Abstract
G protein-gated inward rectifier K+ (GIRK) channels allow neurotransmitters, via G protein-coupled receptor stimulation, to control cellular electrical excitability. In cardiac and neuronal cells this control regulates heart rate and neural circuit activity. We present the 3.5 Å resolution crystal structure of the mammalian GIRK2 channel in complex with βγ G protein subunits, the central signaling complex that links G protein-coupled receptor stimulation to K+ channel activity. Short-range atomic and long-range electrostatic interactions stabilize four βγ G protein subunits at the interfaces between four K+ channel subunits, inducing a pre-open state of the channel. The pre-open state exhibits a conformation that is intermediate between the closed and constitutively active mutant, open conformations. The resultant structural picture is compatible with “membrane delimited” activation of GIRK channels by G proteins and the characteristic burst kinetics of channel gating. The structures also permit a conceptual understanding of how the signaling lipid PIP2 and intracellular Na+ ions participate in multi-ligand regulation of GIRK channels.
Introduction
In 1921 Otto Loewi established the existence of chemical synaptic transmission by showing that vagus nerve stimulation slows the heart rate through release of a chemical substance he called vagusstoff1, 2. Vagusstoff was later shown to be acetylcholine, the major neurotransmitter of the parasympathetic nervous system2, 3. Once released from the vagus nerve, acetylcholine binds to the m2 muscarinic receptor, a G protein-coupled receptor (GPCR) in heart cell membranes, and causes the release of G protein subunits Gα and Gβγ from the receptor’s intracellular surface4. The Gβγ subunits activate G protein-gated Inward Rectifier K+ (GIRK) channels, causing them to open5–10. Open GIRK channels drive the membrane voltage towards the resting (Nernst K+) potential, which slows the rate of membrane depolarization, as depicted (figure 1a). In atrial pacemaker cells of the heart, this directly decreases firing frequency and thus heart rate11. Isoforms of the GIRK channel also exist in neurons, which permit G protein-mediated regulation of neuronal electrical excitability12.
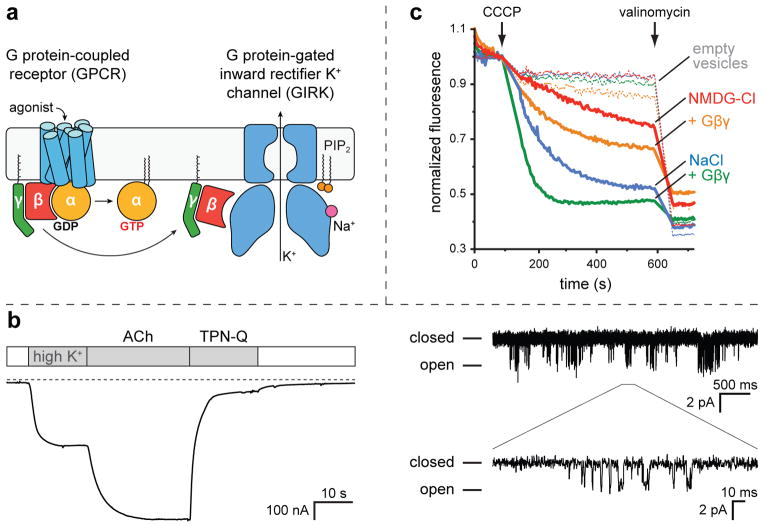
Figure 1. Function and crystal lattice arrangement of GIRK.
a) Schematic of GPCR activation of GIRK channels. Agonist binding to a GPCR promotes the exchange of GDP for GTP on a bound G protein. This causes the G protein to dissociate from the receptor. The Gα and Gβγ subunits subsequently dissociate from each other where they can then interact with effector proteins. Gβγ binding to the cytoplasmic domain of a GIRK channel in the presence of PIP2 causes the channel to open. GIRK channels are also activated by elevated levels of intracellular Na+ ions. b) Example of GPCR-activation of GIRK. The truncated GIRK construct used for crystallography was co-expressed with the M2 muscarinic receptor in Xenopus laevis oocytes. (left) Whole cell current was measured using two-electrode voltage-clamp while holding the cell at −60 mV. The white bars indicate a physiological extracellular solution, while the gray bars represent a solution containing 98 mM KCl. The application of 10 μM acetylcholine (ACh, a M2R agonist), or 1 μM of tertiapin-Q (TPN-Q, a specific GIRK2 blocker) is also indicated. The traces under the dashed line represent negative, inward currents. (right) Single channel recordings in the on-cell patch-clamp configuration. The patch pipette contained 96 mM KCl and 10 μM ACh. The patch was held at −100 mV. A closeup of one of the burst openings is shown on the bottom. c) Activation of purified GIRK channels reconstituted into lipid vesicles. Channel activity was monitored in the presence of either NMDG-Cl or NaCl using a fluorescence-based assay described in detail in the Methods section. Purified Gβγ was added to some of the samples in either the NMDG-Cl or NaCl buffers, as indicated. The initiation of K+ flux by the addition of the H+ ionophore CCCP is indicated. The addition of the K+ ionophore valinomycin to measure total flux capacity of the vesicles is also indicated. The dashed lines represent the same experimental conditions, except that the vesicles do not contain any GIRK.
For several decades electrophysiological and biochemical methods have been applied to understand how G protein subunits activate GIRK channels. Specific mutations on the Gβγ subunit13–17 and on the channel18, 19 were shown to alter G protein-mediated activation of GIRK channels. Biochemical and NMR studies identified components of both the G protein and channel that appear to interact with each other20–22. Together these studies point to a direct interaction between the G protein subunits and the channel to achieve channel activation. Here we present the crystal structure of a GIRK channel bound to Gβγ subunits, a key signaling complex in the G protein-mediated control of electrical excitability.
GIRK2 activation by G protein subunits
Our study addresses GIRK2 (Kir3.2), a neuronal GIRK channel that is able to function as a tetramer of identical subunits23. Activation of GIRK2, which from here on we refer to as GIRK, by GPCR stimulation is shown using an assay in which the m2 muscarinic GPCR is co-expressed together with GIRK channels in Xenopus oocytes24 (figure 1b, left). Initial replacement of Na+ by K+ in the extracellular solution causes some current to flow into the oocyte, measured using two-electrode voltage clamp. When acetylcholine is then applied, a larger inward K+ current is turned on. Inhibition of current by tertiapin-q, a bee venom toxin derivative, establishes the current as mediated by the GIRK channel, a fraction of which is active in the absence of acetylcholine25. The fraction of current activated by acetylcholine is variable, depending on the oocyte. Isolated membrane patches show the characteristic gating of single GIRK channels (figure 1b, right). These channels display “burst kinetics”, during which time an activated channel flickers rapidly between conducting (open) and non-conducting (closed) states, an interesting property that we will consider later. These electrophysiological recordings and other functional studies here were carried out with the identical construct used for crystallization and structural analysis. Hereafter we refer to this construct, which consists of residues 52–380, as the wild-type channel. We emphasize that removal of the disordered N- and C-termini does not appear to alter the functional properties of the channel in any of the electrophysiological and flux measurements we have made.
All studies of G protein-mediated GIRK channel activation to date have been carried out with native cells or with cell lines in which components were expressed heterologously (as in figure 1b). Because we now have in hand individual isolated components – namely the GIRK channel, Gβγ subunits, and the signaling lipid phosphatidylinositol 4,5 bisphosphate (PIP2) – we tested whether these alone (i.e. in the absence of other cellular components) are sufficient to produce a competent signaling complex. Using a flux assay in which the isolated components are reconstituted into synthetic lipid vesicles we find that they are indeed sufficient: baseline K+ flux observed in the absence of Gβγ is strongly enhanced in the presence of Gβγ (figure 1c). GIRK is also activated by intracellular Na+, which accounts for the greater flux observed in Na+ than in NMDG+26–29. However, even in the presence of Na+, Gβγ still causes significant enhancement of flux. These measurements with purified, reconstituted components confirm the conclusion reached through electrophysiological studies, that Gβγ in the presence of membranes containing PIP2 is sufficient to increase the open probability of GIRK channels. These experiments do not exclude a possible role for the Gα subunit in regulating the GIRK channel or in conferring G protein specificity, that is, explaining why GIRK normally is activated by Gβγ subunits associated with “inhibitory” Gαi/o subunits and not by Gβγ subunits associated with stimulatory Gαs subunits8. While many questions remain, these reconstitution experiments show that Gβγ by itself is sufficient to activate the GIRK channel.
Role of membrane in complex formation
Efforts to purify a stable GIRK-Gβγ protein complex in detergent solutions were unsuccessful. We therefore attempted to grow crystals of the complex in dodecylmaltoside by combining individually purified GIRK and Gβγ proteins at a 2–3 fold molar excess of Gβγ in the presence of a 10-fold molar excess of PIP2. Crystals containing both GIRK and Gβγ grew and diffracted to 3.5 Å resolution. These were of space group I422 with one GIRK monomer and one Gβγ subunit per asymmetric unit. Phases were solved by molecular replacement using previously determined structures of GIRK and Gβγ as search models30, 31. A model of the complex was built and refined to working and free residuals, Rw and Rf, of 22.8% and 26.5%, respectively (Table S1). The biological unit consists of one channel tetramer, four Gβγ subunits, four PIP2 molecules and four Na+ ions bound to regulatory sites in addition to K+ ions in the selectivity filter. Intracellular Na+, PIP2 and Gβγ are all physiological regulators of GIRK channel gating5–8, 26–29, 32.
The arrangement of protein molecules within the crystal lattice is notable in light of a functional phenomenon known as “membrane delimited” activation of GIRK channels by G protein stimulation (figure 2a and figure S1a,b). Electrophysiological studies showed that in reaching the channel, Gβγ subunits behave as if to diffuse while attached to the membrane’s cytoplasmic surface (i.e. membrane delimited) 5–10. In the crystal we observe pseudo-membrane layers consisting of transmembrane channel domains (TMDs) and aqueous layers consisting of cytoplasmic channel domains (CTDs) and Gβγ subunits (figure 2a). The Gβγ subunits are oriented such that the C-terminus of the Gγ subunit, which contains a covalent lipid molecule, a geranylgeranyl group, is pointed directly at the membrane layer as if to function as an anchor (figure 2a,b)33, 34. We note that a similar arrangement of the G protein subunits was observed in the crystal of the β2 adrenergic GPCR in complex with the Gαβγ heterotrimer, which was determined in lipid cubic phases35. Thus, the GIRK-Gβγ crystal is compatible with physiological membrane delimited Gβγ activation of GIRK. We also note that our ability to achieve a complex in a crystal with membrane-like layers, but not in detergent solutions, implies that a membrane is important in the formation of the complex between GIRK and Gβγ.
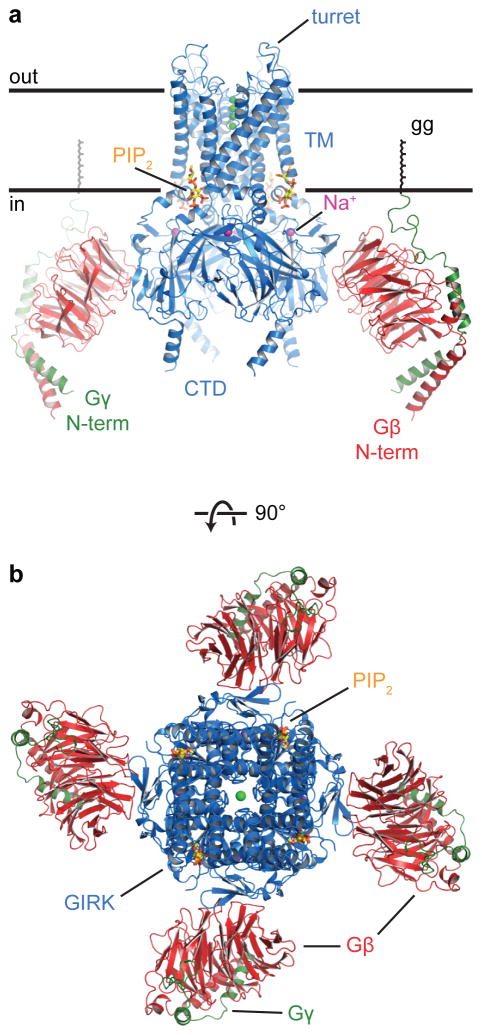
Figure 2. Overall structure of the GIRK-Gβγ complex.
a) A side view of the complex. The front Gβγ dimer was removed for clarity. The approximate extent of the phospholipid bilayer is shown by the thick black lines. The “gg” label points out the geranylgeranyl lipid modification at the C-terminus of Gγ. Bound Na+ ions are shown as purple spheres. c) Top-down view of the complex from the extracellular side of the cell. The GIRK, Gβ, and Gγ proteins are colored blue, red, and green respectively. The PIP2 molecules are shown as sticks. K+ ions are shown as green spheres.
The protein complex
Two views of the GIRK channel show the gating regulators Na+, PIP2, and Gβγ subunits bound (figure 2b,c). On the extracellular side of the membrane the channel’s turrets, which surround the pore entryway, project approximately 10 Å beyond the membrane surface. Previously we speculated that GIRK channels are more susceptible to pore-blocking toxins than are some other Kir channels, because the turrets are more widely spaced in GIRK and thus allow toxins to access the pore31. The structure of the turrets in this crystal are better defined than in the previous GIRK structures31, and indeed support this hypothesis (figure S2).
On the intracellular side, large CTDs project beyond the membrane surface, approximately 50 Å into the cell (figure 2b). These domains provide an extensive surface through which molecules inside the cell can bind in order to regulate channel gating. The Gβγ subunits interact directly with the CTD via Gβ and to the membrane via the covalent lipid attached to Gγ. Although we do not see the lipids in the crystal, the inferred covalent lipid interaction with the membrane is depicted. The entire GIRK channel-Gβγ complex forms a 120 Å × 120 Å square against the intracellular surface of the membrane (figure 2c).
The contact surface between GIRK and Gβ is approximately 700 Å2 (figure 3a–c). On GIRK the contact surface is formed by secondary structure elements βK, βL, βM and βN from one channel subunit and by elements βD and βE from an adjacent channel subunit (figure 3d). The occurrence of the binding site at the interface between two channel subunits is likely to be important for mechanistic reasons, discussed below. On Gβ the contact surface is formed by β sheet elements that form blades 1 and 7 on one edge of the β propeller (figure 3c,e).
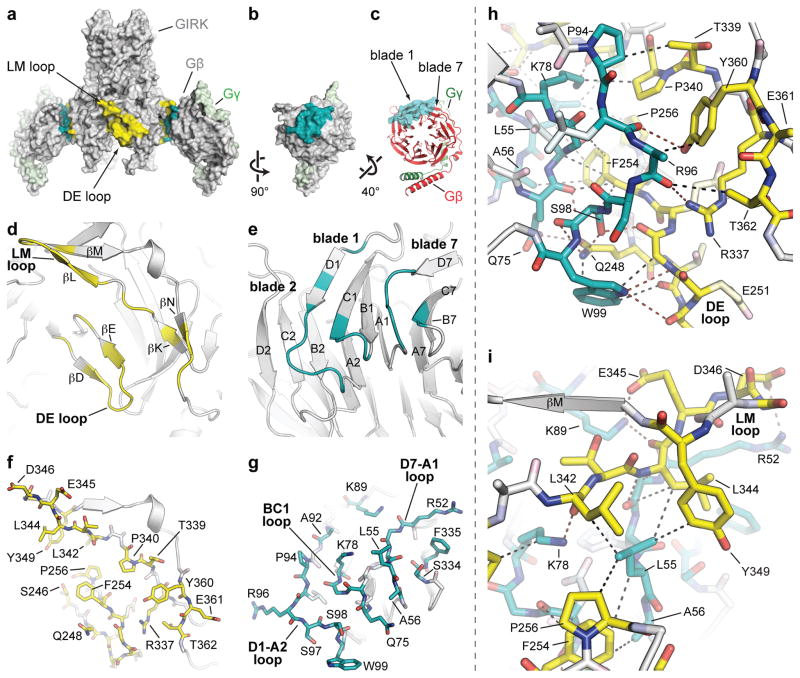
Figure 3. The GIRK-Gβγ binding interface.
a) Surface representation of the GIRK-Gβγ complex. The binding site on GIRK is colored yellow, and the binding site on Gβγ is colored cyan. The front Gβγ dimer is removed for clarity. The overall orientation is the same as in Figure 2a. b) A 90°-rotated view of a Gβγ dimer from panel a to more clearly show the binding interface. c) The Gβγ dimer is rotated upwards to orient the central axis of the β-propeller orthogonal to the page. d,e) A cartoon rendering of the binding interface on GIRK (d) and Gβ (e). Residues involved in the binding site are colored yellow on GIRK (d) and cyan on Gβ (e), and respectively correspond to the highlighted regions in panels a and b. In the d–e and f–g pairs of panels, the binding site can be approximately recapitulated by rotating each panel 90° towards each other, like making a sandwich. f,g) The same view as in panels d and e, except now the residues involved in the binding site are shown as sticks. h, i) A closeup of the GIRK-Gβγ interaction, focused on the DE loop, βK, and βN region (h) or the LM loop region (i) of GIRK. Selected hydrogen bond and van der Waals interactions are shown as dashed lines as a visual aid.
How do the contact surfaces observed in the crystal structure compare with inferences drawn using other biophysical methods? Transferred cross saturation and chemical shift perturbation NMR experiments have been used to identify amino acids on the GIRK CTD that interact with Gβγ or change upon its binding20. These amino acids, colored purple (and orange for L344), fall mainly within or near the perimeter of the surface on GIRK that contacts Gβγ in the crystal structure (figure S3a). Mutational studies also identified numerous amino acids that affect GIRK activation by G protein stimulation13–19. These amino acids are colored orange on the surface of GIRK and Gβγ (figure S3a,b). There are some outliers that may influence function indirectly or alternatively may disrupt protein structure, but most lie within or near the GIRK-Gβγ contact surface. Thus, both NMR and mutagenesis studies lend support to a biologically relevant signaling complex formed in the crystal structure.
Molecular determinants of Gβγ binding
All members of the inward rectifier K+ (Kir) channel family share the same general molecular architecture, but as far as we know only the GIRKs (Kir3s) are directly regulated by G protein subunits36. Many amino acids that compose the Gβγ binding surface on GIRK are also conserved among the G protein-independent Kir channels, but a small set are unique to GIRK (figure S4). This unique set includes Gln248 and Phe254 on the βD-βE loop and the sequence Leu-Thr/Ser-Leu (342–344) on the βL-βM loop (figure 3f). Gln248 forms contacts with Gln75, Ser98, and Trp99 on the Gβ subunit; and mutations at Ser98 and Trp99 diminish Gβγ activation of GIRK (figure 3f,g,h)13, 15. The Leu-Thr/Ser-Leu sequence contacts Leu55 and Lys78 on Gβ (figure 3f,g,i). Mutations involving these residues also affect Gβγ activation of GIRK13. Thus, we can begin to understand Gβγ recognition in terms of short-range interactions afforded by a relatively small set of residues on the surface of the GIRK channel to which the Gβγ subunits bind.
Long-range electrostatic interactions between GIRK and Gβγ also appear significant (figure 4a,b). Multiple acidic residues on the βL-βM loop of the GIRK CTD complement an electropositive swath on the binding surface of the Gβ subunit (figure 4a,b,d). By comparison, a G protein-independent Kir channel contains several lysine amino acids on its βL-βM loop that render its surface potential less electronegative (figure 4c,e). Thus, electrostatic complementarity probably plays a role in binding affinity and specificity. In addition, by acting over longer distances, electrostatic forces are able to guide diffusing molecules into the formation of an encounter complex, where then short-range interactions are able to take hold37. Such long-range guidance would seem to make sense here by directing to the K+ channel the diffusion of Gβγ once it is released from an activated GPCR.
Figure 4. The role of electrostatics at the GIRK-Gβγ interface.
a–c) Surface representations are shown for Gβγ (a), GIRK2 (b), and IRK247 (c), and are colored according to calculated electrostatic surface potentials (red: −100mV, blue: +100mV). The proteins are shown in the same orientation as in Figure 3d–g, except slightly zoomed out. A black outline of a cartoon representation of GIRK (or IRK2 in panel c) is overlaid to help the viewer match interacting surfaces. d,e) A closeup view of LM loop region on GIRK2 (d) or IRK2 (e). The Cα atom for aspartate or glutamate residues are shown as red balls, and arginines and lysines are shown as blue balls. f) (top) Cartoon representations of the GIRK-Gβγ complex and the β2AR-Gαsβγ complex35 are shown. The black lines highlight the difference in the relative orientation of the two Gβγ dimers to the membrane (gray rectangle). (bottom) Isocontour representations of the electrostatic potential for an isolated GIRK channel (left) or a Gβγ dimer (middle and right) are shown (red: −25 mV, blue: +25 mV).
Charged lipids on the membrane’s inner leaflet dominate the electric field close to the membrane where G protein signaling occurs38. With this fact in mind, figure 4f illustrates another potentially important role for electrostatic interactions between GIRK and Gβγ. In the β2 adrenergic GPCR-Gαβγ complex, the Gβγ subunits appear oriented to maximize positive protein charge contact with negative charges on the membrane surface35. In the GIRK-Gβγ complex, while Gβγ resides at the same level with respect to the plane of the membrane, it is tilted roughly 35°. The tilt should reduce favorable electrostatic interactions between Gβγ and the membrane, while new favorable interactions with GIRK presumably compensate. Thus, favorable electrostatic interactions between GIRK and Gβγ may help to reorient Gβγ with respect to the membrane.
Figure S5 compares the different contact surfaces that Gβγ utilizes to interact with other proteins, including Gα and four other effector and regulatory proteins, including GIRK. These comparisons support three conclusions. First, the β propeller of Gβ creates a large sticky surface that enables a multitude of unique interactions. Second, the GIRK binding site on Gβγ overlaps the Gα binding site. This observation, while anticipated, underscores the necessity of receptor activation and G protein subunit dissociation (into Gα and Gβγ) in order to achieve channel activation4–8. Third, the contact surface of RGS9 (Regulator of G protein Signaling 9) on Gβ5 is essentially non-overlapping with that of GIRK on Gβ1γ2, although a conformational change would be required in RGS9 to allow it to bind to a GIRK-Gβγ complex39. RGS9 in the nervous system suppresses the activity of opioid- and dopamine-mediated G protein signaling40–42 . Further studies will be needed to determine whether these signaling pathways intersect.
Gating control by Gβγ
What does the complex structure tell us about the regulation of GIRK channel gating by the Gβγ subunits? With the exception of the C-terminal half of Gγ, which is displaced by a crystal contact, Gβγ is structurally unchanged whether bound to Gα or to GIRK. The GIRK structure on the other hand is altered by the presence of Gβγ. Most notably the CTD is rotated about the channel axis 4° counterclockwise (viewed from the membrane) relative to the TMD (figure 5a,b and Supplementary video 1). The CTD rotation is associated with an unwrapping and splaying of the right-handed bundle of four inner helices that form the closed “inner helical gate” in the absence of Gβγ. Four Phe192 side chains on the inner helices come together to form the narrowest constriction in the closed inner helical gate. These Phe side chains are partially disordered in the slightly splayed structure with Gβγ subunits bound, but still, the pathway seems too narrow to conduct hydrated K+ ions: other clearly open K+ channels, such as the Kv1.2 voltage-dependent K+ channel, have a minimum diameter of 10 Å (distance between van der Waals surfaces), but here it is only 6 – 7 Å43–46. But should we expect an open conformation in the crystal structure? The single channel recordings suggest perhaps not (figure 1b). G protein subunits are most likely bound to the GIRK channel during the duration of an activity burst; however, during the burst the channel flickers rapidly with a relatively low open probability. This might suggest that the GIRK structure we observe in the presence of Gβγ, which adopts a distinctly different conformation than the structure without Gβγ, represents a G protein activated, pre-open conformation, corresponding to the channel part way along the reaction pathway from closed to open.
Figure 5. Gβγ-induced conformational changes in GIRK.
a–e) The structure of wild-type (WT) GIRK in complex with PIP2 (PDB id: 3SYA) is shown (gray) and is compared to the GIRK-Gβγ complex (blue) and the R201A mutant GIRK in complex with PIP2 (PDB id: 3SYQ) (green). All structures are aligned by a structurally inert region around the selectivity filter at the top of the transmembrane domain to show the relative twisting of the cytoplasmic domains. Panels a and c show a Cα ribbon trace of a side view of the transmembrane domain, with Phe192 shown as sticks for reference. Panels b and d show a top-down view of the cytoplasmic domain, with the degree and direction of twisting indicated. Panel e is a 70° rotated view of panel c to highlight the conformational changes of the inner helicies of the two subunits that bound PIP2 in this structure. f) A Cα ribbon trace of part of the cytoplasmic domain from the WT GIRK + Gβγ structure. Red coloring reflects the rmsd between the WT GIRK and WT GIRK + Gβγ structures when they are aligned by their cytoplasmic domains. This highlights the additional conformational changes that happen in the cytoplasmic domain apart from the rigid-body twisting shown in panels b and d. The most intense red represents a rmsd of 0.8 Å.
In a previous study, we determined the crystal structure of a constitutively-open point mutant of the GIRK channel, R201A, which is conductive in the absence of G protein stimulation31. The mutant structure is indeed open, and its comparison to the wild type channels in the absence and presence of Gβγ is suggestive of a mechanism (figure 5c–e, figure 6). In the mutant channel the CTD is rotated an additional 4° beyond the rotation caused by Gβγ, and the CTD subunits have undergone an internal conformational change associated with widening of the membrane-facing apex of the CTD. This widening further opens the inner helical gate to a diameter of 9 Å. One caveat is that only two PIP2 molecules are bound to the tetramer in the mutant channel – to diagonally opposed subunits – so that opening is 2-fold rather than 4-fold symmetric. Packing in the mutant crystal appears to have prevented the binding of PIP2 molecules to all four subunits, which is observed in the wild type structures31. We suspect that had four PIP2 molecules bound to the mutant channel, then opening would be symmetric. Despite the asymmetry of the R201A mutant, the conformation of GIRK in the Gβγ complex is clearly intermediate between the closed (Gβγ-free) and opened (R201A) structures. A morph between these conformations shows that binding of Gβγ causes a 4° rotation of the CTD and a slight splaying of the inner helices. The R201A mutation produces a further 4° rotation, a conformational change within the CTD subunits, and an opening of the inner helical gate (Supplementary video 2). These conformations could account for the burst kinetic behavior of single GIRK channels if binding of the Gβγ subunits produces a pre-open conformation in the membrane, from which the channel flickers rapidly between open (conductive) and pre-open (nonconductive) conformations (figure 6, highlighted pathway). This hypothesis would predict that the R201A mutant channels should exhibit a higher open probability. Unfortunately, due to reduced expression levels, we have been unable to characterize the single channel behavior of this mutant in either Xenopus oocytes or chinese hamster ovary cells.
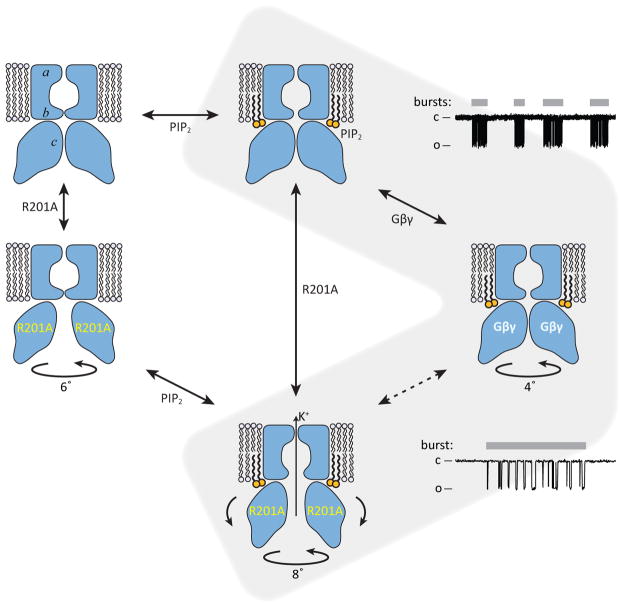
Figure 6. A model of gating regulation of GIRK channels.
The blue shapes depict a GIRK channel with a selectivity filter (a) and two gates: the inner helix gate (b) and the G loop gate (c). The structures correspond to WT GIRK without PIP2 or Gβγ (3SYO), WT GIRK with PIP2 only (3SYA), R201A GIRK without PIP2 or Gβγ (3SYP), WT GIRK with PIP2 and Gβγ (4KFM), and R201A GIRK with PIP2 (3SYQ). Circular arrows with degrees indicate CTD rotation about the pore axis with respect to the TMD, relative to WT structures without Gβγ. Curved arrows in the R201A GIRK with PIP2 reflect the outward rocking of CTD subunits observed in this structure. Idealized single-channel recordings are shown on the right (expanded time scale on the bottom) to illustrate our current hypothesis regarding the gating transitions that the channel undergoes. Inter-burst periods correspond to a channel with only PIP2 bound (top); bursts (gray bars) correspond to the channel with PIP2 and Gβγ bound, which fluctuates rapidly (indicated by dashed arrows) between non-conducting (right) and conducting (bottom) conformations.
Discussion
GIRK2 channels are regulated by PIP2, G protein subunits and intracellular Na+ ions5–8, 26–29, 32. We show in reconstitution experiments, using purified components, that these regulators individually activate the channel partially and in combination activate it to a greater extent. We present a crystal structure of GIRK with all three regulators bound. Together with previously determined crystal structures of GIRK and an R201A mutant of GIRK, both determined in the presence and absence of PIP2, we have pieced together a structural description of conformational states that might underlie the sequential activation of GIRK channels (figure 6, highlighted pathway). The binding of Gβγ subunits to GIRK causes a rotation of the CTD with respect to the TMD and a partial splaying of the inner helices. This conformation is intermediate between the closed and R201A open conformations. Full opening is associated with a further rotation of the CTD and splaying open of the inner helical gate.
Together the structures permit conceptual explanations for multi-ligand regulation. PIP2 is required for full gate opening in the R201A mutant channel31. Thus, PIP2 seems to play a facilitative role; under conditions that favor opening, PIP2 helps, presumably by strengthening the interface between the CTD and TMD where PIP2 is bound. The Na+ ion is bound to the CTD at a position that undergoes a conformational change when the channel opens. Thus, we should expect Na+ binding to be thermodynamically coupled to channel gating, allowing Na+ to function as a regulator.
The GIRK-Gβγ complex structure raises the important question of stoichiometry: how many Gβγ subunits are required to open the GIRK channel? We do not know the answer to this question, but based on the structure we are compelled to speculate. Gβγ binding causes a rotation of the CTD associated with splaying of the inner helices to open the gate. The rotation no doubt occurs because Gβγ binds at the interface between two adjacent CTD subunits, which produces detectable relative motions of the subunits, and inferred strain between them (figure 5f and Supplementary video 3). This, we believe, is the source of the rotation. One Gβγ subunit causing strain across one of four interfaces is probably not enough. Four is undoubtedly better. Can a single GPCR in the neighborhood of a GIRK channel catalyze a sufficient number GDP to GTP exchange reactions and release Gβγ subunits to activate the channel, or do multiple GPCRs surround a GIRK channel? If multiple surround, are GIRK channels and GPCRs randomly distributed or are they organized in a stoichiometric cluster or array? The GIRK-Gβγ complex provides a starting point for addressing these questions.
Methods
Molecular Biology
A truncated GIRK2 construct (containing residues 52–380) was cloned into the pPICZ, or pGEM vectors for high-level expression or electrophysiology, respectively, as previously described48. The full-length human G protein β1 and γ2 subunits were cloned into pFastbac vectors. The γ2 construct also included an N-terminal His10 tag, followed by a yellow fluorescent protein (YFP), and then a PreScission protease site (LEVLFQ/GP). Individual baculoviruses were made from these pFastbac vectors using the Bac-to-Bac system (Invitrogen).
Protein expression and purification
GIRK2 was expressed in P. pastoris as previously described48. GIRK2 was extracted and purified from P. pastoris cells essentially as previously described48, with a few exceptions: Dodecyl-β-D-maltopyranoside (DDM) was used in all steps instead of decyl-β-D-maltopyranoside (DM). DDM was used at 4 % for extraction, 0.4 % during the Talon purification, and 0.05 % on the Superdex-200 column. 10 mM imidazole was included during the batch binding to the Talon resin. PreScission protease-cleaved protein was loaded onto the Superdex-200 column at a sufficiently high concentration such that the 1 mL peak was at least 1 mg/mL. This was done to reduce the final detergent concentration in the concentrated protein, which was necessary for growing large, thick crystals. The protein was concentrated to 30–40 mg/mL in a 50K MWCO, and was typically used immediately.
The G proteins were expressed in High Five insect cells (Invitrogen). High Five cells were grown at 27° C in Express Five serum-free media (Invitrogen), supplemented with L-glutamine. The cells were grown to a density of 1–2 million cells/mL and then infected with a volume of baculovirus for each protein empirically determined to give optimal expression (~30 mL). After 48 hrs, the cells were harvested by centrifugation at 4000 g × 15 min. The cell pellets were resuspended in a small volume of the supernatant and this slurry was transferred to 50 mL conical tubes (approximately 1 L of cells per 50 mL tube). After another centrifugation at 4000 g × 15 min, the supernatant was removed and the cell pellets (~15 mL) were frozen in liquid N2 and then stored at −80° C until needed.
A typical Gβγ prep involved purifying protein from 8 L worth of cells. All procedures were performed at 4° C unless indicated. Frozen cell pellets were added to 480 mL of room temperature (RT) buffer comprised of 50 mM Hepes pH 8, 65 mM NaCl, 1 mM EDTA, 5 mM β-mercaptoethanol (βME), and protease inhibitors (PIs) (0.1 mg/mL pepstatin, 1 mg/mL leupeptin, 1 mg/mL aprotinin, 0.1 mg/mL soy trypsin inhibitor, 1 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride). This solution was stirred at RT in a stainless steel beaker until the pellets melted. Then the beaker was transferred to ice and the solution was sonicated using a probe sonicator (Branson) for 6 × 1 min, with a 1 min cool down in between. The lysed cells were then spun at 35000 g × 35 min to pellet the membranes. The supernatant was poured off and the pellets were each briefly rinsed with ~5 mL of 50 mM Hepes pH 8, 50 mM NaCl, 0.1 mM MgCl2, 5 mM βME, and PIs. The pellets were then resuspended in the same buffer using a dounce homogenizer to a final volume of 350 mL. Na-cholate was added to a final concentration of 1.5 %, and the solution was stirred for 40 min. The solubilized membranes were spun again at 35000 g × 35 min to pellet insoluble material. The supernatant was diluted with two volumes of dilution buffer (20 mM Hepes pH 8, 300 mM NaCl, 5 mM βME, 7.5 mM imidazole, 0.5 % anzergent 3–12 (Anatrace), and PIs) and then added to 32 mL of Talon resin (Clontech) pre-equilibrated in dilution buffer. This suspension was stirred for 1 hr, then spun at 1000 g × 5 min. The resin was transferred to a column and washed with 4 column volumes (cv) of 20 mM Hepes pH 8, 300 mM NaCl, 5 mM βME, 5 mM imidazole, and 0.5 % anzergent 3–12; then, 4 cv of 20 mM Hepes pH 8, 50 mM NaCl, 5 mM βME, 10 mM imidazole, and 0.5 % anzergent 3–12. Then the protein was eluted from the column with 20 mM Hepes pH 8, 40 mM NaCl, 5 mM βME, 200 mM imidazole, and 0.5 % anzergent 3–12. DTT and EDTA were added to 5 mM and 1 mM, respectively. PreScission protease was also added at 1:20 protease:total protein, and incubated over night. The next day, an additional amount of PreScission (1:40 protease:total protein) was added and incubated at RT for 2 hrs. This solution was then diluted down to a conductivity of ~5 mS/cm with 20 mM Hepes pH 8, 5 mM βME, 1 % anzergent 3–12. A white precipitate usually formed at this step, which was mostly comprised of contaminants. This was pelleted by a brief centrifugation, and the supernatant was further filtered through a 0.22 μm filter before loading onto a Mono Q 16/10 column, equilibrated with buffer A (20 mM Hepes pH 8, 40 mM NaCl, 5mM βME, and 0.7 % 3-[(3-Cholamidopropyl)-Dimethylammonio]-1-Propane Sulfonate/N,N-Dimethyl-3-Sulfo-N-[3-[[3α,5β,7α,12α)-3,7,12-Trihydroxy-24-Oxocholan-24-yl]Amino]propyl]-1-Propanaminium Hydroxide (CHAPS; Anatrace). The column was washed with 15 cv buffer A, then the protein was eluted with a 50 cv gradient from 0 – 20 % buffer B (buffer A with 1 M NaCl). The Gβγ protein eluted as a major peak as well as several minor peaks that were assumed to be unprenylated or differentially phosphorylated species. The major peak was collected and concentrated in a 30K MWCO concentrator to at least 5 mg/mL. This was then run on a Superdex-200 column equilibrated in 20 mM Tris pH 7.5, 100 mM KCl, 5 mM DTT, 0.7 % CHAPS) in multiple runs of ~2.5 mg protein per run. This helped to remove trace smaller molecular weight Gβγ protein, which was assumed to be an unprenylated species. Peak fractions were again concentrated in a 30K MWCO concentrator to ~10 mg/mL. Glycerol was added to 20 % and the protein was frozen in liquid N2 in 150 μL aliquots and stored at −80° C until needed. Approximately 8–10 mg worth of these aliquots were thawed as needed and the detergent was then exchanged to DDM while bound to a 1 mL Q Sepharose column (HiTrap, GE Healthcare) at RT. This was done to ensure complete detergent exchange as well as to get a high protein concentration without a high detergent concentration, which was necessary for growing large, thick crystals. DDM (anagrade) was added to 1 % final from a 10 % stock. This was then slowly diluted with two volumes of 20 mM Tris pH 7.5, 5 mM DTT, and 0.05 % DDM, and then loaded onto the HiTrap column equilibrated with buffer C (20 mM Tris pH 7.5, 30 mM KCl, 5 mM DTT, and 0.05 % DDM). The column was washed with 25 mL buffer C, then 3 mL 10 % buffer D (buffer C with 300 mM KCl), 3 mL 20 % buffer D, 9 mL 30 % buffer D, then the protein was eluted with 100 % buffer D. The middle of this elution peak yielded about 0.75 mL of 8–10 mg/mL. This was then further concentrated in a 30K MWCO concentrator. After one centrifugation spin of the concentrator, the protein was diluted with the appropriate volume of 20 mM Tris, 5 mM DTT, including an appropriate concentration of EDTA to bring the [KCl] down to 150 mM and the EDTA concentration up to 1 mM. The protein was then further concentrated to 40–50 mg/mL and stored on ice until needed.
Crystallization
A typical crystallization experiment involved mixing concentrated GIRK2, Gβγ, and 1,2-dioctanoyl-sn-glycero-3-phospho-(1′-myo-inositol-4′,5′-bisphosphate) (C8-PIP2, Avanti Polar Lipids) at a final concentration of 200 μM, 400 μM, and 2 mM, respectively. The mixture was incubated at RT for at least 2 hrs before mixing 1:1 (0.2 μL + 0.2 μL) with the crystallization solution (600 mM NaK tartrate, 50 mM Na-ADA (N-(2-Acetamido)iminodiacetic acid) pH 5.7 – 5.9). The crystals were grown at 20° C using the hanging drop vapor diffusion method. The crystals appeared after a few days and grew as thin square plates or plate clusters to full size within a week. The crystals were cryoprotected by first adding 1 μL of a solution containing 20 mM Tris pH 7.5, 150 mM KCl, 1 mM EDTA, 0.5 % DDM, 1 mM DTT, 720 mM NaK Tartrate, 50 mM Na-ADA pH 5.7, and 1 mM C8-PIP2 directly to the drop. Crystals were gently broken off of the clusters and then briefly transferred to a new solution containing 20 mM Tris pH 7.5, 0.5 % DDM, 1 mM DTT, 50 mM Na-ADA pH 5.7, 1 mM C8-PIP2, and 2.35 – 2.4 M NaK Tartrate, depending on the crystal size. The crystals were then flash-frozen in liquid N2.
Structure determination
Diffraction data were collected at the Advanced Photon Source 23ID-B beamline (λ=1.033 Å). Diffraction images were processed with the HKL2000 program suite49. The crystals diffracted anisotropically (3.45 × 3.45 × 3.8 Å along the a*, b*, and c* axes, respectively), so integrated diffraction data were truncated to these diffraction limits using a script available from the UCLA Diffraction Anisotropy Server50. The crystals were highly sensitive to radiation damage, so data from three sites on one crystal and one site on another crystal were scaled together in HKL2000 to form the most complete dataset. Rmerge and Rpim diffraction data statistics were calculated using the RMERGE program51.
The structure was solved with Phaser52 by sequentially searching for a GIRK2 monomer (PDB id: 3SYA) and then a Gβ1γ2 dimer (PDB id: 1GP2, chains B and G). Initial rigid-body refinement of the molecular replacement solution with REFMAC53, 54 identified the twist between the cytoplasmic and transmembrane domains of GIRK2. The model was then further modified in Coot55 and refined with REFMAC, using jelly-body restraints. TLS refinement in REFMAC was used in the final rounds of refinement.
Because of the poor quality of electron density in the K+ selectivity filter, distance restraints were used during refinement between the K+ ions in the selectivity filter to constrain their positions based on known properties from high-resolution K+ channel structures. In the final stages of refinement, a strong electron density feature near the interfacial helix of the channel was modeled as a DDM maltose headgroup with a five-carbon aliphatic chain. This was based on bilobal shape of the density, the location of this density at the presumed boundary of the DDM micelle, and the presence of 15mM DDM in the crystallization condition. PIP2 and Na+ ligands were carried over from the original search model and their presence was confirmed using simulated annealing omit maps (figure S6a,b).
Comprehensive model validation was performed with MolProbity56 (as embedded within PHENIX57), with 94.5/5.5 % of residues falling within the favored and allowed region of the Ramachandran plot, respectively. Simulated annealing omit maps were calculated using PHENIX. Data collection and refinement statistics are shown in Table S1. All figures were made using PyMOL (www.pymol.org). Videos were made in PyMOL using intermediate structures interpolated with the CNS58, 59 script from the Yale Morph Server60, 61. Electrostatics were calculated using APBS and visualized using the APBS plugin in PyMOL62. Disordered side chains were added back to the model in the most common rotamer conformation to make the calculations more accurate.
Electrophysiology
Two-electrode voltage-clamp recordings of GIRK2 currents in Xenopus laevis oocytes were performed as previously described48. For patch clamp recordings of GIRK2 currents, X. laevis oocytes were injected with cRNA for the M2 muscarinic receptor and GIRK2 as previously described. The patch pipettes typically had a resistance of 4 MΩ and were filled with 96 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 10 mM K-Hepes pH 7.5, and 10 μM acetylcholine. The bath solution contained 96 mM KCl, 5 mM EGTA, and 10 mM K-Hepes pH 7.5. The recordings were made in the on-cell configuration and the patch was held at −100 mV. The currents were recorded with Axon Axopatch 200B (Molecular Devices), filtered at 1 kHz and sampled at 10 kHz using an analog-to-digital converter (Digidata 1440A, Axon Instruments). pClamp10.1 software (Axon Instruments) was used for controlling the amplifier and data acquisition. Uninjected oocytes showed no detectable currents.
Reconstitution into lipid vesicles
Purified GIRK2 was reconstituted into lipid vesicles by first mixing chloroform solutions of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG), and L-α-phosphatidylinositol-4,5-bisphosphate (PI(4,5)P2; from porcine brain; predominant acyl chains are 18:0 and 20:4) at mass ratios of 3:1:0.04, and then drying this solution under an argon gas stream. The dried lipid film was placed in a vacuum desiccator for a few hours and then resuspended in 20 mM K-Hepes pH 7.35, 150 mM KCl, 1 mM EDTA, 35 mM CHAPS (at 10 mg/mL) by incubating for 2 hrs at RT with periodic sonication. Purified GIRK2 was concentrated to ~2 mg/mL and then added to 100 μL of the solubilized lipids, typically at a 1:300 protein:lipid mass ratio, and incubated at RT for 30 min. Dehydrated 1 mL Sephadex G-50 (Sigma) columns were prepared by loading 1 mL of hydrated Sephadex G-50 resin (equilibrated in 20 mM K-Hepes pH 7.35, 150 mM KCl, 1 mM EDTA) on to a small plastic spin column, and then briefly centrifuging at 1500 g. The protein-lipid mixture was then gently pipetted onto the resin bed and the columns were briefly spun up to 1000 g to remove the detergent and form the proteoliposomes.
Flux assay
10 μL of vesicles were added to 190 μL of flux buffer (20 mM K-Hepes pH 7.35, 150 mM NaCl, 1 mM EDTA, and 2 μM of the pH-sensitive dye 9-Amino-6-Chloro-2-Methoxyacridine (ACMA)) in a 96-well black plate (in some experiments, the NaCl was replaced with N-methyl-d-glucamine HCl (NMDG-Cl)). This creates a K+ gradient across the vesicle membrane and a negative potential inside the vesicle if there are open K+ channels. Fluorescence was monitored every 5 s at 410/490 nm (excitation/emission, 20 nm bandwidth). After a baseline was established, the H+ ionophore carbonyl cyanide m-chlorophenyl hydrazone (CCCP) is added to 1 μM from a 400 μM stock and briefly mixed. This allows protons to enter the vesicles, drawn in the by negative potential, which causes quenching of ACMA. After 10 min, the K+ ionophore valinomycin is added to 20 nM from a 8 μM stock and briefly mixed. This acts as a shunt to indicate the total capacity of the vesicles. All fluorescence time-courses were normalized to the fluorescence prior to CCCP addition to account for slight differences in fluorescence between the wells. In some experiments, a concentrated stock of CHAPS-solubilized purified Gβγ was added to the vesicles before diluting them in the flux buffer to give a final concentration of 180 nM.
Supplementary Material
Acknowledgments
We thank P. Hoff and members of D. Gadsby’s laboratory (Rockefeller University) for assistance with oocyte preparation; Y. Hsiung for assistance with insect cell culture; R. Sanishvili, N. Venugopalan, and S. Corcoran (GM/CA, Advanced Photon Source, Argonne National laboratory) for assistance at the synchrotron; and members of the MacKinnon Lab. R.M. is an investigator in the Howard Hughes Medical Institute.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions M.R.W. performed the experiments. M.R.W and R.M. analyzed the data and wrote the paper.
Author Information Atomic coordinates and structure factors for the reported crystal structure have been deposited into the Protein Data Bank under accession code 4KFM.
The authors declare no competing financial interest.
References
- 1.Loewi O. Über humorale Übertragbarkeit der Herznervenwirkung. Pflügers Arch. 1921;189:239–242. [Google Scholar]
- 2.Loewi O. The Chemical Transmission of Nerve Action (Nobel Lecture) 1936. [Google Scholar]
- 3.Loewi O, Navratil E. Über humorale Übertragbarkeit der Herznervenwirkung. Pflügers Arch. 1926;214:678–688. [Google Scholar]
- 4.Gilman AG. G proteins: transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- 5.Logothetis DE, Kurachi Y, Galper J, Neer EJ, Clapham DE. The beta gamma subunits of GTP-binding proteins activate the muscarinic K+ channel in heart. Nature. 1987;325:321–326. doi: 10.1038/325321a0. [DOI] [PubMed] [Google Scholar]
- 6.Reuveny E, et al. Activation of the cloned muscarinic potassium channel by G protein beta gamma subunits. Nature. 1994;370:143–146. doi: 10.1038/370143a0. [DOI] [PubMed] [Google Scholar]
- 7.Wickman KD, et al. Recombinant G-protein beta gamma-subunits activate the muscarinic-gated atrial potassium channel. Nature. 1994;368:255–257. doi: 10.1038/368255a0. [DOI] [PubMed] [Google Scholar]
- 8.Pfaffinger PJ, Martin JM, Hunter DD, Nathanson NM, Hille B. GTP-binding proteins couple cardiac muscarinic receptors to a K channel. Nature. 1985;317:536–538. doi: 10.1038/317536a0. [DOI] [PubMed] [Google Scholar]
- 9.Breitwieser GE, Szabo G. Uncoupling of cardiac muscarinic and beta-adrenergic receptors from ion channels by a guanine nucleotide analogue. Nature. 1985;317:538–540. doi: 10.1038/317538a0. [DOI] [PubMed] [Google Scholar]
- 10.Soejima M, Noma A. Mode of regulation of the ACh-sensitive K-channel by the muscarinic receptor in rabbit atrial cells. Pflügers Arch. 1984;400:424–431. doi: 10.1007/BF00587544. [DOI] [PubMed] [Google Scholar]
- 11.Trautwein W, Dudel J. Zum Mechanismus der Membranwirkung des Acetylcholin an der Herzmuskelfaser. Pflügers Arch. 1958;266:324–334. doi: 10.1007/BF00416781. [DOI] [PubMed] [Google Scholar]
- 12.Luscher C, Slesinger PA. Emerging roles for G protein-gated inwardly rectifying potassium (GIRK) channels in health and disease. Nat Rev Neurosci. 2010;11:301–315. doi: 10.1038/nrn2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ford CE, et al. Molecular basis for interactions of G protein betagamma subunits with effectors. Science. 1998;280:1271–1274. doi: 10.1126/science.280.5367.1271. [DOI] [PubMed] [Google Scholar]
- 14.Albsoul-Younes AM, et al. Interaction sites of the G protein beta subunit with brain G protein-coupled inward rectifier K+ channel. J Biol Chem. 2001;276:12712–12717. doi: 10.1074/jbc.M011231200. [DOI] [PubMed] [Google Scholar]
- 15.Mirshahi T, Robillard L, Zhang H, Hebert TE, Logothetis DE. Gbeta residues that do not interact with Galpha underlie agonist-independent activity of K+ channels. J Biol Chem. 2002;277:7348–7355. doi: 10.1074/jbc.M109999200. [DOI] [PubMed] [Google Scholar]
- 16.Zhao Q, et al. Interaction of G protein beta subunit with inward rectifier K(+) channel Kir3. Mol Pharmacol. 2003;64:1085–1091. doi: 10.1124/mol.64.5.1085. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Q, et al. Dominant negative effects of a Gbeta mutant on G-protein coupled inward rectifier K+ channel. FEBS Lett. 2006;580:3879–3882. doi: 10.1016/j.febslet.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 18.He C, Zhang H, Mirshahi T, Logothetis DE. Identification of a potassium channel site that interacts with G protein betagamma subunits to mediate agonist-induced signaling. J Biol Chem. 1999;274:12517–12524. doi: 10.1074/jbc.274.18.12517. [DOI] [PubMed] [Google Scholar]
- 19.Finley M, Arrabit C, Fowler C, Suen KF, Slesinger PA. betaL-betaM loop in the C-terminal domain of G protein-activated inwardly rectifying K(+) channels is important for G(betagamma) subunit activation. J Physiol. 2004;555:643–657. doi: 10.1113/jphysiol.2003.056101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yokogawa M, Osawa M, Takeuchi K, Mase Y, Shimada I. NMR analyses of the Gbetagamma binding and conformational rearrangements of the cytoplasmic pore of G protein-activated inwardly rectifying potassium channel 1 (GIRK1) J Biol Chem. 2011;286:2215–2223. doi: 10.1074/jbc.M110.160754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slesinger PA, Reuveny E, Jan YN, Jan LY. Identification of structural elements involved in G protein gating of the GIRK1 potassium channel. Neuron. 1995;15:1145–1156. doi: 10.1016/0896-6273(95)90102-7. [DOI] [PubMed] [Google Scholar]
- 22.Huang CL, Slesinger PA, Casey PJ, Jan YN, Jan LY. Evidence that direct binding of G beta gamma to the GIRK1 G protein-gated inwardly rectifying K+ channel is important for channel activation. Neuron. 1995;15:1133–1143. doi: 10.1016/0896-6273(95)90101-9. [DOI] [PubMed] [Google Scholar]
- 23.Kofuji P, Davidson N, Lester HA. Evidence that neuronal G-protein-gated inwardly rectifying K+ channels are activated by G beta gamma subunits and function as heteromultimers. Proc Natl Acad Sci U S A. 1995;92:6542–6546. doi: 10.1073/pnas.92.14.6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kubo Y, Reuveny E, Slesinger PA, Jan YN, Jan LY. Primary structure and functional expression of a rat G-protein-coupled muscarinic potassium channel. Nature. 1993;364:802–806. doi: 10.1038/364802a0. [DOI] [PubMed] [Google Scholar]
- 25.Jin W, Lu Z. Synthesis of a stable form of tertiapin: a high-affinity inhibitor for inward-rectifier K+ channels. Biochemistry. 1999;38:14286–14293. doi: 10.1021/bi991205r. [DOI] [PubMed] [Google Scholar]
- 26.Ho IH, Murrell-Lagnado RD. Molecular mechanism for sodium-dependent activation of G protein-gated K+ channels. J Physiol 520 Pt. 1999;3:645–651. doi: 10.1111/j.1469-7793.1999.00645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho IH, Murrell-Lagnado RD. Molecular determinants for sodium-dependent activation of G protein-gated K+ channels. J Biol Chem. 1999;274:8639–8648. doi: 10.1074/jbc.274.13.8639. [DOI] [PubMed] [Google Scholar]
- 28.Sui JL, Petit-Jacques J, Logothetis DE. Activation of the atrial KACh channel by the betagamma subunits of G proteins or intracellular Na+ ions depends on the presence of phosphatidylinositol phosphates. Proc Natl Acad Sci U S A. 1998;95:1307–1312. doi: 10.1073/pnas.95.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sui JL, Chan KW, Logothetis DE. Na+ activation of the muscarinic K+ channel by a G-protein-independent mechanism. J Gen Physiol. 1996;108:381–391. doi: 10.1085/jgp.108.5.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wall MA, et al. The structure of the G protein heterotrimer Gi alpha 1 beta 1 gamma 2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- 31.Whorton MR, MacKinnon R. Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell. 2011;147:199–208. doi: 10.1016/j.cell.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang CL, Feng S, Hilgemann DW. Direct activation of inward rectifier potassium channels by PIP2 and its stabilization by Gbetagamma. Nature. 1998;391:803–806. doi: 10.1038/35882. [DOI] [PubMed] [Google Scholar]
- 33.Mumby SM, Casey PJ, Gilman AG, Gutowski S, Sternweis PC. G protein gamma subunits contain a 20-carbon isoprenoid. Proc Natl Acad Sci U S A. 1990;87:5873–5877. doi: 10.1073/pnas.87.15.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamane HK, et al. Brain G protein gamma subunits contain an all-trans-geranylgeranylcysteine methyl ester at their carboxyl termini. Proc Natl Acad Sci U S A. 1990;87:5868–5872. doi: 10.1073/pnas.87.15.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hibino H, et al. Inwardly rectifying potassium channels: their structure, function, and physiological roles. Physiol Rev. 2010;90:291–366. doi: 10.1152/physrev.00021.2009. [DOI] [PubMed] [Google Scholar]
- 37.Sheinerman FB, Norel R, Honig B. Electrostatic aspects of protein-protein interactions. Curr Opin Struct Biol. 2000;10:153–159. doi: 10.1016/s0959-440x(00)00065-8. [DOI] [PubMed] [Google Scholar]
- 38.McLaughlin S. The electrostatic properties of membranes. Annu Rev Biophys Biophys Chem. 1989;18:113–136. doi: 10.1146/annurev.bb.18.060189.000553. [DOI] [PubMed] [Google Scholar]
- 39.Cheever ML, et al. Crystal structure of the multifunctional Gbeta5-RGS9 complex. Nat Struct Mol Biol. 2008;15:155–162. doi: 10.1038/nsmb.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zachariou V, et al. Essential role for RGS9 in opiate action. Proc Natl Acad Sci U S A. 2003;100:13656–13661. doi: 10.1073/pnas.2232594100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rahman Z, et al. RGS9 modulates dopamine signaling in the basal ganglia. Neuron. 2003;38:941–952. doi: 10.1016/s0896-6273(03)00321-0. [DOI] [PubMed] [Google Scholar]
- 42.Kovoor A, et al. D2 dopamine receptors colocalize regulator of G-protein signaling 9-2 (RGS9-2) via the RGS9 DEP domain, and RGS9 knock-out mice develop dyskinesias associated with dopamine pathways. J Neurosci. 2005;25:2157–2165. doi: 10.1523/JNEUROSCI.2840-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang Y, et al. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 44.Jiang Y, et al. The open pore conformation of potassium channels. Nature. 2002;417:523–526. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- 45.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 46.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 47.Hansen SB, Tao X, MacKinnon R. Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature. 2011;477:495–498. doi: 10.1038/nature10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whorton MR, MacKinnon R. Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell. 2011;147:199–208. doi: 10.1016/j.cell.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otwinowski Z, Minor W, Carter Charles W., Jr . Methods in Enzymology. Vol. 276. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 50.Strong M, et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiss MS. Global indicators of X-ray data quality. J Appl Crystallogr. 2001;34:130–135. [Google Scholar]
- 52.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta crystallographica. Section D, Biological crystallography. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.The CCP4 suite: programs for protein crystallography. Acta crystallographica. Section D, Biological crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 55.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D, Biological crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 59.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 60.Echols N, Milburn D, Gerstein M. MolMovDB: analysis and visualization of conformational change and structural flexibility. Nucleic Acids Res. 2003;31:478–482. doi: 10.1093/nar/gkg104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krebs WG, Gerstein M. The morph server: a standardized system for analyzing and visualizing macromolecular motions in a database framework. Nucleic Acids Res. 2000;28:1665–1675. doi: 10.1093/nar/28.8.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.