Abstract
The Drosophila mushroom body (MB) is a key associative memory center that has also been implicated in the control of sleep. However, the identity of MB neurons underlying homeostatic sleep regulation, as well as the types of sleep signals generated by specific classes of MB neurons, has remained poorly understood. We recently identified two MB output neuron (MBON) classes whose axons convey sleep control signals from the MB to converge in the same downstream target region: a cholinergic sleep-promoting MBON class and a glutamatergic wake-promoting MBON class. Here we deploy a combination of neurogenetic, behavioral, and physiological approaches to identify and mechanistically dissect sleep-controlling circuits of the MB. Our studies reveal the existence of two segregated excitatory synaptic microcircuits that propagate homeostatic sleep information from different populations of intrinsic MB “Kenyon cells” (KCs) to specific sleep-regulating MBONs: sleep-promoting KCs increase sleep by preferentially activating the cholinergic MBONs, while wake-promoting KCs decrease sleep by preferentially activating the glutamatergic MBONs. Importantly, activity of the sleep-promoting MB microcircuit is increased by sleep deprivation and is necessary for homeostatic rebound sleep (i.e., the increased sleep that occurs after, and in compensation for, sleep lost during deprivation). These studies reveal for the first time specific functional connections between subsets of KCs and particular MBONs and establish the identity of synaptic microcircuits underlying transmission of homeostatic sleep signals in the MB.
INTRODUCTION
The Drosophila MB is an associative memory network of evolutionary origin likely conserved with mammalian cerebral cortex that has also been implicated in the control of sleep [1–12]. Fly sleep exhibits all of the key features of vertebrate sleep, including circadian regulation and homestatic rebound, and can be operationally identified as periods of locomotor quiescence lasting five minutes or longer [13, 14]. The MB contains ~2000 KCs divided into 7 anatomical classes based on the specific projections of their axons into the α/β, α'/β', or γ lobes (Figure 1A)[15]. KCs receive synaptic inputs from sensory systems and in turn synapse convergently onto 34 MBONs of 21 cell types, whose dendrites contiguously tile the MB lobes, defining 15 non-overlapping compartments [15, 16]. MBON cell types are uniquely named by their stereotyped arborizations in these compartments, numbered consecutively within each lobe starting from the intersection of the horizontal and vertical lobes [16]. Interestingly, the dendrites of some MBONs extend into two neighboring compartments residing in different lobes, thus providing a potential mechanism for convergence of multiple KC classes onto a common MBON class [16]. Because of the lack of tools enabling specific genetic targeting of each of these individual KC and MBON cell types for functional manipulation and imaging, it has remained poorly understood which particular MB cell types are involved in controlling sleep, in which MB cell types homeostatic sleep signals are generated, and whether and how they are assembled into sleep-controlling circuits.
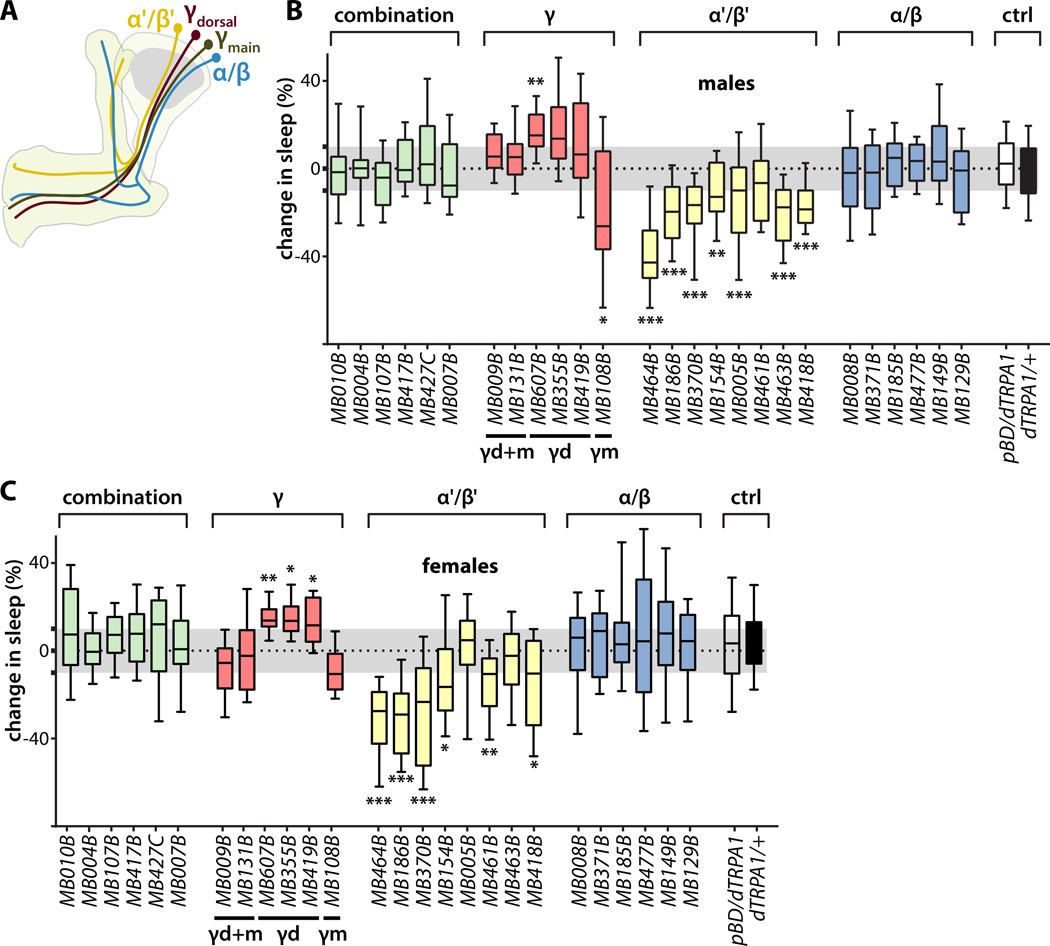
Figure 1. Identification of sleep-promoting and wake-promoting KCs.
(A) Schematic representation of the MB showing the lobe-specific axon projections of the indicated classes of KCs. Axons of α'/β' and α/β KCs bifurcate to innervate the vertical α' and α lobes and horizontal β' and β lobes, while the axons of γ KCs do not bifurcate and innervate only the horizontal γ lobe.
(B) Change in sleep of male flies induced by activation of KC subsets targeted by the indicated split-GAL4 driver lines to express the dTRPA1 temperature-gated depolarizing cation channel. Percent change in sleep is defined as (sleep on day 1 – sleep on day 2) / sleep on day 1, where the ambient temperature is increased on day 2 from 21.5 °C to 28.5 °C to activate the neurons expressing dTRPA1. Activation of either α'/β' or γmain (γm) KCs promoted wake, while activation of γdorsal (γd) KCs promoted sleep. Midline, box boundaries, and whiskers represent median, quartiles, and 10th and 90th percentiles, respectively. Split-GAL4 lines are grouped by the indicated lobe-specific projections of their targeted KC subsets. Each split-GAL4 line was compared to an enhancerless GAL4 (pBDPGAL4U, indicated in the figures as pBDG4U or pBD) by Kruskal-Wallis non-parametric one-way ANOVA and Dunn's post-hoc correction for multiple comparisons (*, p<0.05; **, p<0.01; ***p<0.001; n=24–46 flies per genotype).
(C) Change in sleep of female flies, as in (B).
To comprehensively identify the KCs and MBONs that control sleep, we performed a neuronal activation screen using a new library of intersectional “split-GAL4” fly lines, each of which drives expression of an effector transgene under the control of the upstream activating sequence (UAS) to defined MB cell classes [16–19]. We recently reported the results of an initial thermogenetic activation screen of the subset of this library that targets MBONs, which revealed several classes of sleep-controlling MBONs with dendrites in distinct lobe compartments, including two whose axons converge in the same downstream target region: cholinergic sleep-promoting MBON-γ2α'1 and glutamatergic wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral [16, 19]. Here we report the extension of this thermogenetic activation screen to each of the KC cell types, revealing that α'/β' and γmain (γm) KCs are wake-promoting and γdorsal (γd) KCs are sleep-promoting.
We then used behavioral genetic and physiological techniques to identify synaptic microcircuits connecting these sleep-controlling KC classes to the previously identified sleep-controlling MBONs. We determined that wake-promoting α'/β' and γm KCs preferentially excite wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral, and sleep-promoting γd KCs preferentially excite sleep-promoting MBON-γ2α'1. To identify a potential role for these microcircuits in the generation and/or propagation of homeostatic sleep signals, we used optical imaging of a genetically encoded fluorescent voltage indicator to determine that sleep deprivation increases spontaneous electrical activity in the sleep-promoting γd KCs and MBON-γ2α'1 while inhibiting it in wake-promoting α'/β' KCs and MBON-γ5β′2a/β′2mp/β′2mp_bilateral. Importantly, acute thermogenetic silencing of the synaptic outputs of sleep-promoting MBON-γ2α'1 severely attenuates homeostatic rebound sleep occurring in response to sleep deprivation, establishing the essential role played by the sleep-promoting synaptic microcircuit in homeostatic sleep regulation. Taken together these studies reveal for the first time the necessary and sufficient cellular and synaptic mechanisms underlying transmission of homeostatic sleep signals by microcircuits of the Drosophila MB. Our studies also reveal for the first time a role for functionally segregated synaptic connectivity between particular KC classes and MBONs in the control of behavior, an issue of great general importance given the diversity of neurobehavioral contexts in which the MB plays a central role.
RESULTS
Control of sleep by KCs
To determine how intrinsic signals are generated in the MB to set the activity of the previously identified sleep-controlling MBONs, we screened split-GAL4 lines targeting KCs [16] in male and female flies (Figure 1B,C; S1A). Each of these split-GAL4 lines (or a negative control "empty" split-GAL4 line, pBDPGAL4U, lacking active genomic enhancer sequences) was crossed to a UAS-dTRPA1 line to drive expression of the temperature-gated depolarizing cation channel dTRPA1 [20]. Baseline sleep-wake cycles at 21.5°C and the effects of dTRPA1-mediated neuronal activation induced by a shift to 28.5°C were measured using the automated infrared beam-break locomotor assay that is the widely accepted standard in the field (Trikinetics, Waltham, MA) [21, 22]. Change in sleep is defined as the difference in sleep amount between the first 24 hours at 28.5°C and the baseline 24 hours of sleep at 21.5°C, divided by the baseline sleep amount. Transient activation of α'/β' KCs strongly decreased sleep (i.e., increased wake) (Figure 1B,C; 2A,B), consistent with a recent report that a GABAergic interneuron, MB-APL, promotes sleep by inhibiting α'/β' KCs [7]. Activation of γd KCs increased sleep, while activation of γm KCs decreased sleep (Figure 1B,C; 2C,D). Interestingly, however, simultaneous activation of γd and γm KCs had no effect on sleep (Figure 1B,C), consistent with integration of their opposing effects. While activation of γd KCs using each of the three γd split-GAL4 drivers increases sleep in female flies, only MB607B significantly increases sleep in male flies. This is consistent with the greater strength of MB607B as reflected in fluorescence intensity when used to drive GFP expression [16] and the fact that male flies sleep substantially more than females at baseline, making increases in sleep more difficult to detect in males than in females. In contrast to α'/β' and γ KCs, activation of α/β KCs, either all together or in subsets, had no effect on sleep (Figure 1B,C). This is in contrast to the previous suggestion of a role for α/β KCs in regulating sleep [3], and is perhaps explained by differences between the traditional GAL4 driver used there and the split-GAL4 drivers used here. Simultaneous activation of combinations of KCs encompassing both the sleep-promoting and wake-promoting classes had no net effect on sleep, suggesting counterbalancing influences on downstream targets (Figure 1B,C). To distinguish specific sleep effects from possible confounding locomotor deficits we also quantified locomotor activity when flies are awake. Activation of sleep-promoting KCs did not suppress locomotor activity while awake, nor did activation of wake-promoting KCs increase locomotor activity while awake, ruling out nonspecific locomotor effects that could be misconstrued as changes in sleep (Figure S1B). Furthermore, as expected for neurons that regulate sleep and not just locomotor activity, transient activation of α'/β' KCs to suppress sleep for a single night results in homeostatic rebound sleep the subsequent day (Figure 2E,F,G). Detailed sleep parameters for all lines tested are presented in Table S1, and screening results reanalyzed using the aggregate of all lines as a reference control for each individual line are presented in Table S2. These results reveal the identities of distinct classes of sleep-promoting and wake-promoting KCs.
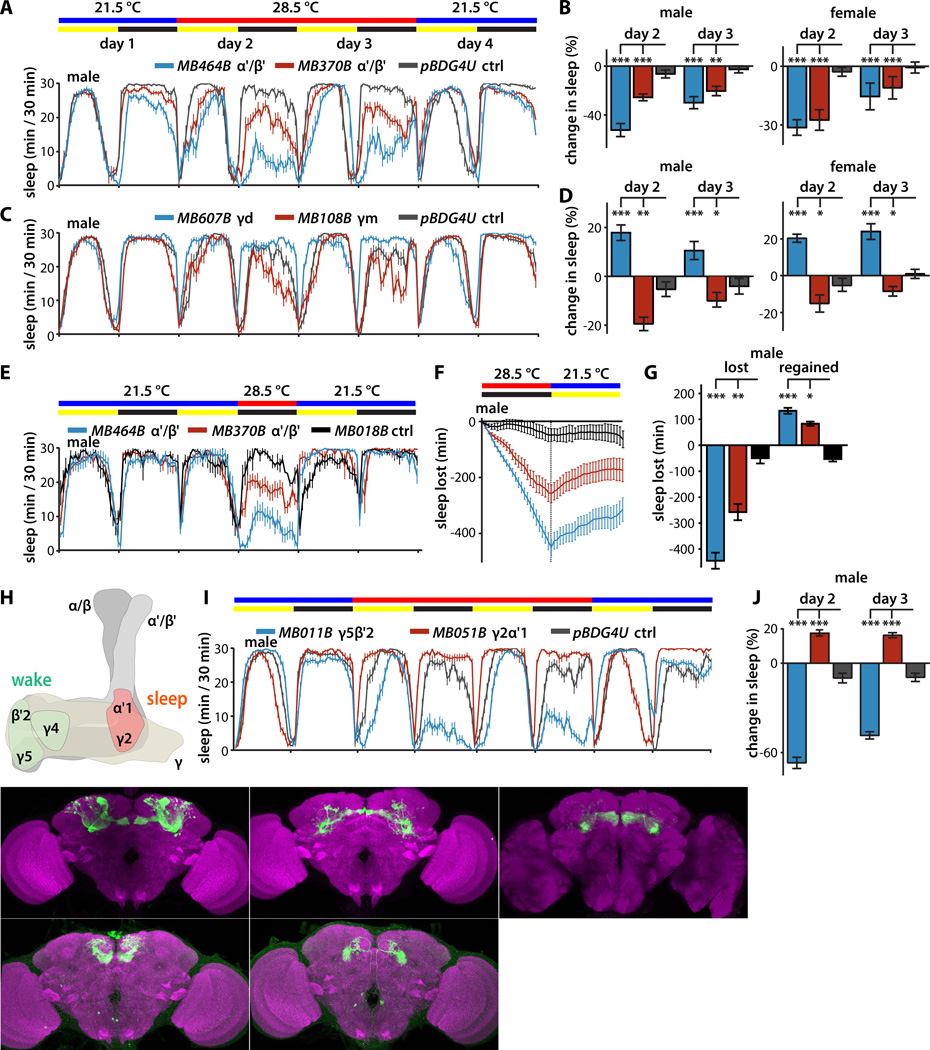
Figure 2. Sleep profiles of flies following thermogenetic activation of KCs and MBONs.
(A) Sleep profiles of male flies expressing dTRPA1 in α'/β' KCs driven by the indicated split-GAL4 lines (mean±sem). Flies were maintained in 12 hr:12 hr light:dark (LD) conditions, and the temperature was increased for days 2 and 3 of the four day experiment.
(B) Quantification of change in sleep relative to day 1 of male and female flies expressing dTRPA1 in α'/β' KCs. Genotypes are color coded as in (A). Statistical analysis was by one-way ANOVA and Dunnett's paired-comparison test.
(C) Sleep profiles of male flies expressing dTRPA1 in γd or γm KCs under the control of the indicated split-GAL4 lines, treated as in (A).
(D) Quantification of change in sleep of male and female flies expressing dTRPA1 in γd or γm KCs. Genotypes are color coded as in (C); statistical analysis as in (B).
(E) Sleep profiles of male flies expressing dTRPA1 in α'/β' KCs driven by the indicated split-GAL4 lines (mean±sem). Flies were maintained in 12 hr:12 hr light:dark (LD) conditions, and the temperature was increased for 12 hr during the night of day 2 of this three day experiment.
(F) Cumulative sleep lost during thermogenetic sleep suppression at 28.5°C and regained during subsequent recovery for 12 hours at 21.5°C (mean±sem). Genotypes are color coded as in (E).
(G) Quantification of sleep lost during thermogenetic sleep suppression and regained during the 12 hours of recovery. Sleep suppression induced by dTRPA1-mediated activation of α'/β' KCs using either of two split-GAL4 drivers resulted in homeostatic rebound compared to control flies expressing dTRPA1 in a cholinergic MBON that does not influence sleep (MB018B). Genotypes are color coded as in (D); statistical analysis was by one-way ANOVA and Dunnett's paired-comparison test. (*, p<0.05; **, p<0.01; ***p<0.001; n=22–24 flies per genotype.)
(H) Schematic representation of the MB lobes indicating the compartment-specific dendritic projections of sleep-regulating MBON-γ5β′2a/β′2mp/β′2mp_bilateral, MBON-γ4>γ1γ2, and MBON-γ2α'1.
(I) Sleep profiles of male flies expressing dTRPA1 in MBONs under the control of the indicated split-GAL4 lines, treated as in (A).
(J) Quantification of change in sleep of male flies expressing dTRPA1 in MBONs. Genotypes color coded as in (F); statistical analysis as in (B).
(K) Whole brains of flies expressing GFP under the control of the indicated MBON and KC split-GAL4 driver lines, immunostained for GFP (green) and the synaptic neuropil marker BRUCHPILOT (magenta).
Synaptic microcircuits connecting KCs with MBONs
To address how the sleep control signals from KCs propagate to downstream sleep control centers outside the MB, we next turned our attention to the connections between KCs and MBONs. We recently reported that transient activation of cholinergic MBON-γ2α'1, whose dendrites arborize in the γ2 and α'1 compartments (Figure 2H) [16], increases sleep in female flies [19]. Conversely, activation of the glutamatergic MBON-γ4>γ1γ2 or a cluster of MBONs whose dendrites arborize in the γ5 and/or β'2 compartments decreases sleep in female flies [19]. Dendrites of MBON-γ4>γ1γ2 arborize in the γ4 compartment and project their axons to the γ1 and γ2 compartments (Figure 2H) [16]. Three MBON cell types arborize dendrites in the γ5 and/or β'2 compartments: one with dendrites in both γ5 and β'2 (MBON-γ5β'2a, also known as MB-M6), and two with dendrites solely in β'2 (MBON-β'2mp, also known as MB-M4, and MBON-β'2mp-bilateral) [16]. Because the available split-GAL4 lines used to target these MBONs are active in all three cell types, we cannot functionally distinguish between them and will refer to them collectively as MBON-γ5β′2a/β′2mp/β′2mp_bilateral. The axon terminals of wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral and sleep-promoting MBON-γ2α'1 converge in the same target regions outside the MB in the superior medial protocerebrum (SMP) and crepine (CRE) [16].
We repeated the MBON transient activation screen in male flies and found that, as in females, glutamatergic MBON-γ5β′2a/β′2mp/β′2mp_bilateral and MBON-γ4>γ1γ2 are wake-promoting, and cholinergic MBON-γ2α'1 is sleep-promoting (Figure 2I,J; S2A). Importantly, our previous extensive analysis using video monitoring of flies in an open arena rules out primary locomotor effects of MBON activation with dTRPA1 [19]. Specifically, acute optogenetic activation of MBON-γ5β′2a/β′2mp/β′2mp_bilateral or MBON-γ2α'1 had no effect on walking speed, and acute thermogenetic activation of MBON-γ5β′2a/β′2mp/β′2mp_bilateral or MBON-γ2α'1 had no effect on the fraction of time spent performing any of more than one dozen distinct behaviors detected and quantified using automated computer vision algorithms [19]. As with the KCs, assessment of activity while awake also rules out non-specific locomotor effects (Figure S3). In addition, dTRPA1-mediated activation of MBON-γ2α'1 or γd KCs does not prevent arousal in a vial tapping assay [see, e.g., 7]; rather, as expected for activation of sleep-promoting neurons, arousal threshold appears greater, arousal response is attenuated, and return to locomotor quiescence is accelerated (Movie S1, S2). Representative images of the expression patterns of sleep-regulating KCs and MBONs are shown in Figure 2K. As an additional independent genetic control for the split-GAL4 screening results, we compared each KC and MBON split-GAL4/UAS-dTRPA1 screen hit to each split-GAL4/+ negative control genotype that possesses the two split-GAL4 driver transgenes but lacks the UAS-dTRPA1 effector transgene (Figure S2B). Previous detailed anatomical analysis of MBON-γ5β′2a/β′2mp/β′2mp_bilateral and MBON-γ2α'1 indicates that their dendrites are each contacted by all three of the sleep-controlling KC classes: sleep-promoting γd, and wake-promoting γm and α'/β' [16]. This pattern of shared anatomical connectivity between KCs and MBONs raises the question how sleep control information of opposite sign propagates from KCs to particular MBONs.
To test the hypothesis that sleep control signals flow from KCs to MBONs through parallel segregated sleep-promoting and wake-promoting synaptic microcircuits, we examined whether blocking the synaptic outputs of a particular MBON class could prevent the effect on sleep of activating a KC class. We focused our attention on MBON-γ5β′2a/β′2mp/β′2mp_bilateral and MBON-γ2α'1, as these MBONs convey sleep control signals to what is likely a common downstream target. To probe functional connectivity of α'/β' KCs to MBONs, we activated these KCs with dTRPA1 targeted using the orthogonal LexA-LexAop binary expression system [23, 24], while simultaneously silencing the synaptic outputs of either MBON-γ5β′2a/β′2mp/β′2mp_bilateral or MBON-γ2α'1 with Shibirets1 temperature-sensitive inhibitor of synaptic vesicle recycling [25] targeted using split-GAL4 (Figure 3A). To simultaneously activate dTRPA1 and Shibirets1, the ambient temperature was shifted from 18.5°C to 31.5°C for one night, and change in sleep was assessed relative to the previous night at 18.5°C. Key controls for these experiments include no-driver negative control flies that express neither dTRPA1 nor Shibirets1, flies that express Shibirets1 in MBONs but not dTRPA1 in KCs, and flies that express dTRPA1 in the α'/β' KCs but not Shibirets1 in MBONs. No-driver control flies slept ~10% less at 31.5°C, consistent with known intrinsic effects of elevated temperature on nighttime sleep [see, e.g., 26]. As expected from our split-GAL4 screen results (Figure 1B,C), activation of α'/β' KCs in male flies expressing dTRPA1 under the control of R35B12-LexA driver significantly decreased night sleep (Figure 3B,C,D). Silencing synaptic outputs of MBON-γ2α'1 or MBON-γ5β′2a/β′2mp/β′2mp_bilateral had no effect on nighttime sleep on its own (Figure 3B). However, silencing wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral completely blocked the decrease in sleep induced by activation of α'/β' KCs, but silencing synaptic outputs of sleep-promoting MBON-γ2α'1 had no effect (Figure 3B,C). Again, measurement of activity while awake rules out non-specific locomotor effects (Figure S4A). As expected for manipulation of homeostatic sleep-regulating circuits, sleep suppression induced by combined thermogenetic activation/silencing induces subsequent homeostatic rebound sleep (Figure 3E). These results support a segregated microcircuit in which α'/β' KCs promote wake by synaptic excitation of wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral, and establish MBON-γ5β′2a/β′2mp/β′2mp_bilateral as the sole non-redundant output pathway by which α'/β' KCs control sleep.
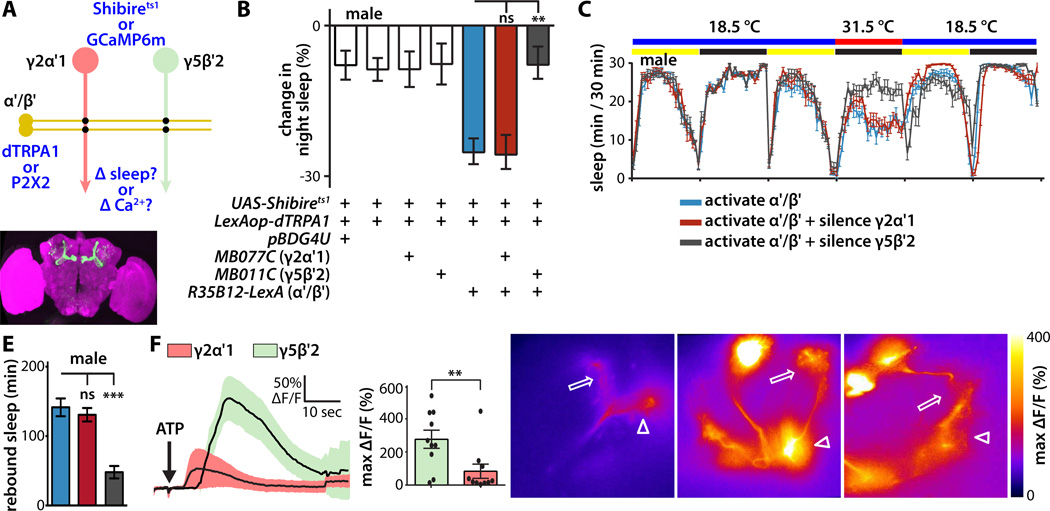
Figure 3. Segregated synaptic information flow from wake-promoting KCs to wake-promoting MBONs.
(A) Schematic of experimental design to probe synaptic connections between wake-promoting α'/β' KCs and sleep-regulating MBONs in male flies. For behavioral genetic experiments, dTRPA1 was expressed in α'/β' KCs using the R35B12-LexA driver, and Shibirets temperature-gated silencer of synaptic release in MBONs using split-GAL4. For physiological experiments, P2X2 ATP-gated depolarizing cation channel was expressed in α'/β' KCs, and GCaMP6m fluorescent Ca 2+ indicator in MBONs.
(B) Change in night sleep compared to the previous night of male flies bearing the indicated transgenes after temperature shift from 18.5°C to 31.5°C to simultaneously activate dTRPA1 and Shibirets1. Synaptic silencing of wake-promoting MBON-γ5β′ 2a/β′ 2mp/β′ 2mp_bilateral, but not sleep-promoting MBON-γ2α'1, prevented the wake-promoting effect of temperature-induced activation of α'/β' KCs. Statistical comparisons between indicated groups were by one-way ANOVA with Dunnett's paired-comparison test (n=48–62 flies per genotype).
(C) Sleep profiles of male flies expressing dTRPA1 in α'/β' KCs and Shibirets1 in the indicated MBONs. Flies were maintained in LD conditions, and the temperature was increased during the second night of the experiment.
(D) Whole brain of fly expressing GFP under the control of the R35B12-LexA driver, immunostained for GFP (green) and the synaptic neuropil marker BRUCHPILOT (magenta).
(E) Quantification of sleep regained during the 12 hr recovery after combined thermogenetic activation/silencing in the experiment shown in (B, C). The quantity of rebound sleep is defined as the difference in sleep amount between the 12 hr recovery period and the previous baseline 12 hr light period. Genotypes are color coded as in (B, C); statistical analysis was by one-way ANOVA and Dunnett's paired-comparison test (n = 25–32 flies per genotype).
(F) Ca2+ responses of GCaMP6m-expressing MBON-γ5β′2a/β′2mp/β′2mp_bilateral or MBON-γ2α'1 following ATP-mediated activation of P2X2-expressing α'/β' KCs. Line graph depicts ΔF/F time course (mean±sem); bar graph depicts peak ΔF/F (mean±sem). MBON-γ5β′2a/β′2mp/β′2mp_bilateral responded much more strongly to α'/β' KC activation than MBON-γ2α'1. Statistical comparison of peak responses was by unpaired t-test (n=10 brains per genotype).
(G) Heat map showing peak Ca2+ responses of representative GCaMP6m-expressing MBON-γ2α'1, MBON-γ5β'2a, or MBON-β'2mp in response to α'/β' KC activation. Arrowheads indicate MBON dendrites, arrows indicate axon terminals, and asterisks indicate cell bodies. Panel width is 80µm.
To probe physiological connectivity of α'/β' KCs to MBONs, we activated α'/β' KCs with P2X2 ATP-gated depolarizing cation channel targeted using LexA-LexAop [27, 28], while simultaneously measuring postsynaptic MBON responses optically using GCaMP6m fluorescent Ca 2+ indicator targeted using split-GAL4 [29] (Figure 3A). Transient activation of P2X2-expressing α'/β' KCs by local delivery of ATP using a picospritzer induced a robust sustained Ca 2+ response in the dendrites of wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral, but only a small response in sleep-promoting MBON-γ2α' 1 (Figure 3F,G). This Ca2+ signal propagated from the dendrites to the axon, axon terminals, and cell bodies of MBON-γ5β′2a/β′2mp/β′2mp_bilateral (Figure 3G; Movie S3). This much stronger excitatory synaptic connection between wake-promoting α'/β' KCs and wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral than with sleep-promoting MBON-γ2α'1 establishes the underlying physiological mechanism for functional segregation of the wake-promoting microcircuit defined by the simultaneous activation-silencing experiments described above (Figure 3B,C).
We next employed the same strategies to probe connectivity between γd and γm KCs and MBONs (Figure 4A). Because of the lack of available LexA driver lines specific for either γd or γm KCs, we used the R14H06-LexA line that is active in γd, γm, and α/β KCs, but not in α'/β' KCs [30]. Simultaneous activation of γd, γm, and α/β KCs induced a net decrease in sleep in male flies (Figure 4B,C,D), which could potentially be explained by a stronger influence of wake-promoting γm KCs than sleep-promoting γd KCs given possible relative dTRPA1 expression levels driven by this particular LexA driver line. When activated alone α/β KCs did not affect sleep (Figure 1A; Figure S4C). While synaptic silencing of wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral suppressed the wake-promoting effect of simultaneous activation of γd, γm, and α/β KCs, synaptic silencing of sleep-promoting MBON-γ2α'1 increased it (Figure 4B,C). Furthermore, as expected for manipulation of homeostatic sleep-regulating circuits, sleep suppression induced by combined thermogenetic activation/silencing induces subsequent homeostatic rebound sleep (Figure 4E). We also tested female flies, which sleep less than males, and thus provide additional head room to detect increases in sleep. Simultaneous activation of γd, γm, and α/β KCs while silencing wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral in female flies increases sleep during the day compared to silencing sleep-promoting MBON-γ2α'1 (Figure 4F,G). Simultaneous activation of γd, γm, and α/β KCs while silencing sleep-promoting MBON-γ2α'1 in female flies suppresses sleep at night compared to silencing wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral (Figure 4F,G). Taken together, these functional results in male and female flies suggest that simultaneous activation of γd and γm KCs activates both MBON-γ5β′2a/β′2mp/β′2mp_bilateral and MBON-γ2α'1. Consistent with this conclusion, P2X2-mediated simultaneous activation of γd, γm, and α/β KCs induces robust Ca2+ increases in both MBON-γ5β′2a/β′2mp/β′2mp_bilateral and MBON-γ2α'1 (Figure 4H,I), while activation of only α/β KCs has no effect (Figure 4J). Moreover, the Ca2+ increase in MBON-γ5β′2a/β′2mp/β′2mp_bilateral occurs only in MBON-γ5β'2a dendrites and not in MBON-β'2mp, indicating that KC-to-MBON synapses are compartmentalized (Figure 4I; Movie S4). These results support the existence of a second segregated microcircuit in which γd KCs promote sleep by synaptic excitation of sleep-promoting MBON-γ2α'1, and that γm KCs promote wake by exciting wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral.
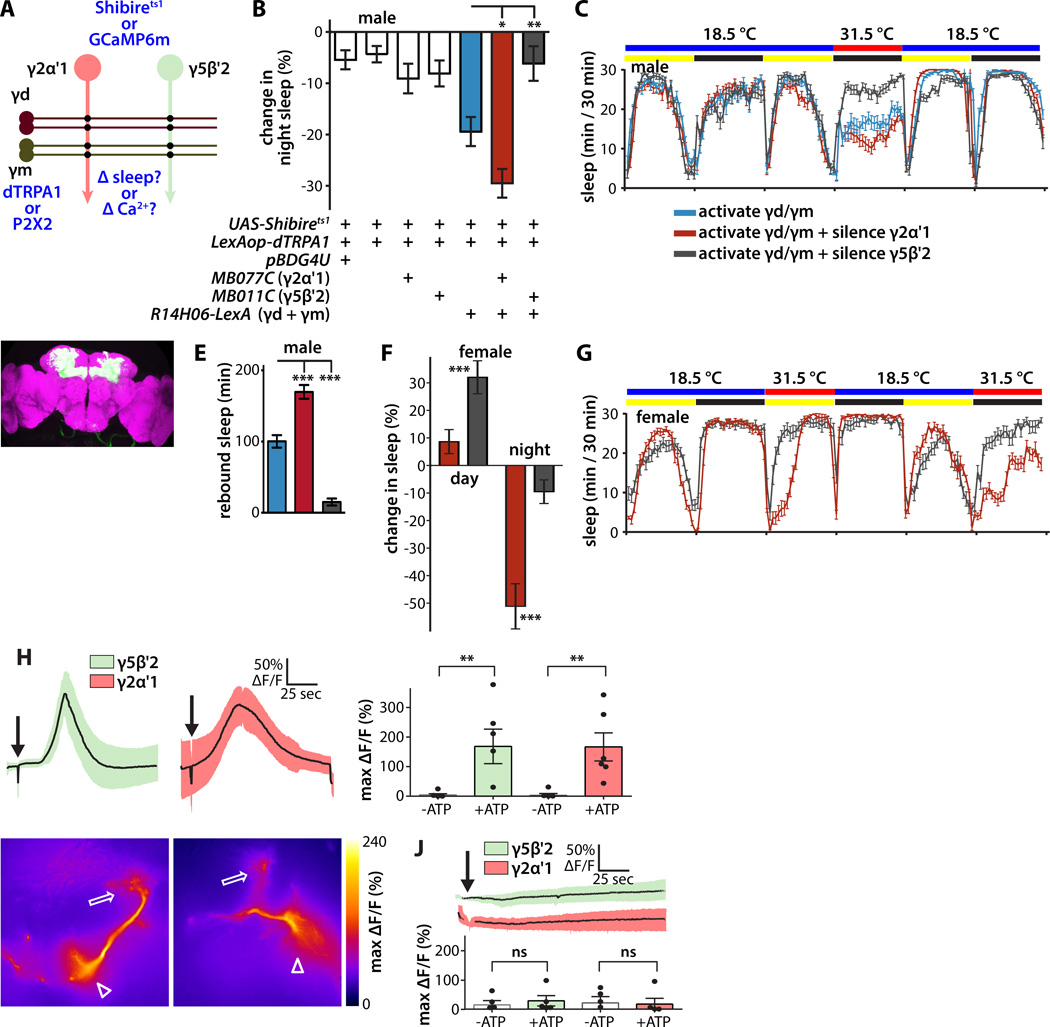
Figure 4. Segregated synaptic information flow from sleep-promoting KCs to sleep-promoting MBONs.
(A) Schematic of experimental design to probe synaptic connections between γ KCs and sleep-regulating MBONs in male flies. For behavioral genetic experiments, dTRPA1 was expressed in γd, γm, and α/β KCs using R14H06-LexA, and Shibirets in MBONs using split-GAL4. For physiological experiments, P2X2 ATP-gated depolarizing cation channel was expressed in γd, γm, and α/β KCs, and GCaMP6m fluorescent Ca2+ indicator in MBONs.
(B) Synaptic silencing of wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral suppressed the net wake-promoting effect of simultaneous activation of γd, γm, and α/β KCs in male flies at night. Conversely, synaptic silencing of sleep-promoting MBON-γ2α'1 augmented the wake-promoting effect of activation of γd, γm, and α/β KCs. Experimental procedure and statistical comparisons were as in Figure 3B (n=48–79 flies per genotype).
(C) Sleep profiles of male flies expressing dTRPA1 in γd, γm, and α/β KCs and Shibirets1 in MBONs as indicated.
(D) Whole brain of fly expressing GFP under the control of R14H06-LexA, immunostained for GFP (green) and the synaptic neuropil marker BRUCHPILOT (magenta).
(E) Quantification of sleep regained during the 12 hours of recovery after combined thermogenetic activation/silencing in the experiment shown in (B, C). Genotypes are color coded as in (B, C); statistical analysis was by one-way ANOVA and Dunnett's paired-comparison test (n=28–31 flies per genotype).
(F) Simultaneous activation of γd, γm, and α/β KCs while silencing wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral in female flies increases sleep during the day compared to silencing sleep-promoting MBON-γ2α'1. Simultaneous activation of γd, γm, and α/β KCs while silencing sleep-promoting MBON-γ2α'1 in female flies suppresses sleep at night compared to silencing wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral. Experimental procedure and statistical comparisons were as in (B); genotypes are color coded as in (B, C) (n=16–18 flies per genotype).
(G) Sleep profiles of female flies expressing dTRPA1 in γd, γm, and α/β KCs and Shibirets1 in MBONs as indicated. Genotypes are color coded as in (B, C).
(H) Ca2+ responses of GCaMP6m-expressing MBON-γ5β′2a/β′2mp/β′2mp_bilateral or MBON-γ2α'1 following ATP-mediated activation of γd, γm, and α/β KCs. Line graph depicts ΔF/F time course; bar graph depicts peak ΔF/F following either ATP or vehicle application. MBON-γ5β'2a and MBON-γ2α'1 each responded strongly to simultaneous activation of γd, γm, and α/β KCs. Statistical comparison of peak responses was by unpaired t-test (n=5–6 brains per genotype and condition).
(I) Heat map showing peak Ca2+ responses of representative GCaMP6m-expressing MBON-γ2α'1 or MBON-γ5β′2a/β′2mp/β′2mp_bilateral in response to combined γd, γm, and α/β KC activation using R14H06-LexA. Arrowheads indicate MBON dendrites, arrows indicate axon terminals, and asterisks indicate cell bodies; panel width is 60µm.
(J) Ca2+ responses of GCaMP6m-expressing MBON-γ5β′2a/β′2mp/β′2mp_bilateral or MBON-γ2α'1 following ATP-mediated activation of α/β KCs using R44E04-LexA. Neither respond to activation of α/β KCs. Graphs and statistical comparison as in (E) (n=5–6 brains per genotype and condition).
Propagation of homeostatic sleep signals through MB microcircuits
The absence of any detectable effect on night sleep of silencing synaptic outputs of sleep-controlling MBONs except when sleep-controlling KCs are exogenously activated (Figure 3B; Figure 4B) suggests that these microcircuits are relatively inactive under basal conditions when flies are able to sleep as much or as little as they choose. We thus hypothesized that the MB sleep-control microcircuits are endogenously activated under physiological or environmental conditions that alter sleep pressure, such as sleep deprivation. Interestingly, recent studies reveal changes in KC Ca2+ levels correlated to quiescent or active states of the fly [31], but the identity of these KCs and their relationship to homeostasis remains uncertain. To test this hypothesis, we compared the electrical activity of sleep-controlling KCs and MBONs in sleep-deprived and – replete conditions, using a standard method for intermittent mechanical perturbation of sleep (see Experimental Procedures). For direct optical measurement of neuronal electrical activity we genetically targeted expression of the “ArcLight” genetically encoded fluorescent voltage indicator [32, 33] using MB split-GAL4 drivers. We measured electrical activity in sleep-regulating MB neurons in fly brain explants acutely dissected from flies immediately following twelve hours of sleep deprivation during the night (or non-deprived flies collected at the same time of day), as acute dissection is much faster than preparation of the fly for tethered in vivo measurements, and previous studies establish that phase-dependent network state and spontaneous activity of circadian clock neurons is maintained in this acute explant preparation [32, 34–36].
These optical electrophysiology measurements revealed significantly greater membrane electrical activity of sleep-promoting MBON-γ2α' 1 dendrites in the brains of sleep-deprived flies than in those that are sleep replete, measured at the same time of day (Figure 5A,B,C,D). In contrast, electrical activity in wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral dendrites was decreased following sleep deprivation (Figure 5E,F,G,H). To determine whether these changes in MBON spontaneous electrical activity induced by sleep deprivation are intrinsic to the MBONs or, rather, are due to altered activity in the presynaptic KCs, we imaged electrical activity in the KCs themselves. Sleep deprivation increased the spontaneous activity of sleep-promoting γd KCs (Figure 5I,J,K,L) and decreased the spontaneous activity of wake-promoting α'/β' KCs (Figure 5M,N,O,P), but had no effect on wake-promoting γm KCs (Figure 5Q,R,S,T). The specificity of these opposite effects of sleep deprivation on the sleep-promoting and wake-promoting MBONs and KCs excludes general non-specific alteration of neural activity by sleep deprivation, and suggests that their relative activity levels could encode sleep need. These results establish for the first time that the MB sleep control microcircuits not only are engaged by exogenous activation, but also are modulated by sleep deprivation, with the encoding (although not necessarily the generation) of sleep need originating in KCs and propagating to MBONs.
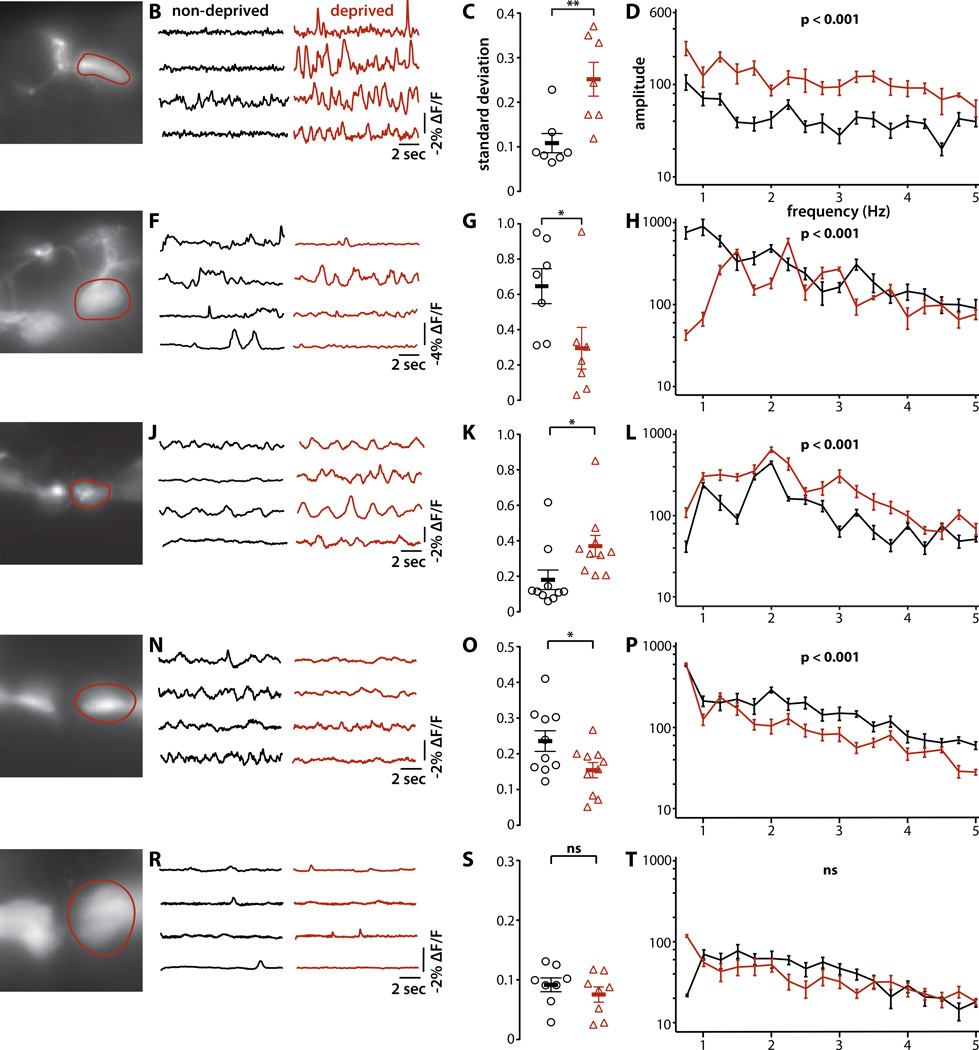
Figure 5. Sleep deprivation activates the sleep-promoting microcircuit and inhibits the wake-promoting microcircuit.
(A, E, I, M, Q) Representative images of ArcLight genetically encoded fluorescent voltage indicator expressed in the indicated sleep-regulating MBONs and KCs. Spontaneous membrane electrical activity was measured in the MBON dendrites or KC axons innervating the MB lobes (outlined in red). We employed the following split-GAL4 drivers, respectively, for the indicated cell types: MB077C, MB011C, MB607B, MB464B, MB108B.
(B, F, J, N, R) Optical recordings of electrical activity of the indicated cell types in brains explanted from four flies sleep-deprived for 16 hours (red) and four non-deprived flies (black), representative of recordings from at least seven flies for each cell type.
(C, G, K, O, S) Quantification of standard deviation of ArcLight optical signals (mean±sem) of sleep-deprived (red) and non-deprived (black) flies. Variation of MBON-γ2α'1 and γd KC optical signals was significantly greater in sleep-deprived than in non-deprived flies, while variation of MBON-γ5β′2a/β′2mp/β′2mp_bilateral and α'/β' KCs was significantly smaller in sleep-deprived than in non-deprived flies. Variation of γm KCs was unaffected by sleep deprivation. Statistical comparisons were by unpaired t-test; **, p<0.01; *, p<0.05; n ≥ 7 flies per cell type and condition).
(D, H, L, P, T) Power spectrum of MBON and KC optical signals computed using fast Fourier transform with 0.2Hz bin width. Powers at each frequency were averaged (±sem) across flies. Power of MBON-γ2α'1 and γd KC optical signals was significantly greater in sleep-deprived flies, while power of MBON-γ5β′2a/β′2mp/β′2mp_bilateral and α'/β' KCs was significantly smaller. Power of γm KCs was unaffected by sleep deprivation. Statistical comparisons were by two-way ANOVA with repeated measures; n ≥ 7 flies per genotype and condition).
These optical electrophysiology experiments are consistent with altered spontaneous neural activity in the sleep-regulating MB microcircuits encoding homeostatic sleep need in response to sleep deprivation. Further evidence in support of this hypothesis is provided by our observation that synaptic silencing of MBON-γ5β′2a/β′2mp/β′2mp_bilateral significantly suppressed the wake-promoting effect of the drug carbamazepine (CBZ) (Figure 6A,B,C). CBZ suppresses fly sleep at least in part by decreasing GABAergic tone [37], although CBZ may also have other unidentified targets. Under normal conditions, GABAergic pathways might regulate sleep in part by inhibiting the MBON-γ5β′2a/β′2mp/β′2mp_bilateral wake-promoting microcircuit; for example, it has recently been shown that dorsal paired medial neurons increase sleep by GABAergic inhibition of wake-promoting α'/β' KCs [7].
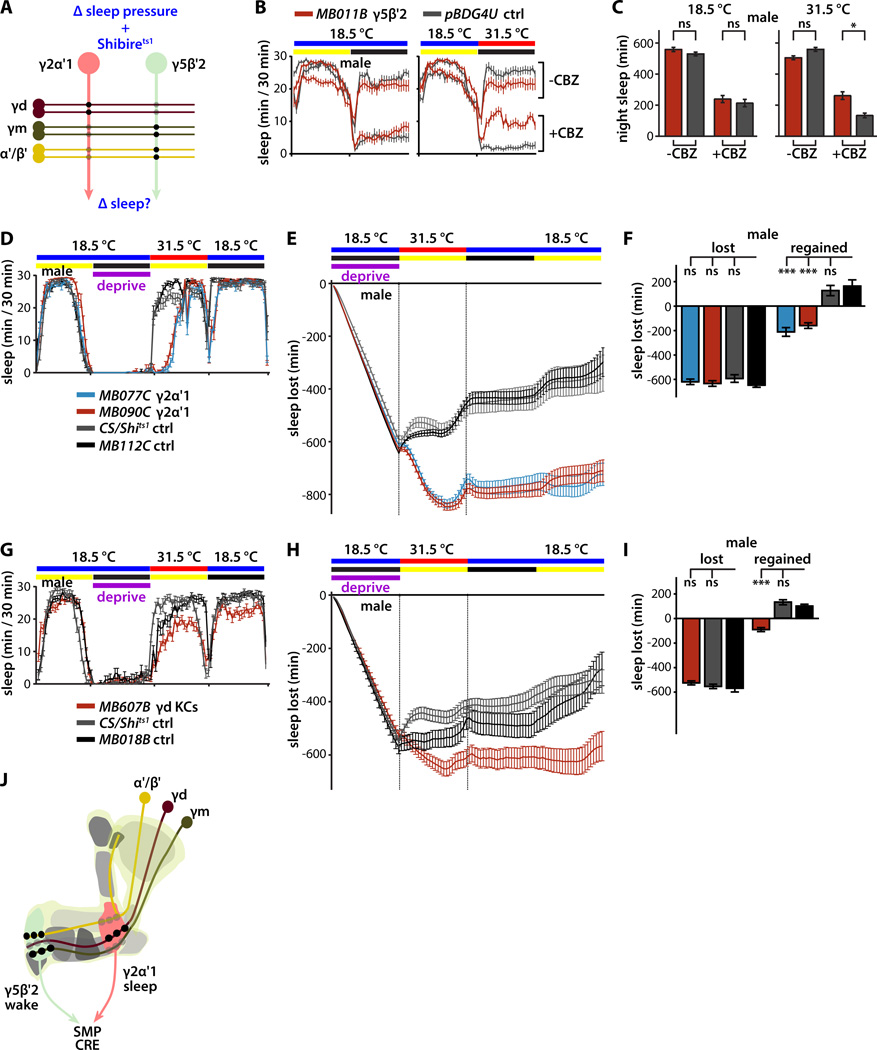
Figure 6. Requirement for propagation of homeostatic sleep signals through MB microcircuits.
(A) Schematic of experimental design to test hypothesis that MB microcircuits propagate essential homeostatic sleep signals. Effects of synaptic silencing of sleep-regulating MBONs on drug-induced waking or sleep deprivation-induced homeostatic rebound sleep were measured.
(B) Sleep profiles of male flies either on control food or food containing carbamazepine (CBZ; 0.1 mg/ml) wake-promoting drug. Synaptic silencing of Shibirets1-expressing MBON-γ5β′2a/β′2mp/β′2mp_bilateral by temperature shift to 31.5°C suppressed the wake-promoting effect of CBZ.
(C) Quantification of change in night sleep of flies in (B). Genotypes color coded as in (B); statistical analysis by ANOVA and Dunnett's paired-comparison test (n=46–55 flies per condition and genotype).
(D) Sleep profiles of male flies expressing Shibirets1 in sleep-promoting MBON-γ2α'1 deprived of sleep for one night at 18.5°C and then shifted to 31.5°C for the following day. Synaptic silencing of MBON-γ2α'1 using either of two split-GAL4 drivers potently suppressed homeostatic rebound sleep induced by sleep deprivation, in comparison to control flies expressing Shibirets1 in a GABAergic MBON that doesn't influence sleep (MB112C) or control flies bearing the UAS-Shits1 transgene but lacking any split-GAL4 driver.
(E) Cumulative sleep lost during sleep deprivation at 18.5°C and regained during subsequent recovery for 12 hours at 31.5°C and then 24 hours 18.5°C (mean±sem). Genotypes are color coded as in (D).
(F) Quantification of sleep lost during deprivation and regained during the 12 hours of recovery at 31.5°C. Homeostatic rebound was completely suppressed by Shibirets1-mediated synaptic silencing of sleep-promoting MBON-γ2α'1 using either the MB077C or MB090C split-GAL4 drivers. Genotypes are color coded as in (D); statistical analysis was by one-way ANOVA and Dunnett's paired-comparison test (n=25–32 flies per genotype).
(G) Sleep profiles of male flies expressing Shibirets1 in sleep-promoting γd KCs deprived of sleep for one night at 18.5°C and then shifted to 31.5°C for the following day. Synaptic silencing of γd KCs potently suppressed homeostatic rebound sleep induced by sleep deprivation, in comparison to control flies expressing Shibirets1 in a cholinergic MBON that doesn't influence sleep (MB018B) or control flies bearing the UAS-Shits1 transgene but lacking any split-GAL4 driver.
(H) Cumulative sleep lost during sleep deprivation at 18.5°C and regained during subsequent recovery for 12 hours at 31.5°C and then 24 hours 18.5°C (mean±sem). Genotypes are color coded as in (G).
(I) Quantification of sleep lost during deprivation and regained during the 12 hours of recovery at 31.5°C. Homeostatic rebound in male flies was completely suppressed by Shibirets1-mediated synaptic silencing of γd KCs. Genotypes are color coded as in (G); statistical analysis was by one-way ANOVA and Dunnett's paired-comparison test (n=26–30 flies per genotype).
(J) Schematic model of synaptic microcircuits of the Drosophila MB that control sleep. γm and α'/β' KC activity promotes wake by synaptic activation of MBON-γ5β′2a/β′2mp/β′2mp_bilateral, while γd KC activity promotes sleep by synaptic activation of MBON-γ2α'1. Sleep deprivation induces homeostatic rebound sleep by activating the γd KC -> MBON-γ2α'1 sleep-promoting microcircuit. MBON-γ5β′2a/β′2mp/β′2mp_bilateral and MBON-γ2α'1 microcircuits oppositely regulate sleep via convergent connections to target neurons in SMP and/or CRE.
We also examined whether synaptic silencing of sleep-promoting MBON-γ2α'1 suppresses homeostatic rebound sleep induced by sleep deprivation. Consistent with an essential role in physiological control of sleep, synaptic silencing of MBON-γ2α'1 mediated by Shibirets1 targeted using either of two independent split-GAL4 lines severely reduced the homeostatic rebound sleep that occurs during the day after a night of sleep deprivation in male flies (Figure 6D,E,F). This effect is specific to Shibirets1-mediated silencing and not a non-specific effect of elevated temperature, as control flies lacking Shibirets1 expression or expressing Shibirets1 in a GABAergic MBON that doesn't influence sleep regain a substantial fraction of the sleep lost (Figure 6D,E,F). Importantly, assessment of activity while awake rules out non-specific locomotor effects (Figure S5). Similar results were obtained with female flies (Figure S6A,B,C). Interestingly, dTRPA1-mediated activation of MBON-γ5β′2a/β′2mp/β′2mp_bilateral suppresses homeostatic rebound sleep (Figure S6D,E,F). To assess whether the necessary homeostatic sleep signal propagates to MBON-γ2α'1 from γd KCs—as is suggested by the physiological data of Figure 5—we silenced γd KCs after sleep deprivation, and found a similar suppression homeostatic rebound sleep (Figure 6G,H,I). In control experiments, we confirmed the absence of any effects of Shibirets1 on rebound sleep in the absence of a temperature shift (Figure S6G,H,I), and no or minimal effects of Shibirets1-mediated synaptic silencing on basal daytime sleep in the absence of mechanical sleep deprivation (Figure S6J,K). Interestingly, even after termination of Shibirets1-mediated silencing of γd KCs or MBON-γ2α'1, lost rebound sleep was not recovered (Figure 6E,H). This suggests the possibility that activation of the sleep-promoting microcircuit eventually discharges homeostatic sleep debt even when its synaptic outputs are silenced, which prevents actual recovery of lost sleep.
DISCUSSION
We have used a combination of sophisticated cell-specific genetic manipulations with behavioral sleep analysis and optical electrophysiology to provide an unprecedented level of detailed understanding of the propagation of homeostatic sleep signals through microcircuits of the Drosophila MB. Specifically, we have identified two parallel segregated compartment-specific microcircuits that regulate sleep: a wake-promoting microcircuit that originates in α'/β' and γm KCs and converges onto MBON-γ5β′2a/β′2mp/β′2mp_bilateral, and a sleep-promoting microcircuit that originates in γd KCs and converges onto MBON-γ2α'1 (Figure 1,2,3,4). Importantly, we have shown not only that exogenous activation of these microcircuits is sufficient to regulate sleep, but also that physiological manipulation of sleep need by sleep deprivation alters their endogenous neural activity (Figure 5), and propagation of these neural signals to downstream targets outside the MB is essential for the generation of homeostatic rebound sleep (Figure 6).
Previous studies using broadly expressed traditional GAL4 drivers have implicated the MB in the control of sleep [3, 4, 6, 8–12], but due to lack of appropriate cell-specific drivers, were unable to resolve specific sleep-regulating MB cell types, although very recent studies have specifically implicated α'/β' KCs and MB-MV1/PPL1 dopaminergic MB neurons in regulating sleep [7, 38]. Moreover, previous studies have not established a role for the MB in the generation and/or propagation of homeostatic sleep signals necessary for rebound following sleep deprivation. While a mutation of the amnesiac gene, which is expressed in a pair of neurons innervating the MB lobes, was shown to impair homeostatic sleep rebound [39], rebound was not found to be strongly affected by very broad synaptic inactivation of the MB KCs that comprise the lobes [4]. In this report, we have established a comprehensive catalog of the KCs and MBONs that control sleep, making use of a novel library of split-GAL4 lines targeting each of the cell types of the MB. Combining sophisticated behavioral genetic and optical electrophysiology approaches has allowed us to determine the roles of specific MB cell types in encoding homeostatic sleep signals under physiological conditions. We then identified specific synaptic microcircuits linking sleep-controlling KCs to sleep-controlling MBONs, revealing the synaptic mechanisms underlying the propagation of homeostatic sleep signals through the MB associative network.
Based on these results, we propose a detailed mechanistic model for homeostatic control of sleep by excitatory microcircuits in the Drosophila MB (Figure 6J). Wake-promoting MBON-γ5β′2a/β′2mp/β′2mp_bilateral and sleep-promoting MBON-γ2α'1 each receive anatomical inputs from both wake-promoting γm and α'/β' KCs and sleep-promoting γd KCs [16]. However, functional segregation of sleep control information into separate microcircuits is enforced by greater synaptic weights between γm and α'/β' KCs and MBON-γ5β′2a/β′2mp/β′2mp_bilateral, and between γd KCs and MBON-γ2α'1. Anatomical studies indicate that the axons of MBON-γ5β′2a/β′2mp/β′2mp_bilateral and MBON-γ2α'1 exit the MB and terminate convergently in the SMP and CRE neuropils [16, 40]. Intriguingly, dendrites of some CX neurons—a brain region involved in locomotor control [41] and implicated in sleep and sleep homeostasis [42–45]—arborize in SMP and CRE. We thus speculate that segregated homeostatic sleep-promoting and wake-promoting signals are generated in the KC-to-MBON microcircuits of the MB and propagate to the CX, where they are integrated to ultimately control sleep. Future studies are needed to further refine our understanding of the neurochemistry and physiology of the MB sleep control microcircuits, explore the mechanisms by which sleep deprivation alters microcircuit activity, and elucidate the connections between the MB and its downstream sleep control targets. In light of the relationship between sleep, learning, and synaptic homeostasis [see, e.g., 42, 46] and our recent discovery that sleep-regulating MBON-γ5β′2a/β′2mp/β′2mp_bilateral and MBON-γ2α'1 are also important for some forms of associative learning [19], it will be of great interest to determine how activity of the sleep-controlling MB synaptic microcircuits influences memory formation and consolidation.
EXPERIMENTAL PROCEDURES
Additional details concerning all experimental procedures are found in the Supplemental Experiment Procedures.
Molecular and genetic methods/fly stocks
LexA lines used in the experiments—R35B12-LexA, R14H06-LexA, and R44E04-LexA—are described in [23, 30]. Detailed genotype information of split-GAL4 strains—including p65ADZp-ZpGAL4DBD combinations—is as described [16]. p65ADZp split-half transgenes were inserted at attP40 or VK00027 while ZpGAL4DBD split-half transgenes were inserted at attP2. Split-GAL4 lines with the terminal letter "B", such as MB077B, contain p65ADZp in attP40 and ZpGAL4DBD in attP2, while "C" lines, such as MB112C, contain the p65ADZp in attVK00027 and ZpGAL4DBD in attP2, recombined on the third chromosome. We chose particular split-GAL4 lines as internal controls for particular experiments subsequent to the initial screen based on (1) showing no detectable sleep phenotype in the screen, (2) having the two split-half transgenes inserted in the same attP sites as the experimental groups in that experiment, and (3) being active in a different MB cell type than the experimental groups in that experiment, specifically a cell type in which no split-GAL4 lines active in that cell type induced sleep phenotypes.
Sleep assays
Total 24-hour sleep quantity (daytime plus nighttime sleep) was extracted from DAM system locomotor activity data as described [47]: sleep is defined as a contiguous period of inactivity lasting five minutes or more [13, 14]. For all screen hits, waking activity was calculated as the number of beam crossings/min when the fly was awake.
Carbamazepine feeding
CBZ was dissolved in 45% (2-hydroxypropyl)-beta-cyclodextrin (Sigma) as described in [37] to prepare a stock solution. For CBZ experiments, flies were loaded in tubes containing 5% agarose and 2% sucrose with 0.1mg/ml CBZ.
Sleep deprivation
Flies were sleep deprived by the intermittent mechanical perturbation method for 12 hours at night while housed in TriKinetics DAM monitors. Flies received mechanical perturbations on a horizontal shaker with a total duty cycle of 15 seconds per minute, delivered in 8 pulses of 1–3 seconds each occurring intermittently at random times. Sleep lost during deprivation and regained afterwards was calculated as described [48]. Sleep loss was calculated by subtracting the amount of sleep occurring during each thirty minute period of sleep deprivation from the amount of sleep occurring during the corresponding period of the previous unperturbed day:night cycle. Sleep rebound was calculated by subtracting the amount of sleep occurring during each thirty minute period following mechanical deprivation from the amount of sleep occurring during the corresponding period of the previous unperturbed day:night cycle.
Stimulation of KCs by ATP/P2X2 and simultaneous GCaMP6m imaging of MBONs
LexAop2-dsRed in attp18 (X); LexAop2-P2X2 in su(Hw)attp5(II); UAS-GCaMP6m in VK0005(III) flies were generated using standard molecular and genetic methods, with the original transgenes as described [18, 28, 29, 49]. These flies were crossed to flies bearing appropriate LexA and split-GAL4 driver transgenes.
ArcLight optical electrophysiology
ArcLight imaging of spontaneous neural activity in fly brain explants was performed as previously described [32]. Fly brains were acutely dissected from flies after twelve hours of sleep deprivation at night or from non-deprived flies at the same time of day and imaged within five minutes of dissection.
Immunohistochemistry
Dissection and immunohistochemistry of fly brains were performed as previously described with minor modifications [30].
Tap-response Arousal Assay
We probed the quiescence induced by activation of sleep promoting neuron by dTRPA1 by tapping vials housing flies as previously described [7]. Genotypes were hidden with tape, and recordings were thus carried out blind to which vial contained experimental and which contained control flies.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Heather Dionne, Ming Wu, Tomoko Ohyama, Austin Pavin, and Romain Franconville for many helpful discussions, as well as fly stocks. We thank Leslie Vosshall, Nelson Spruston, Tanya Wolff, and Marina Picciotto for comments on the manuscript. The Janelia Fly facility (Karen Hibbard, Todd Laverty and other members of the fly core) helped in fly husbandry, and the FlyLight Project Team performed brain dissections, histological preparations, and confocal imaging. We thank members of the Nitabach lab for helpful comments on experimental approaches and conceptual frameworks, specifically Davide Raccuglia, Christina Paquin, Karl Barber, and Michael Kunst. Work in the laboratory of M.N.N. at Yale University was supported in part by the National Institute of Neurological Disorders and Stroke, National Institutes of Health (NIH) (R01NS055035, R01NS056443, R01NS091070), the National Institute of General Medical Sciences, NIH (R01GM098931), and the Kavli Institute for Neuroscience. D.S. and M.N.N. performed portions of this work as participants in the Janelia Visiting Scientist Program. D.S. was also supported by start up funds from the College of Arts and Science, University of San Diego.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Tomer R, Denes AS, Tessmar-Raible K, Arendt D. Profiling by image registration reveals common origin of annelid mushroom bodies and vertebrate pallium. Cell. 2010;142:800–809. doi: 10.1016/j.cell.2010.07.043. [DOI] [PubMed] [Google Scholar]
- 2.Heisenberg M, Borst A, Wagner S, Byers D. Drosophila mushroom body mutants are deficient in olfactory learning. Journal of neurogenetics. 1985;2:1–30. doi: 10.3109/01677068509100140. [DOI] [PubMed] [Google Scholar]
- 3.Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441:757–760. doi: 10.1038/nature04811. [DOI] [PubMed] [Google Scholar]
- 4.Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441:753–756. doi: 10.1038/nature04739. [DOI] [PubMed] [Google Scholar]
- 5.Zars T, Fischer M, Schulz R, Heisenberg M. Localization of a short-term memory in Drosophila. Science. 2000;288:672–675. doi: 10.1126/science.288.5466.672. [DOI] [PubMed] [Google Scholar]
- 6.Guo F, Yi W, Zhou M, Guo A. Go signaling in mushroom bodies regulates sleep in Drosophila. Sleep. 2011;34:273–281. doi: 10.1093/sleep/34.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haynes PR, Christmann BL, Griffith LC. A single pair of neurons links sleep to memory consolidation in Drosophila melanogaster. eLife. 2015;4 doi: 10.7554/eLife.03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seugnet L, Suzuki Y, Merlin G, Gottschalk L, Duntley SP, Shaw PJ. Notch signaling modulates sleep homeostasis and learning after sleep deprivation in Drosophila. Current biology : CB. 2011;21:835–840. doi: 10.1016/j.cub.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueno T, Kume K. Functional characterization of dopamine transporter in vivo using Drosophila melanogaster behavioral assays. Front Behav Neurosci. 2014;8:303. doi: 10.3389/fnbeh.2014.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu M, Robinson JE, Joiner WJ. SLEEPLESS is a bifunctional regulator of excitability and cholinergic synaptic transmission. Current biology : CB. 2014;24:621–629. doi: 10.1016/j.cub.2014.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yi W, Zhang Y, Tian Y, Guo J, Li Y, Guo A. A subset of cholinergic mushroom body neurons requires Go signaling to regulate sleep in Drosophila. Sleep. 2013;36:1809–1821. doi: 10.5665/sleep.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan Q, Joiner WJ, Sehgal A. A sleep-promoting role for the Drosophila serotonin receptor 1A. Current biology : CB. 2006;16:1051–1062. doi: 10.1016/j.cub.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 13.Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI. Rest in Drosophila is a sleep-like state. Neuron. 2000;25:129–138. doi: 10.1016/s0896-6273(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 14.Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–1837. doi: 10.1126/science.287.5459.1834. [DOI] [PubMed] [Google Scholar]
- 15.Tanaka NK, Tanimoto H, Ito K. Neuronal assemblies of the Drosophila mushroom body. The Journal of comparative neurology. 2008;508:711–755. doi: 10.1002/cne.21692. [DOI] [PubMed] [Google Scholar]
- 16.Aso Y, Hattori D, Yu Y, Johnston RM, Iyer NA, Ngo TT, Dionne H, Abbott LF, Axel R, Tanimoto H, et al. The neuronal architecture of the mushroom body provides a logic for associative learning. eLife. 2014;3:e04577. doi: 10.7554/eLife.04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52:425–436. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aso Y, Sitaraman D, Ichinose T, Kaun KR, Vogt K, Belliart-Guerin G, Placais PY, Robie AA, Yamagata N, Schnaitmann C, et al. Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. eLife. 2014;3:e04580. doi: 10.7554/eLife.04580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, Garrity PA. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454:217–220. doi: 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfeiffenberger C, Lear BC, Keegan KP, Allada R. Processing sleep data created with the Drosophila Activity Monitoring (DAM) System. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5520. pdb prot5520. [DOI] [PubMed] [Google Scholar]
- 22.Pfeiffenberger C, Lear BC, Keegan KP, Allada R. Locomotor activity level monitoring using the Drosophila Activity Monitoring (DAM) System. Cold Spring Harb Protoc. 2010 doi: 10.1101/pdb.prot5518. pdb prot5518. [DOI] [PubMed] [Google Scholar]
- 23.Liu C, Placais PY, Yamagata N, Pfeiffer BD, Aso Y, Friedrich AB, Siwanowicz I, Rubin GM, Preat T, Tanimoto H. A subset of dopamine neurons signals reward for odour memory in Drosophila. Nature. 2012;488:512–516. doi: 10.1038/nature11304. [DOI] [PubMed] [Google Scholar]
- 24.Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nature neuroscience. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- 25.Kitamoto T. Conditional modification of behavior in Drosophila by targeted expression of a temperature-sensitive shibire allele in defined neurons. Journal of neurobiology. 2001;47:81–92. doi: 10.1002/neu.1018. [DOI] [PubMed] [Google Scholar]
- 26.Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJ, Kang K, Liu X, Garrity PA, Rosbash M, et al. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron. 2008;60:672–682. doi: 10.1016/j.neuron.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lima SQ, Miesenbock G. Remote control of behavior through genetically targeted photostimulation of neurons. Cell. 2005;121:141–152. doi: 10.1016/j.cell.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Yao Z, Macara AM, Lelito KR, Minosyan TY, Shafer OT. Analysis of functional neuronal connectivity in the Drosophila brain. Journal of neurophysiology. 2012;108:684–696. doi: 10.1152/jn.00110.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenett A, Rubin GM, Ngo TT, Shepherd D, Murphy C, Dionne H, Pfeiffer BD, Cavallaro A, Hall D, Jeter J, et al. A GAL4-driver line resource for Drosophila neurobiology. Cell reports. 2012;2:991–1001. doi: 10.1016/j.celrep.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bushey D, Tononi G, Cirelli C. Sleep- and wake-dependent changes in neuronal activity and reactivity demonstrated in fly neurons using in vivo calcium imaging. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:4785–4790. doi: 10.1073/pnas.1419603112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao G, Platisa J, Pieribone VA, Raccuglia D, Kunst M, Nitabach MN. Genetically targeted optical electrophysiology in intact neural circuits. Cell. 2013;154:904–913. doi: 10.1016/j.cell.2013.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin L, Han Z, Platisa J, Wooltorton JR, Cohen LB, Pieribone VA. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron. 2012;75:779–785. doi: 10.1016/j.neuron.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cao G, Nitabach MN. Circadian control of membrane excitability in Drosophila melanogaster lateral ventral clock neurons. J Neurosci. 2008;28:6493–6501. doi: 10.1523/JNEUROSCI.1503-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunst M, Hughes ME, Raccuglia D, Felix M, Li M, Barnett G, Duah J, Nitabach MN. Calcitonin gene-related peptide neurons mediate sleep-specific circadian output in Drosophila. Current biology : CB. 2014;24:2652–2664. doi: 10.1016/j.cub.2014.09.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheeba V, Gu H, Sharma VK, O'Dowd DK, Holmes TC. Circadian-and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. Journal of neurophysiology. 2008;99:976–988. doi: 10.1152/jn.00930.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agosto J, Choi JC, Parisky KM, Stilwell G, Rosbash M, Griffith LC. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nature neuroscience. 2008;11:354–359. doi: 10.1038/nn2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berry JA, Cervantes-Sandoval I, Chakraborty M, Davis RL. Sleep Facilitates Memory by Blocking Dopamine Neuron-Mediated Forgetting. Cell. 2015;161:1656–1667. doi: 10.1016/j.cell.2015.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu W, Guo F, Lu B, Guo A. amnesiac regulates sleep onset and maintenance in Drosophila melanogaster. Biochem Biophys Res Commun. 2008;372:798–803. doi: 10.1016/j.bbrc.2008.05.119. [DOI] [PubMed] [Google Scholar]
- 40.Ito K, Shinomiya K, Ito M, Armstrong JD, Boyan G, Hartenstein V, Harzsch S, Heisenberg M, Homberg U, Jenett A, et al. A systematic nomenclature for the insect brain. Neuron. 2014;81:755–765. doi: 10.1016/j.neuron.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 41.Strauss R, Heisenberg M. A higher control center of locomotor behavior in the Drosophila brain. J Neurosci. 1993;13:1852–1861. doi: 10.1523/JNEUROSCI.13-05-01852.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Donlea JM, Thimgan MS, Suzuki Y, Gottschalk L, Shaw PJ. Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science. 2011;332:1571–1576. doi: 10.1126/science.1202249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Q, Liu S, Kodama L, Driscoll MR, Wu MN. Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Current biology : CB. 2012;22:2114–2123. doi: 10.1016/j.cub.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Donlea JM, Pimentel D, Miesenbock G. Neuronal machinery of sleep homeostasis in Drosophila. Neuron. 2014;81:860–872. doi: 10.1016/j.neuron.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ueno T, Tomita J, Tanimoto H, Endo K, Ito K, Kume S, Kume K. Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nature neuroscience. 2012;15:1516–1523. doi: 10.1038/nn.3238. [DOI] [PubMed] [Google Scholar]
- 46.Bushey D, Tononi G, Cirelli C. Sleep and synaptic homeostasis: structural evidence in Drosophila. Science. 2011;332:1576–1581. doi: 10.1126/science.1202839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Donelson NC, Kim EZ, Slawson JB, Vecsey CG, Huber R, Griffith LC. High-resolution positional tracking for long-term analysis of Drosophila sleep and locomotion using the "tracker" program. PloS one. 2012;7:e37250. doi: 10.1371/journal.pone.0037250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417:287–291. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- 49.Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.