latent membrane protein 1 requires localization at activation sites and TRAF6, but not TRAF2 or TRAF3. Proc. Natl. Acad. Sci. USA 105: 18448–18453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Konno H., Yamamoto T., Yamazaki K., Gohda J., Akiyama T., Semba K., Goto H., Kato A., Yujiri T., Imai T., et al. 2009. TRAF6 establishes innate immune responses by activating NF-κB and IRF7 upon sensing cytosolic viral RNA and DNA. PLoS One 4: e5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xia Z. P., Sun L., Chen X., Pineda G., Jiang X., Adhikari A., Zeng W., Chen Z. J. 2009. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 461: 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi U. Y., Hur J. Y., Lee M. S., Zhang Q., Choi W. Y., Kim L. K., Lee W. B., Oh G. T., Kim Y. J. 2014. Tripartite motif-containing protein 30 modulates TCR-activated proliferation and effector functions in CD4+ T cells. PLoS One 9: e95805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.