Abstract
NRG-1β (neuregulin-1β) serves multiple functions during embryonic heart development by signalling through ErbB family receptor tyrosine kinases (ErbB2, ErbB3 and ErbB4). Previous studies reported that NRG-1β induces cardiomyogenesis of mESCs (mouse embryonic stem cells) at the later stages of differentiation through ErbB4 receptor activation. In the present study we systematically examined NRG-1β induction of cardiac myocytes in mESCs and identified a novel time window, the first 48 h, for NRG-1β-based cardiomyogenesis. At this time point ErbB3, but not ErbB4, is expressed. In contrast with the later differentiation of mESCs in which NRG-1β induces cardiomyogenesis via the ErbB4 receptor, we found that knocking down ErbB3 or ErbB2 with siRNA during the early differentiation inhibited NRG-1β-induced cardiomyogenesis in mESCs. Microarray analysis of RNA expression at this early time point indicated that NRG-1β treatment in mESCs resulted in gene expression changes important to differentiation including upregulation of components of PI3K (phosphoinositide 3-kinase), a known mediator of the NRG-1β/ErbB signalling pathway, as well as activation of CREB (cAMP-response-element-binding protein). Further study demonstrated that the NRG-1β-induced phosphorylation of CREB was required for cardiomyogenesis of mESCs. In summary, we report a previously unrecognized role for NRG-1β/ErbB3/CREB signalling at the pre-mesoderm stage for stem cell cardiac differentiation.
Keywords: cardiac differentiation, ErbB3, ErbB4, mouse embryonic stem cell, neuregulin-1β (NRG-1β), signalling
INTRODUCTION
PSCs (pluripotent stem cells), such as ESCs (embryonic stem cells) and induced PSCs, hold tremendous promise for repairing damaged heart tissues because of their capability to differentiate into all body cell types, including cardiovascular cells. However, the mechanisms of PSC cardiomyogenesis are largely unknown, and generating sufficient quantities of cardiomyocytes remains a formidable challenge. Our laboratory and others are currently investigating the potential for small molecules and growth factors to facilitate cardiomyogenesis and thereby improve the outcome of currently existing stem cell therapies [1–3].
NRG-1β (neuregulin-1β), a member of the epidermal growth factor family, is required for cardiac development as well as homoeostasis and repair of the postnatal heart. The biological function of NRG-1β is exerted via tyrosine kinase receptors of the ErbB family (ErbB2, ErbB3 and ErbB4) [4–11]. NRG-1β directly binds to the tyrosine kinase receptors ErbB3 or ErbB4, inducing heterodimerization with ErbB2 as well as homodimerization of ErbB4 with subsequent stimulation of tyrosine kinase activity of the ErbB2 and ErbB4 receptors leading to activation of intracellular signalling. The importance of NRG-1β/ErbB signalling in cardiac development is highlighted by the facts that mice with disrupted expression of NRG-1, ErbB2, ErbB3 or ErbB4 die in utero with failure of ventricular trabeculation and cardiac cushion formation [6,7,12,13]. Additionally, we and others have demonstrated important effects of NRG-1β on postnatal and adult myocytes, such as regulation of cell survival, growth, proliferation and stress responses in vitro [14–20]. Moreover, activation of NRG-1β/ErbB signalling using recombinant NRG-1β resulted in improved left ventricular function in rats after myocardial infarction, and clinical trials with recombinant forms of NRG-1β have shown positive effects on the cardiac function of patients with heart failure [21,22].
Previously, NRG-1β has been shown to promote cardiac induction in mESCs (mouse ESCs) when cells were treated at the later differentiation stages [23–25]. These studies demonstrated that NRG-1β induces cardiomyocyte formation in mESCs by signalling through the ErbB4 receptor [24,25]. We sought to examine more systematically the effects of NRG-1β on cardiomyogenic ability in mESCs at various time windows, and found that NRG-1β can actually induce two waves of cardiomyocyte differentiation from mESCs at two distinct treatment time windows, namely early pre-mesoderm stage (day 0–2) and the later differentiation stage (day 3–7). We demonstrated further that NRG-1β-induced cardiomyogenesis at the early treatment time frame occurs via the ErbB3 receptor and activation of CREB (cAMP-response-element-binding protein).
EXPERIMENTAL
Cell culture and reagents
CGR8 and R1 mESC lines were grown in 0.2% gelatine-coated dishes as monolayers in growth medium composed of GMEM (Glasgow minimal essential medium; Sigma–Aldrich) supplemented with 10% FBS (Gibco), 2 mM l-glutamine (Cellgro), 0.05 mM 2-mercaptoethanol (Sigma–Aldrich) and 200 units/ml murine LIF (leukaemia inhibitory factor; Millipore). Every 24 h, cells were washed in 1 × PBS and the culture medium was replaced. Cells were passaged when confluence reached 50–60% to preserve the undifferentiated phenotype. NRG-1β was purchased from R&D Systems.
mESC cardiac differentiation
mESCs were trypsinized and EBs (embryonic bodies) were generated by the hanging-drop method at day 0. In brief, 500 cells in 20 µl of EB differentiation medium were used to make EBs in hanging drops for 2 days (day 0–2). The EB differentiation medium was composed of IMDM (Iscove’s modified Dulbecco’s medium; Gibco) supplemented with 20% FBS, 1.6 mM l-glutamine, 1 × non-essential amino acids, 0.08 mM 2-mercaptoethanol, and either 50 ng/ml NRG-1β or a water vehicle. At day 2 of differentiation (day 2), treatment with NRG-1β was discontinued. The EBs were transferred on to uncoated Petri dishes and suspended in differentiation medium for 2 days (day 2–4). On day 4, the EBs were moved on to gelatine-coated six-well plates, allowed to attach and then incubated in differentiation medium until day 12. Throughout this time, the medium was replaced every 48–72 h. Each day, differentiating cell clusters were microscopically examined for the presence of contracting cardiomyocytes.
In addition, uncoated 96-well round-bottomed plates was used to form EBs, which allowed us to quantify the number of beating EBs. Aliquots of 500 ESCs in 100 µl of EB differentiation medium were distributed in each well of the 96-well plates in the presence of NRG-1β or a vehicle control. Beginning on day 2, the medium was replaced every 48–72 h with differentiation medium. EBs were microscopically examined for contracting cardiomyocytes on days 8–12.
Immunofluorescence and confocal microscopy
EBs treated with NRG-1β or a vehicle control (day 0~2) were plated at day 4 on glass coverslip culture chambers coated with 1% gelatin. At day 10, EBs were fixed in 5% formaldehyde at room temperature (23°C) for 30 min, and then permeabilized with 0.2%Triton X-100 in PBS. After blocking with 1 mg/ml BSA in PBS, cells were incubated with mouse monoclonal anti-α-actinin (Sigma) or mouse anti-cTnT (cardiac troponin T; Santa Cruz Biotechnology) antibodies at concentrations recommended by the manufacturers. After overnight incubation, cells were washed several times with PBS and then incubated with Alexa Fluor™ 488-conjugated rabbit anti-(mouse IgG) (Molecular Probes) and 5 µM DAPI. Immunofluorescence images were obtained using both a Leica inverted microscope (× 10) and a Zeiss inverted LSM 510 confocal microscope (× 40).
qPCR (quantitative real-time PCR)
CGR8 cells were harvested on days 0, 2, 3, 4, 6, 8, 10 and 12 of EB differentiation and stored at −80°C in the cell lysis buffer RLT (Qiagen). Three independent samples were collected for each time point studied. Total RNA was extracted using the RNeasy Mini kit according to the manufacturer’s instructions and treated with RNase-free DNase I (Qiagen). First-strand cDNA was synthesized with the SuperScript III First-Strand Synthesis SuperMix for qPCR (Invitrogen). Using cDNA as the template, TaqMan real-time PCR assays were performed in triplicate using the ABI Prism 7900 HT sequence detection system (Applied Biosystems) according to the manufacturer’s instructions. Data were normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase), and the levels of gene expression were normalized to that of day 0 DMSO-treated cells. The following TaqMan probe and primer sets (Applied Biosystems) were used: nkx2.5 (Mm00657783_m1), myh6 (Mm00440354_m1), brachyury T (Mm00436877_m1) gapdh (Mm99999915_g1), erbB2 (Mm00658541_m1), erbB3 (Mm01159990_g1) and erbB4 (Mm01256793_m1).
Gene expression microarrays
CGR8 cells were harvested at day 2 of EB differentiation and total RNA was extracted as described above. RNA quality measurements using a 2100 Bioanalyzer (Agilent) and further sample processing were performed in the GSR (Genome Sciences Resource) core at Vanderbilt University. Processed and labelled samples were subsequently hybridized to the mouse Gene 1.0 ST whole-transcriptome array (Affymetrix). Three biological replicates were used for each sample type (cells treated with either 50 ng/ml NRG-1β or a water vehicle), for a total of six arrays. Raw data files were RMA (robust multi-chip average) normalized followed by statistical analysis using Partek Genomics Suite version 6.6 [26]. Quality assessment was performed based on Affymetrix internal controls, box whisker plots, histograms, PCA (principal components analysis) and the S.D. of biological replicates. Following normalization and quality assessment, Partek was used to perform pairwise comparisons of average group values and one-way ANOVA. Only transcripts that resulted in a fold-change of at least 1.5 and a multiple hypothesis [B-H (Benjamini–Hochberg)]-corrected P value of less than 0.05 were considered as statistically significantly.
Gene functions, as shown in the Supplementary Online Data (http://www.biochemj.org/bj/458/bj4580335add.htm), were determined using publicly available records of NCBI Entrez Gene, Stanford SOURCE, Aceview, and the PubMed databases. Statistical analyses (including correction for multiple hypothesis testing) for identification of overrepresented ontologies and functions were performed using IPA (Ingenuity Pathway Analysis) software (Ingenuity Systems).
A putative NRG-responsive signalling pathway was inferred from the gene expression results, qPCR, Western blot analysis, siRNA experiments and a variety of publically available data resources using the 3D pathway builder drawing tool, available online via the Protein Lounge (http://www.proteinlounge.com/PathwayBuilder.aspx). A combination of microarray and other experimental results, literature searches, online gene database information and functional analysis results was used to construct the pathway based on known signalling pathways and reported research findings.
Western blotting
EB protein lysates were collected on day 10, separated by SDS/PAGE (10% gel) and transferred on to PVDF membranes. Mouse cTnT expression was detected using the Odyssey system (Li-Cor Bioscience) following incubation with a mouse anticTnT antibody (1:200 dilution) and IRDye 800CW-conjugated goat anti-(mouse IgG) (1:5000 dilution; Li-Cor Bioscience). Rabbit anti-phospho-CREB (Ser133) antibody (1:1000 dilution; Cell Signaling Technology) and mouse anti-CREB antibody (1:1000 dilution; Cell Signaling Technology) were used to detect phospho-CREB and total CREB respectively. Mouse α-tubulin antibody (1:2000 dilution; Abcam) was used as a loading control.
ErbB3 and CREB knockdown
CGR8 cells were transfected with siRNA ErbB3 (Santa Cruz Biotechnology) or siRNA CREB (Cell Signaling Technology) as well as their corresponding siRNA negative controls by following the manufacturer’s instructions. After overnight incubation, ESCs were subjected to differentiation as described above. Protein extracts at day 2 were subjected to Western blotting and RNA from EBs at day 10 were then collected, purified and subjected to qPCR.
Statistical analysis
All values are expressed as means ± S.E.M. for at least three independent experiments. Comparison of means was conducted using Student’s t test and results were considered statistically significant at P <0.05.
RESULTS
NRG-1β induces two waves of cardiomyogenesis in mESCs
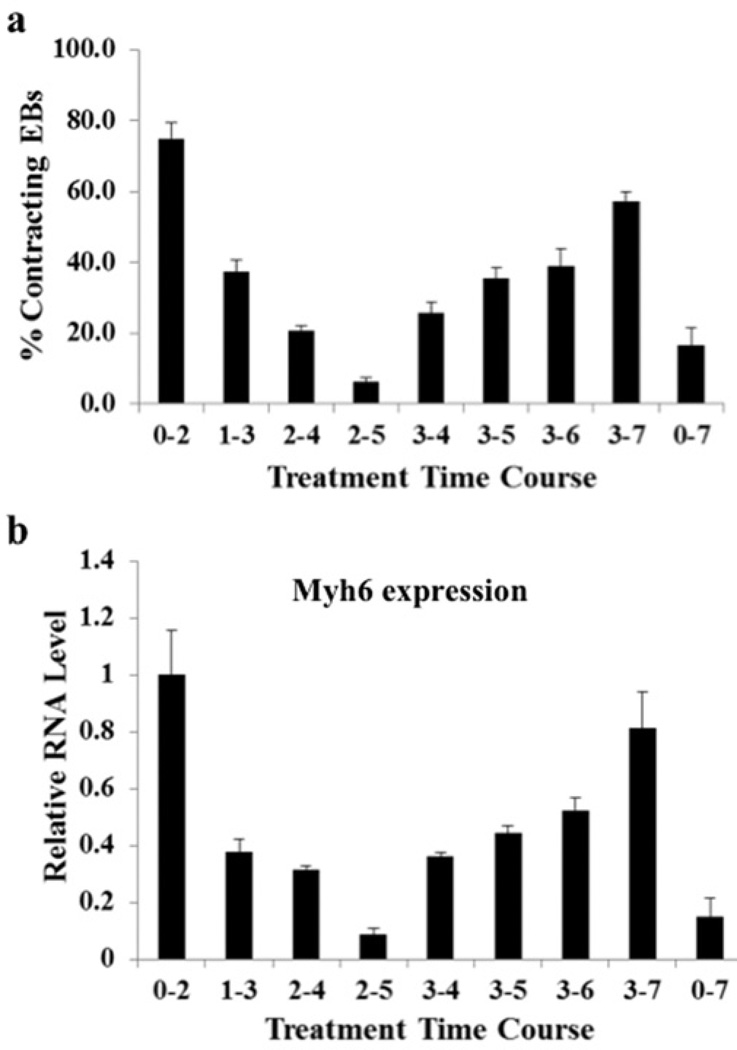
To assess the effect of NRG-1β on mESC cardiomyogenesis in a quantitative manner, we utilized our previously reported method of forming EBs in 96-well microtitre plates [1]. We administered 50 ng/ml NRG-1β at various time intervals during EB differentiation. PBS vehicle control typically resulted in approximately 4–6% contracting EBs at day 10 in all scenarios. NRG-1β treatment from day 0 to 2 led to 75% contracting EBs at day 10, whereas the contracting EBs dropped significantly to 37.5% when NRG-1β was administrated in mESCs during day 1–3. Interestingly, NRG-1β treatment from day 2 to 5 only resulted in approximately 6%contracting EBs, whereas treatment from day 3 to 5 led to 35% contracting EBs, implying that treatment during day 2–3 may have a detrimental effect on ESC cardiomyogenesis. Consistent with previous reports that NRG-1β treatment during later stage induction, day 3–7, resulted in robust ESC cardiac formation [23–25], our quantification method showed contracting EBs were elevated to approximately 57% during this time frame. Similar cardiac induction was also observed in R1 ESCs (results not shown), suggesting that the cardiomyogenic effects of NRG-1β were not cell line-restricted. In summary, our initial observation indicated that NRG-1β can induce two waves of cardiac formation in mESCs at two distinct time windows (Figure 1a and Supplementary Movie S1 at http://www.biochemj.org/bj/458/bj4580335add.htm). To confirm further our finding, we formed EBs from CGR8 ESCs using the hanging-drop method on day 0 and administrated 50 ng/ml NRG-1β at various time intervals. RNA samples were then collected on day 10 to examine cardiac marker myosin heavy chain [Myh6 (myosin, heavy polypeptide 6, cardiac muscle, α)] gene expression. The qPCR results closely mirrored the contracting EB counting, and confirmed that NRG-1β treatment during day 0–2 and day 3–7 significantly elevated Myh6 expression at day 10 (Figure 1b). As early NRG-1β treatment (day 0–2) induced a greater cardiac formation than later treatment (day 3–7) as reported previously [24,25], we focused the present study on the early time window of NRG-1β cardiomyogenesis in mESCs.
Figure 1. Recombinant NRG-1β promotes formation of beating EBs at critical time windows.
(a) Time windows for cardiac induction in mESCs by NRG-1β treatment were systematically examined from day 0 to 2, day 1 to 3, day 2 to 4, day 2 to 5, day 3 to 4, day 3 to 5, day 3 to 6, day 3 to 7 and day 0 to 7. The percentages of contracting EBs at day 10 of differentiation demonstrated that NRG-1β treatment during day 0–2 and day 3–7 can induce two waves of cardiac differentiation in mESCs. Results were obtained from at least 48 EBs for each time point in three individual experiments (P <0.001). (b) qPCR analysis confirmed that NRG-1β treatment during day 0–2 and day 3–7 significantly induced the cardiac marker Myh6 gene expression in mESCs. CGR8 ESCs were treated with 50 ng/ml NRG-1β at the different time intervals, and RNA samples collected on day 10 were subjected to qPCR to determine Myh6 gene expression. Myh6 RNA expression induced by NRG-1β at day 0~2 was set to 1, and the relative RNA expression level was calculated by normalizing Myh6 RNA expression at each specific NRG-1β treatment time window with its RNA expression induced by NRG-1β at day 0~2. qPCR results were obtained from two independent experiments.
Characterization of NRG-1β-induced cardiomyocytes
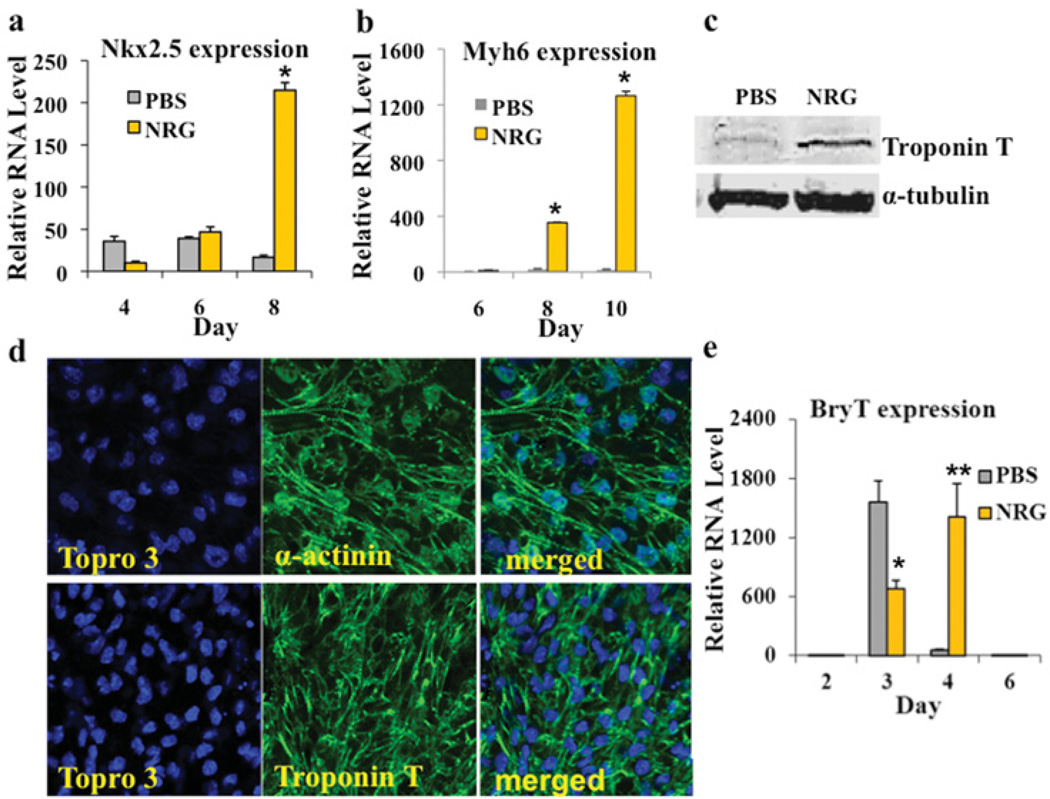
To gauge further the extent of cardiomyogenesis, we administrated 50 ng/ml NRG-1β to CGR8 cells when hanging drops were prepared at day 0. At day 2 of EB formation, NRG-1β was washed out and EBs were resuspended in differentiation medium for an additional 2 days before they were transferred on to gelatin-coated six-well plates (at day 4). The vehicle PBS was used as a negative control. At day 10, CGR8 cells treated with NRG-1β formed large synchronously beating areas, whereas relatively fewer beating foci were observed in the vehicle controls. Consistent with this finding, NRG-1β-treated CGR8 cells from day 0 to 2 of differentiation led to large increases in the expression of several cardiac genes as measured by qPCR. For instance, NRG-1β treatment resulted an approximate 12-fold increase in the expression of the cardiac marker Nkx2.5 (NK2 homeobox 5) at day 8 and an 83-fold increase in the expression of cardiac Myh6 at day 10, when compared with the vehicle controls (Figures 2a and 2b). In addition, Western blotting indicated sarcomeric troponin T protein levels were highly up-regulated after NRG-1β treatment in contrast with the vehicle controls (Figure 2c). Increased cardiac formation was confirmed further by immunofluorescence for cardiac proteins α-actinin and cTnT (Figure 2d).
Figure 2. NRG-1β treatment from day 0 to 2 of ESC differentiation strongly induces cardiomyogenesis.
(a and b) NRG-1β treatment from day 0 to 2 increases the expression of the cardiac markers Nkx2.5 and Myh6. The qPCR results represent the relative expression normalized to that of PBS vehicle-treated cells at day 0. Measurements were obtained from at least three independent experiments for each time point (*P <0.01). (c) Western blotting showing induction of cTnT protein in NRG-1β-treated mESCs on day 10 in comparison with the PBS-treated controls. An antibody against α-tubulin was used as a loading control. (d) mESCs treated with NRG-1β differentiated to cardiomyocytes that expressed the sarcomere proteins α-actinin (upper panels) and cTnT (lower panels). Confocal images were taken using a Zeiss inverted LSM 510 confocal microscope (× 40). Topro 3, TO-PRO-3™ (Molecular Probes). (e) NRG-1β treatment from day 0 to 2 down-regulated BryT+ at day 3 (*P <0.01), but led to a higher expression of BryT+ at day 4, in comparison with vehicle controls (**P <0.05). The relative RNA expression level of each marker was calculated by normalizing the marker RNA expression at each specific day with the RNA expression at day 0.
Bry-T (brachyury T) is an early mesoderm marker whose expression is typically initiated at day 3 in mESCs. Previous studies have demonstrated that two distinct pools of Bry-T+ mesoderm cells exist sequentially during mESC differentiation. Bry-T+ mesoderm cells isolated at approximately day 3.25 are committed to become haemangioblasts and Bry-T+ mesoderm cells isolated around day 4.25 are committed to cardiovascular lineages including cardiomyocytes [1,27]. To understand how NRG-1β treatment from day 0 to 2 induces cardiac cell differentiation of mESCs, we extracted mRNA from EB samples treated with NRG-1β or the vehicle control at day 3 and day 4 respectively. The qPCR result indicated that early NRG-1β treatment (day 0~2) suppressed Bry-T expression at day 3 and up-regulated Bry-T expression at day 4, suggesting that the early pre-mesoderm NRG-1β treatment promotes cardiac formation in mESCs by suppressing the haemangioblast-committed mesoderm and up-regulating the cardiac-committed mesoderm pool (Figure 2e).
NRG-1β-induced mESCs cardiomyogenesis at day 0–2 requires ErbB3
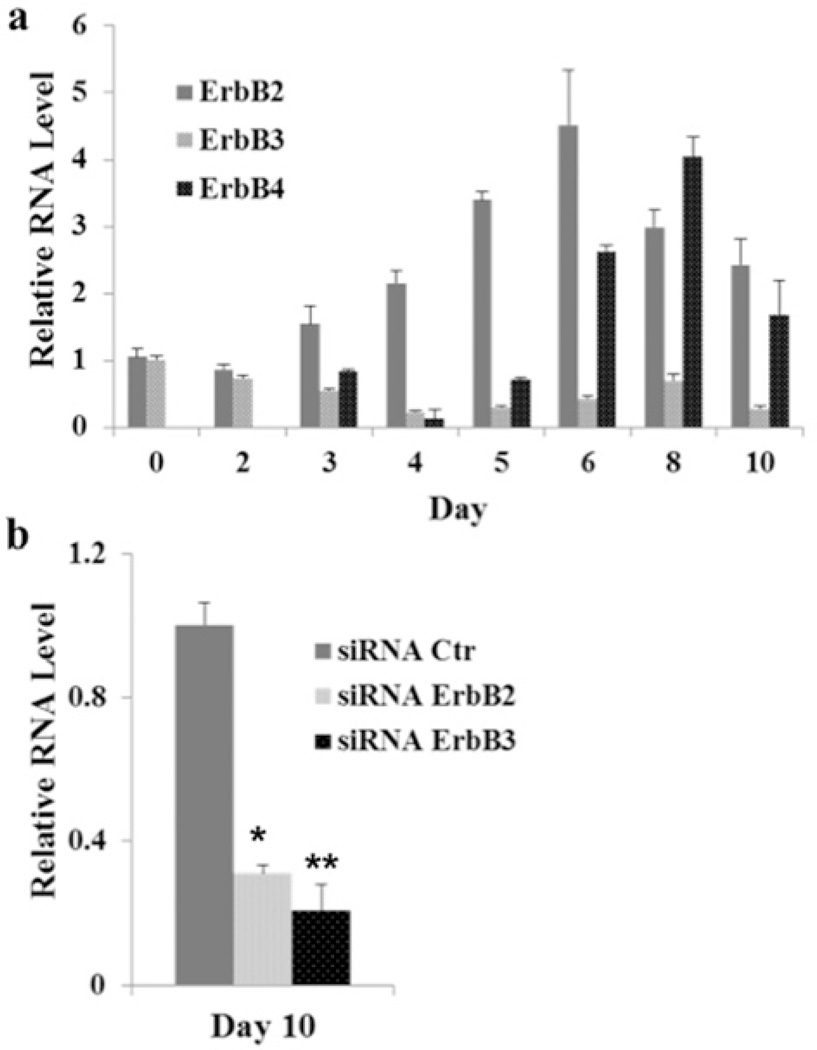
As the biological effects of NRG-1β are mediated through tyrosine kinase ErbB receptors, including ErbB2, ErbB3 and ErbB4, we quantified the mRNA expression levels of the ErbB receptors during the differentiation of CGR8 cells (Figure 3a). Interestingly, ErbB2 and ErbB3 were expressed during the first 48 h of mESC differentiation, but ErbB4 was not detected until day 3, suggesting that NRG-1β induces mESC cardiomyogenesis during the early time window (day 0–2) via the ErbB3 receptor.
Figure 3. ErbB3 receptor is required for NRG-1β-induced mESC cardiomyogenesis.
(a) mRNA expression of the ErbB receptors was quantified during mESC differentiation. mRNA expression of ErbB2, ErbB3 and ErbB4 was measured by qPCR during differentiation at day 0, 2, 4, 6, 8 and 10. Measurements were obtained from at least three independent experiments for each time point. (b) Transient knock down of ErbB3 or ErbB2 with siRNA at the early differentiation stage significantly blunted the NRG-1β-induced cardiac formation in mESCs at day 10. The qPCR results were obtained from at least three independent experiments (*P <0.01 and **P <0.05).
To test this hypothesis, we transfected CGR8 cells with ErbB3 siRNA overnight at day −1, and then initiated NRG-1β-induced differentiation of EBs in hanging droplets from day 0 to 2. EBs were harvested at day 2 and day 10 for RNA isolation. The qPCR results showed that ErbB3 was effectively knocked down by siRNA (results not shown), and cardiac gene expression of Myh6 at day 10 was dramatically down-regulated in ErbB3 siRNA-treated CGR8 cells (Figure 3b). In addition, as ErbB3 lacks an active kinase domain, and it has to form a heterodimer with ErbB2 to exert the biological function of NRG-1β, we knocked down ErbB2 further with siRNA in the presence of NRG-1β. The qPCR results show that ErbB2 knockdown reduced significantly NRG-1β-induced Myh6 expression at day 10 (Figure 3b). Taken together, these results indicate that NRG-1β induces early mESC cardiomyogenesis after 48 h of treatment via the ErbB3 receptor. This is in contrast with NRG-1β-induced cardiac induction of mESCs at the later differentiation stage treatment, which occurs via the ErbB4 receptor [23–25].
NRG-1β promotes global gene expression changes important for cardiomyogenesis
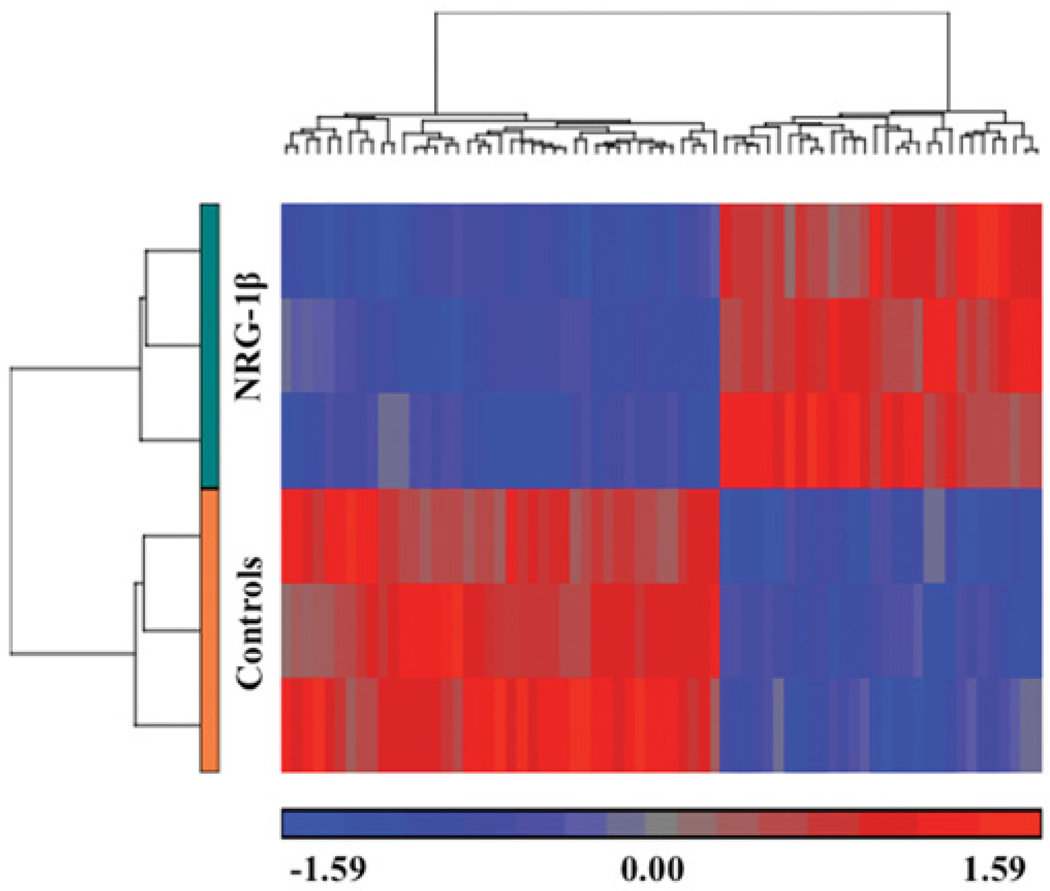
To assess transcriptional responses to NRG-1β that might account for the observed cardiomyogenesis, microarrays were used to measure early gene expression changes in NRG-1β-treated CGR8 cells compared with the vehicle-treated controls. NRG-1β treatment resulted in the alteration of 1058 probes representing 767 characterized genes (Figure 4 and Supplementary Online Data). Although the fold differences were generally subtle (ranging from 10% to 60% induction), functional analysis indicated that these genes were highly related to one another and thus might represent an early indication of the transcriptional process involved in cardiomyogenesis. On the basis of this functional analysis, the most statistically significant biological processes were embryonic development (B-H P value = 1.9 × 10−5), cell morphogenesis involved in cell differentiation (B-H P value = 1.4 × 10−4), and regulation of cell adhesion and migration (B-H P value = 3.4 × 10−4).
Figure 4. Hierarchical clustering of genes differentially expressed between the NRG-1β- and vehicle-treated mESCs.
Hierarchical clustering of the 125 most significantly differential probes (at least 1.5-fold change; P <0.05) between vehicle- and NRG-1β-treated mESCs at day 2. Bright red, bright blue and grey indicate the highest, lowest and median normalized signal values respectively as indicated below the heat map. For each sample type, control (orange) and NRG treatment (teal) as labelled, there were three biological replicates. Columns represent individual genes. Rows and vertical dendrograms indicate individual sample clustering.
Signalling pathway analysis identified several important pathways involved in self-renewal and differentiation of ESCs treated with NRG-1β. Differential genes in these categories were researched further to identify interrelated associations with each other and in known signalling cascades. When up- and down-regulated genes were displayed based on their known locations within these pathways, two major processes were readily apparent. Multiple genes encoding proteins that promote stem cell renewal were down-regulated in ESCs in response to NRG-1β treatment (Supplementary Figure S1, shown in green, at http://www.biochemj.org/bj/458/bj4580335add.htm). Conversely, genes important for cardiomyogenesis, cytoskeletal stability and contraction were generally up-regulated (Supplementary Figure S1, shown in red).
Activation of CREB is involved in mESC cardiomyogenesis
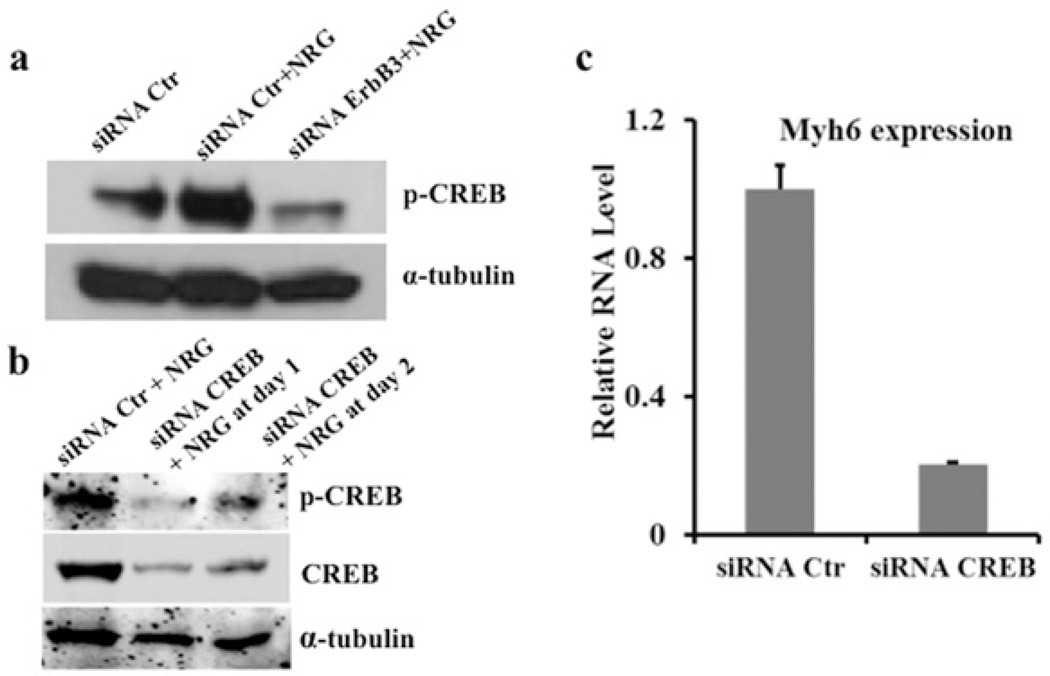
NRG-1β treatment induced up-regulation of the genes that encode the catalytic and regulatory subunits of PI3K (phosphoinositide 3-kinase) (Pi3kcg and Pi3kr1 respectively), which is a known component of the ErbB signalling pathway leading to activation of Akt [also known as PKB (protein kinase B)]. Akt has been shown to directly activate the transcription factor CREB by phosphorylation at Ser133, and phosphorylated CREB regulates a variety of cellular processes related to cell fate, including proliferation, differentiation and survival [28–31]. More recently, CREB was identified as a regulatory target of Akt that is important for the differentiation of pancreatic cells [32]. We therefore probed NRG-1β-treated ESC protein lysates at day 2 for CREB phosphorylated at Ser133 (phospho-CREB). Immunoblots demonstrated that NRG-1β increased phospho-CREB levels, whereas knocking down ErbB3 significantly attenuated phospho-CREB levels at day 2, leading to a dramatic reduction in NRG-1β-induced Myh6 expression at day 10 (Figures 3b and 5a). These results suggest that NRG-1β/ErbB3 signalling in the context of early cardiomyogenesis may involve activation of the transcription factor CREB.
Figure 5. Activation of CREB is involved in NRG-1β-induced cardiomyogenesis of mESC differentiation.
NRG-1β treatment from day 0 to 2 significantly induced active phospho-CREB (p-CREB) and transient knockdown of ErbB3 with siRNA dramatically attenuated active phospho-CREB in the presence of NRG-1β (a). CREB siRNA effectively reduced CREB expression and levels of phospho-CREB during the initial 48 h of mESC treatment with 50 ng/ml NRG-1β (b) and prevented cardiomyogenesis as indicated by reduced Myh6 expression on day 10 (c). Myh6 RNA expression in the siRNA control (Ctr) samples was set to 1, and the relative RNA expression level was calculated by normalizing Myh6 RNA expression with its RNA expression in the siRNA control samples. The qPCR results were obtained from two independent experiments.
CREB knockdown significantly reduces NRG-1β-induced ESC cardiomyogenesis
Because the phospho-CREB level is dramatically elevated upon NRG-1β induction at the early pre-mesoderm stage during ESC cardiomyogenesis, we next examined whether CREB activation is indispensable for cardiomyocyte formation. We transiently reduced CREB using siRNA during the early stages of ESC differentiation in the presence of NRG-1β, and then examined the expression of the cardiac marker Myh6 on day 10. Western blotting confirmed that CREB was effectively reduced by siRNA during the first 48 h of differentiation (Figure 5b), and the qPCR results demonstrated that Myh6 was significantly down-regulated on day 10 when CREB expression was reduced (Figure 5c), suggesting that CREB activation was required for NRG-1β/ErbB3-induced ESC cardiomyogenesis.
DISCUSSION
NRG-1β/ErbB signalling plays critical roles in cardiac development, homoeostasis and heart repair. Previously it has been shown that NRG-1β can induce the cardiomyogenesis of mESCs at late-stage differentiation by signalling through the ErbB4 receptor [24,25]. In the present study, we add to this literature by demonstrating that NRG-1β can actually induce two waves of cardiomyocyte formation from mESCs at two distinct treatment time windows, namely a previously unidentified early pre-mesoderm stage (day 0–2) and a later differentiation stage (day 3–7). In contrast with the later stage of differentiation in which NRG-1β signals through the ErbB4 receptor for cardiomyogenesis of mESCs, we found that NRG-1β can induce substantial cardiomyocyte formation in mESCs via the ErbB3/ErbB2 receptors at the early pre-mesoderm stage of treatment, presumably by suppressing haemangioblast-committed mesoderm cells and up-regulating the cardiac-committed mesoderm pool [1,27].
The observed NRG-1β/ErbB3 signalling-mediated induction of cardiomyogenesis is an interesting and, to some extent, unexpected finding. Previous studies demonstrated that ErbB3 is not absolutely required for early cardiogenesis in mouse embryos including formation of ventricular myocytes [13], and NRG-1β signalling in fully differentiated cardiomyocytes occurs via ErbB2/ErbB4 heterodimers [23–25]. These facts, together with the report that adult cardiomyocytes do not typically express ErbB3 at appreciable levels [33], raise an interesting question: whether NRG-1β/ErbB3 signalling at the pre-mesoderm stage can induce context-dependent pre-cardiac mesoderm progenitor cell formation. This question warrants further investigation as it may lead to identification of a novel population of progenitor cells for potential cell therapies.
Moreover, CREB activation by phosphorylation at Ser133 (phospho-CREB) is known to protect the myocardium from apoptosis by up-regulating the anti-apoptotic proteins of the Bcl-2 family [34–36]. However, the role of activated CREB in cardiac differentiation has not been reported. On the basis of our microarray analysis and subsequent verification experiments, we found that NRG-1β/ErbB3 signalling induced CREB phosphorylation and increased mESC development to cardiomyocytes. In addition, transient knockdown of CREB with siRNA during early differentiation significantly reduced NRG-1β-induced cardiomyogenesis in mESCs, suggesting that CREB activation by NRG-1β/ErbB3 signalling is indispensable during ESC cardiomyogenesis. Further elucidation of the role of phospho-CREB in ESC cardiac commitment and dissection of the molecular mechanism of NRG-1β/ErbB3/phospho-CREB regulation may uncover new biological insights relevant to cardiomyogenesis.
Previously NRG-1β/ErbB4 signalling was reported to regulate cardiac subtype specification in hESCs (human ESCs) [37]. This raises an interesting question of whether subtypes of cardiomyocytes (i.e. pacemaker nodal cells and atrial and ventricular cardiomyocytes) generated through NRG-1β/ErbB3 signalling at the early pre-mesoderm stage are the same as those generated through NRG-1β/ErbB4 signalling at the later differentiation stage. Further investigation to address this question in ESCs would provide important insights into cardiac subtype cell development in early embryonic heart formation, providing practical approaches to derive homogenous cardiomyocyte populations for future clinical translation of stem cell-based therapy.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Holly M. Smith for technical support. Images of confocal microscopy were performed through the use of the VUMC Cell Imaging Shared Resource.
FUNDING
This work was supported by the National Institutes of Health [grant number U01 HL100398] and a seed fund of the College of Veterinary Medicine at Western University of Health Sciences (to J.H).
Abbreviations
- B-H
Benjamini–Hochberg
- Bry-T
brachyury T
- CREB
cAMP-response-element-binding protein
- cTnT
cardiac troponin T
- EB
embryonic body
- ESC
embryonic stem cell
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- mESC
mouse ESC
- Myh6
myosin, heavy polypeptide 6, cardiac muscle, α
- Nkx2.5
NK2 homeobox 5
- NRG
neuregulin
- PI3K
phosphoinositide 3-kinase
- PSC
pluripotent stem cell
- qPCR
quantitative real-time PCR
Footnotes
AUTHOR CONTRIBUTION
Jijun Hao, Cristi Galindo and Truc-Linh Tran performed the experiments; Jijun Hao and Douglas Sawyer designed the experiments. Jijun Hao, Cristi Galindo and Douglas Sawyer wrote the paper.
REFERENCES
- 1.Hao JJ, Daleo MA, Murphy CK, Yu PB, Ho JN, Hu JY, Peterson RT, Hatzopoulos AK, Hong CC. Dorsomorphin, a selective small molecule inhibitor of BMP signaling, promotes cardiomyogenesis in embryonic stem cells. PLoS ONE. 2008;3:e2904. doi: 10.1371/journal.pone.0002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ao A, Hao JJ, Hopkins CR, Hong CC. DMH1, a novel BMP small molecule inhibitor, increases cardiomyocyte progenitors and promotes cardiac differentiation in mouse embryonic stem cells. PLoS ONE. 2012;7:e41627. doi: 10.1371/journal.pone.0041627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang HM, Hao JJ, Hong CC. Cardiac induction of embryonic stem cells by a small molecule inhibitor of Wnt/β-catenin signaling. ACS Chem. Biol. 2011;6:192–197. doi: 10.1021/cb100323z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brutsaert DL. Cardiac endothelial—myocardial signaling: its role in cardiac growth, contractile performance, and rhythmicity. Physiol. Rev. 2003;83:59–115. doi: 10.1152/physrev.00017.2002. [DOI] [PubMed] [Google Scholar]
- 5.Cote GM, Miller TA, Lebrasseur NK, Kuramochi Y, Sawyer DB. Neuregulin-1α and β isoform expression in cardiac microvascular endothelial cells and function in cardiac myocytes in vitro. Exp. Cell Res. 2005;311:135–146. doi: 10.1016/j.yexcr.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 6.Gassmann M, Casagranda F, Orioli D, Simon H, Lai C, Klein R, Lemke G. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature. 1995;378:390–394. doi: 10.1038/378390a0. [DOI] [PubMed] [Google Scholar]
- 7.Meyer D, Birchmeier C. Multiple essential functions of neuregulin in development. Nature. 1995;378:386–390. doi: 10.1038/378386a0. [DOI] [PubMed] [Google Scholar]
- 8.Lee KF, Simon H, Chen H, Bates B, Hung MC, Hauser C. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature. 1995;378:394–398. doi: 10.1038/378394a0. [DOI] [PubMed] [Google Scholar]
- 9.Rentschler S, Zander J, Meyers K, France D, Levine R, Porter G, Rivkees SA, Morley GE, Fishman GI. Neuregulin-1 promotes formation of the murine cardiac conduction system. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10464–10469. doi: 10.1073/pnas.162301699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeisberg EM, Tarnavski O, Zeisberg M, Dorfman AL, McMullen JR, Gustafsson E, Chandraker A, Yuan X, Pu WT, Roberts AB, et al. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat. Med. 2007;13:952–961. doi: 10.1038/nm1613. [DOI] [PubMed] [Google Scholar]
- 11.Pentassuglia L, Sawyer DB. The role of Neuregulin-1β/ErbB signaling in the heart. Exp. Cell Res. 2009;315:627–637. doi: 10.1016/j.yexcr.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee KF, Simon H, Chen H, Bates B, Hung MC, Hauser C. Requirement for neuregulin receptor Erbb2 in neural and cardiac Development. Nature. 1995;378:394–398. doi: 10.1038/378394a0. [DOI] [PubMed] [Google Scholar]
- 13.Erickson SL, O’Shea KS, Ghaboosi N, Loverro L, Frantz G, Bauer M, Lu LH, Moore MW. ErbB3 is required for normal cerebellar and cardiac development: a comparison with ErbB2- and heregulin-deficient mice. Development. 1997;124:4999–5011. doi: 10.1242/dev.124.24.4999. [DOI] [PubMed] [Google Scholar]
- 14.Zhao YY, Sawyer DR, Baliga RR, Opel DJ, Han X, Marchionni MA, Kelly RA. Neuregulins promote survival and growth of cardiac myocytes. Persistence of ErbB2 and ErbB4 expression in neonatal and adult ventricular myocytes. J. Biol. Chem. 1998;273:10261–10269. doi: 10.1074/jbc.273.17.10261. [DOI] [PubMed] [Google Scholar]
- 15.Fukazawa R, Miller TA, Kuramochi Y, Frantz S, Kim YD, Marchionni MA, Kelly RA, Sawyer DB. Neuregulin-1 protects ventricular myocytes from anthracycline-induced apoptosis via erbB4-dependent activation of PI3-kinase/Akt. J. Mol. Cell. Cardiol. 2003;35:1473–1479. doi: 10.1016/j.yjmcc.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 16.Baliga RR, Pimental DR, Zhao YY, Simmons WW, Marchionni MA, Sawyer DB, Kelly RA. NRG-1-induced cardiomyocyte hypertrophy. Role of PI-3-kinase, p70(S6K), and MEK-MAPK-RSK. Am. J. Physiol. 1999;277:H2026–H2037. doi: 10.1152/ajpheart.1999.277.5.H2026. [DOI] [PubMed] [Google Scholar]
- 17.Kuramochi Y, Cote GM, Guo X, Lebrasseur NK, Cui L, Liao R, Sawyer DB. Cardiac endothelial cells regulate reactive oxygen species-induced cardiomyocyte apoptosis through neuregulin-1β/erbB4 signaling. J. Biol. Chem. 2004;279:51141–51147. doi: 10.1074/jbc.M408662200. [DOI] [PubMed] [Google Scholar]
- 18.Lemmens K, Fransen P, Sys SU, Brutsaert DL, De Keulenaer GW. Neuregulin-1 induces a negative inotropic effect in cardiac muscle: role of nitric oxide synthase. Circulation. 2004;109:324–326. doi: 10.1161/01.CIR.0000114521.88547.5E. [DOI] [PubMed] [Google Scholar]
- 19.Bersell K, Arab S, Haring B, Kuhn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138:257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- 20.Wadugu B, Kuhn B. The role of neuregulin/ErbB2/ErbB4 signaling in the heart with special focus on effects on cardiomyocyte proliferation. Am. J. Physiol. Heart Circ. Physiol. 2012;302:H2139–H2147. doi: 10.1152/ajpheart.00063.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jabbour A, Hayward CS, Keogh AM, Kotlyar E, McCrohon JA, England JF, Amor R, Liu X, Li XY, Zhou MD, et al. Parenteral administration of recombinant human neuregulin-1 to patients with stable chronic heart failure produces favourable acute and chronic haemodynamic responses. Eur. J. Heart Fail. 2011;13:83–92. doi: 10.1093/eurjhf/hfq152. [DOI] [PubMed] [Google Scholar]
- 22.Gao R, Zhang J, Cheng L, Wu X, Dong W, Yang X, Li T, Liu X, Xu Y, Li X, Zhou M. A Phase II, randomized, double-blind, multicenter, based on standard therapy, placebo-controlled study of the efficacy and safety of recombinant human neuregulin-1 in patients with chronic heart failure. J. Am. Coll. Cardiol. 2010;55:1907–1914. doi: 10.1016/j.jacc.2009.12.044. [DOI] [PubMed] [Google Scholar]
- 23.Suk Kim H, Hidaka K, Morisaki T. Expression of ErbB receptors in ES cell-derived cardiomyocytes. Biochem. Biophys. Res. Commun. 2003;309:241–246. doi: 10.1016/s0006-291x(03)01521-3. [DOI] [PubMed] [Google Scholar]
- 24.Sun M, Yan X, Bian Y, Caggiano AO, Morgan JP. Improving murine embryonic stem cell differentiation into cardiomyocytes with neuregulin-1: differential expression of microRNA. Am. J. Physiol. Cell. Physiol. 2011;301:C21–C30. doi: 10.1152/ajpcell.00141.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z, Xu G, Wu Y, Guan Y, Cui L, Lei X, Zhang J, Mou L, Sun B, Dai Q. Neuregulin-1 enhances differentiation of cardiomyocytes from embryonic stem cells. Med. Biol. Eng. Comput. 2009;47:41–48. doi: 10.1007/s11517-008-0383-2. [DOI] [PubMed] [Google Scholar]
- 26.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 27.Kattman SJ, Adler ED, Keller GM. Specification of multipotential cardiovascular progenitor cells during embryonic stem cell differentiation and embryonic development. Trends Cardiovasc. Med. 2007;17:240–246. doi: 10.1016/j.tcm.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 28.Du KY, Montminy M. CREB is a regulatory target for the protein kinase Akt/PKB. J. Biol. Chem. 1998;273:32377–32379. doi: 10.1074/jbc.273.49.32377. [DOI] [PubMed] [Google Scholar]
- 29.Giachino C, De Marchis S, Giampietro C, Parlato R, Perroteau I, Schutz G, Fasolo A, Peretto P. cAMP response element-binding protein regulates differentiation and survival of newborn neurons in the olfactory bulb. J. Neurosci. 2005;25:10105–10118. doi: 10.1523/JNEUROSCI.3512-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng JC, Kinjo K, Judelson DR, Chang J, Wu WS, Schmid I, Shankar DB, Kasahara N, Stripecke R, Bhatia R, et al. CREB is a critical regulator of normal hematopoiesis and leukemogenesis. Blood. 2008;111:1182–1192. doi: 10.1182/blood-2007-04-083600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bleckmann SC, Blendy JA, Rudolph D, Monaghan AP, Schmid W, Schutz G. Activating transcription factor 1 and CREB are important for cell survival during early mouse development. Mol. Cell. Biol. 2002;22:1919–1925. doi: 10.1128/MCB.22.6.1919-1925.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li XY, Zhan XR, Liu XM, Wang XC. CREB is a regulatory target for the protein kinase Akt/PKB in the differentiation of pancreatic ductal cells into islet β-cells mediated by hepatocyte growth factor. Biochem. Biophys Res. Commun. 2011;404:711–716. doi: 10.1016/j.bbrc.2010.12.048. [DOI] [PubMed] [Google Scholar]
- 33.Zhao YY, Sawyer DR, Baliga RR, Opel DJ, Han XQ, Marchionni MA, Kelly RA. Neuregulins promote survival and growth of cardiac myocytes: persistence of ErbB2 and ErbB4 expression in neonatal and adult ventricular myocytes. J. Biol. Chem. 1998;273:10261–10269. doi: 10.1074/jbc.273.17.10261. [DOI] [PubMed] [Google Scholar]
- 34.Fentzke RC, Korcarz CE, Lang RM, Lin H, Leiden JM. Dilated cardiomyopathy in transgenic mice expressing a dominant-negative CREB transcription factor in the heart. J. Clin. Invest. 1998;101:2415–2426. doi: 10.1172/JCI2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng FJ, Jiao SM, Yu B. Picroside II protects cardiomyocytes from hypoxia/reoxygenation-induced apoptosis by activating the PI3K/Akt and CREB pathways. Int. J. Mol. Med. 2012;30:263–270. doi: 10.3892/ijmm.2012.987. [DOI] [PubMed] [Google Scholar]
- 36.Watson PA, Reusch JEB, McCune SA, Leinwand LA, Luckey SW, Konhilas JP, Brown DA, Chicco AJ, Sparagna GC, Long CS, Moore RL. Restoration of CREB function is linked to completion and stabilization of adaptive cardiac hypertrophy in response to exercise. Am. J. Physiol. Heart Circ. Physiol. 2007;293:H246–H259. doi: 10.1152/ajpheart.00734.2006. [DOI] [PubMed] [Google Scholar]
- 37.Zhu WZ, Xie Y, Moyes KW, Gold JD, Askari B, Laflamme MA. Neuregulin/ErbB signaling regulates cardiac subtype specification in differentiating human embryonic stem cells. Circ. Res. 2010;107:776–786. doi: 10.1161/CIRCRESAHA.110.223917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.