Abstract
The population structure, virulence, and antimicrobial resistance of uropathogenic E. coli (UPEC) from cats are rarely characterized. The aim of this study was to compare and characterize the UPEC isolated from cats in four geographic regions of USA in terms of their multilocus sequence typing (MLST), virulence profiles, clinical signs, antimicrobial resistance and phylogenetic grouping. The results showed that a total of 74 E. coli isolates were typed to 40 sequence types with 10 being novel. The most frequent phylogenetic group was B2 (n = 57). The most frequent sequence types were ST73 (n = 12) and ST83 (n = 6), ST73 was represented by four multidrug resistant (MDR) and eight non-multidrug resistant (SDR) isolates, and ST83 were significantly more likely to exhibit no drug resistant (NDR) isolates carrying the highest number of virulence genes. Additionally, MDR isolates were more diverse, and followed by SDR and NDR isolates in regards to the distribution of the STs. afa/draBC was the most prevalent among the 29 virulence-associated genes. Linking virulence profile and antimicrobial resistance, the majority of virulence-associated genes tested were more prevalent in NDR isolates, and followed by SDR and MDR isolates. Twenty (50%) MLST types in this study have previously been associated with human isolates, suggesting that these STs are potentially zoonotic. Our data enhanced the understanding of E. coli population structure and virulence association from cats. The diverse and various combinations of virulence-associated genes implied that the infection control may be challenging.
Introduction
Urinary tract infection (UTI) is one of the most frequent bacterial infection in both human and companion animals. Uropathogenic Escherichia coli (UPEC), belonging to extraintestinal pathogenic E. coli (ExPEC), is the most common bacterium isolated from canine and feline UTIs [1]. Considering the physical closeness in which many humans live with their pet companions, these organisms are responsible for significant social and economic costs for both communities and public health resources as previous study indicate that there may be cross transmission of ExPEC between animals and humans [2]. The ability of UPEC to cause symptomatic UTIs is associated with expression of numerous virulence-associated genes. This includes a broad array of adhesins, invasins, toxins and proteins, which are responsible for pathogenesis outside the gastrointestinal tract [3,4]. Virulence-associated genes are present in increased numbers in UPEC compared to E. coli that remain in the gastrointestinal tract. The presence of some genes, such as papA, papC, sfa/foc, afa/draBC, iutA and kpsMT II are often responsible for acute UTIs [5,6]. Previous surveys have demonstrated similarities among clinical E. coli isolates from humans, dogs, and cats with respect to genomic background and virulence genes, suggesting possible zoonotic transmission [7].
Transmission of antimicrobial resistance among bacterial isolates is an increasing problem in infectious diseases [8]. Antimicrobial susceptible isolates mostly derive from phylogenetic group B2 and are associated with higher virulence-associated genes prevalence than antimicrobial resistant isolates, which are typically associated with shifts toward groups D and A [9]. Some studies have shown that virulence of E.coli isolates are sometimes associated with antimicrobial resistance, whereas other studies have reported that antimicrobial resistance and virulence-associated genes are only weakly linked [8,10].
Knowledge of the molecular pathogenicity of ExPEC infections has improved markedly over the last decade, however, the genetic background and virulence profiles of feline UPEC have been studied to a much lesser extent or in much smaller sample populations in the E. coli isolates from cats. The aim of the current study was to compare and characterize the UPEC isolated from cats in the United States in terms of their virulence, antimicrobial resistance profiles, phylogenetic grouping, and multilocus sequence typing (MLST) in order to obtain a comprehensive understanding of the population structure of UPEC.
Materials and Methods
Bacterial Isolates
We have a primary surveillance study population consisting of 740 E. coli isolates acquired from cats located in four geographic regions in the United States: West (California), South (North Carolina), Midwest (Ohio and Illinois), and Northeast (Massachusetts) from January 2008 through January 2013. Organisms isolated from the urine of cats with suspected UTI had been submitted to a nationally recognized veterinary diagnostic laboratory that receives samples throughout the United States. Organisms had been isolated and identified as E. coli isolates. Upon receipt by Clinical Pharmacology Laboratory (CPL) at Auburn University, identification as E. coli was reconfirmed based on reculture overnight on BBL CHROMagar Orientation (BD Diagnostics, Franklin Lakes, NJ) at 37°C. And then, the isolates were harvested and stored in trypticase soy broth containing 30% glycerol at -80°C for further analysis.
Antimicrobial Susceptibility Testing and Extended-spectrum β-lactamases (ESBLs) Detection
Antimicrobial susceptibility testing was performed for all isolates using custom microdilution susceptibility plates according to the manufacturer’s protocol (Trek Diagnostic Systems, Inc., Cleveland, OH). A panel of sixteen drugs was studied, representing six drug classes, classified into 12 antimicrobial categories: penicillins, penicillins + β-lactam inhibitors, antipseudomonal + β-lactam inhibitor, non-extended spectrum cephalosporins (1st generation cephalosporins), extended-spectrum cephalosporins (3rd and 4th generation cephalosporins), cephamycins, carbapenems, tetracyclines, phenicols, fluoroquinolones, folate pathway inhibitors, and aminoglycosides was tested [11,12], the antibacterial agents and the corresponding breakpoints were listed in S1 Table), and the MICs were recorded using the Sensititre Vizion system (Trek Diagnostic Systems) (The breakpoint MICs of the drugs tested in S1 Table). All MIC determinations were performed in triplicate and reference strain E. coli ATCC 25922 was used for quality control. The results were interpreted by the guidelines of CLSI [13]. Each isolate was categorized as no drug (NDR), non-multidrug (SDR), multidrug (MDR) resistance. SDR was defined as resistance to 1 or 2 of the previously described 12 antimicrobial categories. MDR was defined as resistance to three or more categories [11].
Additionally, the E. coli isolates were tested for extended-spectrum β-lactamase (ESBL) production using microdilution-based Sensititre (Trek Diagnostic Systems, Inc., Cleveland, OH) with ESBL Confirmatory MIC plates (ESB1F) according to previous study of our laboratory [14].
Seventy four (10% of primary surveillance study population isolates, including all of NDR [n = 12], with the remaining comparised of isolates expression SDR [n = 24] and MDR [n = 36]). The SDR and MDR isolates represent 9% of isolates in the entire population expressing SDR and MDR respectively and were randomly selected. Among the 74 patients, 15 (20.3%) were male and 59 (79.7%) were female, and 58 (78.4%) of them are more than 10 years old. Additionally, each isolate was classified in terms of the severity of clinical signs as to absent, mild, moderate, severe or life-threating with the latter generally reflecting pyelonephritis with urosepsis. The scoring system reflected the clinical impression of the veterinarian in consultation with the pet owner. Further, each isolate was also designated as either absent (asymptomatic bacteriuria [ABU]) versus non-ABU (mild, moderate, severe, and life-threatening).
Phylogenetic Grouping and Virulence Genotyping
The distribution of phylogenetic groups amongst UPEC isolates was determined as recently improved phylotyping PCR approach described by Clermont and colleagues [15]. PCR virulence typing was performed in the triplex and duplex PCR reactions as previously described [16] or conventional PCR reaction. All E. coli isolates tested were screened for 29 virulence-associated genes of extraintestinal E. coli, including a pathogenicity associated island (PAI), representing six categories: adhesins (fimH, papA, papC, papE, papG, papG I, papG II, papG III, sfa/focDE, afa/draBC, sfaS, focA, focG, and bmaE), toxins (hlyA, hlyD and cnf1), capsule synthesis (kpsMT II, kpsMT K1, kpsMT K5 and rfc), siderophores (fyuA, iroN, ireA and iutA), invasin (ibeA) and miscellaneous genes (traT, PAI and cvaC). The primer sequences are listed in Table 1.
Table 1. The Oligonucleotide primers of virulence-associated genes used in this study.
| Target gene | Accession No. | Primer sequence (5’-3’) | Fragment size (bp) | Reference |
|---|---|---|---|---|
| fimH | AJ225176 | TGCAGAACGGATAAGCCGTGG/ GCAGTCACCTGCCCTCCGGTA | 508 | [16] |
| papA | X61239 | ATGGCAGTGGTGTCTTTTGGTG/ CGTCCCACCATACGTGCTCTTC | 717 | [16] |
| papC | X61239 | GTGGCAGTATGAGTAATGACCGTTA/ ATATCCTTTCTGCAGGGATGCAATA | 205 | [16] |
| papE | X61239 | GCAACAGCAACGCTGGTTGCATCAT/ AGAGAGAGCCACTCTTATACGGACA | 336 | [16] |
| papG | X61239 | CTGTAATTACGGAAGTGATTTCTG/ ACTATCCGGCTCCGGATAAACCAT | 1070 | [16] |
| papG I | X61239 | TCGTGCTCAGGTCCGGAATTT/ TCCAGAAATAGCTCATGTAACCCG | 479 | [40] |
| papG II | M20181 | GGGATGAGCGGGCCTTTGAT/ CGGGCCCCCAAGTAACTCG | 190 | [41] |
| papG III | X61238 | GGCCTGCAATGGATTTACCTGG/ CCACCAAATGACCATGCCAGAC | 258 | [41] |
| sfa/focDE | Unpublished | CTCCGGAGAACTGGGTGCATCTTAC/CGGAGGAGTAATTACAAACCTGGCA | 410 | [42] |
| afa/draBC | X76688 | GGCAGAGGGCCGGCAACAGGC/CCCGTAACGCGCCAGCATCTC | 559 | [41] |
| sfaS | S53210 | GTGGATACGACGATTACTGTG/CCGCCAGCATTCCCTGTATTC | 240 | [16] |
| focA | DQ301498 | ATGCGTCYGCTGTCACCACGG/ GGCGTCGGCGTTGGCAATAC | 458 | This study |
| focG | DQ301498 | CAGCACAGGCAGTGGATACGA/GAATGTCGCCTGCCCATTGCT | 364 | [16] |
| bmaE | M15677 | ATGGCGCTAACTTGCCATGCTG/ AGGGGGACATATAGCCCCCTTC | 507 | [16] |
| hlyA | M10133 | AACAAGGATAAGCACTGTTCTGGCT/ACCATATAAGCGGTCATTCCCGTCA | 1177 | [43] |
| hlyD | AM690759 | CTCCGGTACGTGAAAAGGAC/ GCCCTGATTACTGAAGCCTG | 904 | [16] |
| cnfI | U42629 | ATCTTATACTGGATGGGATCATCTTGG/ GCAGAACGACGTTCTTCATAAGTAT | 974 | [43] |
| kpsMT II | X53819 | GCGCATTTGCTGATACTGTTG/ CATCCAGACGATAAGCATGAGCA | 272 | [16] |
| kpsMT K1 | M57382 | TAGCAAACGTTCTATTGGTGC/ CATCCAGACGATAAGCATGAGCA | 153 | [16] |
| kpsMT K5 | X53819 | CAGTATCAGCAATCGTTCTGTA/ AACCATACCAACCAATGCGAG | 159 | [16] |
| rfc | U39042 | ATCCATCAGGAGGGGACTGGA/ CATCCAGACGATAAGCATGAGCA | 788 | [16] |
| fyuA | Z38064 | TGATTAACCCCGCGACGGGAA/ CGCAGTAGGCACGATGTTGTA | 880 | [16] |
| iroN | CP001671 | AAGTCAAAGCAGGGGTTGCCCG/ GACGCCGACATTAAGACGCAG | 667 | This study |
| ireA | AF320691 | GATGACTCAGCCACGGGTAA/ CCAGGACTCACCTCACGAAT | 254 | This study |
| iutA | X05874 | GGCTGGACATCATGGGAACTGG/ CGTCGGGAACGGGTAGAATCG | 302 | [44] |
| ibeA | L42624 | AGGCAGGTGTGCGCCGCGTAC/ TGGTGCTCCGGCAAACCATGC | 170 | [16] |
| traT | J01769 | GGTGTGGTGCGATGAGCACAG/ CACGGTTCAGCCATCCCTGAG | 290 | [16] |
| PAI | AF003742 | GGACATCCTGTTACAGCGCGCA/ TCGCCACCAATCACAGCCGAAC | 930 | [16] |
| cvaC | X57525 | CACACACAAACGGGAGCTGTT/ CTTCCCGCAGCATAGTTCCAT | 680 | [16] |
Multilocus Sequence Typing
All 74 UPEC isolates were assigned to multilocus sequence types as described previously [17]. PCR amplification and sequencing of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA and recA) were performed following the protocols specified at the E. coli MLST website (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli). All the primer sequences of seven genes are available at http://mlst.warwick.ac.uk/mlst/dbs/Ecoli/documents/primersColi_html. The 50 μl amplification reaction mixture comprised 2 μl of template DNA, 1.5 μl of each primer (25 pmol/μl), 25 μl 2×PCR Super Master Mix (Biotool LLC., TX) and 20 μl sterilized distilled water. The reaction conditions were an initial denaturation step at 94°C for 2 min, followed by 30 cycles of the following conditions: denaturation at 94°C for 1 min, 1 min primer annealing at 54–60°C, and extension at 72°C for 2 min, with a final extension step at 72°C for 5 min.
Amplicons from seven housekeeping genes were purified using a QIAquick PCR Purification Kit (Qiagen, Inc., Valencia, CA). Sequencing of the PCR products was performed using the services of Macrogen (Macrogen Inc., Rockville, MD), and then alignment between the sequences reference sequences of E. coli MG1655 were done using MEGA version 6.0 software. Allele numbers for seven gene fragments of each isolate were obtained by comparing with corresponding allele available in MLST E. coli database (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli), and Sequence type (ST) of each isolate was determined by combining seven allelic profiles.
Statistical Analysis
Significance was determined by Pearson’s Chi-squared test with Yates continuity correction using ‘R’ software (version 3.0.1). The threshold for statistical significance was a P values of <0.05.
Results
Antimicrobial Susceptibility
The antimicrobial susceptibility results showed that 72.4% (62/74) expressed resistance to at least one antimicrobial drug, including 26 (35.1%) SDR phenotype and 36 (48.6%) MDR phenotype isolates. Only 12 (16.2%) isolates were fully susceptible to all antimicrobials tested (NDR). Regarding resistance to drugs, the highest rates of resistance was expressed toward doxycycline with100% (62/62), and followed by cephalothin at 98.4% (61/62), and ampicillin (62.9%), and the least prevalence was meropenem (4.8%). The percentages of severity were moderate (39.2%; 29/74), followed by mild (18.9%), severe (24.3%) and life-threatening (4.1%). The ABU isolates represented 13.5% (10/74) of total feline UPEC isolates.
Phylogenetic Groups
As is present in Table 2, the predominant phylogenetic group was B2 (74.3%, 55/74), followed by B1 (9.5%, 7/74), D (6.8%, 6/74), A (2.7%, 2/74), E (2.7%, 2/74), and F (2.7%, 2/74) (P < 0.001, B2 vs. B1, D, A, E and F). Phylogenetic group B2 also accounted for 91.7%, 76.9% and 66.7% of the NDR, SDR and MDR phenotype isolates, respectively. Additionally, the isolates from groups A and E expressed MDR phenotype. Phylogenetic analysis in relation to severity of clinical signs revealed that the majority of isolates from patients were considered clinically severe, and belonged to group B2 (53.8%; 10/18). While all the group D isolates belonged to ABU isolates.
Table 2. The prevalence of virulence-associated genes among 74 uropathogenic E. coli isolates from cats.
| Prevalence [no. (%)] of virulence genes | P value | ||||||
|---|---|---|---|---|---|---|---|
| Total number(n = 74) | NDR (n = 12) | SDR (n = 26) | MDR (n = 36) | NDRvsSDR | NDRvsMDR | SDRvsMDR | |
| Phylogenetic group | |||||||
| B2 | 55 (74.3) | 11 (91.7) | 20 (76.9) | 24 (66.7) | 0.009 | 0.007 | 0.092 |
| B1 | 7 (9.5) | 1 (8.3) | 4 (15.4) | 2 (5.6) | 0.001 | 0.008 | 0.054 |
| D | 6 (8.1) | 0 (0) | 0 (0) | 6 (13.9) | 1 | <0.0001 | <0.0001 |
| A | 2 (2.7) | 0 (0) | 0 (0) | 2 (5.6) | 1 | <0.0001 | <0.0001 |
| E | 2 (2.7) | 0 (0) | 0 (0) | 2 (5.6) | 1 | <0.0001 | <0.0001 |
| F | 2 (2.7) | 0 (0) | 2 (7.7) | 0 (0) | <0.0001 | 1 | <0.0001 |
| Virulence determinants | |||||||
| Adhesins | |||||||
| afa/draBC | 68 (91.9) | 12 (100) | 24 (92.3) | 32 (88.9) | 0.465 | 0.587 | 0.309 |
| fimH | 64 (86.5) | 11 (91.7) | 23 (88.5) | 30 (83.3) | 0.302 | 0.302 | 0.204 |
| papA | 47 (63.5) | 11 (91.7) | 16 (61.5) | 20 (55.6) | 0.000 | 0.000 | 0.112 |
| papE | 47 (63.5) | 9 (75.0) | 16 (61.5) | 22 (61.1) | 0.017 | 0.044 | 0.178 |
| sfa/focDE | 47 (63.5) | 10 (83.3) | 20 (76.9) | 17 (47.2) | 0.068 | 0.008 | 0.004 |
| papC | 45 (60.8) | 10 (83.3) | 18 (69.2) | 17 (47.2) | 0.038 | 0.008 | 0.018 |
| focG | 44 (59.5) | 10 (83.3) | 16 (61.5) | 18 (50.0) | 0.000 | 0.006 | 0.048 |
| papG | 21 (28.4) | 9 (75.0) | 11 (42.3) | 1 (2.8) | 0.000 | 0.000 | 0.000 |
| focA | 31 (41.9) | 5 (41.7) | 13 (50.0) | 13 (36.1) | 0.143 | 0.068 | 0.038 |
| papG III | 30 (40.5) | 11 (91.7) | 8 (30.8) | 11 (30.6) | 0.000 | 0.000 | 0.957 |
| sfaS | 9 (12.2) | 1 (8.3) | 5 (19.2) | 3 (8.3) | 0.002 | 1 | 0.002 |
| papG II | 2 (2.7) | 0 (0) | 2 (7.7) | 0 (0) | 0.086 | 1 | 0.086 |
| papG I | 1 (1.4) | 1 (8.3) | 0 (0) | 0 (0) | 0.092 | 0.092 | 1 |
| bmaE | 1 (1.4) | 1 (8.3) | 0 (0) | 0 (0) | 0.092 | 0.092 | 1 |
| Toxins | |||||||
| hlyD | 56 (75.7) | 11 (91.7) | 19 (73.1) | 26 (72.2) | 0.027 | 0.024 | 0.915 |
| hlyA | 45 (60.8) | 11 (91.7) | 17 (65.4) | 17 (47.2) | 0.011 | 0.005 | 0.027 |
| cnf1 | 45 (60.8) | 9 (75) | 18 (69.2) | 18 (50.0) | 0.068 | 0.013 | 0.024 |
| Capsule synthesis | |||||||
| kpsMT II | 41 (55.4) | 8 (66.7) | 15 (57.7) | 18 (50.0) | 0.051 | 0.027 | 0.059 |
| rfc | 12 (16.2) | 3 (25.0) | 5 (9.2) | 4 (11.1) | 0.032 | 0.038 | 0.107 |
| kpsMTK 5 | 11 (14.9) | 3 (25.0) | 4 (15.4) | 4 (11.1) | 0.051 | 0.038 | 0.077 |
| kpsMTK 1 | 3 (4.1) | 2 (16.7) | 1 (3.8) | 0 (0) | 0.000 | 0.000 | 0.002 |
| Siderophores | |||||||
| fyuA | 62 (83.8) | 12 (100) | 24 (92.3) | 26 (72.2) | 0.025 | 0.001 | 0.015 |
| iroN | 45 (60.8) | 8 (66.7) | 17 (65.4) | 20 (55.6) | 0.166 | 0.064 | 0.075 |
| iutA | 45 (60.8) | 6 (50.0) | 13 (50.0) | 26 (72.2) | 1 | 0.015 | 0.015 |
| ireA | 21 (28.4) | 4 (33.3) | 10 (38.5) | 7 (19.4) | 0.409 | 0.009 | 0.006 |
| Invasin | |||||||
| ibeA | 13 (17.6) | 4 (33.3) | 4 (15.4) | 5 (13.9) | 0.029 | 0.039 | 0.132 |
| Others | |||||||
| traT | 50 (67.6) | 12 (100) | 15 (57.7) | 23 (63.9) | 0.004 | 0.005 | 0.131 |
| PAI | 44 (59.5) | 10 (83.3) | 15 (57.7) | 19 (52.8) | 0.020 | 0.009 | 0.138 |
| cvaC | 6 (8.1) | 4 (33.3) | 0 (0) | 2 (5.6) | 0.000 | 0.000 | 0.128 |
The 29 virulence-associated genes analysed were: afa/draBC, Dr-binding adhesins; fimH, mannose-specific adhesin of type 1 fimbriae; papA, P fimbriae structural subunit; papE, fimbriae tip pilins; papC, p fimbriae assembly; papG, p fimbriae adhesin (and alleles I, II, and III); sfa/focDE, S and F1C fimbriae; sfaS, S fimbriae; focG, focA, F1C fimbriae; bmaE, blood group M fimbriae; hlyD, hlyA, α-haemolysin; cnf1, cytotoxic necrotizing factor type 1; kpsM II, group 2 capsule in addition to specifically targeting K1 and K5 genes; rfc, O antigen polymerase; fyuA, ferric yersiniabactin receptor; iutA, aerobactin receptor; iroN, almochelin receptor; ireA, iron-responsive element gene; ibeA, invasion of brain endothelium; traT, serum-resistance associated; PAI, pathogenicity island; cvaC, Colicin-V.
Distribution of Virulence-associated Genes
The frequencies of the 29 virulence-associated genes were summarized in Table 2. The overall prevalence of these genes ranged from 1.4% (papG I and bmaE) to 91.9% (afa/draBC). The profiles of virulence-associated genes were extremely diverse, with each isolate characterized by a different profile. All of the 74 E. coli isolates harbored 2 to 24 virulence-associated genes studied. The isolates of phylogenetic groups B2 possessed averages of 14.9 virulence-associated genes, which was higher than among those in groups F, E, D, B1 and A (11.5, 10.5, 8.3, 7.7 and 4.5, respectively). Moreover, a number of virulence-associated genes such as hlyD, hlyA, cnf1 and iroN were detected significantly more frequently in phylogenic group B2 compared with other groups (P < 0.01). While fimH, sfa/focDE, afa/draBC, traT, iutA and fyuA genes were widely distributed among all groups at different percentages.
Linking the virulence profile and clinical signs, a statistically significant difference could not be detected for the distribution of each virulence-associated genes among the five levels of severity (P > 0.05) with the exception of the hlyA, which was more prevalent in severe and life-threatening isolates. The proportion was various as isolates were categorized as either ABU vs non-ABU, the virulence-associated genes present in a greater proportion of non-ABU isolates were papG III, focG and cnf1 (P < 0.05). The discriminator among the virulence-associated genes tested between ABU and non-ABU was cnf1, which occurred in 35.3% of ABU vs 68.4% non-ABU (P < 0.001). Moreover, there was a strong correlation between the distribution of the virulence-associated genes and the resistant phenotype. Overall, 100% (12/12) NDR isolates, 84.6% (22/26) SDR isolates and 61.1% (22/36) MDR isolates encoded ten or more of the 29 virulence-associated genes tested. Furthermore, the distribution rates of most genes tested were higher in NDR isolates, followed by significant descending gradients in SDR and MDR isolates with the exception of iutA, sfaS, focA, papG II and ireA (Table 2), the NDR, SDR and MDR phenotype isolates harbored an average of 17.4, 13.5 and 10.9 virulence-associated genes, respectively. It is noteworthy that papG III was significantly higher in NDR isolates (91.7%) than in SDR (30.8%) and MDR phenotype (30.6%) isolates (P < 0.001). Among the vast majority of the SDR or MDR phenotype isolates, resistance to more antimicrobial or antimicrobial classes possessed less virulence-associated genes. Moreover, eight ESBL positive E. coli isolates were detected in MDR phenotype, and they were distributed in phylogenetic groups B2, D and E, and the number of virulence-associated genes present in these isolates varied between 2 and 15. However, it is difficult to link the virulence profiles with phylogenetic groups among these ESBL positive isolates as the limited isolates.
MLST Analysis and Phylogenetic Relationships of the UPEC Isolates
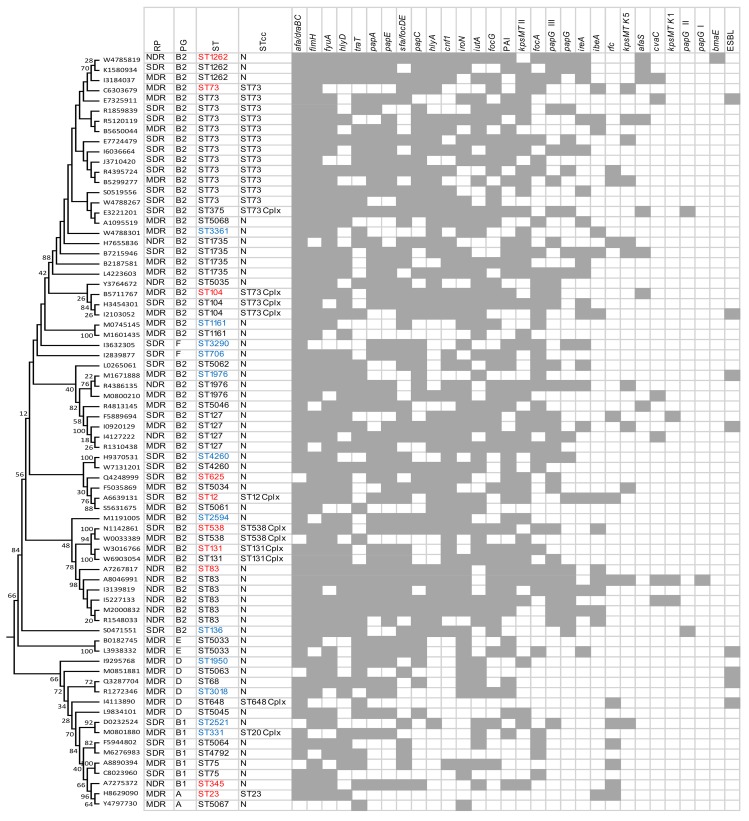
We used MLST to determine the diversity and phylogenetic relationships of the UPEC isolates. Sequences were concatenated for each isolate and aligned using ClustalW in MEGA 6.0. The evolutionary history of 74 E. coli isolates tested was inferred using the maximum likelihood method based on the Tamura-Nei model and concatenated sequences of all seven genes (Fig 1). The 74 UPEC isolates analyzed were assigned to 40 distinct sequence types (STs) with ten (25%) being novel. The most frequent ST was ST73 (n = 12, 16.2%) followed by ST83 (n = 6, 8.1%), ST73 were represented by four multidrug MDR and eight SDR isolates, the ST83 isolates were significantly associated with NDR but carried the highest number of virulence-associated genes. The majority of (94.4%, 17/18) of ST73 and ST83 isolates were assigned to groups B2, and they were also associated with the severe and life-threatening situations. There were 7, 18, and 26 STs in the NDR, SDR and MDR isolates, with the Simpson's diversity index 77.3%, 91.1% and 97.9%, respectively. Two MDR isolates (2.7%) belonging to phylogenetic group B2 were ST131. Additionally, some STs or clonal complexes identified in this study have also been found to be associated with both humans and animals according to the MLST database (Fig 1, red font), and some ones were identified in humans or animals, even in water (Fig 1, blue font). Eight ESBL-producing isolates belonged to eight different STs (Fig 1), and this is the first report on ESBL positive E. coli of ST5033, ST5063 and ST104 in cats.
Fig 1. Maximum likelihood tree constructed using MEGA 6.0 based on the nucleotide sequences of seven housekeeping genes: adk, gyrB, fumC, icd, mdh, purA and recA, and depicting infrerred phylogency of 74 uropathogenic E. coli (UPCE) from cats.
Resistant phenotype (RP), phylogenetic group (PG), sequence type (ST), ST clonal complex (STcc; “N” indicates No STcc), virulence-associated genes and the prevalence of ESBL were displayed the right of the dendrogram. Virulence-associated genes were arranged in descending order according their corresponding prevalence. Gray square indicates the presence of the virulence-associated genes and ESBL. The sequence types highlighted in red were also found to be associated with both humans and other animals, and sequence types highlighted in blue were identified in humans or animals, or in water.
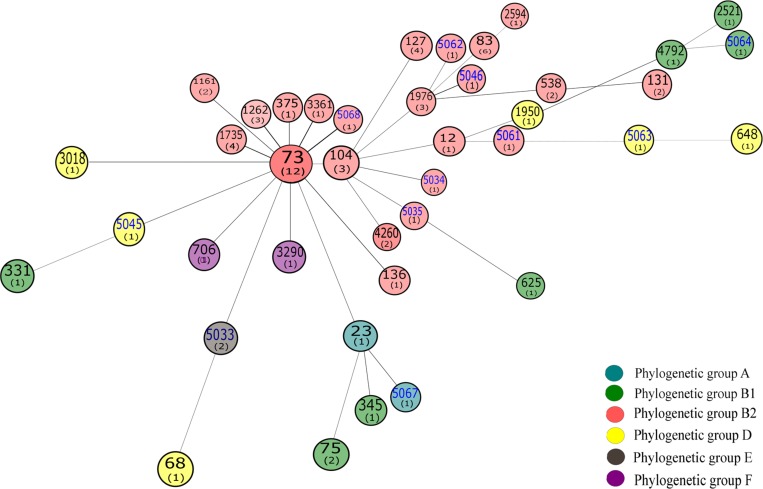
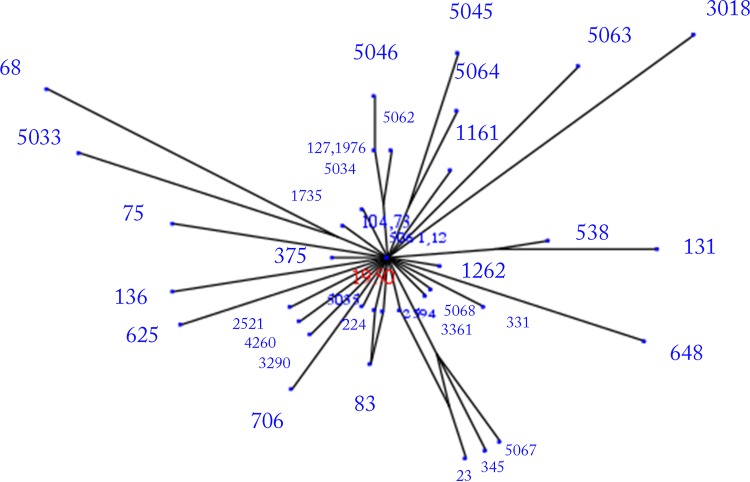
The 40 STs identified in this study were compared with all identified E. coli MLST types (In May, 2015, 7629 isolates in the E. coli MLST database belonging to 4613 STs) using the eBURST. It is clear from this analysis that these E. coli isolates were distributed widely among multiple clonal complexes (S1 Fig). In order to further evaluate the relationship between STs, the Minimum spanning (MS) tree was generated from the allelic profiles of the tested isolates using a web version of MS Tree (http://pubmlst.org/analysis/). MS tree showed that the tested E. coli mainly classed into five clonal complexes, which is represented by ST73, ST104, ST1976, ST23 and ST12, respectively. ST73 served as the predicted founder in the MS tree (Fig 2). Meanwhile, Splits tree decomposition demonstrated a similar network among E. coli STs (Fig 3). As evident from Fig 3, most group A and B1 isolates had shorter branches, suggesting that they were closely related. This is consistent with previous studies that group A and B1 isolates were considered as sister groups [18,19].
Fig 2. Minimum spanning (MS) tree was generated from the allelic profiles of seven housekeeping genes: adk, gyrB, fumC, icd, mdh, recA and purA.
Each ST is represented by a circle named with its ST, and the number in the brackets of each circle represent the number of each ST in our isolates tested. The blue fonts are the novel ST identified in this study.
Fig 3. Splits tree decomposition network was obtained using distance matrix obtained from allelic profiles using a web version of Splits-Tree (http://pubmlst.org/analysis/).
Most groups A and B1 isolates had shorter branches, suggesting that they were closely related as the group A and B1 isolates were considered as sister groups.
Discussion
In the current study, we investigated the association between phylogenetic background, antimicrobial resistance, virulence profiles, clinical signs as well as the genetic relatedness of the UPEC isolated from cats in four geographic regions of the United States. Additionally, we demonstrated that UPEC from cats exhibited distinctive virulence profiles and phylogenetic background based on different clinical sign levels.
It is reported that antimicrobial susceptible and resistant ExPEC isolates are fundamentally different in bacterial populations [9,20,21]. In general, more than 94% of isolates belonged to phylogenetic groups B2, B1 and D. A similar observation has been made with ExPEC human clinical isolates [22–24]. Our data also showed that the phylogenetic profile differed between the NDR, SDR and MDR isolates. It is notable that almost all NDR isolates derived from group B2, compared with the proportion of 84.6% and 66.7%, respectively, for SDR and MDR isolates (P < 0.01). This was consistent with the previous study that demonstrated susceptible UPEC isolates to predominantly belong to the B2 phylogenetic group [25]. Moreover, consistently in human medicine, the proportion of group B2 is higher for severe disease, and group D was high in ABU isolates. Previous studies have found that UPEC isolates from cats share similarities with E. coli isolates that cause serious extraintestinal infections in humans. As such, E. coli also might have zoonotic and reverse zoonotic potential due to the considerable commonality observed between human and [14]animal E. coli isolates from UTIs [26,27].
Virulence profile analysis (29 genes) revealed that isolates belonging to phylogenetic groups B2 more frequently carry virulence-associated genes than that of group B1, D, A, C, E and F isolates. A high prevalence of afa/draBC, fimH, fyuA and traT (≥ 67.6%) reinforced the premise that adhesins, toxins and iron acquisition systems were more prevalent among ExPEC. Usually, fimbrial adhesins are the most common factors associated with virulent E. coli in UTI [28]. Our results firstly agreed in that afa/draBC (Dr-binding adhesins), a key virulence marker of ExPEC [29] was the most prevalent virulence gene. P fimbriae are the second common virulence genes of UPEC, with pap genes being associated with pyelonephritis [30]. In the current study, papA, papC and papE were present in high percentages (60.8%-63.5%), suggesting that the isolates from the urine of cats have greater capabilities to colonize kidneys and generate pyelonephritis [31]. PapG adhesin included papG I, papG II and papG III, and their prevalence have host specificity. According to our data, papG I and papG II were only observed in one and two isolates, respectively. It is in accordance with previous literature for which papG I has been observed in a lower frequency (0%–6.0%) in E. coli isolated from UTIs in cats, dogs and humans, while papG III was present in 95% cat isolates [32]. Our findings showed that the prevalence of papG III in NDR isolates (91.7%) was significantly higher prevalent than in SDR isolates (30.8%) and MDR (30.6%) isolates. These results indicated that papG III was the predominant papG allele in the pathogenesis of cat UPEC, and the isolates carrying papG III or not might correlate with the susceptibility or resistance of E. coli. In regards to resistance and virulence, our results revealed a strong inverse correlation between the distribution of the virulence-associated genes and the resistant phenotype. The distribution of virulence-associated genes statistically differed between and within different resistant phenotype. The vast majority of the genes tested were significantly more common in NDR isolates, followed by SDR and MDR isolates. Moreover, the prevalence of virulence-associated genes is inversely associated with antibiotic resistance, videlicet, the prevalence of virulence-associated genes was highest for NDR isolates, followed by significant descending gradients to SDR, and then MDR isolates. The exceptions were iutA, sfaS, focA, papG II and ireA. iutA was universally detected at a higher frequency in MDR isolates, and four other genes were more prevalent in SDR isolates. Meanwhile, within the majority of SDR or MDR phenotype isolates, resistance to more antimicrobial or antimicrobial categories harbored less virulence-associated genes. These findings further reinforced the previous studies that MDR E. coli from urinary tract infections tend to be associated with a decrease in the presence of virulence compared with the susceptible isolates [25,33].
The MLST was developed as a scalable typing system to determine the diversity and phylogenetic relationships of the isolates based on seven housekeeping genes, and it provides reproducibility, comparability, and transferability between laboratories [34]. Previous studies identified four major ST complexes, ST14, ST69, ST73 and ST95, associated with UPEC [35,36]. We found a highly diverse population representing 40 ST, with 10 being novel, clustered into ST73, ST104, ST1976, ST23 and ST12 clonal complexes, and 75% of STs were firstly reported in E. coli isolates from cats. The MDR phenotype isolates had a richer ST diversity (26 STs) than the SDR counterparts (18 STs) and NDR (7 STs), and MDR isolates also showed higher genetic diversity than SDR and NDR isolates with the corresponding ratio of the MLST types to the isolate number in MDR, SDR and NDR phenotype were 72.2% (26/36), 69.2% (18/26) and 58.3% (7/12). Whether or not it may imply that the genetic diversity of UPEC will gradually increase as the resistant phenotypes change from NDR, to SDR, to MDR must be further elucidated in the future studies. An interesting finding was that ST83 occurred in 50% (6/12) fully susceptible isolates, which possessed averages of 18.7 virulence-associated genes. However, it was not found in any resistant isolate. Thus far, there are four ST83 voluntarily submitted to the publicly accessible E. coli MLST database, including isolates from a cat, horse and Celebese ape. This is the first report of a relatively high prevalence of ST83 found in fully susceptible UPEC from cats. The relationship between the ST83 and resistance of E. coli needs further investigation in the future. It is worthy to mention that twenty (50%) MLST types showed relatedness to STs commonly associated with bacteremia in human patients, which provides further evidence that UPEC from cats and humans are shared.
Notably, ST131 were identified in two (2.7%) non-ESBL-producing MDR isolates associated with severe clinical sign level. However, neither isolate had a high virulence gene content nor serious resistance compared with other isolates. Previous studies have demonstrated that most ST131 are strongly associated with CTX-M-15 ESBL-producing, high virulence and fluoroquinolone resistance [37,38]. It is suggested that the isolates studied here are not a part of the pandemic clade. Nonetheless, ongoing surveillance for ST131 isolates is necessary as it is now rapidly and globally disseminated. The role in companion animals as a source or as a site of reverse zoonosis in ST131 transmission dynamics is not clear, as most companion animal ST131 isolates showed a high degree of relatedness to human ST131 isolates [39].
Conclusion
Taken together, our results discovered a clear association between the phylogenetic groups, resistant phenotypes, virulence profiles, MLST types and different levels of clinical signs. Generally, the resistant isolates harbored less virulence-associated genes than susceptible ones, and phylogenetic groups B2 and D isolates were associated more frequently with virulence-associated genes than those belonging to other groups. MLST analysis revealed a highly diverse population representing 40 STs including 10 novel STs and two ST131 isolates, and twenty (50%) MLST types showed relatedness to STs commonly associated with humans. The most frequent MLST types in susceptible and resistant isolates were ST83 and ST73, respectively, and the genetic diversities statistically significant ascending gradients from NDR, through SDR, to MDR. Additionally, the UPEC isolates also exhibited distinctive patterns of association with virulence profiles and phylogenetic background based on different levels of clinical signs. The results significantly advance our understanding of the distinctive UPEC in feline, and may help for controlling the UTIs in future.
Supporting Information
Blue nodes represent predicted founder STs and sub-founders are indicated in yellow, and all other STs marked as black dots.
(DOCX)
(DOCX)
Acknowledgments
The authors wish to acknowledge the assistance of laboratory staff in the clinical pharmacology laboratory of Auburn University.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by the Fundamental Research Funds for the Central Universities (no. QN2013029), National Science Foundation of Shaanxi province (no. 2014JM3071), and partially supported by grant D07-MS006 from Morris Animal Foundation.
References
- 1. Osugui L, de Castro AF, Iovine R, Irino K, Carvalho VM (2014) Virulence genotypes, antibiotic resistance and the phylogenetic background of extraintestinal pathogenic Escherichia coli isolated from urinary tract infections of dogs and cats in Brazil. Vet Microbiol 171: 242–247. 10.1016/j.vetmic.2014.03.027 [DOI] [PubMed] [Google Scholar]
- 2. Johnson JR, Clabots C (2006) Sharing of virulent Escherichia coli clones among household members of a woman with acute cystitis. Clin Infect Dis 43: e101–108. [DOI] [PubMed] [Google Scholar]
- 3. Mulvey MA, Lopez-Boado YS, Wilson CL, Roth R, Parks WC, Heuser J, et al. (1998) Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli . Science 282: 1494–1497. [DOI] [PubMed] [Google Scholar]
- 4. Bien J, Sokolova O, Bozko P (2012) Role of Uropathogenic Escherichia coli Virulence Factors in Development of Urinary Tract Infection and Kidney Damage. Int J Nephrol 2012: 681473 10.1155/2012/681473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yuri K, Nakata K, Katae H, Yamamoto S, Hasegawa A (1998) Distribution of uropathogenic virulence factors among Escherichia coli strains isolated from dogs and cats. J Vet Med Sci 60: 287–290. [DOI] [PubMed] [Google Scholar]
- 6. Johnson JR, Kaster N, Kuskowski MA, Ling GV (2003) Identification of urovirulence traits in Escherichia coli by comparison of urinary and rectal E. coli isolates from dogs with urinary tract infection. J Clin Microbiol 41: 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnson JR, Johnston B, Clabots CR, Kuskowski MA, Roberts E, DebRoy C (2008) Virulence genotypes and phylogenetic background of Escherichia coli serogroup O6 isolates from humans, dogs, and cats. J Clin Microbiol 46: 417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boerlin P, Travis R, Gyles CL, Reid-Smith R, Janecko N, Lim H, et al. (2005) Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl Environ Microbiol 71: 6753–6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cooke NM, Smith SG, Kelleher M, Rogers TR (2010) Major differences exist in frequencies of virulence factors and multidrug resistance between community and nosocomial Escherichia coli bloodstream isolates. J Clin Microbiol 48: 1099–1104. 10.1128/JCM.02017-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang XM, Jiang HX, Liao XP, Liu JH, Zhang WJ, Zhang H, et al. (2010) Antimicrobial resistance, virulence genes, and phylogenetic background in Escherichia coli isolates from diseased pigs. FEMS Microbiol Lett 306: 15–21. 10.1111/j.1574-6968.2010.01917.x [DOI] [PubMed] [Google Scholar]
- 11. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. (2012) Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18: 268–281. 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 12. Thungrat K, Price SB, Carpenter DM, Boothe DM (2015) Antimicrobial susceptibility patterns of clinical Escherichia coli isolates from dogs and cats in the United States: January 2008 through January 2013. Vet Microbiol 179: 287–295. 10.1016/j.vetmic.2015.06.012 [DOI] [PubMed] [Google Scholar]
- 13.CLSI (2013) Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacterial Isolated from Animals; Approved Standard. Fourth Edition and Supplement, CLSI document VET01-A4 (standard) and VET01-S2 (supplement). Clinical and Laboratory Standards Institute, Wayne, PA USA. pp. ISBN 1-56238- 58770.
- 14. Aly SA, Debavalya N, Suh SJ, Oryazabal OA, Boothe DM (2012) Molecular mechanisms of antimicrobial resistance in fecal Escherichia coli of healthy dogs after enrofloxacin or amoxicillin administration. Can J Microbiol 58: 1288–1294. 10.1139/w2012-105 [DOI] [PubMed] [Google Scholar]
- 15. Clermont O, Christenson JK, Denamur E, Gordon DM (2013) The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Env Microbiol Rep 5: 58–65. [DOI] [PubMed] [Google Scholar]
- 16. Johnson JR, Stell AL (2000) Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis 181: 261–272. [DOI] [PubMed] [Google Scholar]
- 17. Wirth T, Falush D, Lan RT, Colles F, Mensa P, Wieler LH, et al. (2006) Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60: 1136–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jaureguy F, Landraud L, Passet V, Diancourt L, Frapy E, Guigon G, et al. (2008) Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains . BMC Genomics 9: 560 10.1186/1471-2164-9-560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu G, Ehricht R, Mafura M, Stokes M, Smith N, Pritcharg, et al. (2012) Escherichia coli isolates from extraintestinal organs of livestock animals harbour diverse virulence genes and belong to multiple genetic lineages. Vet Microbiol 160: 197–206. 10.1016/j.vetmic.2012.05.029 [DOI] [PubMed] [Google Scholar]
- 20. Zhao L, Chen X, Zhu X, Yang W, Dong L, Xu X, et al. (2009) Prevalence of virulence factors and antimicrobial resistance of uropathogenic Escherichia coli in Jiangsu province (China). Urology 74: 702–707. 10.1016/j.urology.2009.01.042 [DOI] [PubMed] [Google Scholar]
- 21. Jaureguy F, Carbonnelle E, Bonacorsi S, Clec'h C, Casassus P, Bingen E, et al. (2007) Host and bacterial determinants of initial severity and outcome of Escherichia coli sepsis. Clin Microbiol Infect 13: 854–862. [DOI] [PubMed] [Google Scholar]
- 22. Moreno E, Planells I, Prats G, Planes AM, Moreno G, Andreu A (2005) Comparative study of Escherichia coli virulence determinants in strains causing urinary tract bacteremia versus strains causing pyelonephritis and other sources of bacteremia. Diagn Microbiol Infect Dis 53: 93–99. [DOI] [PubMed] [Google Scholar]
- 23. Rijavec M, Muller-Premru M, Zakotnik B, Zgur-Bertok D (2008) Virulence factors and biofilm production among Escherichia coli strains causing bacteraemia of urinary tract origin. J Med Microbiol 57: 1329–1334. 10.1099/jmm.0.2008/002543-0 [DOI] [PubMed] [Google Scholar]
- 24. Piatti G, Mannini A, Balistreri M, Schito AM (2008) Virulence factors in urinary Escherichia coli strains: phylogenetic background and quinolone and fluoroquinolone resistance. J Clin Microbiol 46: 480–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wagner S, Gally DL, Argyle SA (2014) Multidrug-resistant Escherichia coli from canine urinary tract infections tend to have commensal phylotypes, lower prevalence of virulence determinants and ampC-replicons. Vet Microbiol 169: 171–178. 10.1016/j.vetmic.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Freitag T, Squires RA, Schmid J, Elliott J (2005) Feline uropathogenic Escherichia coli from Great Britain and New Zealand have dissimilar virulence factor genotypes. Vet Microbiol 106: 79–86. [DOI] [PubMed] [Google Scholar]
- 27. Johnson JR, Delavari P, Stell AL, Whittam TS, Carlino U, Russo TA (2001) Molecular comparison of extraintestinal Escherichia coli isolates of the same electrophoretic lineages from humans and domestic animals. J Infect Dis 183: 1546–1546. [DOI] [PubMed] [Google Scholar]
- 28. Tarchouna M, Ferjani A, Ben-Selma W, Boukadida J (2013) Distribution of uropathogenic virulence genes in Escherichia coli isolated from patients with urinary tract infection. Int J Infect Dis 17: e450–453. 10.1016/j.ijid.2013.01.025 [DOI] [PubMed] [Google Scholar]
- 29. Karisik E, Ellington MJ, Livermore DM, Woodford N (2008) Virulence factors in Escherichia coli with CTX-M-15 and other extended-spectrum beta-lactamases in the UK. J Antimicrob Chemother 61: 54–58. [DOI] [PubMed] [Google Scholar]
- 30. Jacobsen SM, Stickler DJ, Mobley HL, Shirtliff ME (2008) Complicated catheter-associated urinary tract infections due to Escherichia coli and Proteus mirabilis. Clin Microbiol Rev 21: 26–59. 10.1128/CMR.00019-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Antao EM, Wieler LH, Ewers C (2009) Adhesive threads of extraintestinal pathogenic Escherichia coli . Gut Pathog 1: 22 10.1186/1757-4749-1-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Feria C, Machado J, Duarte Correia J, Goncalves J, Gaastra W (2001) Distribution of papG alleles among uropathogenic Escherichia coli isolated from different species. FEMS Microbiol Lett 202: 205–208. [DOI] [PubMed] [Google Scholar]
- 33. Koczura R, Mokracka J, Barczak A, Krysiak N, Kaznowski A (2013) Association between the presence of class 1 integrons, virulence genes, and phylogenetic groups of Escherichia coli isolates from river water. Microb Ecol 65: 84–90. 10.1007/s00248-012-0101-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schierack P, Steinruck H, Kleta S, Vahjen W (2006) Virulence factor gene profiles of Escherichia coli isolates from clinically healthy pigs. Appl Environ Microbiol 72: 6680–6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lau SH, Reddy S, Cheesbrough J, Bolton FJ, Willshaw G, Cheasty T, et al. (2008) Major uropathogenic Escherichia coli strain isolated in the northwest of England identified by multilocus sequence typing. J Clin Microbiol 46: 1076–1080. 10.1128/JCM.02065-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tartof SY, Solberg OD, Manges AR, Riley LW (2005) Analysis of a uropathogenic Escherichia coli clonal group by multilocus sequence typing. J Clin Microbiol 43: 5860–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M (2010) Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis 51: 286–294. 10.1086/653932 [DOI] [PubMed] [Google Scholar]
- 38. Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, et al. (2014) Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A 111: 5694–5699. 10.1073/pnas.1322678111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Platell JL, Cobbold RN, Johnson JR, Heisig A, Heisig P, et al. (2011) Commonality among Fluoroquinolone-Resistant Sequence Type ST131 Extraintestinal Escherichia coli Isolates from Humans and Companion Animals in Australia. Antimicrob Agents Chemothe 55: 3782–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mitsumori K, Terai A, Yamamoto S, Yoshida O (1998) Identification of S, F1C and three PapG fimbrial adhesins in uropathogenic Escherichia coli by polymerase chain reaction. FEMS Immunol Med Microbiol 21: 261–268. [DOI] [PubMed] [Google Scholar]
- 41. Johnson JR, Brown JJ (1996) A novel multiply primed polymerase chain reaction assay for identification of variant papG genes encoding the Gal(alpha 1–4)Gal-binding PapG adhesins of Escherichia coli . J Infect Dis 173: 920–926. [DOI] [PubMed] [Google Scholar]
- 42. Le Bouguenec C, Archambaud M, Labigne A (1992) Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J Clin Microbiol 30: 1189–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamamoto S, Terai A, Yuri K, Kurazono H, Takeda Y, Yoshida O (1995) Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol Med Microbiol 12: 85–90. [DOI] [PubMed] [Google Scholar]
- 44. Johnson JR, Brown JJ, Carlino UB, Russo TA (1998) Colonization with and acquisition of uropathogenic Escherichia coli as revealed by polymerase chain reaction-based detection. J Infect Dis 177: 1120–1124. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Blue nodes represent predicted founder STs and sub-founders are indicated in yellow, and all other STs marked as black dots.
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.