Abstract
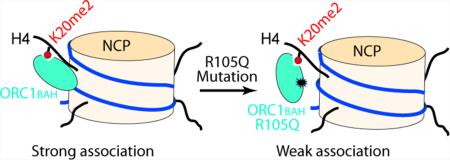
Recent studies have identified several genetic mutations within the BAH domain of human Origin Recognition Complex subunit 1 (hORC1BAH), including the R105Q mutation, implicated in Meier-Gorlin Syndrome (MGS). However, the pathological role of the hORC1 R105Q mutation remains unclear. In this study, we have investigated the interactions of the hORC1BAH domain with histone H4K20me2, DNA, and the nucleosome core particle labeled with H4Kc20me2, a chemical analog of H4K20me2. Our study revealed a nucleosomal DNA binding site for hORC1BAH. The R105Q mutation reduces the hORC1BAH–DNA binding affinity, leading to impaired hORC1BAH–nucleosome interaction, which likely influences DNA replication initiation and MGS pathogenesis. This study provides an etiologic link between the hORC1 R105Q mutation and MGS.
Eukaryotic DNA replication is initiated by assembling the prereplication complex (pre-RC) to chromosomal sites, termed origins of replication.1,2 Assembly of pre-RC is nucleated by the heterohexameric origin recognition complex (ORC, comprised of subunits ORC1–6),3 which is further joined by CDT1, CDC6, and hexameric minichromosome maintenance (MCM) helicase.4 Throughout the entire cell cycle, the subunits of ORC2 through ORC5 form a stable core complex (ORC(2–5)) that interacts dynamically with ORC1 and ORC6.5 During the G1 phase, the interaction of ORC1 with ORC(2–5) helps recruit ORC to nuclear structures, which is essential for subsequent chromatin loading of pre-RC.6,7 After cells enter the S phase, ORC1 is selectively degraded by a ubiquitin-dependent mechanism,6 which thereby prevents reinitiation of DNA replication during a single cell division cycle.
Emerging evidence has revealed that the DNA replication initiation machinery is linked to a variety of human diseases.8 For instance, germline mutations in five genes from pre-RC (ORC1, ORC4, ORC6, CDT1, and CDC6) have recently been identified to be implicated in Meier-Gorlin syndrome (MGS), an autosomal recessive primordial dwarfism characterized by absent or hypoplastic kneecaps, markedly small ears, and short statue.9 Several lines of evidence have indicated that the MGS-associated ORC mutations may lead to impaired DNA replication: the ORC6 mutation (Y232S), located in the C-terminal helix of ORC6, has been shown to impede the assembly of ORC;10 the ORC4 mutation (Y174C) is found to be pathogenic in functional assays of cell growth.11 In the case of ORC1, the MGS-associated mutations (e.g., F89S, R105Q, and E127G) mainly fall in its N-terminal BAH (Bromo Adjacent Homology) domain,12,13 a domain that mediates the association of ORC with chromatin.14,15 Furthermore, a recent study by Kuo et al. has demonstrated that the metazoan ORC1 BAH domain (ORC1BAH) specifically binds to histone H4 dimethylated at lysine 20 (H4K20me2), an epigenetic mark that is enriched at replication origins.14 This recognition appears to be important for association of ORC1 with chromatin and DNA replication initiation, as wild type human ORC1 (hORC1), but not the H4K20me2 interaction-defective mutants of hORC1, was able to rescue the MGS-like proportionate dwarfism phenotype in an ORC1-knockdown zebrafish model.14
The work by Kuo et al. has also revealed that two of the MGS-associated ORC1 mutations, F89S and E127G, both led to significantly reduced binding affinity of the hORC1 BAH (hORC1BAH) domain for H4K20me2,14 suggesting that the hORC1 F89S and E127G mutations may affect the hORC1 function through impairing the hORC1–H4K20me2 interaction. In addition to the hORC1 F89S and E127G mutations, the hORC1 R105Q is a recurrent mutation that was identified in a cohort of MGS patients with severe development defects.12,13,16 A recent study by Hossain and Stillman has indicated that this mutation abolishes the ability of hORC1 to inhibit the kinase activity of cyclin E-CDK2, which consequently affects centrosome copy number control in cells.17 On the basis of this observation, it has been proposed that residue R105 of hORC1 is located in its cyclin E-CDK2 binding surface; the R105Q mutation disrupts the hORC1-mediated inhibition of cyclin E-CDK2, which therefore contributes to centrosome defects and the MGS phenotype.17 While this study has uncovered a link between the hORC1 R105Q mutation and centrosome duplication, whether the ORC1 R105Q mutation directly influences the hORC1-chromatin interaction and DNA replication initiation remains unclear.
In this study, we have investigated the effect of the hORC1 R105Q mutation on the hORC1BAH–H4K20me2 interaction using a combined chemical, biochemical, and cellular approach. First, through isothermal titration calorimetry (ITC) analysis, we demonstrate that wild type hORC1BAH and its R105Q mutant bind to the histone H4(14–25)K20me2 peptide with similar affinity. Second, our electrophoretic mobility shift assay (EMSA) and protein sequence analysis identified a DNA binding site for wild type ORC1BAH, which is largely disrupted by the R105Q mutation. On this basis, we performed in vitro reconstitution of the nucleosome core particle (NCP) labeled with H4Kc20me2, a chemical analog of H4K20me2, and compared the interactions of wild type and R105Q hORC1BAH with the H4Kc20me2-labeled NCP. Through quantitative EMSA analysis, we show that the hORC1BAH R105Q mutation significantly decreases the binding affinity of the hORC1BAH domain for the H4Kc20me2-labeled NCP. Finally, we performed the chromatin fractionation experiment to show that the R105Q mutation reduces the ORC1-chromatin association in vivo. Taken together, this study provides first evidence that the hORC1 R105Q mutation reduces the chromatin loading of hORC1 through impairing the interaction between hORC1 and the nucleosomal DNA, which may consequently affect initiation of DNA replication and contribute to MGS pathogenesis.
Interactions of Wild Type and R105Q hORC1BAH with H4K20me2
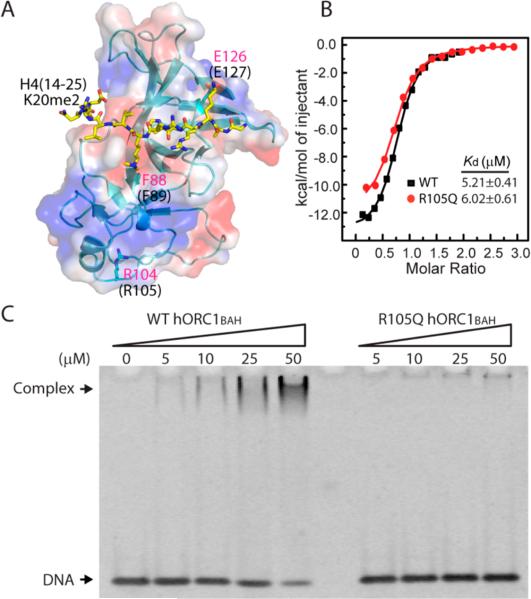
Cellular evidence has indicated that the MGS-associated ORC1 mutations lead to reduced pre-RC assembly and impaired DNA replication licensing.13 Accordingly, structural analysis of the mouse ORC1BAH (mORC1BAH) domain in complex with the histone H4(14–25)K20me2 peptide indicates that two of these mutations, F89S and E127G, are located in regions that impinge on the H4K20me2 binding (Figure 1A).14 This observation has been confirmed by ITC analysis, which showed that the F89S and E127G mutations lead to reduced binding affinity of the hORC1BAH domain for the H4(14–25)K20me2 peptide by 2- and 4-fold, respectively.14 By contrast, the R105Q mutation is located in a region that is distant from the H4K20me2 binding site, implying that this mutation may have a negligible impact on the H4K20me2 binding (Figure 1A). To clarify the functional consequence of the hORC1 R105Q mutation, we have performed the ITC assay to measure the binding affinity of the hORC1BAH R105Q mutant for the H4(14–25)K20me2 peptide. As shown in Figure 1B, the hORC1BAH R105Q mutant binds to the H4K20me2 peptide with a dissociation constant (Kd) of 6.02 μM, equivalent to what was previously determined for the wild type hORC1BAH domain (Kd = 5.21 μM)14 within experimental error. This observation, consistent with the pull-down assay in a previous study,17 suggests that the hORC1 R105Q mutation does not affect the binding of hORC1 to histone H4K20me2.
Figure 1.
hORC1 R105Q mutant retention of the binding affinity of hORC1BAH for histone H4K20me2, but not for DNA. (A) Structure of the mORC1BAH–H4(14–25)K20me2 complex (PDB 4DOW), with residues of H4(14–25)K20me2 shown in yellow stick representation, and mORC1 F88, R104, and E126 (equivalent to F89, R105, and E127 in hORC1BAH, respectively) shown in blue stick representation. (B) ITC analysis of the bindings of wild type hORC1BAH and its R105Q mutant to the histone H4(14–25)K20me2 peptide. (C) Titration of wild type hORC1BAH (left) and its R105Q mutant (right) against a 14-mer DNA duplex using EMSA. The protein–DNA complex and free DNA are indicated by arrows.
Interactions of Wild Type and R105Q hORC1BAH with DNA
Given the fact that the interaction between ORC1 and H4K20me2 occurs in the context of a nucleosome in vivo, we ask whether the hORC1BAH domain interacts with DNA and, if it does, whether the hORC1 R105Q mutation disrupts such an interaction. To address these issues, we have titrated the hORC1BAH domain against a 14-mer DNA duplex and analyzed DNA binding using the EMSA method. Under a salt concentration of 100 mM NaCl, we observed that the 14-mer DNA is continuously shifted by increasing the amount of wild type hORC1BAH (Figure 1C), indicative of formation of the hORC1BAH–DNA complex. By contrast, the hORC1BAH R105Q mutant in the same concentration range failed to shift the DNA significantly (Figure 1C), suggesting that the hORC1BAH R105Q mutant largely loses its DNA binding affinity. When we further lowered the salt concentration to 20 mM NaCl and used a 153-mer Widom DNA18 as a probe, we observed that the DNA binding affinity of the hORC1BAH R105Q mutant is slightly increased, albeit still much weaker than that of wild type hORC1BAH (Figure S1). It is noted that the other two MGS-associated mutants, hORC1BAH F89S and E127G, both bind to DNA with a similar affinity to that of wild type hORC1BAH (Figure S2). Consistent with these observations, analysis of surface electrostatic potentials of the structure of the mORC1BAH–H4K20me2 complex reveals that the R105 equivalent residue in mORC1 (R104) is located in a region that is abundant with basic residues, forming a potential DNA binding surface (Figure 1A and Figure S3A). We then investigated this potential DNA binding region through site-directed mutagenesis and DNA binding assay. We observed that two mutations within this area, the K99A/R100A double mutation and K163A mutation, both dramatically reduced the hORC1BAH–DNA interaction, supporting that the R105Q surrounding area is the DNA binding surface of hORC1BAH (Figure S3B). It is worth noting that this DNA binding site does not appear to be conserved in all BAH domains (Figure S3C,D). For instance, structure-based sequence alignment of the ORC1BAH domains from diverse species indicates that five residues that participate in formation of this potential DNA binding site are highly conserved in metazoan ORC1BAH domains, but not in their yeast counterparts (Figure S3D). Further, in the structures of the yeast Sir3 BAH domain in complex with a nucleosome,19–21 the corresponding region does not make any appreciable contact with nucleosomal DNA (Figure S3C). Taken together, these observations identified a DNA binding site for the metazoan ORC1BAH domain. The R105Q mutation presumably alters the DNA binding site of the hORC1BAH domain, resulting in reduced binding of the hORC1BAH domain with DNA.
Interactions of Wild Type and R105Q hORC1BAH with the NCP Labeled with H4Kc20me2
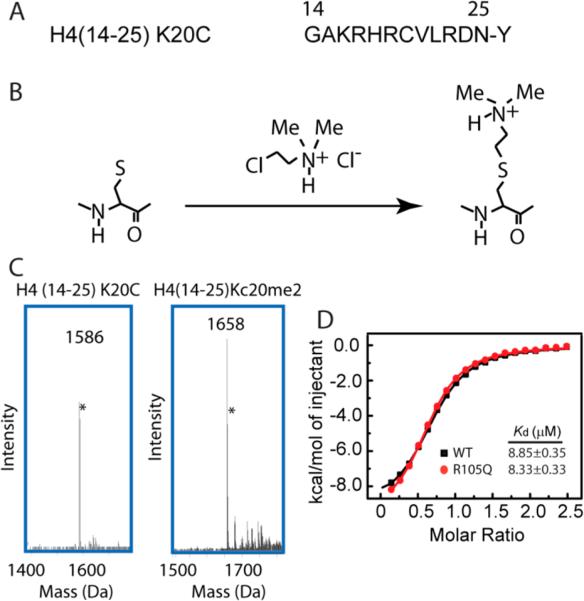
On the basis of the ITC and EMSA analysis, we postulated that the hORC1 R105Q mutation, through disruption of the hORC1BAH–DNA interaction, might impair the association of the hORC1BAH domain with the H4K20me2-modified NCP. To test this, we set out to investigate the effect of the hORC1 R105Q mutation on the hORC1BAH–H4K20me2 interaction at the nucleosomal level. Considering that preparation of the epigenetically modified histone proteins using the traditional semisynthetic approach suffers from low efficiency,22 we have resorted to a chemical approach, in which the “epigenetically” modified histone protein is prepared by covalently linking the chemical analog of an epigenetic mark (e.g., methyl group) to the recombinant protein.23,24 This method, taking advantage of the fact that the cysteine residues can be alkylated with haloethylamines to produce amino-ethylcysteine residues,23 has been used to introduce mono-, di-, or trimethyl-lysine analogs to specific sites of recombinantly expressed histones.24 To test the feasibility of this approach in the study of the hORC1BAH–H4K20me2 interaction, we first generated the H4(14–25)Kc20me2 peptide, analogue of the H4(14–25)K20me2 peptide, by chemical reaction of (2-cholorethyl)-dimethylammonium chloride with residue C20 of the H4(14–25) K20C peptide (Figure 2A,B), which was subsequently verified by mass spectrometry (Figure 2C). Next, we performed an ITC assay to measure the binding affinity of the hORC1BAH domain for the H4(14–25)Kc20me2 peptide. In agreement with the measurements for the hORC1BAH–H4K20me2 bindings (Figure 1B), our ITC analysis indicated that both wild type hORC1BAH and its R105Q mutant interact with the H4(14–25)Kc20me2 peptide, with Kds of 8.85 μM and 8.33 μM, respectively (Figure 2D). These values are comparable with those determined for the H4(14–25)K20me2 peptide (Figure 1B), therefore suggesting that the H4Kc20me2 mark is capable of recapitulating the biochemical property of H4K20me2 in this study.
Figure 2.
Generation of the H4Kc20me2 mark, a chemical analog of H4K20me2. (A) Amino acid sequence of the H4(14–25) K20C peptide, with an extra tyrosine introduced to the C-terminus for spectrophotometric measurement. (B) Schematic depicting the generation of the H4Kc20me2 modification.24 (C) Mass spectra of the H4(14–25) K20C and H4(14–25)Kc20me2 peptides. The peaks indicated by the asterisks arise from the isotope effects. (D) ITC analysis of the bindings of wild type hORC1BAH and its R105Q mutant to the histone H4(14–25)Kc20me2 peptide.
Next, we proceeded to perform in vitro reconstitution of the NCP labeled with H4Kc20me2. In essence, we first introduced the K20C mutation on the recombinant histone H4 protein by site-directed mutagenesis, then we obtained histone H4Kc20me2 by chemical reaction of the H4 K20C mutant with (2-cholorethyl)-dimethylammonium chloride, as described above. The reaction product was again verified by mass spectrometry (Figure S4A,B). This success has allowed us to generate the NCP labeled with H4Kc20me2, using recombinant histone H3, H4 Kc20me2, H2A, and H2B proteins, and Widom 601-DNA,18 following a reported protocol.25 On this basis, we have evaluated the binding specificities of the hORC1BAH domain with H4Kc20me2 at both histone octamer and nucleosomal levels using pull down assays (Figure S5A,B). Our results show that the hORC1BAH domain preferentially binds to the H4Kc20me2-modified histone octamer and NCP over their unmodified counterparts (Figure S5A,B), consistent with the previously observed H4K20me2-binding specificity of hORC1BAH.14
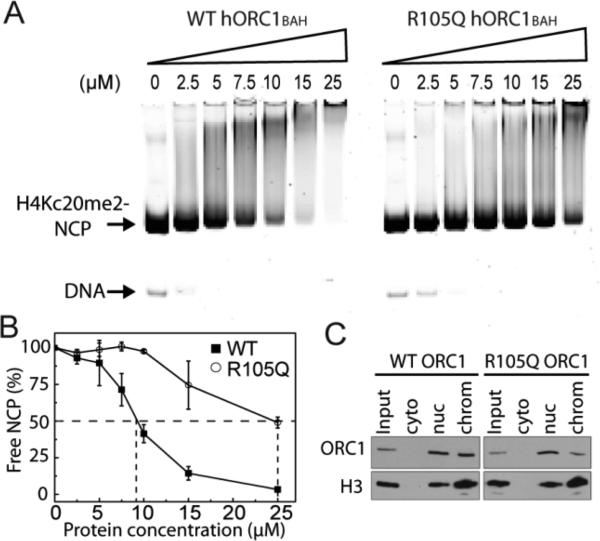
We then performed the EMSA analysis to compare the bindings of wild type hORC1BAH and its R105Q mutant to the H4Kc20me2-labeled NCP. As shown in Figure 3A, the free H4Kc20me2-modified NCP is strongly shifted by increasing the amount of wild type hORC1BAH, confirming a tight association between wild type hORC1BAH and the H4Kc20me2-modified NCP. By contrast, gel shift of the H4Kc20me2-modified NCP is significantly reduced in the presence of the same amount of the R105Q hORC1BAH mutant. Quantitative analysis of the amount of free NCP over various concentrations of the hORC1BAH domain reveals that the R105Q mutation reduces the affinity of the hORC1BAH domain for the H4Kc20me2-modified NCP by at least a factor of 2 (Figure 3B), indicating that the R105Q mutation impairs the interaction between the hORC1BAH domain and the H4K20me2-modified NCP.
Figure 3.
hORC1 R105Q mutation reduction of the binding affinity of hORC1BAH for the H4Kc20me2-labeled NCP. (A) Titrations of wild type hORC1BAH (left) and its R105Q mutant (right) against the H4Kc20me2-labeled NCP using EMSA. The free H4Kc20me2-labeled NCP and DNA are indicated by arrows. (B) Quantification of hORC1BAH domain binding to H4Kc20me2-labeled NCP from A. The mean value (±s.d.) for the percentage of the unbound NCPs, derived from two independent experiments, is plotted against hORC1BAH concentration. The concentration of hORC1BAH required for 50% binding to NCP is indicated by dashed lines. (C) Western blot analysis of lysates from U2OS cells stably expressing FLAG-ORC1 (WT or R105Q) biochemically separated into soluble and chromatin-bound fractions and probed with anti-Flag and anti-H3 to detect ORC1 and H3 as indicated. Input (33%), whole cell extract; cyto, cytoplasmic fraction; nuc, nuclear protein fraction extracted in 420 mM salt; chrom, chromatin-bound fraction.
Chromatin Loading of Wild Type and R105Q ORC1 in vivo
To examine the effect of R105Q mutation on ORC1-chromatin binding in vivo, we stably expressed in U2OS cells flag-tagged wild type and R105Q ORC1, respectively, followed by chromatin fractionation and Western blotting of Flag-ORC1. Our results show that, although the overall expression levels of wild type and R105Q ORC1 in the nucleus are similar, the level of chromatin-bound R105Q ORC1 is significantly lower than that of wild type ORC1 (Figure 3C). This observation is consistent with a previous study,13 which showed that the chromatin-bound hORC1 and human ORC2 proteins were evidently reduced in skin fibroblasts harboring the hORC1 R105Q mutation.
In summary, our study reveals that the hORC1BAH domain binds to both histone H4K20me2 and DNA components of the H4K20me2-modified nucleosome. Our observations complement the previous study by Kuo et al. that the H4K20me2 binding is essential for chromatin loading of ORC and likely attributes to the pathological effects of two of the MGS-associated ORC1 mutations (F89S and E127G)14 and suggest that the interaction between the hORC1BAH domain and DNA is also required for proper chromatin loading of hORC1. Impairment of the hORC1–chromatin interaction by the hORC1 R105Q mutation will probably affect the assembly of ORC on chromatin and subsequent DNA replication initiation, which, along with the R105Q-caused defect in centrosome number control,17 contributes to the pathogenesis of MGS.
METHODS
For protein expression and purification, nucleosome core particle reconstitution, histone octamer pull-down assay, nucleosome pull-down assay, and mass spectrometry, please see the Supporting Information.
ITC Measurements
Wild type or R105Q hORC1BAH domain (~0.08 mM each) and the histone peptides (~1 mM) were dialyzed against a buffer containing 20 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 2 mM DTT. The peptide was titrated into the hORC1BAH domain at 5 °C using the microCal ITC200 instrument (GE healthcare). The ITC data were analyzed using the Origin 7.0 software.
EMSA (Electrophoretic Mobility Shift Assay)
A random 14-bp DNA duplex (upper strand: 5′-GGAGGCCTCCTGCT-3′) was used to characterize DNA binding of wild type hORC1BAH and its F89S, K99A/R100A, R105Q, E127G, and K163A mutants. Binding reactions contained 3–5 μM dsDNA and various concentrations of protein, dissolved in 10 μL of binding buffer (10 mM Tris-HCl, pH 7.5, 100 mM NaCl). Binding reaction mixtures were electrophoresed in 6% DNA retardation gel (Invitrogen) in 0.25× or 0.5× TBE (89 mM Tris-borate, pH 8.4) buffer at 4 °C under 100 V for ~1 h. DNA was visualized by ethidium bromide staining, or SYBR green staining scanned by a Typhoon imager (GE Healthcare).
To perform EMSA for Widom 601 DNA or H4Kc20me2-labeled NCP, 1.0 μM Widom 601 DNA or 1.0–1.5 μM H4Kc20me2-labeled NCP was mixed with various concentrations of wild type or R105Q hORC1BAH in a 10 μL buffer containing 20 mM Tris-HCl, pH 7.5, 20 mM NaCl, 0.8 mM EDTA, and 1.2 mM DTT. The samples were loaded to a 6% DNA retardation gel in 0.25× TBE buffer and run at 4 °C under 100 V for 1 h and 10 min. The gel was stained with 10 000× diluted SYBR green I for 1 h, scanned by Typhoon imager. The intensity of the gel shift was integrated by the ImageJ software (http://imagej.nih.gov/ij/). Then the ratio of the intensity of free NCP in each lane over the intensity of the NCP in the hORC1BAH-free lane was calculated and used for normalization of the percentage of free nucleosome in each lane.
Chromatin Fraction of ORC1
U2OS cells were cultured in DMEM (Life Technologies) supplemented with 10% fetal bovine serum (Gibco/Life Technologies), penicillin-streptomycin (Life Technologies), and l-glutamine (Life Technologies). Retroviral transduction was performed as previously described14 to generate cells stably expressing pBabe-puro-3xFLAG-ORC1 WT or R105Q.14 Cells were selected under puromycin (Sigma) at 2 μg mL−1 for 72 h before collection.
Biochemical fractionation was adapted from a protocol previously described.26 Briefly, 1 × 107 cells per cell line were collected, washed in PBS, and lysed in Buffer A (10 mM HEPES pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10%glycerol, 1 mM DTT, cOmplete protease inhibitor tablet (Roche)), containing Triton X-100 at a final concentration of 0.1%, for 8 min on ice. Cytoplasmic proteins were separated from nuclei by centrifugation at 1300g for 5 min at 4 °C. The first supernatant was recovered and clarified by centrifugation at 21 000g for 10 min at 4 °C to generate the cytoplasmic fraction. Nuclei pellets were washed once in Buffer A and lysed in Buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, cOmplete protease inhibitor) for 30 min on ice, and soluble proteins were separated from chromatin by centrifugation at 1700g for 5 min at 4 °C. Nuclear proteins were extracted by incubating chromatin pellets in Buffer B with 420 mM NaCl for 20 min on ice, followed by centrifugation at 1700g for 5 min at 4 °C, and the supernatant was collected as a nuclear protein fraction. Chromatin pellets were resuspended in the SDS sample buffer and solubilized by sonication for 10 min in a Bioruptor (Diagenode). For whole cell extracts, 1 × 107 cells per cell line were collected, washed in PBS, and lysed in a RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 0.1% SDS, 1 mM DTT, cOmplete protease inhibitor tablet) for 10 min on ice followed by sonication for 10 min in a Bioruptor. Samples were clarified by centrifugation at 21 000g for 10 min at 4 °C.
Supplementary Material
ACKNOWLEDGMENTS
J.S. is supported by the Basil O'Connor Starting Scholar Research Award, March of Dimes foundation, and University of California, Riverside start-up fund. This work was supported in part by grants from the NIH to O.G. (R01 GM079641). We thank R. New from UCR ACIF for help in the mass spectrometry (NSF CHE-0541848).
Footnotes
Supporting Information
Supplementary methods, Figures S1–S5. This material is available free of charge via the Internet at http://pubs.acs.org.
O.G. is a co-founder of EpiCypher, Inc.
REFERENCES
- 1.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 2.Mendez J, Stillman B. Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. BioEssays. 2003;25:1158–1167. doi: 10.1002/bies.10370. [DOI] [PubMed] [Google Scholar]
- 3.Bell SP. The origin recognition complex: from simple origins to complex functions. Genes Dev. 2002;16:659–672. doi: 10.1101/gad.969602. [DOI] [PubMed] [Google Scholar]
- 4.Nishitani H, Lygerou Z, Nishimoto T, Nurse P. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature. 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- 5.Dhar SK, Delmolino L, Dutta A. Architecture of the human origin recognition complex. J. Biol. Chem. 2001;276:29067–29071. doi: 10.1074/jbc.M103078200. [DOI] [PubMed] [Google Scholar]
- 6.DePamphilis ML, Blow JJ, Ghosh S, Saha T, Noguchi K, Vassilev A. Regulating the licensing of DNA replication origins in metazoa. Curr. Opin. Cell Biol. 2006;18:231–239. doi: 10.1016/j.ceb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Ohta S, Tatsumi Y, Fujita M, Tsurimoto T, Obuse C. The ORC1 cycle in human cells: II. Dynamic changes in the human ORC complex during the cell cycle. J. Biol. Chem. 2003;278:41535–41540. doi: 10.1074/jbc.M307535200. [DOI] [PubMed] [Google Scholar]
- 8.Shen Z. The origin recognition complex in human diseases. Bioscience reports. 2013;33:475–483. doi: 10.1042/BSR20130036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorlin RJ, Cervenka J, Moller K, Horrobin M, Witkop CJ., Jr. Malformation syndromes. A selected miscellany. Birth Defects, Orig. Artic. Ser. 1975;11:39–50. [PubMed] [Google Scholar]
- 10.Bleichert F, Balasov M, Chesnokov I, Nogales E, Botchan MR, Berger JM. A Meier-Gorlin syndrome mutation in a conserved C-terminal helix of Orc6 impedes origin recognition complex formation. eLife. 2013;2:e00882. doi: 10.7554/eLife.00882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guernsey DL, Matsuoka M, Jiang H, Evans S, Macgillivray C, Nightingale M, Perry S, Ferguson M, LeBlanc M, Paquette J, Patry L, Rideout AL, Thomas A, Orr A, McMaster CR, Michaud JL, Deal C, Langlois S, Superneau DW, Parkash S, Ludman M, Skidmore DL, Samuels ME. Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nat. Genet. 2011;43:360–364. doi: 10.1038/ng.777. [DOI] [PubMed] [Google Scholar]
- 12.Bicknell LS, Bongers EM, Leitch A, Brown S, Schoots J, Harley ME, Aftimos S, Al-Aama JY, Bober M, Brown PA, van Bokhoven H, Dean J, Edrees AY, Feingold M, Fryer A, Hoefsloot LH, Kau N, Knoers NV, Mackenzie J, Opitz JM, Sarda P, Ross A, Temple IK, Toutain A, Wise CA, Wright M, Jackson AP. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat. Genet. 2011;43:356–359. doi: 10.1038/ng.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bicknell LS, Walker S, Klingseisen A, Stiff T, Leitch A, Kerzendorfer C, Martin CA, Yeyati P, Al Sanna N, Bober M, Johnson D, Wise C, Jackson AP, O'Driscoll M, Jeggo PA. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat. Genet. 2011;43:350–355. doi: 10.1038/ng.776. [DOI] [PubMed] [Google Scholar]
- 14.Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, Chen JK, Patel DJ, Gozani O. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature. 2012;484:115–119. doi: 10.1038/nature10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noguchi K, Vassilev A, Ghosh S, Yates JL, DePamphilis ML. The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo. EMBO J. 2006;25:5372–5382. doi: 10.1038/sj.emboj.7601396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Munnik SA, Otten BJ, Schoots J, Bicknell LS, Aftimos S, Al-Aama JY, van Bever Y, Bober MB, Borm GF, Clayton-Smith J, Deal CL, Edrees AY, Feingold M, Fryer A, van Hagen JM, Hennekam RC, Jansweijer MC, Johnson D, Kant SG, Opitz JM, Ramadevi AR, Reardon W, Ross A, Sarda P, Schrander-Stumpel CT, Sluiter AE, Temple IK, Terhal PA, Toutain A, Wise CA, Wright M, Skidmore DL, Samuels ME, Hoefsloot LH, Knoers NV, Brunner HG, Jackson AP, Bongers EM. Meier-Gorlin syndrome: growth and secondary sexual development of a microcephalic primordial dwarfism disorder. Am. J. Med. Genet. Part A. 2012;158A:2733–2742. doi: 10.1002/ajmg.a.35681. [DOI] [PubMed] [Google Scholar]
- 17.Hossain M, Stillman B. Meier-Gorlin syndrome mutations disrupt an Orc1 CDK inhibitory domain and cause centrosome reduplication. Genes Dev. 2012;26:1797–1810. doi: 10.1101/gad.197178.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 19.Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 A resolution. Science. 2011;334:977–982. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnaudo N, Fernandez IS, McLaughlin SH, Peak-Chew SY, Rhodes D, Martino F. The N-terminal acetylation of Sir3 stabilizes its binding to the nucleosome core particle. Nat. Struct. Mol. Biol. 2013;20:1119–1121. doi: 10.1038/nsmb.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang D, Fang Q, Wang M, Ren R, Wang H, He M, Sun Y, Yang N, Xu RM. Nalpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain. Nat. Struct Mol. Biol. 2013;20:1116–1118. doi: 10.1038/nsmb.2637. [DOI] [PubMed] [Google Scholar]
- 22.Flavell RR, Muir TW. Expressed protein ligation (EPL) in the study of signal transduction, ion conduction, and chromatin biology. Acc. Chem. Res. 2009;42:107–116. doi: 10.1021/ar800129c. [DOI] [PubMed] [Google Scholar]
- 23.Kenyon GL, Bruice TW. Novel sulfhydryl reagents. Methods Enzymol. 1977;47:407–430. doi: 10.1016/0076-6879(77)47042-3. [DOI] [PubMed] [Google Scholar]
- 24.Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 26.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.