Abstract
Background
Silver nanoparticles (AgNPs) have attracted much interest and have been used for antibacterial, antifungal, anticancer, and antiangiogenic applications because of their unique properties. The increased usage of AgNPs leads to a potential hazard to human health. However, the potential effects of AgNPs on animal models are not clear. This study was designed to investigate the potential impact of AgNPs on pregnant mice.
Methods
The synthesis of AgNPs was performed using culture extracts of Bacillus cereus. The synthesized AgNPs were characterized by X-ray diffraction, Fourier transform infrared spectroscopy, and transmission electron microscopy. AgNPs were administrated into pregnant mice via intravenous infusion at 1.0 mg/kg doses at 6.5 days postcoitum (dpc). At 13.5, 15.5, and 17.5 dpc, the pregnant mice were euthanized, and the embryo and placenta were isolated. The meiotic status of oocytes was evaluated. DNA methylation studies were performed, and aberrant imprinting disrupted fetal, placental, and postnatal development. Quantitative real-time polymerase chain reaction analysis and Western blot were used to analyze various gene expressions.
Results
The synthesized AgNPs were uniformly distributed and were spherical in shape with an average size of 8 nm. AgNPs exposure increased the meiotic progression of female germ cells in the fetal mouse ovaries, and maternal AgNP exposure significantly disrupted imprinted gene expression in 15.5 dpc embryos and placentas, such as Ascl2, Snrpn, Kcnq1ot1, Peg3, Zac1, H19, Igf2r, and Igf2; DNA methylation studies revealed that AgNPs exposure significantly altered the methylation levels of differentially methylated regions of Zac1.
Conclusion
The results from this study indicated that early exposure to AgNPs has the potential to disrupt fetal and postnatal health through epigenetic changes in the embryo and abnormal development of the placenta. These results can contribute to research involved in the safe use of various biomedical applications of AgNPs and improves the understanding of the development of AgNPs in biomedical applications.
Keywords: silver nanoparticles, methylation, meiosis, gene expression
Introduction
Nanoparticles are used in biomedical applications such as therapeutics, catalysis, antimicrobial agents, transfection vectors, biosensing devices, and fluorescent labels.1–5 Silver nanoparticles (AgNPs), as one of the most commonly used metal nanoparticles, have recently gained popularity. AgNPs, with a broad antibacterial activity against both gram-negative and gram-positive microorganisms, has been employed for antimicrobial properties, and thus they are involved in the production of several medical products like catheters or implants6 and are also used for their antifungal,7 antiangiogenic,8 and anticancer properties.9 Despite their potential benefits to society, biological adverse effects of AgNPs must be carefully evaluated. Several studies have demonstrated that AgNPs can enter into cells in fish, mice, and humans and be distributed throughout the body.10–13 AgNPs can be systemically distributed throughout and may be accumulated and redistributed between various organs, leading to decreased body weight, changes in blood biochemical parameters, and inflammation.14–16 Previous studies have indicated that AgNPs induce cytotoxicity via the generation of reactive oxygen species (ROS), leading to increases in intracellular oxidative stress or Jun amino-terminal kinases (JNK) activation, which leads to the release of cytochrome C into the cytosol and translocation of Bax into mitochondria.17,18 These releases induce cell apoptosis, autophagy, and necrosis.19,20 Previous studies have shown that AgNP-treated cells cause different cellular responses that may be important for their use in medicine and their toxicity.20–22
In mice, the cells migrate to the nascent gonads and proliferate at ~10.5–11.5 days postcoitum (dpc) and then migrate from the gonads to the testes or ovaries at 13.5 dpc.23,24 Female germ cells, or oogonia, stop dividing and enter meiosis prophase I at 13.5 dpc through the leptotene, zygotene, pachytene, and diplotene stages to become oocytes in ovaries and are finally arrested at the diplotene stage.25 Synaptonemal complex protein 3 (SCP3), synaptonemal complex protein 1 (SCP1), stimulated by retinoic acid 8 (Stra8), deleted in azoospermia-like (Dzal), and DNA meiotic recombinase 1 (Dmc1) are meiotic markers.26 In mice, retinoic acid, an extrinsic meiosis-initiating signal, induces the transcription and expression of Stra8, which in turn governs the meiotic program.27,28 Dazl, an intrinsic factor, is required for meiotic DNA replication and the subsequent events of meiotic prophase.29 SCP3 plays a critical role in pairing and recombination of homologous chromosomes in meiosis 1.30 Exposure to environmental factors in pregnant mice affects the expression of meiosis genes and meiotic progression.31,32
The placenta is an important organ for material exchange between fetal and maternal blood and is the main source of nutrients for the fetus.33 Imprinted genes play important roles in placental differentiation, growth, and function, with profound effects on fetal development.34,35 It has been suggested that changes in imprinted gene dosage in the placentae may compromise the prenatal control of nutritional resources.36 Epigenetic events are crucial for early development but can be affected by environmental factors and diets.37,38 Gallou-Kabani et al39 showed that a high-fat diet during pregnancy modified the expression of imprinted genes and local and global DNA methylation patterns in the placenta, and global DNA methylation resulted in sex- and diet-specific differences. Martha et al40 demonstrated that bisphenol A (BPA) exposure may perturb fetal and postnatal health through epigenetic changes in the embryo and alterations in placental development. Abnormal expression of an imprinted gene will affect the development of the fetus, its health later in life, and the child’s behavior.41,42 Igf2 is a paternal allele gene and encodes a fetal growth factor; deletion of the Igf2 gene caused a significant decrease in fetal and placental weight compared with a control group.43 Loss of function alleles on insulin-like growth factor 2 receptor (Igf2r) growth functions resulted in overgrowth of the embryo heart and placenta with partial perinatal lethality.44 In addition, low expression of H19 messenger RNA (mRNA) may contribute to the development of macrosomia.45 Exposure to AgNPs caused mitochondrial dysfunction, which leads to the induction of ROS, DNA damage, and chromosomal aberrations, apoptosis, and autophagy.46 AgNPs increased ROS production in cells, leading to apoptosis through the activation of p42/p44, JNKs, or p38 mitogen-activated protein kinases or p53-dependent processes.47,48
Because of the extensive use of AgNPs in consumer products and increasing potential of human and environmental exposure, safety and regulatory concerns regarding AgNPs are growing.49,50 The primary objective of this study was to synthesize smaller AgNPs using biological systems. The secondary objective of this study was to investigate the effect of the smaller-size AgNPs on fetal development, including imprinted gene expression and on imprinted gene DNA methylated regions (DMRs) in fetuses during pregnancy. Finally, we investigated the effect of AgNPs on the expression of meiosis genes and meiosis progression during pregnancy.
Materials and methods
Materials
Luria–Bertani agar was purchased from USB Corporation (Santa Clara, CA, USA). Mueller–Hinton broth and agar, silver nitrate, and crystal violet were purchased from Sigma-Aldrich (St Louis, MO, USA). All the other chemicals were purchased from Sigma-Aldrich unless otherwise stated.
Synthesis and characterization of AgNPs
Synthesis and characterization of AgNPs was carried out according to the method described earlier.8 Briefly, Bacillus mycoides strain was inoculated into flasks containing sterile Luria–Bertani broth and incubated for 24 hours at 37°C with agitation (200 rpm). After incubation, the culture was centrifuged at 10,000 rpm for 10 minutes, and the supernatant was used for AgNP synthesis. In a typical reaction, the culture supernatant was mixed with 5 mM aqueous AgNO3 solution (add 50 μL of 5 M aqueous AgNO3 solution in 50 mL culture supernatant, aqueous AgNO3 dissolved in deionized water to prepare aqueous AgNO3 solution) and incubated at 60°C for 6 hours in a water bath. This liquid was then centrifuged at 3,000 rpm for 10 minutes, the supernatant was the AgNPs solution. The as-prepared particles were characterized as previously described.51 X-ray diffraction (XRD) analyses were performed using an X-ray diffractometer (Bruker D8 DISCOVER; Bruker AXS GmBH, Karlsruhe, Germany). The high-resolution XRD measurements were performed at 3 kW with a Cu target using a scintillation counter (λ =1.5406 Å) at 40 kV and 40 mA and were recorded in the range of 2θ =5°–80°. Further characterization of the AgNPs surface changes and composition was performed using Fourier transform infrared (FT-IR) spectroscopy (Perkin Elmer Spectroscopy GX; Perkin Elmer, Waltham, MA, USA). Transmission electron microscopy (TEM; Hitachi H-7500, Seoul National University, Seoul, Republic of Korea) was used to determine the size and morphology of the AgNPs. TEM images of bio-AgNPs were obtained at an accelerating voltage of 300 kV.
Animals, treatment, and experimental design
All the experiments were performed with approval from the Institutional Animal Care and Use Committee at Konkuk University (IACUC approval number KU11035), Seoul, Republic of Korea. Virgin, female, 8-week-old ICR (imprinting control region) mice were purchased for this study and were housed in temperature-controlled (21°C–22°C) and light-controlled (12-hour light, 12-hour dark cycle) environments with 70% humidity and fed under ad libitum conditions. The females were mated with male mice and inspected daily for a mating plug. At 0.5 dpc, vaginal plugs were detected. Each female was housed individually.
The mice were divided into two groups (20 mice in each group) as follows: group 1, control; group 2, 1.0 mg/kg AgNP-treated group. The control group received phosphate-buffered saline without AgNPs using the same method used to administer AgNPs suspensions; the 1.0 mg AgNPs/kg was administrated to the mice via intravenous infusion one time at 1.0 mg/kg doses at 6.5 dpc. At 13.5, 15.5, and 17.5 dpc, the pregnant mice were sacrificed through cervical dislocation euthanasia, and then embryo and placenta were isolated for further study.
Evaluation of oocyte meiotic prophase I stages
The pregnant mice in the control and experimental groups were sacrificed with spinal dislocation death at 13.5 and 17.5 dpc, respectively, for collection of developing fetal oocytes. The meiotic status of the oocytes was confirmed by chromosome condition examination.31 The oocytes were treated in 1% sodium citrate for 20 minutes at room temperature, fixed in 4% paraformaldehyde, spread on polylysine-coated slides for 8 hours, blocked in tris-buffered saline with 1% donkey serum and 3% bovine serum albumin for 30 minutes at room temperature, and then incubated overnight at 4°C with SCP3 antibody (Novus, Littleton, CO, USA) at a dilution of 1:200. The next day, sections were incubated with Cy3-labeled goat anti-rabbit IgG at a dilution of 1:50 (Beyotime, Nantong, People’s Republic of China) at 37°C for 1.5 hours, followed by incubation with Hoechst33342 at a dilution of 1:1,000 for 5 minutes at room temperature. The slides were analyzed under a fluorescence microscope (Olympus BX51; Olympus Corporation, Tokyo, Japan), and the meiotic prophase stages were determined based on the SCP3 immunostaining characteristic patterns within the germ cells.32
Methylation studies
DNA was isolated using a micro DNA isolation kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. One microgram of DNA was treated with sodium bisul-fate using a Methyl amp™ DNA modification kit (Epigentek, Farmingdale, NY, USA) according to the manufacturer’s instructions. The bisulfite-treated DNA was subject to nested (seminested) polymerase chain reaction (PCR) amplification for pleiomorphic adenoma gene-like 1 (Zac1) and Igf2r (primer sequences are given in Table 1). The PCR products were separated by electrophoresis on 1% agarose gel, and bands of appropriate size were isolated from the gel and purified with the Wizard SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA). Purified DNA was cloned into a pGEM-T easy Vector (Promega) according to the manufacturer’s instructions. Positive clones were obtained by antibiotic selection, and the insert was DNA sequenced.
Table 1.
Primers used for quantitative RT-PCR
| Genes | Sequences of primers | GenBank |
|---|---|---|
| GAPDH | F: 5′-AGGTCGGTGTGAACGGATTTG-3′ | NC_000072.6 |
| R: 5′-TGTAGACCATGTAGTTGAGGTCA-3′ | ||
| Stra8 | F: 5′-CTCCTCCTCCACTCTGTTGC-3′ | XM_004046 273 |
| R: 5′-GCGGCAGAGACAATAGGAAG-3′ | ||
| Dazl | F: 5′-TGACGTGGATGTGCAGAAGAT-3′ | NM_010021 |
| R: 5′-AGGAGGATATGCCTGAACATACT-3′ | ||
| Scp1 | F: 5′-AGAAGCAAAAGCCCTTCACACT-3′ | NC_000069 |
| R: 5′-ACCTTTATCAATGTTTTCCCGACT-3′ | ||
| Scp3 | F: 5′-CCGCTGAGCAAACATCTAAAG-3′ | NC_000076 |
| R: 5′-ATCAGCAACATCTTCTTCTGAACC-3′ | ||
| Dmc1 | F: 5′-CCCTCTGTGTGACAGCTCAAC-3′ | NM_010059 |
| R: 5′-GGTCAGCAATGTCCCGAAG-3′ | ||
| Cx37 | F: 5′-GTCAGCGTGCTGTGTAAGAGTG-3′ | NC_000070.6 |
| R: 5′-GCAGGTTGAGCACCAGGGAGATGAC-3′ | ||
| Cx43 | F: 5′-TGTGGGCAAGACACGAATATG-3′ | NC_000076.6 |
| R: 5′-GACAAGGTCCAAGCCTACTCCC-3′ | ||
| Gdf9 | F: 5′-CTGATAGGCGAGGTGAGACC-3′ | NC_000077.6 |
| R: 5′-GAGCCGGACGGTATTGTAGA-3′ | ||
| Bmp15 | F: 5′-TTCTTAGAATTCTTTTGTGGGGAGT-3′ | NC_000086.7 |
| R: 5′-TGGTATAACTTGAGCATGTACCTCA-3′ | ||
| Zp1 | F: 5′-GTCCGACTCCTGCAGAGAAC-3′ | NC_000085.6 |
| R: 5′-TGATGGTGAAGCGCTGATAG-3′ | ||
| Zp2 | F: 5′-AAGGTCTTGAGCAGGAACGA-3′ | NC_000073.6 |
| R: 5′-GGGTGGAAAGTAGTGCGGTA-3′ | ||
| Zp3 | F: 5′-CCGAGCTGTGCAATTCCCAGA-3′ | NC_000071.6 |
| R: 5′-AACCCTCTGAGCCAAGGGTGA-3′ | ||
| Nobox | F: 5′-CTATCCTGACAGTGACAAACGCC-3′ | NC_000072.6 |
| R: 5′-CACCCTCTCAGCACCCTCATTAT-3′ | ||
| Figla | F: 5′-ACAGAGCAGGAAGCCCAGTA-3′ | NC_000072.6 |
| R: 5′-TGGGTAGCATTTCCCAAGAG-3′ | ||
| Bax | F: 5′-ATGCGTCCACCAAGAAGCTGAG-3′ | NC_000073.6 |
| R: 5′-CCCCAGTTGAAGTTGCCATCAG-3′ | ||
| Bcl2 | F: 5′-ACTGAATGTAGATAATGG-3′ | NC_000067.6 |
| R: 5′-TAACTGTAACTGATAAGG-3′ | ||
| Caspase3 | F: 5′-GATAATGTCTTAGAACTTGAATCC-3′ | NC_000074.6 |
| R: 5′-CTTCCATAAATCAGGTCCAA-3′ | ||
| Ascl2 | F: 5′-TGAGCATCCCACCCCCCTA-3′ | NC_000073.6 |
| R: 5′-CCAAACATCAGCGTCAGTATAG-3′ | ||
| Snrpn | F: 5′-CTGCAGCACATTGACTATAGGATG-3′ | NC_000073 |
| R: 5′-ACAGTCATTGAAACCAAGTTCTCC-3′ | ||
| Kcnq1ot1 | F: 5′-TCCATGGTGAGGTCTTATCTTGTT-3′ | NC_000073.6 |
| R: 5′-GAAAAGAAGCGTGTTGAAGAAAAA-3′ | ||
| Peg3 | F: 5′-AAGGCTCTGGTTGACAGTCGTG-3′ | NC_000073.6 |
| R: 5′-TTCTCCTTGGTCTCACGGGC-3′ | ||
| Zac1 | F: 5′-TCACCATTCACAATTATTCCCACT-3′ | NC_000076 |
| R: 5′-CTTCCGGTTGAATGTCTTCTCAC-3′ | ||
| H19 | F: 5′-ACCCCGGGATGACTTCATC-3′ | NC_000073.6 |
| R: 5′-TATCTCCGGGACTCCAAACC-3′ | ||
| Igf2r | F: 5′-TTACACATGGGAAGCTGTTGACT-3′ | NC_000083.6 |
| R: 5′-CGGCAGTTCTCTGTCTTTAGGTC-3′ | ||
| Igf2 | F: 5′-GGGGAGCTATGGGTAGGAAG-3′ | NC_000073.6 |
| R: 5′-GGCATAAAATTTGGGGTCCT-3′ |
Abbreviation: RT-PCR, real-time polymerase chain reaction.
RNA extraction and RT-PCR
Total RNA was prepared using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. Reverse transcription was performed using a QuantiTect® Reverse Transcription kit (Qiagen), and quantitative real-time PCR (qRT-PCR) was performed using Maxima SYBR Green/ROX qPCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA), all according to the manufacturers’ instructions. Samples were analyzed in triplicate for each gene, and the housekeeping gene of β-actin was used as a control gene. The expression level was evaluated by 2−ΔΔCt. The primers are listed in Table 1.
Western blot
Western blot analysis was performed according to the procedure previously described.26 The samples were harvested and washed with ice-cold phosphate-buffered saline followed by treatment with ice-cold radioimmunoprecipitation assay lysis buffer. Loading buffer was added, and the samples were boiled for 5 minutes. The proteins were loaded and run on 8%–12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Subsequently, the proteins were transferred to polyvinylidene difluoride membranes (Millipore Corp, Atlanta, GA, USA). The membranes were blocked with 6% nonfat dry milk at room temperature for 1 hour before being incubated with the indicated primary antibodies overnight at 4°C, followed by horseradish peroxidase-conjugated secondary antibodies at room temperature for 1 hour. The blots were developed using enhanced chemiluminescence. The tested proteins included anti-SCP3 (Abcam, ab15093, Abcam, Cambridge, UK) and anti-actin (Abcam, ab8226, Abcam).
Statistical methods
For each set of results, independent trials were repeated at least three times; the data are represented as means ± standard deviation. The differences among the means were statistically tested by Student’s t-test or one-way analysis of variance followed by Tukey’s test for multiple comparisons using Graph-Pad Prism analysis software (GraphPad Software, Inc., La Jolla, CA, USA). Comparisons were considered significant at P<0.05 and P<0.01.
Results and discussion
Characterization of AgNPs
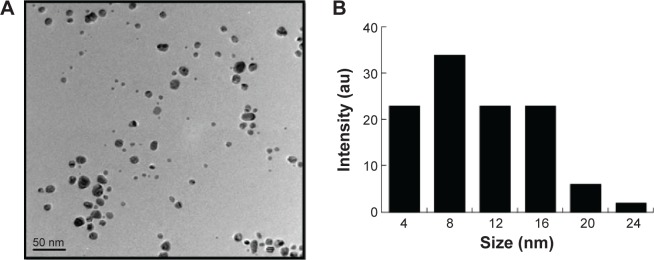
The AgNPs were synthesized using culture filtrate of B. mycoides. Synthesis of AgNPs using the biological method appears to be simple, cost-effective, and environmentally friendly. The as-prepared AgNPs were characterized using ultraviolet spectrophotometry, XRD, FT-IR, and TEM. The absorption spectra of the AgNPs were characteristic of highly symmetric single-band absorption, with a peak maxima at 426 nm (Figure 1), indicating excitation of surface plasmons typical for AgNPs. In addition, a strong broad peak was observed between 370 and 445 nm, indicating AgNP formation. Further characterization was performed to confirm the crystalline nature of the AgNPs. The XRD spectra for the AgNPs revealed three intense peaks over the entire spectrum of 2θ values, ranging from 10° to 80° (Figure 2). A comparison of our XRD spectra with the standard confirmed that the AgNPs formed in our experiments were nanocrystals, as evidenced by the peaks at 2θ values of 32.3°, 46.3°, 57.5°, and 77.0° corresponding to the (111), (200), (220), and (311) planes for silver, respectively (Figure 2). FT-IR spectra were used to identify the presence of biomolecules that could potentially contribute to the reduction of Ag+ ions or capping of the bioreduced AgNPs. As observed in Figure 3, the band at 1,389.10 cm−1 can be assigned to the C–N stretching vibrations of the aromatic and aliphatic amines. The FT-IR spectra of AgNPs synthesized from bacterial supernatant showed peaks at 3,434.78 and 1,628.34 cm−1, which are characteristic of the stretching modes for the O–H and C=O groups (Figure 3). AgNPs of different sizes exhibited similar spectra without any significant differences. The characterization of nanoparticles using TEM is a necessary step before assessing their in vitro toxicity.52 The physical and chemical properties of nanoparticles such as their particle size, size distribution, morphology, composition, surface area, surface chemistry, and reactivity in solution are important factors that are essential to assess nanoparticle toxicity.52 As seen in Figure 4A, the synthesized AgNPs were distributed uniformly and were spherical with a size of 4–24 nm, the average size was 8 nm. The mean size and standard deviation were calculated in random fields of various numbers of particles (Figure 4B).
Figure 1.
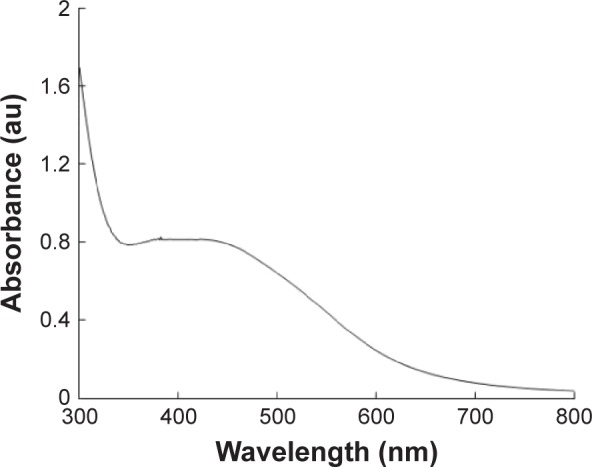
The absorption spectrum of AgNPs synthesized by Bacillus mycoides culture supernatant.
Note: The absorption spectrum of AgNPs exhibited a strong broad peak at 426 nm, which is assigned to surface plasmon resonance of the particle.
Abbreviations: AgNPs, silver nanoparticles; au, arbitrary unit.
Figure 2.
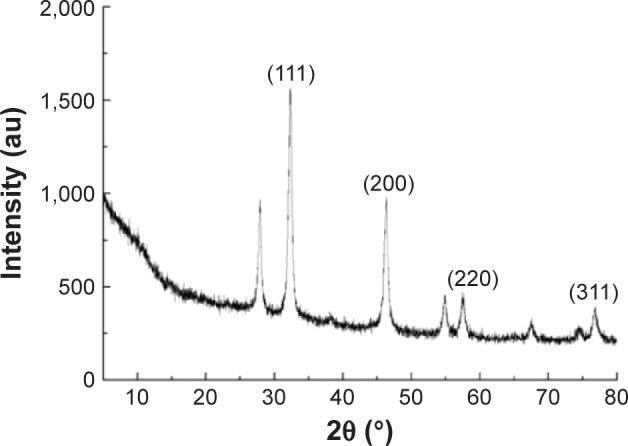
XRD spectra of AgNPs.
Note: There are three intense peaks across the spectrum of 2θ values ranging from 10° to 80°.
Abbreviations: XRD, X-ray diffraction; AgNPs, silver nanoparticles; au, arbitrary unit.
Figure 3.
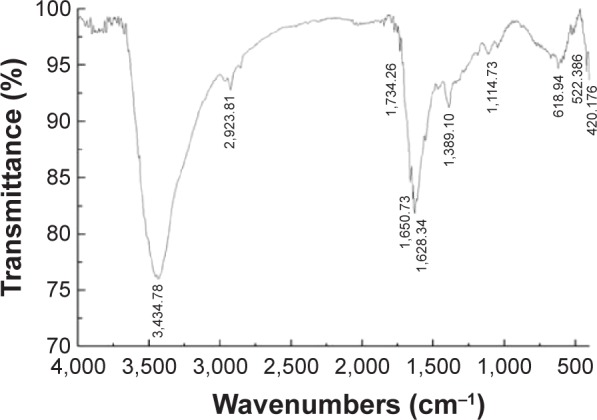
FT-IR spectra of AgNPs.
Abbreviations: FT-IR, Fourier transform infrared; AgNPs, silver nanoparticles.
Figure 4.
TEM images of AgNPs.
Notes: (A) TEM images of several fields were used to measure the AgNP particle size; micrograph (B) size distributions based on TEM images of AgNPs ranging from 4 to 24 nm.
Abbreviations: TEM, transmission electron microscopy; AgNPs, silver nanoparticles; au, arbitrary unit.
Previous studies reported the use of a biological system for the synthesis of AgNPs using supernatant from Bacillus licheniformis53 and Escherichia coli cultures,8 in which the particles had an average size of 40 and 50 nm, respectively. Shankar et al54 produced AgNPs between 16 and 40 nm in size from geranium leaf extract. Mukherjee et al55 synthesized monodispersed spherical nanoparticles (20–60 nm) along with very few bigger particles (~90 nm).55 Rodríguez-León et al56 synthesized two different populations of nanoparticles: one small in size with an average diameter around 3–5 nm and another larger in size, between 10 and 20 nm.56 Several fungi produce various sizes of AgNPs, including spent mushroom substrate, Pleurotus florida, Volvariella volvacea, and Ganoderma neo-japonicum, with an average size of 30.5, 20±5, 15, and 5 nm.57–60 Together with these previous reports, our TEM data suggest that using a biological system can produce a defined and controlled size and that biomolecules stabilize AgNPs and prevent their aggregation, which are useful in several biomedical applications.
Maternal AgNPs exposure increased the meiotic progression of female germ cells in fetal mouse ovary
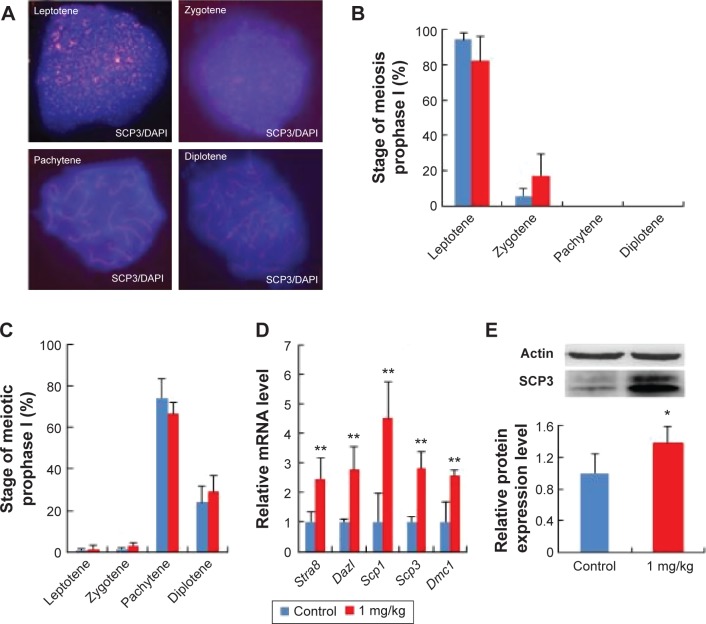
To investigate the effects of AgNPs on the meiotic progression of female germ cells within fetal ovaries after the exposure of pregnant mice to AgNPs, we prepared chromosome spreads from female fetal germ cell suspensions and immunostained them for the marker of meiotic prophase I, namely SCP3, a structural component of the axial and lateral parts of the synaptonemal complex. The results are presented in Figure 5A and B. At 17.5 dpc, the percentages of oocytes at the pachytene and diplotene stage in the treated group were 66.56% and 29.17%, respectively. In the control group, the percentages of oocytes at the pachytene and diplotene stage were 74.4% and 23.76%, respectively (Figure 5C). These results indicate that AgNP exposure promotes the progression of meiotic prophase I of oocytes.
Figure 5.
Effects of AgNPs on progress of meiosis prophase I.
Notes: (A) The characteristics of chromosomes in oocytes at meiosis I stained with SCP3 antibody (red) and Hoechst33342 (blue). (B) The percentage of fetal mouse oocytes at different phases of meiosis prophase I with or without exposure of pregnant mice to DEHP at 13.5 dpc. (C) The percentage of fetal mouse oocytes at different phases of meiosis prophase I with or without the exposure of pregnant mice to DEHP at 17.5 dpc. (D) The relative mRNA expression of meiosis-specific genes, including Stra8, Dazl, Scp1, Scp3, and Dmc1 at 13.5 dpc in the ovaries of fetal mice with or without maternal AgNPs exposure. Each sample for RNA extraction contains four ovaries, and the expression of these genes was normalized to β-actin. (E) The relative protein expression of Scp3 gene at 13.5 dpc in the ovaries of fetal mice with or without maternal AgNPs exposure. Every three ovaries were harvested at 13.5 dpc. All the experiments were repeated at least three times. The results are presented as means ± SD. *P<0.05; **P<0.01.
Abbreviations: AgNPs, silver nanoparticles; DEHP, diethylhexyl phthalate; dpc, days postcoitum; SD, standard deviation; mRNA, messenger RNA; SCP, synaptonemal complex protein.
To further confirm the effects of AgNPs on meiosis, quantitative RT-PCR and Western blotting were used to examine the expression of meiosis-specific genes at both the transcriptional and translational levels after AgNP exposure. As observed in Figure 5D–E, the relative mRNA expression levels of the meiosis-specific genes, Stra8, Dazl, Scp1, Scp3, and Dmc1, were increased significantly in the AgNP-treated groups compared with the control groups at 13.5 dpc (Figure 5D). In accordance with the expression of the transcription level, a similar protein expression pattern was observed in the SCP3 expression (Figure 5E).
BPA and diethylhexyl phthalate, two endocrine-disrupting chemicals, decreased the expression of meiosis gene Stra8 by promoting DNA methylation and inhibited meiotic progression of female germ cells in fetal mouse ovary.31,32 After AgNP treatment, the expression of meiosis genes were upregulated: meiosis progression was promoted by AgNPs (Figure 5). Further studies are required to prove whether AgNPs could promote the development of the fetus.
Effect of AgNPs on various gene expressions of fetal ovaries
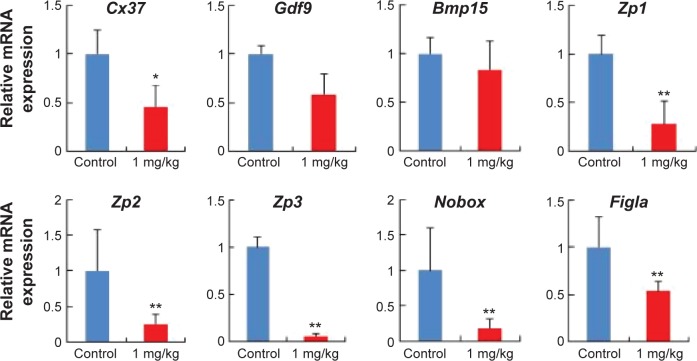
To analyze the effects of AgNPs in pregnant mice, we investigated the mRNA expression of the development-related genes of oocytes (Figure 6). It is well known that the Cx37 gene is thought to play a more critical role in ovarian function because the absence of either connexins causes a loss of oocyte–granulosa cell coupling and disruption of folliculogenesis.61,62 Zona pellucida (ZP) is an extracellular coating synthesized and secreted by the oocyte during follicular development and surrounds the plasma membrane of mammalian eggs and is involved in oogenesis.63,64 Several studies have suggested that growth differentiation factor 9 (Gdf9) and bone morphogenetic protein 15 (Bmp15) are required for early oocyte differentiation.65,66 Newborn ovary homeobox (Nobox), a homologous gene expression of a specific oocyte, is expressed in oocytes and plays an important role in oocyte survival and primordial follicle formation, which mutates, leading to the formation of syncytial follicles instead of primordial follicles.67,68 Factor in the germline alpha (Figla), an oocyte-specific basic helix-loop-helix transcription factor, plays critical dual roles in the activation of oocyte-associated genes and is required for primordial follicle formation and the expression of many genes required for folliculogenesis, fertilization, and early embryonic survival.69 As observed in Figure 6, after AgNP treatment, the relative mRNA levels of Cx37, ZP glycoprotein 1, 2, and 3 (Zp1, Zp2, and Zp3), and Figla genes were significantly reduced in the experimental group compared with those in the control group (P<0.05). In addition, Gdf9 and Bmp15 expression decreased in the AgNP-treated group.
Figure 6.
AgNP exposure altered total mRNA expression of the development-related genes of oocytes.
Notes: Relative mRNA levels of genes related to the development of oocytes at 15.5 dpc. n=15 in control group; n=14 in AgNPs-treated group. All of the experiments were repeated at least three times. The results are presented as means ± SD. *P<0.05; **P<0.01.
Abbreviations: AgNPs, silver nanoparticles; dpc, days postcoitum; mRNA, messenger RNA.
Aberrant expression of imprinted genes is associated with significant changes in total expression in placenta
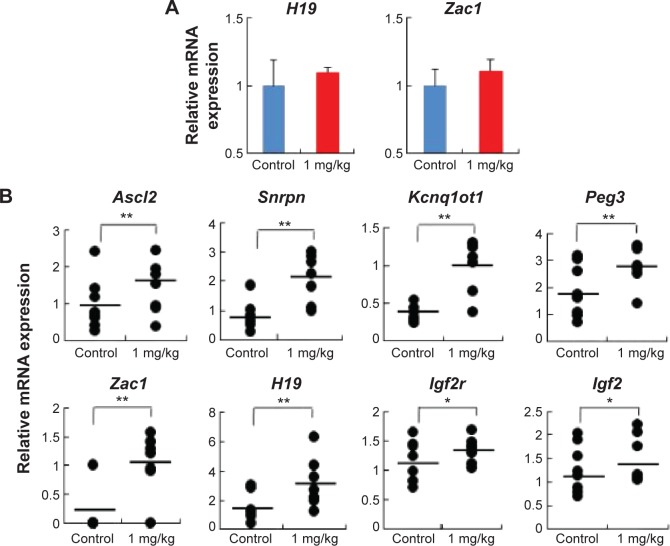
To address whether the total expression of imprinted genes was affected by AgNP exposure, we analyzed the embryonic and placental tissues from the offspring of pregnant mice treated with and without AgNPs. We investigated the expression of the imprinted genes H19 and Zac1 in embryonic tissues (Figure 7A). RT-PCR analysis of H19 and Zac1 revealed that the mean total RNA of the embryos from mice exposed to AgNPs increased compared with that of the control embryos (1.09±0.04 versus 1.00±0.27 and 1.12±0.09 versus 1.00±0.11, respectively; n=15 in control group, n=14 in AgNPs-treated group; Figure 7A). In addition, we investigated the expression of eight imprinted genes, Ascl2, Snrpn, Kcnq1ot1, Peg3, Zac1, H19, Igf2r, and Igf2, relative to the reference gene Gapdh at 15.5 dpc. qRT-PCR analysis of the imprinted genes revealed that placentas from mice exposed to AgNPs showed a significant increase of mean total RNA compared with the control placentas. In female placentas, eight genes (Ascl2, Snrpn, Kcnq1ot1, Peg3, Zac1, H19, Igf2r, and Igf2) displayed changes in their expression patterns when the mother was treated with AgNPs. The expression of eight genes increased significantly in the AgNP-treated group compared with that of the control (P<0.01, Figure 7B). A similar pattern was also observed in male placentas (Figure S1).
Figure 7.
AgNP exposure altered total mRNA expression of imprinted genes relative to reference genes in embryos and placentas of female mouse at 15.5 dpc.
Notes: (A) Relative mRNA expression of H19 and Zac1 genes in embryos (n=15 in control group; n=14 in AgNPs-treated group). (B) Relative mRNA expression of imprinted genes (Ascl2, Snrpn, Kcnq1ot1, Peg3, Zac1, H19, Igf2r, and Igf2) in placentas (n=8 in control group; n=7 in AgNP-treated group). The results are presented as means ± SD. *P<0.05; **P<0.01.
Abbreviations: AgNPs, silver nanoparticles; SD, standard deviation; dpc, days postcoitum; mRNA, messenger RNA.
Exposure of AgNPs disrupted the expression of imprinted genes in a tissue-specific manner (Figure 7A and B). Exposure of AgNPs resulted in significantly more placentas exhibiting a high expression of the paternally expressed Snrpn gene (Figure 7B). Loss of imprinting (LOI) occurred in placentas from AgNP-exposed mice compared with the control (P<0.01; Figure 7B). Analysis of the placentas exhibiting LOI in the AgNP-treated group indicated that the normally repressed maternal Snrpn allele contributed a proportion of the total expression (Figure 7B). At the Zac1 locus, AgNP exposure significantly resulted in LOI in the treated-group placentas compared with the controls (Figure 7B; P<0.05), whereas no effects were observed in the embryos (Figure 7A). However, the same expression pattern was also observed in H19 of the embryos (Figure 7A).
Imprinted genes are dosage-sensitive. They encode proteins involved in common pathways and play multiple roles in the placenta, including regulation of the embryonic growth and transport capacity, thereby controlling the supply of nutrients to the fetus.70–73 Placental expression profile of imprinted genes also impacts birth weight.74 Several lines of evidence have suggested that DNA methylation status can be altered by maternal diet.75 Maternal BPA exposure significantly disrupted imprinted gene expression in embryos and placentas, and the majority of affected genes were expressed abnormally in the placenta.40 Also, a high-fat diet during pregnancy modified the expression of imprinted genes and local and global DNA methylation patterns in the placenta.39 Sferruzzi-Perri et al38 also founded that feeding high-sugar and high-fat diets affected several growth regulatory imprinted genes (Igf2, Dlk1, Snrpn, Grb10, and H19) independently of changes in DNA methylation.
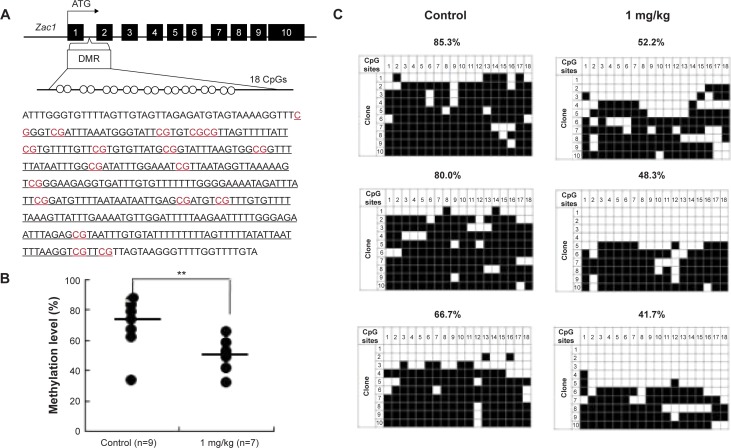
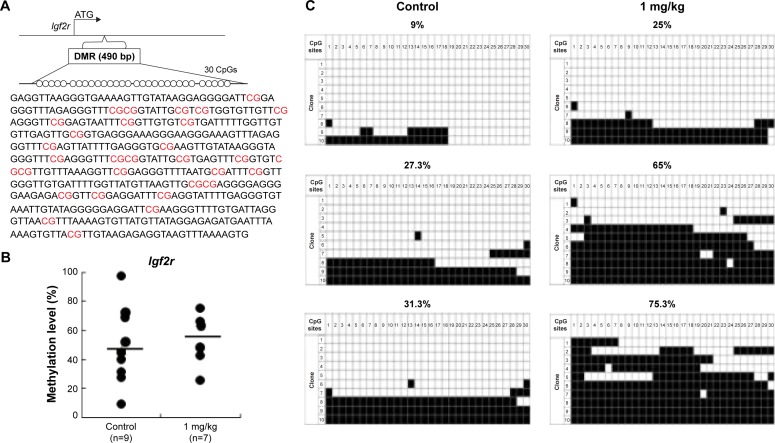
AgNP exposure alters average DNA methylation levels of the Zac1 DMR and Igf2r DMR
To determine whether the LOI observed in the tissues from the AgNP-exposed mice was linked to abnormal patterns of DNA methylation, we performed DNA methylation analysis in 15.5 placentas. The Zac1 imprint control element, which is a DMR containing 18CpG, plays a crucial role in regulating many imprinted genes in this cluster.63 We therefore investigated whether adaptation of the AgNP supply to fetal demand in pregnant mice involved the imprint control element/DMR regulating these important placental transporter systems. Pyrosequencing analysis of a subset of placentas (that of the Zac1 gene) revealed a significant reduction in the mean methylation levels in AgNP-treated placentas (73.61%; n=9) compared with controls (48.75%; n=7; P<0.01 through analysis of variance; Figure 8). However the methylation of Igf2r revealed a slight increase in the mean methylation levels in AgNP-treated placentas (52.7%; n=9) compared with the controls (45.5%; n=7; Figure 9). Three placentas from both control and AgNP exposure groups were analyzed by bisulfite sequencing, and the methylation status of ICR is shown in Figures 8C and 9C.
Figure 8.
Analysis of control and AgNP-exposed placentas indicated that exposure significantly reduced methylation of Zac1 gene.
Notes: The Zac1 DMRs are depicted in (A), including the sequences analyzed in the (B) pyrosequencing and (C) bisulfite sequencing assays. (A) In total, 18 CpG sites (highlighted in red) located in a 383 bp region of the Zac1 DMR were assayed by bisulfite sequencing. The 18 CpGs were analyzed by pyrosequencing. (B) Pyrosequencing data in control and AgNPs exposure are shown with samples exhibiting black circles of Zac1. The y-axis represents the percentage of total methylation. The black horizontal line in each exposure group indicates the average methylation. The sample sizes analyzed in each exposure group are indicated; P<0.01 when analyzed using one-way ANOVA. The results are presented as means ± SD. **P<0.01. (C) Three placentas from both the control and AgNP exposure groups were analyzed by bisulfite sequencing, and the methylation status of Zac1 is shown. The CpG sites within DMRs are shown; filled squares denote methylated and open squares denote unmethylated. The percentages of methylation at all the CpGs are indicated. The average CpG methylation levels are 72.82% in the control group and 50.07% in the AgNPs-treated group (P<0.01).
Abbreviations: AgNPs, silver nanoparticles; DMR, DNA methylated region; ANOVA, analysis of variance.
Figure 9.
Analysis of control and AgNP-exposed placentas indicated that exposure increased methylation of the Igf2r gene.
Notes: The Igf2r DMRs are depicted in (A), including the sequences analyzed in the (B) pyrosequencing and (C) bisulfite sequencing assays. (A) In total, 30 CpG sites (highlighted in red) located in a 490 bp region of the Igf2r DMR were assayed by bisulfite sequencing. The 30 CpGs were analyzed by pyrosequencing. (B) Pyrosequencing data for the control and AgNPs exposure are presented with the samples exhibiting black circles of Igf2r. The y-axis represents the percentage of total methylation. The black horizontal line in each exposure group indicates the average methylation. The sample sizes analyzed in each exposure group are indicated. (C) Three placentas from both the control and AgNP exposure groups were analyzed by bisulfite sequencing, and the methylation status of Igf2r is indicated. The CpG sites within DMRs are shown; filled squares denote methylated and open squares denote unmethylated. The percentages of methylation at all CpGs are indicated. The average CpG methylation levels are 45.5% in the control group and 52.7% in the AgNPs-treated group (P<0.01).
Abbreviations: AgNPs, silver nanoparticles; DMR, DNA methylated region.
Zac1 and Igf2r genes are two important imprinted genes. Imprinted genes involved in the control of embryonic growth. Zac1, a member of coregulated gene, alters the expression of several of these imprinted genes, including H19, Igf2, Cdkn1c, and regulates the Igf2/H19 locus through binding to a shared enhancer.70 Igf2r gene, a member insulin-like growth factor (IGF) system. Fetal growth is largely controlled by the complex IGF system.75 The expression of the Zac1 and Igf2r genes are controlled by a germline differentially methylated regions. Upregulated expression of Zac1 gene is correlated with the downregulation of methylation of Zac1 (Figure 8). But there was only a slight increase in the mean methylation of the Igf2r DMR (Figure 9). Our findings suggest that upregulation of Igf2r genes is independent of imprinting mechanisms. Some studies have shown that the altered expression was not associated with imprinting dysregulation or LOI.76 Further detailed studies are warranted to determine the effects and precise role of AgNPs on the embryonic growth.
Conclusion
This study describes the synthesis of a controlled size of AgNPs using a biological system, which is simple, convenient, cost-effective, and environmentally friendly. We prepared smaller AgNPs with an average size of 8 nm and investigated the effect of the AgNPs in pregnant mice. The results indicated that the AgNPs disturbed the first meiosis progression and expression of imprinted genes in placental tissue. In addition, the epigenetic methylation was affected, the methylation of Zac1 gene was significantly reduced in AgNP-treated placentas, and the mean methylation of Igf2r revealed a slight increase in AgNP-treated group. These changes in placental gene expression could potentially explain the accelerated fetal growth observed during the third trimester. Upregulation of maternally expressed imprinted genes induces primordial germ cells to prematurely stop proliferation and enter into meiotic prophase. This study could facilitate the production of nanoparticles with improved biocompatibility and the creation of safer nanomaterials.
Supplementary material
Samples from the control and AgNP-treated group placentas on the male mouse at 15.5 dpc were analyzed for total expression of the imprinted genes.
Notes: The relative mRNA expression of the imprinted genes (Ascl2, Snrpn, Kcnq1ot1, Peg3, Zac1, H19, Igf2r, and Igf2) in placentas (n=8 in control group; n=7 in AgNPs-treated group). The results are presented as means ± SD. *P<0.05; **P<0.01.
Abbreviations: AgNPs, silver nanoparticles; SD, standard deviation; dpc, days postcoitum; mRNA, messenger RNA.
Acknowledgments
This paper was supported by the KU – Research Professor Program of the Konkuk University. Dr Sangiliyandi Gurunathan received support from a Konkuk University KU – Full-time Professorship. This work was also supported by the Science Research Center (2015R1A5A1009701) from the National Research Foundation of Korea and the Woo Jang-Choon project (PJ007849) from the Rural Development Administration, Republic of Korea.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Shin KS, Choi JY, Park CS, Jang HJ, Kim K. Facile synthesis and catalytic application of silver-deposited magnetic nanoparticles. Catal Lett. 2009;133(1–2):1–7. [Google Scholar]
- 2.Zhou L, He X, He D, Wang K, Qin D. Biosensing technologies for Mycobacterium tuberculosis detection: status and new developments. Clin Dev Immunol. 2011;2011:193963. doi: 10.1155/2011/193963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kreuter J, Gelperina S. Use of nanoparticles for cerebral cancer. Tumori. 2008;94:271–277. doi: 10.1177/030089160809400220. [DOI] [PubMed] [Google Scholar]
- 4.Yoon KY, Hoon BJ, Park JH, Hwang J. Susceptibility constants of Escherichia coli and Bacillus subtilis to silver and copper nanoparticles. Sci Total Environ. 2007;373:572–575. doi: 10.1016/j.scitotenv.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Su J, Zhang J, Liu L, Huang Y, Mason RP. Exploring feasibility of multicolored CdTe quantum dots for in vitro and in vivo fluorescent imaging. J Nanosci Nanotechnol. 2008;8:1174–1177. [PubMed] [Google Scholar]
- 6.Kim JS, Kuk E, Yu KN, et al. Antimicrobial effects of silver nanoparticles. Nanomedicine. 2007;3:95–101. doi: 10.1016/j.nano.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Khatami M, Pourseyedi S. Phoenix dactylifera (date palm) pit aqueous extract mediated novel route for synthesis high stable silver nanoparticles with high antifungal and antibacterial activity. IET Nanobiotechnol. 2015;9(4):184–190. doi: 10.1049/iet-nbt.2014.0052. [DOI] [PubMed] [Google Scholar]
- 8.Gurunathan S, Lee KJ, Kalishwaralal K, et al. Antiangiogenic properties of silver nanoparticles. Biomaterials. 2009;30(31):6341–6350. doi: 10.1016/j.biomaterials.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 9.Sriram MI, Kanth SBM, Kalishwaralal K, Gurunathan S. Antitumor activity of silver nanoparticles in Dalton’s lymphoma ascites tumor model. Int J Nanomedicine. 2010;5(1):753–762. doi: 10.2147/IJN.S11727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarhan OM, Hussein RM. Effects of intraperitoneally injected silver nanoparticles on histological structures and blood parameters in the albino rat. Int J Nanomedicine. 2014;24(9):1505–1517. doi: 10.2147/IJN.S56729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Massarsky A, Trudeau VL, Moon TW. Predicting the environmental impact of nanosilver. Environ Toxicol Pharmacol. 2014;38(3):861–873. doi: 10.1016/j.etap.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Bhalli JA, Ding W, et al. Cytotoxicity and genotoxicity assessment of silver nanoparticles in mouse. Nanotoxicology. 2014;8(1):36–45. doi: 10.3109/17435390.2013.855827. [DOI] [PubMed] [Google Scholar]
- 13.Vrček IV, Zuntar I, Petlevski R, et al. Comparison of in vitro toxicity of silver ions and silver nanoparticles on human hepatoma cells. Environ Toxicol. 2014 Nov 28; doi: 10.1002/tox.22081. Epub. [DOI] [PubMed] [Google Scholar]
- 14.Panyala NR, Pena-Mendez EM, Havel J. Silver or silver nanoparticles: a hazardous threat to the environment and human health? J App Biomed. 2008;6:117–129. [Google Scholar]
- 15.Kim YS, Kim JS, Cho HS, et al. Twenty-eight-day oral toxicity genotoxicity, and gender-related tissue distribution of silver nanoparticles in Sprague-Dawley rats. Inhal Toxicol. 2008;20:575–583. doi: 10.1080/08958370701874663. [DOI] [PubMed] [Google Scholar]
- 16.Dziendzikowska K, Gromadzka-Ostrowska J, Lankoff A, et al. Time-dependent biodistribution and excretion of silver nanoparticles in male Wistar rats. J Appl Toxicol. 2012;32(11):920–928. doi: 10.1002/jat.2758. [DOI] [PubMed] [Google Scholar]
- 17.Hsin YH, Chen CF, Huang S, et al. The apoptotic effect of nanosilver is mediated by a ROS- and JNK-dependent mechanism involving the mitochondrial pathway in NIH3T3 cells. Toxicol Lett. 2008;179(3):130–139. doi: 10.1016/j.toxlet.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 18.Miethling-Graff R, Rumpker R, Richter M, et al. Exposure to silver nanoparticles induces size- and dose-dependent oxidative stress and cytotoxicity in human colon carcinoma cells. Toxicol In Vitro. 2014;28(7):1280–1289. doi: 10.1016/j.tiv.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Chan WH. Citrinin induces apoptosis via a mitochondria-dependent pathway and inhibition of survival signals in embryonic stem cells, and causes developmental injury in blastocysts. Biochem J. 2007;404:317–326. doi: 10.1042/BJ20061875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sur I, Altunbek M, Kahraman M, Culha M. The influence of the surface chemistry of silver nanoparticles on cell death. Nanotechnology. 2012;23(37):375102. doi: 10.1088/0957-4484/23/37/375102. [DOI] [PubMed] [Google Scholar]
- 21.Ito K, Hirao A, Arai F, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12:446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- 22.Choi YJ, Kim NH, Lim MS, et al. Geldanamycin attenuates 3 nitropropionic acid induced apoptosis and JNK activation through the expression of HSP 70 in striatal cells. Int J Mol Med. 2014;34(1):24–34. doi: 10.3892/ijmm.2014.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saffman EE, Lasko P. Germline development in vertebrates and invertebrates. Cell Mol Life Sci. 1999;55:1141–1163. doi: 10.1007/s000180050363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLaren A. Primordial germ cells in the mouse. Dev Biol. 2003;262:1–15. doi: 10.1016/s0012-1606(03)00214-8. [DOI] [PubMed] [Google Scholar]
- 25.McLaren A, Southee D. Entry of mouse embryonic germ cells into meiosis. Dev Biol. 1997;187(1):107–113. doi: 10.1006/dbio.1997.8584. [DOI] [PubMed] [Google Scholar]
- 26.Feng YM, Liang GJ, Pan B, et al. Notch pathway regulates female germ cell meiosis progression and early oogenesis events in fetal mouse. Cell Cycle. 2014;13(5):782–791. doi: 10.4161/cc.27708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koubova J, Menke DB, Zhou Q, et al. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. Proc Natl Acad Sci U S A. 2006;103:2474–2479. doi: 10.1073/pnas.0510813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson EL, Baltus AE, Roepers-Gajadien HL, et al. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc Natl Acad Sci U S A. 2008;105:14976–14980. doi: 10.1073/pnas.0807297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y, Gill ME, Koubova J, Page DC. Germ cell-intrinsic and -extrinsic factors govern meiotic initiation in mouse embryos. Science. 2008;322:1685–1687. doi: 10.1126/science.1166340. [DOI] [PubMed] [Google Scholar]
- 30.Syrjänen JL, Pellegrini L, Davies OR. A molecular model for the role of SYCP3 in meiotic chromosome organisation. Elife. 2014;3:e02963. doi: 10.7554/eLife.02963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang XF, Zhang T, Han ZH, et al. Transgenerational inheritance of ovarian development deficiency induced by maternal diethyl-hexyl phthalate exposure. [June 12, 2014];Reprod Fertil Dev. doi: 10.1071/RD14113. Epub. [DOI] [PubMed] [Google Scholar]
- 32.Zhang HQ, Zhang XF, Zhang LJ, et al. Fetal exposure to bisphenol A affects the primordial follicle formation by inhibiting the meiotic progression of oocytes. Mol Biol Rep. 2012;39(5):5651–5657. doi: 10.1007/s11033-011-1372-3. [DOI] [PubMed] [Google Scholar]
- 33.Larqué E, Pagán A, Prieto MT, et al. Placental fatty acid transfer: a key factor in fetal growth. Ann Nutr Metab. 2014;64(3–4):247–253. doi: 10.1159/000365028. [DOI] [PubMed] [Google Scholar]
- 34.Fowden AL, Sibley C, Reik W, Constancia M. Imprinted genes, placental development and fetal growth. Horm Res. 2006;65:50–58. doi: 10.1159/000091506. [DOI] [PubMed] [Google Scholar]
- 35.Peters J. The role of genomic imprinting in biology and disease: an expanding view. Nat Rev Genet. 2014;15(8):517–530. doi: 10.1038/nrg3766. [DOI] [PubMed] [Google Scholar]
- 36.Charalambous M, da Rocha ST, Ferguson-Smith AC. Genomic imprinting, growth control and the allocation of nutritional resources: consequences for postnatal life. Curr Opin Endocrinol Diabetes Obes. 2007;14:3–12. doi: 10.1097/MED.0b013e328013daa2. [DOI] [PubMed] [Google Scholar]
- 37.Loke YJ, Galati JC, Morley R, et al. Association of maternal and nutrient supply line factors with DNA methylation at the imprinted IGF2/H19 locus in multiple tissues of newborn twins. Epigenetics. 2013;8(10):1069–1079. doi: 10.4161/epi.25908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sferruzzi-Perri AN, Vaughan OR, Haro M, et al. An obesogenic diet during mouse pregnancy modifies maternal nutrient partitioning and the fetal growth trajectory. FASEB J. 2013;27(10):3928–3937. doi: 10.1096/fj.13-234823. [DOI] [PubMed] [Google Scholar]
- 39.Gallou-Kabani C, Gabory A, Tost J, et al. Sex- and diet-specific changes of imprinted gene expression and DNA methylation in mouse placenta under a high-fat diet. PLoS One. 2010;5(12):e14398. doi: 10.1371/journal.pone.0014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martha S, Isaac S, Clementina M, Marisa SB. Bisphenol a exposure disrupts genomic imprinting in the mouse. PLoS Genet. 2013;9(4):e1003401. doi: 10.1371/journal.pgen.1003401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nielsen CH, Larsen A, Nielsen AL. DNA methylation alterations in response to prenatal exposure of maternal cigarette smoking: a persistent epigenetic impact on health from maternal lifestyle? [December 6, 2014];Arch Toxicol. doi: 10.1007/s00204-014-1426-0. Epub. [DOI] [PubMed] [Google Scholar]
- 42.Hillman SL, Finer S, Smart MC, et al. Novel DNA methylation profiles associated with key gene regulation and transcription pathways in blood and placenta of growth-restricted neonates. Epigenetics. 2015;10(1):50–61. doi: 10.4161/15592294.2014.989741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Constancia M, Hemberger M, Hughes J, et al. Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature. 2002;417:945–948. doi: 10.1038/nature00819. [DOI] [PubMed] [Google Scholar]
- 44.Hughes J, Frago S, Bühnemann C, Carter EJ, Hassan AB. Maternal transmission of a humanised Igf2r allele results in an Igf2 dependent hypomorphic and non-viable growth phenotype. PLoS One. 2013;8(2):e57270. doi: 10.1371/journal.pone.0057270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang H, Yu Y, Xun P, et al. Maternal mRNA expression levels of H19 are inversely associated with risk of macrosomia. Arch Med Sci. 2014;10(3):525–530. doi: 10.5114/aoms.2014.43746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee YH, Cheng FY, Chiu HW, et al. Cytotoxicity, oxidative stress, apoptosis and the autophagic effects of silver nanoparticles in mouse embryonic fibroblasts. Biomaterials. 2014;35(16):4706–4715. doi: 10.1016/j.biomaterials.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 47.Satapathy SR, Mohapatra P, Preet R, et al. Silver-based nanoparticles induce apoptosis in human colon cancer cells mediated through p53. Nanomedicine (Lond) 2013;8(8):1307–1322. doi: 10.2217/nnm.12.176. [DOI] [PubMed] [Google Scholar]
- 48.Inkielewicz-Stepniak I, Santos-Martinez MJ, Medina C, Radomski MW. Pharmacological and toxicological effects of co-exposure of human gingival fibroblasts to silver nanoparticles and sodium fluoride. Int J Nanomedicine. 2014;9:1677–1687. doi: 10.2147/IJN.S59172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Lima R, Seabra AB, Durán N. Silver nanoparticles: a brief review of cytotoxicity and genotoxicity of chemically and biogenically synthesized nanoparticles. J Appl Toxicol. 2012;32(11):867–879. doi: 10.1002/jat.2780. [DOI] [PubMed] [Google Scholar]
- 50.Schrand AM, Dai L, Schlager JJ, Hussain SM. Toxicity testing of nanomaterials. Adv Exp Med Biol. 2012;745:58–75. doi: 10.1007/978-1-4614-3055-1_5. [DOI] [PubMed] [Google Scholar]
- 51.Gurunathan S, Kalishwaralal K, Vaidyanathan R, et al. Biosynthesis, purification and characterization of silver nanoparticles using Escherichia coli. Colloids Surf B Biointerfaces. 2009;74(1):328–335. doi: 10.1016/j.colsurfb.2009.07.048. [DOI] [PubMed] [Google Scholar]
- 52.Murdock RC, Braydich-Stolle L, Schrand AM, Schlager JJ, Hussain SM. Characterization of nanomaterial dispersion in solution prior to in vitro exposure using dynamic light scattering technique. Toxicol Sci. 2008;101(2):239–353. doi: 10.1093/toxsci/kfm240. [DOI] [PubMed] [Google Scholar]
- 53.Kalimuthu K, Suresh Babu R, Venkataraman D, Bilal M, Gurunathan S. Biosynthesis of silver nanocrystals by Bacillus licheniformis. Colloids Surf B Biointerfaces. 2008;65(1):150–153. doi: 10.1016/j.colsurfb.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 54.Shankar SS, Ahmad A, Sastry M. Geranium leaf assisted biosynthesis of silver nanoparticles. Biotechnol Prog. 2003;19(6):1627–1631. doi: 10.1021/bp034070w. [DOI] [PubMed] [Google Scholar]
- 55.Mukherjee S, Chowdhury D, Kotcherlakota R, et al. Potential theranostics application of bio-synthesized silver nanoparticles (4-in-1 system) Theranostics. 2014;4:316–335. doi: 10.7150/thno.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodríguez-León E, Iniguez-Palomares R, Navarro RE, et al. Synthesis of silver nanoparticles using reducing agents obtained from natural sources (Rumex hymenosepalus extracts) Nanoscale Res Lett. 2013;8(1):318. doi: 10.1186/1556-276X-8-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vigneshwaran N, Kathe KA, Varadarajan PV, Nachane RP, Balasubramanya RH. Silver-protein (core-shell) nanoparticle production using spent mushroom substrate. Langmuir. 2007;23:7113–7117. doi: 10.1021/la063627p. [DOI] [PubMed] [Google Scholar]
- 58.Philip D. Biosynthesis of Au, Ag and Au–Ag nanoparticles using edible mushroom extract. Spectrochim Acta A Mol Biomol Spectrosc. 2009;73:374–381. doi: 10.1016/j.saa.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 59.Bhat R, Deshpande R, Ganachari SV, Huh DS, Venkataraman A. Photo-irradiated biosynthesis of silver nanoparticles using edible mushroom Pleurotus florida and their antibacterial activity studies. Bioinorg Chem Appl. 2011;2011:650979. doi: 10.1155/2011/650979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gurunathan S, Raman J, Abd Malek SN, John PA, Vikineswary S. Green synthesis of silver nanoparticles using Ganoderma neo-japonicum Imazeki: a potential cytotoxic agent against breast cancer cells. Int J Nanomedicine. 2013;8:4399–4413. doi: 10.2147/IJN.S51881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simon AM, Goodenough DA, Li E, Paul DL. Female infertility in mice lacking connexin 37. Nature. 1997;385(6616):525–529. doi: 10.1038/385525a0. [DOI] [PubMed] [Google Scholar]
- 62.Veitch GI, Gittens JE, Shao Q, Laird DW, Kidder GM. Selective assembly of connexin37 into heterocellular gap junctions at the oocyte/granulosa cell interface. J Cell Sci. 2004;117:2699–2707. doi: 10.1242/jcs.01124. [DOI] [PubMed] [Google Scholar]
- 63.Izquierdo-Rico MJ, Gimeno L, Jiménez-Cervantes C, Ballesta J, Avilés M. Biosynthesis of hamster zona pellucida is restricted to the oocyte. Theriogenology. 2011;75(3):463–472. doi: 10.1016/j.theriogenology.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 64.Gook DA, Edgar DH, Borg J, Martic M. Detection of zona pellucida proteins during human folliculogenesis. Hum Reprod. 2008;23(2):394–402. doi: 10.1093/humrep/dem373. [DOI] [PubMed] [Google Scholar]
- 65.Yan C, Wang P, DeMayo J, et al. Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol Endocrinol. 2001;15:854–866. doi: 10.1210/mend.15.6.0662. [DOI] [PubMed] [Google Scholar]
- 66.Bristol-Gould SK, Kreeger PK, Selkirk CG, et al. Postnatal regulation of germ cells by activin: the establishment of the initial follicle pool. Dev Biol. 2006;298:132–148. doi: 10.1016/j.ydbio.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 67.Rajkovic A, Pangas SA, Ballow D, Suzumori N, Matzuk MM. NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science. 2004;305:1157–1159. doi: 10.1126/science.1099755. [DOI] [PubMed] [Google Scholar]
- 68.Lechowska A, Bilinski S, Choi Y, et al. Premature ovarian failure in nobox-deficient mice is caused by defects in somatic cell invasion and germ cell cyst breakdown. J Assist Reprod Genet. 2011;28:583–589. doi: 10.1007/s10815-011-9553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Soyal SM, Amleh A, Dean J. FIGalpha, a germ cell-specific transcription factor required for ovarian follicle formation. Development. 2000;127:4645–4654. doi: 10.1242/dev.127.21.4645. [DOI] [PubMed] [Google Scholar]
- 70.Varrault A, Gueydan C, Delalbre A, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006;11(5):711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 71.Tycko B, Morison IM. Physiological functions of imprinted genes. J Cell Physiol. 2002;192:245–258. doi: 10.1002/jcp.10129. [DOI] [PubMed] [Google Scholar]
- 72.Charalambous M, Cowley M, Geoghegan F, et al. Maternally-inherited Grb10 reduces placental size and efficiency. Dev Biol. 2010;337:1–8. doi: 10.1016/j.ydbio.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 73.Gabory A, Ripoche MA, Le Digarcher A, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136:3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- 74.Kappil MA, Green BB, Armstrong DA, et al. Placental expression profile of imprinted genes impacts birth weight. Epigenetics. 2015;10(9):842–849. doi: 10.1080/15592294.2015.1073881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kwong WY, Miller DJ, Ursell E, et al. Imprinted gene expression in the rat embryo–fetal axis is altered in response to periconceptional maternal low protein diet. Reproduction. 2006;132:265–277. doi: 10.1530/rep.1.01038. [DOI] [PubMed] [Google Scholar]
- 76.Diplas AI, Lambertini L, Lee MJ, et al. Differential expression of imprinted genes in normal and IUGR human placentas. Epigenetics. 2009;4:235–240. doi: 10.4161/epi.9019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Samples from the control and AgNP-treated group placentas on the male mouse at 15.5 dpc were analyzed for total expression of the imprinted genes.
Notes: The relative mRNA expression of the imprinted genes (Ascl2, Snrpn, Kcnq1ot1, Peg3, Zac1, H19, Igf2r, and Igf2) in placentas (n=8 in control group; n=7 in AgNPs-treated group). The results are presented as means ± SD. *P<0.05; **P<0.01.
Abbreviations: AgNPs, silver nanoparticles; SD, standard deviation; dpc, days postcoitum; mRNA, messenger RNA.