The discovery of microRNAs (miRNAs) is one of the most significant breakthroughs in recent decades. miRNAs are a class of small, endogenous, non-coding RNAs (19–24 nt) that negatively regulate the expression of target genes by binding to their 3′ untranslated region and are involved in various biological processes. Three main approaches that are globally used for the detection of miRNA include direct cloning, forward genetics and computational approaches using bioinformatics.1 The bioinformatic approach uses the available transcriptomic and genomic data for mining miRNAs using a homology search with the existing miRNAs. In the current study, a computational approach was employed to identify potent miRNAs involved in the inflammatory response of the liver. The rat model of lipopolysaccharide (LPS)-induced liver inflammation was used, and the expression of the candidate miRNAs in hepatocytes was quantified by real-time polymerase chain reaction (qRT-PCR).
Inflammation is the first step toward combating antigens and toward tissue recovery, and in some instances, it proceeds to a chronic state associated with debilitating autoimmune diseases, such as multiple sclerosis or even cancer. Chronic inflammation of liver tissues may lead to fibrosis, steatosis and cirrhosis, and may end with cancer. The response of liver tissue to chronic injury is stimulated by pro-inflammatory factors, such as bacterial endotoxin or interleukin-1.2 There is an increasing paradigm that miRNAs regulate different biological processes in various cell types within the liver, including liver pathology and cancer.3 The expression of MiR-517a, miR-892a and miR-106a* in bile was increased in patients with Ischemic type biliary lesions, a biliary complication encountered after liver transplantation.4 Although several miRNAs have been reported, miR-155, miR-146a and miR-125b are considered potent miRNAs involved in inflammation-mediated hepatocyte damage at multiple levels by targeting a pro-inflammatory cytokine production cascade where Toll-like receptor and intracellular signaling pathways determine pro- and anti-inflammatory cytokines.5
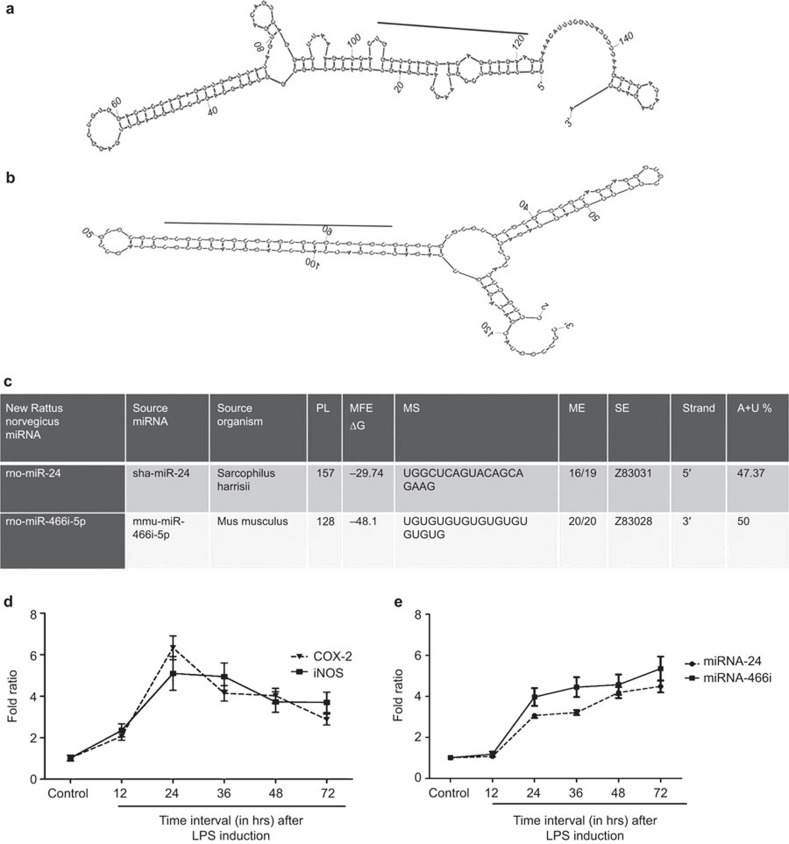
An increasing body of evidence has clearly shown that a differential expression of miRNAs is associated with liver diseases, such as hepatitis C and hepatitis B, metabolic disorders and drug abuse.6 Different approaches have been used to identify novel miRNAs that are involved in liver disorders. In our study, we used a computational approach that enabled us to identify two inflammatory responsive liver miRNAs from 89 liver EST sequences: rno-miR-24 and rno-miR-466i-5p. In the current study, the MiRBase database was used to identify miRNAs and their conserved homologues from liver ESTs under inflammatory conditions. In brief, the EST sequences (89, February 2013) of hepatic transcripts preferentially expressed during acute inflammation were downloaded from the NCBI database (http://www.ncbi.nlm.nih.gov/nuccore) by searching ‘liver inflammation', and the inflammatory-responsive miRNAs were predicted as described by Thirugnanasambantham et al.1 The careful analysis of 89 liver EST sequences revealed 10 non-protein-encoding ESTs homologous to the miRNA sequences. A secondary structural analysis of the pre-miRNA-related sequence from the 10 non-coding ESTs revealed the presence of 2 miRNA encoding transcripts in the liver inflammatory EST sequences (Figure 1a and b). The source of the sequences, length of the precursor sequences and their minimum folding free energies and A+U content for the predicated miRNAs are shown in Figure 1c. Among the predicted miRNAs from the analyzed liver ESTs, rno-miR-24 was observed in the indirect strand and rno-miR-466i-5p was observed in the direct strand. rno-miR-466i-5p and rno-miR-24 had a minimum free energy of −29.74 and −48.1 kcal/mol, respectively, with an average of −38.92 kcal/mol. While considering the A+U percentage in the predicted miRNAs, it was approximately 50% and 47.4% in rno-miR-466i-5p and rno-miR-24, respectively.
Figure 1.
Inflammatory responsive miRNA identified using a computational approach and its stem-loop RT-PCR expression analysis in rat hepatocytes with LPS induction. (a) Hairpin structure of predicted Rattus norvegicus miRNA rno-miR-24; (b) hairpin structure of predicted Rattus norvegicus miRNA rno-miR-466i-5p; (c) details of predicted Rattus norvegicus miRNA; (d) RT-PCR expression analysis of pro-inflammatory gene (COX2 and iNOS) expression fold ratio with time dependence; (e) stem-loop RT-PCR expression analysis of miRNA (rno-miR-24 and rno-miR-466i-5p) expression fold ratio with time dependence. LPS, lipopolysaccharide; microRNA, miRNA; RT-PCR, reverse transcription polymerase chain reaction.
To further confirm the expression of the identified miRNAs in response to inflammation in liver cells, the hepatocytes were isolated from rat livers by the collagenase perfusion method,7 and inflammation was induced with LPS (1 µg/ml; Sigma-Aldrich, St Louis, MO, USA). Total RNA was extracted for the quantitative determination of COX-2, iNOS and the housekeeping gene GAPDH using real-time PCR analysis from stimulated and unstimulated cells at 4, 8, 12, 24, 36 and 48 h after LPS challenge. The forward and reverse primers used for inflammatory marker gene expressions were as follows: iNOS forward: 5′-ACAACAGGAACCTACCAGCTCA-3′, reverse: 5′-GATGTTGTAGCGCTGTGTGTCA-3; COX-2 forward: 5′-TGTATGCTACCATCTGGCTTCGG-3′, reverse: 5′-GTTTGGAAC AGTCGCTCGTCATC-3′, and GAPDH forward: 5′- GTATTGG GCGCCTGGTCACC-3′, reverse: 5′-CGCTCCTGGAAGATGGTGATGG-3′). The cycling condition was 35 cycles of 94 °C for 30 s, 60 °C for 45 s and 72 °C for 1 min followed by a final extension step at 72 °C for 7 min. Reverse transcription and quantitative expression analyses of rno-miR-24, rno-miR-466i-5p, and the endogenous control U6 were performed using real-time PCR as described previously.8 The primer sequences used for stem-loop RT-PCR were as follow: rno-miR-24 stem loop RT primer 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTTCTG-3′ forward primer 5′-GCGGCGGTGGCTCAGTACAGC-3′ rno-miR-466i-5p stem loop RT primer 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCACACA-3′ forward primer 5′-GCGGCGGTGTGTGTGTGTGTG-3′ Universal reverse primer 5′-GTGCAGGGTCCGAGGT-3′ U6 RT primer 5′-AACGCTTCACGAATTTGCGTG-3′ U6 forward primer 5′-GCTCGCTTCGGCAGCACA-3′ U6 reverse primer 5′-GAGGTATTCGCACCAGAGGA-3′. The relative expression of target genes was stated as a fold ratio using the (ΔΔCT check) 2−ΔΔCT method.
The expression patterns of the genes encoding inflammatory markers (Figure 1d) and identified miRNAs (Figure 1e) were analyzed in control and LPS-stimulated hepatocytes using real-time PCR. The expression of mRNA encoding COX-2 and iNOS were upregulated in proportion with LPS stimulation up to 24 h. We observed that the inflammatory markers COX-2 and iNOS reached the maximum level at 24 h (6.33- and 5.09-fold, respectively) and started to decline from 36 h (4.14- and 4.94-expression fold, respectively) after LPS treatment. However, the expression of both miRNAs was detected from 12 h after LPS treatment, in parallel with inflammation progression, as denoted by the expression of inflammatory markers. The maximum expression of both miR-24 and miR-466i-5p was reached 72 h (5.38 and 4.48 expression fold, respectively) after LPS treatment.
In in vitro studies, increases in mRNA expression of COX-2 and iNOS levels upon LPS stimulation of hepatocytes were observed in this study. A similar expression pattern of iNOS was noticed from earlier studies in liver slices, where the expression was observed after 5 h of incubation with LPS and increased until 24 h.9 We found that the expression levels of both miRNAs were upregulated with LPS treatment. A similar result was observed with miR-155, a regulator of inflammation.5 Simultaneously, as denoted by the expression of inflammatory markers (COX-2 and iNOS), the inflammation was suppressed with the increase in expression of rno-miR-24 and rno-miR-466i-5p. Song et al.10 reported the upregulation of miR-466i in SST-treated chronic liver disease. In conclusion, our data identify two new miRNAs, miR-24 and miR-466i, which are upregulated in hepatocytes in response to inflammatory stimuli. This upregulation was reciprocally related with the inflammatory markers COX-2 and iNOS, suggesting that they are target genes in liver inflammation. The anti-inflammatory miRNAs identified with a computational approach are known to be involved in the regulation of inflammatory reactions. Furthermore, the analysis of inflammatory markers revealed that the level of inflammation was reduced with an increase in the expression level of the identified miRNA. Therefore, data from this study may open intriguing possibilities for the therapeutic value of the identified miRNAs as anti-inflammatory factors for liver injury.
Acknowledgments
All of the authors are thankful to the Pondicherry Centre for Biological Sciences for providing the necessary facility to carry out the work. Financial support, from the State Bank of India (RASMECC), Pondicherry, India, in the form of a start-up loan to establish the institute, is also gratefully acknowledged. KT is a recipient of the Young Scientist grant (SB/FT/LS-382/2012), Science and Engineering Research Board, Department of Science and Technology, Government of India, and their support is duly acknowledged.
References
- 1Thirugnanasambantham K, Hairul-Islam VI, Saravanan S, Subasri S, Subastri A. Computational approach for identification of Anopheles gambiae miRNA involved in modulation of host immune response. Appl Biochem Biotechnol 2013; 170: 281–291. [DOI] [PubMed] [Google Scholar]
- 2Koj A, Jura J. Complex analysis of genes involved in the inflammatory response: interleukin-1-induced differential transcriptome of cultured human hepatoma HepG2 cells. Acta Biochim Pol 2003; 50: 573–582. [PubMed] [Google Scholar]
- 3Zhang X, Zhang ZQ, Dai FH, Shi BS, Chen L, Zhang XX et al. Comparison of circulating, hepatocyte specific messenger RNA and microRNA as biomarkers for chronic hepatitis B and C. PLoS One 2014; 9: e92112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Lankisch TO, Voigtländer T, Manns MP, Holzmann A, Dangwal S, Thum T. MicroRNAs in the bile of patients with biliary strictures after liver transplantation. Liver Transpl 2014; 20: 673–688. [DOI] [PubMed] [Google Scholar]
- 5Bala S, Petrasek J, Mundkur S, Catalano D, Levin I, Ward J et al. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology 2012; 56: 1946–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Bala S, Marcos M, Szabo G. Emerging role of microRNAs in liver diseases. World J Gastroenterol 2009; 15: 5633–5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Webster CR, Usechak P, Anwer MS. cAMP inhibits bile acid-induced apoptosis by blocking caspase activation and cytochrome c release. Am J Physiol Gastrointest Liver Physiol 2002; 283: G727–G738. [DOI] [PubMed] [Google Scholar]
- 8Kramer MF. Stem-loop RT-qPCR for miRNAs. Curr Protoc Mol Biol 2011; Chapter 15: Unit 15.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Olinga P, Merema MT, de Jager MH, Derks F, Melgert BN, Moshage H et al. Rat liver slices as a tool to study LPS-induced inflammatory response in the liver. J Hepatol 2001; 35: 187–194. [DOI] [PubMed] [Google Scholar]
- 10Song KH, Kim YH, Kim BY. Sho-saiko-to, a traditional herbal medicine, regulates gene expression and biological function by way of microRNAs in primary mouse hepatocytes. BMC Complement Altern Med 2014; 14: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]