Abstract
Increased IFN-α production contributes to the pathogenesis of infectious and autoimmune diseases. Plasmacytoid dendritic cells (pDCs) from females produce more IFN-α upon TLR7 stimulation than pDCs from males, yet the mechanisms underlying this difference remain unclear. In this article, we show that basal levels of IFN regulatory factor (IRF) 5 in pDCs were significantly higher in females compared with males and positively correlated with the percentage of IFN-α–secreting pDCs. Delivery of recombinant IRF5 protein into human primary pDCs increased TLR7-mediated IFN-α secretion. In mice, genetic ablation of the estrogen receptor 1 (Esr1) gene in the hematopoietic compartment or DC lineage reduced Irf5 mRNA expression in pDCs and IFN-α production. IRF5 mRNA levels furthermore correlated with ESR1 mRNA levels in human pDCs, consistent with IRF5 regulation at the transcriptional level by ESR1. Taken together, these data demonstrate a critical mechanism by which sex differences in basal pDC IRF5 expression lead to higher IFN-α production upon TLR7 stimulation in females and provide novel targets for the modulation of immune responses and inflammation.
Introduction
Differences in immune responses between females and males, including responsiveness to vaccination (1), have been reported (2–5) but often remain overlooked in immunological studies and particularly in human viral infections (6), because most studies have been carried out in rodents (7–10). As a general rule, females exhibit more robust humoral and cell-mediated immune responses to antigenic challenges compared with males (1, 11–13). Furthermore, females are also often more prone to immune-related pathology and autoimmunity (14). The heightened inflammatory immune responses observed in females have been suggested to contribute to sex differences in the clinical manifestations, immune responses and outcome of viral diseases, including influenza A virus (15), hantavirus (16), hepatitis C virus (17, 18), and HIV-1 (19, 20).
The pathways underlying these sex differences in the manifestations of viral and autoimmune diseases are not well understood, but increasing data suggest a critical role of the TLR7 pathway and resulting type I IFN production in the outcome of these diseases (21–23). Our group and others have previously shown that plasmacytoid dendritic cells (pDCs) derived from females produced significantly more IFN-α in response to TLR7 ligands than pDCs derived from males, resulting in stronger immune activation (24, 25), and that sex hormones can regulate the IFN-α response to TLR7 stimulation (26, 27). However, the mechanisms underlying this sex difference in TLR7-induced IFN-α production by pDCs remain unknown.
IFN-α induction is regulated primarily at the transcriptional level by the IFN regulatory factors (IRF) family (28–30). In response to stimulation, these transcription factors are phosphorylated on serine residues, a modification that stimulates protein dimerization, nuclear translocation, and interaction with transcriptional coactivators (31, 32). pDCs constitutively express high levels of IRF5 and IRF7 (33–35). TLR7 activation of pDCs leads to the activation and phosphorylation of both IRF5 and IRF7 (31, 36, 37). IRF7 is widely recognized as the “master regulator” of type I IFN production (32), whereas IRF5 has been shown to be a central mediator of TLR7 signaling (33, 38). In addition, IRF5 polymorphisms have been associated with multiple autoimmune diseases, and in particular systemic lupus erythematous and rheumatoid arthritis (39–42), two autoimmune diseases characterized by overproduction of type I IFN and by significant sex differences in prevalence. Autoimmune-risk haplotypes exhibit higher IRF5 levels (43) and are associated with increased levels of IFN-α (44–46), suggesting that expression of IRF5 contributes to the development of autoimmune diseases (47).
In this study, we investigated the role of IRF5 and IRF7 for the difference in IFN-α production observed between females and males. Our results demonstrate that IRF5 levels are regulated by the estrogen receptor α (ERα) in mice, and that sex difference in IRF5 expression in human pDCs can lead to higher IFN-α production in females compared with males after TLR7 stimulation, providing potential novel targets for the modulation of inflammation and immune responses in both chronic viral and autoimmune diseases.
Materials and Methods
Study subjects and samples
Human samples were collected from individuals recruited and enrolled at Massachusetts General Hospital, and all subjects gave written, informed consent for participation in these studies. The study was approved by the Partners Human Research Committee. Characteristics of the patient cohort are available in Supplemental Table I. No significant differences in age (p = 0.18, two-tailed Mann–Whitney U test) or ethnicity were noticed (p = 0.1, Fischer Exact test) between the 53 females and 37 males included in this study. When available (n = 26), information on the use of oral contraceptives containing sex hormones was collected. The female study subjects included 18.9% (n = 10) of postmenopausal or surgically sterile females; 50% (n = 8) and 18% (n = 3) of premenopausal females reported using oral contraceptives and using an intrauterine device, respectively. Subgroups were used in the different analyses performed, with some donors being tested across multiple assays. Blood was collected in lithium heparin tubes, and PBMCs were separated from whole blood by Ficoll-Histopaque density centrifugation (Sigma-Aldrich, St. Louis, MO). Cells were resuspended in R-10 [RPMI 1640 (Sigma-Aldrich) containing 10% heat-inactivated FBS (Sigma-Aldrich), 2500 U/ml penicillin, 2500 μg/ml streptomycin, 100 mM l-glutamine (Corning, Lowell, MA)] and counted. Blood was processed within 5 h after venipuncture to prevent the loss of pDC responsiveness to TLR ligands (48).
Mice
Mice selectively lacking ERα in the hematopoietic compartment or in the DC lineage were generated by crossing B6 mice carrying an estrogen receptor 1 (Esr1) gene in which exon 2 was flanked by loxP sites (ERαfl/fl) with B6 mice expressing the Cre recombinase under the control of the Tie2 promoter-enhancer (Tie2-ERαKO) or the CD11c promoter (CD11c-ERαKO) as described elsewhere (49). Littermate wild-type (WT) mice were used as controls. Mice were bred and maintained in a specific pathogen-free animal facility. Eight- to 12-wk-old female mice were used in all experiments. The INSERM U1043 Institutional Review Board for animal experimentation approved protocols.
Measurement of single-cell cytokine production by flow cytometry
Intracellular cytokine staining assays were carried out as previously described (25). In brief, freshly isolated PBMCs were resuspended in R-10 at a concentration of 1.5 million cells/ml, and 1 ml PBMCs was stimulated in FACS tubes with 1 μg/ml CL097, a synthetic TLR7 ligand (imidazoquinoline; Invivogen, San Diego, CA). A total of 5 μg/ml brefeldin A (Sigma-Aldrich) was added to each tube immediately after addition of the stimulant to inhibit cellular cytokine release. Unstimulated cells with 5 μg/ml brefeldin A added served as a negative control. Intracellular cytokine content of pDCs was determined after 20 h of stimulation as previously described (50). PBMCs were stained for surface markers using anti-CD3 Alexa Fluor 700, anti-CD19 Alexa Fluor 700, anti-CD56 Alexa Fluor 700, anti-CD11c PE, anti-CD14 allophycocyanin-Cy7, anti–HLA-DR Pacific blue, and anti-CD123 PE-Cy5 (all from BD Biosciences, San Jose, CA). pDCs were defined as CD3negCD19negCD56negHLA-DRposCD14negCD11cnegCD123bright cells. Cells were fixed and permeabilized using Fix&Perm Medium A and B (Invitrogen, Carlsbad, CA) and stained intracellularly with anti–IFN-α FITC (PBL IFN Source, Piscataway, NJ), anti–IL-12 allophycocyanin, and anti–TNF-α PE-Cy7 (BD Biosciences). Flow cytometry data were acquired within 2 h of staining on a BD Biosciences LSRII equipped with four lasers. Spectral overlap was corrected by appropriate compensation, and rainbow beads were used to maintain the consistency of the fluorescence intensity between experiments. The frequency of cytokine-producing pDCs was determined by subsequent analysis using FlowJo software (version 8.5.2; Tree Star, Ashland, OR). Unstimulated cells were used to define background cytokine production level and subtracted from the frequency in stimulated samples.
Measurement of ex vivo protein levels of transcription factors by flow cytometry
Freshly isolated PBMCs were fixed by adding cold 4% paraformaldehyde (PFA) directly into the culture medium to obtain a final concentration of 2% PFA. Cells were incubated in fixative for 30 min at 37°C, then washed and pelleted. Cells were permeabilized by vortexing to resuspend in 500 μl ice-cold methanol and incubated for 10 min at −20°C, and subsequently washed twice. For IRF5 staining, fixed and permeabilized cells were incubated for 10 min at room temperature and in the dark with the unconjugated rabbit monoclonal IRF5 Ab (Abcam, Cambridge, MA), washed, and then stained with a secondary goat anti-rabbit Alexa Fluor 700 Ab (Invitrogen). Unconjugated rabbit IgG (Cell Signaling Technology, Danvers, MA) was used as isotype control. Cells were stained for surface markers for 30 min at room temperature as described earlier. For IRF7 staining, anti-IRF7 Alexa Fluor 488 (BD Biosciences) was also added to the surface stain mix. Finally, the cells were washed, pelleted, and resuspended in 100 μl PBS containing 2% heat-inactivated FBS. All washes were performed with PBS containing 2% heat-inactivated FBS at 4°C. The IFN-α secretion assay (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) was used in combination with this protocol as per manufacturer’s instructions. Samples were acquired on the BD Biosciences LSRII within 2 h of staining. The mean fluorescence intensities (MFIs) of IRF5 and IRF7 in pDCs, CD3+ T cells, and monocytes/conventional dendritic cells (cDCs) and the frequencies of IFN-α–secreting pDCs were determined by subsequent analysis using FlowJo software.
Subcellular quantification of IRF5 protein levels in pDCs using the TissueFAXS slide scanning system
Two million fresh PBMCs were seeded in 50 μl R-10 on poly-d-lysine–coated plates and simultaneously stimulated with 1 μg/ml CL097 for 2 h. Cells were then fixed with 4% PFA for 20 min at room temperature and permeabilized with ice-cold methanol (10 min at −20°C). Unconjugated anti-IRF5 (Abcam) or rabbit IgG (Cell Signaling Technology) was added and the plates were incubated at 4°C overnight. The slides were subsequently washed in PBS supplemented with 2% normal goat serum and incubated in goat anti-rabbit IgG Alexa Fluor 546 (Invitrogen) for 30 min at room temperature. Cells were successively stained at room temperature with CD123 allophycocyanin (BD Biosciences) for 1 h, goat anti-mouse IgG2a Alexa Fluor 647 (Invitrogen) for 30 min, HLA-DR Alexa Fluor 488 for 1 h (Exbio, Vestec, Czech Republic), goat anti-mouse IgG1 Alexa Fluor 488 (Invitrogen) for 30 min, with three washes in PBS supplemented with 2% normal goat serum between each stain. All slides were mounted in Prolong Gold Antifade reagent with DAPI (Invitrogen). The sample slides were scanned using the TissueFAXS (TissueGnostics GmbH, Vienna, Austria) slide scanning system based on a Zeiss Axio Imager Z2 upright epifluorescence microscope. Images were captured using a Zeiss EC Plan-Neofluar 100× 1.3NA objective in combination with a PCO (Kelheim, Germany) monochrome 12-bit CCD camera. This slide scanning system uses a Maerzhaeuser motorized scanning stage with eight-slide holder to permit scanning and stitching together of many fields of view into one image. In this way, all of the plated cells could be scanned on each coverslip and then the fluorescence intensity of the different markers evaluated on a per-cell basis using TissueQuest imaging analysis software (TissueGnostics GmbH). pDCs were identified as HLA-DR+CD123+ cells. Each fluorescence channel was thresholded to visually segment the cells based on average per-cell fluorescence intensity. Cytoplasm and nuclear IRF5 fluorescence was then separately determined using a cytoplasm mask for IRF5, a nuclear mask for IRF5, and a whole-cell mask for IRF5. For each sample, a minimum of 150 pDCs was imaged to determine the mean intensity of IRF5.
IRF5 recombinant protein delivery using a vector-free microfluidic platform
pDCs were enriched from PBMCs using the pDC Enrichment Kit (Stemcell, Vancouver, Canada) following manufacturer’s instructions. Cells were resuspended in R-10 at 1000 cells/μl and were mixed with Cascade Blue–labeled 3-kDa dextran molecules, for control of delivery, and with either 0.03–0.06 μg/μl IRF5 recombinant protein (Abcam or Origene, Rockville, MD) or 0.05–0.1 μg/μl of the control TUBA1A recombinant protein (Abcam), and subsequently placed in the device’s inlet reservoir. Delivery was performed using a vector-free microfluidic platform as previously described (51, 52) and illustrated in Supplemental Fig. 1A. In brief, cells were mechanically deformed while passing through the microfluidic device (SQZ Biotechnologies, USA) at a pressure of 80 or 120 psi, resulting in the transient formation of holes in the cell membrane allowing content from the surrounding buffer to diffuse into the cytosol. Cells were incubated at room temperature in the delivery solution for 5 min after treatment to ensure closure of membrane holes before being subjected to any further treatment, as previously described (52). Delivery efficiency was assessed using FITC-labeled, 70-kDa dextran probes and/or Cascade blue–labeled, 3-kDa dextran molecules mimicking protein and small interfering RNA deliveries, respectively. Appropriate controls were included to correct for reduced IFN-α–producing capacity of pDCs subjected to mechanical deformation. Delivery of TUBA1A protein, which is not involved in the IFN-α production pathway, was used as control for determining any nonspecific effect on IFN-α production. IRF5 levels were subsequently measured by flow cytometry as described earlier. Cells were then stimulated for 2 h with 1 μg/ml CL097. Supernatants were collected and run on the Milliplex Human 29 cytokine/chemokine magnetic bead panel kit (Millipore, Billerica, MA). Viability was assessed by using the Aqua LIVE/DEAD staining (Invitrogen) as per manufacturer’s instructions. IFN-α secretion was measured using Miltenyi’s IFN-α secretion assay following manufacturer’s instructions.
Intracellular cytokine staining of murine pDCs
Murine bone marrow (BM) cell suspensions were activated with a preparation of oligonucleotides PolyU (Sigma-Aldrich) with 1,2-dioleoyloxy-3-trimethylammonium-propane (DOTAP) [8 μl of a cationic liposome preparation (DOTAP; Roche) mixed with 1 μg PolyU in 150 μl RPMI, in a polystyrene tube]. BM cells were stimulated with PolyU-DOTAP preparation for 4 h, and 5 μg/ml brefeldin A (eBioscience) was added for the last 2 h of culture. Nonspecific staining was blocked with 5 μg/ml anti-CD16/CD32 (2.4G2; American Type Culture Collection). BM cell suspensions were then stained with PE-Cy7–labeled anti-CD11c (N418) and allophycocyanin-labeled mouse plasmacytoid dendritic cell Ag-1 (all from eBioscience). Intracellular cytokine staining was performed with mixed FITC-labeled IFN-α/β–specific Abs (RMMA-1/RMMB-1; PBL). Unstimulated cells and isotypes were used as control staining. Data were acquired on a Fortessa (BD Biosciences).
Quantification of IRF5 protein expression in mouse splenic B cells
B cells were individually purified from mouse spleens by positive selection using anti-CD19 beads (Miltenyi). Cells were lysed in lithium dodecyl sulfate sample buffer (Invitrogen) and analyzed by immunoblotting. Membranes were probed with anti-IRF5 (polyclonal rabbit IgG; Cell Signaling 4950) or anti–β-actin (monoclonal mouse IgG1; Sigma A1978) Abs, followed by incubation with appropriate HRP-conjugated secondary Abs. Densitometric analysis was performed using Image Lab software v5.0 (Bio-Rad).
Quantification of IRF5 and IRF7 mRNA expression in mouse pDCs
BM cell suspensions were stained with allophycocyanin-labeled mouse plasmacytoid dendritic cell Ag-1 and PE-Cy7–labeled anti-CD11c (all from eBioscience) for 30 min at 4°C, and double-positive cells were sorted using a FACSAria (BD Biosciences). RNA from purified pDCs was extracted using the NucleoSpin RNA XS and treated with DNase I following manufacturer’s instructions (Macherey-Nagel). RNA samples were retrotranscribed into cDNA using oligo-dT, random primers, and the SuperScript III Reverse Transcriptase (Life Technologies). Quantitative PCRs were performed using Irf5 and Irf7 QuantiTect Primer Assays with SYBR green PCR Mastermix (QIAGEN). Gene transcripts were normalized to Hprt gene abundance, and relative mRNA levels were calculated by the expression 2−∆Ct.
In situ IRF5 mRNA expression assay by flow cytometry
Five million PBMCs were pelleted and surface stained on ice for 30 min. Cells were subjected to the QuantiGene FlowRNA assay (eBioscience, San Diego, CA) as per manufacturer’s instructions with type6-B2M probe, type1-ESR1 probe, and a customized ultrasensitive type4-IRF5 probe (probes are all from eBioscience). To control for nonspecific probe interaction, we replaced type4-IRF5 probe and type1-Esr1 probe by type4-TLR7 probe and type1-TLR9 probe. The bacterial DapB probes were used as a control. To gain sensitivity, we increased target incubation time from 2 to 3 h. Similarly, preamplification and amplification incubation times were increased from 1.5 to 2 h. Samples were run in duplicates and acquired on the BD Biosciences Fortessa within 2 h of staining. The MFIs of IRF5, ESR1, and B2M probes were determined by subsequent analysis using FlowJo software. Values were excluded if the duplicates exhibit >20% difference.
Statistical analysis
Comparison between females and males was calculated using Wilcoxon rank tests (Mann–Whitney) or unpaired t tests. Comparison of IRF5 MFI between IFN-α–secreting pDCs and nonsecreting pDCs was calculated using the paired Wilcoxon rank tests. Linear regression was calculated using Spearman rank-based correlation. For IRF5 protein delivery experiments, we used Wilcoxon signed rank for comparison of the increase in the percentage of IFN-α secretion relative to the control therefore normalized to 1. Comparison between WT mice and ERαKO mice was calculated using the unpaired t tests.
Results
Sex differences in the IFN-α/TLR7 pathway in pDCs
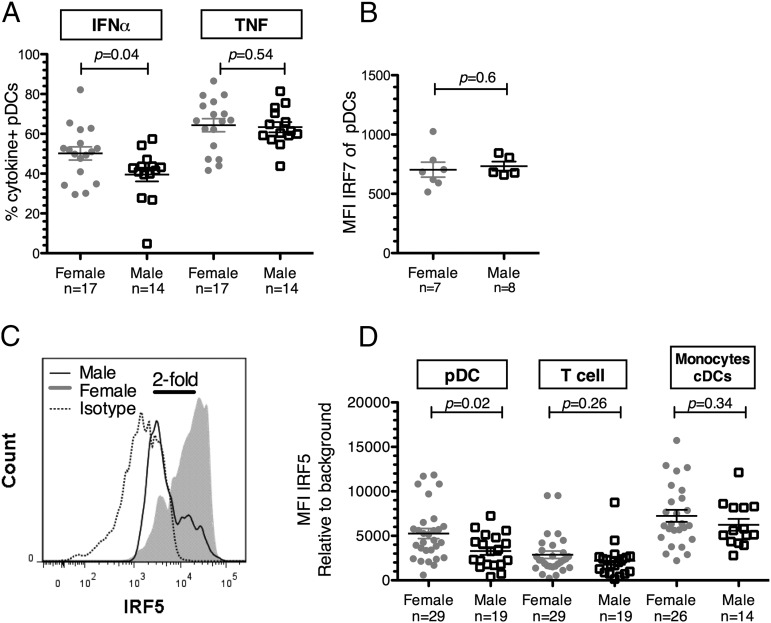
We and others have previously reported that pDCs isolated from females produce markedly more IFN-α in response to TLR7 ligands than pDCs derived from males (24–26). These results were confirmed in this study by measuring the frequency of IFN-α–producing pDCs in a first group of 31 healthy individuals (17 females, 14 males) (Supplemental Table I). A significantly higher percentage of IFN-α–producing pDCs after 20 h of stimulation with the synthetic TLR7/8 ligand CL097 was observed in females than in males (p = 0.04, two-tailed Mann–Whitney U test; Fig. 1A). Neither age nor ethnicity influenced IFN-α production by pDCs (p = 0.1, r = 0.3, Spearman rank-based correlation; p = 1.0, Fisher Exact test). The mean frequency of IFN-α–producing pDCs was 50.15% in females and 39.53% in males, in line with previous reports (25). In contrast, no sex difference was noticed in the percentage of TNF-α–producing pDCs (p = 0.54, two-tailed Mann–Whitney U test; Fig. 1A).
FIGURE 1.
Sex differences in TLR7 signaling in pDCs. (A) The percentage of IFN-α–secreting pDCs was significantly higher in females (n = 17) than in males (n = 14), whereas the percentage of TNF-α–secreting pDCs did not differ between females and males after 20 h of stimulation with 1 μg/ml CL097. Brefeldin A was added at 5 μg/ml at time of stimulation to block cytokine secretion. (B) MFI of IRF7 was determined ex vivo in pDCs from females (n = 7) or age-matched male donors (n = 5) using FlowJo software. No difference between sex in IRF7 expression in pDCs was observed. (C) Flow cytometry histogram overlays show the MFI of the IRF5 in pDCs from a male representative of the male average (empty curve) and in pDCs from a female representative of the female average (filled gray curve). There was a 2-fold difference in the MFI from those two samples. Isotype control is represented with dotted curve. (D) MFI of IRF5 was determined ex vivo in pDCs, T cells, and monocytes/cDCs from females (n = 29 for pDCs and T cells, and n = 26 from monocytes/cDCs) or age-matched male donors (n = 19 for pDCs and T cells, and n = 14 from monocytes/cDCs) using FlowJo software. pDCs derived from females exhibited significantly higher IRF5 levels than pDCs derived from males. IRF5 levels in T cells did not differ between females and males. Comparison between sexes was calculated using the Wilcoxon rank tests. Error bars indicate the mean and SEM.
Mechanisms underlying sex difference in IFN-α production remain to be elucidated. IRF7 and IRF5 are two crucial transcription factors activated upon TLR7 stimulation that modulate IFN-α production (33). Ex vivo levels of IRF5 and IRF7 in pDCs were measured subsequently by flow cytometry in a second group of healthy donors (Supplemental Table I). No sex difference was observed in the ex vivo levels of IRF7 in pDCs (p = 0.64, two-tailed Mann–Whitney U test; Fig. 1B). In contrast, pDCs derived from females contained 1.6 times more IRF5 than pDCs derived from age- and ethnicity-matched males, as measured by the MFI level of ex vivo IRF5 expression (females: n = 29, males: n = 19; p = 0.02, two-tailed Wilcoxon Rank test; Fig. 1D). IRF5 expression among pDCs appeared heterogeneous with some pDCs expressing no or very low levels of IRF5 as determined by the use of an isotype control (Fig. 1C). Notably, no difference in IRF5 protein levels was noticed between premenopausal females under hormonal birth control (n = 8) and those without hormonal birth control (n = 11). Ex vivo expression levels of IRF5 protein were also examined in CD3+ T cells and in monocytes/cDCs, in which the MFI of IRF5 was significantly lower and higher than in pDCs (p < 0.0001, Wilcoxon matched-pairs signed rank test), respectively. Ex vivo expression levels of IRF5 in CD3+ T cells and monocytes/cDCs did not significantly differ between females and males (p = 0.26 and p = 0.34, respectively, two-tailed Mann–Whitney U test; Fig. 1D), suggesting that the sex difference in ex vivo levels of IRF5 is cell type dependent. These results suggest a potential role of IRF5, but not IRF7, levels in the observed sex differences in IFN-α production.
Sex difference in basal IRF5 protein levels can influence pDC responses to TLR7 stimulation
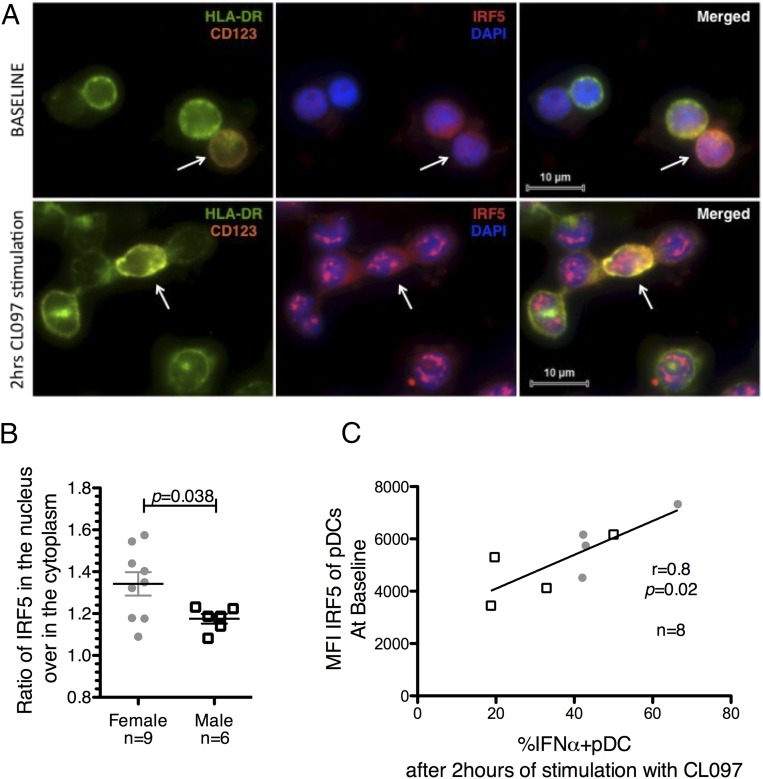
To gain a better understanding of the biological consequences of the sex difference in basal protein levels of IRF5 in pDCs, we measured IRF5 subcellular localization after TLR7 stimulation using the TissueFAXS slide scanning system. PBMCs were stimulated for 2 h with CL097 before staining for IRF5. pDCs were identified among all of the plated cells as HLA-DR+CD123+ (Fig. 2A), and the levels of IRF5 in both the cytoplasm and the nucleus from all pDCs on the slide were subsequently quantified. The ratio of nuclear IRF5 to cytoplasmic IRF5 was calculated to determine the changes in IRF5 translocation to the nucleus. Higher ratios of IRF5 in the nucleus after stimulation were observed in females than in males (p = 0.038; unpaired t test; Fig. 2B). The total amount of IRF5 among pDCs did not vary between baseline and after 2 h of stimulation with CL097 (p = 0.19, paired Mann–Whitney U test). This suggests that the sex difference in IRF5 levels observed before stimulation might translate to higher nuclear IRF5 content after TLR7 stimulation.
FIGURE 2.
IRF5 in pDCs is activated upon TLR7 stimulation and associated with IFN-α secretion. (A and B) PBMCs were stimulated with CL097 at 1 μg/ml for 2 h and then stained for IRF5–Alexa Fluor 546, HLA-DR–Alexa Fluor 488, and CD123–Alexa Fluor 647. The sample slides were scanned in Prolong Gold using the slide scanning system based on a Zeiss Axio Imager Z2 upright epifluorescence microscope. Images were captured using a Zeiss EC Plan-Neofluar 100× 1.3NA objective in combination with a PCO monochrome 12-bit CCD camera. pDCs were identified among all of the plated cells as HLA-DR+CD123+ cells. Quantification was performed using the TissueQuest analysis software. (A) Representative images at baseline and after 2 h of stimulation are shown with CD123 in orange, HLA-DR in green, DAPI staining of the nucleus in blue, and IRF5 in red. (B) The dot plot represents the ratio of nuclear IRF5 on cytoplasmic IRF5 in pDCs after 2 h of stimulation with CL097. Higher IRF5 ratio in the nucleus in females (n = 9) than in males (n = 6). Error bars indicate the mean and SEM. Comparison between sexes was calculated using unpaired t tests. (C) IRF5 and IFN-α expression were measured in pDCs at baseline and after 2 h of stimulation with CL097, respectively. A significant correlation between the basal levels of IRF5 in pDCs and the percentage of IFN-α–secreting pDCs after 2 h of stimulation with CL097 was observed. Linear regression was calculated in eight healthy donors (females: n = 4, close round shapes; males: n = 4, open squares) with Spearman rank-based correlation.
The consequences of the sex difference in basal levels of IRF5 in pDCs on IFN-α production by pDCs were subsequently examined by measuring ex vivo IRF5 levels and the percentage of IFN-α–secreting pDCs by flow cytometry. Ex vivo IRF5 levels before stimulation positively correlated with the percentage of IFN-α–secreting pDCs after 2 h of stimulation with CL097 (r = 0.8, p = 0.02, Spearman rank-based correlation; Fig. 2C). Nevertheless, a subset of IFN-α–secreting pDCs expressed no or low IRF5 levels, suggesting that IRF5 may not be the sole factor involved in IFN-α production. Altogether, these data demonstrate a link between the basal quantity of IRF5 in pDCs and the production of IFN-α.
Delivery of exogenous IRF5 protein increases IFN-α secretion in response to TLR7 stimulation in pDCs
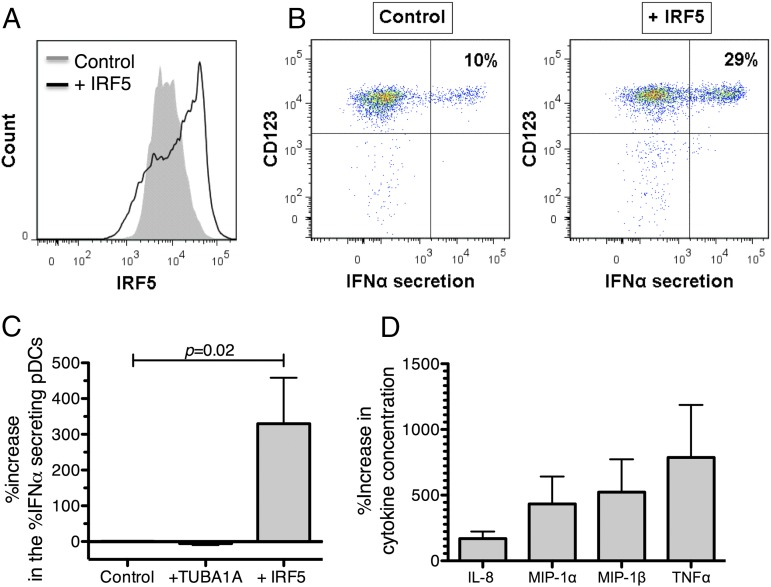
We further examined the direct impact of IRF5 protein levels on IFN-α production. Given that pDCs left in culture for >6 h have significantly reduced capabilities to produce IFN-α in response to TLR7 stimulation (48), techniques involving long incubation period such as small interfering RNA or transfection of vectors containing IRF5 cannot be applied. Thus, we used a technique recently described by Sharei and colleagues (51) for the direct delivery of IRF5 recombinant protein into human primary pDCs. A microfluidic device injects cells in narrow lanes inducing cell constriction, creating transient holes in plasma membranes, and enabling passive entry of molecules. Different microfluidic devices (described in Supplemental Table II) were tested as the size of the constriction and the number of constrictions were previously shown to influence delivery efficiency (51). The best delivery efficiency without significant loss of cell viability was obtained using the 10-4 × 5iS chip where 10 is the length of constriction in micrometers, 4 is the width of the constriction in micrometers, and 5 is the number of times the constriction is repeated through each channel (Supplemental Fig. 1C, 1D). pDCs subjected to this optimized delivery method exhibited decreased IFN-α secretion in response to TLR7 ligand compared with untreated pDCs, whereas no unspecific IFN-α production was induced in the absence of TLR7 ligand. The 10-4 × 5iS device enabled efficient delivery of IRF5 protein into live primary pDCs (Fig. 3A, Supplemental Fig. 1B), allowing us to assess the impact of IRF5 protein delivery into primary pDCs on IFN-α production. Fig. 3B shows representative plots of IFN-α–secreting pDCs with or without IRF5 protein delivery. IRF5 delivery resulted in a significant increase (3-fold) in the percentage of IFN-α–secreting pDCs (n = 7, p = 0.02, Wilcoxon signed rank test; Fig. 3C). In contrast, delivery of TUBA1A protein, a protein irrelevant for the pDC TLR7/IFN-α pathway, did not lead to changes in IFN-α production (Fig. 3C), validating that the increase of IFN-α secretion was not due to unspecific stimulation by the protein delivery method.
FIGURE 3.
Exogenous IRF5 enhance IFN-α production after TLR7 stimulation. Enriched pDC population was subjected to the microfluidic device with or without (control) recombinant IRF5 protein in the surrounding milieu. IRF5 levels were measured at baseline. pDCs were then stimulated with CL097 for 2 h and IFN-α secretion was measured. (A) Flow cytometry histogram overlays show the mean intensities of the IRF5 in pDCs after IRF5 delivery (empty curve) and in the control (filled curve) after treatment with the microfluidic device. A 2-fold increase in the MFI of IRF5 in pDCs is observed upon IRF5 delivery. (B) Primary flow data of IFN-α secretion in pDCs after 2 h of TLR7 stimulation in the control sample and after IRF5 delivery. (C) The percentage of IFN-α–secreting pDCs increased specifically upon delivery of IRF5 protein. The percentage of IFN-α–secreting pDCs upon delivery of IRF5 was normalized to the control. In contrast, no significant changes occurred in the percentage of IFN-α–secreting pDCs upon delivery of TUBA1A, a protein irrelevant to the IFN-α pathway. Data for IRF5 delivery are representative of seven independent experiments. Wilcoxon signed rank for comparison of the percentage increase in the percentage of IFN-α secretion relative to the control therefore normalized to 1. (D) Supernatants were collected after 2 h of stimulation and run on the Milliplex Human 29 cytokine/chemokine magnetic bead panel kit (Millipore). Increase in the concentration of TNF-α, MIP-1α, MIP-1β, and IL-8 is observed upon delivery of IRF5 protein. Error bars indicate the mean and SEM.
Apart from its role in IFN-α production, IRF5 has been described to also promote transcription of proinflammatory cytokines such as TNF-α upon TLR stimulation (38). Supernatants were therefore collected after 2 h of CL097 stimulation from control pDCs and pDCs in which IRF5 protein was overexpressed, and cytokines were measured using the Milliplex Human 29 cytokine/chemokine magnetic bead panel kit. Consistent with previous work (53), 2 h of CL097 stimulation did not induce the production of IL-2, IL-4, IL-5, IL-7, IL-10, IL-13, IL-15, IL-17, IFN-γ, MIG, and eotaxin, whereas TNF-α, IL-8, MIP-1α, and MIP-1β protein secretion were induced. pDCs in which IRF5 protein was overexpressed exhibited increased TNF-α, IL-8, MIP-1α, and MIP-1β production compared with control pDCs (Fig. 3D). Overall, we showed increased production of IFN-α and other inflammatory cytokines after delivery of exogenous IRF5 protein into primary human pDCs using a novel method of protein delivery, confirming the role of IRF5 in mediating TLR7 signaling and cytokine production in human pDCs (54).
Sex difference in IRF5 levels in pDCs is associated with ERα signaling
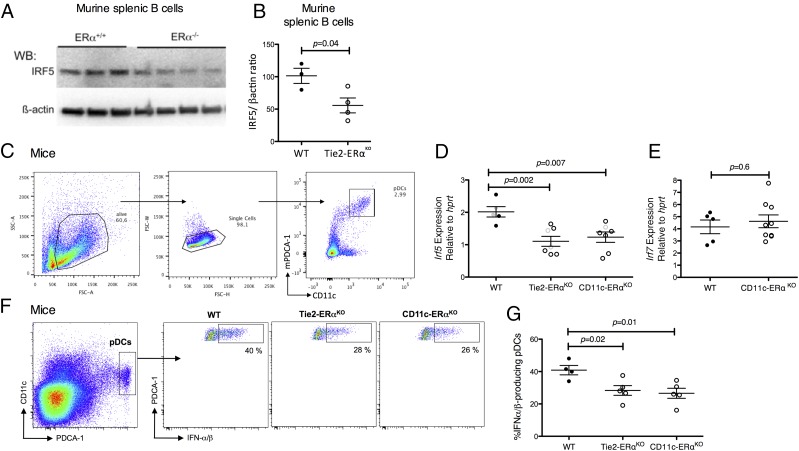
Previous reports have shown that estrogens can modulate IFN-α production by pDCs both in mice (26) and in humans (55). Studies have also shown higher Irf5 mRNA levels in splenic B cells derived from female than in age-matched male mice and from WT mice as compared with ERα knockout mice (56). We therefore investigated ERα-dependent regulation of IRF5 in female mice using tissue-specific ERα-deficient mice (26, 49). Before assessing levels of Irf5 mRNA in pDCs, and in agreement with previous works (56), we observed a lower expression of IRF5 protein in splenic B cells from Tie2-ERαKO mice, lacking ERα in the hematopoietic compartment, as compared with ERαflox/flox WT mice (p = 0.04, unpaired t test; Fig. 4A, 4B) (56). We then sorted BM pDCs from ERαflox/flox WT, Tie2-ERαKO, and CD11c-ERαKO female mice, which specifically lack ERα in dendritic cells (Fig. 4C). We observed that BM pDCs derived from unmanipulated Tie2-ERαKO female mice and CD11c-ERαKO female mice exhibited significantly less Irf5 mRNA expression than in pDCs from WT mice (p = 0.003 and p = 0.007, respectively, unpaired t test; Fig. 4D). By contrast, Irf7 mRNA levels were similar between WT and CD11c-ERαKO pDCs (p = 0.6, unpaired t test; Fig. 4E). We further demonstrated that reduced Irf5 expression in BM-derived pDCs from Tie2-ERαKO mice and from CD11c-ERαKO mice was associated with an impaired capacity of pDCs to produce IFN-α in response to TLR7 engagement. Indeed, significantly less IFN-α was produced in response to TLR7 stimulation by pDCs from Tie2-ERαKO or CD11c-ERαKO mice than from WT littermate controls (p = 0.02 and p = 0.01, respectively, unpaired t test; Fig. 4G). These results demonstrate that Irf5 expression correlates with the level of TLR7-mediated IFN-α production in pDCs and is, at least partially, regulated by ERα signaling.
FIGURE 4.
Defective estrogen signaling in mice is associated with reduced IRF5 levels and diminished TLR7-mediated production of IFN-α by pDCs. (A and B) B cells were individually purified from WT (n = 3) and Tie2-ERαKO (n = 4) mice spleens by positive selection using anti-CD19 beads (Miltenyi). Protein lysates of B cells were analyzed by immunoblotting for IRF5 and β-actin protein expression. (A) Representative Western blot of the results is shown. (B) Densitometry results normalized to β-actin show decreased IRF5 protein levels in splenic B cells from Tie2-ERαKO as compared with WT mice. (C–E) pDCs from the BM of female WT Tie2-ERαKO and CD11c-ERαKO mice were sorted based on the expression of CD11c and PDCA-1 from BM cells. RNA samples were retrotranscribed into cDNA, and quantitative PCRs were performed using Irf5 and Irf7 QuantiTect Primer Assays with SYBR green PCR Mastermix (Qiagen). Gene transcripts were normalized to Hprt gene abundance, and relative mRNA levels were calculated by the expression 2−∆Ct. Comparison between mice groups was calculated using the unpaired t tests. (C) Gating strategy for pDC sorting is shown. (D) RNA was isolated for each group from one to two pools of four to five mice (in gray) and from three to six individual mice (in black). Irf5 mRNA levels were significantly lower in pDCs from Tie2-ERαKO (n = 14) and CD11c-ERαKO (n = 10) than in pDCs from WT mice (n = 12). (E) RNA was isolated from five individual WT mice and nine individual CD11c-ERαKO (n = 9) mice. Irf7 mRNA levels in pDCs did not differ between WT and CD11c-ERαKO mice. (F and G) BM cells from WT, Tie2-ERαKO, and CD11c-ERαKO mice were stimulated with the TLR7 ligand PolyU for 4 h, in the presence of brefeldin A for the last 2 h. (F) Representative flow cytometry plots showing IFN-α production by pDCs (CD11c+PDCA1+). (G) ERα deletion in hematopoietic compartment (Tie2-ERαKO) or in the DC lineage (CD11c-ERαKO) was associated with diminished frequency of IFN-α/β–producing pDCs from mice upon TLR7 stimulation. Comparison between mice groups was calculated using the unpaired t tests. Error bars indicate the mean and SEM.
To address the question of ERα-dependent regulation of the IRF5 gene in humans, we quantified ESR1 (ERα gene) and IRF5 mRNA transcripts in the same pDCs purified from humans using a novel flow cytometry–based in situ hybridization assay, the QuantiGene FlowRNA Assay. This assay based on the use of specific oligonucleotide probes coupled with branched DNA signal amplification offers the advantage over standard quantitative RT-PCR techniques to control for the potential heterogeneity in the expression among a defined cell type. Fig. 5A shows histograms of the detection of IRF5 mRNA and ESR1 mRNA. A significant correlation between the ESR1 and IRF5 transcripts was observed in pDCs from females, but not in pDCs from males (females: n = 10, r = 0.81, p = 0.04; males: n = 7, r = 0.45, p = 0.31, Spearman correlation; Fig. 5B). In addition, IRF5 mRNA and ESR1 mRNA in the pDC population correlated at the single-cell levels in both male and female healthy donors (Fig. 5C). Potential nonspecific interactions between the type1-Esr1 and type4-IRF5 probes were controlled for by simultaneously testing two different probes (type4-TLR7 and type1-TLR9) coupled to the same fluorophores as used for ESR1 and IRF5 probes. Overall, these data demonstrate that estrogen-dependent regulation of IRF5 transcription via ERα can result in sex differences in IRF5 levels of pDCs and downstream IFN-α production.
FIGURE 5.
Sex difference in IRF5 levels in human pDCs is associated with estrogen signaling. mRNA levels of IRF5 and ESR1 were measured in human pDCs using the QuantiGene FlowRNA assay. (A) Flow cytometry histogram overlays show the mean intensities of IRF5 or Esr1 mRNA (empty curve) compared with control with the irrelevant dapB probes (filled gray curve) in pDCs. (B) A significant correlation between IRF5 and ESR1 mRNA levels is observed in human pDCs derived from females (closed round shapes, n = 10, p = 0.04, r = 0.84), but not in pDCs derived from males (open squares, n = 7, p = 0.31, r = 0.45). Samples were run in duplicates. Linear regression was calculated with Spearman rank-based correlation. Error bars indicate the mean and SEM. (C) Flow cytometry contour plots show correlated ESR1 mRNA and IRF5 mRNA expressions in the pDC population at the single-cell level, respectively, in representative male (left panel) and female (right panel) healthy donors. Control with irrelevant type 1 and 6 probes is shown in gray in both plots.
Discussion
Important differences exist between males and females in the outcome of infectious diseases and occurrence of autoimmune diseases for which the pDC IFN-α response has been implicated in the pathology. Sex differences in IFN-α production by pDCs upon TLR7 stimulation have been previously described by our group and others (24–26). In this study, we investigated the potential role of two key regulators of IFN-α production, IRF7 and IRF5, in sex differences in IFN-α production by pDCs. Although no sex-based difference in IRF7 expression was observed, significantly higher protein levels of IRF5 were detected in pDCs purified from females than in pDCs derived from males. Basal protein levels of IRF5 were positively associated with the level of IFN-α secretion by TLR7-stimulated pDCs, and experimental increase in IRF5 protein levels resulted in increased IFN-α secretion by human pDCs. BM-derived pDCs from female mice with conditional ERα knockout exhibited impaired capacity to produce IFN-α in response to TLR7 engagement and significantly less Irf5 mRNA expression than pDCs from WT mice. Altogether, these results identify sex hormone–mediated regulation of IRF5 levels in pDCs as an underlying mechanism for the higher percentage of IFN-α–secreting pDCs in females as compared with males after TLR7 stimulation.
Previous studies have reported conflicting results regarding the role of IRF5 in IFN-α transcription (33, 38, 57, 58). These studies differed in cell type and stimulations used, suggesting that the impact of IRF5 on type I IFNs transcription may be cell type and stimulus dependent. In our study, the basal levels of IRF5 were found to be significantly higher in pDCs from females than in those from males. IRF5 was furthermore observed to be functional upon TLR7 stimulation, with IRF5 levels before stimulation being correlated with the percentage of IFN-α–secreting pDCs after 2 h of TLR7 stimulation. However, it should be noted that a subset of IFN-α–secreting pDCs with undetectable IRF5 levels was also present. This observation suggests that IRF5 alone does not account for all the IFN-α produced by pDCs and may reflect the well-known role of IRF7 in IFN-α production. Altogether, these data demonstrate that sex difference in basal levels of IRF5 can drive sex differences in IFN-α production upon TLR7 stimulation.
Although sex-based differences were observed in pDCs, no sex difference in IRF5 expression was observed in T cells, confirming a cell-specific regulation of IRF5 expression. A potential explanation for the cell-specific differences in IRF5 might be the presence of multiple alternatively spliced isoforms of IRF5. These different IRF5 isoforms have been shown to have distinct cell type–specific expression, regulation, cellular localization, and function (59). Human pDCs express four distinct alternatively spliced isoforms (V1, V2, V3, and V4), with V3 and V4 being the most predominant transcripts expressed in unstimulated pDCs. In contrast, the V4 (and also V1) isoforms are not detected in unstimulated T cells (59). In addition, cell type–specific posttranscriptional regulation, for example, by the expression of cell-specific microRNAs, may be involved in differences in IRF5 expression (60, 61). Basal IRF5 expression among pDCs is heterogeneous, with some pDCs expressing no or low levels of IRF5. Although the Ab against IRF5 used in this study is recognizing all IRF5 isoforms, it is possible that differences in the affinity for the various isoforms may account for the low IRF5 levels reported in some pDCs. Overall, our data show that the role of IRF5 in IFN-α transcription is cell type specific and results in differential sex-based expression of IRF5 as well as IFN-α expression in pDCs.
To clarify the impact of IRF5 on IFN-α transcription in primary human pDCs, we used a novel technique previously described to deliver exogenous recombinant protein into human primary pDCs (51). The delivery of recombinant IRF5 protein led to an increase in the percentage of IFN-α–secreting pDCs, confirming the ex vivo association observed between IRF5 levels and TLR7-induced IFN-α production. In addition to IFN-α production, IRF5 is also involved in the transcription of proinflammatory cytokines such as IL-6 and TNF-α (38). In agreement with this, we observed an increase in the production of TNF-α, IL-8, MIP-1α, and MIP-1β upon IRF5 delivery in cells and after 2 h of TLR7 stimulation. Previous studies by our group and others did not detect sex differences in TNF-α production after overnight stimulation with TLR7 ligand (24, 25). In contrast, Seillet and colleagues (26) recently reported an increased frequency in TNF-α–producing pDCs in women as compared with men upon 5 h of stimulation with a TLR7 ligand. Such discrepancy could be explained by the shorter stimulation times used in Seillet et al.’s study (26), which may have unmasked the sex differences by limiting cytokine-specific feedback regulatory mechanisms. Therefore, although increased IRF5 may lead to increased TNF-α production by pDCs upon short stimulation time, feedback regulatory mechanisms may later be dampening this production so that no difference is observed with longer stimulation time.
Biological sex differences in the human immune responses to infections or autoimmune diseases can be caused by genetic factors linked to sex chromosomes and/or the modulation of immunity by sex hormones. The precise functional mechanisms by which sex hormones might regulate the IFN-α response of pDCs are unknown but are thought to involve ERα signaling (26, 27). ERs are expressed on all PBMCs including pDCs (26, 55, 62–64). The interaction of ERα with target gene promoters can occur either directly, through specific estrogen response elements, or indirectly through contacts with other DNA-bound transcription factors such as the specificity protein 1, but also as the AP-1 or the NF-κB, both involved in the transcription of type I IFNs and proinflammatory cytokines (65–68). A previous study in mice showed higher Irf5 mRNA levels in splenic B cells from female than from age-matched male mice and lower levels of Irf5 mRNA in ERα knockout mice as compared with WT mice (56). pDCs from Tie2-ERαKO and CD11c-ERαKO intact female mice lack ERα in the hematopoietic compartment or in CD11c+ DC lineage, respectively, and have been shown to be impaired in their capacity to produce IFN-α in response to TLR9 engagement (26). We further show in this article that pDCs from Tie2-ERαKO and CD11c-ERαKO mice are also altered in their ability to produce IFN-α after ex vivo TLR7 stimulation, and that this was associated with decreased Irf5 mRNA expression levels as compared with their littermate controls.
Species-specific differences, in particular in splice patterns of IRF5, may bias the translation of our results obtained from mice to humans. Indeed, spliced variants of IRF5 were identified only in human cells, whereas in inbred strains of mice Irf5 encodes for a dominant unspliced transcript (59). Interestingly, an ERα binding site has been identified using the University of California Santa Cruz Genome Browser (http://genome.ucsc.edu) at position 128561334–128561609 on human chromosome 7, which is 16,385 bp upstream the IRF5 gene (IRF5 chr7:128577994–128590088), suggesting that IRF5 may also be regulated by sex hormones in humans. Furthermore, one of the described IRF5 polymorphism in humans, the CGGGG indel, is associated with increased expression of IRF5 itself because of the presence of an additional SP1 binding site, an ERα cofactor (47, 69). In this study, we found a significant correlation between IRF5 and ESR1 mRNA levels in pDCs from females but not in pDCs from males in humans, highlighting the dependency of IRF5 mRNA on estrogen signaling in females. Although we were not able to simultaneously measure IRF5 mRNA and ERα protein in the same cells, previous reports suggested that ESR1 mRNA and ERα protein expression correlate (70–72). Importantly, the ERαKO mice models used in this study do not exclude that nonhormonal pathways and particularly X-chromosome–linked factors participate in sex-specific regulation of IRF5 in pDCs. Recently, it was shown that X chromosome dosage contributed independently from sex hormones to the sex bias in the pDC TLR7-mediated IFN-α response (55). Accordingly, we observe a trend toward higher IRF5 levels persisting in postmenopausal females as compared with age-matched males (p = 0.06). This might also explain why we did not observe significant difference in IRF5 levels between premenopausal females under hormonal birth control and premenopausal females having regular menstrual cycle.
In conclusion, this study demonstrates that pDCs from healthy females exhibit higher basal levels of IRF5 than pDCs from healthy males. Furthermore, higher levels of IRF5 in pDCs are directly associated with higher IFN-α responses to TLR7 stimulation. This sex-based difference appeared to be partially due to ERα signaling-mediated modulation, because Irf5 mRNA expression was significantly reduced in female mice with a conditional knockout for ERα and correlated, in humans, to ESR1 mRNA expression in pDCs from females. These data provide new insights into the mechanisms underlying the higher inflammation observed in females in infectious and autoimmune diseases, and identify IRF5 as an attractive target for specific modulation of the IFN-α pathway.
Supplementary Material
Acknowledgments
We are grateful to the healthy volunteers for blood donations and to Pamela Richtmyer, who provided tremendous support for the recruitment. We thank Mike Waring and Adam Chicoine for excellent assistance with flow cytometry through the Harvard University Center for AIDS Research (HU CFAR) Immunology Core at the Ragon Institute. We thank Véronique Morin, Anne Oudin, and Rima Zoorob for excellent assistance at the Centre d’Immunologie et des Maladies Infectieuses.
This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases Grants R01 AI078784 and P01 AI078897, fellowships from the National Health and Medical Research Council of Australia (Grant 519578 to J.J.C.), a Ragon Fellowship from The Phillip T. and Susan M. Ragon Foundation (to J.J.C.), a fellowship from the French National Agency for Research on AIDS and Viral Hepatitis (Grant 2013-219 to M.G.), the Fondation pour la Recherche Médicale (Grant DEQ2000329169 to J.-C.G.), the Conseil Régional Midi-Pyrénées (J.-C.G.), the Arthritis Fondation Courtin (J.-C.G.), and the Fondation ARC pour la recherche sur le cancer (J.-C.G.).
The online version of this article contains supplemental material.
- BM
- bone marrow
- cDC
- conventional dendritic cell
- DOTAP
- 1,2-dioleoyloxy-3-trimethylammonium-propane
- ERα
- estrogen receptor α
- Esr1
- estrogen receptor 1
- IRF
- IFN regulatory factor
- MFI
- mean fluorescence intensity
- pDC
- plasmacytoid dendritic cell
- PFA
- paraformaldehyde
- WT
- wild-type.
Disclosures
A.S. has personal financial interest in SQZ Biotechnologies. A.S. had no influence on the results and discussion presented in this article. M.G. was funded by eBioscience to present her work using the FlowRNA QuantiGene Assay at the meeting organized by the French Society of Immunology in Lille, France on November 5, 2014 (conference fees, second-class rail travel from Paris to Lille, and two nights’ accommodation). S.Z. received a grant from Gilead to study the impact of pregnancy on the IFN-α pathway in HIV-1–infected women.
References
- 1.Klein S. L., Jedlicka A., Pekosz A. 2010. The Xs and Y of immune responses to viral vaccines. Lancet Infect. Dis. 10: 338–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grossman C. J. 1985. Interactions between the gonadal steroids and the immune system. Science 227: 257–261. [DOI] [PubMed] [Google Scholar]
- 3.Olsen N. J., Kovacs W. J. 1996. Gonadal steroids and immunity. Endocr. Rev. 17: 369–384. [DOI] [PubMed] [Google Scholar]
- 4.Mitchell L. A. 1999. Sex differences in antibody- and cell-mediated immune response to rubella re-immunisation. J. Med. Microbiol. 48: 1075–1080. [DOI] [PubMed] [Google Scholar]
- 5.Green M. S., Shohat T., Lerman Y., Cohen D., Slepon R., Duvdevani P., Varsano N., Dagan R., Mendelson E. 1994. Sex differences in the humoral antibody response to live measles vaccine in young adults. Int. J. Epidemiol. 23: 1078–1081. [DOI] [PubMed] [Google Scholar]
- 6.Klein S. L. 2000. The effects of hormones on sex differences in infection: from genes to behavior. Neurosci. Biobehav. Rev. 24: 627–638. [DOI] [PubMed] [Google Scholar]
- 7.Boissier J., Chlichlia K., Digon Y., Ruppel A., Moné H. 2003. Preliminary study on sex-related inflammatory reactions in mice infected with Schistosoma mansoni. Parasitol. Res. 91: 144–150. [DOI] [PubMed] [Google Scholar]
- 8.Cao W., Manicassamy S., Tang H., Kasturi S. P., Pirani A., Murthy N., Pulendran B. 2008. Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat. Immunol. 9: 1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hannah M. F., Bajic V. B., Klein S. L. 2008. Sex differences in the recognition of and innate antiviral responses to Seoul virus in Norway rats. Brain Behav. Immun. 22: 503–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinstein Y., Ran S., Segal S. 1984. Sex-associated differences in the regulation of immune responses controlled by the MHC of the mouse. J. Immunol. 132: 656–661. [PubMed] [Google Scholar]
- 11.Villacres M. C., Longmate J., Auge C., Diamond D. J. 2004. Predominant type 1 CMV-specific memory T-helper response in humans: evidence for gender differences in cytokine secretion. Hum. Immunol. 65: 476–485. [DOI] [PubMed] [Google Scholar]
- 12.Hewagama A., Patel D., Yarlagadda S., Strickland F. M., Richardson B. C. 2009. Stronger inflammatory/cytotoxic T-cell response in women identified by microarray analysis. Genes Immun. 10: 509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Lunzen J., Altfeld M. 2014. Sex differences in infectious diseases-common but neglected. J. Infect. Dis. 209(Suppl. 3): S79–S80. [DOI] [PubMed] [Google Scholar]
- 14.Voskuhl R. 2011. Sex differences in autoimmune diseases. Biol. Sex Differ. 2: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein, S. L. 2012. Sex influences immune responses to viruses, and efficacy of prophylaxis and treatments for viral diseases. BioEssays 34: 1050–1059. [DOI] [PMC free article] [PubMed]
- 16.Klein S. L., Marks M. A., Li W., Glass G. E., Fang L. Q., Ma J. Q., Cao W. C. 2011. Sex differences in the incidence and case fatality rates from hemorrhagic fever with renal syndrome in China, 2004-2008. Clin. Infect. Dis. 52: 1414–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grebely J., Page K., Sacks-Davis R., van der Loeff M. S., Rice T. M., Bruneau J., Morris M. D., Hajarizadeh B., Amin J., Cox A. L., et al. InC3 Study Group 2014. The effects of female sex, viral genotype, and IL28B genotype on spontaneous clearance of acute hepatitis C virus infection. Hepatology 59: 109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Berg C. H., Grady B. P., Schinkel J., van de Laar T., Molenkamp R., van Houdt R., Coutinho R. A., van Baarle D., Prins M. 2011. Female sex and IL28B, a synergism for spontaneous viral clearance in hepatitis C virus (HCV) seroconverters from a community-based cohort. PLoS One 6: e27555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farzadegan H., Hoover D. R., Astemborski J., Lyles C. M., Margolick J. B., Markham R. B., Quinn T. C., Vlahov D. 1998. Sex differences in HIV-1 viral load and progression to AIDS. Lancet 352: 1510–1514. [DOI] [PubMed] [Google Scholar]
- 20.Addo M. M., Altfeld M. 2014. Sex-based differences in HIV type 1 pathogenesis. J. Infect. Dis. 209(Suppl. 3): S86–S92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson E. B., Brooks D. G. 2013. Decoding the complexity of type I interferon to treat persistent viral infections. Trends Microbiol. 21: 634–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganguly D., Chamilos G., Lande R., Gregorio J., Meller S., Facchinetti V., Homey B., Barrat F. J., Zal T., Gilliet M. 2009. Self-RNA-antimicrobial peptide complexes activate human dendritic cells through TLR7 and TLR8. J. Exp. Med. 206: 1983–1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teijaro J. R., Ng C., Lee A. M., Sullivan B. M., Sheehan K. C., Welch M., Schreiber R. D., de la Torre J. C., Oldstone M. B. 2013. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science 340: 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berghöfer B., Frommer T., Haley G., Fink L., Bein G., Hackstein H. 2006. TLR7 ligands induce higher IFN-alpha production in females. J. Immunol. 177: 2088–2096. [DOI] [PubMed] [Google Scholar]
- 25.Meier A., Chang J. J., Chan E. S., Pollard R. B., Sidhu H. K., Kulkarni S., Wen T. F., Lindsay R. J., Orellana L., Mildvan D., et al. 2009. Sex differences in the Toll-like receptor-mediated response of plasmacytoid dendritic cells to HIV-1. Nat. Med. 15: 955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seillet C., Laffont S., Trémollières F., Rouquié N., Ribot C., Arnal J. F., Douin-Echinard V., Gourdy P., Guéry J. C. 2012. The TLR-mediated response of plasmacytoid dendritic cells is positively regulated by estradiol in vivo through cell-intrinsic estrogen receptor α signaling. Blood 119: 454–464. [DOI] [PubMed] [Google Scholar]
- 27.Seillet C., Rouquié N., Foulon E., Douin-Echinard V., Krust A., Chambon P., Arnal J. F., Guéry J. C., Laffont S. 2013. Estradiol promotes functional responses in inflammatory and steady-state dendritic cells through differential requirement for activation function-1 of estrogen receptor α. J. Immunol. 190: 5459–5470. [DOI] [PubMed] [Google Scholar]
- 28.Ozato K., Tailor P., Kubota T. 2007. The interferon regulatory factor family in host defense: mechanism of action. J. Biol. Chem. 282: 20065–20069. [DOI] [PubMed] [Google Scholar]
- 29.Taniguchi T., Takaoka A. 2002. The interferon-alpha/beta system in antiviral responses: a multimodal machinery of gene regulation by the IRF family of transcription factors. Curr. Opin. Immunol. 14: 111–116. [DOI] [PubMed] [Google Scholar]
- 30.Honda K., Takaoka A., Taniguchi T. 2006. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25: 349–360. [DOI] [PubMed] [Google Scholar]
- 31.Barnes B. J., Moore P. A., Pitha P. M. 2001. Virus-specific activation of a novel interferon regulatory factor, IRF-5, results in the induction of distinct interferon alpha genes. J. Biol. Chem. 276: 23382–23390. [DOI] [PubMed] [Google Scholar]
- 32.Honda K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., Shimada N., Ohba Y., Takaoka A., Yoshida N., Taniguchi T. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434: 772–777. [DOI] [PubMed] [Google Scholar]
- 33.Schoenemeyer A., Barnes B. J., Mancl M. E., Latz E., Goutagny N., Pitha P. M., Fitzgerald K. A., Golenbock D. T. 2005. The interferon regulatory factor, IRF5, is a central mediator of toll-like receptor 7 signaling. J. Biol. Chem. 280: 17005–17012. [DOI] [PubMed] [Google Scholar]
- 34.Izaguirre A., Barnes B. J., Amrute S., Yeow W. S., Megjugorac N., Dai J., Feng D., Chung E., Pitha P. M., Fitzgerald-Bocarsly P. 2003. Comparative analysis of IRF and IFN-alpha expression in human plasmacytoid and monocyte-derived dendritic cells. J. Leukoc. Biol. 74: 1125–1138. [DOI] [PubMed] [Google Scholar]
- 35.Coccia E. M., Severa M., Giacomini E., Monneron D., Remoli M. E., Julkunen I., Cella M., Lande R., Uzé G. 2004. Viral infection and Toll-like receptor agonists induce a differential expression of type I and lambda interferons in human plasmacytoid and monocyte-derived dendritic cells. Eur. J. Immunol. 34: 796–805. [DOI] [PubMed] [Google Scholar]
- 36.Sato M., Suemori H., Hata N., Asagiri M., Ogasawara K., Nakao K., Nakaya T., Katsuki M., Noguchi S., Tanaka N., Taniguchi T. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13: 539–548. [DOI] [PubMed] [Google Scholar]
- 37.Marié I., Smith E., Prakash A., Levy D. E. 2000. Phosphorylation-induced dimerization of interferon regulatory factor 7 unmasks DNA binding and a bipartite transactivation domain. Mol. Cell. Biol. 20: 8803–8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takaoka A., Yanai H., Kondo S., Duncan G., Negishi H., Mizutani T., Kano S., Honda K., Ohba Y., Mak T. W., Taniguchi T. 2005. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature 434: 243–249. [DOI] [PubMed] [Google Scholar]
- 39.Nordang G. B., Viken M. K., Amundsen S. S., Sanchez E. S., Flatø B., Førre O. T., Martin J., Kvien T. K., Lie B. A. 2012. Interferon regulatory factor 5 gene polymorphism confers risk to several rheumatic diseases and correlates with expression of alternative thymic transcripts. Rheumatology (Oxford) 51: 619–626. [DOI] [PubMed] [Google Scholar]
- 40.Dieguez-Gonzalez R., Calaza M., Perez-Pampin E., de la Serna A. R., Fernandez-Gutierrez B., Castañeda S., Largo R., Joven B., Narvaez J., Navarro F., et al. 2008. Association of interferon regulatory factor 5 haplotypes, similar to that found in systemic lupus erythematosus, in a large subgroup of patients with rheumatoid arthritis. Arthritis Rheum. 58: 1264–1274. [DOI] [PubMed] [Google Scholar]
- 41.Graham R. R., Kozyrev S. V., Baechler E. C., Reddy M. V., Plenge R. M., Bauer J. W., Ortmann W. A., Koeuth T., González Escribano M. F., Pons-Estel B., et al. Argentine and Spanish Collaborative Groups 2006. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat. Genet. 38: 550–555. [DOI] [PubMed] [Google Scholar]
- 42.Sigurdsson S., Padyukov L., Kurreeman F. A., Liljedahl U., Wiman A. C., Alfredsson L., Toes R., Rönnelid J., Klareskog L., Huizinga T. W., et al. 2007. Association of a haplotype in the promoter region of the interferon regulatory factor 5 gene with rheumatoid arthritis. Arthritis Rheum. 56: 2202–2210. [DOI] [PubMed] [Google Scholar]
- 43.Feng D., Stone R. C., Eloranta M. L., Sangster-Guity N., Nordmark G., Sigurdsson S., Wang C., Alm G., Syvänen A. C., Rönnblom L., Barnes B. J. 2010. Genetic variants and disease-associated factors contribute to enhanced interferon regulatory factor 5 expression in blood cells of patients with systemic lupus erythematosus. Arthritis Rheum. 62: 562–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niewold T. B., Rivera T. L., Buyon J. P., Crow M. K. 2008. Serum type I interferon activity is dependent on maternal diagnosis in anti-SSA/Ro-positive mothers of children with neonatal lupus. Arthritis Rheum. 58: 541–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hedl M., Abraham C. 2012. IRF5 risk polymorphisms contribute to interindividual variance in pattern recognition receptor-mediated cytokine secretion in human monocyte-derived cells. J. Immunol. 188: 5348–5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rullo O. J., Woo J. M., Wu H., Hoftman A. D., Maranian P., Brahn B. A., McCurdy D., Cantor R. M., Tsao B. P. 2010. Association of IRF5 polymorphisms with activation of the interferon alpha pathway. Ann. Rheum. Dis. 69: 611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clark D. N., Read R. D., Mayhew V., Petersen S. C., Argueta L. B., Stutz L. A., Till R. E., Bergsten S. M., Robinson B. S., Baumann D. G., et al. 2013. Four promoters of IRF5 respond distinctly to stimuli and are affected by autoimmune-risk polymorphisms. Front. Immunol. 4: 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meier A., Fisher A., Sidhu H. K., Chang J. J., Wen T. F., Streeck H., Alter G., Silvestri G., Altfeld M. 2008. Rapid loss of dendritic cell and monocyte responses to TLR ligands following venipuncture. J. Immunol. Methods 339: 132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lélu K., Laffont S., Delpy L., Paulet P. E., Périnat T., Tschanz S. A., Pelletier L., Engelhardt B., Guéry J. C. 2011. Estrogen receptor α signaling in T lymphocytes is required for estradiol-mediated inhibition of Th1 and Th17 cell differentiation and protection against experimental autoimmune encephalomyelitis. J. Immunol. 187: 2386–2393. [DOI] [PubMed] [Google Scholar]
- 50.Meier A., Alter G., Frahm N., Sidhu H., Li B., Bagchi A., Teigen N., Streeck H., Stellbrink H. J., Hellman J., et al. 2007. MyD88-dependent immune activation mediated by human immunodeficiency virus type 1-encoded Toll-like receptor ligands. J. Virol. 81: 8180–8191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharei A., Zoldan J., Adamo A., Sim W. Y., Cho N., Jackson E., Mao S., Schneider S., Han M. J., Lytton-Jean A., et al. 2013. A vector-free microfluidic platform for intracellular delivery. Proc. Natl. Acad. Sci. USA 110: 2082–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharei A., Cho N., Mao S., Jackson E., Poceviciute R., Adamo A., Zoldan J., Langer R., Jensen K. F. 2013. Cell squeezing as a robust, microfluidic intracellular delivery platform. J. Vis. Exp. DOI: 10.3791/50980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Birmachu W., Gleason R. M., Bulbulian B. J., Riter C. L., Vasilakos J. P., Lipson K. E., Nikolsky Y. 2007. Transcriptional networks in plasmacytoid dendritic cells stimulated with synthetic TLR 7 agonists. BMC Immunol. 8: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steinhagen F., McFarland A. P., Rodriguez L. G., Tewary P., Jarret A., Savan R., Klinman D. M. 2013. IRF-5 and NF-κB p50 co-regulate IFN-β and IL-6 expression in TLR9-stimulated human plasmacytoid dendritic cells. Eur. J. Immunol. 43: 1896–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laffont S., Rouquié N., Azar P., Seillet C., Plumas J., Aspord C., Guéry J. C. 2014. X-Chromosome complement and estrogen receptor signaling independently contribute to the enhanced TLR7-mediated IFN-α production of plasmacytoid dendritic cells from women. J. Immunol. 193: 5444–5452. [DOI] [PubMed] [Google Scholar]
- 56.Shen H., Panchanathan R., Rajavelu P., Duan X., Gould K. A., Choubey D. 2010. Gender-dependent expression of murine Irf5 gene: implications for sex bias in autoimmunity. J. Mol. Cell Biol. 2: 284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin R., Yang L., Arguello M., Penafuerte C., Hiscott J. 2005. A CRM1-dependent nuclear export pathway is involved in the regulation of IRF-5 subcellular localization. J. Biol. Chem. 280: 3088–3095. [DOI] [PubMed] [Google Scholar]
- 58.Yanai H., Chen H. M., Inuzuka T., Kondo S., Mak T. W., Takaoka A., Honda K., Taniguchi T. 2007. Role of IFN regulatory factor 5 transcription factor in antiviral immunity and tumor suppression. Proc. Natl. Acad. Sci. USA 104: 3402–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mancl M. E., Hu G., Sangster-Guity N., Olshalsky S. L., Hoops K., Fitzgerald-Bocarsly P., Pitha P. M., Pinder K., Barnes B. J. 2005. Two discrete promoters regulate the alternatively spliced human interferon regulatory factor-5 isoforms. Multiple isoforms with distinct cell type-specific expression, localization, regulation, and function. J. Biol. Chem. 280: 21078–21090. [DOI] [PubMed] [Google Scholar]
- 60.Kriegel A. J., Liu Y., Liu P., Baker M. A., Hodges M. R., Hua X., Liang M. 2013. Characteristics of microRNAs enriched in specific cell types and primary tissue types in solid organs. Physiol. Genomics 45: 1144–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taniguchi K., Ito Y., Sugito N., Kumazaki M., Shinohara H., Yamada N., Nakagawa Y., Sugiyama T., Futamura M., Otsuki Y., et al. 2015. Organ-specific PTB1-associated microRNAs determine expression of pyruvate kinase isoforms. Sci. Rep. 5: 8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mao A., Paharkova-Vatchkova V., Hardy J., Miller M. M., Kovats S. 2005. Estrogen selectively promotes the differentiation of dendritic cells with characteristics of Langerhans cells. J. Immunol. 175: 5146–5151. [DOI] [PubMed] [Google Scholar]
- 63.Phiel K. L., Henderson R. A., Adelman S. J., Elloso M. M. 2005. Differential estrogen receptor gene expression in human peripheral blood mononuclear cell populations. Immunol. Lett. 97: 107–113. [DOI] [PubMed] [Google Scholar]
- 64.Yakimchuk K., Jondal M., Okret S. 2013. Estrogen receptor α and β in the normal immune system and in lymphoid malignancies. Mol. Cell. Endocrinol. 375: 121–129. [DOI] [PubMed] [Google Scholar]
- 65.Kushner P. J., Agard D. A., Greene G. L., Scanlan T. S., Shiau A. K., Uht R. M., Webb P. 2000. Estrogen receptor pathways to AP-1. J. Steroid Biochem. Mol. Biol. 74: 311–317. [DOI] [PubMed] [Google Scholar]
- 66.Stein B., Yang M. X. 1995. Repression of the interleukin-6 promoter by estrogen receptor is mediated by NF-kappa B and C/EBP beta. Mol. Cell. Biol. 15: 4971–4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Björnström L., Sjöberg M. 2005. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol. Endocrinol. 19: 833–842. [DOI] [PubMed] [Google Scholar]
- 68.Safe S., Kim K. 2008. Non-classical genomic estrogen receptor (ER)/specificity protein and ER/activating protein-1 signaling pathways. J. Mol. Endocrinol. 41: 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sigurdsson S., Göring H. H., Kristjansdottir G., Milani L., Nordmark G., Sandling J. K., Eloranta M. L., Feng D., Sangster-Guity N., Gunnarsson I., et al. 2008. Comprehensive evaluation of the genetic variants of interferon regulatory factor 5 (IRF5) reveals a novel 5 bp length polymorphism as strong risk factor for systemic lupus erythematosus. Hum. Mol. Genet. 17: 872–881. [DOI] [PubMed] [Google Scholar]
- 70.Du X., Li X. Q., Li L., Xu Y. Y., Feng Y. M. 2013. The detection of ESR1/PGR/ERBB2 mRNA levels by RT-QPCR: a better approach for subtyping breast cancer and predicting prognosis. Breast Cancer Res. Treat. 138: 59–67. [DOI] [PubMed] [Google Scholar]
- 71.Pentheroudakis G., Kotoula V., Eleftheraki A. G., Tsolaki E., Wirtz R. M., Kalogeras K. T., Batistatou A., Bobos M., Dimopoulos M. A., Timotheadou E., et al. 2013. Prognostic significance of ESR1 gene amplification, mRNA/protein expression and functional profiles in high-risk early breast cancer: a translational study of the Hellenic Cooperative Oncology Group (HeCOG). PLoS One 8: e70634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wirapati P., Sotiriou C., Kunkel S., Farmer P., Pradervand S., Haibe-Kains B., Desmedt C., Ignatiadis M., Sengstag T., Schütz F., et al. 2008. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 10: R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.