Abstract
Upon virus infection, retinoic acid–inducible gene I–like receptors in host cells recognize viral RNA and activate type I IFN expression. Previously, we identified WD repeat domain (WDR) 5 as one positive regulator for pathway activation. In this study, we report that WDR82, a homolog protein of WDR5, acts opposite to WDR5 and inhibits the activation of the retinoic acid–inducible gene I signaling pathway. WDR82 overexpression inhibits virus-triggered pathway activation, whereas its knockdown enhances induced IFN-β expression. WDR82 is localized on the mitochondria, and its first N-terminal WD40 domain is critical for localization. WDR82 interacts with TNFR-associated factor (TRAF) 3, and its overexpression promotes K48-linked, but not K63-linked, polyubiquitination on TRAF3. Furthermore, WDR82 knockdown inhibits viral replication in the cell, whereas its overexpression has the opposite effect. Interestingly, WDR82 regulates Sendai virus–induced IFNB1 expression in a cell type–specific manner. Taken together, our findings demonstrate that WDR82 is a negative regulator of virus-triggered type I IFNs pathway through mediating TRAF3 polyubiquitination status and stability on mitochondria.
Introduction
The innate immune system is the first line for host to defend the invasion of microbial pathogens (1–3). The host cells use pattern recognition receptors to recognize various pathogen-associated molecular patterns, such as nucleic acids, proteins, lipids, and carbohydrates (2, 4–6). The recognition induces a series of signaling events and leads to production of type I IFNs and proinflammatory cytokines (2, 3, 7, 8). The synthesized IFNs further activate a wide range of antiviral genes and subsequent antiviral response. Several families of pattern recognition receptors have been identified, including TLRs, retinoic acid–inducible gene I (RIG-I)–like receptors, cyclic GMP–AMP synthase–like receptors, NOD-like receptors, and C-type lectin receptors (4, 5). RIG-I–like receptors, including RIG-I, melanoma differentiation-associated gene 5, and laboratory of genetics and physiology 2, have been identified as sensors for RNA viruses (1, 7, 8). RIG-I recognizes short dsRNA or 5-triphosphate panhandle RNA produced during viral infection and replication (9). Upon recognition, the conformation of RIG-I is changed and recruited to mitochondria by virus-induced signaling adaptor (VISA, also known as MAVS, IPS-1, and Cardif) (10–13). The activated VISA then recruits mediator of IFN regulatory factor (IRF) 3 activation (MITA), TNFR-associated factor (TRAF) 3, and TRAF6 to the mitochondria and forms a huge signaling complex (1, 7, 8, 14–16). TRAF6 activates the IKK complex, which phosphorylates IκBα and activates NF-κB. TRAF3 activates the TBK1/IKKε/NEMO complex to phosphorylate IRF3/7, which then translocate to the nucleus and drive the transcription of IFNs (1, 7, 8, 17).
Recently, we identified that WD repeat domain (WDR) 5, a WD40 repeat protein involved in epigenetic regulation, translocates to mitochondria upon virus treatment and promotes the activation of virus-triggered type I IFNs signaling (18, 19). WDR5 was initially identified as one common subunit of all six histone H3K4 methyltransferase complexes, namely SET domain containing 1A/B (SETD1A/B) and myeloid/lymphoid or mixed-lineage leukemia 1–4 (18, 20, 21). WDR82 is also a WD40 repeat protein and one subunit of the SETD1A/B complexes (22, 23). WDR5 and WDR82 have close molecular masses and share high similarity in protein sequence (22). It is proposed that their WD40 repeat domains provide platforms for protein–protein interactions (18, 24). SETD1A/B are the major enzymes responsible for global histone H3K4 trimethylation in mammalian cells (22), which is highly associated with active transcription and considered a hallmark for active transcribed genes. Both WDR5 and WDR82 are critical for the constitution of complexes. In the absence of either protein, SETD1A/B complexes fall apart and global H3K4 trimethylation is impaired (22). Despite their high similarity, WDR5 and WDR82 interact with the different domains of SETD1A, which suggest they have different interaction partners (23).
Because WDR5 plays an important role in the virus-triggered type I IFNs signaling pathway, we were interested whether WDR82 also regulates the pathway. In this study, our data demonstrate that WDR82 inhibits the activation of the virus-triggered IFNB signaling pathway through regulating TRAF3 polyubiquitination and stability, which is opposite to the role of WDR5.
Materials and Methods
Cell lines and reagents
HEK293 cell lines were cultured in DMEM supplemented with 10% FBS and 1% penicillin and streptomycin. U2OS cells were cultured in RPMI 1640 supplemented with 10% FBS and 1% penicillin and streptomycin.
MitoTracker Red (Molecular Probes); mAbs against Flag (Sigma-Aldrich), hemagglutinin (HA; OriGene), Myc-tag (ABclonal), enhanced GFP (eGFP; ABclonal), β-actin (ABclonal), and phospho–TBK1-172 (Epitomics); and polyclonal Abs against TRAF3 (Santa Cruz Biotechnology), WDR5 (Upstate Biotechnology), phospho–IRF3-Ser396 (Cell Signaling Technology), and IRF3 (Epitomics) were purchased from the indicated manufacturers. The antisera for WDR82 was raised from mice by the human full length of WDR82 purified from Escherichia coli.
Immunoprecipitation
The cells were harvested and lysed in Nonidet P-40 lysis buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 0.5% Nonidet P-40) or high-salt lysis buffer (20 mM HEPES [pH 7.4], 10% glycerol, 0.35 M NaCl, 1 mM MgCl2, 0.5% Triton X-100, 1 mM DTT) with proteinase inhibitors. The supernatant was then incubated with protein G beads (GE Healthcare) and desired Ab at 4°C for 4 h. The beads were spun down and washed three times with lysis buffer. The final drop of wash buffer was vacuumed out and SDS loading buffer was added to the beads, followed by Western blot.
RNA interference, reverse transcription, and quantitative PCR
The indicated cells were transfected with small interfering RNA (siRNA) and were scraped down and collected by centrifugation. Total RNA was extracted with an RNA extraction kit (Yuanpinghao) according to the manufacturer’s instructions. Approximately 1 μg total RNA was used for reverse transcription with a first-strand cDNA synthesis kit (Toyobo). The amount of mRNA was assayed by quantitative PCR. β-Actin was used to normalize the amount of each sample. Assays were repeated at least three times. Data shown are average values ± SD of one representative experiment. Primer and siRNA sequences are as follows: IFNB1, forward, 5′-CAACTTGCTTGGATTCCTAC-3′, reverse, 5′-CTGTCCTTGAGGCAGTATTC-3′; WDR82, forward, 5′-CTCCATCGTGCTCTATGACT-3′, reverse, 5′-GATGAGGTCCACACCATATT-3′; Actin, forward, 5′-CAGCACAATGAAGATCAAGA-3′, reverse, 5′-GATCCACATCTGCTGGAAG-3′; Sendai virus (SeV) genomic primers, forward, 5′-AAACGCATCACGTCTCTTCC-3′, reverse, 5′-TTCTCAGCTCTGCTTAGGGG-3′; WDR82 siRNA, no. 1, 5′-AGAGAACCCUGUACAGUAA-3′, no. 2, 5′-UGCCAAACAUUUACAGAAA-3′, no. 3, 5′-GGCCUGAAAGGGAGGGAAA-3′.
Immunofluorescence staining
Cells were cultured on coverslips and fixed with freezing methanol after washing twice in PBS. The coverslips were then washed three times by PBS and blocked in PBS with 1% BSA for 10 min. The coverslips were hybridized with first and second Abs for 1 h. Then the coverslips were mounted with a ProLong antifade kit (Invitrogen) and observed with fluorescence microscopy.
Immunoprecipitation
The cells were harvested and lysed in Nonidet P-40 lysis buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 0.5% Nonidet P-40) or high-salt lysis buffer (20 mM HEPES [pH 7.4], 10% glycerol, 0.35 M NaCl, 1 mM MgCl2, 0.5% Triton X-100, 1 mM DTT) with proteinase inhibitors. The supernatant was then incubated with protein G beads (GE Healthcare) and desired Ab at 4°C for 4 h. The beads were spun down and washed three times with lysis buffer. The final drop of wash buffer was vacuumed out and SDS loading buffer was added to the beads followed by Western blot.
Luciferase reporter assay
HEK293T cells (∼1 × 105) were seeded on 24-well plates and transfected by calcium phosphate precipitation. The reporter plasmid of pRL-TK or pRL-SV40 Renilla luciferase was added to each transfection to normalize transfection efficiency. A dual-specific luciferase assay kit (Promega) was used for luciferase assays. Assays were repeated at least three times. Data shown are average values ± SD of one representative experiment.
Vesicular stomatitis virus–GFP replication assays
Cells were transfected with siRNAs for 48 h, which was followed by vesicular stomatitis virus (VSV)-GFP infection for 1 h, and then cells were washed with PBS three times and replaced by new medium. Twelve to 24 h later, cells were observed by fluorescence microscopy and lysated for further immunoblot analysis with anti-GFP.
Preparation of mouse peritoneal macrophages
Peritoneal exudate cells from C57BL/6 mice were obtained by washing the peritoneal cavity with 5 ml fresh ice-cold DMEM. Peritoneal cells were counted in a hemocytometer, plated in 12-well plates, and allowed to adhere in for 1 h at 37°C with 5% CO2. Then supernatants were removed and, after washing with ice-cold PBS, the adherent cells were incubated in fresh DMEM containing 10% FBS and 1% penicillin/streptomycin at 37°C with 5% CO2.
Isolation of mouse embryo fibroblasts
Mouse embryo fibroblasts (MEFs) were isolated from C57BL/6 mice 13–15 d after coitus. Each embryo was dissected into 5 ml sterile PBS, voided of internal organs, and sheared through scissors in the presence of 0.25% trypsin-EDTA. After 30 min incubation with gentle shaking at 37°C, DMEM with 10% FBS was added to inactivate trypsin. The cells were filtered through a 74-μm nylon mesh and washed by fresh DMEM twice. Then cells were plated onto 100-mm cell culture dishes and incubated for 24 h at 37°C with 5% CO2. Adherent cells were used as MEFs. All MEFs used in this study were within five passages.
Results
WDR82 regulates the activation of virus-triggered type I IFNs signaling pathway
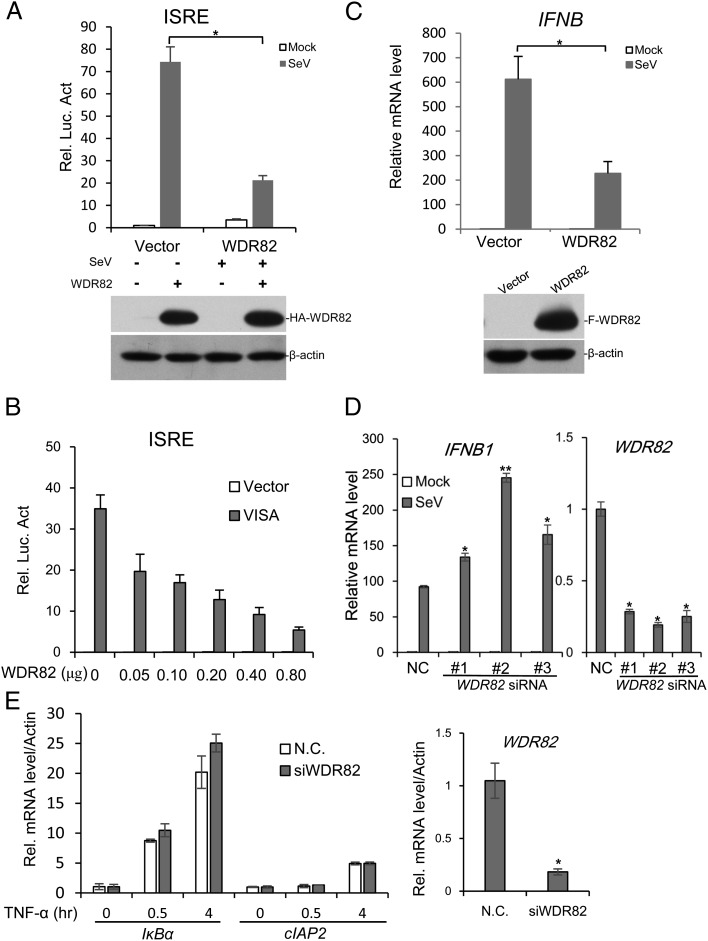
To investigate whether WDR82 is involved in the regulation of virus-triggered type I IFNs signaling, an expression plasmid containing WDR82 cDNA was transfected into the cell and SeV was used to activate the signaling pathway. The result of IFN-stimulated response element (ISRE) luciferase assay indicated that exogenous expressed WDR82 inhibited the activation of virus-triggered signaling (Fig. 1A). The further experiment showed that WDR82 inhibited ISRE luciferase activity in a dose-dependent manner (Fig. 1B). The RT-PCR result showed that WDR82 overexpression indeed repressed the inducible expression of IFN-β mRNA by virus (Fig. 1C). Three independent siRNAs were used to knock down WDR82 in HEK293 cells, and the activation of IFN-β mRNA was greatly elevated 12 h after virus treatment (Fig. 1D). Similar results were also obtained in ACHN and 769-P cell lines (data not shown). In the contrast, WDR82 knockdown does not affect the expression of target genes induced by TNF-α (Fig. 1E). Collectively, these findings indicate that WDR82 inhibits the activation of the type I IFNs pathway by SeV specifically.
FIGURE 1.
WDR82 inhibits SeV-induced ISRE activation and IFNB transcription. (A) WDR82 overexpression inhibits activated ISRE report gene. HEK293 cells (1 × 105) were transfected with ISRE reporter plasmid (0.1 μg) together with control (1 μg) or WDR82 plasmid (1 μg). Twelve hours later, cells were infected with SeV and luciferase assays were performed. Lysates were immunoblotted with anti-HA and anti–β-actin. (B) WDR82 inhibits ISRE activation in a dose-dependent manner. HEK293 cells (1 × 105) were cotransfected with ISRE reporter (0.1 μg) and the indicated amounts of WDR82 plasmid, together with control (0.2 μg) or VISA plasmid (0.2 μg). Cell lysates were performed with luciferase assays 24 h after transfection. (C) WDR82 inhibits IFNB gene transcription. WDR82 stably expressed HEK293 cells, and control cells were infected with SeV for 12 h and RT-PCR was performed with IFNB primers. Cell lines were also immunoblotted with anti-Flag and anti–β-actin. (D) WDR82 knockdown promotes IFNB transcription. HEK293 cells were transfected with WDR82 and with three different siRNAs. Forty-eight hours later, cells were infected with SeV for 12 h before performing RT-PCR experiments. The efficiency of WDR82 knockdown was measured with RT-PCR. (E) HEK293 cells were transfected with WDR82 siRNA and treated with TNF-α. The mRNA levels of IκBα and cIAP2 were measured by quantitative PCR. *p < 0.05, **p < 0.01.
WDR82 regulates the activation of key signaling molecules opposite to WDR5
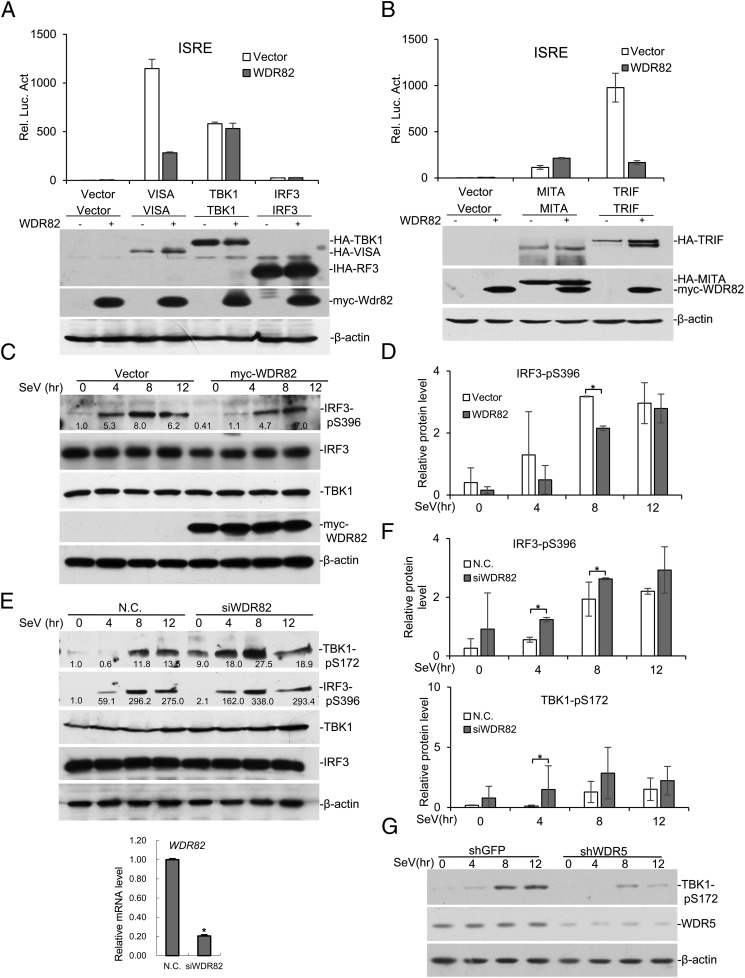
Although WDR82 shares high homology with WDR5, and our data suggest that it plays an opposite role to WDR5 in regulating virus-triggered signaling. To study the underlying mechanisms, we first assessed the roles of WDR82 on pathway activation by different molecules. WDR82 nicely inhibits ISRE-luciferase activity induced by VISA, but not TBK1 or IRF3 (Fig. 2A). Moreover, WDR82 inhibits ISRE-luciferase activity induced by Toll/IL-1R domain-containing adapter inducing IFN-β (TRIF), but not by MITA/STING (Fig. 2B). VISA and TRIF activate IRF3 dependent on TRAF3, but MITA/STING usually do not (25). The data suggest that WDR82 may regulate RIG-I signaling through TRAF3. To further confirm this, we examined its effects on the phosphorylation of TBK1 and IRF3, the hallmarks of type I IFNs pathway activation. When WDR82 was exogenously expressed in the cell, the induced phosphorylation of TBK1 and IRF3 was impaired, whereas the total IRF3 remained the same (Fig. 2C, 2D). WDR82 knockdown by siRNA caused increased phosphorylation of IRF3 and TBK1 (Fig. 2E, 2F). In comparison, WDR5 knockdown caused a decrease in TBK1 phosphorylation (Fig. 2G). These data suggest that WDR82 regulates the virus-triggered type I IFNs signaling pathway in a manner different from that of WDR5, which is consistent with our previous observations.
FIGURE 2.
WDR82 inhibits the phosphorylation of key signaling molecules. (A) VISA, TBK1, and IRP3 were transfected into HEK293 cells with or without WDR82. ISRE luciferase activity was assayed to measure pathway activation. (B) MITA/STING and TRIF were transfected into HEK293 with or without WDR82. Then ISRE luciferase activity was measured. (C and D) HEK293 cells were transfected with WDR82 and treated with SeV for indicated times. TBK1-pS172, IRF3-pS396, IRF3, Myc-WDR82, and β-actin were measured with Western blotting in the lysates. Quantification of Western blotting bands from three experiments are shown in (D). (E and F) HEK293 cells transfected with WDR82 siRNA were infected with SeV and the efficiency of WDR82 knockdown was confirmed by RT-PCR experiments. Western blots and protein quantification were carried out as in (C) and (D). (G) WDR5 was knocked down by shRNA and experiments were performed as in (C). Lysates were immunoblotted with anti–TBK1-pS172, anti-WDR5, and anti–β-actin. *p < 0.05.
The induction of IFN-β by viral infection is also reported to be regulated at the transcriptional level (26). Because WDR82 was previously characterized as a transcription regulator, it might regulate the transcription of IRF3 target genes, rather than the signaling pathway. However, WDR82 and WDR5 are required for the normal function of SETD1A/B complexes, which catalyze H3K4 methylation and are globally associated with active transcription. If the inhibitory effect on IFN-β activation is due to the function of WDR82 in transcription, WDR82 then should activate IFN-β expression rather than inhibition, which is opposite to our discovery. Therefore, our data suggest the regulation of IFN-β activation by WDR82 is not at the level downstream of IRF3 activation.
Localization of WDR82 to mitochondria
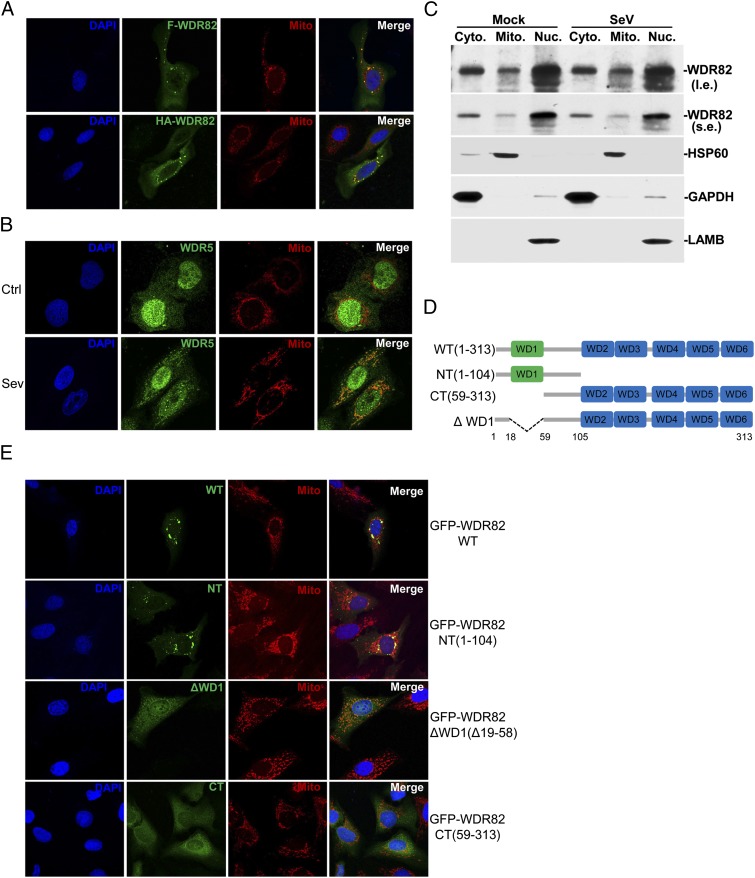
Our previous report showed that GFP-tagged WDR5 is translocated to mitochondria in response to viral treatment (19). We compared the subcellular localization of WDR82 and WDR5. Because we lack a good WDR82 Ab for staining the endogenous protein, we expressed WDR82 with different tags in HEK293 cells. Portions of both Flag- and HA-tagged WDR82 were observed to be localized on mitochondria, even without SeV treatment (Fig. 3A). We used a WDR5 Ab and found the endogenous WDR5 distributed both in nucleus and cytoplasm; upon viral treatment, a portion of WDR5 is translocated to mitochondria (Fig. 3B), which is consistent with our previous report (19). Moreover, we fractionated the cell components and confirmed by Western blot that WDR82 is localized on mitochondria (Fig. 3C).
FIGURE 3.
WDR82 is localized on mitochondria. (A) Confocal microscopy of WDR82 subcellular localization. U2OS cells grown on slides were transfected with Flag- or HA-tagged WDR82 plasmids. Twenty-four hours later, cells were fixed and blocked with 0.1% BSA and followed by staining with DAPI, anti-Flag or anti-HA, and MitoTracker Red. (B) The subcellular localization of endogenous WDR5. Cells were infected with or without SeV, and endogenous WDR5 was stained with the corresponding Ab. (C) Cells were fractionated and endogenous WDR82 was blotted. (D and E) The first N-terminal WD domain is essential for mitochondria localization of WDR82. Multiple truncations of C-terminal EGFP-tagged WDR82 was transfected into U2OS, followed by staining with DAPI and MitoTracker Red. (A), (B), and (E) Original magnification ×600.
A series of GFP-tagged truncations were generated to investigate which domain is responsible for the mitochondrial localization of WDR82 (Fig. 3D). The results indicated that the N-terminal containing the first WD40 domain is sufficiently localized to mitochondria (Fig. 3E). When the first WD40 domain was deleted from the full-length protein, the mutants abolished the localization on mitochondria and were distributed over the whole cell (Fig. 3E), which suggested that the first WD40 domain (residues 19–58) is critical for the specific localization of WDR82 on mitochondria.
Interaction between WDR82 and TRAF3
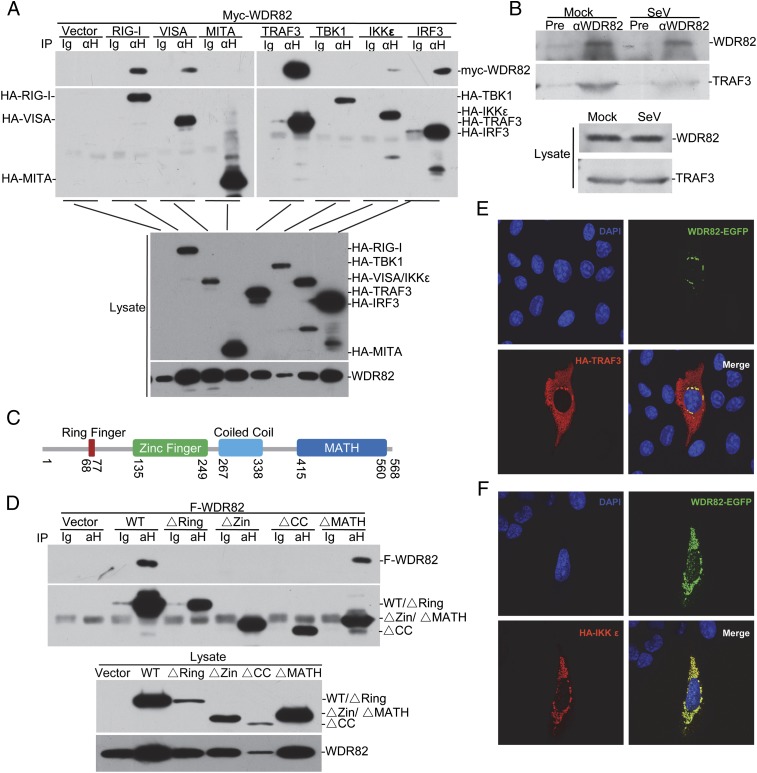
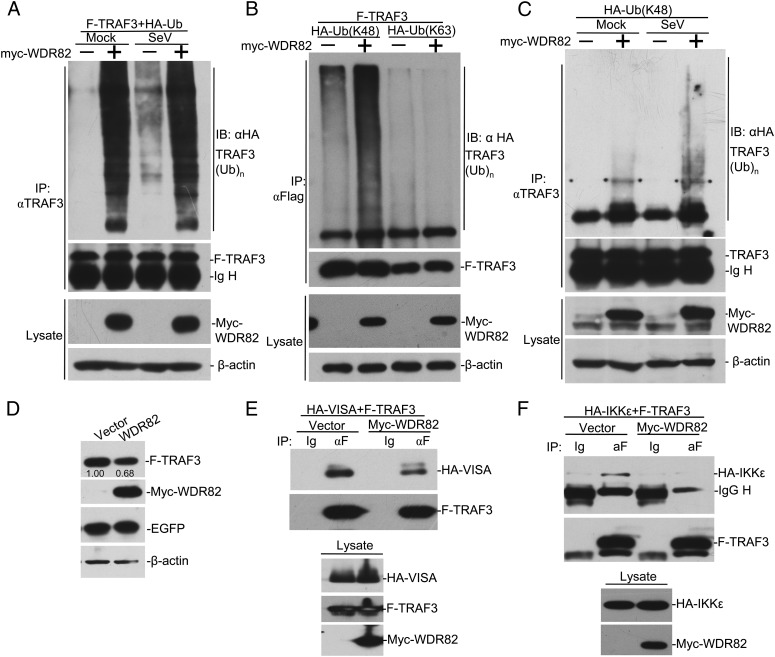
The above data clearly show that WDR82 is involved in the regulation of virus-triggered type I IFNs signaling through a different mechanism from WDR5. To further elucidate the underlying biochemical mechanisms, we performed coimmunoprecipitation assays between WDR82 and other key molecules. The results showed that exogenous WDR82 was associated with multiple key molecules in the pathway, including RIG-I, VISA, TRAF3, IKKε, and IRF3 (Fig. 4A). Upon viral stimuli, these signaling molecules are recruited to the outer membrane of mitochondria and together form a huge complex (1, 7). We noticed that the interaction between WDR82 and TRAF3 was the strongest and speculated that WDR82 interacts with the complex through TRAF3. The coimmunoprecipitation assays between endogenous proteins proved that WDR82 indeed interacts with TRAF3 in the cell (Fig. 4B). We did not observe its interaction with other signaling molecules at the endogenous level (data not shown). To further characterize TRAF3 domains interacting with WDR82, we made a series of TRAF3 truncations. The data for immunoprecipitation assays indicated that the N-terminal of TRAF3 is critical, and deletion of either ring finger, zinc finger, or coiled-coil domains abolished its interaction with WDR82 (Fig. 4C, 4D). The results of immunostaining also exhibited that exogenous expressed WDR82 colocalized with TRAF3 and IKKε (Fig. 4E, 4F). Collectively, these data indicate that WDR82 is associated with the RIG-I signaling complex, probably through interaction with TRAF3.
FIGURE 4.
Interaction between WDR82 and TRAF3. (A) Overexpressed WDR82 interacts with TRAF3. HEK293 cells were cotransfected with Myc-tagged WDR82 plasmids and HA-tagged RIG-I, VISA, MITA, TRAF3, TBK1, IKKε, and IRF3. Lysates were coimmunoprecipitated with anti-HA and Western blotted with anti-HA and anti-Myc. (B) Endogenous interaction between TRA3 and WDR82. HEK293 cells were infected with SeV, and coimmunoprecipitation and Western blotting were performed with the indicated Ab. (C) Sketch map of TRAF3 domains. (D) Mapping of TRAF3–WDR82 interacting domain. HA-tagged TRAF3 truncations and Flag-tagged WDR82 were cotransfected into HEK293 cells. Coimmunoprecipitation and immunoblot assays were performed as indicated. (E and F) WDR82 is colocalized with TRAF3 and IKKε. C-terminal EGFP-tagged WDR82 and HA-tagged TRAF3 or IKKε were cotransfected into U2OS cells. Immunofluorescence staining was performed as indicated. (E and F) Original magnification ×600.
WDR82 enhances K48-linked polyubiquitination of TRAF3
TRAF3 is a key molecule in the cascade of RIG-I signaling pathway, and its K63-linked polyubiquitination is required for TBK1 recruitment and downstream events; additionally, the regulation of different forms of polyubiquitination chains emerged as critical to the regulation of innate immunity pathways (14, 27–31). To further elucidate the biochemical mechanisms, we investigate the impact of WDR82 in regulating TRAF3 polyubiquitination. Myc-WDR82, Flag-TRAF3, and HA-ubiquitin (Ub) were coexpressed in the cell. An in vivo ubiquitination assay was performed with anti-Flag IP and anti-HA Western blotting. The data suggested that WDR82 promotes TRAF3 polyubiquitination (Fig. 5A). Viral treatment enhanced TRAF3 polyubiquitination, but it had little effect on the WDR82-triggered signal (Fig. 5A). Further characterization indicated that WDR82 promotes the K48-linked, but not the K63-linked, polyubiquitination chain on TRAF3 (Fig. 5B). WDR82 overexpression also enhanced the K48 chain on the endogenous TRAF3, which was further elevated upon viral treatment (Fig. 5C). K48-linked polyubiquitination is often related with protein degradation by proteasomes. To further verify our discovery, we measured the TRAF3 level in the lysate and the result indicated that the exogenous TRAF3 was at a lower level when coexpressed with WDR82 (Fig. 5D). These results suggest that WDR82 regulates K48-linked polyubiquitination on TRAF3.
FIGURE 5.
WDR82 regulates TRAF3 stability by promoting its K48-linked polyubiquitination. (A) WDR82 promotes the polyubiquitination of TRAF3. HEK293 cells were transfected with HA-tagged ubiquitin plasmids and Myc-WDR82. Twenty-four hours later, cells were infected with SeV and cell lysates were immunoprecipitated with anti-TRAF3. The immunoprecipitates were then immunoblotted with anti-HA or anti-TRAF3. The lysate was assayed with anti-Myc and anti–β-actin. (B and C) WDR82 promotes the K48-linked polyubiquitination of TRAF3. The experiments were performed as in (A), except K48 only or K63 only ubiquitin was used instead of wild-type. (D) HEK293 cells were transfected with the indicated plasmids. Twenty-four hours later, cell lysates were immunoblotted with indicated Abs. Relative protein level was labeled below the corresponding bands. (E and F) WDR82 overexpression impairs the interaction between TRAF3 with VISA or IKKε. HEK293 cells were transfected with the indicated plasmids. Twenty-four hours later, lysates were immunoprecipitated with anti-Flag. The immunoprecipitates and cell lysates were immunoblotted with indicated Abs.
WDR82 inhibits TRAF3 interaction with other signaling molecules
Based on the above discovery, we speculated that WDR82 should regulate the interaction between TRAF3 and other molecules. We studied the interaction of TRAF3 with VISA or IKKε with or without WDR82 overexpression. The results showed that when coexpressed with WDR82, TRAF3 pulled down much less VISA and IKKε, whereas their protein levels in lysates were equal (Fig. 5E, 5F). These results further confirmed that WDR82 inhibits RIG-I signaling activation through regulating TRAF3.
WDR82 enhances virus replication in the cell
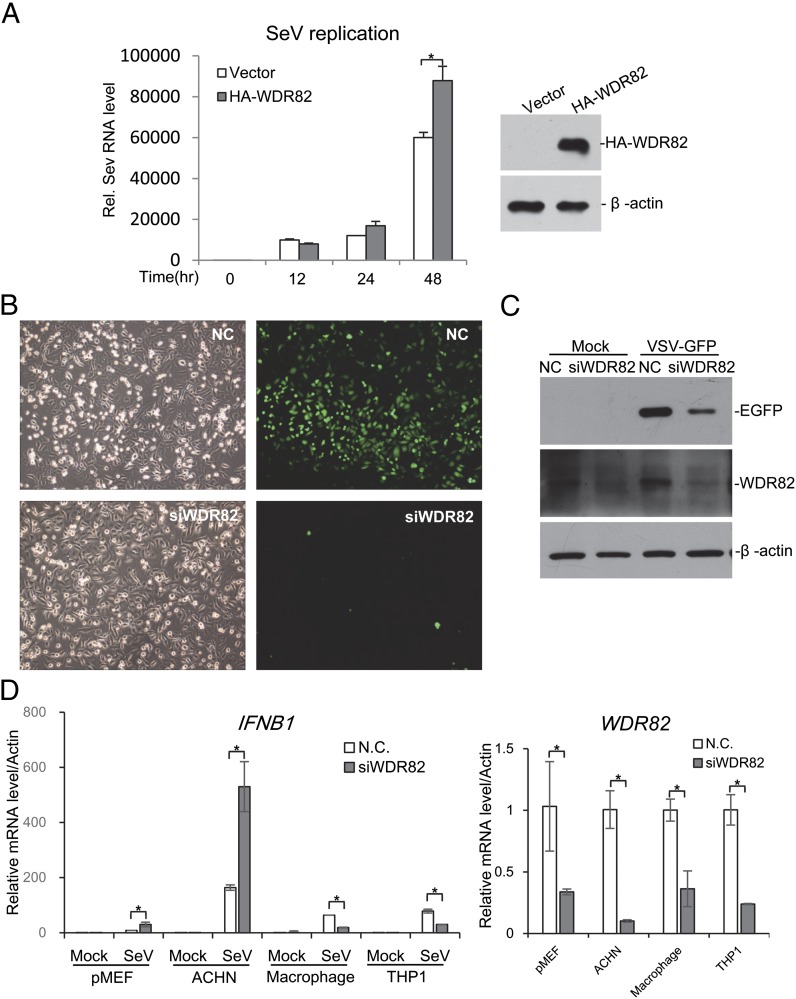
To investigate whether WDR82 affects virus replication in the cell, we used SeV to infect cells and measured virus genomic RNA via RT-PCR. The result indicated that more viral genomic RNA existed in WDR82 overexpression cells (Fig. 6A). Meanwhile, we used one engineered VSV strain containing GFP cDNA. WDR82 was knocked down in the cell and the cells were infected with VSV-GFP. The microscopy images showed that the green fluorescence of GFP was much less in WDR82 deficient cells, although the cell numbers were similar (Fig. 6B). The expressed GFP protein was further examined by Western blotting, which again showed that less GFP protein was expressed in WDR82 knockdown cells (Fig. 6C). These data indicated that WDR82 promotes the replication of RNA viruses through inhibiting RIG-I pathway activation.
FIGURE 6.
WDR82 is critical for the replication of VSV and SeV. (A) Overexpression of WDR82 promotes SeV replication. HEK293 cells were transfected with WDR82 for 12 h followed by SeV infection for the indicated time. Then, SeV genomic RNA in cells was extracted for further real-time PCR tests by specific genomic primers. Parts of cells were lysated and immunoblotted with the indicated Ab. (B and C) WDR82 knockdown impairs VSV-GFP replication. U2OS cells were transfected with WDR82 or control siRNAs for 48 h followed by VSV-GFP infection for 1 h, and then cells were replaced with new medium for another 24 h. The cells were observed by fluorescence microscopy and lysated for further immunoblot analysis with the indicated Ab. (B) Original magnification ×200. (D) Primary MEFs and macrophages were isolated as described in Materials and Methods. The indicated cell lines were transfected with WDR82 siRNA and treated with SEV. IFNB1 mRNA was measured by quantitative PCR. *p < 0.05.
WDR82 regulates RIG-I signaling in multiple cell lines
To investigate whether WDR82 regulates antiviral signaling in a tissue-specific manner, we knocked down WDR82 and measured IFNB1 activation in multiple cell lines. Interestingly, WDR82 deficiency enhanced IFNB1 expression in primary MEF and ACHN cell lines, but it was repressed in primary mouse macrophages and THP1 cell lines (Fig. 6D). These findings suggest thatWDR82 regulates RIG-I signaling in a tissue-dependent manner, which fits its dual roles in the cell, a positive regulator for gene transcription and a repressor for RIG-I signaling,
Discussion
Similar to WDR5, WDR82 was initially identified as one subunit of SETD1A/B complexes, which play important roles in histone methylation and transcription regulation (18, 22, 23). Our studies extended their roles in the cytoplasm to regulate antiviral signaling pathways (18, 19). Mixed-lineage leukemia 1, one of the H3K4 methyltransferases, was also shown to be localized in cytoplasm and involved in regulating NF-κB signaling pathway (32). This and our previous work have shown that a number of key factors functioning in transcription regulation also exist in cytoplasm and have important roles in signaling pathways. Recently, several groups have reported that key factors involved in epigenetic regulation also play important roles in signaling pathways in a way that is not dependent on their enzyme activity. The complexes of histone H3K4 methyltransferases consist of many common and unique subunits (20, 33). It will be interesting to investigate whether other related proteins have similar effects in antiviral signaling and other pathways.
The formation of the polyubiquitination chain on TRAF3 has been proven to be one of the key steps in the activation of the virus-triggered type I IFN signaling pathway (14). Moreover, the formation of different types of Ub chains emerged to be another important way to regulate protein functions in the cell. In this study, we found that WDR82 promotes the K48-linked Ub chain on TRAF3, and we provided important clues for the study of Ub-regulated pathway activation. However, further study is still required to demonstrate the detailed mechanisms.
Although WDR82 is among the closest homologs of WDR5, it plays opposite roles in the type I IFN signaling pathway. Both proteins are localized on mitochondria: WDR5 positively regulates pathway activation and WDR82 inhibits it. Considering their high similarity, it is possible that the two proteins compete with each other on mitochondria. WDR82 is localized on mitochondria to ensure pathway inactivation in the absence of virus, whereas WDR5 is floating around in the cell. Upon infection, WDR5 is recruited to mitochondria together with other signaling factors, and it competes with WDR82 for protein interaction and promotes pathway activation. It will be interesting to investigate how the two WD40 repeat proteins, which collaborate in the nucleus to regulate transcription, compete in the cytoplasm to regulate the virus-triggered type I IFN signaling pathway. The cell type–specific effects also reflect the complexity of the antiviral RIG-I signaling regulated by WDR82. It will be interesting to investigate whether it is associated with tissue-specific antiviral response and diseases.
Acknowledgments
We thank Dr. Hong-Bing Shu of Wuhan University for sharing reagents and for discussions on the manuscript.
This work was supported by National Basic Research Program of China 973 Program Grants 2011CB504206 and 2012CB518700, National Natural Science Foundation of China Grants 30971502 and 91019013 (to M.W.) and 31221061, 31200653, and 31370866 (to L.-Y.L.), and Program for New Century Excellent Talents in University Grant NCET-11-0410 (to M.W.).
- EGFP
- enhanced GFP
- HA
- hemagglutinin
- IRF
- IFN regulatory factor
- ISRE
- IFN-stimulated response element
- MEF
- mouse embryo fibroblast
- MITA
- mediator of IFN regulatory factor 3 activation
- RIG-I
- retinoic acid–inducible gene I
- SETD1A/B
- SET domain containing 1A/B
- SeV
- Sendai virus
- siRNA
- small interfering RNA
- TRAF
- TNFR-associated factor
- TRIF
- Toll/IL-1R domain–containing adapter inducing IFN-β
- Ub
- ubiquitin
- VISA
- virus-induced signaling adaptor
- VSV
- vesicular stomatitis virus
- WDR
- WD repeat domain.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Loo Y. M., Gale M., Jr. 2011. Immune signaling by RIG-I-like receptors. Immunity 34: 680–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun L., Liu S., Chen Z. J. 2010. SnapShot: pathways of antiviral innate immunity. Cell 140: 436–436.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seth R. B., Sun L., Chen Z. J. 2006. Antiviral innate immunity pathways. Cell Res. 16: 141–147. [DOI] [PubMed] [Google Scholar]
- 4.Kugelberg E. 2014. Pattern recognition receptors: curbing gut inflammation. Nat. Rev. Immunol. 14: 583. [DOI] [PubMed] [Google Scholar]
- 5.Takeuchi O., Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140: 805–820. [DOI] [PubMed] [Google Scholar]
- 6.Ran Y., Shu H. B., Wang Y. Y. 2014. MITA/STING: a central and multifaceted mediator in innate immune response. Cytokine Growth Factor Rev. 25: 631–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoneyama M., Onomoto K., Jogi M., Akaboshi T., Fujita T. 2015. Viral RNA detection by RIG-I-like receptors. Curr. Opin. Immunol. 32: 48–53. [DOI] [PubMed] [Google Scholar]
- 8.Fitzgerald M. E., Rawling D. C., Vela A., Pyle A. M. 2014. An evolving arsenal: viral RNA detection by RIG-I-like receptors. Curr. Opin. Microbiol. 20: 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5: 730–737. [DOI] [PubMed] [Google Scholar]
- 10.Xu L. G., Wang Y. Y., Han K. J., Li L. Y., Zhai Z., Shu H. B. 2005. VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol. Cell 19: 727–740. [DOI] [PubMed] [Google Scholar]
- 11.Sun Q., Sun L., Liu H. H., Chen X., Seth R. B., Forman J., Chen Z. J. 2006. The specific and essential role of MAVS in antiviral innate immune responses. Immunity 24: 633–642. [DOI] [PubMed] [Google Scholar]
- 12.Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437: 1167–1172. [DOI] [PubMed] [Google Scholar]
- 13.Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K. J., Takeuchi O., Akira S. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6: 981–988. [DOI] [PubMed] [Google Scholar]
- 14.Corn J. E., Vucic D. 2014. Ubiquitin in inflammation: the right linkage makes all the difference. Nat. Struct. Mol. Biol. 21: 297–300. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y. Y., Ran Y., Shu H. B. 2012. Linear ubiquitination of NEMO brakes the antiviral response. Cell Host Microbe 12: 129–131. [DOI] [PubMed] [Google Scholar]
- 16.Liu S., Chen J., Cai X., Wu J., Chen X., Wu Y. T., Sun L., Chen Z. J. 2013. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. eLife 2: e00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lei C. Q., Zhong B., Zhang Y., Zhang J., Wang S., Shu H. B. 2010. Glycogen synthase kinase 3β regulates IRF3 transcription factor-mediated antiviral response via activation of the kinase TBK1. Immunity 33: 878–889. [DOI] [PubMed] [Google Scholar]
- 18.Wu M., Shu H. B. 2011. MLL1/WDR5 complex in leukemogenesis and epigenetic regulation. Chin J Cancer 30: 240–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y. Y., Liu L. J., Zhong B., Liu T. T., Li Y., Yang Y., Ran Y., Li S., Tien P., Shu H. B. 2010. WDR5 is essential for assembly of the VISA-associated signaling complex and virus-triggered IRF3 and NF-κB activation. Proc. Natl. Acad. Sci. USA 107: 815–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eissenberg J. C., Shilatifard A. 2010. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev. Biol. 339: 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wysocka J., Swigut T., Milne T. A., Dou Y., Zhang X., Burlingame A. L., Roeder R. G., Brivanlou A. H., Allis C. D. 2005. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121: 859–872. [DOI] [PubMed] [Google Scholar]
- 22.Wu M., Wang P. F., Lee J. S., Martin-Brown S., Florens L., Washburn M., Shilatifard A. 2008. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol. Cell. Biol. 28: 7337–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J. H., Skalnik D. G. 2008. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol. Cell. Biol. 28: 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trievel R. C., Shilatifard A. 2009. WDR5, a complexed protein. Nat. Struct. Mol. Biol. 16: 678–680. [DOI] [PubMed] [Google Scholar]
- 25.Ishikawa H., Ma Z., Barber G. N. 2009. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461: 788–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo W.-W., Lian H., Zhong B., Shu H. B., Li S. 2014. Krüppel-like factor 4 negatively regulates cellular antiviral immune response. Cell. Mol. Immunol. DOI: 10.1038/cmi.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tseng P. H., Matsuzawa A., Zhang W., Mino T., Vignali D. A., Karin M. 2010. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat. Immunol. 11: 70–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhong B., Zhang Y., Tan B., Liu T. T., Wang Y. Y., Shu H. B. 2010. The E3 ubiquitin ligase RNF5 targets virus-induced signaling adaptor for ubiquitination and degradation. J. Immunol. 184: 6249–6255. [DOI] [PubMed] [Google Scholar]
- 29.Li S., Zheng H., Mao A. P., Zhong B., Li Y., Liu Y., Gao Y., Ran Y., Tien P., Shu H. B. 2010. Regulation of virus-triggered signaling by OTUB1- and OTUB2-mediated deubiquitination of TRAF3 and TRAF6. J. Biol. Chem. 285: 4291–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong B., Zhang L., Lei C., Li Y., Mao A. P., Yang Y., Wang Y. Y., Zhang X. L., Shu H. B. 2009. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 30: 397–407. [DOI] [PubMed] [Google Scholar]
- 31.Qin Y., Zhou M. T., Hu M. M., Hu Y. H., Zhang J., Guo L., Zhong B., Shu H. B. 2014. RNF26 temporally regulates virus-triggered type I interferon induction by two distinct mechanisms. PLoS Pathog. 10: e1004358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X., Zhu K., Li S., Liao Y., Du R., Zhang X., Shu H. B., Guo A. Y., Li L., Wu M. 2012. MLL1, a H3K4 methyltransferase, regulates the TNFα-stimulated activation of genes downstream of NF-κB. J. Cell Sci. 125: 4058–4066. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi Y. H., Westfield G. H., Oleskie A. N., Trievel R. C., Shilatifard A., Skiniotis G. 2011. Structural analysis of the core COMPASS family of histone H3K4 methylases from yeast to human. Proc. Natl. Acad. Sci. USA 108: 20526–20531. [DOI] [PMC free article] [PubMed] [Google Scholar]