Abstract
The biological activity of the multifunctional cytokine interleukin-1 (IL-1) is mediated by its receptors. The aim of this study was to determine if an association exists between single nucleotide polymorphisms (SNPs) in the IL-1 type 1 and 2 receptor genes (IL1R1 and IL1R2) and the expression level of membrane-bound IL1Rs on subpopulations of mononuclear cells or serum levels of soluble IL-1 receptors. It was observed that healthy individuals with the genotype TT in SNP rs2234650:C>T had a lower percentage of intact CD14+ monocytes expressing IL1R1 on their surface. The SNP rs4141134:T>C in IL1R2 has also been associated with the percentage of intact CD3+ T cells expressing IL1R2. Furthermore, individuals carrying the CC allele of SNP rs4141134:T>C and the TT allele of SNP rs2071008:T>G in IL1R2 had a lower density of IL1R2s on the surface of CD14+ monocytes in lipopolysaccharide (LPS)-stimulated PBMC cultures. In summary, this study demonstrated that IL-1 receptor gene polymorphisms could be one of the factors influencing the expression of membrane-bound IL-1 receptors (IL1R) on immunocompetent cells.
Keywords: interleukin-1, membrane-bound receptor, SNPs, soluble receptor
Introduction
Interleukin-1 (IL-1) is a cytokine involved in a wide range of physiological processes, including having a central role in the regulation of acute and chronic inflammation.1,2 The biological effects of IL-1 (IL-1α and IL-1β) are achieved by the binding of the cytokine to the membrane-bound IL-1 type 1 receptor (IL1R1).3,4 IL1R1 is a glycoprotein with a molecular mass of 80 kDa that is predominantly expressed on endothelial cells, smooth muscle cells, epithelial cells, hepatocytes, fibroblasts, keratinocytes, epidermal dendritic cells and T lymphocytes.5 IL-1 binding to the extracellular domain of IL1R1 promotes the recruitment of the IL-1 receptor accessory protein, which leads to signal transduction mediated by the cytoplasmic domains of IL1R1 and IL1R2 receptors.6
The IL-1 type 2 receptor (IL1R2) is unable to initiate signaling and only acts as a ‘decoy' receptor.3,7 IL1R2 is a glycoprotein with a molecular mass of 60 kDa that is expressed on monocytes, neutrophils, T and B lymphocytes.8,9 Studies have also provided evidence for the existence of soluble receptors for IL-1 (sIL1Rs),10,11 which are derived via proteolytic cleavage of membrane-bound receptors12 or by alternative splicing.13 The sIL1Rs act as natural inhibitors of IL-1 activity by preventing the interaction between IL-1 and membrane-bound IL1Rs.6,14 The IL1R1 and IL1R2 genes are located on the long arm of chromosome 2 (2q12–13) and both appear to be driven by multiple promoters. The IL1R1 gene has three distinct promoters which have no TATA or CAAT box motifs and belong to a group of housekeeping gene promoters. These three promoters (referred to as exons 1A, 1B and 1C) generate transcripts with different 5′-untranslated regions.15,16 It is known that the expression of cytokines and their receptors can be influenced by genetic polymorphisms.19,20 Furthermore, polymorphisms in the regulatory regions of cytokines genes can also alter mRNA splicing, mRNA stability and the level of gene transcription.17,18
Functional cellular responses to cytokines depend on the expression level of the specific receptors. It is therefore important to know not only the percentage of cells expressing these membrane-bound receptors but also the density of these receptors on the cells.21,22 Differing expression levels of IL-1 receptors type 1 and 2 have previously been shown in T-lymphocyte populations.23 Moreover, prior studies have examined the association of polymorphisms in the IL-1 receptor genes with different diseases. However, data on association of these polymorphisms with receptor protein expression levels have not yet been published.
The aim of this study was to determine if there is an association between polymorphisms in the IL-1 type 1and type 2 receptor genes and the expression levels of these membrane-bound receptors on different subpopulations of mononuclear cells. We also examined the serum levels of sIL1Rs in healthy individuals.
Material and methods
Blood samples
Blood samples were obtained from Blood Procurement Station No. 1 at the Novosibirsk Blood Center and collected in sterile VACUETTE ethylenediaminetetraacetic acid and serum clot activator tubes (Greiner Bio-One, Kremsmünster, Austria). Samples (n=150) were obtained from individuals from Novosibirsk (Western Siberia, Russia) and included 83 males and 67 females between the ages of 19 and 55 years (mean: 34±1 years). Inclusion in the study required that all individuals belong to the Russian ethnic group, were healthy and unrelated. The main exclusion criteria were those standard for blood donors in the Russian Federation: age (<18, >60 years of age), body weight (<50 kg), blood pressure (systolic <90, >160 mmHg; diastolic <60, >100 mmHg), body temperature (>37 °C), pulse (<50, >100 BPM), diseases (URTI, quinsy, flu, virus hepatitis, tuberculosis, brucellosis, typhus, tularemia, leprosy, HIV, syphilis, oncological diseases), allergy, drug treatment (antibiotics, analgesics), pregnancy and lactation periods. Research was performed in accordance with the Declaration of Helsinki and was approved by the local ethics committee of the FSBI ‘Research Institute of Clinical Immunology'. All individuals provided informed consent before the study was performed.
Isolation of mononuclear cells and stimulation conditions
Peripheral blood mononuclear cells (PBMCs) were isolated from peripheral blood according to standard methods using a Ficoll–Urografin density gradient (ρ=1.077 g/cm3).24 The isolated PBMCs were cultured in RPMI-1640 medium containing 10% fetal calf serum, 2 mM L-glutamine, 10 mM HEPES buffer, 0.5 mm 2-mercaptoethanol, 80 µg/ml gentamicin and 100 µg/ml benzylpenicillin for 24 h under 5% SO2 at 37 °S. The cells were cultured at a concentration of 2×106 cells/ml in 96-well plates (TPP, Trasadingen, Switzerland) and in the presence or absence of lipopolysaccharide (LPS) from Escherichia coli serotype 055:B5 (Sigma-Aldrich, St Louis, MO, USA) at a final concentration of 200 ng/ml. The LPS stimulation index was calculated as a simple ratio of the number of membrane-bound IL1Rs on CD14+ cells in cultures with and without LPS stimulation.
Determination of expression levels of membrane-bound IL1 type 1 and 2 receptors
The levels of membrane-bound IL1R1 and IL1R2 on various subpopulations of mononuclear cells was determined by flow cytometry using antibodies from eBioscience (San Diego, CA, USA), including anti-human CD3 allophycocyanin (cat. no. 17-0037-42), anti-human CD14 fluorescein isothiocyanate (cat. no. 11-0149-42) and anti-human CD19 phycoerythrin-cyanine 7 (cat. no. 25-0199-42), as well as antibodies from R&D Systems (Minneapolis, MN, USA), including anti-human IL1R1 phycoerythrin (cat. no. FAB269P) and anti-human IL1R2 phycoerythrin (cat. no. FAB663). To convert the fluorescence intensity values of cells expressing an appropriate marker into absolute values of receptor level, BD QuantiBRITE PE calibration particles were used (cat. no. 340495; BD Biosciences, San Jose. CA, USA).25
Determination of serum levels of soluble IL1 type 1 and 2 receptors
Serum levels of soluble IL-1 type 1 and 2 receptors were determined via enzyme-linked immunosorbent assay (ELISA) using the Human IL1sR1 ELISA and Human IL1sR2 ELISA kits (RayBiotech, Norcross, GA, USA) according to the manufacturer's instructions. The sensitivity of the assays was 6 pg/ml (sIL1R1) and 5 pg/ml (sIL1R2).
Genotyping methods
single nucleotide polymorphisms (SNPs) located within IL1R1 and IL1R2 genes were selected from the SNP database at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/snp). Specific SNPs were chosen for study using the following criteria: (i) location within the promoter region of the IL1R1 and IL1R2 genes; (ii) high frequency of minor allele expression (>10%); and (iii) potential for association with pathological states.
Genomic DNA was isolated by phenol-chloroform extraction.26 Genotyping was performed using restriction fragment length polymorphism analysis of polymerase chain reaction (PCR)-amplified fragments. Primer sequences for SNP IL1R1 rs2234650:C>T have been described previously.27 Primers for SNP IL1R1 rs3917225:A>G, IL1R2 rs4141134:T>C and IL1R2 rs2071008:T>G were designed using NCBI/Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast). Primers (Table 1) were synthesized by BIOSAN (Novosibirsk, Russia).
Table 1. Primers and restriction endonucleases used for genotyping SNPs in the IL1R1 and IL1R2 genes.
| SNP | Primer sequences | Restriction endonucleases | Restriction products |
|---|---|---|---|
| IL1R1 rs3917225:A>G | 5′-TCT GGG GCA TAC TCA CAG GGGT-3′5′-AGC TGG GTT GTG GTA GCC TTA CTG-3′ | AsuHP I | AA: 411 bpGG: 172/239 bp |
| IL1R1 rs2234650:C>T | 5′-TTGGAGGATGGCCCATGAAGACC-3′5′-CTGTTACGCGCCCGGATGAAAAA-3′ | Pst I | TT: 350 bpCC: 253/97 bp |
| IL1R2 rs4141134:T>C | 5′-CCA TGC CAT CTG CTC TTG GCC AT-3′5′-GAC CAG ACT TTG GAA AGG CCT CC-3′ | Msp I | TT: 448 bpCC: 258/190 bp |
| IL1R2 rs2071008:T>G | 5′-CTT ACA TGG CTG GTG CCT TT-3′5′-TAT CTC CCA TCC CAC ATG GT-3′ | Dra III | TT: 357 bpGG: 194/163 bp |
IL1R, IL-1 receptor; SNP, single nucleotide polymorphism.
PCR was performed using a PTC-200 DNA thermocycler (MJ Research Inc., Watertown, MA, USA). The 20 µl reaction volume contained 1–2 units Taq DNA polymerase (SibEnzyme, Novosibirsk, Russia), 0.5 µM of each primer, 0.25 mM of each deoxyribonucleotide-triphosphate and 50–200 ng of genomic DNA. The reaction buffer added to the DNA polymerase contained 60 mM Tris-HCl (pH 8.5, 25 °C), 1.5 mM MgCl2, 25 mM KCl, 10 mM 2-mercaptoethanol and 0.1% Triton X-100. PCR conditions were as follows: initial denaturation for 3 min at 95 °C; followed by 30 cycles (rs3917225:A>G, rs2234650:C>T, and rs2071008:T>G) or 38 cycles (rs4141134:T>C) as follows: denaturation at 94 °C for 20 s; annealing at 57 °C for 15 s (rs3917225:A>G); 70 °C for 15 s (rs2234650:C>T); 60 °C for 15 s (rs4141134:T>C); and 58 °C for 15 s (rs2071008:T>G); extension at 72 °C for 20 s, with a final extension at 72 °C for 2 min.
PCR products were incubated with restriction enzymes from SibEnzyme (Table 1) at 37 °C. Restriction products were analyzed by electrophoresis of 2% agarose (Medigen, Novosibirsk, Russia) in Tris-acetate-ethylenediaminetetraacetic acid buffer. The pUC19 plasmid digested with Msp I served as a molecular weight marker (SibEnzyme). Restriction products were visualized under ultraviolet light and the molecular weights of fragments were evaluated with the aid of a Videodensitometer ImageMaster VDS (Pharmacia Biotech, Uppsala, Sweden).
Statistical analysis
Statistical analysis was performed using the software STATISTICA 7.0 (StatSoft Inc. Moscow, Russia). Compliance of genotype frequency distributions with the Hardy-Weinberg equilibrium was established by the χ2 test (online calculator provided by Tufts University, Boston, MA, USA (http://www.tufts.edu/∼mcourt01/Documents/Court%200l lab%20-%20HW%20calculator.xls)). Data were tested for normality by the Shapiro–Wilk's W test and the Kolmogorov–Smirnov Z test. The hypothesis that the respective distribution was normal was rejected in most cases. More stringent non-parametric tests were used for subsequent analysis and all data were expressed as median values with quartiles. Correlation analysis was performed using Spearman's rank correlation. Comparison of all sample values and the determination of genotypic relationships with protein expression levels were performed using the Kruskal–Wallis H test, and the examination between the medians using the Mann–Whitney U test. All differences were considered significant at P≤0.05. After performing Bonferroni's correction for the pairwise comparisons of genotypes, P-values <0.017 were considered to indicate statistical significance.
Results
Expression levels of membrane-bound IL1 type 1 and 2 receptors on subpopulations of mononuclear cells
Expression levels of membrane-bound IL-1 type 1 and 2 receptors on subpopulations of mononuclear cells are shown in Table 2. In each subpopulation of mononuclear cells (CD3+ T lymphocytes, CD19+ B lymphocytes and SD14+ monocytes), a significantly higher percentage of cells was estimated to express IL1R1 vs. IL1R2 (P<0.05). In addition, a greater proportion of SD14+ monocytes was IL1R1 positive (IL1R1+), compared to CD3+ T lymphocytes and CD19+ B lymphocytes (P<0.001).
Table 2. Levels of expression of membrane-bound IL-1 type 1 and 2 receptors on intact CD3+, CD19+, and CD14+ PBMC subpopulations and on CD14+ cells from both unstimulated and LPS-stimulated PBMC cultures from 150 healthy individuals.
| Median (quartiles) | ||||
|---|---|---|---|---|
| Percent of cells expressing receptors | Number of receptors per cell | |||
| IL1R1 | IL1R2 | IL1R1 | IL1R2 | |
| Intact CD3+ cells | 10.1 (7.1–13.9)1 | 2.9 (1.5–5.7) | 1148.9 (931.8–1537.5)1 | 611.3 (485.4–750.6)3 |
| Intact CD19+ cells | 3.4 (2.6–5.2)2 | 2.8 (2.0–4.8) | 1597.0 (1222.6–1908.9)2 | 1128.1 (960.4–1369.4) |
| Intact CD14+ cells | 61.0 (40.0–74.6)4 | 2.9 (1.5–7.4)4 | 920.6 (731.5–1226.9)4 | 1035.4 (801.3–1389.5)4 |
| Unstimulated CD14+ cells | 46.1 (28.6–70.3)5 | 14.5 (8.9–59.4)5 | 1568.7 (1306.0–1981.8)5 | 1621.3 (1341.3–2155.2)5 |
| LPS-stimulated CD14+ cells | 75.7 (64.2–82.0) | 8.4 (4.3–28.7) | 3519.9 (2865.0–4473.6) | 1507.0 (1155.3–1915.4) |
Abbreviations: IL1R, IL-1 receptor; LPS, lipopolysaccharide; PBMC, peripheral blood mononuclear cell.
Data expressed as medians (interquartile range).
Mononuclear cells (2×106/ml) were cultivated in the absence and presence of LPS (E. coli serotype 055:B5) at a concentration of 200 ng/ml for 24 h.
Significant difference from intact CD14+ cells (P<0.001).
Significant difference from intact CD3+ and CD14+ cells (P<0.001).
Significant difference from intact CD19+ and CD14+ cells (P<0.001).
Significant difference from CD14+ cells from unstimulated PBMC cultures (P<0.001).
Significant difference from CD14+ cells from LPS-stimulated PBMC cultures (P<0.05).
The number of membrane-bound IL1Rs per cell was determined using Quantibrite PE Calibration Beads, and the least average amounts of IL1R1s and IL1R2s were determined for SD14+ monocytes and CD3+ T lymphocytes (P<0.001), respectively. CD3+ T lymphocytes and CD19+ B lymphocytes showed higher levels of IL1R1 per cell than IL1R2 (P<0.001), while in SD14+ monocytes, the number of IL1R2s per cell was greater than the number of IL1R1s (P<0.05).
The comparison of IL1R expression levels on SD14+ monocytes in unstimulated versus LPS-stimulated PBMC revealed a significant increase in both the percentage of cells expressing IL1R1 and the absolute number of IL1R1s molecules in response to mitogen stimulation (P<0.001). In addition, a significant decrease in the percentage of cells expressing IL1R2 and a decrease in the absolute number of receptors were observed in response to LPS activation (P<0.05). Statistically significant differences (P<0.001) were also observed for the percentage of IL1R+ cells and the absolute number of these receptors on cells in both the total intact CD14+ cell populations and the CD14+ cells from the unstimulated PBMC culture.
Serum levels of sIL1 type 1 and 2 receptors
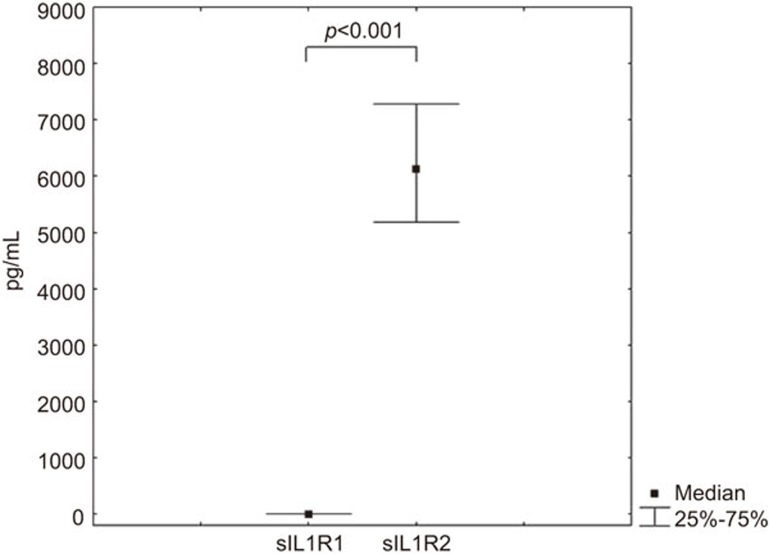
The serum levels of sIL1R1 and sIL1R2 were also examined (Figure 1). Overall, the serum levels of sIL1R2 were significantly higher than sIL1R1 (P<0.001), but there was no correlation between sIL1R1 and sIL1R2 levels.
Figure 1.
Levels of soluble IL-1 type 1 and 2 receptors in the serum of healthy individuals (n=150). The median and quartiles are shown in the histogram. The P-valuesindicated were calculated by the Mann–Whitney test. IL-1, interleukin-1.
Association of SNPs rs3917225:A>G and rs2234650:C>T in the promoter region of the IL1R1 gene with IL-1R expression levels
The genotypes and allele frequencies of SNPs in the promoter regions of the IL1R1 (rs3917225:A>G, rs2234650:C>T) and IL1R2 (rs4141134:T>C, rs2071008:T>G) genes in healthy individuals from Novosibirsk (Western Siberia, Russia) are shown in Table 3. The genotypes and allele frequencies of all four examined polymorphisms corresponded with the Hardy–Weinberg equilibrium (HWE) (P>0.05). No statistically significant differences were found in genotypes and allele frequencies between groups of men and women.
Table 3. Genotypes and allele frequencies of SNPs in the IL1R1 and IL1R2 genes.
| SNPs | Genotype | Genotype frequency, % (n) | Allele | Allele frequency, % |
|---|---|---|---|---|
| IL1R1 rs3917225:A>G | AA | 38.1 (56) | A | 62 |
| AG | 46.9 (69) | G | 38 | |
| GG | 15 (22) | |||
| IL1R1 rs2234650:C>T | CC | 21.6 (27) | C | 50 |
| CT | 57.6 (72) | T | 50 | |
| TT | 20.8 (26) | |||
| IL1R2 rs4141134:T>C | TT | 38.9 (58) | T | 64 |
| TC | 51 (76) | C | 36 | |
| CC | 10.1 (15) | |||
| IL1R2 rs2071008:T>G | TT | 78.6 (114) | T | 88 |
| TG | 19.3 (28) | G | 12 | |
| GG | 2.1 (3) |
Abbreviations: IL1R, IL-1 receptor; SNP, single nucleotide polymorphism.
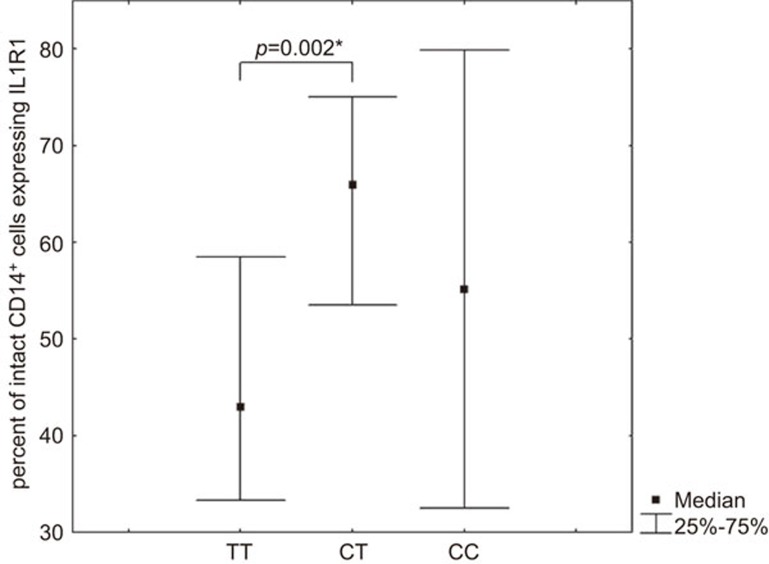
There was no statistically significant relationship between the SNPs rs3917225:A>G and rs2234650:C>T in the promoter regions of the IL1R1 gene and the level of soluble IL-1 type 1 receptors. Furthermore, SNP rs3917225:A>G, in the promoter of the IL1R1 gene, did not show any association with the expression level of IL1R1 as the P-value was no longer significant after performing the Bonferroni correction. The analysis of IL1R1 expression in terms of SNP rs2234650:C>T genotypes in the IL1R1 gene indicated a statistically significant difference in the percentage of cells expressing IL1R1 in intact CD19+ V lymphocytes (P=0.011, Mann–Whitney test) and CD14+ monocytes (P=0.005, Mann–Whitney test) subpopulations, as well as in the amount of membrane-bound IL1R1 on intact CD14+ monocytes (P=0.049, Mann–Whitney test). However, pairwise analysis of the expression of IL1R1 in carriers of different genotypes at SNP rs22344650:C>T showed significant differences only in the percentage of intact CD14+ monocytes. Specifically, individuals with the TT genotype have a lower percentage of intact CD14+ monocytes expressing IL1R1 than carriers of the CT genotype (Figure 2)
Figure 2.
Percentage of intact CD14+ cells expressing IL1R1 in carriers of different IL1R1 SNP rs2234650:C>T genotypes. The P-values indicated were calculated by the Mann–Whitney test. *Significant differences after performing Bonferroni corrections (TT vs. CT). IL1R, IL-1 receptor; SNP, single nucleotide polymorphism.
Association of SNPs rs4141134:T>C and rs2071008:T>G in the promoter region of the IL1R2 gene with the expression level of membrane-bound IL1R2
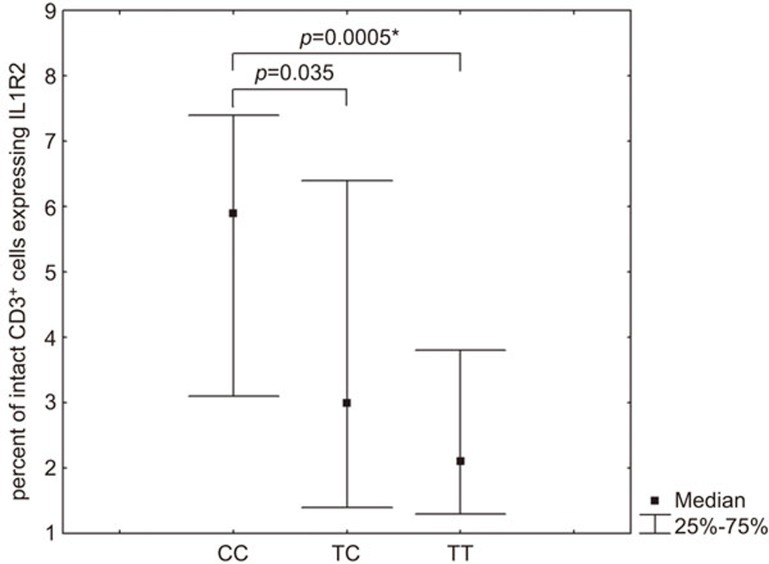
The analysis of expression level and genetic variation in SNP rs4141134:T>C in the promoter of the IL1R2 gene indicated a statistically significant difference in the percentage of cells expressing IL1R2 in subpopulations of intact CD3+ T cells, as well as on CD14+ monocytes in LPS-stimulated PBMC cultures (P<0.05 using Mann–Whitney and Kruskal–Wallis tests). The pairwise analysis demonstrated that individuals with the CC genotype have a higher percentage of cells expressing IL1R2 in the intact CD3+ T-cell population (Figure 3).
Figure 3.
Percentage of intact CD3+ cells expressing IL1R2 in carriers of different IL1R2 SNP rs4141134:T>C genotypes. The P-values indicated were calculated by the Mann–Whitney test. *Significant differences after performing Bonferroni corrections (CC vs. TT). IL1R, IL-1 receptor; SNP, single nucleotide polymorphism.
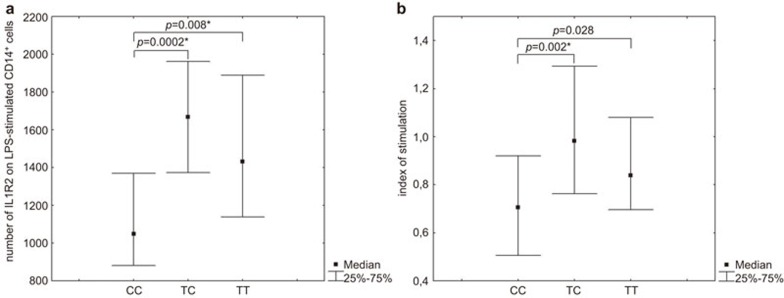
Our data also indicates that individuals with the CC genotype in SNP rs4141134:T>C have a lower density (i.e., absolute number) of IL1R2 on CD14+ monocytes in LPS-stimulated PBMC cultures (Figure 4a) and have a lower stimulation index of IL1R2 expression on CD14+ monocytes cultured in the presence of LPS (Figure 4b).
Figure 4.
Association of different IL1R2 SNP rs4141134:T>C genotypes with the expression levels of membrane-bound IL1R2. (a) Number of membrane-bound IL1R2s on CD14+ monocytes from LPS-stimulated PBMC cultures. (b) LPS stimulation index. The P-values indicated were calculated by the Mann–Whitney test. *Significant differences after performing Bonferroni corrections. IL1R, IL-1 receptor; LPS, lipopolysaccharide; PBMC, peripheral blood mononuclear cell; SNP, single nucleotide polymorphism.
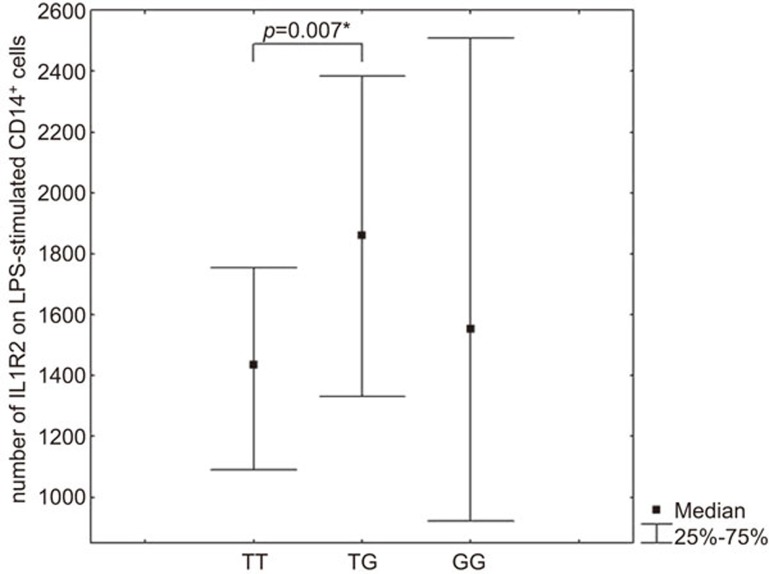
We also found an association of the SNP rs2071008:T>G in the IL1R2 gene with the expression level of membrane-bound IL1R2. Individuals with the TT genotype showed a lower level of membrane-bound IL-1 type 2 receptors on CD14+ monocytes in LPS-stimulated PBMC cultures, as opposed to individuals with the TG genotype (Figure 5). The frequency of the GG genotype was extremely low (2.1%); therefore, the above analysis was not able to be carried out in this group.
Figure 5.
Number of membrane-bound IL1R2s on CD14+ monocytes from LPS-stimulated PBMC cultures in carriers of different IL1R2 SNP rs2071008:T>G genotypes. The P-values indicated were calculated by the Mann–Whitney test. *Significant differences after performing Bonferroni corrections (TT vs. TG). IL1R, IL-1 receptor; LPS, lipopolysaccharide; PBMC, peripheral blood mononuclear cell; SNP, single nucleotide polymorphism.
Discussion
In this study, we examined the expression of membrane-bound IL-1 type 1 and 2 receptors on different subpopulations of mononuclear cells from the peripheral blood of healthy individuals. We have demonstrated quantitative differences in expression levels, both in the percentage of cells carrying the receptors, and in the absolute number of these receptors expressed on the surface of CD3+ T lymphocytes, CD19+ B lymphocytes and CD14+ monocytes. It is known that IL1R1 (as opposed to the ‘decoy' receptor IL1R2) is capable of promoting the signal transduction that leads to its effector functions. Thus, it is possible that the functional response of a cell population to IL-1 may depend on the balance between the levels of IL1R1 and IL1R2 on the cells. An investigation of intact subpopulations of mononuclear cells has shown that within the population of CD14+ monocytes, the percentage of IL1R1+ cells was 20 times higher than percentage of IL1R2+ cells. However, no significant differences in absolute numbers of IL1R1s and IL1R2s on CD14+ cells were observed. The percentage of CD3+ lymphocytes expressing IL1R1 was three times higher than percentage of cells expressing IL1R2. The absolute amount of IL-1 type 1 receptors on CD3+ cells was also two times higher than absolute amount of type 2 receptors. Our data are fully consistent with the literature on the level of IL1R1 and IL1R2 mRNAs in T cells.23 Additionally, this comparison of mononuclear cell subpopulations has shown that a higher percentage of the CD14+ monocytes population were also IL1R1+. The CD19+ B lymphocytes showed the lowest percentage of IL1R1+ cells. However, no differences have been observed in the absolute number of IL1R1s per cell within the CD3+, CD19+ and CD14+ subpopulations. Therefore, the percentage of receptor positive cells and the expression density of these receptors on individual cells are not always related.
This study also investigated changes in the expression levels of IL1R1 and IL1R2 as a result of LPS stimulation. Saccani et al.28 and Penton-Rol et al.29 found that incubating cells in presence of LPS led to an increase in IL1R1 mRNA levels and a decrease in IL1R2 mRNA levels. Our data examining the changes in protein levels are consistent with these previous studies and reflective of those changes in mRNA levels during cell stimulation. A reproducible increase in the percentage of IL1R1+ cells and in the absolute amount of IL1R1 molecules was observed when we compared the expression of IL1R1 on CD14+ monocytes from unstimulated versus LPS-stimulated PBMC cultures. Analysis of IL-1 type 2 receptor expression after LPS stimulation also has revealed a decrease in the percentage of IL1R2+ cells and a decrease in the average number of IL1R2s on CD14+ cells. When we compared IL1R1 and IL1R2 expression between intact CD14+ monocytes with CD14+ monocytes that had been cultivated for 24 h, we observed a similar increase in the number of IL1R1s and IL1R2s by approximately 600 receptors. Thereby, a decrease in the percentage of IL1R1+ cells and an increase in the percentage of IL1R2+ cells occurred, suggesting that these changes in the expression levels of the IL1Rs were due to changes in the culture microenvironment.
Therefore, we demonstrated differences in the expression levels of membrane-bound IL-1 type 1I and 2II receptors on immunocompetent cells. It is known that differences in expression of cytokine receptors can be caused by gene polymorphisms.20,22 Polymorphisms in cytokine genes and their receptors have been the focus of intensive investigations for the last 15 years. Prior studies have found associations of specific polymorphisms with the predisposition to certain diseases, with their clinical course and outcome, as well as with appropriate therapies.30,31 Polymorphisms within the promoter regions of genes may directly influence the structure of transcription factor binding sites and mRNA stability,17,18 thereby leading to differences in protein expression.
A number of studies have been performed to examine the role of the SNP rs3917225:A>G in the IL1R1 gene. Nakki et al.32 showed that the A allele is associated with severe hand osteoarthritis. Park et al.33 found that the rs3917225:A>G polymorphism did not associated with papillary thyroid carcinoma. One of the most investigated polymorphisms in the IL1R1 gene is the SNP rs2234650:C>T (also known as 1970 C/T PstI). Previous has shown an association of the T allele of SNP rs2234650:C>T with diabetes mellitus type I,34 AIDS progression35 and the C allele has been associated with asthma.36 It is known that the activity of specific mediators (i.e., cytokines) depends on the density of their receptor on cells.21,22 Thus, it may be assumed that individuals with the above SNP genotypes in the IL1R1 gene may be more susceptible to the action of IL-1 due to their increased expression of the signal transduction-capable IL-1 type 1 receptors. Our study demonstrated that individuals with the homozygous TT genotype in SNP rs2234650:C>T in the IL1R1 gene have a lower percentage of cells expressing IL1R1 on the intact CD14+ monocyte population. These data provide evidence that the T allele is associated with lower expression of membrane-bound IL1R1s on populations of CD14+ cells. Thus, it may be assumed that the population of monocytes in individuals with the TT genotype may have a lowered functional response to IL-1 mediators. However, these assumptions will require direct experimental confirmation.
Polymorphisms in the IL1R2 gene are not well understood. We examined two polymorphisms (SNPs rs4141134:T>C and rs2071008:T>G) in the promoter region of the IL1R2 gene. Individuals with the CC genotype in SNP rs4141134:T>C showed an increase in the percentage of CD3+ T cells expressing membrane-bound IL1R2. The percentage of cells carrying specific surface receptors may predetermine the potential reaction to certain mediators. It is likely that the biological effects of a given mediator may be stronger in a population of cells carrying increased percentage of the mediator's receptor. In this case, however, the higher percentage of IL1R2+ cells exerts the reverse effect, i.e., blocking signal transduction by competing with IL1R1 for binding of the mediator (IL-1).37 Thus, an increase in the percentage of cells expressing the ‘decoy' receptor, IL1R2, limits the functional effect of IL-1 in these subpopulations.
The individuals with the CC genotype in SNP rs4141134:T>C and the TT genotype in SNP rs2071008:T>G have lower levels of membrane-bound IL1R2 on CD14+ cells isolated from LPS-stimulated PBMC cultures. It is well known that LPS activates the release of the IL-1 cytokine.38 Hence, it may be expected that those individuals homozygous for the C allele in rs4141134:T>C and the T allele in rs2071008:T>G individuals should show stronger effects in response to LPS stimulation, as they have a lower number of ‘decoy' IL1R2 receptors on the surface of CD14+ monocytes. Most likely the T allele in IL1R2 rs4141134:T>C has the greater potential for stimulation of IL-1 type 2 receptor expression.
The modification of binding sites for transcription factors is the most possible mechanism linking SNPs and gene expression activity. The AliBaba2.1 software was used to examine the potential for transcription factors to bind to specific allele sequences (http://www.gene-regulation.com/pub/programs/alibaba2/index.html). The presence of the G allele in SNPs rs3917225:A>G and rs2071008:T>G presents possible binding sites for transcription factors Pit-1a and TAF-1, respectively. TAF1 (RNA polymerase II, TATA box binding protein-associated factor, 250 kDa) is an essential protein required for progression of the cell cycle and repression of apoptosis in mammalian cells.39 The TAF1 has intrinsic protein kinase activity,40 histone acetyltransferase activity41 and ubiquitin-activating activity (E1/E2).42 TAF1 binds to and modulates the transcriptional activity of proteins such as c-Jun,43 Mdm244 and cyclin D145 and, therefore, participates in cancer progression.
The presence of the C allele in SNP rs2234650 provides the ability to bind the transcription factor Yin Yang 1, whereas the T allele sequence can bind the transcription factor activating protein 1. Yin Yang 1 is a ubiquitous and multifunctional zinc-finger transcription factor that interacts with histone acetyltransferases and deacetylases for its transcriptional activity and is also involved in inflammation and tumorigenesis.46 Yin Yang 1has been shown to enhance the cyclooxygenase-2 protein expression and prostaglandin D2 production elicited by LPS treatment.47 Activating protein 1 is a dimeric transcription factor, implicated in the regulation of a variety of cellular processes including proliferation and survival, differentiation, growth, inflammation, apoptosis, cell migration and transformation.48 It is possible that the differences in IL1R expression are due to changes in the binding of these transcription factors. However, the involvement of these transcription factors in the regulation of IL1R genes expression requires direct experimental confirmation at the level of mRNA and protein synthesis.
There are some potential limitations to this study. The sample size was relatively small and collected from a homogeneous ethnic group. Gene expression was tested at the protein level, but mRNA levels not were assessed. Furthermore, only a limited number of SNPs were investigated. However, the presented results provide a basis for further study of the functionality these polymorphisms and potential molecular mechanisms of the revealed associations.
Conclusion
This study has shown quantitative differences in the expression levels of membrane-bound IL-1 type 1 and 2 receptors on subpopulations of immunocompetent cells from healthy individuals. Moreover, differences in the expression levels of two types of membrane-bound IL1Rs on cells cultivated in the presence of the polyclonal activator LPS were estimated. Furthermore, it was established that differences in the expression of the IL1Rs may be related to single nucleotide polymorphisms in promoter regions of these genes. Therefore, polymorphisms of the IL1R genes may be one of the factors influencing the level of expression of membrane-bound IL1Rs on PBMC.
Acknowledgments
This study was supported by the Federal Target Program ‘Scientific and scientific-pedagogical personnel of innovative Russia' for 2009–2013 (state contract no. 02.740.11.0707).
The authors declare no conflicts of interest.
References
- 1Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol 2009; 27: 519–550. [DOI] [PubMed] [Google Scholar]
- 2Gabay C, Lamacchia C, Palmer G. IL-1 pathways in inflammation and human diseases. Nat Rev Rheumatol 2010; 6: 232–241. [DOI] [PubMed] [Google Scholar]
- 3Colotta F, Re F, Muzio M, Bertini R, Polentarutti N, Sironi M et al. Interleukin-1 type II receptor: a decoy target for IL-1 that regulated by IL-4. Science 1993; 261: 472–475. [DOI] [PubMed] [Google Scholar]
- 4Sims JE, Gayle MA, Slack JL, Alderson MR, Bird TA, Giri JG et al. Interleukin 1 signaling occurs exclusively via the type I receptor. Proc Natl Acad Sci USA 1993; 90: 6155–6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Dinarello CA. IL-1 receptor type I. In: Oppenheim JJ, Feldmann M, Durum SK, Hirano T, Vilcek J, Nicola NA (ed.) Cytokine Reference: A Compendium of Cytokines and Other Mediators of Host Defense. London: Academic Press, 2001: 1587–1600. [Google Scholar]
- 6Arend WP, Palmer G, Gabay C. IL-1, IL-18, and IL-33 families of cytokines. Immunol Rev 2008; 223: 20–38. [DOI] [PubMed] [Google Scholar]
- 7Mantovani A, Locati M, Vecchi A, Sozzani S, Allavena P. Decoy receptors: a strategy to regulate inflammatory cytokines and chemokines. Trends Immunol 2001; 22: 328–336. [DOI] [PubMed] [Google Scholar]
- 8Wang M, Windgassen D, Papoutsakis ET. Comparative analysis of transcriptional profiling of CD3+, CD4+ and CD8+ T cells identifies novel immune response players in T-cell activation. BMC Genomics 2008; 9: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Matsushima K, Akahoshi T, Yamada M, Furutani Y, Oppenheim JJ. Properties of a specific interleukin 1 (IL 1) receptor on human Epstein Barr virus-transformed B lymphocytes: identity of the receptor for IL 1-alpha and IL 1-beta. J Immunol 1986; 136: 4496–4502. [PubMed] [Google Scholar]
- 10Symons JA, Eastgate JA, Duff GW. Purification and characterization of a novel soluble receptor for interleukin 1. J Exp Med 1991; 174: 1251–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Arend WP, Malyak M, Smith MF Jr, Whisenand TD, Slack JL, Sims JE et al. Binding of IL-1 alpha, IL-1 beta, and IL-1 receptor antagonist by soluble IL-1 receptors and levels of soluble IL-1 receptors in synovial fluids. J Immunol 1994; 153: 4766–4774. [PubMed] [Google Scholar]
- 12Orlando S, Sironi M, Bianchi G, Drummond AH, Boraschi D, Yabes D et al. Role of metalloproteases in the release of the IL-1 type II decoy receptor. J Biol Chem 1997; 272: 31764–31769. [DOI] [PubMed] [Google Scholar]
- 13Liu C, Hart RP, Liu XJ, Clevenger W, Maki RA, de Souza EB. Cloning and characterization of an alternatively processed human type II interleukin-1 receptor mRNA. J Biol Chem 1996; 271: 20965–20972. [DOI] [PubMed] [Google Scholar]
- 14Symons JA, Young PR, Duff GW. Soluble type II interleukin 1 (IL-1) receptor binds and blocks processing of IL-1 beta precursor and loses affinity for IL-1 receptor antagonist. Proc Natl Acad Sci USA 1995; 92: 1714–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Ye K, Vannier E, Clark BD, Sims JE, Dinarello CA. Three distinct promoters direct transcription of different 5′ untranslated regions of the human interleukin 1 type I receptor: a possible mechanism for control of translation. Cytokine 1996; 8: 421–429. [DOI] [PubMed] [Google Scholar]
- 16Dale M, Nicklin MJ. Interleukin-1 receptor cluster: gene organization of IL1R2, IL1R1, IL1RL2 (IL-1Rrp2), IL1RL1 (T1/ST2), and IL18R1 (IL-1Rrp) on human chromosome 2q. Genomics 1999; 57: 177–179. [DOI] [PubMed] [Google Scholar]
- 17Bidwell J, Keen L, Gallagher G, Kimberly R, Huizinga T, McDermott MF. Cytokine gene polymorphism in human disease: on-line databases. Genes Immun 1999; 1: 3–19. [DOI] [PubMed] [Google Scholar]
- 18Shastry BS. SNPs: impact on gene function and phenotype. Methods Mol Biol 2009; 578: 3–22. [DOI] [PubMed] [Google Scholar]
- 19Brookes AJ. The essence of SNPs. Gene 1999; 234: 177–186. [DOI] [PubMed] [Google Scholar]
- 20Sennikov SV, Vasilyev FF, Lopatnikova JA, Shkaruba NS, Silkov AN. Polymorphisms in the tumor necrosis factor receptor genes affect the expression levels of membrane-bound type i and type ii receptors. Mediators Inflamm 2014; 2014: 745909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Reynes J, Portales P, Segondy M, Baillat V, André P, Réant B et al. CD4+ T cell surface CCR5 density as a determining factor of virus load in persons infected with human immunodeficiency virus type 1. J Infect Dis 2000; 181: 927–933. [DOI] [PubMed] [Google Scholar]
- 22Moraga I, Harari D, Schreiber G, Uzé G, Pellegrini S. Receptor density is key to the alpha2/beta interferon differential activities. Mol Cell Biol 2009; 29: 4778–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Bebes A, Kovács-Sólyom F, Prihoda J, Kui R, Kemény L, Gyulai R. Interleukin-1 receptors are differentially expressed in normal and psoriatic T cells. Mediators Inflamm 2014; 2014: 472625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Böyum A. Separation of leukocytes from blood and bone marrow. Introduction. Scand J Clin Lab Invest 1968; Suppl 97: 7. [PubMed] [Google Scholar]
- 25Vasilyev FF, Lopatnikova JA, Sennikov SV. Optimized flow cytometry protocol for analysis of surface expression of interleukin-1 receptor types I and II. Cytotechnology 2013; 65: 795–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Maniatis T, Sambrook J, Fritsch EF. Molecular Cloning—A Laboratory Manual. New York: Cold Spring Harbor Press, 1989: 76–85. [Google Scholar]
- 27Bergholdt R, Karlsen AE, Johannesen J, Hansen PM, Dinarello CA, Nerup J et al. Characterization of polymorphisms of an interleukin 1 receptor type 1 gene (IL1RI) promotor region (P2) and their relation to insulin-dependent diabetes mellitus (IDDM). The Danish Study Group of Diabetes in Childhood. Cytokine 1995; 7: 727–733. [DOI] [PubMed] [Google Scholar]
- 28Saccani S, Polentarutti N, Penton-Rol G, Sims JE, Mantovani A. Divergent effects of LPS on expression of IL-1 receptor family members in mononuclear phagocytes in vitro and in vivo. Cytokine 1998; 10: 773–780. [DOI] [PubMed] [Google Scholar]
- 29Penton-Rol G, Orlando S, Polentarutti N, Bernasconi S, Muzio M, Introna M et al. Bacterial lipopolysaccharide causes rapid shedding, followed by inhibition of mRNA expression, of the IL-1 type II receptor, with concomitant up-regulation of the type I receptor and induction of incompletely spliced transcripts. J Immunol 1999; 162: 2931–2938. [PubMed] [Google Scholar]
- 30Smith AJ, Humphries SE. Cytokine and cytokine receptor gene polymorphisms and their functionality. Cytokine Growth Factor Rev 2009; 20: 43–59. [DOI] [PubMed] [Google Scholar]
- 31Hollegaard MV, Bidwell JL. Cytokine gene polymorphism in human disease: on-line databases, Supplement 3. Genes Immun 2006; 7: 269–276. [DOI] [PubMed] [Google Scholar]
- 32Näkki A, Kouhia ST, Saarela J, Harilainen A, Tallroth K, Videman T. Allelic variants of IL1R1 gene associate with severe hand osteoarthritis. BMC Med Genet 2010; 11: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Park SW, Kim MK, Kwon KH, Kim J. Association between a promoter polymorphism (rs2192752, -1028A/C) of interleukin 1 receptor, type I (IL1R1) and location of papillary thyroid carcinoma in a Korean population. Int J Immunogenet 2012; 39: 501–507. [DOI] [PubMed] [Google Scholar]
- 34Metcalfe KA, Hitman GA, Pociot F, Bergholdt R, Tuomilehto-Wolf E, Tuomilehto J et al. An association between type 1 diabetes and the interleukin-1 receptor type 1 gene. The DiMe Study Group. Childhood Diabetes in Finland. Hum Immunol 1996; 51: 41–48. [DOI] [PubMed] [Google Scholar]
- 35Do H, Vasilescu A, Carpentier W, Meyer L, Diop G, Hirtzig T et al. Exhaustive genotyping of the interleukin-1 family genes and associations with AIDS progression in a French cohort. J Infect Dis 2006; 194: 1492–1504. [DOI] [PubMed] [Google Scholar]
- 36Mahdaviani SA, Rezaei N, Moradi B, Dorkhosh S, Amirzargar AA, Movahedi M. Proinflammatory cytokine gene polymorphisms among Iranian patients with asthma. J Clin Immunol 2009; 29: 57–62. [DOI] [PubMed] [Google Scholar]
- 37Lang D, Knop J, Wesche H, Raffetseder U, Kurrle R, Boraschi D et al. The type II IL-1 receptor interacts with the IL-1 receptor accessory protein: a novel mechanism of regulation of IL-1 responsiveness. J Immunol 1998; 161: 6871–6877. [PubMed] [Google Scholar]
- 38Cavaillon JM, Fitting C, Caroff M, Haeffner-Cavaillon N. Dissociation of cell-associated interleukin-1 (IL-1) and IL-1 release induced by lipopolysaccharide and lipid A. Infect Immun 1989; 57: 791–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Wassarman DA, Sauer F. TAF(II)250: a transcription toolbox. J Cell Sci 2001; 114(Pt 16): 2895–902. [DOI] [PubMed] [Google Scholar]
- 40Dikstein R, Ruppert S, Tjian R. TAFII250 is a bipartite protein kinase that phosphorylates the base transcription factor RAP74. Cell 1996; 84: 781–790. [DOI] [PubMed] [Google Scholar]
- 41Mizzen CA, Yang XJ, Kokubo T, Brownell JE, Bannister AJ, Owen-Hughes T et al. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell 1996; 87: 1261–1270. [DOI] [PubMed] [Google Scholar]
- 42Pham AD, Sauer F. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science 2000; 289: 2357–2360. [DOI] [PubMed] [Google Scholar]
- 43Lively TN, Ferguson HA, Galasinski SK, Seto AG, Goodrich JA. c-Jun binds the N terminus of human TAF(II)250 to derepress RNA polymerase II transcription in vitro. J Biol Chem 2001; 276: 25582–25588. [DOI] [PubMed] [Google Scholar]
- 44Allende-Vega N, Saville MK, Meek DW. Transcription factor TAFII250 promotes Mdm2-dependent turnover of p53. Oncogene 2007; 26: 4234–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45Kloet SL, Whiting JL, Gafken P, Ranish J, Wang EH. Phosphorylation-dependent regulation of cyclin D1 and cyclin A gene transcription by TFIID subunits TAF1 and TAF7. Mol Cell Biol 2012; 32: 3358–3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene 2006; 25: 1125–1142. [DOI] [PubMed] [Google Scholar]
- 47Joo M, Wright JG, Hu NN, Sadikot RT, Park GY, Blackwell TS et al. Yin Yang 1 enhances cyclooxygenase-2 gene expression in macrophages. Am J Physiol Lung Cell Mol Physiol 2007; 292: L1219–L1226. [DOI] [PubMed] [Google Scholar]
- 48Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J Cell Sci 2004; 117(Pt 25): 5965–5973. [DOI] [PubMed] [Google Scholar]