Abstract
Fibroblast growth factor 23 (FGF23) is a potent regulator of phosphate (Pi) and vitamin D homeostasis. The transcription factor, early growth response 1 (egr-1), is a biomarker for FGF23-induced activation of the ERK1/2 signaling pathway. We have shown that ERK1/2 signaling blockade suppresses renal egr-1 gene expression and prevents FGF23-induced hypophosphatemia and 1,25-dihydroxyvitamin D (1,25(OH)2D) suppression in mice. To test whether egr-1 itself mediates these renal actions of FGF23, we administered FGF23 to egr-1 -/- and wild-type (WT) mice. In WT mice, FGF23 induced hypophosphatemia and suppressed expression of the renal Na/Pi cotransporters, Npt2a and Npt2c. In FGF23-treated egr-1 -/- mice, hypophosphatemic response was greatly blunted and Na/Pi cotransporter expression was not suppressed. In contrast, FGF23 induced equivalent suppression of serum 1,25(OH)2D concentrations by suppressing renal cyp27b1 and stimulating cyp24a1 mRNA expression in both groups of mice. Thus, downstream of receptor binding and ERK1/2 signaling, we can distinguish the effector pathway that mediates FGF23-dependent inhibition of Pi transport from the pathway that mediates inhibition of 1,25(OH)2D synthesis in the kidney. Furthermore, we demonstrate that the hypophosphatemic effect of FGF23 is significantly blunted in Hyp/egr-1 -/- mice; specifically, serum Pi concentrations and renal Npt2a and Npt2c mRNA expression are significantly higher in Hyp/egr-1 -/- mice than in Hyp mice. We then characterized the egr-1 cistrome in the kidney using ChIP-sequencing and demonstrate recruitment of egr-1 to regulatory DNA elements in proximity to several genes involved in Pi transport. Thus, our data demonstrate that the effect of FGF23 on Pi homeostasis is mediated, at least in part, by activation of egr-1.
Introduction
Fibroblast growth factor 23 (FGF23) is a bone-derived circulating hormone that is critical for phosphate (Pi) and vitamin D homeostasis. Excess circulating FGF-23 is responsible for the pathogenesis of hypophosphatemic syndromes; specifically, X-linked hypophosphatemia (XLH), autosomal recessive and dominant hypophosphatemic rickets, and tumor induced osteomalacia [1–3]. FGF-23 excess induces renal Pi wasting with resultant hypophosphatemia, 1,25 dihydroxyvitamin D (1,25(OH)2D) deficiency, rickets and osteomalacia. The kidney is the primary physiologic target for FGF23 action. FGF23 decreases serum Pi concentrations by directly inhibiting renal Pi reabsorption via the sodium-dependent Pi cotransporters, Npt2a and Npt2c in the renal proximal tubule [4–8]. FGF23 suppresses renal 1,25(OH)2D synthesis by directly suppressing cyp27b1 and stimulating cyp24a1 gene expression [6,7]; cyp27b1 and cyp24a1 encode for the enzymes responsible for the synthesis and degradation of 1,25(OH)2D, respectively. In a mouse model of XLH (Hyp mouse), we showed that serum FGF23 concentrations are 17-fold higher than in normal mice [7] and mitogen activated protein kinase (MAPK)/extracellular signal regulated kinase 1/2 (ERK1/2) signaling is constitutively activated in the kidney. The transcription factor, early growth response 1 (egr-1), is downstream of ERK1/2 and is a biomarker for activation of ERK1/2 signaling by FGF23 [4,9]. Egr-1 mRNA expression is two-fold higher in Hyp mice than in normal mice [10,11]. Blockade of ERK1/2 signaling in the kidney in Hyp mice is associated with reduction in egr-1 gene expression and improvements in the hypophosphatemia, 1,25(OH)2D deficiency and skeletal mineralization defects [10,11]. Thus, our studies demonstrated that activation of the ERK1/2 signaling pathway by FGF23 is critical to the pathogenesis of hypophosphatemia and 1,25(OH)2D deficiency in Hyp mice.
The role of egr-1 in the kidney is not well studied, and whether it directly mediates FGF23-dependent regulation of renal Pi and vitamin D metabolism is unknown. To test whether egr-1 itself mediates the renal actions of FGF23, we determined the effects of FGF23 on renal Pi transport and 1,25(OH)2D synthesis in egr-1 null (egr-1 -/-) and Hyp/egr-1 -/- mice. To obtain a comprehensive view of the genes regulated by egr-1 in the kidney, we employed chromatin-immunoprecipitation (ChIP) coupled with genome-wide sequencing (ChIP-seq) analysis to characterize the FGF23-dependent egr-1 cistrome in the kidney. We integrated the ChIP-seq data with gene expression profiling data to identify direct targets that are both bound by egr-1 and regulated by the FGF23/egr-1 signaling axis.
Methods
Animals
We purchased egr-1 -/- mice and their wild-type littermates, 50–76 days of age, from Jackson Laboratory (Bar Harbor, ME). Phenotypic features of mice with global egr-1 gene deletion have been described [12]. All mice were fed a constant diet containing 0.6% phosphorus and 1% calcium (Teklad diet 98243, Harlan Laboratories, Madison, WI) starting 4 days before the experiment. Mice were injected intraperitoneally (IP) with vehicle or human recombinant FGF-23 (Genzyme, MA) (150ng/g body weight Q12 x 3 doses) and sacrificed 1 hour after the 3rd dose (n = 8–10 mice per group). Female Hyp mice purchased from Jackson Laboratories were crossed with egr-1 +/- heterozygous males to generate Hyp/egr-1 -/- mice that were studied at 50–80 days of age. For ChIP-seq and microarray experiments, 6 week old C57/Bl6J wild-type mice were administered a single IP dose of FGF23 (150ng/g body weight) or vehicle and sacrificed after 1 and 2 hrs. Blood was obtained for determination of serum biochemistries. The kidneys were removed and frozen for subsequent preparation of RNA and protein samples. All procedures used in this study were approved by the Committee on Animal Research, University of California San Francisco.
Serum Biochemistry
Serum phosphorus and calcium concentrations were determined using kits from Stanbio Laboratories (San Antonio, TX). Serum 1,25(OH)2D concentrations were determined using an enzyme immunoassay (EIA) kit from Immunodiagnostic Systems, Inc. (Scottsdale, AZ). Serum intact parathyroid hormone (PTH) concentrations were determined using EIA kits from Immutopics International (San Clemente, CA).
Real-Time PCR
Total RNA was isolated from kidney using TRIzol reagent (Invitrogen). Probes and primer sets were custom designed as previously described [11]. The mRNA abundance of the gene of interest, expressed relative to that of gus mRNA, was quantitated by real-time PCR using the ABI 7900 HT Sequence Detection System (Applied Biosystems) [6].
Western Blot Analysis
Mouse kidney total protein (25μg), and renal brush border membrane vesicles (BBMV)(25μg) were isolated [13,14] and fractionated on 8% SDS-polyacrylamide gel as previously described [11]. For detection of Npt2a and Npt2c proteins, membranes were probed with a rabbit anti-Npt2a polyclonal antibody (1:300) (Alpha Diagnostic International, Inc., San Antonio, TX) and rabbit anti-Npt2c polyclonal antibody (1:1000) (Sigma-Aldrich, St. Louis, MO). Equal protein loading was determined using a rabbit anti-β-actin polyclonal antibody (1:5000) (Cell Signaling Technology, Danvers, MA). The membranes were subsequently blotted using an infrared (IR) labeled secondary antibody (Li-Cor Biosciences, Lincoln, NE). The bound complex was detected using Odyssey Infrared Imaging System (LiCor Biotechnology).
Chromatin-immunoprecipitation and genome wide sequencing
Chromatin was prepared from mouse kidney as per standard protocol (2 kidneys from each mouse were used to prepare one chromatin sample). 30μg chromatin and 3.4 μg of rabbit monoclonal anti-Egr-1 antibody (Cell Signaling Technology, Danvers, MA) were used for the ChIP experiments. Vehicle-treated mouse kidneys were used as a biological negative control. Prior to sequencing, we validated the egr-1 ChIP samples by ChIP-QPCR using Nab1 primers (positive control for egr-1 binding) and normalized to ChIP samples with control IgG and untreated chromatin (negative controls). Induction of Egr-1 binding to Nab1 was 10-fold higher in FGF23-treated samples compared to vehicle-treated samples (data not shown). DNA libraries were subsequently prepared and amplified after ChIP and submitted for sequencing on the Illumina Hi-Seq platform (Active Motif, CA). Genomic regions with local enrichments in signal intensity (“peaks”) were identified using the MACS algorithm method [15] that looks for significant enrichments in the ChIP data file when compared to the Input data file (i.e. random background). MACS is suitable to identify the binding sites of transcription factors with known binding motifs. Peaks are reported as intervals that have a start and end coordinate. The length of each interval varies from 500 bp to 1500 bp. Intervals present in Input or IgG control samples (i.e. false peaks) were used to identify false positives in egr-1 ChIP samples. To compare peak metrics between 2 samples, we grouped overlapping intervals into “Active Regions”, which are defined by the start and end coordinate of the most upstream and downstream interval, respectively (= union of overlapping intervals). In locations where only one sample has an interval, this interval defines the active region. After defining the active regions, their exact locations along with their proximities to gene annotations are reviewed using the data visualization tools on the UCSC (http://genome.ucsc.edu) and IGB genome browsers (http://bioviz.org/igb/download.html). False discovery rates were calculated and were noted to be low at less than 0.1%. Approximately 9 million sequence tags were uniquely mapped to the genome per sample. After normalization, we obtained ~ 9,000 and 7,000 unique active regions for FGF23-treated, 1 and 2 hr samples, respectively. To refine the in vivo consensus egr-1 motif, we queried MEME (a web-based tool to identify transcription factor binding motifs) [16,17] with the top 1,200 enriched regions corresponding to the most statistically significant binding sites identified by ChIP-seq. Additional transcription factor motifs that were identified in the vicinity of egr-1 binding are shown in S1 Table. We validated the results of the ChIP-seq dataset using the Nab1 gene as a positive control. This active region (Nab 1-AR) was further shown to contain an enhancer element that is responsive to FGF23 in HEK293 cells transfected with the Nab 1-AR sequence upstream of the luciferase reporter gene (S1 Fig).
Microarray and Pathway Analysis
Microarray analysis was performed on kidneys from 6 week-old mice treated with FGF23 or vehicle for 1 hr. Total RNAs were isolated using TRIreagent (Molecular Research Center, Cincinnati, OH, USA). The expression of 45,000 genes was tested on the kidney samples using the Illumina.SingleColor. Mouse WG-6_V2_0_R1_11278593_A chip (Illumina, San Diego, CA, USA) at the DNA Discovery Core of University of Tennessee Health Science Center. Data were analyzed using GeneSpring GX software (Agilent Technologies, Santa Clara, CA, USA). The Robust Multichip Averaging probe summarization algorithm was used to perform background correction, normalization, and probe summarization. Microarray data were normalized per chip and per gene to the median. Genes were filtered to include only those that were expressed in at least one of the samples. The statistical analysis was performed using Mann-Whitney unpaired test with a p value cutoff of p<0.05 followed by Benjamini-Hochberg multiple test correction to minimize the false positive discovery rate (FDR). The resulting data were compared with our ChIP-seq data and previously published data reflecting the renal transcriptome in Col4a3-/- [18] and FGF23 transgenic mice [19] with chronic FGF23 excess. Pathway analysis was performed using the Ingenuity program (Ingenuity Systems, Redwood City, CA, USA) to match the identified genes of interest to already known broader networks of genes contained in the literature database.
Cell Culture and Transfection
Human embryonic kidney (HEK-293) cells (a kind gift from Makoto Kuro-o, University of Texas Southwestern, Dallas, TX) were stably transfected with the transmembrane (Tm) form of mouse klotho-pEF1 expression vector and maintained as previously described [5]. Klotho is an obligatory co-factor for FGF-23 and confers tissue specificity for it actions in target tissues [4]. It is well established that FGF-23-dependent signal activation in HEK-293 cells is dependent on Tm klotho [4,5]. HEK-293 cells were plated at 130,000/well in 24-well plates, in DMEM H-21 with 10% FBS (Hyclone, Waltham, MA). At 80% confluence, HEK293 cells were transfected with a thymidine kinase (TK) promoter-driven luciferase plasmid containing Nab 1-active region (AR) (930bp in length) and treated with FGF23 (100ng/ml) or vehicle for 24hrs. Luciferase activity was normalized to the empty vector and expressed as relative luciferase units (RLU).
Statistical Analysis
Data are expressed as means ± SEM. The significance of differences between vehicle and treatment groups was analyzed by ANOVA or t-test when appropriate, using Sigma Stat statistical software (Jandel Scientific, San Rafael, CA).
Results
Effects of egr-1 gene deletion on Pi homeostasis in mice
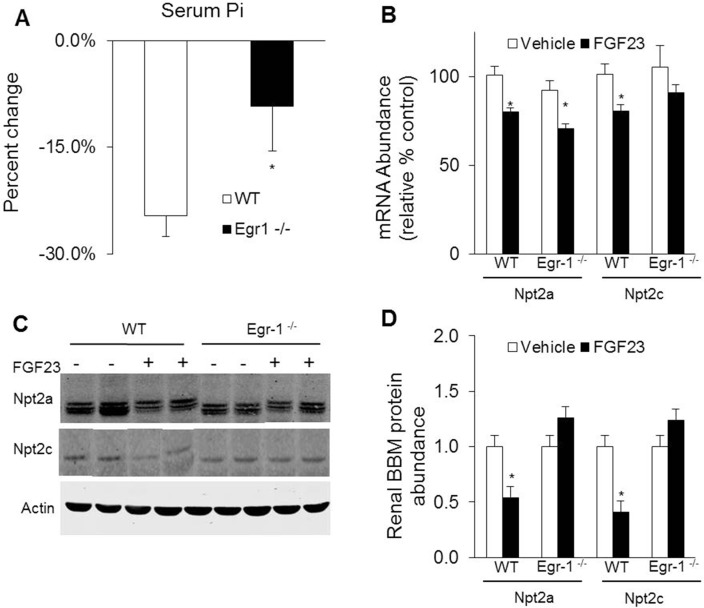
To determine whether egr-1 mediates the phosphaturic effect of FGF23, we administered FGF23 intraperitoneally to egr-1 -/- and WT mice. At baseline, the mean serum Pi concentrations were not significantly different in egr-1 -/- mice when compared to WT mice. After treatment with FGF23, serum Pi concentration decreased by 27% in WT mice (8.3 ± 0.4 vs 6.1 ± 0.3 mg/dl, P<0.05) and by 12% in egr-1 -/- mice (7.4 ± 0.5 vs 6.5 ± 0.3 mg/dl, P = 0.06). Thus, the hypophosphatemic response to FGF23 was blunted by 56% in egr-1 -/- mice (Fig 1A). We then determined the renal mRNA and protein abundance of the NaPi cotransporters, Npt2a and Npt2c. At baseline, renal abundance of Npt2a and Npt2c mRNA and protein were not significantly different between the two groups. After treatment with FGF23, Npt2a mRNA expression decreased significantly in both groups of mice by approximately 22% (P<0.05). However, Npt2a protein abundance in renal brush border membrane (BBM) preparations decreased only in WT mice and was unchanged in egr-1 -/- mice. The renal abundance of both Npt2c mRNA and protein decreased significantly only in FGF23-treated WT mice but were unchanged in FGF23-treated egr-1 -/- mice (Fig 1B–1D).
Fig 1. Effects of egr-1 gene deletion on phosphate homeostasis.
Egr-1 -/- and wild-type (WT) mice were treated with vehicle or FGF23. A. Serum phosphorus concentration. Bars depict percent change when compared to vehicle treated group (n = 8–10 mice/group). B. Renal Npt2a and Npt2c mRNA abundance were quantitated by real-time PCR, normalized to that of gus mRNA, and expressed as a percent relative to vehicle-treated mice. Bars depict mean ± SEM (n = 8–10 mice/group) * P<0.05, compared to egr-1 -/- and WT mice treated with vehicle. C and D. Renal Npt2a and Npt2c protein abundance in renal brush border membrane vesicle preparations normalized to β-actin.
Effects of egr-1 gene deletion on vitamin D homeostasis in mice
To determine the effect of egr-1 gene deletion on vitamin D homeostasis, we measured serum 1,25(OH)2D concentrations and the renal abundance of cyp27b1 and cyp24a1 mRNA (Fig 2A–2C). At baseline, the mean serum 1,25(OH)2D concentration in egr-1 -/- mice was not significantly different from that in WT mice (Fig 3A). FGF23 treatment induced significant suppression of serum 1,25(OH)2D concentrations in both groups of mice (Fig 2A). At baseline, abundance of cyp27b1 and cyp24a1 mRNA in egr-1 -/- mice was not significantly different from those in WT mice. With FGF23 treatment, cyp27b1 expression was greatly suppressed and cyp24a1 mRNA was greatly increased in both groups of mice (Fig 2B and 2C). The mean serum calcium and PTH concentrations did not differ between egr-1 -/- and WT mice in the vehicle-treated group or after FGF23 treatment (data not shown). Thus, FGF23 can regulate vitamin D metabolism normally even in the absence of egr-1 expression.
Fig 2. Effects of egr-1 gene deletion on renal 1,25(OH)2D metabolism.
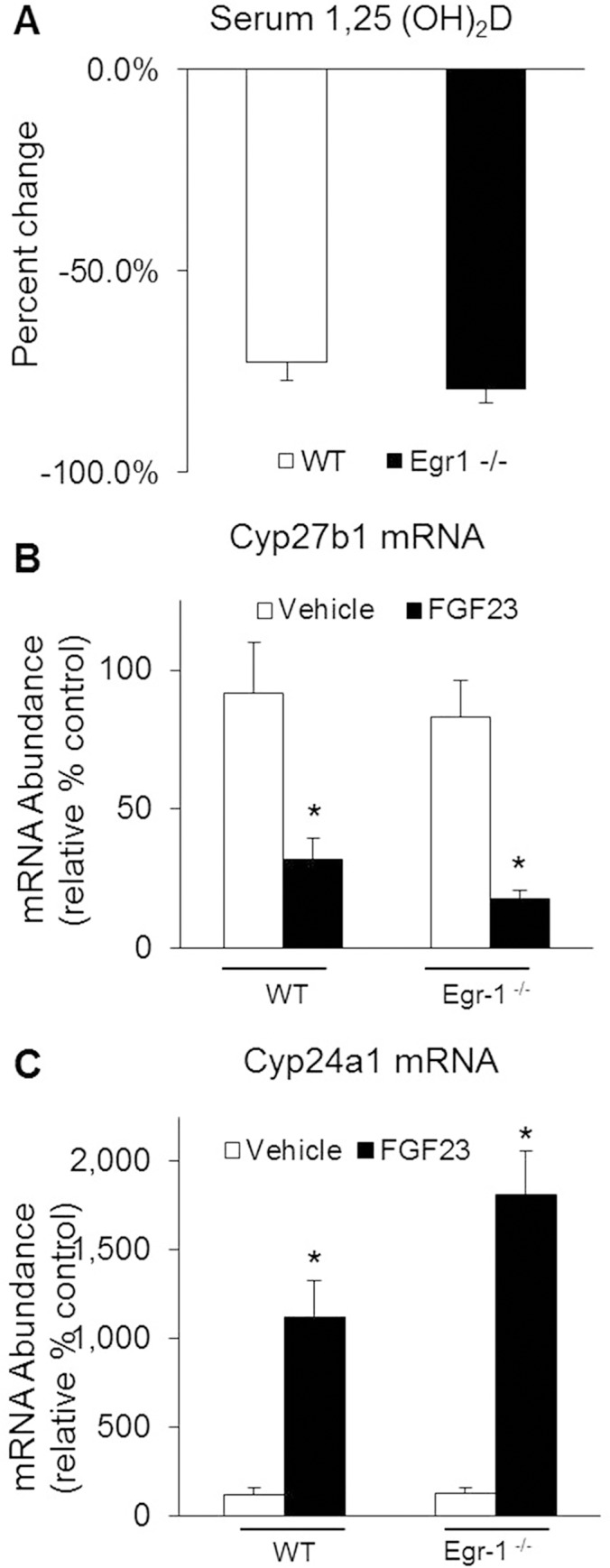
Egr-1 -/- and wild-type (WT) mice were treated with vehicle or FGF23. A. Serum 1,25(OH)2D concentrations. B. Renal cyp27b1 and, C. Renal cyp24a1 mRNA abundance were quantitated by real-time PCR, normalized to that of gus mRNA, and expressed as a percent relative to vehicle-treated WT mice. Bars depict mean ± SEM (n = 8–10 mice/group) * P<0.05, compared to egr-1 -/- and WT mice treated with vehicle.
Fig 3. Effects of egr-1 gene deletion on phosphate homeostasis in Hyp mice.
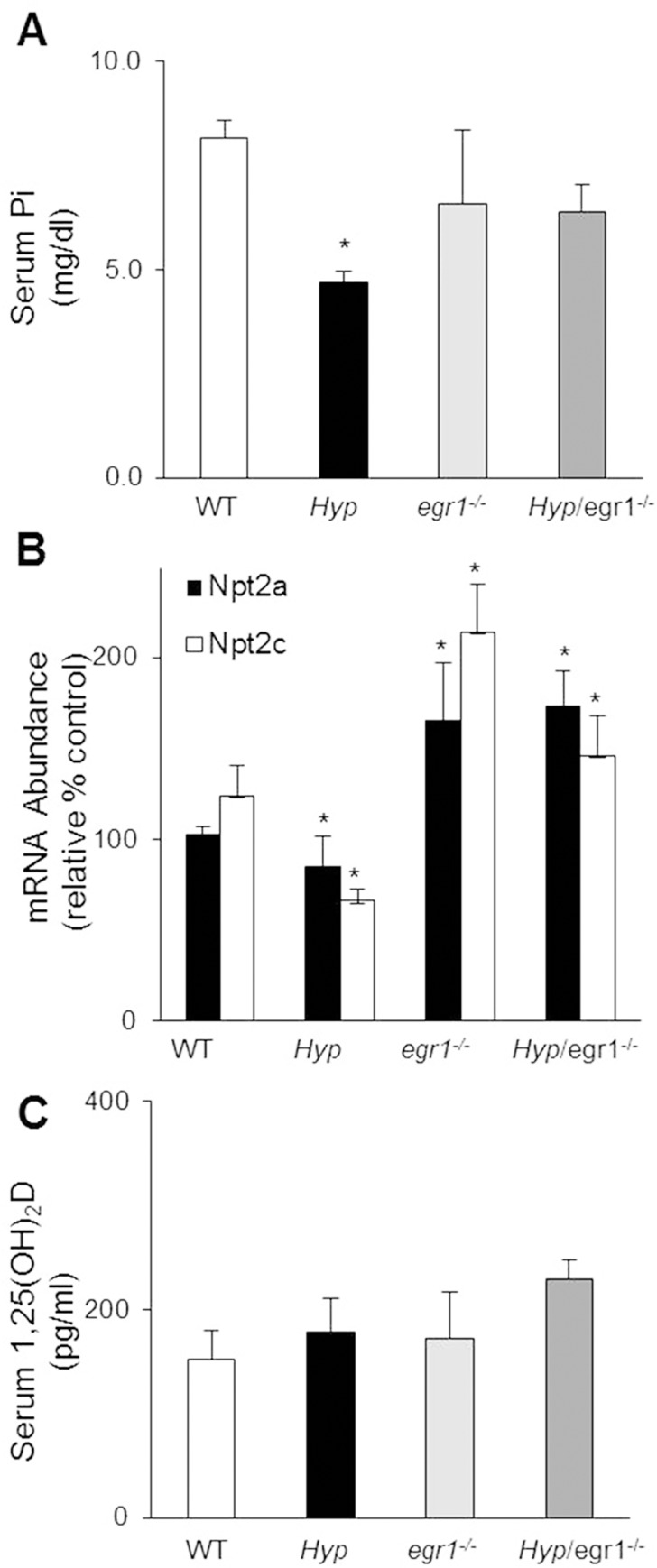
WT, Hyp, egr-1 -/- and Hyp/egr-1 -/- mice were sacrificed at 7–10 weeks of age to determine the following: A. Serum phosphorus concentration. B. Renal Npt2a and Npt2c mRNA abundance were quantitated by real-time PCR, normalized to that of gus mRNA, and expressed as a percent relative to WT mice. C. Serum 1,25 (OH)2D concentration. Bars depict mean ± SEM (n = 5 mice/group) * P<0.05, compared to WT mice.
Effects of egr-1 gene deletion on Pi homeostasis in Hyp mice
Hyp mice exhibit phosphaturia and hypophosphatemia due to excess circulating FGF23 and this hypophosphatemic effect is completely rescued when the fgf23 gene is deleted in Hyp/fgf23 -/- mice [20,21]. To determine whether egr-1 mediates the hypophosphatemia induced by FGF23 in Hyp mice, we generated the double mutant Hyp/egr-1 -/- mice. In Hyp/egr-1 -/- mice, serum Pi concentrations were not significantly different from egr-1 -/- or WT mice (Fig 3A). As expected, serum Pi concentrations were 42% lower in Hyp mice than in WT mice, consistent with previous reports. Renal mRNA abundance of Npt2a and Npt2c were lower in Hyp mice when compared to WT mice and this effect was completely reversed by ablation of the egr-1 gene in Hyp/egr-1 -/- mice (Fig 3B). The mean serum 1,25(OH)2D, calcium and PTH concentrations in Hyp/egr-1 -/- mice were not significantly different from those in egr-1 -/- or WT mice (Fig 3C and S2 Table).
Characterization of the egr-1 cistrome in mice kidney
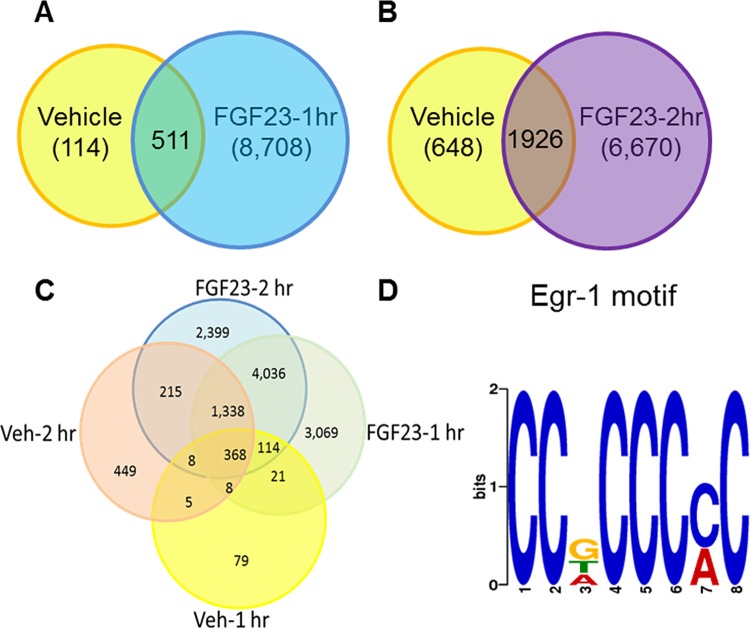
To further explore the molecular mechanisms responsible for the blunted hypophosphatemic response to FGF23 treatment in mice lacking the egr-1 gene, we sought to obtain a comprehensive view of the genes regulated by FGF23 downstream of egr-1. To define the egr-1 cistrome (i.e genome-wide location of transcription factor (egr-1) binding-sites) in the kidney, we performed ChIP-seq analysis in WT mice treated with FGF23 or vehicle. We obtained unique DNA sequences or “active regions” across the mouse genome (Fig 4A–4C). We constructed an Euler diagram to display the active regions identified in the 4 groups of mice. 43% of the unique active regions were identified in both treatment groups, 32% and 25% of the active regions were unique to the 1hr and 2hr treatment groups, respectively (Fig 4C). The top consensus motif in our dataset identified using MEME software tool was indeed the egr-1 consensus sequence 5'-CCG CCC CC-3' (Fig 4D), thereby validating the ChIP-seq experiment. This motif is identical to the most overrepresented sequence identified in a recent egr-1 ChIP-seq study of mouse brain [22], and is a close match to the canonical egr-1 DNA response element previously characterized [23].
Fig 4. Schematic representation of the active regions identified by ChIP-seq analysis in normal mice kidney after treatment with FGF23 or vehicle.
A-C. Venn and Euler diagrams showing overlap of egr-1 binding sites between different treatment groups. D. Motif discovery using MEME. Consensus motif for egr-1, as determined by querying the 1,200 most enriched ChIP-seq loci.
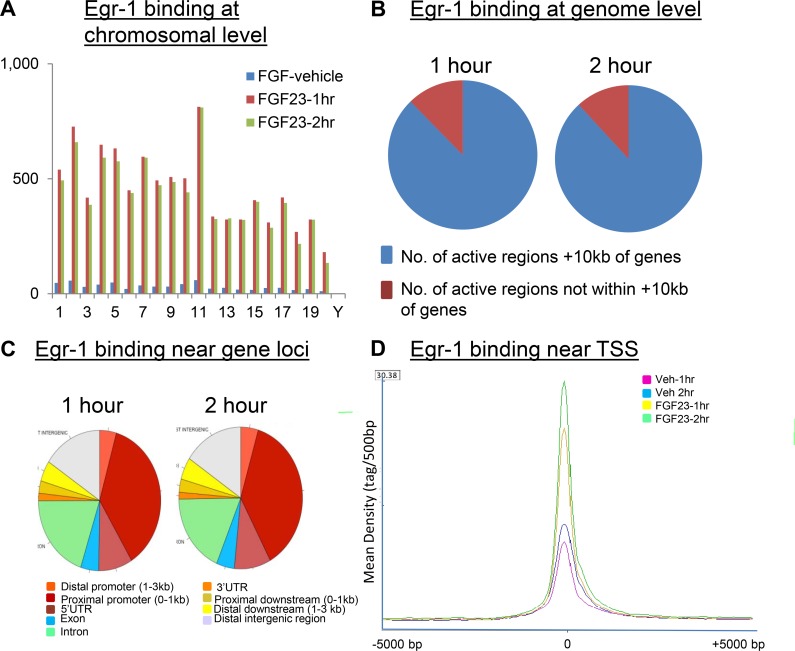
We next examined the distribution of egr-1 binding sites across the mouse genome in FGF23-treated mice. The locations and amplitudes of peaks were strikingly similar between 1 hr and 2 hr samples in FGF23-treated mice; similarity in egr-1 binding was seen at the chromosomal level (Fig 5A) and genome wide level (Fig 5B and 5C). The majority (88%) of egr-1-bound regions mapped within 5 kb of transcription start sites (TSS) (Fig 5D), which is consistent with reports showing that transcription factors prefer to bind in the vicinity of genes [24]. The distribution of the egr-1 binding sites near individual gene loci was further analyzed and is depicted in Fig 5C. We discovered a striking enrichment for egr-1-binding sites (41 and 42% for the FGF23-treated 1 hr and 2 hr samples, respectively) in the proximal promoter (0–1kb) region of individual gene loci. A significant number of binding sites were also observed in the introns and distant intergenic region (Fig 5C).
Fig 5. Analysis of the egr-1 cistrome.
Genome-wide distribution of egr-1 binding sites in normal mice kidney after treatment with FGF23 or vehicle. A. Visualization of egr-1-binding sites across all mouse chromosomes. B. Spatial distribution of egr-1 binding sites at the genome level and, C. near individual genes. D. The ChIP-seq and input data files were analyzed by seqMINER, which analyzes the signal profiles across mouse promoters (TSS), and clusters the profiles according to binding patterns. The result shows that the 3 clusters with the strongest signals (vehicle, FGF23 -1hr and FGF23-2hr) contained 1328, 3288, and 4811 genes (total = 9427), respectively. Vehicle and FGF23-treated samples did not show a difference in signal profile but differed markedly in signal intensity.
Characterization of egr-1 binding to active regions near genes responsible for renal Pi transport
To further analyze the role of egr-1 in FGF23-mediated inhibition of renal Pi transport, we queried the egr-1 ChIP seq database for genes encoding proteins reported to participate in intracellular Pi transport in the kidney (i.e. cotransporters, scaffold and trafficking proteins). In mice treated with FGF23, we found a 2- to 16- fold increase in egr-1 binding near genes encoding for Na/Pi cotransporters (Npt2a and Npt2c), scaffold proteins (NHERF1-3, ezrin and GABARAP), and trafficking proteins (megalin and vacuolar ATPase). Several of the active regions we identified were found in the core promoter region which strongly suggests that these genes are transcriptionally regulated by FGF23 (Table 1). Although egr-1 gene ablation did not alter the suppressive effect of FGF23 on Npt2a mRNA in egr-1-/- mice, we identified four active regions bound by egr-1 near the Npt2a gene. Of note, PDZK2, Pex-19, Shank2E and cortactin were not bound by egr-1.
Table 1. List of genes present in the CHIP-seq datasets and reported in the literature to be involved in renal Pi transport.
| Gene Name | Known Function | Location of AR (Distance from TSS) | Length of AR (bp) | Average Fold Change* |
|---|---|---|---|---|
| Slc34a1 (Npt2a) | Co transporter | -7746,159, 9,260,15434 | 777, 691, 1092,752 | 2 |
| Slc34a3 (Npt2c) | Co transporter | 5576, 9505,10401 | 2268, 881, 521 | 3 |
| Slc9a3r1 (NHERF-1) | Scaffolding protein | 53,1641, 15829, 18715,24077 | 910,1577, 884,848, 481 | 3 |
| Slc9a3r1 (NHERF-2) | Scaffolding protein | 259,17633 | 1223, 499 | 16 |
| PDZK1 (NHERF-3) | Scaffolding protein | -24 | 482 | 2 |
| MAP17 (PDZK1P1) | Scaffolding protein | 2209 | 617 | 2 |
| Ezrin (Vil2) | Scaffolding protein | -295, 40534, 51001, 54448 | 808, 791, 1377,713 | 8 |
| GABARAP | Scaffolding protein | -66,5101,9248 | 1172, 2573, 729 | 3 |
| Megalin (LRP2) | Protein trafficking | -65 | 735 | 6 |
| Vacuolar ATPase (ATP6V1A) | Protein trafficking | -426 | 1620 | 2 |
| Pex-19 | Scaffolding protein | - | - | Not detected |
| PDZK2 (NHERF-4) | Scaffolding protein | - | - | Not detected |
| Shank2E | Scaffolding protein | - | - | Not detected |
| Cortactin | Scaffolding protein | - | - | Not detected |
*Fold change represents egr-1 binding events in FGF23 treated 1-hr sample when compared to vehicle-treated sample. AR-Active Region, TSS-Transcription start site
Identification of direct Egr-1 target genes
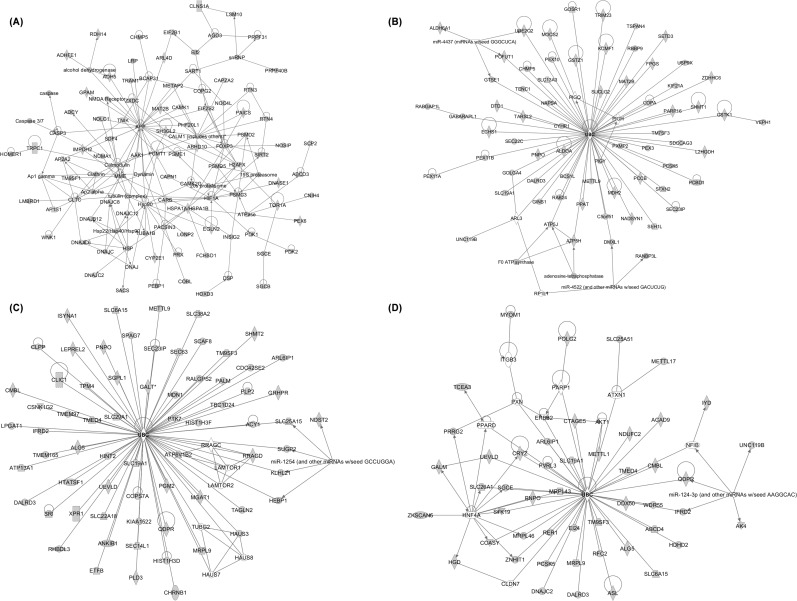
We next determined “direct” egr-1 target genes; i.e., genes that are both bound by egr-1 and actively transcribed in the kidney after FGF23 treatment. To this end, we performed RNA microarray analyses on kidney tissue from mice treated with FGF23 for 1hr and intersected the microarray and the ChIP-seq data for the same time point. The list of genes with the highest fold change in the microarray analysis is provided in S3 Table. Based on the expression patterns from the microarray analysis and using a cutoff value of p<0.05, we found 1,425 genes common to both datasets; of these, 120 (8%) were up-regulated and 1,305 (92%) were down-regulated by egr-1 after FGF23 injection. The list of genes with the highest fold change in the intersection analysis is shown in S4 Table. We then performed an Ingenuity Pathway Analysis (IPA) on 1,425 genes. IPA categorized the genes into 5 top molecular and cell functions and 3 top regulatory networks of equal importance (S5 Table). “Cellular assembly and organization” was identified in both networks and function categories, and connections between genes from this network were mapped in Fig 6A. In this category, several members of the calmodulin family were among the most interconnected genes in the network and therefore represented valid candidates for FGF23-regulated gene targets via egr-1. Consistent with this finding, previous studies have shown that the calmodulin-dependent protein kinase pathway is involved in Pi transport [25,26]. Our findings suggest that FGF23 activates the calmodulin pathway to inhibit renal Pi transport.
Fig 6. Ingenuity Pathway analysis (IPA)-Top regulatory networks representing genes involved in cellular assembly and organization.
A. Network of common genes found in CHIP-seq and microarray datasets. Ubiquitin C (UBC) shown as the central regulator. B. Network of common genes found in CHIP-seq, microarray and Col4a3-/- mice datasets, C. Network of common genes found in CHIP-seq and FGF23Tg datasets and, D. Network of common genes found in microarray and FGF23Tg datasets. The networks are built according to the identified interconnected pathways involving the majority of genes displaying direct interactions. Genes represented in gray belong to the dataset. Genes represented in white are intermediary regulators that do not belong to the cluster.
To further elaborate the genes regulated by FGF23/egr-1 signaling axis in conditions of both acute and chronic elevations of serum FGF23, we did the following bioinformatics analyses; we intersected the set of genes common to Chip-Seq and microarray datasets with available microarray expression profiles reported previously from kidney samples of mice with FGF23 excess (Alport’s chronic kidney mouse model (Col4a3-/-), and FGF23 transgenic (FGF23Tg) mice) [18,19]. Intersection with the Col4a3-/- dataset identified 222 genes in common with the Chip-Seq & Microarray dataset. Down-stream IPA analyses of the 222 genes, revealed a single top regulatory network out of 4 significant networks computed. This network was formed by genes involved in “cellular assembly and organization” (S6 Table), with ubiquitin C as a central network regulator. This rare occurrence, of a single central network organizing gene, reflects that protein ubiquitinylation by ubiquitin C is the only known common mediator between all the regulated genes (Fig 6B). Individual intersection analysis identified 1,406 and 501 genes in common between Col4a3-/- and Chip-Seq, and Col4a3-/- and microarray datasets, respectively. Separate IPAs were performed on each group of genes and “cellular assembly and organization” was identified again as the top regulatory network. A similar approach was used to intersect the common genes from the Chip-seq and microarray datasets with the FGF23Tg database. Intersection of the FGF23Tg dataset identified 468 and 131genes in common with the Chip-Seq and microarray datasets, respectively. Ubiquitin C was consistently found as a single factor connecting the gene network (Fig 6C and 6D). Thus, cellular assembly and organization network, and ubiquitin C were consistently identified in the intersection of four genomic datasets associated with FGF23 excess.
FGF23/Egr-1 signaling down-regulates vesicle trafficking proteins in the kidney
In the mouse brain, a significant number of egr-1 target genes are strongly associated with vesicle transport [22]. Since vesicle trafficking is critical for movement of ion transporters to and from the proximal tubule brush border membrane, we next sought to determine whether vesicle trafficking proteins in the kidney are regulated by FGF23/egr-1 signaling. We queried the human vesicle trafficking protein database [27] (a mouse database is currently not available). Using a 2-fold cut off for increased egr-1 binding, we identified 102 genes from our ChIP-seq dataset that overlap with the vesicle trafficking database. We also performed a Gene Set Enrichment Analysis (GSEA) with a cutoff of p<0.05 and a false discovery rate (FDR) q-value of p<0.1, and found that 47 vesicle trafficking genes were bound by egr-1 following FGF23 treatment (data not shown). We then intersected the egr-1 ChIP-seq data with the microarray data for the same time point (1hr) to obtain a list of vesicle trafficking genes that are both bound by egr-1 and whose gene expression is regulated by FGF23. Using GSEA with the same established parameters, we found that 14 vesicle genes were down-regulated in both databases (Table 2) including two lectins (ERGIC-1 and -3), SNX9, a membrane remodeling protein which interacts with actin-regulating proteins [28], the lysosomal trafficking protein VPS41 and synaptophysin.
Table 2. List of vesicle trafficking genes that are present in both CHIP-seq and microarray datasets.
| Gene Name | Symbol | Fold Change* |
|---|---|---|
| DownRegulated Genes | ||
| B-cell receptor-associated protein 31 | BCAP31 | -1.7 |
| sorting nexin 9 | SNX9 | -1.7 |
| golgi SNAP receptor complex member 1 | GOSR1 | -1.6 |
| transmembrane emp24 protein transport domain containing 4 | TMED4 | -1.6 |
| endoplasmic reticulum-golgi intermediate compartment 1 | ERGIC1 | -1.5 |
| SH3-domain GRB2-like 2 | SH3GL2 | -1.5 |
| syntaxin 18 | STX18 | -1.4 |
| vacuolar protein sorting 41 | VPS41 | -1.4 |
| adaptor protein complex AP-2, alpha 2 subunit | AP2A2 | -1.3 |
| adaptor-related protein complex AP-4, epsilon 1 | AP4E1 | -1.3 |
| endoplasmic reticulum-golgi intermediate compartment 3 | ERGIC3 | -1.3 |
| synaptophysin | SYP | -1.3 |
| S. cerevisiae vesicle trafficking protein-like C 22 | SEC22C | -1.2 |
| AP2 associated kinase 1 | AAK1 | -1.1 |
*Fold change represents change in gene expression in microarray datasets in FGF23 treated 1-hr sample when compared to vehicle-treated sample. Fold change in ChIP-seq datasets not shown.
DNase I: A novel mechanism for FGF23 to regulate renal Pi reabsorption
By intersecting all 4 databases that reflect FGF23 excess (Chip-seq, microarray, Col4a3-/- and FGF23Tg) using IPA, we identified 18 down-regulated genes (Table 3), among which DNase I was one of the top candidates. Interestingly, DNase I was previously found to be one of the top down-regulated genes in the kidney of Hyp, FGF23Tg and Col4a3-/- mice [18,19,29]. Here, we demonstrate that egr-1 binds to an active region ~ -7700 bp from the transcription start site of DNase I gene (Fig 7A). To study the functional role of DNase I, we determined whether DNase I gene expression is regulated by FGF23 and by dietary Pi intake. Normal mice were either injected with FGF23 or fed a high (1.65%), or low (0.02%) Pi diet for 5 days. DNase I mRNA expression was significantly suppressed by FGF23 injection (47%) and by high Pi diet (20%), whereas a low Pi diet induced a 200% increase in DNase I mRNA expression (Fig 7B and 7C). Furthermore, FGF23 failed to suppress renal DNase I mRNA expression in egr-1-/- mice (Fig 7B). These studies demonstrate that DNase I is indeed regulated by FGF23 and dietary Pi intake, and may play a physiologic role in Pi homeostasis.
Table 3. List of genes down regulated in Chip-seq, microarray, Col4a3-/- and FGF23Tg datasets.
| Gene Name | Symbol | *Fold Change |
|---|---|---|
| DownRegulated Genes | ||
| Adenylate kinase isoenzyme 4 | AK3L1 | -2.0 |
| Deoxyribonuclease I | DNASE1 | -1.6 |
| Estrogen-related receptor alpha | ESRRA | -1.6 |
| Zinc finger with KRAB and SCAN domains 5 | ZKSCAN5 | -1.6 |
| Prostasin | PRSS8 | -1.5 |
| Proprotein convertase subtilisin/kexin type 5 | PCSK5 | -1.5 |
| Autocrine motility factor receptor, E3 ubiquitin protein ligase | AMFR | -1.5 |
| Nucleosome assembly protein 1-like 1 | NAP1L1 | -1.5 |
| Peroxisomal biogenesis factor 11 alpha | PEX11A | -1.5 |
| Solute carrier family 19 (folate transporter), member 1 | SLC19A1 | -1.5 |
| Homogentisate 1,2 dioxygenase | HGD | -1.4 |
| Glutathione S-transferase zeta 1 | GSTZ1 | -1.4 |
| Lon peptidase 1 | LONP1 | -1.4 |
| DALR anticodon binding domain containing 3 | DALRD3 | -1.4 |
| Unc-119 Homolog B | UNC119B | -1.2 |
| Pyridoxamine 5'-phosphate oxidase | PNPO | -1.1 |
| Androgen receptor | AR | -1.1 |
| 5-azacytidine-induced protein 2 | AZI2 | -1.1 |
*Fold change represents change in gene expression in microarray datasets in FGF23 treated 1-hr sample when compared to vehicle-treated sample.
Fig 7. Renal DNase I gene expression.
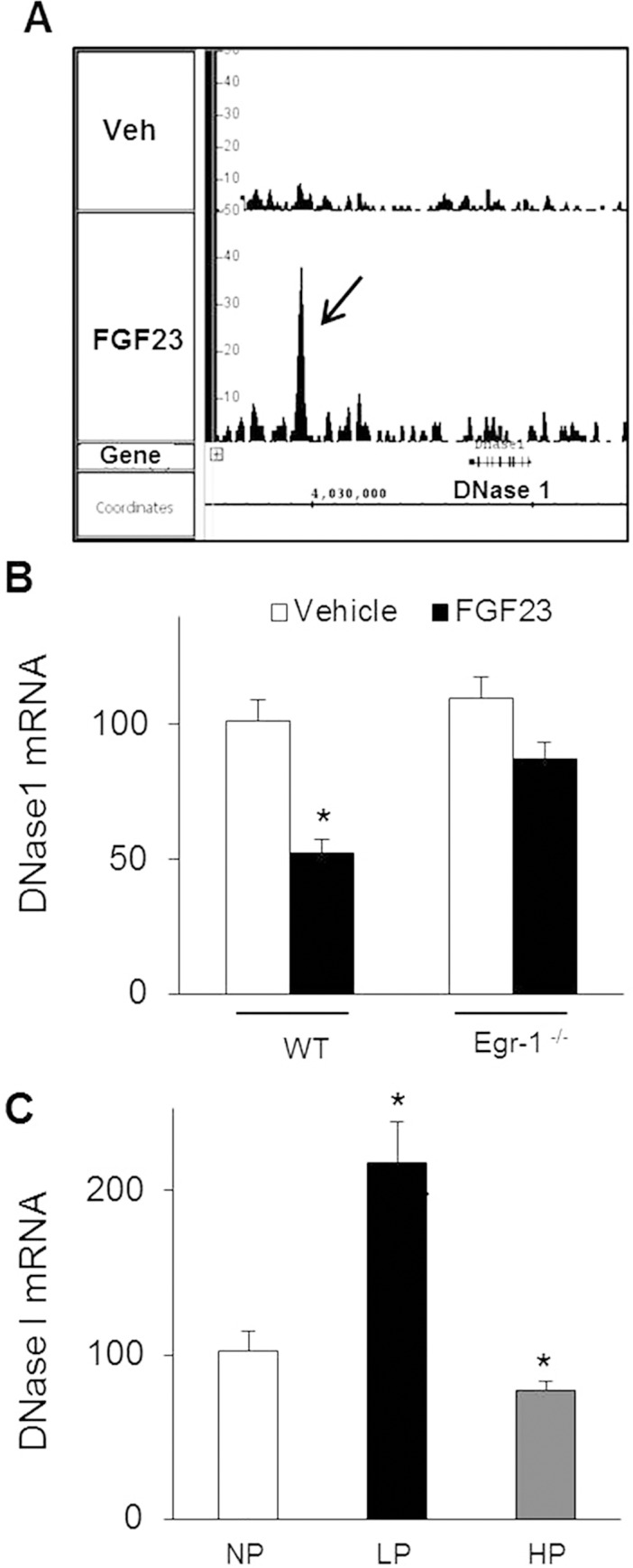
A. Egr-1 ChIP-seq analysis showed recruitment of egr-1 to DNase I. Peak signal intensity for egr-1 binding up-stream (-7700 bp) of DNase I gene in FGF23-treated mice. A low signal is noted in the vehicle-treated sample. B. Renal DNase I mRNA abundance in WT and egr-1 -/- mice treated with vehicle or FGF23. C. Renal DNase I mRNA abundance in normal mice fed a normal (NP), low (LP) or high (HP) Pi diet for 5 days. Bars depict mean ± SEM (n = 5 mice/group). * P < 0.05 compared to control group.
Discussion
In the present study, we provide evidence that egr-1 mediates FGF23-dependent inhibition of renal Pi transport but plays no apparent role in FGF23-mediated inhibition of 1,25(OH)2D synthesis. Further, we characterize the FGF23-dependent egr-1 cistrome in the kidney. To our knowledge ours is the first study to identify a transcription factor, egr-1, that mediates the hypophosphatemic effect of FGF23 and to differentiate the effector pathways that regulates inhibition of renal Pi reabsorption from that of 1,25(OH)2D synthesis. Using genome-wide ChIP-sequencing, we have identified components in the Pi transport network that are regulated by the FGF23/egr-1 signaling axis, including putative novel targets with protein trafficking function.
Previously, we showed that systemic inhibition of MEK/ERK1/2 signaling in Hyp mice improved hypophosphatemia by 70% [10,11]. In the present study, we demonstrate that deletion of the egr-1 gene, which is downstream of MEK/ERK1/2 signaling, blunts the hypophosphatemic response to acute administration of FGF23 by approximately 56% in egr-1 -/- mice. In Hyp/egr-1 -/- mice which have chronically high circulating FGF23 concentrations, deletion of the egr-1 gene improves hypophosphatemia by 50% when compared to Hyp mice. We also observed that suppression of Na/Pi cotransporter abundance was blocked in egr-1 -/- mice treated with FGF23 and in Hyp/egr-1 -/- mice. Furthermore, using bioinformatics we demonstrate that egr-1 transcriptionally regulates the gene expression of scaffold and trafficking proteins that mediate the insertion and retrieval of Npt2a from the BBM as discussed further below.
It is a currently held view that Pi reabsorption is regulated by changing the number of Na/Pi cotransporters that reside in the BBM rather than by changing their intrinsic transport activity [30]. BBM abundance of Npt2a and Npt2c is dependent on their gene transcription, and their insertion and retrieval from the BBM, which in turn is dependent on their interaction with scaffolding and trafficking proteins [31–34]. However, little is known about FGF23-induced inhibition of renal Pi reabsorption at the cellular level. We and others have shown that FGF23 inhibits BBM Pi uptake via suppression of Npt2a and Npt2c mRNA and protein abundance [6,8]. The specific role for NHERF-1 in FGF23-induced inhibition of Pi transport was also recently demonstrated [35,36]. To investigate the interaction of the cotransporters with intracellular scaffolding and trafficking proteins, we utilized a novel bioinformatics approach to expand our limited knowledge of the Pi transport network in the context of FGF23 action. We identified several novel putative gene targets for FGF23, including those that encode a subset of scaffold and vesicle trafficking proteins in the kidney (Tables 1 and 2). Several of the active regions identified were within the proximal promoter region (0–1kb) including binding sites for ezrin, GABARAP, megalin, vacuolar ATPase and NHERF-3. Interestingly, egr-1 bound to genes encoding scaffolding proteins, NHERF1, 2 and 3 of which NHERF-2 showed the strongest binding at 16-fold when compared to controls. Of note, several of the genes encoding vesicle trafficking proteins were significantly down regulated by FGF23. Given the rapid transcriptional regulation of these vesicle trafficking genes by FGF23 in an egr-1-dependent manner, it is possible that the direct repression of the vesicle genes may be responsible for the reduced insertion and/or endocytic removal of Npt2a and Npt2c from the apical BBM and the consequent phosphaturic effect of FGF23. Further study is needed to precisely map the intracellular Pi transport network that is regulated by FGF23/egr-1 signaling. Interestingly, a recent ChIP-seq study described the role of egr-1 in mouse brain tissue. A significant number of egr-1 target genes in the brain were strongly associated with protein targeting and localization, endocytosis, and vesicle transport [22]. Thus, the similarities in the egr-1 ChIP-seq datasets in mouse kidney and brain suggest that the cellular functions of egr-1 are highly conserved.
We have demonstrated that blockade of MEK/ERK1/2 signaling corrects the aberrant renal 1,25(OH)2D synthesis in Hyp mice [10,11]. In the present study however, suppression of renal 1,25(OH)2D synthesis was intact in egr-1-/- mice treated with FGF23. These findings provide evidence that downstream of FGF receptor binding and ERK1/2 signaling, the pathways diverge such that egr-1 is required for FGF23-dependent inhibition of Pi transport but not for inhibition of 1,25(OH)2D synthesis. Identification of the different signaling pathways for these two actions of FGF23 may have important clinical significance in patients with chronic kidney disease (CKD). In pre-dialysis CKD, excess circulating FGF23 induces phosphaturia, an action thought to be helpful in maintaining Pi homeostasis as the glomerular filtration rate declines [37–40]. However, excess FGF23 induces suppression of renal 1,25(OH)2D synthesis and reduction in circulating 1,25(OH)2D [39,41], actions thought to be critical to the development of secondary hyperparathyroidism [37,38,42,43]. Thus, distinguishing the effector pathways by which FGF23 regulates Pi reabsorption from those of 1,25(OH)2D synthesis maybe important to formulate future therapy for patients with CKD.
Our study has the following limitations. We utilized whole kidney samples and thus, interpretation of the data can be confounded by potential cell type-specific differences in egr-1-binding patterns. To overcome this limitation, we analyzed the expression of known genes within the Na/Pi cotransport network whose expression is largely restricted to the proximal tubule. Secondly, we focused our investigations on the kidney but whether egr-1 regulates intestinal Pi transport is unknown. However, FGF23 does not directly regulate intestinal Pi transport; indirect effects are observed due to its suppressive effect on vitamin D metabolism [44–46]. Thus, it is unlikely that egr-1 plays a direct role in the regulation of intestinal Pi transport.
Prior to the present study, few direct gene targets of FGF23 signaling were known. By integrating ChIP-seq and gene expression data, we identified a set of genes regulated by FGF23/egr-1 signaling. Through intersections of multiple genomic datasets we have identified novel genes and regulatory networks for FGF23; specifically, ubiquitin C and DNase I. The occurrence of a single central network regulator (i.e ubiquitin C) emerging from the intersection of 4 large genomic data sets is very rare, and suggests that ubiquitinylation by ubiquitin C is the common mediator between those genes regulated by FGF23. Previous studies have reported that ubiquitinylation of Npt2a in the kidney and Npt2b in the intestine determine their protein abundance in the apical BBM [47,48]. Further studies are needed to elucidate the specific role of ubiquitin C in renal Pi transport. We identified DNase I as another novel candidate gene for regulation of renal Pi transport. In conditions where serum FGF23 concentrations are increased, DNase I gene expression is significantly down regulated as seen in Hyp mice and in WT mice fed a high dietary Pi intake or treated with exogenous FGF23. A low Pi diet elicits the opposite effect; serum FGF23 concentrations are lower [7] and DNase I gene expression is 2-fold higher when compared to a normal Pi diet. These findings demonstrate that DNase I is regulated by both FGF23 and dietary Pi intake in normal mice. DNase I bind to the monomeric (G) form of actin and prevent filamentous (F) actin formation. Actin cytoskeleton is known to participate in the endocytic retrieval of Na/Pi cotransporters [49,50] and it is possible that DNase I modulates the stability of the actin cytoskeleton. Further studies are needed to elucidate the specific role of DNase I in renal Pi transport.
In summary, our data demonstrate that the effect of FGF23 on Pi homeostasis in mice is mediated, at least in part, by activation of transcription factor, egr-1. Thus we have distinguished the signaling pathway that mediates FGF23-dependent inhibition of Pi reabsorption from that which mediates inhibition of 1,25(OH)2D synthesis. Using genome wide ChIP-sequencing, we have characterized the egr-1 cistrome in the mouse kidney and identified several gene targets downstream of FGF23/egr-1 signaling. This study provides a large genomic database suitable for further exploration and understanding of the molecular basis for disorders of Pi homeostasis associated with FGF23 excess.
Supporting Information
A. Signal histogram of the peaks visualized for the positive control gene, Nab1. Integrated Genome Browser (IGB) shots showing egr-1 binding sites near the Nab1 gene. Binding of egr-1 to the regulatory region up-stream (-1500 bp) of Nab1gene. A low signal is noted in the vehicle-treated sample. B. Peak signal intensity for Nab1 gene in FGF23-treated (1 and 2hr) mice, represented as fold change over vehicle-treated sample. C. HEK 293 cells transfected with Nab 1 active region (930bp)—Luc plasmid and treated with FGF23 (100ng/ml) or vehicle for 24hrs. Luciferase activity was normalized to the empty vector and expressed as relative luciferase units (RLU). Data expressed as Mean ± SEM, n = 3 replicates, *P<0.05.
(JPG)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by University of California San Francisco Clinical and Translational Science Institute-Strategic Opportunity Support grant (to FP), and the UCSF Pediatric Nephrology Innovative Research Fund (to AAP).
References
- 1. Jonsson KB, Zahradnik R, Larsson T, White KE, Sugimoto T, Imanishi Y, et al. Fibroblast growth factor 23 in oncogenic osteomalacia and X-linked hypophosphatemia. N Engl J Med 2003. April 24;348(17):1656–63. [DOI] [PubMed] [Google Scholar]
- 2. White KE, Jonsson KB, Carn G, Hampson G, Spector TD, Mannstadt M, et al. The autosomal dominant hypophosphatemic rickets (ADHR) gene is a secreted polypeptide overexpressed by tumors that cause phosphate wasting. J Clin Endocrinol Metab 2001. February;86(2):497–500. [DOI] [PubMed] [Google Scholar]
- 3. Feng JQ, Ward LM, Liu S, Lu Y, Xie Y, Yuan B, et al. Loss of DMP1 causes rickets and osteomalacia and identifies a role for osteocytes in mineral metabolism. Nat Genet 2006. November;38(11):1310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Urakawa I, Yamazaki Y, Shimada T, Iijima K, Hasegawa H, Okawa K, et al. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature 2006. December 7;444(7120):770–4. [DOI] [PubMed] [Google Scholar]
- 5. Kurosu H, Ogawa Y, Miyoshi M, Yamamoto M, Nandi A, Rosenblatt KP, et al. Regulation of fibroblast growth factor-23 signaling by klotho. J Biol Chem 2006. March 10;281(10):6120–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Perwad F, Zhang MY, Tenenhouse HS, Portale AA. Fibroblast growth factor 23 impairs phosphorus and vitamin D metabolism in vivo and suppresses 25-hydroxyvitamin D-1alpha-hydroxylase expression in vitro. Am J Physiol Renal Physiol 2007. November;293(5):F1577–F1583. [DOI] [PubMed] [Google Scholar]
- 7. Perwad F, Azam N, Zhang MY, Yamashita T, Tenenhouse HS, Portale AA. Dietary and serum phosphorus regulate fibroblast growth factor 23 expression and 1,25-dihydroxyvitamin D metabolism in mice. Endocrinology 2005. December;146(12):5358–64. [DOI] [PubMed] [Google Scholar]
- 8. Shimada T, Hasegawa H, Yamazaki Y, Muto T, Hino R, Takeuchi Y, et al. FGF-23 is a potent regulator of vitamin D metabolism and phosphate homeostasis. J Bone Miner Res 2004. March;19(3):429–35. [DOI] [PubMed] [Google Scholar]
- 9. Farrow EG, Davis SI, Summers LJ, White KE. Initial FGF23-mediated signaling occurs in the distal convoluted tubule. J Am Soc Nephrol 2009. May;20(5):955–60. 10.1681/ASN.2008070783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang MY, Ranch D, Pereira RC, Armbrecht HJ, Portale AA, Perwad F. Chronic Inhibition of ERK1/2 Signaling Improves Disordered Bone and Mineral Metabolism in Hypophosphatemic (Hyp) Mice. Endocrinology 2012. April 1;153(4):1806–16. 10.1210/en.2011-1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ranch D, Zhang MYH, Portale AA, Perwad F. Fibroblast Growth Factor-23 Regulates Renal 1,25(OH)2D and Phosphate Metabolism via the MAP Kinase Signaling Pathway in Hyp Mice. J Bone Miner Res 2011;26(8):1883–90. 10.1002/jbmr.401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee SL, Tourtellotte LC, Wesselschmidt RL, Milbrandt J. Growth and differentiation proceeds normally in cells deficient in the immediate early gene NGFI-A. J Biol Chem 1995. April 28;270(17):9971–7. [DOI] [PubMed] [Google Scholar]
- 13. Zhang MYH, Wang X, Wang JT, Compagnone NA, Mellon SH, Tenenhouse HS, et al. Dietary phosphorus transcriptionally regulates 25-hydroxyvitamin D-1a-hydroxylase gene expression in the proximal renal tubule. Endocrinology 2002;143:587–95. [DOI] [PubMed] [Google Scholar]
- 14. Biber J, Stieger B, Stange G, Murer H. Isolation of renal proximal tubular brush-border membranes. Nat Protoc 2007;2(6):1356–9. [DOI] [PubMed] [Google Scholar]
- 15. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008;9(9):R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 2011. June 15;27(12):1696–7. 10.1093/bioinformatics/btr189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 2009. July;37(Web Server issue):W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dai B, David V, Martin A, Huang J, Li H, Jiao Y, et al. A comparative transcriptome analysis identifying FGF23 regulated genes in the kidney of a mouse CKD model. PLoS One 2012;7(9):e44161 10.1371/journal.pone.0044161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marsell R, Krajisnik T, Goransson H, Ohlsson C, Ljunggren O, Larsson TE, et al. Gene expression analysis of kidneys from transgenic mice expressing fibroblast growth factor-23. Nephrol Dial Transplant 2008. March;23(3):827–33. [DOI] [PubMed] [Google Scholar]
- 20. Sitara D, Razzaque MS, Hesse M, Yoganathan S, Taguchi T, Erben RG, et al. Homozygous ablation of fibroblast growth factor-23 results in hyperphosphatemia and impaired skeletogenesis, and reverses hypophosphatemia in Phex-deficient mice. Matrix Biol 2004. November;23(7):421–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu S, Zhou J, Tang W, Jiang X, Rowe DW, Quarles LD. Pathogenic role of Fgf23 in Hyp mice. Am J Physiol Endocrinol Metab 2006. July;291(1):E38–E49. [DOI] [PubMed] [Google Scholar]
- 22. Koldamova R, Schug J, Lefterova M, Cronican AA, Fitz NF, Davenport FA, et al. Genome-wide approaches reveal EGR1-controlled regulatory networks associated with neurodegeneration. Neurobiol Dis 2014. March;63:107–14. 10.1016/j.nbd.2013.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Christy B, Nathans D. DNA binding site of the growth factor-inducible protein Zif268. Proc Natl Acad Sci U S A 1989. November;86(22):8737–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. O'Geen H, Lin YH, Xu X, Echipare L, Komashko VM, He D, et al. Genome-wide binding of the orphan nuclear receptor TR4 suggests its general role in fundamental biological processes. BMC Genomics 2010;11:689 10.1186/1471-2164-11-689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Friedlaender MM, Darmon D, Wald H, Popovtzer MM. The in vivo and in vitro effect of calmodulin antagonists on the renal actions of 25(OH) vitamin D3 in the rat. Pflugers Arch 1989. December;415(3):372–80. [DOI] [PubMed] [Google Scholar]
- 26. Lu M, Barber LE, Renfro JL. Renal transepithelial phosphate secretion: luminal membrane voltage and Ca2+ dependence. Am J Physiol 1994. October;267(4 Pt 2):F624–F631. [DOI] [PubMed] [Google Scholar]
- 27. Pietrosemoli N, Pancsa R, Tompa P. Structural disorder provides increased adaptability for vesicle trafficking pathways. PLoS Comput Biol 2013;9(7):e1003144 10.1371/journal.pcbi.1003144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shin N, Ahn N, Chang-Ileto B, Park J, Takei K, Ahn SG, et al. SNX9 regulates tubular invagination of the plasma membrane through interaction with actin cytoskeleton and dynamin 2. J Cell Sci 2008. April 15;121(Pt 8):1252–63. 10.1242/jcs.016709 [DOI] [PubMed] [Google Scholar]
- 29. Meyer MH, Dulde E, Meyer RA Jr. The genomic response of the mouse kidney to low-phosphate diet is altered in X-linked hypophosphatemia. Physiol Genomics 2004. June 17;18(1):4–11. [DOI] [PubMed] [Google Scholar]
- 30. Biber J, Hernando N, Forster I. Phosphate transporters and their function. Annu Rev Physiol 2013. February 10;75:535–50. 10.1146/annurev-physiol-030212-183748 [DOI] [PubMed] [Google Scholar]
- 31. Weinman EJ, Steplock D, Shenolikar S, Blanpied TA. Dynamics of PTH-induced disassembly of Npt2a/NHERF-1 complexes in living OK cells. Am J Physiol Renal Physiol 2011. January;300(1):F231–F235. 10.1152/ajprenal.00532.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weinman EJ, Steplock D, Zhang Y, Biswas R, Bloch RJ, Shenolikar S. Cooperativity between the phosphorylation of Thr95 and Ser77 of NHERF-1 in the hormonal regulation of renal phosphate transport. J Biol Chem 2010. August 13;285(33):25134–8. 10.1074/jbc.M110.132423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cunningham R, Biswas R, Steplock D, Shenolikar S, Weinman E. Role of NHERF and scaffolding proteins in proximal tubule transport. Urol Res 2010. August;38(4):257–62. 10.1007/s00240-010-0294-1 [DOI] [PubMed] [Google Scholar]
- 34. Weinman EJ, Biswas RS, Peng G, Shen L, Turner CL, E X, et al. Parathyroid hormone inhibits renal phosphate transport by phosphorylation of serine 77 of sodium-hydrogen exchanger regulatory factor-1. J Clin Invest 2007. November;117(11):3412–20. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35. Weinman EJ, Steplock D, Shenolikar S, Biswas R. Fibroblast growth factor-23-mediated inhibition of renal phosphate transport in mice requires sodium-hydrogen exchanger regulatory factor-1 (NHERF-1) and synergizes with parathyroid hormone. J Biol Chem 2011. October 28;286(43):37216–21. 10.1074/jbc.M111.288357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Andrukhova O, Zeitz U, Goetz R, Mohammadi M, Lanske B, Erben RG. FGF23 acts directly on renal proximal tubules to induce phosphaturia through activation of the ERK1/2-SGK1 signaling pathway. Bone 2012. September;51(3):621–8. 10.1016/j.bone.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hasegawa H, Nagano N, Urakawa I, Yamazaki Y, Iijima K, Fujita T, et al. Direct evidence for a causative role of FGF23 in the abnormal renal phosphate handling and vitamin D metabolism in rats with early-stage chronic kidney disease. Kidney Int 2010. November;78(10):975–80. 10.1038/ki.2010.313 [DOI] [PubMed] [Google Scholar]
- 38. Isakova T, Wahl P, Vargas GS, Gutierrez OM, Scialla J, Xie H, et al. Fibroblast growth factor 23 is elevated before parathyroid hormone and phosphate in chronic kidney disease. Kidney Int 2011. June;79(12):1370–8. 10.1038/ki.2011.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Levin A, Bakris GL, Molitch M, Smulders M, Tian J, Williams LA, et al. Prevalence of abnormal serum vitamin D, PTH, calcium, and phosphorus in patients with chronic kidney disease: results of the study to evaluate early kidney disease. Kidney Int 2007. January;71(1):31–8. [DOI] [PubMed] [Google Scholar]
- 40. Portale AA, Wolf M, Juppner H, Messinger S, Kumar J, Wesseling-Perry K, et al. Disordered FGF23 and Mineral Metabolism in Children with CKD. Clin J Am Soc Nephrol 2014. February 9;9(2):344–53. 10.2215/CJN.05840513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pitts TO, Piraino BH, Mitro R, Chen TC, Segre GV, Greenberg A, et al. Hyperparathyroidism and 1,25-dihydroxyvitamin D deficiency in mild, moderate, and severe renal failure. J Clin Endocrinol Metab 1988;67:876–81. [DOI] [PubMed] [Google Scholar]
- 42. Nagano N, Miyata S, Abe M, Kobayashi N, Wakita S, Yamashita T, et al. Effect of manipulating serum phosphorus with phosphate binder on circulating PTH and FGF23 in renal failure rats. Kidney Int 2006. February;69(3):531–7. [DOI] [PubMed] [Google Scholar]
- 43. Gutierrez O, Isakova T, Rhee E, Shah A, Holmes J, Collerone G, et al. Fibroblast growth factor-23 mitigates hyperphosphatemia but accentuates calcitriol deficiency in chronic kidney disease. J Am Soc Nephrol 2005. July;16(7):2205–15. [DOI] [PubMed] [Google Scholar]
- 44. Marks J, Debnam ES, Unwin RJ. Phosphate homeostasis and the renal-gastrointestinal axis. Am J Physiol Renal Physiol 2010. August;299(2):F285–F296. 10.1152/ajprenal.00508.2009 [DOI] [PubMed] [Google Scholar]
- 45. Barthel TK, Mathern DR, Whitfield GK, Haussler CA, Hopper HA, Hsieh JC, et al. 1,25-Dihydroxyvitamin D3/VDR-mediated induction of FGF23 as well as transcriptional control of other bone anabolic and catabolic genes that orchestrate the regulation of phosphate and calcium mineral metabolism. J Steroid Biochem Mol Biol 2007. March;103(3–5):381–8. [DOI] [PubMed] [Google Scholar]
- 46. Miyamoto K, Ito M, Kuwahata M, Kato S, Segawa H. Inhibition of intestinal sodium-dependent inorganic phosphate transport by fibroblast growth factor 23. Ther Apher Dial 2005. August;9(4):331–5. [DOI] [PubMed] [Google Scholar]
- 47. Lederer ED, Ameen M, Smith H, Merchant ML, Khundmiri SJ. The role of ubiquitination of the type IIa sodium phosphate cotransporter (Npt2a) in protein trafficking. FASEB J 22, 1158.23. 3-1-2008. [Google Scholar]
- 48. Palmada M, Dieter M, Speil A, Bohmer C, Mack AF, Wagner HJ, et al. Regulation of intestinal phosphate cotransporter NaPi IIb by ubiquitin ligase Nedd4-2 and by serum- and glucocorticoid-dependent kinase 1. Am J Physiol Gastrointest Liver Physiol 2004. July;287(1):G143–G150. [DOI] [PubMed] [Google Scholar]
- 49. Lanzano L, Lei T, Okamura K, Giral H, Caldas Y, Masihzadeh O, et al. Differential modulation of the molecular dynamics of the type IIa and IIc sodium phosphate cotransporters by parathyroid hormone. Am J Physiol Cell Physiol 2011. October;301(4):C850–C861. 10.1152/ajpcell.00412.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Blaine J, Okamura K, Giral H, Breusegem S, Caldas Y, Millard A, et al. PTH-induced internalization of apical membrane NaPi2a: role of actin and myosin VI. Am J Physiol Cell Physiol 2009. December;297(6):C1339–C1346. 10.1152/ajpcell.00260.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Signal histogram of the peaks visualized for the positive control gene, Nab1. Integrated Genome Browser (IGB) shots showing egr-1 binding sites near the Nab1 gene. Binding of egr-1 to the regulatory region up-stream (-1500 bp) of Nab1gene. A low signal is noted in the vehicle-treated sample. B. Peak signal intensity for Nab1 gene in FGF23-treated (1 and 2hr) mice, represented as fold change over vehicle-treated sample. C. HEK 293 cells transfected with Nab 1 active region (930bp)—Luc plasmid and treated with FGF23 (100ng/ml) or vehicle for 24hrs. Luciferase activity was normalized to the empty vector and expressed as relative luciferase units (RLU). Data expressed as Mean ± SEM, n = 3 replicates, *P<0.05.
(JPG)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.