Abstract
To investigate how cells sense stiffness in settings structurally similar to native extracellular matrices (ECM), we designed a synthetic fibrous material with tunable mechanics and user-defined architecture. In contrast to flat hydrogel surfaces, these fibrous materials recapitulated cell-matrix interactions observed with collagen matrices including stellate cell morphologies, cell-mediated realignment of fibers, and bulk contraction of the material. While increasing the stiffness of flat hydrogel surfaces induced mesenchymal stem cell spreading and proliferation, increasing fiber stiffness instead suppressed spreading and proliferation depending on network architecture. Lower fiber stiffness permitted active cellular forces to recruit nearby fibers, dynamically increasing ligand density at the cell surface and promoting the formation of focal adhesions and related signaling. These studies demonstrate a departure from the well-described relationship between material stiffness and spreading established with hydrogel surfaces, and introduce fiber recruitment as a novel mechanism by which cells probe and respond to mechanics in fibrillar matrices.
Keywords: Extracellular matrix, fibers, hydrogels, remodeling, mechanotransduction, mechanosensing, focal adhesions, rigidity, stiffness, electrospinning, biomechanics, cell shape
Main
The adhesion of cells to the extracellular matrix (ECM) regulates many cellular functions including spreading, migration, proliferation, and differentiation, and thus plays a major role in embryonic development, adult tissue homeostasis, and disease pathogenesis1–3. Because the biophysical and biochemical properties of native ECMs are difficult to experimentally modulate, synthetic materials have been crucial towards isolating the contributions of specific matrix properties in regulating cell adhesion and function. In particular, elastic hydrogel surfaces (e.g. polyacrylamide gels) have proven indispensable in demonstrating that substrate stiffness or rigidity can itself modulate cell behavior4–6, as these materials afford precise control of stiffness, independent of cell adhesive ligand density7. However, it is unclear how findings on these smooth, flat surfaces extend to in vivo settings, where cells reside in or on complex three-dimensional (3D) ECMs consisting of meshworks of fibers with diameters typically on the order of micrometers8–10. These networks of fibers vary widely in density and organization depending on the tissue (e.g. dense, aligned collagen bundles in tendon versus loose, less organized networks in glandular organs). The micrometer-scale architecture of these fibrous networks constrains spatially where cells can form adhesions and imparts complex mechanical characteristics due to non-linear stiffening in response to loading and differential rigidity in axial versus transverse directions with respect to fiber orientation - all features that cannot be captured with existing isotropic, linear elastic hydrogel surfaces. Given the lack of mechanically tunable synthetic materials possessing fibrous structure at physiologic length scales, an understanding of how cells sense and respond to the mechanics of fibrillar microenvironments remains an open challenge. Here, we establish a novel material system that incorporates fibrillar structure while still maintaining synthetic control over mechanical and adhesive features and apply this system to elucidate mechanisms of how cells interpret ECM stiffness in fibrous networks.
Fabrication of a synthetic fibrillar ECM with controllable architecture and mechanics
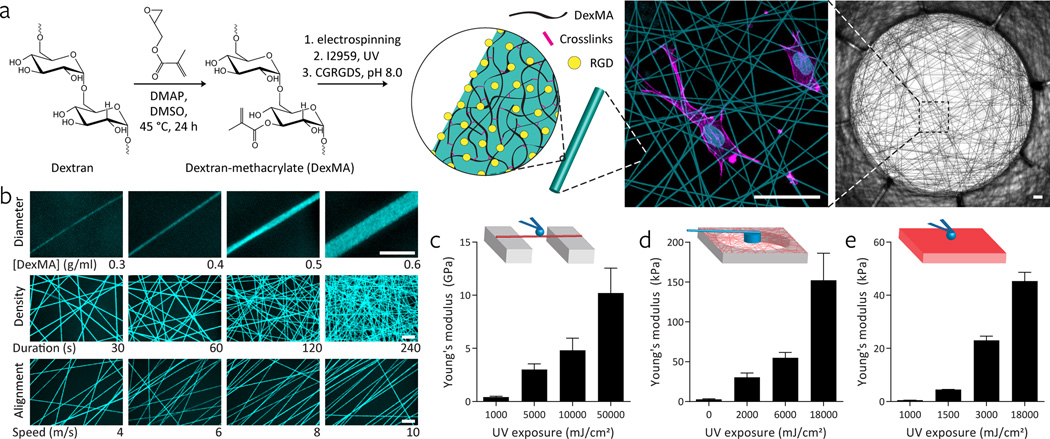
To develop a material system for studying fibrillar mechanosensing, we combined polymer chemistry, electrospinning, and soft lithography. As a base material, we formulated a protein-resistant, methacrylated dextran (DexMA, Fig. 1a, Supplementary Fig. 1)11 that could be functionalized with cell adhesive moieties following substrate fabrication (Fig. 1a, Supplementary Fig. 3–4). Fiber networks with controllable architecture and mechanics were fabricated by electrospinning the polymer onto collection substrates such that fibers were suspended across microfabricated wells. The geometry of the wells defined boundary conditions and elevated networks to exclude a mechanical contribution from the underlying rigid surface. Numerous structural parameters were tuned in this system, including fiber diameter (via solution concentration, Supplementary Fig. 7), fiber density (via fiber collection durations), and fiber anisotropy (via rotational speed of the collection surface) (Fig. 1b). Exposure to UV light crosslinked DexMA, rendering fibers insoluble and allowing stiffness to be modulated through the extent of light exposure. To measure the mechanics of individual DexMA fibers as a function of UV exposure, we performed micro-scale three-point bending tests using AFM (Supplementary Fig. 2)12,13. The Young’s modulus of individual fibers was tunable between 140 MPa and 10 GPa (Fig. 1c), approximating the range of reported values for various fibrous biopolymers such as collagen (0.5–10 GPa)12,14. As cells probe the mechanics of not just a single fiber, but a network composed of many fibers, a macroscale measurement of network mechanics was also developed (Supplementary Fig. 2). Increasing UV exposure to increase fiber modulus without altering other network parameters (Supplementary Figs. 3, 9) led to an increase in network stiffness as expected (Fig. 1d). A salient feature of the DexMA polymer is that in addition to fibrous networks, we can generate smooth hydrogel surfaces lacking fibrous topography from the same material to serve as a direct comparator in our studies. Tuning mechanics by UV photocrosslinking yielded hydrogels with moduli between 450 Pa and 45 kPa as determined by AFM nanoindentation and Hertz contact mechanics (Fig. 1e).
Figure 1. A novel approach to engineering fibrillar microenvironments with tunable mechanical and architectural features.
a, Hierarchical overview of fabricating cell-adhesive suspended networks of dextran methacrylate (DexMA) fibers. Dextran is reacted with glycidyl methacrylate to generate DexMA. Following addition of a photoinitiator, DexMA is electrospun onto microfabricated substrates to define networks of suspended fibers. Networks are photo-crosslinked to tune fiber stiffness and the RGD is incorporated to enable cell attachment (scale bars, 100 µm). b, Through modulation of the electrospinning fabrication process, networks with varying fiber diameter (polymer solution concentration), density (fiber collection duration), and alignment (collection surface translation speed) can be generated, enabling the modeling of diverse fibrillar ECMs present in different tissue systems throughout the body (scale bars: 10 µm). c, Young’s modulus of individual fibers isolated over PDMS troughs and measured by three point bending AFM; n ≥ 12, mean ± s.d. d, Young’s modulus of DexMA fiber networks measured by cylindrical indentation with a calibrated cantilever; n ≥ 5, mean ± s.d. e, Young’s modulus of DexMA flat hydrogels determined by AFM spherical probe nanoindentation and Hertz contact mechanics; n ≥ 6, mean ± s.d.
Upon processing DexMA into hydrogel or fibrillar form, the adhesive peptide CGRGDS (RGD) was coupled to substrates via Michael-type addition with unreacted methacrylates (Supplementary Fig. 3–4). Although functionalization with other peptides or even full-length proteins is possible, we chose a non-fibrillar adhesion moiety to exclude the confounding mechanical contribution of a superimposed meshwork of ECM proteins, which has previously been shown to influence cellular mechanosensing15. Thus, RGD coupled directly to DexMA fiber networks or hydrogel surfaces ensured that the ECM stiffness experienced by seeded cells was defined entirely by the structure of the material.
Synthetic fiber networks recapitulate collagen matrices at multiple length scales
Several distinctions exist between this material and in vivo ECMs or even purified fibrous biopolymers such as type I collagen. Structural features including persistence length, tortuosity, fiber diameter, and 3D organization likely diverge from natural fibrillar ECMs. Biochemically, the exclusive use of RGD differs from the more complex ligand-receptor interactions mediating adhesion to native fibrillar proteins, such as the additional GFOGER and GVMGFO adhesion sequences on collagens or tension-induced exposure of cryptic binding sites in fibronectin16–19. Given these disparities, we assessed how faithfully our synthetic fibrillar material recapitulated topographical and mechanical interactions between cells and a bona fide fibrous ECM. For these studies, we chose type I collagen, the most abundant fibrillar protein in mammals and a well-studied model in mechanobiology over several decades8,20. As a comparator possessing chemistry identical to fiber networks but lacking fibrous topography, cells were also cultured on the surface of DexMA hydrogels.
We began by examining cytoskeletal organization in cells cultured on these materials. All studies presented here used human mesenchymal stem cells (hMSCs), a model cell type for mechanosensing studies6,15,21,22, although similar trends were observed with human dermal mouse fibroblasts. A high degree of similarity was observed between DexMA fiber networks and type I collagen matrices (Fig. 2a); cells in both conditions adopted a spindly morphology possessing thin, elongated processes that terminated in branched protrusions, with punctate vinculin-rich focal adhesions (FA) sequestered to the tips of these protrusions. Centrally located FAs were not visible due to high cytosolic signal, and hMSCs possessed relatively few actin stress fibers traversing the cytoplasm, although actin was enriched at the hulls of processes. This shift in vinculin from FAs to the cytosol and paucity of stress fibers has been suggested to be representative of cells in 3D matrices or in vivo tissues20,23–26. In contrast, cells seeded on stiff hydrogels adopted a well-spread, lamellar morphology with larger, elongated FAs distributed throughout the cell and numerous stress fibers similar to cells cultured on glass or plastic24. Taken together, the surprising similarity between type I collagen matrices and fiber networks in contrast to flat hydrogel surfaces suggests that fibrillar topography had a stronger influence on cell morphology than the biochemical nature of these interactions.
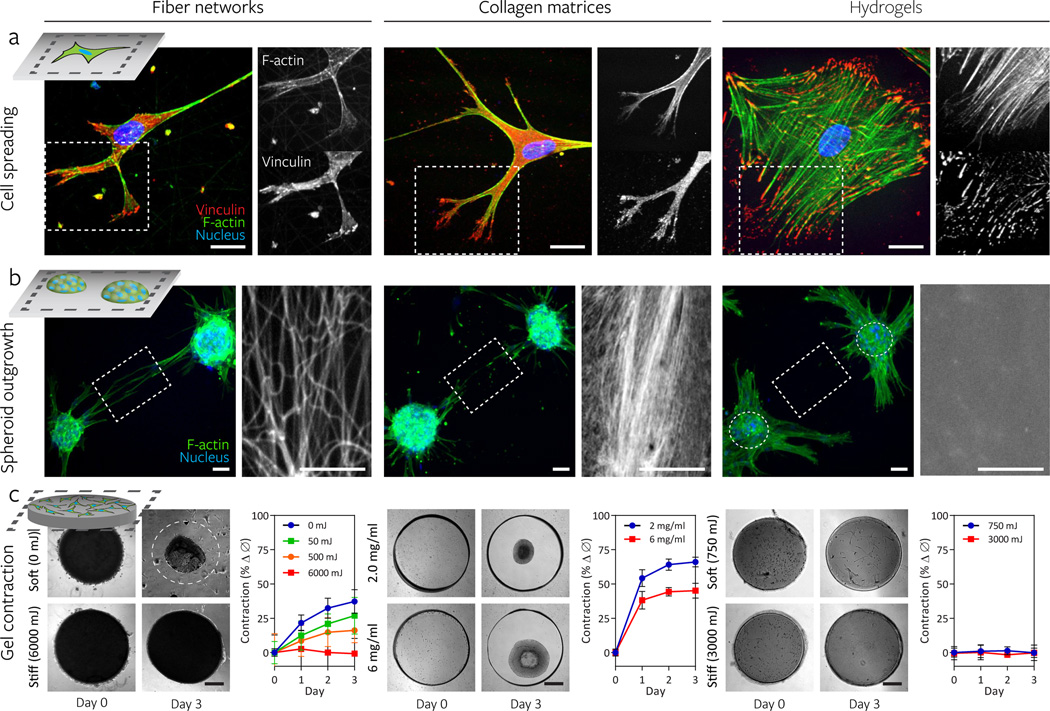
Figure 2. Synthetic fiber networks induce similar topographical and mechanical interactions with cells as collagen matrices at multiple length scales.
Comparison of cell spreading (a), spheroid outgrowth (b), and gel contraction (c) across hMSC-seeded DexMA fiber networks (left column), type I collagen matrices (middle column), and flat DexMA hydrogels (right column). a, Cytosolic and focal adhesion-localized vinculin (red), counter-stained for F-actin (green) and cell nuclei (blue) with phalloidin and Hoechst 33342, respectively. Dashed box designates region shown with F-actin or vinculin channel isolated (scale bars, 50 µm). b, Outgrowth of hMSC spheroids (100 cells/spheroid) stained for F-actin (green) and cell nuclei (blue). Rhodamine methacrylate-coupled DexMA fibers and hydrogel surface and Picrosirius Red-stained collagen are shown in the region spanning two spheroids, as indicated by the dashed box (scale bars, 50 µm). c, DexMA fibers and hydrogels and collagen were processed into thick circular slabs and seeded at high density with hMSCs. Initial and final states of soft (top) and stiff (bottom) constructs. Cell-mediated contraction was determined by normalizing construct diameter to the initial diameter; mean ± s.d., n ≥ 6 (scale bars, 500 µm).
Stopak and Harris demonstrated how cellular forces could physically reorganize ECM, culturing tissue explants within collagen matrices and observing the alignment of fibers in fields of high tension that directed cell migration between adjacent explants27. To test whether DexMA fiber networks could capture this phenomenon, multicellular hMSCs spheroids were seeded on the synthetic fiber networks. Indeed, cell forces pulled fibers into alignment and cells began to directionally migrate towards adjacent spheroids (Fig. 2b), mimicking the response on collagen matrices. This response did not occur on DexMA hydrogels of a range of different stiffnesses, highlighting the comparatively limited range of deformations in non-fibrous materials28. These results reinforce a high degree of mechanical similarity between our synthetic fibrillar ECM and collagen, and provide evidence of the potential for ECM remodeling and long range force transmission in this material29,30, a process relevant to, for example, cancer cell escape from primary tumors during metastasis31.
In addition to local remodeling, macroscale contraction has been observed in fibroblast-populated collagen matrices and used to assess how tensile forces generated by cells elicit morphogenetic changes32–34. When we seeded DexMA fiber networks unattached to any boundary constraints, compaction occurred over a three day time course, comparable to collagen (Supplementary Fig. 5), whereas seeded DexMA hydrogels of a range of stiffnesses and unseeded DexMA fiber samples did not change in diameter (Fig. 2c). Altering the concentration of collagen can tune the contraction response over a limited range, but such perturbation simultaneously alters fibril and adhesive ligand density as well as mechanics. Importantly, in contrast to biologic ECMs, we can independently tune fiber stiffness, density, and architecture to dictate the ability of cells to deform the material.
Increasing fiber stiffness suppresses cell spreading and proliferation
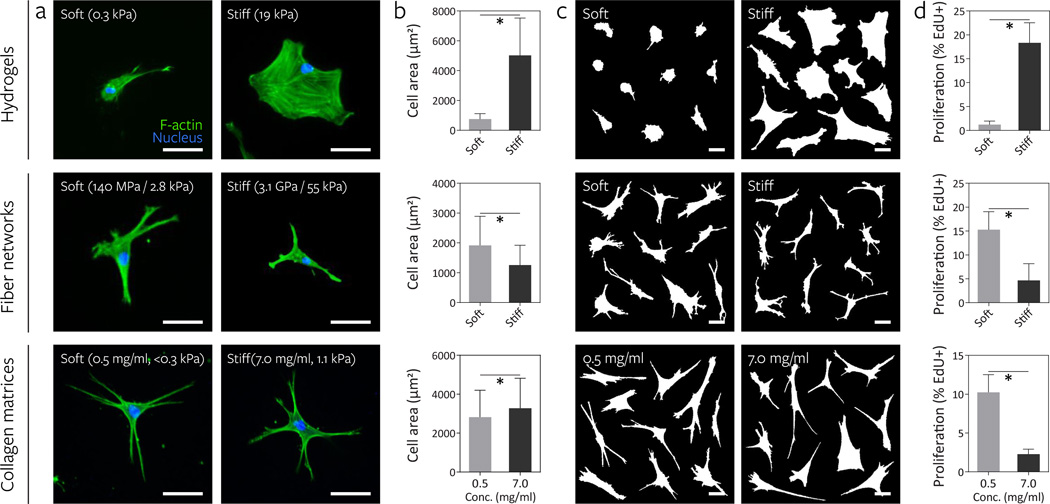
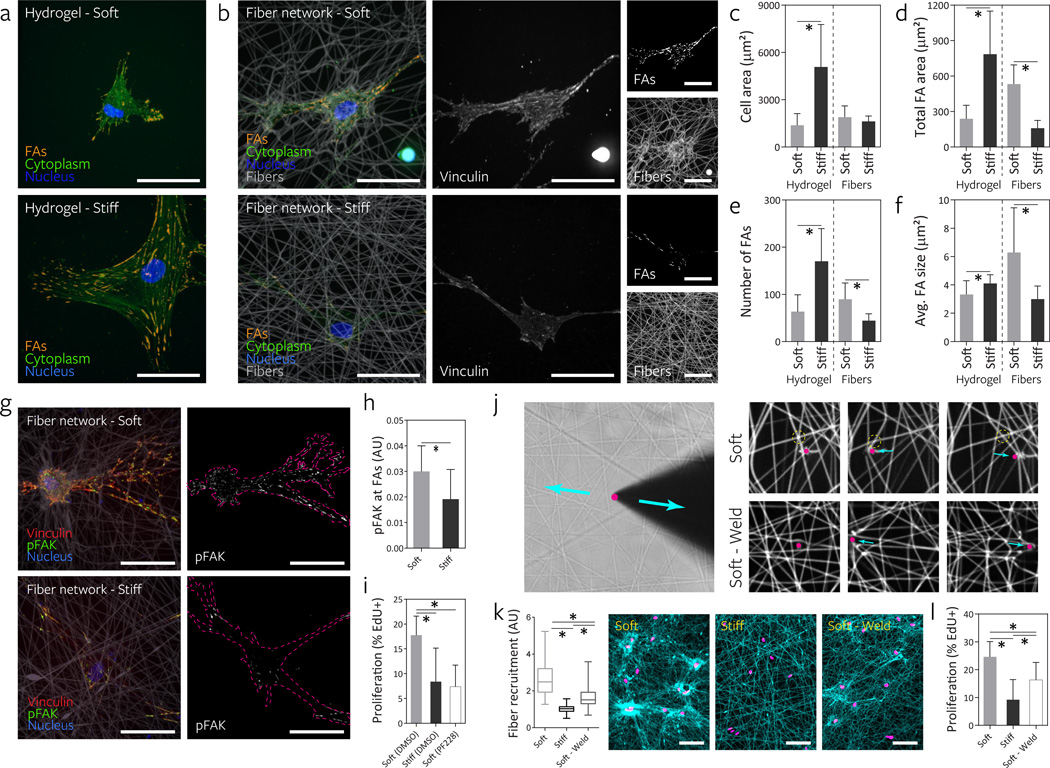
Having validated our synthetic material, we next tested whether cells respond to an increase in material stiffness in fiber networks in similar fashion to hydrogel surfaces. To assay a cell response thoroughly described in the literature, we first examined the relationship between material stiffness and cell spreading. Tuning DexMA crosslinking to define soft (290 Pa) and stiff (19.1 kPa) hydrogels, we confirmed previous observations4–6,35: at low stiffness cells failed to spread; with increasing stiffness, spreading and the formation of large lamellae occurred (Fig. 3a–c). To examine a longer term, functional outcome, we measured cell proliferation over a two day time course. In these experiments, we observed uniformly low cell death across all conditions, suggesting cytocompatibility and no direct effects of stiffness on cell viability (Supplementary Fig. 8). With stiffness-induced increases in cell area from soft to stiff hydrogels, we observed a commensurate increase in cell proliferation (Fig. 3d), again in line with numerous previous reports35–37. Surprisingly, when cells were cultured on soft (fiber: 140 MPa, network: 2.8 kPa) and stiff (fiber: 3.1 GPa, network: 55 kPa) fiber networks with all other parameters maintained constant, cell area did not increase with fiber stiffness as observed on hydrogel surfaces, but instead showed a modest but significant decrease (Fig. 3a–c). Moreover, in stark contrast to hydrogel surfaces, cell proliferation in soft networks was three-fold higher than in stiff networks fabricated at the same fiber density (Fig. 3d).
Figure 3. Increasing fiber stiffness suppresses cell spreading and proliferation.
The effect of altering material stiffness on hMSC spreading and proliferation was examined on DexMA hydrogels (top row, soft: 290 Pa, stiff: 19.1 kPa) and fiber networks (middle row, soft: 140 MPa fiber, 2.8 kPa network; stiff: 3.1 GPa fiber, 55 kPa network). Low (0.5 mg/mL, <0.3 kPa) and high (7.0 mg/mL, 1.1 kPa) concentration type I collagen matrices where bulk stiffness and adhesive ligand density increase in tandem were included for comparison (bottom row). a, Actin cytoskeletal organization of representative hMSCs 16 h after seeding, stained for F-actin (green) and cell nuclei (blue) (scale bars, 50 µm). b, Quantification of cell area; mean ± s.d., n ≥ 64 cells, * P < 0.05. c, Cell outlines of ten representative cells (scale bars, 50 µm). d, Proliferation of hMSCs over two days as determined by EdU incorporation; mean ± s.d., n ≥ 13 ROI with totals of 750–1500 cells analyzed, * P < 0.05.
We then examined whether this inverted proliferation response occurs in an established fibrillar setting and excluded an RGD-specific response by repeating the experiment with collagen matrices, in which stiffness and ligand density were simultaneously modulated by altering collagen concentration. Although increasing either stiffness or ligand density alone increases cell area and proliferation on synthetic hydrogels7,35–38, we found that the soft, low concentration collagen matrices promoted proliferation compared to the stiffer, high concentration matrices (Fig. 3a–d). Taken together, these data suggest that in fibrous ECMs, the relationship between substratum stiffness and cell function is inverted as compared to on hydrogel surfaces.
Fiber recruitment by cells increases local adhesive ligand density, apparent stiffness, and adhesion signaling
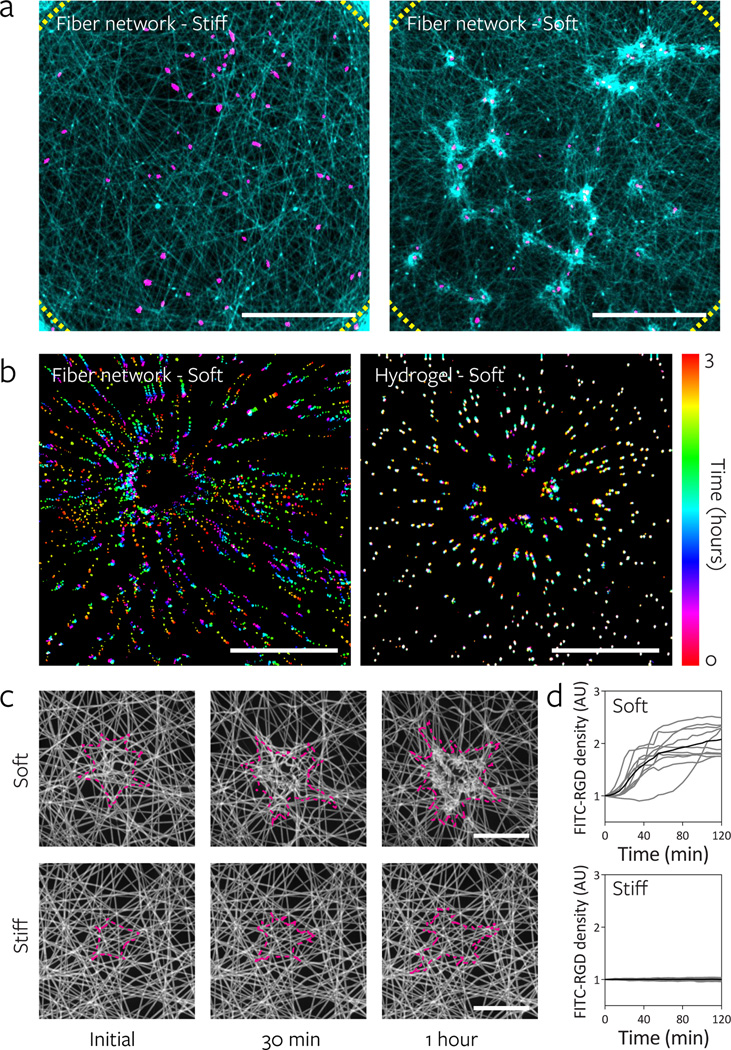
Modulating fiber stiffness had a significant effect on the ability of cells to reorganize fibers. In soft networks, cells pulled and deformed the networks, resulting in significant recruitment of fibers to the cell and the formation of numerous densely compacted clusters of fibers (Fig. 4a, Supplementary Video 3). Conversely, stiff networks presented a rigid and essentially immobile ECM that underwent negligible architectural remodeling. The range of deformations in soft networks is illustrated by the displacement of fluorescent microspheres embedded within soft DexMA fibers upon cell attachment (Fig. 4b), and qualitatively by movies of cells migrating on soft versus stiff fibers (Supplementary Video 1 & 2). Microsphere (and fiber) motion was unidirectional and displacements were high in soft networks (Supplementary Fig. 6, Supplementary Videos 4 & 5). In contrast, displacements in soft hydrogels were an order of magnitude smaller and underwent cyclic motions, suggestive of “load-and-fail” dynamics described previously by Chan et al39.
Figure 4. Lower fiber and network stiffness enables cell-mediated reorganization of the material and clustering of adhesive ligands local to the cell.
a, Reorganization of stiff and soft fiber networks 16 h after hMSC seeding. Fibers imaged by coupling with rhodamine methacrylate (cyan) and thresholded cell nuclei labeled with Hoechst 33342 (magenta). Dotted lines indicate the periphery of the suspended network (scale bars, 500 µm). b, Temporally color-coded overlays capturing the motion of beads embedded within soft fibers (fiber: 140 MPa, network: 2.8 kPa) and soft hydrogels (290 Pa) over a 3 h time course following hMSC seeding. c, Time-lapse images of FITC-RGD coupled fiber recruitment during the first two hours of hMSCs spreading on soft (top) and stiff (bottom) networks. Cell outlines shown in magenta (scale bars, 50 µm). d, Quantification of FITC-RGD fluorescence intensity in a 50 µm diameter circular region centered on the cell’s nucleus. Intensity was normalized to adjacent acellular areas; n = 10 cells.
The considerable physical reorganization and clustering of fibers in soft networks resulted in increased adhesive ligand density adjacent to the cell. Time lapse confocal imaging of fiber networks functionalized with fluorescently tagged RGD demonstrated a rapid 2–3 fold increase of local RGD density following cell attachment to soft networks, with little change in stiff networks (Fig. 4c–d, Supplementary Video 3). Consistent with the conclusion that increased matrix contact with cells due to fiber recruitment provides the mechanism for enhanced response, we observed that differences in proliferation between soft and stiff fibers decreased and ultimately were eliminated by increasing the initial fiber density of the network (Supplementary Fig. 10). Interestingly, we also observed decreased fiber recruitment with increasing fiber density, likely due to the distribution of cellular contractile work across more adhesion – a phenomenon that has previously been described40 – deforming each fiber to a lesser degree.
Increases in ligand density could enhance FA assembly38,41. Indeed, soft networks were found to promote FA formation (vinculin localization) over stiff networks (Fig. 5b), even when restricting analysis to cells with similar spread areas (as spreading is correlated with enhanced FA formation4,42) (Fig. 5c). While in both conditions FAs localized to the tips of extended cell processes, hMSCs on soft networks possessed markedly more centrally located FAs, directly localized to underlying clustered fibers. Average FA size, number of FAs, and total FA area were all higher in cells on soft fiber networks, and consistently inverted as compared to the response to stiffness on hydrogel surfaces (Fig. 5a, d–f). Focal adhesion kinase (FAK) plays a central role in transducing adhesion into biochemical signals43, and markedly more phosphorylated FAK was detected at FA sites in soft fiber networks (Fig. 5g–h). This increased FAK activation was functionally important to the proliferation induced on soft networks, as inhibition of FAK with a selective inhibitor (PF-57322844) abrogated the increase in proliferation on soft networks (Fig. 5i).
Figure 5. Fibrillar ECM remodeling promotes focal adhesion (FA) formation and FAK phosphorylation to increase proliferation.
a, FA formation of representative hMSCs seeded on DexMA hydrogels of low and high stiffness as visualized by cytosol extraction, vinculin immunostaining, and subsequent image analysis to identify FAs (orange). The cell’s cytoplasm (green) and nucleus (blue) are also shown (scale bars, 50 µm). b, FA formation of representative hMSCs on DexMA fiber networks of low and high fiber stiffness 16 h after seeding. Composite images (left) showing FAs (orange), cytoplasm (green), nuclei (blue) and fibers (grey). Single channel images of vinculin (middle) and fibers (right, bottom) as well as identified FAs (right, top) (scale bars, 50 µm). Cell area (c), total FA area (d), total number of FAs (e), and average FA size (f); mean ± s.d., n = 12 cells, * P < 0.05. g, Merged images of representative hMSCs 16 h after seeding co-stained for vinculin (red) and phospho-FAK (green). Single channel images of phospho-FAK with cells outlined in magenta (scale bars, 50 µm). h, Quantification of phospho-FAK localization to FAs determined by fluorescence intensity; mean ± s.d., n = 10 cells, * P < 0.05. i, Effect of FAK phosporylation inhibition on proliferation of hMSCs over two days, as determined by EdU incorporation; mean ± s.d., n ≥ 9 ROI with totals of 450–1500 cells analyzed, * P < 0.05. j, To test fiber-fiber connectivity, a diamond sharpened blade was placed adjacent to individual fibers and reciprocated via micromanipulator. Soft networks as fabricated in all previous studies possess limited connectivity as demonstrated by free sliding of fibers (top row) in contrast to “welded” networks with high fiber-fiber connectivity (bottom row). k, Remodeling of fiber networks 16 h after MSC seeding. Fiber recruitment in 50 µm diameter circular regions centered on the cell nucleus. Fibers imaged by coupling with rhodamine methacrylate (cyan) and thresholded cell nuclei labeled with Hoechst 33342 (magenta) (scale bars, 100 µm). Fluorescence intensity was normalized to adjacent acellular areas. l, Effect of altering fiber-fiber connectivity on proliferation of hMSCs over two days as determined by EdU incorporation; mean ± s.d., n ≥ 9 ROI with totals of 450–1500 cells analyzed, * P < 0.05.
While these studies implicate an important role for fiber recruitment in the cellular response to fibrous matrices, one alternative explanation for the results is that decreasing fiber stiffness also alters the nanoscale mobility or mechanics of RGD on soft fibers, thereby impacting integrin clustering or catch-bond dynamics15,39,45,46. To distinguish fiber recruitment from the effects of RGD mobility or mechanics, we modulated the interconnections or “welding” between fibers as a means to reduce fiber recruitment without altering the stiffness or RGD mobility of individual fibers (Fig. 5j). In standard networks, fiber intersections were a mixture of welded and unwelded states (Supplemental Video 6, 8). To maximize fiber-fiber welding, networks were placed in a controlled humidity environment and provided sufficient moisture prior to light exposure to fuse all juxtaposed fibers (Fig. 5j, Supplementary Videos 7). Indeed, this welding resulted in reduced fiber clustering compared to soft networks, though not to the extent of stiff networks, and resulted in intermediate levels of fiber/RGD recruitment (Fig. 5k). Cell proliferation in these soft welded networks was likewise intermediate between soft and stiff networks (Fig. 5l), supporting the model that fiber recruitment (and not the stiffness of individual fibers per se) is the primary mechanism of mechanotransduction observed in these studies.
Here, we describe a new biomaterial system that recapitulates the fibrous architecture of native ECMs while providing the control offered by synthetic materials. Using this material, we uncovered a previously unrecognized mechanism for transducing matrix stiffness, whereby lower fiber/network stiffness enabled cells to recruit nearby fibers, leading to increased local adhesive ligand density, enhanced adhesion signaling, cell spreading, and proliferative signaling. Notably, we find that multiple structural parameters of fiber networks (ie. fiber stiffness, fiber-fiber welding, and fiber density) converge through cellular fiber recruitment to influence cell function. This link between fiber recruitment and proliferation suggests that spatial rearrangements of ECM are not merely a consequence of cellular forces, but can also feedback to alter cell signaling and function31. Moreover, given that these dramatic structural rearrangements are absent in traditional elastic hydrogel materials, these findings highlight the need for novel materials that can more accurately capture the mechanical behavior of native ECMs. Recent work identifying a role for substrate stress-relaxation in cell spreading through the introduction of viscoelasticity to gels further illustrates this need47. Future innovation to incorporate such properties in addition to dynamic features (ie. cell-mediated matrix degradation, synthesis, and crosslinking) into fibrous materials will be critical in further refining our understanding of the feedback between matrix structure/mechanics and the cellular response.
Most tissues possess bulk moduli in the Pa to kPa range, while the protein fibers that compose these tissues are often in the MPa to GPa range48,49. Our ability to not only recapitulate these features in our synthetic fiber networks, but also independently tune mechanics at different length scales may provide a path to elucidating how cells assimilate mechanic signals from multiple length scales (eg. through integrin clustering and focal adhesion assembly at the nanoscale21,50 and through actomyosin activity between focal adhesions to probe stiffness at the microscale51,52). Integrating insights offered by such materials with theoretical approaches that couple the hierarchical structure and mechanics of fibrous ECM53,54 to models of intracellular dynamics (integrin engagement39,50, stress fiber formation55, focal adhesion assembly56) will be an essential and exciting step towards this goal.
Methods
All reagents were from Sigma-Aldrich (St. Louis, MO) and were used as received, unless otherwise stated.
Synthesis of methacrylated dextran (DexMA)
Dextran (MP Biomedicals, MW 86,000 kDa) was methacrylated by reaction with glycidyl methacrylate, according to a modified, previously described procedure11. In brief, dextran (20 g) and 4-dimethylaminopyridine (2 g) were dissolved in anhydrous dimethyl sulfoxide (100 mL) under vigorous stirring. Glycidyl methacrylate (24.6 mL) was added and the reaction mixture was heated to 45 °C for 24 h. The solution was cooled on ice for 20 min and precipitated into 1 L ice cold 2-isopropanol. The crude product was recovered by centrifugation, redissolved in milli-Q water and dialyzed against milli-Q water for 3 days. The final product was lyophilized and stored at −20 °C until use. DexMA was characterized by1H-NMR (Supplementary Fig. 1). The degree of functionalization was calculated as the ratio of the averaged methacrylate proton integral (6.174 ppm and 5.713 ppm in D2O) and the anomeric proton of the glycopyranosyl ring (5.166 ppm and 4.923 ppm). Since the signal of the anomeric proton of α-1,3 linkages (5.166 ppm) partially overlaps with other protons, a pre-determined ratio11 of 4% α-1,3 linkages was assumed and the total anomeric proton integral was calculated solely based on the integral at 4.923 ppm. A methacrylate:dextran repeat unit ratio of 0.7 was determined.
DexMA fiber network fabrication
3D networks of suspended DexMA fibers were fabricated by a combination of electrospinning and standard soft photolithography. DexMA was dissolved at 0.3–0.6 g/mL in a 1:1 mixture of milli-Q water and dimethylformamide with 0.005% Irgacure 2959 photoinitiator (Ciba, Tarrytown, NY). Electrospinning was accomplished with a custom setup consisting of a high voltage power supply (Gamma High Voltage Research, Ormond Beach, FL), syringe pump (KD Scientific, Holliston, MA), and grounded copper or aluminum collecting surface enclosed within an environmental chamber (Terra Universal, Fullerton, CA). Electrospinning was performed at a flow rate of 0.5 mL/h, voltage of 7.5 kV, and gap distance of 8 cm (Supplementary Fig. 11 a–b). To induce a preferred direction of fiber alignment (Fig. 1), the static grounded surface (6×6×1/8 inch copper sheet) was replaced with a motorized platen rotating at 1200 rpm, a speed sufficient to translate the substrate at a linear velocity of 10 m/s (Supplementary Fig. 11 c–d). Samples were primary crosslinked under UV to stabilize fibers, hydrated in a solution containing 1 mg/mL Irgacure 2959, and then exposed to varying durations of UV (100 mW/cm2) to control the degree of crosslinking and resulting stiffness. Fibers were collected on poly(dimethylsiloxane) (PDMS; Sylgard 184, Dow-Corning, Midland, MI) arrays of circular wells (2 mm diameter) functionalized with methacrylates to promote fiber adhesion. Briefly, silicon wafer masters possessing SU-8 photoresist (Microchem, Westborough, MA) were produced by standard photolithography and used to generate PDMS stamps. Following silanization with trichloro(1H,1H,2H,2H-perfluorooctyl)silane, stamps were used to emboss uncured PDMS onto oxygen plasma-treated coverslips. Well arrays were methacrylated with a 2% v/v solution of 3-(Trimethoxysilyl)propyl methacrylate in ethanol for at least 24 h. To promote fiber-fiber welding, fiber networks were briefly exposed to a humidified environment (60% relative humidity, determined empirically) prior to primary crosslinking. This approach to fuse fibers at overlaying intersection points was predicated on previous work showing the influence of solvent volatility on fiber-fiber welding57. Lateral displacements using a diamond dissecting knife (Type MDL, Electron Microscopy Sciences, Hatfield, PA) controlled by a micromanipulator (HCU-3DM, SmarAct GMBH, Oldenburg, Germany) were applied to networks to confirm the introduction of fiber-fiber welding.
DexMA hydrogel preparation
DexMA was dissolved at 3.75% w/v in M199 media containing sodium bicarbonate (3.5% w/v) and HEPES (10 mM). 10 mg/mL Irgacure 2959 in ethanol was added to a final concentration of 0.02% w/v. Solutions were mixed and spread onto methacrylated coverslips (below). Hydrogel precursors were photo-polymerized using an Omnicure S2000 UV lamp (EXFO, Ontario, Canada) at 100 mW/cm2 (measured at 365 nm). Polymerization times were 7.5 s for soft and 30 s for intermediate stiffness hydrogels. Stiff hydrogels were polymerized for 60 s in an argon chamber. To functionalize coverslips with methacrylates, No. 1 thickness, borosilicate glass coverslips were plasma oxidized using air plasma (K1050X, Emitech) and immersed in a 2% v/v solution of 3-(Trimethoxysilyl)propyl methacrylate in anhydrous toluene for at least 24 h. Prior to use, coverslips were washed in ethanol and water and dried under a stream of nitrogen.
Collagen matrices
Solutions of rat tail collagen I (BD Biosciences) at various concentrations were prepared as in58 and pipetted onto glutaraldehyde functionalized glass coverslips. Coverslips were prepared by exposure to oxygen plasma and immediate treatment with sequential 2 h incubations in 0.1 mg/mL poly-L-lysine and 5% v/v glutaraldehyde.
Mechanical testing
To determine the tensile mechanical properties of individual fibers, three point bending tests were performed using atomic force microscopy (AFM). Single fibers were collected on 200 µm wide by 200 µm tall microfabricated PDMS troughs by electrospinning for short durations (1–3 seconds). Fibers were hydrated and crosslinked to varying degrees by UV exposure as above, and deformed by an AFM tip (0.06 N/m) loaded with a 25 µm diameter bead positioned centrally along the fiber’s length. Young’s modulus was calculated from the resulting load-displacement curves using known equations for a cylindrical rod undergoing three point bending with fixed boundaries (confirmed experimentally, Supplementary Fig. 2). This measurement was performed on a population basis, using a mean fiber diameter to calculate a Young’s modulus from the same population’s mean stiffness. For flat hydrogels, AFM nanoindentation testing was performed with the same probe as above. Young’s modulus was determined by fitting force-indentation curves to known models for Hertzian contact of a spherical indentor on an elastic half space, assuming a Poisson ratio of 0.5. Mechanics of fibrous networks as fabricated for cell studies was determined by indentation with a rigid cylinder. Cylinders (500 µm diameter, 500 µm tall) of SU8 photoresist were microfabricated and affixed to pure tungsten filaments of known mechanical properties. Indentation was performed with a micromanipulator, using a confocal microscope to determine the resulting plane of the fibers at each step of indentation. Young’s modulus was approximated assuming an elastic membrane using the following equation,
where t is the membrane thickness (20 µm), ro is the membrane diameter (1 mm), ri is the indentor diameter, (0.25 mm), and ν is the poisson ratio (0.5), F is the indentation force, δ is the indentation depth, and E is the Young’s modulus.
RGD functionalization of DexMA fiber networks and hydrogels
For cell studies, DexMA fiber networks and hydrogels were functionalized with the cell-adhesive peptide CGRGDS, custom synthesized by Aapptec (Louisville, KY) and supplied as a trifluoroacetate salt. A CGRGDS concentration of 2 mM was used for most cell studies. To couple CGRGDS to available methacrylates via Michael addition, the peptide was dissolved in M199 media containing sodium bicarbonate (3.5% w/v) and HEPES (10 mM) and 1 M NaOH (ca. 10 µL/mL solution) was added to adjust the pH to 7.5–8. The solution was transferred to the substrates and incubated for 1 h at room temperature. Following functionalization, substrates were thoroughly rinsed with PBS prior to cell seeding. In additional studies, quantifying the amount of CGRGDS on fiber substrates, peptides were fluorescently labeled with FITC. CGRGDS (100 mg) was dissolved in 0.15 M sodium bicarbonate buffer (700 µl), the solution was adjusted to pH 8 using 1M sodium hydroxide, and a solution of fluorescein-N-hydroxysuccinimide (Fisher Scientific, Pittsburgh, PA) in dimethylsulfoxide (100 mg in 300 µl) was introduced. The mixture was allowed to react for 3 h at room temperature on a shaker. It is noted that N-hydroxysuccinimide will partially precipitate in aqueous buffer, but will redissolve upon reaction within the first hour of addition. The reaction mixture was desalted using a PD-10 column (Life Technologies, Carlsbad, CA) and lyophilized.
Cell culture
NIH 3T3 fibroblasts were cultured in high glucose DMEM containing 1% penicillin/streptomycin, L-glutamine, and 10% bovine serum. Human MSCs were cultured in low glucose DMEM containing 1% penicillin/streptomycin, and 10% fetal bovine serum (basal media) and expanded to between passage 4 and 6 before use. In typical studies, substrates were seeded at 2,000 cells/cm2 and maintained in basal media. For studies requiring multicellular clusters, pyramid-shaped microwells (AggreWell, Stemcell Technologies, Vancouver, BC) were used to generate spheroids containing 1,000 MSCs. For contraction assays, MSCs were seeded at 10,000 cells/cm2. For studies requiring the inhibition of FAK phosphorylation, PF 573228 (Tocris Biosciences, Bristol, UK) was added to basal media at a concentration of 1 µM.
Fluorescent staining and microscopy
MSCs on fiber and hydrogel substrates were fixed in 4% PFA for 15 min at room temperature. Alternatively, to extract cytoplasmic vinculin, samples were simultaneously fixed and permeabilized in 2% PFA in microtubule stabilizing buffer for 15 min at room temperature. To examine the organization of the actin cytoskeleton, cells were permeabilized with Triton X-100, blocked in 2% BSA, and stained with phalloidin. For immunostaining, samples were permeabilized, blocked for 1 h in 10% FBS containing 0.2% Tween, and incubated with primary (vinculin: 1:500 mouse monoclonal anti-vinculin (Sigma, V9264), phoshoFAK: 1:500 rabbit polyclonal anti-FAK (phospho Y397) (AbCam, ab39967) and secondary (1:1000 Alexa Fluor 488 goat anti-mouse IgG (H+L) or 1:1000 Alexa Fluor 488 donkey anti-rabbit IgG (H+L) (Life Technologies) consecutively for 1 h each at room temperature. For proliferation studies, EdU labeling was performed following the manufacturer’s protocol (ClickIT EdU, Life Technologies). To quantify cell death within fibrous networks, staining with calcein AM/ethidium homodimer (LIVE/DEAD® Cell Viability Assay, Life Technologies) was performed. High resolution confocal imaging of phalloidin-stained samples was performed to identify multinucleated cells, and did not reveal any evidence of incomplete cytokinesis. Samples were imaged at 10 or 40 × on a Zeiss 200M with a spinning disk head (Yokogawa CSU-10 with Borealis), environmental chamber, four laser lines, Photometric Evolve EMCCD camera. Unless otherwise specified, images are presented as maximum intensity projections. Cell area, proliferation, focal adhesion, and fiber recruitment analyses were performed with custom Matlab scripts.
Statistics
Statistical differences were determined by ANOVA or Student's t-test where appropriate, with significance indicated by p<0.05. Sample size is indicated within corresponding figure legends. All data is presented as a mean ± standard deviation.
Supplementary Material
Acknowledgements
This work was supported in part from grants from the National Institutes of Health (grant numbers EB00262, EB001046, HL115553, GM74048, AR056624) and Center for Engineering Cells and Regeneration of the University of Pennsylvania. B.M.B. acknowledges financial support from a Ruth L. Kirschstein National Research Service Award (EB014691) and NIH Pathway to Independence Award (K99HL124322). We would like to thank Ning Wang for helpful discussions. We would also like to acknowledge support from the Penn Regional Nanotechnology Facility and the BME Core Facilities at Boston University.
Footnotes
Contributions
B.M.B., B.T., J.A.B. and C.S.C. designed the materials. B.M.B., B.T., and C.S.C. designed the experiments. B.M.B., B.T., W.Y.W., M.S.S., and I.L.K. conducted experiments and analysed data. V.B.S. helped with analysis and interpretation of mechanical testing data. B.M.B., B.T., and C.S.C. wrote the manuscript.
References
- 1.Discher DE, Janmey P, Wang Y. Tissue Cells Feel and Respond to the Stiffness of Their Substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 2.Wozniak MA, Chen CS. Mechanotransduction in development: a growing role for contractility. Nat Rev Mol Cell Biol. 2009;10:34–43. doi: 10.1038/nrm2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaalouk DE, Lammerding J. Mechanotransduction gone awry. Nat Rev Mol Cell Biol. 2009;10:63–73. doi: 10.1038/nrm2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelham RJ, Wang Y. Cell locomotion and focal adhesions are regulated by substrate flexibility. PNAS. 1997;94:13661–13665. doi: 10.1073/pnas.94.25.13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeung T, et al. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskeleton. 2005;60:24–34. doi: 10.1002/cm.20041. [DOI] [PubMed] [Google Scholar]
- 6.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 7.Engler A, et al. Substrate Compliance versus Ligand Density in Cell on Gel Responses. Biophysical Journal. 2004;86:617–628. doi: 10.1016/S0006-3495(04)74140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pedersen JA, Swartz MA. Mechanobiology in the Third Dimension. Annals of Biomedical Engineering. 2005;33:1469–1490. doi: 10.1007/s10439-005-8159-4. [DOI] [PubMed] [Google Scholar]
- 9.Pathak A, Kumar S. Biophysical regulation of tumor cell invasion: moving beyond matrix stiffness. Integrative Biology. 2011;3:267. doi: 10.1039/c0ib00095g. [DOI] [PubMed] [Google Scholar]
- 10.Baker BM, Chen CS. Deconstructing the third dimension – how 3D culture microenvironments alter cellular cues. J Cell Sci. 2012;125:3015–3024. doi: 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Dijk-Wolthuis WNE, et al. Synthesis, Characterization, and Polymerization of Glycidyl Methacrylate Derivatized Dextran. Macromolecules. 1995;28:6317–6322. [Google Scholar]
- 12.Yang L, et al. Micromechanical bending of single collagen fibrils using atomic force microscopy. J. Biomed. Mater. Res. 2007;82A:160–168. doi: 10.1002/jbm.a.31127. [DOI] [PubMed] [Google Scholar]
- 13.Kluge D, Abraham F, Schmidt S, Schmidt H-W, Fery A. Nanomechanical Properties of Supramolecular Self-Assembled Whiskers Determined by AFM Force Mapping. Langmuir. 2010;26:3020–3023. doi: 10.1021/la904780c. [DOI] [PubMed] [Google Scholar]
- 14.Guthold M, et al. A Comparison of the Mechanical and Structural Properties of Fibrin Fibers with Other Protein Fibers. Cell Biochem Biophys. 2007;49:165–181. doi: 10.1007/s12013-007-9001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trappmann B, et al. Extracellular-matrix tethering regulates stem-cell fate. Nat Mater. 2012;11:642–649. doi: 10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- 16.Knight CG, et al. The Collagen-binding A-domains of Integrins α1β1 and α2β1Recognize the Same Specific Amino Acid Sequence, GFOGER, in Native (Triple-helical) Collagens. J. Biol. Chem. 2000;275:35–40. doi: 10.1074/jbc.275.1.35. [DOI] [PubMed] [Google Scholar]
- 17.Konitsiotis AD, et al. Characterization of High Affinity Binding Motifs for the Discoidin Domain Receptor DDR2 in Collagen. J. Biol. Chem. 2008;283:6861–6868. doi: 10.1074/jbc.M709290200. [DOI] [PubMed] [Google Scholar]
- 18.Zhong C, et al. Rho-mediated Contractility Exposes a Cryptic Site in Fibronectin and Induces Fibronectin Matrix Assembly. J Cell Biol. 1998;141:539–551. doi: 10.1083/jcb.141.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klotzsch E, et al. Fibronectin forms the most extensible biological fibers displaying switchable force-exposed cryptic binding sites. PNAS. 2009;106:18267–18272. doi: 10.1073/pnas.0907518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grinnell F, Ho C-H, Tamariz E, Lee DJ, Skuta G. Dendritic Fibroblasts in Three-dimensional Collagen Matrices. Molecular Biology of the Cell. 2003;14:384–395. doi: 10.1091/mbc.E02-08-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huebsch N, et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khetan S, et al. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nature Materials. 2013;12:458–465. doi: 10.1038/nmat3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking Cell-Matrix Adhesions to the Third Dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 24.Hakkinen KM, Harunaga JS, Doyle AD, Yamada KM. Direct Comparisons of the Morphology, Migration, Cell Adhesions, and Actin Cytoskeleton of Fibroblasts in Four Different Three-Dimensional Extracellular Matrices. Tissue Engineering Part A. 2010;17:713–724. doi: 10.1089/ten.tea.2010.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fraley SI, et al. A distinctive role for focal adhesion proteins in three-dimensional cell motility. Nat. Cell Biol. 2010;12:598–604. doi: 10.1038/ncb2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubow KE, Horwitz AR. Reducing background fluorescence reveals adhesions in 3D matrices. Nat. Cell Biol. 2011;13:3–5. doi: 10.1038/ncb0111-3. author reply 5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stopak D, Harris AK. Connective tissue morphogenesis by fibroblast traction: I. Tissue culture observations. Developmental Biology. 1982;90:383–398. doi: 10.1016/0012-1606(82)90388-8. [DOI] [PubMed] [Google Scholar]
- 28.Ma X, et al. Fibers in the Extracellular Matrix Enable Long-Range Stress Transmission between Cells. Biophysical Journal. 2013;104:1410–1418. doi: 10.1016/j.bpj.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo C-L, et al. Long-range mechanical force enables self-assembly of epithelial tubular patterns. PNAS. 2012;109:5576–5582. doi: 10.1073/pnas.1114781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi Q, et al. Rapid disorganization of mechanically interacting systems of mammary acini. PNAS. 2014;111:658–663. doi: 10.1073/pnas.1311312110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Provenzano P, et al. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Medicine. 2006;4:38. doi: 10.1186/1741-7015-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grinnell F, Lamke CR. Reorganization of hydrated collagen lattices by human skin fibroblasts. J Cell Sci. 1984;66:51–63. doi: 10.1242/jcs.66.1.51. [DOI] [PubMed] [Google Scholar]
- 33.Tomasek JJ, Haaksma CJ, Eddy RJ, Vaughan MB. Fibroblast contraction occurs on release of tension in attached collagen lattices: dependency on an organized actin cytoskeleton and serum. Anat. Rec. 1992;232:359–368. doi: 10.1002/ar.1092320305. [DOI] [PubMed] [Google Scholar]
- 34.Kolodney MS, Elson EL. Correlation of myosin light chain phosphorylation with isometric contraction of fibroblasts. J. Biol. Chem. 1993;268:23850–23855. [PubMed] [Google Scholar]
- 35.Mih JD, Marinkovic A, Liu F, Sharif AS, Tschumperlin DJ. Matrix stiffness reverses the effect of actomyosin tension on cell proliferation. J Cell Sci. 2012;125:5974–5983. doi: 10.1242/jcs.108886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H-B, Dembo M, Wang Y-L. Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. American Journal of Physiology - Cell Physiology. 2000;279:C1345–C1350. doi: 10.1152/ajpcell.2000.279.5.C1345. [DOI] [PubMed] [Google Scholar]
- 37.Wang L-S, Boulaire J, Chan PPY, Chung JE, Kurisawa M. The role of stiffness of gelatin–hydroxyphenylpropionic acid hydrogels formed by enzyme-mediated crosslinking on the differentiation of human mesenchymal stem cell. Biomaterials. 2010;31:8608–8616. doi: 10.1016/j.biomaterials.2010.07.075. [DOI] [PubMed] [Google Scholar]
- 38.Cavalcanti-Adam EA, et al. Cell Spreading and Focal Adhesion Dynamics Are Regulated by Spacing of Integrin Ligands. Biophysical Journal. 2007;92:2964–2974. doi: 10.1529/biophysj.106.089730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan CE, Odde DJ. Traction Dynamics of Filopodia on Compliant Substrates. Science. 2008;322:1687–1691. doi: 10.1126/science.1163595. [DOI] [PubMed] [Google Scholar]
- 40.Yang MT, Sniadecki NJ, Chen CS. Geometric Considerations of Micro- to Nanoscale Elastomeric Post Arrays to Study Cellular Traction Forces. Adv. Mater. 2007;19:3119–3123. [Google Scholar]
- 41.Balaban NQ, et al. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat Cell Biol. 2001;3:466–472. doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- 42.Chen CS, Alonso JL, Ostuni E, Whitesides GM, Ingber DE. Cell shape provides global control of focal adhesion assembly. Biochemical and Biophysical Research Communications. 2003;307:355–361. doi: 10.1016/s0006-291x(03)01165-3. [DOI] [PubMed] [Google Scholar]
- 43.Pasapera AM, Schneider IC, Rericha E, Schlaepfer DD, Waterman CM. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J Cell Biol. 2010;188:877–890. doi: 10.1083/jcb.200906012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Slack-Davis JK, et al. Cellular Characterization of a Novel Focal Adhesion Kinase Inhibitor. J. Biol. Chem. 2007;282:14845–14852. doi: 10.1074/jbc.M606695200. [DOI] [PubMed] [Google Scholar]
- 45.Houseman BT, Mrksich M. The microenvironment of immobilized Arg-Gly-Asp peptides is an important determinant of cell adhesion. Biomaterials. 2001;22:943–955. doi: 10.1016/s0142-9612(00)00259-3. [DOI] [PubMed] [Google Scholar]
- 46.Arnold M, et al. Activation of Integrin Function by Nanopatterned Adhesive Interfaces. Chem Phys Chem. 2004;5:383–388. doi: 10.1002/cphc.200301014. [DOI] [PubMed] [Google Scholar]
- 47.Chaudhuri O, et al. Substrate stress relaxation regulates cell spreading. Nat Commun. 2015;6 doi: 10.1038/ncomms7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fratzl P. Collagen: Structure and Mechanics. Springer Science & Business Media; 2008. [Google Scholar]
- 49.Gautieri A, Vesentini S, Redaelli A, Buehler MJ. Hierarchical Structure and Nanomechanics of Collagen Microfibrils from the Atomistic Scale Up. Nano Lett. 2011;11:757–766. doi: 10.1021/nl103943u. [DOI] [PubMed] [Google Scholar]
- 50.Elosegui-Artola A, et al. Rigidity sensing and adaptation through regulation of integrin types. Nat Mater. 2014;13:631–637. doi: 10.1038/nmat3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choquet D, Felsenfeld DP, Sheetz MP. Extracellular matrix rigidity causes strengthening of integrin-cytoskeleton linkages. Cell. 1997;88:39–48. doi: 10.1016/s0092-8674(00)81856-5. [DOI] [PubMed] [Google Scholar]
- 52.Fu J, et al. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat. Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sander EA, Stylianopoulos T, Tranquillo RT, Barocas VH. Image-based multiscale modeling predicts tissue-level and network-level fiber reorganization in stretched cell-compacted collagen gels. PNAS. 2009;106:17675–17680. doi: 10.1073/pnas.0903716106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abhilash AS, Baker BM, Trappmann B, Chen CS, Shenoy VB. Remodeling of Fibrous Extracellular Matrices by Contractile Cells: Predictions from Discrete Fiber Network Simulations. Biophysical Journal. 2014;107:1829–1840. doi: 10.1016/j.bpj.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walcott S, Sun SX. A mechanical model of actin stress fiber formation and substrate elasticity sensing in adherent cells. PNAS. 2010;107:7757–7762. doi: 10.1073/pnas.0912739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Besser A, Safran SA. Force-Induced Adsorption and Anisotropic Growth of Focal Adhesions. Biophysical Journal. 2006;90:3469–3484. doi: 10.1529/biophysj.105.074377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kidoaki S, Kwon IK, Matsuda T. Structural features and mechanical properties of in situ–bonded meshes of segmented polyurethane electrospun from mixed solvents. J. Biomed. Mater. Res. 2006;76B:219–229. doi: 10.1002/jbm.b.30336. [DOI] [PubMed] [Google Scholar]
- 58.Kuntz RM, Saltzman WM. Neutrophil motility in extracellular matrix gels: mesh size and adhesion affect speed of migration. Biophys J. 1997;72:1472–1480. doi: 10.1016/S0006-3495(97)78793-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.