Summary
Studies examining the mechanisms by which the liver injures and regenerates usually focus on factors and pathways within the liver, neglecting the signaling derived from the gut-liver axis. The intestinal content is rich in microorganisms as well as metabolites generated from both the host and colonizing bacteria. Via the gut-liver axis, this complex “soup” exerts an immense impact on liver integrity and function. This review article summarizes data published in the past 30 years that have demonstrated the signaling derived from the gut-liver axis in relation to liver injury and regeneration. Despite many correlative findings, the intricate networks of pathways involved along with a scarcity of mechanistic data urgently require nutrigenomic, metabolomics, and microbiota profiling approaches to provide a deep understanding of the interplay between nutrition, bacteria, and host response. Such knowledge would better elucidate the molecular mechanisms that link microbiota alteration to host physiological response and vice-versa.
Keywords: gut-liver axis, gut dysbiosis, nuclear receptor, bile acid receptor, FXR, probiotic, prebiotic, partial hepatectomy
Introduction
A unique feature of the liver is its extraordinary regenerative ability. Liver regeneration is crucial for restoration of function following injury and an understanding of the underlying mechanisms would be of therapeutic value in liver disease treatment and transplantation. Liver regeneration is an orchestrated biological process that includes sequential changes in gene expression, growth factor production, and tissue remodeling. Following liver resection, hepatocytes, which are not terminally differentiated, exhibit substantial proliferative capacity. Many mitogens, cytokines, and growth factors, which are involved in liver regeneration, have been identified and extensively reviewed [1–16]. In addition to the presence of growth factors and mitogens, active metabolism is required to generate the energy and precursors for biosynthesis of macromolecules necessary for cell proliferation and tissue remodeling during liver regeneration. Because nuclear receptors play a crucial role in hepatic metabolism, their actions in liver regeneration have been extensively studied in recent years as well [17–27]. However, liver regeneration research has typically focused on signaling pathways intrinsic to the liver, overlooking those derived from the gut. The current review details the signaling within the gut-liver axis and summarizes the interactions between microbiota and bile acids (BAs) in maintaining gastrointestinal health and impacting liver injury and regeneration.
The relationship between gut microbes, liver injury, and liver regeneration
The gut microbiota refers to the 100 trillion bacteria that reside in the human gastrointestinal tract (GI), and is now often referred to as its own organ [28]. Over the past decade, an exponential amount of research into the human microbiome, termed “the forgotten organ”, has shifted our perspective on the influence of the hostmicrobiome relationship in the pathogenesis of human diseases [29]. In addition, gut microbiota affects intestinal signaling and enterohepatic circulation of BAs, a growing body of evidence supports that the gut microbiota may promote liver regeneration and health.
Bacterial endotoxin and liver regeneration
Endotoxin lipopolysaccharides (LPS) are the major components of the outer membrane of Gram-negative bacteria. LPS have three components: O-antigen, core oligosaccharide, and lipid A. O-antigen is exposed on the outer surface of the bacterial and recognized by host antibodies. In contrast, the lipid A is conserved, and those hydrophobic fatty acid chains anchor the LPS into the bacterial membrane. Through toll-like receptor 4, the receptor of LPS, lipid A activates mammalian immune system with production of inflammatory mediators that lead to septic shock [30]. Chemically, LPS do not have O-antigens and only have the lipid A and oligosaccharide core, and LPS administration is frequently used to induce liver injury for in vivo study of hepatic regeneration and function. While this would initially appear to indicate that bacteria negatively influence liver regeneration, evidence indicates that endotoxin is necessary for liver regeneration. Gut-derived endotoxin administered both before and after partial hepatectomy (PHx) induced hepatic DNA synthesis and release of several hepatotrophic factors such as insulin [31]. Conversely, hepatic DNA synthesis in mice was impaired when gut-derived endotoxin was prevented from reaching the liver [32]. In addition, conditions that eliminate bacteria or reduce endotoxin could inhibit DNA synthesis following 67% liver resection. Those conditions include gut sterilization using neomycin and cefazolin, reduction of endotoxin and BAs using cholestyramine, and neutralization of the lipid A portion of circulating endotoxin by polymyxin B [32]. Endotoxin tolerant rats as well as Gram-negative bacteria deficient rats all demonstrated impaired DNA synthesis in response to PHx [32]. Furthermore, LPS could rescue both germ-free and LPS-resistant mice from delayed liver regeneration [33]. The observed LPS-induced hepatocyte proliferation may result from augmentation of hepatocyte growth factor (HGF) activity. Treatment of rats with a combination of LPS and HGF increased JNK and AP-1 DNA binding, possibly through c-JUN and STAT3 up-regulation [34]. LPS-HGF modulation of hepatocyte proliferation indicates potential contribution from the gut microbiota to the liver regeneration program.
Although endotoxin has been shown to induce hepatocyte proliferation, it is important to note that not all endotoxin-releasing bacteria are beneficial for liver regeneration. In mice, orthotopic liver transplantation was associated with increased hepatic inflammation and increased portal endotoxin levels after surgery, often leading to liver injury and rejection [32]. However, when Bifidobacterium, Lactobacillus, Bacteroides, and Eubacterium was increased and Enterobacteriaceae was reduced, portal LPS levels and Kupffer cell activation decreased, which was beneficial for preventing liver injury found in rats after orthotopic liver transplantation [35]. These findings suggest differential effects of specific bacteria on liver regeneration. This is also supported by experiments using antibiotic treatment. It has been shown that norfloxacin treatment did not affect DNA synthesis and hepatic ornithine decarboxylase activity 24 hours after 70% liver resection in a rat model. Thus, selective bowel decontamination with norfloxacin does not seem to change hepatocyte proliferation [36]. A recent study showed that ampicillin-sensitive bacteria were associated with normal liver regeneration [37]. The number of CD1d-dependent natural killer T (NKT) cells was increased in antibiotic-treated hepatectomized mice, and these NKT cells were overly activated to produce elevated interferon-γ. NKT cells deficiency or antibody blockade of the CD1d-NKT interaction increased hepatocyte proliferation, which improved liver regeneration. Moreover, increased Kupffer cells were observed in antibiotic-treated mice, which had elevated interleukin 12 (IL-12) to activate hepatic NKT cells. Interleukin-12p40 deficiency or treatment with anti-IL-12 antibody reduced NKT cell activation and restored liver regeneration in antibiotic-treated mice [37]. Together, mild bacterial translocation with specific bacteria and subsequent endotoxin release is essential to stimulate liver regeneration, but sustained dysbiosis has a negative impact on liver regeneration.
Probiotics
Emerging evidence indicates that the presence of several key bacterial species, mainly Lactobacillus, Bifidobacterium, and Bacteroides species, influences liver injury and regeneration. Carbon tetrachloride-induced cirrhosis was linked to a decreased microbial diversity [38]. In addition, a high proportion of Bifidobacterium animalis was also positively correlated with elevated IL-10 expression, which reinforces the hepatoprotective effects of Bifidobacterium species [38]. Additionally, Bifidobacterium infantis has been implicated in modulating colonic microbial diversity and reducing fecal endotoxin levels [39]. Decreased abundance of these species, particularly Bifidobacterium species, can exacerbate hepatic injury and impede regeneration [40]. Hepatic ischemia/reperfusion (I/R)-induced injury resulted in reduced density of Lactobacillus, Bifidobacterium, and Bacteroides and increased density of Enterococus and Enterobacteriaceae [41]. Probiotic treatment reduces liver injury and examples are listed below. Lactobacillus rhamnosus treatment improved liver function and reduced inflammation in an alcohol-induced liver injury in mice [42, 43]. A combination of Bifidobacterium infantis, Lactobacillus gasseri, and Lactobacillus plantarum relieved colorectal inflammation and tumor-associated hepatic injury [44]. This probiotic treatment in combination with blueberry husks ameliorated dextran sulphate sodium-induced colonic damage to an even greater extent in rats [44]. Lactobacillus salivarius or Pediococcus pentosaceus prevented D-galactosamine-induced rat liver injury as evidenced by reduced total bilirubin as well as colon and liver abnormalities, decreased bacterial translocation and increased IL-10 [45]. Moreover, Pediococcus pentoseceus, Lactococcus raffinolactis, and Lactobacillus paracasei 19 inhibited bacterial translocation after liver resection in rats, and induced hepatocyte mitosis which was delayed by colonic anastomosis [46]. Combined Bifidobacterium longum, Lactobacillus acidophilus, and Enterococcus faecalis treatment in rats that underwent orthotopic liver transplantation protected against liver damage and acute rejection, and altered the intestinal and colonic microbiota by increasing the density of Lactobacillus and Bifidobacterium species [47]. The treatment of synbiotics consisting of four different lactic acid producing bacteria (Pediacoccus pentosaceus, Leuconostoc mesenteroides, Lactobacillus paracasei, and Lactobacillus plantarum) plus four bioactive fibers (β-glucan, inulin, pectin, and resistant starch) improved liver function after liver resection in human [40]. Treatment with the Linex containing Lactobacillus and Bifidobacterium alleviated hepatic injury and restored liver function in chronic liver disease patients by increasing Bifidobacterium, Lactobacillus, and Escherichia abundance [48]. Treatment with Lactobacillus paracasei F19 restored gut microbiota diversity and mitigated liver inflammation and necrosis caused by I/R [41]. Furthermore, Bifidobacterium treatment before I/R reduced endotoxemia, bacterial translocation, and inflammation, and improved intestinal barrier function, which can be potentially beneficial for liver regeneration [42]. Lactobacillus casei treatment has also been shown to mitigate chemical-induced colonic barrier injury and prevent excessive bacterial translocation [49]. Taken together, the conservation of gut microbiota, at least certain species, is consequentially implicated in mucosal homeostasis, which prevents the progression of pathologies.
The effects of bile acids on liver regeneration
There is a great metabolic demand during liver regeneration and BA-mediated intestinal nutrient absorption is essential for proper liver regeneration. However, the concentration of BAs is important in determining whether they are proliferative or cytotoxic. The hydrophobic nature of BAs allows them to act as a detergent for lipid absorption, but the same hydrophobic property can damage cell membranes. The signal transduction of BAs is primarily mediated through G-protein coupled bile salt receptor (TGR5), and farnesoid x receptor (FXR) [50–52].
Bile acid overload impairs liver regeneration
Because there is a fine balance between BAs being cytotoxic or proliferative, much research has focused on factors that alter the ratio of BA to liver volume. Hepatic resection leads to a drastic increase in the ratio of BA to liver volume, overloading the remaining liver with BAs [23, 27, 53]. This sudden spike in hepatic BAs after liver resection can cause devastating cytotoxicity due to increased oxidative stress and cell membrane permeability. There are several hepatoprotective mechanisms, mainly regulated through BA receptors and transporters, in place to prevent BA overload and thus additional liver injury, which are illustrated and explained in Figure 1. It has been shown that TGR5 or FXR knockout (KO) in mice have enlarged total BA pool size, increased inflammation, and impaired liver regeneration [23, 24]. Sequestration of BAs through cholestyramine treatment or suppressed inflammation by Kupffer cell depletion alleviated the delayed liver regeneration seen in TGR5 as well as FXR KO mice [23, 24]. Two-thirds PHx leads to a 2.5-folds increase in BA secretion along with 3-folds increased mRNA expression of multidrug resistant protein 2 (Mdr2), which aids in preventing BA overload [54, 55]. Additionally, the mRNA levels of other BA transporters, bile salt export pump (Bsep) and multidrug resistance protein 3 (Mrp3) are up-regulated in the first 48 hours after PHx, indicating the important cytoprotective effects of tightly regulated BA homeostasis [54]. BAs, through small heterodimer partner (SHP) nuclear receptor, and fibroblast growth factor 15 (FGF15) down-regulate cholesterol 7α-hydroxylase (CYP7A1) and sterol 12α-hydroxylase (CYP8B1) expression, thus prohibiting BA synthesis for 48 hours after liver resection [56]. Consistently, CYP7A1 is inhibited by HGF [57]. Such inhibitory feedback can prevent BA-induced toxicity, hepatocyte apoptosis, and liver damage.
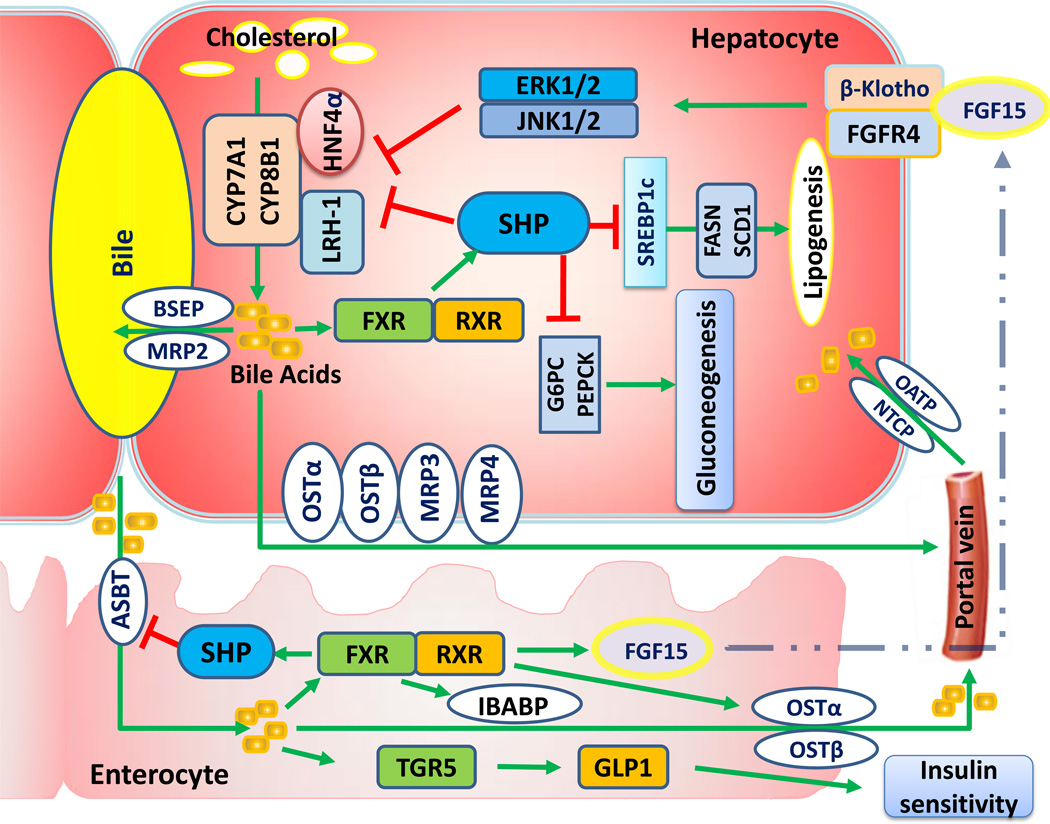
Fig. 1. Bile acid homeostasis and its downstream effects on carbohydrate and lipid metabolism.
Hepatic cholesterol through cholesterol-7α-hydroxylase (CYP7A1) and sterol 12α-hydroxylase (CYP8B1) is converted to bile acids (BAs). BA transporters are involved in the secretion of BAs from the liver into the duodenum, flowing back through the ileum and reabsorbed by the liver. Farnesoid x receptor (FXR) and its targets are involved in the enterohepatic recycling of BAs. FXR-induced small heterodimer partner (SHP) inhibits BA synthesis by down-regulating CYP7A1 and CYP8B1 expression. Additionally, intestinal FXR induces fibroblast growth factor 15 (FGF15) expression, which in turn represses CYP7A1 through fibroblast growth factor receptor 4 (FGFR4) and β-Klotho-mediated signaling. Regarding lipid and carbohydrate metabolism, hepatic FXR activation induces gluconeogenesis and represses lipogenesis through SHP [100, 101]. Intestinal BAs also activate G protein-coupled bile acid receptor (TGR5) to increase production and secretion of glucagon-like peptide-1 (GLP-1), an incretin hormone that promotes insulin sensitivity, and thereby improves glucose disposition [102, 103]. CYP7A1, cholesterol-7α-hydroxylase; CYP8B1, sterol 12α-hydroxylase; BAs, bile acids; FXR, farnesoid x receptor; SHP, small heterodimer partner; FGF15, fibroblast growth factor 15; FGFR4, fibroblast growth factor receptor 4; TGR5, G protein-coupled bile acid receptor; GLP-1, glucagon-like peptide-1.
Bile acid and gut bacteria-derived signaling are essential for liver regeneration
Although excess BAs can cause liver injury and impair liver regeneration, BAs have also been shown to be critical for proper restoration of liver mass and function. Plasma BAs levels were positively correlated with liver regenerative response in rabbits following portal vein embolization [58]. An initial expansion in BA pool size accelerated the regenerative process, which indicates that while excess levels may inhibit regeneration, BAs potentiate hepatocyte proliferation [24]. PHx-induced liver regeneration was markedly delayed in rats when BA pool size was contracted by cholestyramine, a BA sequestrant [59]. Surgical and genetic disruptions of normal BA enterohepatic circulation and influx into the liver severely attenuated liver regeneration after PHx in mice [60]. PHx accompanied by ileal resection resulted in diminished liver regeneration capability, most likely due to loss of BA reabsorption in the ileum [61]. These findings highlight BA circulation through the gut-liver axis as an important regulatory component of the liver regeneration program. Taken together, both the injurious and proliferative effects of BAs on hepatocytes emphasize the importance of appropriately maintaining BA homeostasis to facilitate liver repair.
The role of BA signaling during liver regeneration has been reviewed [62]. For the thoroughness of this review, we briefly cover the role of FXR-associated pathways in regulating liver regeneration. In addition to regulating BA homeostasis, FXR controls lipid and glucose metabolism [63] (Fig.1). FXR whole body KO mice exhibited a delayed liver regeneration due to dysregulated BA synthesis [24]. Intestinal FXR was also found to facilitate liver regeneration through up-regulation of FGF15 in mice (FGF19 in humans) [64]. FGF15 is an ileal-secreted enterokine that is induced by FXR to inhibit BA overproduction [65]. Additionally, intestinal FXR KO impeded liver regeneration as a result of insufficient FGF15 activity which was rescued by administration of exogenous FGF15 [21]. As such, FGF15 KO mice suffered significantly higher lethality rates after liver resection due to hepatic failure relative to wild type mice [64, 66]. Furthermore, hepatocyte-specific FXR KO mice also show delayed liver regeneration from inactivation of CYCLIN D and suppressed HGF-mediated signaling [67]. In addition to the vital role of BA circulation through the gut-liver axis, cytokine and paracrine signaling molecules generated from the liver and intestine including tumor necrosis factor α (TNFα), IL-6, and FGF15/19, and HGF impact liver regeneration as well [68]. HGF treatment reduces inflammation and promotes colonic epithelial regeneration, potentially preventing translocation of harmful microbes and metabolites across the intestinal mucosa [69]. Treating mice with glucagon-like peptide 2 also accelerated PHx-induced liver regeneration [70]. Taken together, liver regeneration is regulated by the enterohepatic circulation of BAs as well as cytokines and growth factors.
The interplay between the gut microbiota and bile acid homeostasis
Gut microbiota modulates bile acid production
Hepatic as well as microbial enzymes are responsible for the synthesis of various BAs (Fig. 2). There is a species difference in BA profiles [71, 72]. In human, cholic acid (CA) and chenodeoxycholic acid (CDCA) are primary BAs [72]. However, in mice, α-muricholic acid (α-MCA) and β-MCA are the major primary BAs [72]. These primary BAs are sterol compounds synthesized from cholesterol and conjugated with mainly glycine in human or taurine in mice [73]. Primary BAs enter the intestinal lumen and undergo deconjugation, dehydroxylation, epimerization, and oxidation using bacterial enzymes [72]. Conjugation increases the aqueous solubility of BAs and renders them largely impermeable to the intestinal epithelium, thus preventing them from exiting the intestinal lumen. The conversion of primary to secondary BAs deoxycholic acid (DCA) and lithocholic acid (LCA) is also mediated via bacterial enzyme 7α-dehydroxylase [73]. Therefore, the composition of BAs in germ-free and conventional rats is drastically different; specifically, germ-free rats have elevated taurine-conjugated BAs and reduced secondary and glycine-conjugated BAs [74]. Among BAs, CDCA has the highest binding affinity to FXR [75]. In mice, tauro-β-MCA (T-β-MCA) is an inhibitor of FXR [76]. These findings point to the possibility that intestinal bacteria not only regulate BA deconjugation, but also BA synthesis through FXR.
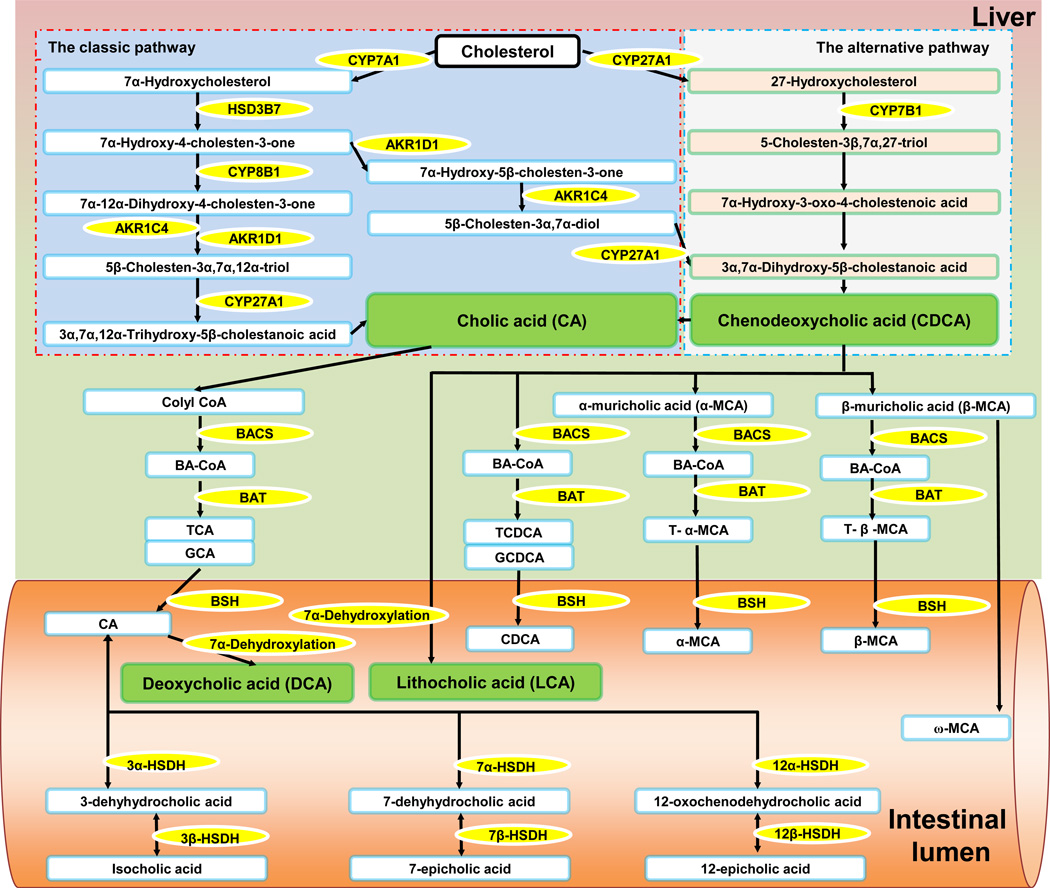
Fig 2. Bile acid synthesis as mediated by hepatic and intestinal bacteria enzymes.
There are two pathways responsible for bile acid (BA) synthesis in the liver. In the classic pathway, the rate-limiting step in BA formation is conversion of cholesterol to 7α-hydroxycholesterol by cholesterol 7α-hydroxylase (CYP7A1). Multiple sequential steps that modify the steroid nucleus and side chain produce two primary BAs, cholic acid (CA) and chenodeoxycholic acid (CDCA). The main enzymes in those modification steps are 3β-hydroxy-Δ5-C27-steroid dehydrogenase (HSD3B7), sterol 12α-hydroxylase (CYP8B1), Δ4-3-oxosteroid-5β-reductase (AKR1D1), 3α-hydroxysteroid dehydrogenase (AKR1C4), and sterol 27-hydroxylase (CYP27A1). Through the alternative pathway, cholesterol is converted into CDCA and the key enzymes involved are CYP27A1 and 25-hydroxycholesterol 7-α-hydroxylase (CYP7B1). Free primary BAs are conjugated through two reactions. First, using BA-CoA synthase (BACS), BA-CoA is generated. Next, BA-CoA:amino acid N-acyltransferase (BAT) amidates BA-CoA with either a taurine or a glycine. In the intestines, 7α-dehydroxylation of CA and CDCA converts the primary BAs into secondary BAs including deoxycholic acid (DCA) and lithocholic acid (LCA), respectively. The known bacterial species possessing 7α-dehydroxylation activity are members of the Firmicutes phylum (Clostridium and Eubacterium) [104]. The taurine or glycine conjugated BAs are catalyzed by bile salt hydrolases (BSHs) to become free BAs. BSH can be detected in bacterial genera including Bacteroides, Bifidobacterium, Clostridium, Lactobacillus, and Listeria [105]. Free BAs can be oxidized by hydroxysteroid dehydrogenases (HSDHs). For example, CA can be converted into 3-dehyhydrocholic acid, 7-dehyhydrocholic acid, and 12-oxochenodehydrocholic acid by 3α-HSDH, 7α-HSDH, and 12α-HSDH, respectively, and further converted into isocholic acid, 7-epicholic acid, and 12-epicholic acid by 3β-HSDH, 7β-HSDH, and 12β-HSDH, respectively. HSDHs are expressed by Firmicutes phylum members, including Eubacterium, Peptostreptococcus, and Ruminococcus [106]. BAs, bile acids; CYP7A1, cholesterol 7α-hydroxylase; CA, cholic acid; CDCA, chenodeoxycholic acid; HSD3B7, 3β-hydroxy-Δ5-C27-steroid dehydrogenase; CYP8B1, sterol 12α-hydroxylase; AKR1D1, Δ4-3-oxosteroid-5β-reductase; AKR1C4, 3α-hydroxysteroid dehydrogenase; CYP27A1, sterol 27-hydroxylase; CYP7B1, 25-hydroxycholesterol 7-α-hydroxylase; BACS, BA-CoA synthase; BAT, BA-CoA:amino acid N-acyltransferase; DCA, deoxycholic acid, LCA, lithocholic acid; HSDH, hydroxysteroid dehydrogenase.
A cross-sectional study of patients with cirrhosis showed elevated primary BAs and Enterobacteriaceae and diminished 7α-dehydroxylating bacteria including Lachonospiraceae, Ruminococcaceae, and Blautia [77]. Mice treated with antibiotics consisting of bacitracin, neomycin, and streptomycin had increased tauro-CA (TCA) and T-β-MCA and reduced secondary BAs, which indicated the diminished intestinal 7α-dehydroxylating bacteria [78]. In addition, antibiotic treatment also suppressed Fgf15 expression and increased Cyp7a1 expression, which indicated the regulation of microbiota on BA synthesis [78]. This modulation of intestinal FXR and BA synthesis carries many potential implications for liver regeneration, and requires further investigation. Additionally, total and fecal secondary BA levels were diminished in patients with cirrhotic livers with Enterobacteriaceae and Ruminococcaceae growth positively correlating with CDCA and DCA levels, respectively [77]. Moreover, in cirrhotic patients who consumed alcohol, analysis of fecal and serum BA levels, serum endotoxin and stool microbiota revealed increased mRNA levels of inflammatory cytokines as well as secondary hydrophobic BAs [79]. Such elevation in cytotoxic secondary BAs may compromise intestinal epithelial integrity and contribute to dysbiosis, which in turn impairs liver regeneration. Taken together, these findings implicate the gut microbiota in modulating the production and composition of BAs.
Bile acids modulate the gut microbiota
While intestinal bacteria modulate BA synthesis, BAs can mutually influence the gut microbial population. In a FXR-dependent manner, conjugated BAs can exert antimicrobial effects in the digestive tract [76]. Consequently, FXR KO mice exhibited higher densities of ileal bacteria and compromised epithelial barrier integrity [80]. This effect was also observed in mice with biliary obstruction and reversible by administration of a FXR agonist [80]. Conversely, hydrophobic, taurine-conjugated BAs enhanced the growth of sulfate-reducing gut bacteria, leading to a “leaky gut” with increased antigen and bacterial translocation, cholelithiasis, carcinoma, inflammatory bowel disease, and colorectal cancer [81, 82]. Moreover, a low-fat diet supplemented with TCA, promoted changes in mouse-host BA composition, which can markedly alter conditions for gut microbial assemblage, resulting in dysbiosis and disrupted immune homeostasis. However, an increase in intestinal T-β-MCA caused by tempol, an antioxidant, reduced the colonic population of Lactobacillus, decreased bile salt hydrolase activity in the feces, and inhibited the intestinal FXR signaling [83]. This evidence suggests that the gut microbiota, as an “organ”, is capable of adapting to dynamic changes in intestinal environment. Exogenous administration of CA up-regulated bacterial 7α-dehydroxylation-mediated DCA production and altered the gut microbiota population with increased abundance of Firmicutes over Bacteroidetes in rat [84]. In addition, exogenous CA increased pathogenic Clostridia and Erysipelotrichi populations, which can lead to colitis and cirrhosis [85]. Overall, it appears that factors influencing either the BA composition or gut microbial diversity may also significantly impact on liver function and regeneration.
The influence of GI disease on liver injury and regeneration as mediated by gut bacteria
Because hepatic regeneration is dependent on signaling mediators derived from the GI tract, diseases or pathologies that disturb the normal intestinal environment, particularly the gut microbiota, could interfere with liver regeneration. Subsequently discussed are studies that have shown a correlation between GI diseases, alterations in the gut microbiota, and hepatic injury as well as regeneration.
Compromised colonic epithelial barrier
Intestinal pathologies are linked to factors involved in liver injury or regeneration. For example, small bowel resection in piglets caused gut microbiota dysbiosis, which resulted in significant BA dysregulation and harmful clinical outcomes including steatorrhoea, persistent diarrhea, liver injury, and impaired regeneration. Small bowel resection also interrupted FXR-mediated signaling pathways, which are essential for liver regeneration [86]. Increased intestinal permeability in alcoholic patients was positively correlated with severity of cirrhosis in alcoholic patients. A “leaky gut” caused endotoxemia in rats and humans and contributed to alcohol-induced hepatic cirrhosis and dysfunction [87]. Furthermore, nonalcoholic fatty liver disease in rats was associated with compromised intestinal barrier integrity and elevated LPS [88]. Knockout toll-like receptor 4, an important modulator of innate immune response to LPS, resulted in aggressive onset of colitis and subsequent bacterial translocation to mesenchymal lymph nodes [89].
Sepsis-induced liver and colonic epithelial damage could be ameliorated by probiotic VSL#3, which restored the diversity of the intestinal microbiota [90]. This study showed that administration of a peroxisome proliferator-activated receptor gamma (PPARγ) inhibitor completely abolished the anticipated probiotic benefits, suggesting that VSL#3 treatment may promote liver regeneration through a PPARγ-mediated pathway. Interestingly, liver regeneration was found to be accelerated in liver-specific PPARγ-null mice on a normal diet, but impaired when mutant mice suffered diet-induced fatty liver, suggesting that PPARγ inhibition may be detrimental in a state of intestinal dysbiosis [91]. Bioactive peptide factors from Bifidobacterium infantis were also shown to improve epithelial cell barrier function and reduce inflammation, implying a potential pathway through which certain beneficial bacteria may enhance liver regeneration by protecting against hepatic damage [92]. Metabolic pathways may also exert a hepatoprotective effect following liver injury. Parenteral administration of glutamine after liver resection dramatically increased liver regeneration by promoting hepatic alanine uptake and intestinal glutamine metabolism. Protein synthesis in colonic epithelium was increased, whereas bacterial translocation and endotoxin levels were greatly reduced [93]. This improvement in intestinal epithelial barrier function may shield the liver from excessive endotoxemia after liver resection.
Liver disease and alterations of gut microbiota
Hepatic diseases have been linked to altered microbial diversity in the intestines that may create a positive feedback cycle that exacerbates hepatic injury and impede liver regeneration. Alcoholic liver disease patients generally had contracted Bacteroides species and expanded Proteobacteria species. [94]. This gut dysbiosis was also correlated with elevated serum endotoxin, likely from excessive bacterial translocation [94]. The presence of endotoxemia along with reduction in Bacteroides density is expected to negatively impact liver regeneration. The study of liver steatosis, alcoholic and non-alcoholic, has proven valuable to illuminating the downstream consequences of gut microbiota alterations. Nonalcoholic steatohepatitis provokes an innate immune response, which stimulates hepatic inflammation through cytokines such as TNFα [95]. Obesity-induced nonalcoholic steatohepatitis also perturbed gut microbiota composition by decreasing total microbial diversity, most likely by Bacteroidetes species contraction [96]. Hepatic lipid contents in patients with choline deficiency have also been shown to affect gut microbial diversity [97]. Treatment with a combination of five Chinese herbs (Compositae : Polygonacease : Zingiberaceae : Clusiaceae : Rubiaceae = 13 : 7 : 7 : 7 : 7) was found to promote growth of short chain fatty acid producer Collinsella while improving steatosis in rats [98]. This altered gut microbiota associated with steatosis, particularly diminished Bacteroidetes abundance, may indicate gut dysbiosis and propagation of further hepatic injury. Other etiologies, such as GI diseases, can also influence hepatic injury through modulation of the gut microbiota. In a rat model of irritable bowel syndrome, administration of Lactobacillus casei and Bifidobacterium lactis either before or after irritable bowel syndrome induction alleviated inflammation and apoptosis in both the colon and liver [99]. Together, there is an intimate relationship between hepatic metabolism, microbiota, and liver injury as well as regeneration.
Summary and future directions
It is well recognized that diet and nutrition play a significant role in the etiology of metabolic diseases and that affects tissue injury and repair. However, the precise mechanisms by which diets affect our health status and outcomes, particularly in the GI system, are poorly understood. Despite the exponential growth in marketing of synbiotics and probiotic products, there is a lack of established mechanistic links between gut microbiota alterations and physiological responses from the host. The current summary provides promising evidence, which indicates intestinal bacteria and BAs cross talk within the gut-liver axis and jointly regulate nutrient absorption, liver metabolism, and inflammatory processes. Thus, BA and bacteria-mediated signaling within the gut-liver axis is crucial for proper execution of injury response and repair, such relationship is summarized in Figure 3. It is critical to gain insights into how nutrient-host and microbiota-host interactions influence an individual’s predisposition to injury and tissue repair. Due to the intricate networks of implicated pathways as well as scarcity of available information, it seems that nutrigenomic, metabolomics, and microbiota profiling approaches are warranted to provide a better understanding regarding the impact of the aforementioned factors in influencing liver function and healing.
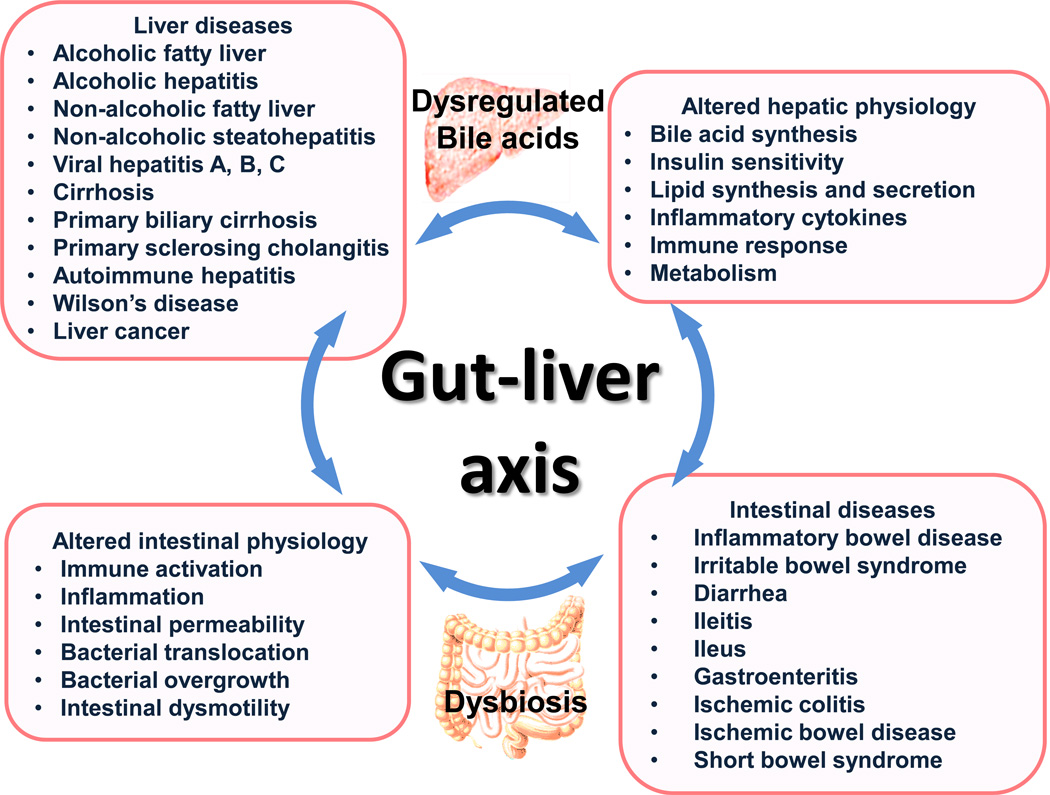
Fig. 3. Overview of the interplay between bile acid dysregulation and gut dysbiosis in the context of liver and GI pathologies.
The gut and the liver are intimately associated, and there is continuous bidirectional communication between these organs through bile acids, hormones, inflammatory mediators, and products of digestion and absorption. A variety of liver diseases affect the bile acid profile, which contributes to gut dysbiosis and intestinal pathogenesis. Similarly, intestinal diseases lead to dysbiosis and change the bile acid profile that in turn affect metabolism and inflammatory response in the liver.
Key Points.
Microbiota and bile acids within the gut-liver axis are crucial in regulating metabolism and inflammatory processes, and thus are important for liver injury and liver regeneration.
There exists a “gut-liver axis” that facilitates bidirectional communication between intestinal microbes and hepatic bile acid metabolism. In one direction, the gut microbiota plays a pivotal role in regulating bile acid homeostasis while on the other end, bile acids influence gut microbiota composition.
Because hepatic regeneration is dependent on signaling mediators derived from the gastrointestinal tract, diseases or pathologies that disturb the normal intestinal environment, particularly the gut microbiota, interfere with liver regeneration.
Despite the exponential growth in marketing of synbiotics and probiotic products, there is a lack of established mechanistic links between gut microbiota alterations and physiological responses from the host. The summarized data provide promising evidence that bile acids and microbiota jointly regulate nutrient absorption, hepatic metabolism, and inflammatory processes thus maintain the health of gut and liver.
Acknowledgement
This study is supported by grants funded by National Institutes of Health CA53596, DK092100, and U01CA179582. The authors thank Thinh Chau and Lisa Teixeira for editing the manuscript.
Abbreviations
- BAs
bile acids
- GI
gastrointestinal tract
- LPS
lipopolysaccharide
- PHx
partial hepatectomy
- HGF
hepatocyte growth factor
- NKT
natural killer T
- IL
interleukin
- I/R
ischemia/reperfusion
- TGR5
G-protein coupled membrane receptor
- FXR
farnesoid x receptor
- KO
knockout
- Mdr2
multidrug resistance 2
- Bsep
bile salt export pump
- Mrp3
multidrug resistance protein 3
- SHP
small heterodimer partner
- FGF15
fibroblast growth factor 15
- CYP7A1
cholesterol 7α-hydroxylase
- CYP8B1
sterol 12α-hydroxylase
- TNFα
tumor necrosis factor α
- CA
cholic acid
- CDCA
chenodeoxycholic acid
- MCA
muricholic acid
- DCA
deoxycholic acid
- LCA
lithocholic acid
- T-β-MCA
tauro-β-muricholic acid
- TCA
tauro-cholic acid
- PPARγ
peroxisome proliferator-activated receptor gamma
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest: The authors who have taken part in this review declared that they do not have any disclosures regarding funding or conflict of interest.
Author’s Contributions: Hui-Xin Liu, literature search, writing and revising manuscript as well as figure illustrations; Ryan Keane, literature search and drafting manuscript. Lili Sheng, writing and editing manuscript; Yu-Jui Yvonne Wan, conception of the paper, revising paper critically for important intellectual content and final approval of the version to be published.
References
- 1.Michalopoulos GK. Liver-Regeneration - Molecular Mechanisms of Growth-Control. Faseb J. 1990;4:176–187. [PubMed] [Google Scholar]
- 2.Michalopoulos GK. Control Mechanisms of Liver-Regeneration. J Gastroenterol. 1994;29:23–29. [PubMed] [Google Scholar]
- 3.Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60–66. doi: 10.1126/science.276.5309.60. [DOI] [PubMed] [Google Scholar]
- 4.Michalopoulos GK, DeFrances M. Liver regeneration. Adv Biochem Eng Biot. 2005;93:101–134. doi: 10.1007/b99968. [DOI] [PubMed] [Google Scholar]
- 5.Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michalopoulos GK. Liver Regeneration. Mol Pathol Lib. 2011;5:261–278. [Google Scholar]
- 7.Michalopoulos GK. Advances in liver regeneration. Expert Rev Gastroent. 2014;8:897–907. doi: 10.1586/17474124.2014.934358. [DOI] [PubMed] [Google Scholar]
- 8.Taub R. Liver regeneration: From myth to mechanism. Nat Rev Mol Cell Bio. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 9.Fausto N, Campbell JS. The role of hepatocytes and oval cells in liver regeneration and repopulation. Mech Develop. 2003;120:117–130. doi: 10.1016/s0925-4773(02)00338-6. [DOI] [PubMed] [Google Scholar]
- 10.Fausto N. Liver regeneration. J Hepatol. 2000;32:19–31. doi: 10.1016/s0168-8278(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 11.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43:S45–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 12.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. J Hepatol. 2012;57:692–694. doi: 10.1016/j.jhep.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 13.Fausto N. New Perspectives on Liver-Regeneration. Hepatology. 1986;6:326–327. doi: 10.1002/hep.1840060229. [DOI] [PubMed] [Google Scholar]
- 14.Fausto N. Liver-Regeneration - Models and Mechanisms. Tr Adv Liv. 1992:1–6. [Google Scholar]
- 15.Monga SP. Role and regulation of beta-catenin signaling during physiological liver growth. Gene expression. 2014;16:51–62. doi: 10.3727/105221614X13919976902138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monga SP. Role of Wnt/beta-catenin signaling in liver metabolism and cancer. The international journal of biochemistry & cell biology. 2011;43:1021–1029. doi: 10.1016/j.biocel.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu HX, Fang YP, Hu Y, Gonzalez FJ, Fang JW, Wan YJY. PPAR beta Regulates Liver Regeneration by Modulating Akt and E2f Signaling. Plos One. 2013;8 doi: 10.1371/journal.pone.0065644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai GL, He L, Bu PL, Wan YJY. Pregnane X receptor is essential for normal progression of liver regeneration. Hepatology. 2008;47:1277–1287. doi: 10.1002/hep.22129. [DOI] [PubMed] [Google Scholar]
- 19.Yang XX, Guo ML, Wan YJY. Hepatocyte retinoid X receptor alpha (RXRalpha) deficiency impairs liver regeneration through multiple pathways. Faseb J. 2009;23 [Google Scholar]
- 20.Liu HX, Ly I, Hu Y, Wan YJY. Retinoic acid regulates cell cycle genes and accelerates normal mouse liver regeneration. Biochem Pharmacol. 2014;91:256–265. doi: 10.1016/j.bcp.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang LS, Wang YD, Chen WD, Wang XC, Lou GY, Liu N, et al. Promotion of Liver Regeneration/Repair by Farnesoid X Receptor in Both Liver and Intestine in Mice. Hepatology. 2012;56:2336–2343. doi: 10.1002/hep.25905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Y, Zhan Q, Liu HX, Chau T, Li YY, Wan YJY. Accelerated Partial Hepatectomy-Induced Liver Cell Proliferation Is Associated with Liver Injury in Nur77 Knockout Mice. Am J Pathol. 2014;184:3272–3283. doi: 10.1016/j.ajpath.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pean N, Doignon I, Garcin I, Besnard A, Julien B, Liu BK, et al. The Receptor TGR5 Protects the Liver From Bile Acid Overload During Liver Regeneration in Mice. Hepatology. 2013;58:1451–1460. doi: 10.1002/hep.26463. [DOI] [PubMed] [Google Scholar]
- 24.Huang WD, Ma K, Zhang J, Qatanani M, Cuvillier J, Liu J, et al. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 2006;312:233–236. doi: 10.1126/science.1121435. [DOI] [PubMed] [Google Scholar]
- 25.Gazit V, Huang JS, Weymann A, Rudnick DA. Analysis of the role of hepatic PPAR? expression during mouse liver regeneration. Hepatology. 2012;56:1489–1498. doi: 10.1002/hep.25880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson SP, Yoon L, Richard EB, Duan CS, Cattley RC, Corton JC. Delayed liver regeneration in peroxisome proliferator-activated receptor-alpha-null mice. Hepatology. 2002;36:544–554. doi: 10.1053/jhep.2002.35276. [DOI] [PubMed] [Google Scholar]
- 27.Liu HX, Hu Y, French SW, Gonzalez FJ, Wan YJ. Forced expression of fibroblast growth factor 21 reverses the sustained impairment of liver regeneration in hPPARalphaPAC mice due to dysregulated bile acid synthesis. Oncotarget. 2015;6:9686–9700. doi: 10.18632/oncotarget.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sommer F, Backhed F. The gut microbiota - masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 29.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. Embo Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 31.Cornell RP. Gut-Derived Endotoxin Elicits Hepatotrophic Factor Secretion for Liver-Regeneration. Am J Physiol. 1985;249:R551–R562. doi: 10.1152/ajpregu.1985.249.5.R551. [DOI] [PubMed] [Google Scholar]
- 32.Cornell RP. Restriction of Gut-Derived Endotoxin Impairs DNA-Synthesis for Liver-Regeneration. Am J Physiol. 1985;249:R563–R569. doi: 10.1152/ajpregu.1985.249.5.R563. [DOI] [PubMed] [Google Scholar]
- 33.Cornell RP, Liljequist BL, Bartizal KF. Depressed Liver-Regeneration after Partial-Hepatectomy of Germ-Free, Athymic and Lipopolysaccharide-Resistant Mice. Hepatology. 1990;11:916–922. doi: 10.1002/hep.1840110603. [DOI] [PubMed] [Google Scholar]
- 34.Gao CH, Jokerst R, Gondipalli P, Cai SR, Kennedy S, Flye MW, et al. Lipopolysaccharide potentiates the effect of hepatocyte growth factor on hepatocyte replication in rats by augmenting AP-1 activity. Hepatology. 1999;30:1405–1416. doi: 10.1002/hep.510300602. [DOI] [PubMed] [Google Scholar]
- 35.Arai M, Mochida S, Ohno A, Arai S, Fujiwara K. Selective bowel decontamination of recipients for prevention against liver injury following orthotopic liver transplantation: Evaluation with rat models. Hepatology. 1998;27:123–127. doi: 10.1002/hep.510270120. [DOI] [PubMed] [Google Scholar]
- 36.MacIntosh EL, Gauthier T, Harding GK, Minuk GY. Selective bowel decontamination does not alter hepatic regeneration in rats. Gastroenterology. 1992;102:1403–1405. [PubMed] [Google Scholar]
- 37.Wu X, Sun R, Chen Y, Zheng X, Bai L, Lian Z, et al. Oral ampicillin inhibits liver regeneration by breaking hepatic innate immune tolerance normally maintained by gut commensal bacteria. Hepatology. 2015;62:253–264. doi: 10.1002/hep.27791. [DOI] [PubMed] [Google Scholar]
- 38.Cuenca S, Sanchez E, Santiago A, El Khader I, Panda S, Vidal S, et al. Microbiome Composition by Pyrosequencing in Mesenteric Lymph Nodes of Rats with CCI4-induced Cirrhosis. J Innate Immun. 2014;6:263–271. doi: 10.1159/000356454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodes L, Saha S, Tomaro-Duchesneau C, Prakash S. Microencapsulated Bifidobacterium longum subsp infantis ATCC 15697 Favorably Modulates Gut Microbiota and Reduces Circulating Endotoxins in F344 Rats. Biomed Res Int. 2014 doi: 10.1155/2014/602832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rayes N, Pilarski T, Stockmann M, Bengmark S, Neuhaus P, Seehofer D. Effect of pre-and probiotics on liver regeneration after resection: a randomised, double-blind pilot study. Benef Microbes. 2012;3:237–244. doi: 10.3920/BM2012.0006. [DOI] [PubMed] [Google Scholar]
- 41.Nardone G, Compare D, Liguori E, Di Mauro V, Rocco A, Barone M, et al. Protective effects of Lactobacillus paracasei F19 in a rat model of oxidative and metabolic hepatic injury. Am J Physiol-Gastr L. 2010;299:G669–G676. doi: 10.1152/ajpgi.00188.2010. [DOI] [PubMed] [Google Scholar]
- 42.Wang YH, Kirpich I, Liu YL, Ma ZH, Barve S, McClain CJ, et al. Lactobacillus rhamnosus GG Treatment Potentiates Intestinal Hypoxia-Inducible Factor, Promotes Intestinal Integrity and Ameliorates Alcohol-Induced Liver Injury. Am J Pathol. 2011;179:2866–2875. doi: 10.1016/j.ajpath.2011.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang YH, Liu YL, Kirpich I, Ma ZH, Wang CL, Zhang M, et al. Lactobacillus rhamnosus GG reduces hepatic TNF alpha production and inflammation in chronic alcohol-induced liver injury. J Nutr Biochem. 2013;24:1609–1615. doi: 10.1016/j.jnutbio.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hakansson A, Branning C, Molin G, Adawi D, Hagslatt ML, Jeppsson B, et al. Blueberry Husks and Probiotics Attenuate Colorectal Inflammation and Oncogenesis, and Liver Injuries in Rats Exposed to Cycling DSS-Treatment. Plos One. 2012;7 doi: 10.1371/journal.pone.0033510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lv LX, Hu XJ, Qian GR, Zhang H, Lu HF, Zheng BW, et al. Administration of Lactobacillus salivarius LI01 or Pediococcus pentosaceus LI05 improves acute liver injury induced by D-galactosamine in rats. Appl Microbiol Biot. 2014;98:5619–5632. doi: 10.1007/s00253-014-5638-2. [DOI] [PubMed] [Google Scholar]
- 46.Seehofer D, Rayes N, Schiller R, Stockmann M, Muller AR, Schirmeier A, et al. Probiotics partly reverse increased bacterial translocation after simultaneous liver resection and colonic anastomosis in rats. J Surg Res. 2004;117:262–271. doi: 10.1016/j.jss.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 47.Xie YR, Chen HZ, Zhu B, Qin N, Chen YB, Li ZF, et al. Effect of Intestinal Microbiota Alteration on Hepatic Damage in Rats with Acute Rejection After Liver Transplantation. Microb Ecol. 2014;68:871–880. doi: 10.1007/s00248-014-0452-z. [DOI] [PubMed] [Google Scholar]
- 48.Kirpich IA, Solovieva NV, Leikhter SN, Shidakova NA, Lebedeva OV, Sidorov PI, et al. Probiotics restore bowel flora and improve liver enzymes in human alcohol-induced liver injury: a pilot study. Alcohol. 2008;42:675–682. doi: 10.1016/j.alcohol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Llopis M, Antolin M, Guarner F, Salas A, Malagelada JR. Mucosal colonisation with Lactobacillus casei mitigates barrier injury induced by exposure to trinitronbenzene sulphonic acid. Gut. 2005;54:955–959. doi: 10.1136/gut.2004.056101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsuei J, Chau T, Mills D, Wan YJY. Bile acid dysregulation, gut dysbiosis, and gastrointestinal cancer. Exp Biol Med. 2014;239:1489–1504. doi: 10.1177/1535370214538743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li TG, Chiang JYL. Bile acids as metabolic regulators. Curr Opin Gastroen. 2015;31:159–165. doi: 10.1097/MOG.0000000000000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li TG, Chiang JYL. Bile Acid Signaling in Metabolic Disease and Drug Therapy. Pharmacol Rev. 2014;66:948–983. doi: 10.1124/pr.113.008201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fukano S, Saitoh Y, Uchida K, Akiyoshi T, Takeda K. Bile-Acid Metabolism in Partially Hepatectomized Rats. Steroids. 1985;45:209–227. doi: 10.1016/0039-128x(85)90071-6. [DOI] [PubMed] [Google Scholar]
- 54.Csanaky IL, Aleksunes LM, Tanaka Y, Klaassen CD. Role of hepatic transporters in prevention of bile acid toxicity after partial hepatectomy in mice. Am J Physiol-Gastr L. 2009;297:G419–G433. doi: 10.1152/ajpgi.90728.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang RX, Sheps JA, Ling V. ABC Transporters, Bile Acids, and Inflammatory Stress in Liver Cancer. Curr Pharm Biotechno. 2011;12:636–646. doi: 10.2174/138920111795163986. [DOI] [PubMed] [Google Scholar]
- 56.Yang F, He YQ, Liu HX, Tsuei J, Jiang XY, Yang L, et al. All-trans retinoic acid regulates hepatic bile acid homeostasis. Biochem Pharmacol. 2014;91:483–489. doi: 10.1016/j.bcp.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song KH, Ellis E, Strom S, Chiang JY. Hepatocyte growth factor signaling pathway inhibits cholesterol 7alpha-hydroxylase and bile acid synthesis in human hepatocytes. Hepatology. 2007;46:1993–2002. doi: 10.1002/hep.21878. [DOI] [PubMed] [Google Scholar]
- 58.Hoekstra LT, Rietkerk M, van Lienden KP, van den Esschert JW, Schaap FG, van Gulik TM. Bile salts predict liver regeneration in rabbit model of portal vein embolization. J Surg Res. 2012;178:773–778. doi: 10.1016/j.jss.2012.06.038. [DOI] [PubMed] [Google Scholar]
- 59.Dong XS, Zhao HL, Ma XM, Wang SM. Reduction in bile acid pool causes delayed liver regeneration accompanied by down-regulated expression of FXR and c-Jun mRNA in rats. J Huazhong U Sci-Med. 2010;30:55–60. doi: 10.1007/s11596-010-0110-8. [DOI] [PubMed] [Google Scholar]
- 60.Naugler WE. Bile Acid Flux Is Necessary for Normal Liver Regeneration. Plos One. 2014;9 doi: 10.1371/journal.pone.0097426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Medeiros AC, Azevedo ACB, Oseas JMD, Gomes MDF, de Oliveira FG, Rocha KBF, et al. The ileum positively regulates hepatic regeneration in rats. Acta Cir Bras. 2014;29:93–98. doi: 10.1590/S0102-86502014000200004. [DOI] [PubMed] [Google Scholar]
- 62.Fan MJ, Wang XC, Xu GY, Yan QF, Huang WD. Bile acid signaling and liver regeneration. Bba-Gene Regul Mech. 2015;1849:196–200. doi: 10.1016/j.bbagrm.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vallim TQD, Tarling EJ, Edwards PA. Pleiotropic Roles of Bile Acids in Metabolism. Cell Metab. 2013;17:657–669. doi: 10.1016/j.cmet.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uriarte I, Fernandez-Barrena MG, Monte MJ, Latasa MU, Chang HCY, Carotti S, et al. Identification of fibroblast growth factor 15 as a novel mediator of liver regeneration and its application in the prevention of post-resection liver failure in mice. Gut. 2013;62:899–910. doi: 10.1136/gutjnl-2012-302945. [DOI] [PubMed] [Google Scholar]
- 65.Schaap FG, Leclercq IA, Jansen PLM, Damink SWO. Prometheus' little helper, a novel role for fibroblast growth factor 15 in compensatory liver growth. J Hepatol. 2013;59:1121–1123. doi: 10.1016/j.jhep.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 66.Kong B, Huang JS, Zhu Y, Li GD, Williams J, Shen S, et al. Fibroblast growth factor 15 deficiency impairs liver regeneration in mice. Am J Physiol-Gastr L. 2014;306:G893–G902. doi: 10.1152/ajpgi.00337.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Borude P, Edwards G, Walesky C, Li F, Ma XC, Kong B, et al. Hepatocyte-Specific Deletion of Farnesoid X Receptor Delays But Does Not Inhibit Liver Regeneration After Partial Hepatectomy in Mice. Hepatology. 2012;56:2344–2352. doi: 10.1002/hep.25918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bohm F, Kohler UA, Speicher T, Werner S. Regulation of liver regeneration by growth factors and cytokines. Embo Mol Med. 2010;2:294–305. doi: 10.1002/emmm.201000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanayama M, Takahara T, Yata Y, Xue F, Shinno E, Nonome K, et al. Hepatocyte growth factor promotes colonic epithelial regeneration via Akt signaling. Am J Physiol-Gastr L. 2007;293:G230–G239. doi: 10.1152/ajpgi.00068.2007. [DOI] [PubMed] [Google Scholar]
- 70.El-Jamal N, Erdual E, Neunlist M, Koriche D, Dubuquoy C, Maggiotto F, et al. Glugacon-like peptide-2: broad receptor expression, limited therapeutic effect on intestinal inflammation and novel role in liver regeneration. Am J Physiol-Gastr L. 2014;307:G274–G285. doi: 10.1152/ajpgi.00389.2012. [DOI] [PubMed] [Google Scholar]
- 71.Hofmann AF, Hagey LR, Krasowski MD. Bile salts of vertebrates: structural variation and possible evolutionary significance. J Lipid Res. 2010;51:226–246. doi: 10.1194/jlr.R000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chiang JYL. Bile acid regulation of gene expression: Roles of nuclear hormone receptors. Endocr Rev. 2002;23:443–463. doi: 10.1210/er.2000-0035. [DOI] [PubMed] [Google Scholar]
- 73.Degirolamo C, Rainaldi S, Bovenga F, Murzilli S, Moschetta A. Microbiota Modification with Probiotics Induces Hepatic Bile Acid Synthesis via Downregulation of the Fxr-Fgf15 Axis in Mice. Cell Rep. 2014;7:12–18. doi: 10.1016/j.celrep.2014.02.032. [DOI] [PubMed] [Google Scholar]
- 74.Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, et al. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. P Natl Acad Sci USA. 2011;108:4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang F, Hu Y, Liu HX, Wan YJY. MiR-22-silenced Cyclin A Expression in Colon and Liver Cancer Cells Is Regulated by Bile Acid Receptor. J Biol Chem. 2015;290:6507–6515. doi: 10.1074/jbc.M114.620369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, et al. Gut Microbiota Regulates Bile Acid Metabolism by Reducing the Levels of Tauro-beta-muricholic Acid, a Naturally Occurring FXR Antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 77.Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58:949–955. doi: 10.1016/j.jhep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Islam S, Felin J, Jantti S, Hyotylainen T, Wahlstrom A, Marschall HU, et al. Gut Microbiota Regulates Bile Acid Metabolism by Reducing the Levels of Tauro-Betamuricholic Acid, a Naturally Occurring Fxr Antagonist. J Hepatol. 2012;56:S556–S556. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 79.Kakiyama G, Hylemon PB, Zhou HP, Pandak WM, Heuman DM, Kang DJ, et al. Colonic inflammation and secondary bile acids in alcoholic cirrhosis. Am J Physiol-Gastr L. 2014;306:G929–G937. doi: 10.1152/ajpgi.00315.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Inagaki T, Moschetta A, Lee YK, Peng L, Zhao GX, Downes M, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. P Natl Acad Sci USA. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ilan Y. Leaky gut and the liver: a role for bacterial translocation in nonalcoholic steatohepatitis. World journal of gastroenterology : WJG. 2012;18:2609–2618. doi: 10.3748/wjg.v18.i21.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.DeMeo MT, Mutlu EA, Keshavarzian A, Tobin MC. Intestinal permeation and gastrointestinal disease. J Clin Gastroenterol. 2002;34:385–396. doi: 10.1097/00004836-200204000-00003. [DOI] [PubMed] [Google Scholar]
- 83.Li F, Jiang CT, Krausz KW, Li YF, Albert I, Hao HP, et al. Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat Commun. 2013;4 doi: 10.1038/ncomms3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Islam KBMS, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, et al. Bile Acid Is a Host Factor That Regulates the Composition of the Cecal Microbiota in Rats. Gastroenterology. 2011;141:1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 85.Shankar V, Hamilton MJ, Khoruts A, Kilburn A, Unno T, Paliy O, et al. Species and genus level resolution analysis of gut microbiota in Clostridium difficile patients following fecal microbiota transplantation. Microbiome. 2014;2:13. doi: 10.1186/2049-2618-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pereira-Fantini PM, Lapthorne S, Joyce SA, Dellios NL, Wilson G, Fouhy F, et al. Altered FXR signalling is associated with bile acid dysmetabolism in short bowel syndrome-associated liver disease. J Hepatol. 2014;61:1115–1125. doi: 10.1016/j.jhep.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 87.Keshavarzian A, Holmes EW, Patel M, Iber F, Fields JZ, Pethkar S. Leaky gut in alcoholic cirrhosis: A possible mechanism for alcohol-induced liver damage. Am J Gastroenterol. 1999;94:200–207. doi: 10.1111/j.1572-0241.1999.00797.x. [DOI] [PubMed] [Google Scholar]
- 88.Su L, Wang JH, Cong X, Wang LH, Liu F, Xie XW, et al. Intestinal immune barrier integrity in rats with nonalcoholic hepatic steatosis and steatohepatitis. Chinese Med J-Peking. 2012;125:306–311. [PubMed] [Google Scholar]
- 89.Fukata M, Michelsen KS, Eri R, Thomas LS, Hu B, Lukasek K, et al. Toll-like receptor-4 is required for intestinal response to epithelial injury and limiting bacterial translocation in a murine model of acute colitis. Am J Physiol-Gastr L. 2005;288:G1055–G1065. doi: 10.1152/ajpgi.00328.2004. [DOI] [PubMed] [Google Scholar]
- 90.Ewaschuk J, Endersby R, Thiel D, Diaz H, Backer J, Ma M, et al. Probiotic bacteria prevent hepatic damage and maintain colonic barrier function in a mouse model of sepsis. Hepatology. 2007;46:841–850. doi: 10.1002/hep.21750. [DOI] [PubMed] [Google Scholar]
- 91.Gazit V, Huang J, Weymann A, Rudnick DA. Analysis of the role of hepatic PPARgamma expression during mouse liver regeneration. Hepatology. 2012;56:1489–1498. doi: 10.1002/hep.25880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ewaschuk JB, Diaz H, Meddings L, Diederichs B, Dmytrash A, Backer J, et al. Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. Am J Physiol-Gastr L. 2008;295:G1025–G1034. doi: 10.1152/ajpgi.90227.2008. [DOI] [PubMed] [Google Scholar]
- 93.Ito A, Higashiguchi T. Effects of glutamine administration on liver regeneration following hepatectomy. Nutrition. 1999;15:23–28. doi: 10.1016/s0899-9007(98)00133-6. [DOI] [PubMed] [Google Scholar]
- 94.Mutlu EA, Gillevet PM, Rangwala H, Sikaroodi M, Naqvi A, Engen PA, et al. Colonic microbiome is altered in alcoholism. Am J Physiol-Gastr L. 2012;302:G966–G978. doi: 10.1152/ajpgi.00380.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tsujimoto T, Kawaratani H, Kitazawa T, Uemura M, Fukui H. Innate Immune Reactivity of the Ileum-Liver Axis in Nonalcoholic Steatohepatitis. Digest Dis Sci. 2012;57:1144–1151. doi: 10.1007/s10620-012-2073-z. [DOI] [PubMed] [Google Scholar]
- 96.de Wit N, Derrien M, Bosch-Vermeulen H, Oosterink E, Keshtkar S, Duval C, et al. Saturated fat stimulates obesity and hepatic steatosis and affects gut microbiota composition by an enhanced overflow of dietary fat to the distal intestine. Am J Physiol-Gastr L. 2012;303:G589–G599. doi: 10.1152/ajpgi.00488.2011. [DOI] [PubMed] [Google Scholar]
- 97.Spencer MD, Hamp TJ, Reid RW, Fischer LM, Zeisel SH, Fodor AA. Association Between Composition of the Human Gastrointestinal Microbiome and Development of Fatty Liver With Choline Deficiency. Gastroenterology. 2011;140:976–986. doi: 10.1053/j.gastro.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yin XC, Peng JH, Zhao LP, Yu YP, Zhang X, Liu P, et al. Structural changes of gut microbiota in a rat non-alcoholic fatty liver disease model treated with a Chinese herbal formula. Syst Appl Microbiol. 2013;36:188–196. doi: 10.1016/j.syapm.2012.12.009. [DOI] [PubMed] [Google Scholar]
- 99.Bellavia M, Rappa F, Lo Bello M, Brecchia G, Tomasello G, Leone A, et al. Lactobacillus Casei and Bifidobacterium Lactis Supplementation Reduces Tissue Damage of Intestinal Mucosa and Liver after 2,4,6-Trinitrobenzenesulfonic Acid Treatment in Mice. J Biol Reg Homeos Ag. 2014;28:251–261. [PubMed] [Google Scholar]
- 100.Zhang YQ, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, et al. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. P Natl Acad Sci USA. 2006;103:1006–1011. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Watanabe M, Houten SM, Wang L, Moschetta A, Mangelsdorf DJ, Heyman RA, et al. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004;113:1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Harach T, Pols TWH, Nomura M, Maida A, Watanabe M, Auwerx J, et al. TGR5 potentiates GLP-1 secretion in response to anionic exchange resins. Sci Rep-Uk. 2012;2 doi: 10.1038/srep00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Potthoff MJ, Potts A, He TT, Duarte JAG, Taussig R, Mangelsdorf DJ, et al. Colesevelam suppresses hepatic glycogenolysis by TGR5-mediated induction of GLP-1 action in DIO mice. Am J Physiol-Gastr L. 2013;304:G371–G380. doi: 10.1152/ajpgi.00400.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 105.Jones BV, Begley M, Hill C, Gahan CGM, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. P Natl Acad Sci USA. 2008;105:13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lepercq P, Gerard P, Beguet F, Raibaud P, Grill JP, Relano P, et al. Epimerization of chenodeoxycholic acid to ursodeoxycholic acid by Clostridium baratii isolated from human feces. Fems Microbiol Lett. 2004;235:65–72. doi: 10.1016/j.femsle.2004.04.011. [DOI] [PubMed] [Google Scholar]