Abstract
Lung and airway epithelial cells generated in vitro from human pluripotent stem cells have applications in regenerative medicine, modeling of lung disease, drug screening and studies of human lung development. Here we describe a strategy for directed differentiation of human pluripotent stem cells into developmental lung progenitors, and their subsequent differentiation into predominantly distal lung epithelial cells. The protocol entails four stages that recapitulate lung development and takes approximately 50 days. First, definitive endoderm is induced in the presence of high concentrations of Activin A. Subsequently, lung-biased anterior foregut endoderm is specified by sequential inhibition of BMP, TGF-β and Wnt signaling. Anterior foregut endoderm is then ventralized by applying Wnt, BMP, FGF and RA signaling to obtain lung and airway progenitors. Finally, these are further differentiated into more mature epithelial cells types using Wnt, FGF, c-AMP and glucocorticoid agonism. This protocol is conducted in defined conditions, does not involve genetic manipulation of the cells, and results in cultures where the majority of the cells express markers of various lung and airway epithelial cells, with a predominance of cells identifiable as functional type II alveolar epithelial cells.
INTRODUCTION
This protocol describes an approach for the directed differentiation of human pluripotent stem cells (hPSCs), either embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs), into lung and airway epithelial cells. This protocol is based on our published work on the generation of anterior foregut endoderm (AFE), from which the lung is derived, and on the subsequent differentiation of AFE into lung and airway epithelial cells1,2.
Directed differentiation of hPSCs involves recapitulating in vivo development to specify desired organ fates in vitro through carefully dosed and timed activation and inhibition of specific signaling pathways3. The lung is derived from lung buds that arise on the anterior ventral aspect of the definitive endoderm (DE), and develop into lung and airways through a complex and coordinated process of branching morphogenesis and lineage specification4. Hence, directed differentiation of hPSCs into pulmonary epithelial cells begins with induction of DE, followed by AFE specification, patterning into a ventral anterior foregut fate, and finally specification of the various airway and lung epithelial cells. Importantly, directed differentiation does not involve introduction of genetic material into the genome.
Applications
This technology has applications in modeling lung diseases affecting the pulmonary epithelia and drug screening, and will provide novel insights into human lung development. For example, this approach can be used to examine the mechanisms and factors that drive lineage and morphogenetic decisions in terminal lung development5. This is important since, whereas the early stages of lung development are fairly well understood in the mouse model4, mechanisms underlying lineage determination and alveolar development are to a large extent unknown. Potential diseases that could be modeled include cystic fibrosis, tracheoesophageal atresia and fistula, surfactant deficiency syndromes, idiopathic pulmonary fibrosis and lung cancer6–9. This approach could also be used to screen for drugs that enhance the production of lung surfactant, failure of which is one the main causes of morbidity and mortality in prematurely born infants10. Ultimately, the ability to generate lung and airway epithelial cells from hPSCs may have applications in regenerative medicine for respiratory diseases.
Comparison with other methods
Directed differentiation of lung and airway tissue has lagged behind other organs. Although several papers have been published in this area11–18, no detailed protocols that would enable easy replication of the findings are available, however11. Early reports used spontaneous differentiation of mouse ESCs11 or drug selection in hESCs, which may lead to generation of cells that underwent undesirable epigenetic or genetic changes12. Using a mouse NKX2.1 reporter ESC line and cell sorting, combined with our published strategy to generate AFE1, Longmire et al. could achieve differentiation of lung progenitors13. Several papers were published using human cells14–18. Ghaedi et al14, employ a variety of differentiation strategies throughout that require serum supplementation14. Our protocol uses only defined conditions, while serum is only briefly applied to inactivate trypsin when cells are passaged. This protocol14 furthermore lacks addition of BMP4 during lung field induction, which we have shown to be indispensable2, a finding that is entirely consistent with studies in mouse lung specification4,19,20. Mou et al.15 only differentiated for 12 days, yielding a purity of early developmental lung progenitors in the single digits, and no evidence of further differentiation15. A third report16 uses air-liquid interphase cultures for terminal differentiation. Using a ventralization strategy similar to our published work2, Gotoh et al.17 generated lung progenitors and successfully identified a surface marker specific for Nkx2.1+ lung cells. These four reports14–16 generated AFE in a fashion similar to our original paper1. Wong et al.18 used air-liquid interphase cultures to generate ciliated proximal airway cells as well, but used a very different strategy to generate AFE. There were no data on efficiency and yield however18. The protocol described here provides a quantitative and efficient system for generating predominantly distal lung, and to a lesser extent, airway epithelial cells.
Experimental design
An overview of the protocol is given in Figure 1. The entire differentiation is conducted in defined serum-free conditions, although serum is used to inactivate trypsin at steps requiring trypsin passaging of the cells. Serum can be substituted with commercially available trypsin inhibitors. The first part involves step-wise, sequential induction of early lung progenitors (Steps 1–26). hPSCs are passaged on Matrigel-coated plates to deplete residual mouse embryonic fibroblasts (MEF) (Steps 1–6). Primitive streak (Steps 7–9) and subsequently DE (Steps 10–13) are induced using established protocols21,22. AFE is then specified based on our previous protocol1 with modifications that specifically improve the yield of NKX2.1+ developmental lung bud progenitors (Steps 14–17)2. More specifically, inhibition of BMP and TGF-β signaling for 24hr is followed by 24hr inhibition of TGF-β and Wnt signaling. Subsequent lung field induction is achieved by activating Wnt, FGF, BMP and retinoic acid signaling (Steps 18–26), pathways critical for lung field specification in the mouse19,23–25. The early lung progenitors generated by this protocol (between days 15 and 25) are capable of in vivo differentiation into the six types of lung and airway epithelial cells after transplantation under the kidney capsule of immunodeficient mice2. The second half of the protocol (Steps 27–30) describes the long-term differentiation of hPSCs-derived lung progenitors in vitro into predominantly distal cells. This step is performed in the presence of Wnt and FGF signaling. Adding factors known to induce alveolar maturation13,26 at this stage leads to a strong enrichment of functional ATII cells in the culture2.
Figure 1. Schematic illustration of the protocol for lung and airway progenitor cell generation.
The schematic illustrates for all sequential steps the media and tissue culture plates used, the timing, and the growth factors and small molecules required.
The cultures can be analyzed using several approaches, including transplantation under the kidney capsule of immune deficient mice, quantitative RT-PCR, immunofluorescence (IF) and uptake and release of fluorescently labeled SP-B. Procedures for this are standard and are described in our original paper2. The antibodies used are shown in Table 1. Appropriate controls are critical however. We use undifferentiated hPSCs as controls for transplantation under the kidney capsule of immune deficient NSG mice. These cells give rise to a teratoma containing cells from all three germ layers. In contrast, cells differentiated into lung progenitors only generate respiratory epithelium2. Controls for SP-B uptake are cells at d15 of differentiation, which do not express any markers of differentiated cells and do not take up SP-B. Positive controls for IF include human adult and fetal lung sections2. Negative controls for IF and qPCR are random differentiation of hPSCs induced by withdrawal of renewal factors, or hPSCs differentiated into alternative endodermal lineages using published strategies. We typically use differentiation into the hepatic lineage27 as a control for IF and qPCR. We believe that such ‘biological negative controls’, i.e., related cells that do not express the markers of interest, should be used as controls for IF. These are frequently more helpful in differentiating non-specific background signal from specific antibody signal, compared with Immunoglobulin (isotype) controls or secondary-only controls.
Table 1.
Antibodies for differentiation culture characterization.
| Antigen | Host | Clone No. | Supplier | Cat. no. | Dilution |
|---|---|---|---|---|---|
| EpCAM | Mouse, APC conjugated | EBA-1 | BD Biosciences | BDB347200 | 1:100 |
| CXCR4 | Mouse, PE conjugated | 12G5 | Invitrogen | MHCXCR404 | 1:200 |
| c-KIT | Mouse, APC conjugated | 104D2 | Invitrogen | CD11705 | 1:100 |
| Foxa2/HNF-3β | Goat | M-20 | Santa Cruz | sc-6554 | 1:50 |
| Nkx2.1/TTF-1 | Mouse | 8G7G3/1 | Invitrogen | 18-0221 | 1:100 |
| Nkx2.1/TTF-1 | Rabbit | Polyclonal | Seven Hills | WRAB-1231 | 1:1000 |
| P63 | Rabbit | H-129 | Santa Cruz | sc-8344 | 1:100 |
| Sox2 | Rabbit | Polyclonal | Stemgent | 09-0024 | 1:100 |
| Sox2 | Goat | Y-17 | Santa Cruz | sc-17320 | 1:100 |
| Mucin1 | Armenian Hamster | MH1 | NeoMarkers | HM-1630-P1ABX | 1:200 |
| Mucin5AC | Mouse (Biotin) | 45M1 | Abcam | ab79082 | 1:100 |
| Mucin5B | Rabbit | H-300 | Santa Cruz | sc-20119 | 1:100 |
| Mucin2 | Rabbit | H-300 | Santa Cruz | sc-15334 | 1:100 |
| Foxj1 | Mouse | 2A5 | e-bioscience | 14-9965-82 | 1:100 |
| CC-10 | Goat | C-20 | Santa Cruz | sc-9770 | 1:100 |
| pro-SP-C | Rabbit | Polyclonal | Seven Hills | WRAB-9337 | 1:2000 |
| SP-C | Rabbit | Polyclonal | Seven Hills | WRAB-76694 | 1:1000 |
| mature SP-B | Rabbit | Polyclonal | Seven Hills | WRAB-48604 | 1:1000 |
| ABCA3 | Rabbit | Polyclonal | Seven Hills | WRAB-70565 | 1:1000 |
| PDPN | Rabbit | FL-162 | Santa Cruz | sc-134482 | 1:100 |
| AQP5 | Goat | G-19 | Santa Cruz | sc-9890 | 1:100 |
| Cleaved caspase 3 | Rabbit | 5A1E | Cell signaling | Asp175 | 1:100 |
Limitations
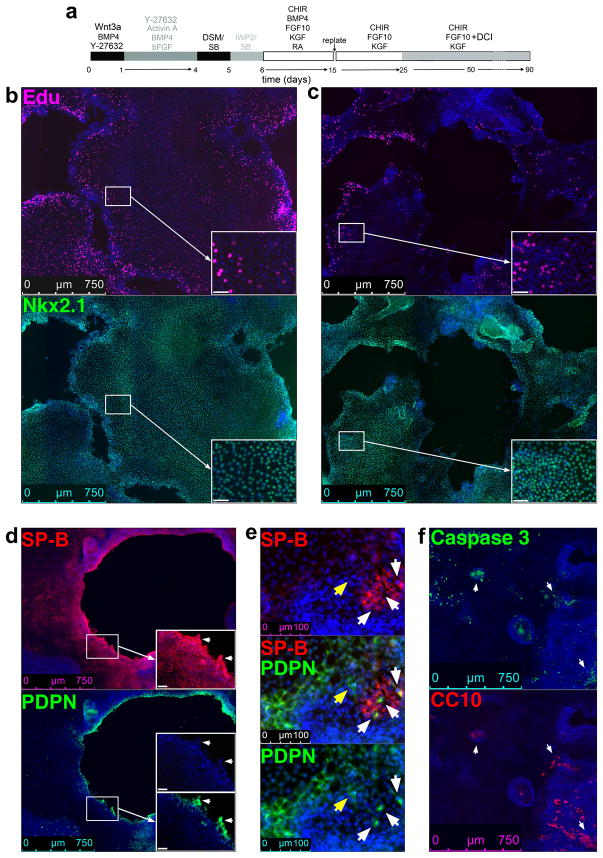
We have shown that after transplantation under the kidney capsule of immunodeficient NSG mice, the progenitors generated in this protocol can generate both proximal airway and distal lung epithelial cells. During further in vitro differentiation according to our protocol, differentiation is clearly biased towards distal lung, however. Furthermore, we believe that the cultures are still at a fetal stage of development. Evidence supporting this notion includes the fact that while cells expressing the basal cell marker, p63, are present, these do not express NFGR, a markers of adult basal cells28 that is absent on p63+ cells in fetal lung2. Furthermore, we show here (see Fig. 7, and ‘Anticipated Results’) that continuous proliferation occurs in areas of the cultures for months, and that cells co-expressing markers for ATI and ATII cells are present in these proliferative areas. Such cells are, based on recent studies in the mouse, considered bipotential alveolar progenitors arising during the saccular stage of alveolar development29. On the other hand, we find apoptosis in areas with cells expressing airway markers. These findings explain the progressive bias of this model towards distal lung.
Figure 7. Dynamics of ~d50 and 3-month cultures of RUES2 cells differentiated according to the protocol shown in panel (a).
(b) 10× tile scan images of expression of NKX2.1 (rabbit) (Table 1) and the uptake of EdU (See Enzymes and other reagents) in ~d50 RUES2 differentiation culture. Cell detachment generates empty spaces in the originally confluent culture, lined by EdU+ cells. (c) Idem as (b) after 3 months of culture. (d) 10× tile scan images of 3 months differentiation cultures stained for SP-B and PDPN (Table 1). Arrows show cells co-expressing SP-B and PDPN lining the empty spaces. (e) Enlarged view showing expression of SP-B and PDPN in 3-months cutlures. Yellow arrow, PDPN-expressing cell; white arrows, cells co-expressing SP-B and PDPN. (f) 10× tile scan images of the expression of CC10 and Cleaved caspase 3 (Table 1). Arrows indicate apoptosis in areas with cells expressing CC10. Apoptosis was not observed in areas with SP-B or PDPN expressing cells (not shown). Scale bar insets = 40 μm
Another limitation is that there is clear variability in the efficiency of this protocol across hES and hiPS lines, with purities of lung progenitors ranging from 70% to nearly 100%. This phenomenon is well established in the field30–32.
Finally, we are cognizant of the fact that for future clinical appliation, hPSCS lines that have not been in contact with any reagents derived from animals are required. This protocol may be the basis of future clinical protocols, however clinical application will require many GMP modifications, of which feeder-free culture of the hPSCs is only one. Long-term maintenance in the absence of feeders compromised DE induction for some hPSCs in our hands, and feeder-independent conditions that maintain hPSCs as competent for differentiation are not yet available. It is hoped that the recently identified methods to convert hESCs to ‘ground state’ or naïve ESCs33,34, similar to mouse ESCs, might solve this issue in the future.
MATERIALS
REAGENTS
Cells
-
hPSCs or hiPSCs. This protocol can be performed with any hPSC line. We use RUES2 (Rockefeller University Embryonic Stem Cell Line 2, NIH approval number NIHhESC-09-0013, Registration number 0013; passage 13–24) and iPSC lines generated from human dermal fibroblasts using Sendai virus (SVhiPS1) or modified mRNA transfection (mRNA hiPS) (Pluripotent Stem Cell Core Facility, Icahn School of Medicine at Mount Sinai, NY). We have also used Sendai Virus generated hiPSC line (SVhiPS2) independently established from a child with influenza.
Caution:
Research involving human stem cells must be conducted in accordance with relevant ethical guidelines and applicable institutional and funding agency regulations.
Irradiated mouse embryonic fibroblasts (MEFs) (Global Stem, Cat. No. GSC-6001G)
Growth media and supplements
1-thioglycerol (MTG, Sigma-Aldrich, cat. no. M6145)
-
2-Mercaptoethanol (Sigma-Aldrich, cat. no. M3148)
CAUTION:
2-Mercaptoethanol is a toxic and corrosive health hazard. Use personal protective equipment while handling and discard waste properly.
-
Activin A (R&D Systems, cat. no. 338-AC)
CRITICAL:
This reagent shows lot-to-lot variation, lot testing for successful DE differentiation is recommended. Alternative replacements need to be tested before use.
All-Trans-Retinoic Acid (ATRA) (Sigma Aldrich, cat. no. R2625)
-
B27 Supplement (Life Technologies, cat. no. 17504-044)
CRITICAL:
This reagent shows lot-to-lot variation, lot testing for successful DE differentiation is recommended.
BMP4 (R&D Systems, cat. no. 314-BP)
Bovine Albumin Fraction V Solution (7.5%) (BSA, Life Technologies, cat. no. 15260-037)
cAMP (Sigma Aldrich, cat. no. B5386)
CHIR99021 (Tocris, cat. no. 04-0004)
Dexamethasone (Sigma Aldrich, cat. no. D4902)
Dorsomorphin dihydrochloride (Tocris, cat. no. 3093)
Dulbecco’s Modified Eagle’s Medium/Ham’s F-12 50/50 Mix (DMEM/F-12) (Cellgro (Corning), cat. no. 10-092-cv)
EGF (R&D Systems, cat. no. 2028-EG-200)
FGF-2 (FGFB) (R&D Systems, cat. no. 233-FB)
FGF10 (R&D Systems, cat. no. 345-FG)
FGF7/KGF (R&D Systems, cat. no. 251-KG)
Glutamax (Life Technologies, cat. no. 35050-061)
Ham’s F-12 Medium (Cellgro (Corning), cat. no. 10-080-CV)
-
hESC-quality FBS (Atlantic Biologicals, cat. no. S10250H)
CRITICAL:
Test for compatibility with hESCs. hESC-quality FBS provides better hPSC maintenance than standard FBS.
3-Isobutyl-1-methylxanthine (IBMX) (Sigma Aldrich, cat. no. I5879)
Iscove’s Modified Dulbecco’s Medium (IMDM) (Life Technologies, cat. no. 12200-036)
IWP2 (Tocris, cat. no. 3533)
-
KnockOut Serum Replacement (KOSR; Life Technologies, cat. no. 10828-028)
CRITICAL:
This reagent shows lot-to-lot variation, lot testing is recommended.
L-Ascorbic Acid (Sigma Aldrich, cat. no. A4544)
MEM Non-Essential Amino Acids Solution (Life Technologies, cat. no. 11140-050)
-
N2 Supplement (Life Technologies, cat. no. 17502-048)
CRITICAL:
This reagent shows lot-to-lot variation, lot testing for successful DE differentiation is recommended.
NOGGIN (R&D Systems, cat. no. 6057)
Penicillin/Streptomycin (Cellgro (Corning), cat. no. 30-002-CI)
Primocin (InvivoGen, cat. no. ANT-PM-2)
SB-431542 (Tocris, cat. no. 1614)
WFI Cell Culture Grade Water (Cellgro (Corning), cat. no. MT-25-055-CM)
Wnt3A (R&D Systems, cat. no. 5036-WN)
Y-27632 dihydrochloride (ROCK inhibitor, Tocris, cat. no. 1245)
Enzymes and other reagents
0.05% Trypsin/0.53 mM EDTA (Fisher Scientific, cat. no. MT-25-051-CI)
10X PBS with Ca2+ and Mg2+ (Cellgro (Corning), cat. no. 46-013-CM)
Accutase/EDTA (Innovative Cell Technologies, cat. no. AT104-100ML)
Click-iT EdU Alexa Fluor 488 Imaging Kit (Life Technologies, cat. no. C10337)
-
DMSO (Sigma-Aldrich, cat. no. D2650-100ML)
CAUTION:
DMSO is toxic. Use personal protective equipment while handling.
Dnase I (VWR, cat. no. 80510-412)
DPBS with Ca2+ and Mg2+ (Cellgro (Corning), cat. no. MT21030CM)
DPBS without Ca2+ and Mg2+ (Cellgro (Corning), cat. no. MT21031CM)
-
Ethanol (Sigma-Aldrich, cat. no. E7023)
CAUTION:
Ethanol is toxic and flammable. Use personal protective equipment while handling.
Fibronectin (Sigma-Aldrich, cat. no. F0895)
Gelatin from bovine skin – Type B, powder (Sigma-Aldrich, cat. no. G9391)
Growth factor-reduced Matrigel (Corning Discovery Labware (BD Biosciences), cat. no. 354230)
-
Hydrochloric acid solution 1.0 M (Sigma-Aldrich, cat. no. 318949)
CAUTION:
Hydrochloric acid is toxic and corrosive. Use personal protective equipment to avoid contact with skin and eyes, collect and discard waste appropriately; do not mix with other waste.
MycoSensor qPCR Assay Kits (Agilent Technologies, cat. no. 302108)
-
Sodium hydroxide standard solution 1mol/L (Sigma-Aldrich, cat. no. 71463)
CAUTION:
Sodium hydroxide is corrosive. Use personal protective equipment to avoid contact with skin and eyes, collect and discard waste appropriately; do not mix with other waste.
EQUIPMENT
Balance (METTLER TOLEDO, model no. AB104-S/PH)
Class II A2 biological safety cabinet (Nuaire, model no. NU-425-400).
CO2 Incubator at 37°C with 95% humidity, 95% air and 5% CO2 (Thermo Scientific, model: Heracell 150i)
CO2 Incubator at 37°C with 95% humidity, 5% O2, 5% CO2, and 90% N2) (Thermo Scientific, model: Forma Series II 3110 Water-Jacketed).
Mini Microcentrifuge
Motorized Leica DMI 6000B fluorescence microscope coupled with Leica DFC365 FX and DFC450 digital cameras and operated by LAS AF 6.2 software (Leica Microsystems GmbH, Wetzlar, Germany)
Tabletop centrifuge (Eppendorf, Centrifuge 5810 R)
Water bath set at 37°C (Fisher Scientific, IsoTemp 210)
1.7 mL Posi-Click Microcentrifuge tubes (Denville Scientific, cat. no. C2170)
6-well Ultra-Low Attachment plate (Corning, cat. no. 3471)
6-well tissue culture-treated plate (Falcon, cat. no. 353046)
24-well tissue culture-treated plate (Falcon, cat. no. 353047)
48-well tissue culture-treated plate (Falcon, cat. no. 353078)
10 cm2 tissue culture-treated plates (Falcon, cat. no. 353003)
15 mL polypropylene conical centrifuge tube (Santa Cruz, cat. no. sc-200249)
50 mL polypropylene conical centrifuge tube (Santa Cruz, cat. no. sc-200252)
Cell passaging tool (Life Technologies, cat. no. 23181-010)
Cell Scraper (Sarstedt, cat. no. 83.1830)
CryoTube vials (Nunc, cat. no. 375418)
P1000 Barrier Pipette Tips (Denville Scientific, cat. no. P1126)
P200 Barrier Pipette Tips (Denville Scientific, cat. no. P1122)
P20 Barrier Pipette Tips (Denville Scientific, cat. no. 1121)
P10 Barrier Pipette Tips (Denville Scientific, cat. no. P1096-FR)
Serological Pipette – 2 mL, individually wrapped (Santa Cruz, cat. no. sc-200276)
Serological Pipette – 5 mL, individually wrapped (Santa Cruz, cat. no. sc-200279)
Serological Pipette – 10 mL, individually wrapped (Santa Cruz, cat. no. sc-200281)
Serological Pipette – 25 mL, individually wrapped (Santa Cruz, cat. no. sc-200283)
Serological Pipette – 50 mL, individually wrapped (Santa Cruz, cat. no. sc-213234)
Sterile Filter Systems, disposable (0.22 uM, PES) (Corning, cat. no. 431097)
Steriflip-GP Filter, 50ml, 0.22μm (EMD Millipore, cat. no. EW-29969-24)
REAGENT SETUP
Human Pluripotent Stem Cell Culture
-
Detailed guidelines for hPSC culture can be found elsewhere, such as ‘ES Cell International Pte Ltd: Methodology Manual Human Embryonic Stem Cell Culture 2005’ and ‘Havard Stem Cell Institute (HSCI) StemBook Protocols for pluripotent cells, URL: http://www.stembook.org/protocols/pluripotent-cells’. Culture hPSCs in a 95% air/5% CO2 tissue culture incubator. We use irradiated MEFs feeders, as we found that this culture system best maintains the endoderm potential of the cells. Plate feeders at a density of ~20,000–25,000 cells/cm2 into gelatin coated 6-well TC plates, and use ~24 hr after plating for hPSC passaging and thawing. Thaw hPSCs at a higher density than that used for passaging. We thaw RUES2 at a density of ~20,000–25,000 cells/cm2. It is optional to add Y-27632 to the hPSC medium. We thaw hiPSCs at a similar density of ~20,000–25,000 cells/cm2, but in the presence of 10mM Y-27632 for 24 hr. Passage hPSCs with Accutase/EDTA every 4 days onto the feeders using a stem cell passaging tool. We passage RUES2 at a density of ~7,000–9,000 cells/cm2 without Rho kinase inhibitor Y-27632, and most iPSCs at a density of ~10,000–12,000 cells/cm2 in the presence of 10mM Y-27632 for 24 hr, respectively. Change hPSC medium daily. For rapidly growing hPSC lines such as RUES2, which has a doubling time of ~9 hr, add fresh hPSC medium on the day of passaging hPSCs, and wait for at least two hours before digesting the cells with Accutase/EDTA. Right before passaging the cells, eliminate prematurely differentiated colonies in hPSC cultures carefully with a 10 or 20 μL pipette tip (there should be very few or none for well-maintained hPSC lines).
Critical: Proper hPSC maintenance is critical for subsequent DE induction. The feeder-to-hPSC ratio (not only the absolute density of feeders) is one of the key factors for proper hPSC maintenance and needs to be optimized for each hPSC line. Too high a ratio causes differentiation (morphological changes) and too low a ratio reduces the capacity of DE specification without noticeable morphological changes (see above for a range of ratios for tested lines). It is recommended to use low passage number (<3) feeder cells. If the DE induction efficiency gradually decreases over sequential passages for a specific cell line, it is worthwhile verifying the hPSC maintenance method.
-
FGF-2. Reconstitute FGF-2 in sterile PBS + 0.1% (wt/vol) final concentration of BSA to obtain a 20 μg/ml solution. Make 1ml aliquots and store them at 20 °C for up to 6 months.
CRITICAL:
For all the cytokines: thaw once. Store thawed aliquots at 4°C for up to a month.
Y-27632. Reconstitute ROCK inhibitor Y-27632 in DMSO to obtain a 10 mM solution. Make 100μL aliquots and store them at 20 °C for up to a year.
50mg/ml Ascorbic acid solution. Prepare a 50mg/ml Ascorbic acid solution by dissolving 500mg of L-Ascorbic acid powder in 10ml of water, then filter with 0.22μM Steriflip-GP filter. Make 100 μl aliquots and store them at 20 °C for up to 6 months. Store thawed aliquot at 4°C and use it within 24 hr.
Dnas i. Reconstitute Dnase I by dissolving 10MU (~167mg) lyophilized powder in 167ml of sterile water to obtain a 1mg/ml (10X) stock solution. Make 1ml aliquots and store them at −20 °C for up to a year. Further dilute 10X stock with sterile PBS to obtain a 1 μg/ml solution, Make 1000μL aliquots and store them at −20 °C for up to 1 year.
Activin A. Reconstitute Activin A in sterile PBS + 0.1% (wt/vol) final concentration of BSA to obtain a 100 μg/ml solution. Make 100μL aliquots and store them at 20 °C for up to 6 months.
Wnt3A. Reconstitute Wnt3A in sterile PBS + 0.1% (wt/vol) final concentration of BSA to obtain a 100 μg/ml solution. Make 100μL aliquots and store them at −20 °C for up to 6 months.
Dorsomorphin dihydrochloride. Reconstitute Dorsomorphin dihydrochloride in DMSO to obtain a 2 mM solution. Make 100μL aliquots and store them at −20 °C for up to a year.
NOGGIN. Reconstitute NOGGIN in sterile PBS + 0.1% (wt/vol) final concentration of BSA to obtain a 100 μg/ml solution. Make 100μL aliquots and store them at −20 °C for up to 6 months.
SB-431542. Reconstitute SB-431542 in DMSO to obtain a 10 mM solution. Make 100μL aliquots and store them at −20 °C for up to a year.
IWP2. Reconstitute IWP2 in DMSO to obtain a 1 mM solution. Make 100μL aliquots and store them at −20 °C for up to a year.
CHIR99021. Reconstitute CHIR99021 in DMSO to obtain a 3 mM solution. Make 100μL aliquots and store them at −20 °C for up to a year.
FGF10. Reconstitute FGF10 in sterile PBS + 0.1% (wt/vol) final concentration of BSA to obtain a 10 μg/ml solution. Make 300μL aliquots and store them at −20 °C for up to 6 months.
FGF7. Reconstitute FGF7 in sterile PBS + 0.1% (wt/vol) final concentration of BSA to obtain a 10 μg/ml solution. Make 300μL aliquots and store them at −20 °C for up to 6 months.
BMP4. Reconstitute BMP4 in sterile PBS + 4 mM final concentration of HCl + 0.1% (wt/vol) final concentration of BSA to obtain a 10 μg/ml solution. Make 200μL aliquots and store them at −20 °C for up to 6 months.
ATRA. Reconstitute All-Trans-Retinoic Acid (ATRA) in DMSO to obtain a 0.5 mM solution. Make 100μL aliquots and store them at −20 °C for up to 3 months. Avoid exposure to light. Store aliquots at 4°C for up to a week.
IBMX. Reconstitute IBMX in DMSO to obtain a 0.5 mM solution. Make 100μL aliquots and store them at −20 °C for up to 6 months.
cAMP. Reconstitute cAMP in sterile water + 0.1N final concentration of Sodium hydroxide to obtain a 0.1 M solution. Make 200μL aliquots and store them at −20 °C for up to 6 months. Avoid exposure to light.
Dexamethasone. Reconstitute Dexamethasone in absolute ethanol to obtain a 1mg/ml (50X) stock solution. Make 200μL aliquots and store them at −20 °C for up to 1 year. Further dilute 50X stock with sterile PBS to obtain a 20 μg/ml solution, Make 200μL aliquots and store them at −20 °C for up to 6 months.
Gelatin. Prepare a 0.1% Gelatin solution in PBS (wt/vol) by first adding 2 grams of lyophilized powder to 1 L of Cellgro H2O, and boiling the mixture for 5–10 minutes, then add 200mL of 10X PBS and additional 800mL of H2O, and cool at 4° C for 1 hour. Filter and aliquot. Store at 4 °C for 6 months.
Matrigel. Prepare ½ diluted Matrigel stocks by adding 1mL of cold DMEM/F12 to 1mL of Matrigel in 15ml conical tubes on ice, stored in −20 °C. Keep all materials coming in contact with the solution cold to avoid polymerization at ~room temperature (22–25 °C). Store thawed ½ Matrigel stocks in 4°C for up to a week. Make 3.3% (v/v) Matrigel solution in DMEM/F12 by diluting the ½ stock with 28mL of cold DMEM/F12, make it right before coating tissue culture-treated plates.
Fibronectin. Aliquot and store Fibronectin at 4 °C for up to 2 years. Make 0.2% or 0.33% (v/v) Fibronectin solution in sterile PBS right before coating 24- or 48- well TC plates.
-
hPSC maintenance media
Combine 400 ml of DMEM/F12, 100 ml of KOSR, 5 ml of GlutaMAX, 5 ml of MEM-NEAA, 3.5 μl of 2-mercaptoethanol and 1ml Primocin to make a total of ~500ml of medium. Filter using a 0.22μm pore size filter unit. This sterile medium can be stored at 2–8 °C for up to 1 month. Add FGF-2 to a final concentration of 20 ng/ml for hPSC culture before use.
MEF conditioned media (CM)
Prepare MEF CM by adding 1.2mL/well of hPSCs maintenance medium to a feeder plate without hPSCs. Ideally CM is collected every 24 hr from d2–5 feeder plates and stored in 4°C, used it within two weeks.
IMDM medium reconstitution
Reconstitute IMDM medium by adding 1 pack of IMDM powder (17.7g) and 3.024g of Sodium Bicarbonate to 1L of WFI Cell Culture Grade Water. Mix well and filter the medium using a 0.22μm pore size filter unit. This sterile medium can be stored at 2–8 °C for up to 1 month.
Medium for MEFs
Combine 425ml of reconstituted IMDM, 75ml of ESC-quality FBS, 20 μl of MTG, 5ml of Glutamax and 5ml of Penicillin/Streptomycin to make 500ml of medium for MEFs. This sterile medium can be stored at 2–8 °C for up to 1 month.
Wash medium
Combine 500ml of reconstituted IMDM, 25ml of ESC-quality FBS, 5ml of Glutamax and 5ml of Penicillin/Streptomycin to make ~500ml of wash medium. This sterile medium can be stored at 2–8 °C for up to 1 month.
Stop medium
Combine 100ml of reconstituted IMDM, 100ml of ESC-quality FBS, 2ml of Glutamax and 2ml of Penicillin/Streptomycin to make ~200ml of medium, this sterile medium can be stored at 2–8 °C for up to 1 month. Add the appropriate amount of Dnase I stock solution immediately before use only to the volume needed, to obtain complete stop medium with a final concentration of 30ng/ml of Dnase I.
Serum-Free Differentation (SFD) medium
For ~1 liter of SFD medium, combine 750ml of reconstituted IMDM, 250ml of F-12, 7.5ml of BSA, 10ml of Glutamax, 5ml of N2, 10ml of B27 and 10ml of Penicillin/Streptomycin. This sterile medium can be stored at 2–8 °C for up to 1 month. On day of use, make fresh SFD+ medium by adding L-Ascorbic acid and MTG to obtain a final concentration of 50μg/ml for L-Ascorbic acid and 0.04μl/ml for MTG, respectively. Make fresh d0–50 differentiation medium as follows:
d0 Medium (Primitive streak formation)
Add 1μl/mL of Rock inhibitor stock, 0.1μl/mL of Wnt3a stock and 0.3μl/mL of BMP4 stock to SFD+ medium to obtain a final concentration of 10 μM for Rock inhibitor, 10 ng/mL for Wnt3a and 3 ng/mL for BMP4, respectively.
d1–3 Medium (DE induction)
Add 1μl/mL of Rock inhibitor stock, 1μl/mL of Activin A stock, 0.05μl/mL of BMP4 stock and 0.25μl/mL of FGF-2 to SFD+ medium to obtain a final concentration of 10 μM for Rock inhibitor, 100ng/ml for Activin A, 0.5 ng/mL for BMP4 and 2.5 ng/mL for FGF-2, respectively.
d4/4.5/5 Medium (AFE induction)
Add 0.75–1μl/mL of Dorsomorphin dihydrochloride stock (as replace of 1μl/mL NOGGIN) and 1μl/mL of SB431542 stock to SFD+ medium to obtain a final concentration of 1.5–2 μM for Dorsomorphin dihydrochloride (equivalent to 100 ng/mL of NOGGIN) and 10 μM of SB431542, respectively.
d5/5.5/6 Medium (AFE induction)
Add 1μl/mL of IWP2 stock and 1μl/mL of SB431542 stock to SFD+ medium to obtain a final concentration of 1 μM for IWP2 and 10 μM of SB431542, respectively.
d6/6.5/7 – d15 Medium
Add 1μl/mL of CHIR99021 stock, 1μl/mL of KGF stock, 1μl/mL of FGF10 stock, 1μl/mL of BMP4 stock, 2μl/mL of EGF stock (optional) and 0.1μl/mL of Retinoic Acid stock to SFD+ medium to obtain a final concentration of 3 μM for CHIR99021, 10ng/ml for KGF, 10ng/ml for FGF10, 10ng/ml for BMP4, 20ng/ml for EGF (optional) and 50 nM for Retinoic Acid, respectively.
d16 – d25 Medium
Add 1μl/mL of CHIR99021 stock, 1μl/mL of KGF stock and 1μl/mL of FGF10 stock to SFD+ medium to obtain a final concentration of 3 μM for CHIR99021, 10ng/ml for KGF and 10ng/ml for FGF10, respectively.
d26 Medium
Add 1μl/mL of CHIR99021 stock, 1μl/mL of KGF stock, 1μl/mL of FGF10 stock, 2.5μl/mL of Dexamethasone stock, 1μl/mL of IBMX stock and 1μl/mL of cAMP stock to SFD+ medium to obtain a final concentration of 3 μM for CHIR99021, 10ng/ml for KGF, 10ng/ml for FGF10, 25 ng/ml for Dexamethasone, 0.1 mM for IBMX and 0.1 mM for cAMP, respectively.
EQUIPMENT SET UP
Fibronectin-coated plates
Coat 24- or 48- well TC plates with freshly made 0.2% or 0.33% (v/v) AcFibronectin solution. Use a volume sufficient to cover the bottom of each well, i.e., 300 μL well for 24-well plates and 150 μL well for 48-well plates. Coat at −4 °C overnight. Store coated plates at −4 °C, use within a week.
Matrigel-coated plates
Coat 10cm2 tissue culture-treated plates with freshly made 3.3% (v/v) Matrigel solution. Use a volume that is enough to cover the bottom of the plate, i.e., 5–6mL per plate. Coat at −4 °C overnight. Store coated plates at −4 °C, use within a week.
PROCEDURE
Matrigel Depletion of MEFs
-
1
Day −3: Thaw ½ diluted Matrigel aliquot overnight on ice.
-
2
Day −2. Prepare Matrigel-coated 10cm2 tissue culture-treated plates (see Reagent set up).
-
3
Day −1. Prewarm accutase to 37° C. Remove medium from hPSC culture and add accutase. Digest hPSC cultures in the incubator. Digest for 2 – 3 min for a near confluent plate. Time varies depending on the density of the hPSC culture. Aspirate Accutase and add wash medium.
-
4
Remove the cells gently with stem cell passaging tool, transfer to a 15 mL conical tube, and pellet at 300 g for 5 minutes at 4°C.
-
5
Remove supernatant with suction, and re-suspend hPSC pellet in 50% fresh hPSC medium + 50% MEF CM. Pay attention to remove the residual wash medium stuck on the wall of the conical tube to avoid residual FBS contamination of the hPSC medium.
-
6
Transfer hPSCs to Matrigel-coated plates after removing non-adhering Matrigel, and place in 95% air/5% CO2incubator. hPSC lines are typically passaged from >75% confluent 6-well plate (~6–9 million cells per plate) onto Matrigel-coated plates in a 1:2 or 1:3 ratio (i.e. 2 to 3 wells per Matrigel plate). The ideal condition to start a differentiation on the following day is to have a 75–90% confluent plate with expansion of distinct, flattened hES colonies (Figure 2a). Ideally, the cells are in the Matrigel-coated plate for less than 24 hr. However, some lines may require more than 24 hours to achieve this morphology. If so, reduce the initial plating ratio to 1:1 or 1:1.5.
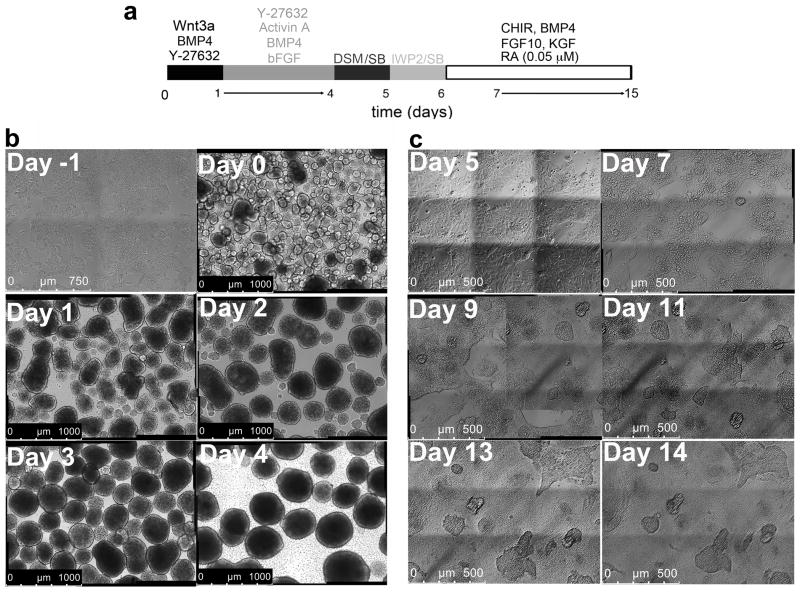
Figure 2. Culture morphology between days −1 and 15.
(a) Differentiation protocol of RUES2 cells shown in panels b–c. (b) 10× tile scan images (3×3) of RUES2 on a Matrigel-coated plate (day −1 of differentiation), showing flattened but distinct colonies, and EBs at days 0–4. (c) Representative bright field 20× images (tile scan, 3×3) at differentiation days 5, 7, 9, 11, 13 and 14. The image position was recorded by motorized Leica DMI 6000B microscope; pictures on the recorded position were taken every two days to demonstrate dynamics of cell proliferation and migration.
Primitive Streak Induction
-
7
On the following day (Day 0), prepare d0 medium (see Reagent Setup)
-
8
Dissociate the hES cells from the Matrigel-coated plates with warm 0.05% trypsin in incubator for 1–1.5 minutes. Tap the plate on the side with medium force, rotate and tap again, repeating this motion 2–3 times. The cells will visibly come off the plate rapidly. Add stop medium to halt the reaction. Cells should be in 3 to 10-cell clusters when visualized under microscope. Transfer to a conical tube. Add an excess of wash medium, and pellet at 300 g for 5 minutes at 4° C.
-
9
Aspirate the supernatant. Gently re-suspend the pellet in a small volume (~3ml) of d0 medium using a 5 mL serological pipette without generating bubbles. Add more d0 medium up to 12 mL per 10 cm2 Matrigel-coated plate, and place the cells in 6-well low-attachment plate(s) to form embryoid bodies (EBs)(2mL per well). Place in a 5% O2/5% CO2 incubator.
Definitive Endoderm Induction
-
10
On the following day (day 1), prepare DE induction medium (see Reagent Setup).
-
11
Collect the EBs in a conical tube, and add 1mL/well of DE medium to the empty low attachment plate(s) to avoid drying of the plate(s). Allow the EBs to settle for 15 minutes at room temperature. Alternatively, pellet at 130 g for 1 minute if the EBs are very small. Aspirate the supernatant, re-suspend the EBs in DE medium (12mL/plate), and place back in the same low-attachment plate(s), in a final volume of 3mL per well. Return to 5% O2/5% CO2 tissue culture incubator.
-
12
Two days later (day 3), prepare 0.2% fibronectin-coated plates (see Equipment Setup), to be used for AFE induction at day 4.5–5.5. 24- or 48-well plates are recommended.
Troubleshooting
-
13
On day 3, prepare DE medium. If cell density is relatively low (1~1.5 million cells per well), remove half of the original “DE medium” added on day 1, add more freshly prepared DE medium to embryoid bodies, 1.5mL per well of low attachment dish. For high-density plates, i.e., 2~3 million cells per well, completely remove DE medium added at day 1 and add fresh DE medium to the embryoid bodies, 3mL per well. In a plate with higher cell density, cells need to be fed on day 2 or day 2.5. Alternatively, split the EBs into one more plate. Return to 5% O2/5% CO2 tissue culture incubator for additional 1–2 days. Representative images of EBs at each day of differentiation into DE are shown in Figure 2b.
Critical step: Verify DE induction kinetics beforehand between days 3.5 and 5 for each hPSC line by flow cytometric analysis of CXCR4, c-KIT and EpCAM (Table 1). Primitive streak and subsequent DE induction from a properly maintained hPSC line should take 4–5 days (duration of Activin A treatment: 3–4 days). It is recommended to start AFE patterning within 6–9 hr after the DE yield maximizes. It is also recommended to work with hPSCs that can generate CXCR4+c-KIT+EpCAM+ cells with ≥80% purity. RUES2 consistently generate ~95% DE. For a given cell line maintained appropriately in defined conditions, the DE induction kinetics should remain stable throughout passages. Change of feeding frequency at Step 13 may affect the kinetics of DE induction however. Always double-check the DE yield at step 16.
Anterior Foregut Patterning of Definitive Endoderm
-
14
On day 4/4.5/5, prepare d4/4.5/5 medium (see Reagent Setup). The optimal concentration of Dorsomorphin dihydrochloride is cell line-dependent, while for some iPS lines, NOGGIN (100ng/ml) appears more efficient.
Troubleshooting
-
15
Collect EBs into a conical tube and add warm 0.05% trypsin (approximately 10mL per plate, total 2.5 – 4.0 min). Flick the tube gently every 15 sec for 1–1.5 min and then flick with medium force constantly for 5–10 sec, while closely watching the EBs. Approximately half should be dissociated as the supernatant becomes “cloudy”. Let the remaining EBs settle for 20 sec and very gently transfer most of the “cloudy” trypsin/supernatant into Stop medium, leave 1–2 ml trypsin, then flick the tube again with medium force for 5–10 sec. Once the rest of the EBs has dissociated completely, halt the reaction with the Stop medium. Add an excess of “Wash” medium, and pellet at 300 g for 5 minutes at 4°C.
Critical step: Dissociate no more than one plate of EBs at a time. Excessive trypsinization causes extensive cell death at step 17. When dissociating a small amount of EBs (~10% of a well) to verify DE induction kinetics, the digestion time should be 30s – 2 min.
-
16
Aspirate supernatant, and resuspend the pellet in d4/4.5/5 medium. Use ~20,000 cells to assess DE induction by flow cytometry for CXCR4, c-KIT and EpCAM (ideally a properly maintained hPSC line should produce 95% CXCR4+c-KIT+EpCAM+ cells). Plate the rest of the cells at a density of 5–7.5×104 cells/cm2 in 0.2% fibronectin-coated 48-well or 24-well plates (prepared at Step 12). Return to 5% O2/5% CO2 incubator. Hold the plate in one hand and gently rock the plate with a few left-to-right motions, place very gently in the incubator (without swilling) to assure even distribution of the cells.
-
17
One day later, prepare d5/5.5/6 medium. Aspirate to remove d4/4.5/5 medium and add d5/5.5/6 medium to the plate. Return to 5% O2/5% CO2 tissue culture incubator.
Lung progenitor induction and expansion
-
18
Prepare d6/6.5/7 medium (see Reagent Setup). It is useful to perform a RA concentration gradient to determine the optimal dosage (0.05–0.1μM for tested hPSCs), as verified by maximal FOXA2 and NKX2.1 expression at day 15.
-
19
Aspirate d5/5.5/6 medium and replace with d6/6.5/7 medium. It is useful to spare a few wells without switching to lung induction medium to assess SOX2 and FOXA2 expression, evidence of appropriate AFE induction, by IF, for example when optimizing the protocol for a new hPSC line.
-
20
Continue culturing for additional 8–10 days, replacing medium every 2nd day. Move the cells to a normoxic tissue culture incubator at a time point between day 8 – 12.
-
21
Use at least 6 wells for RNA extraction and IF staining. Proceed with remaining wells to the next step. Representative bright field images of a selected culture area during differentiation day 5 to 15 are shown in Figure 2c.
-
22
On day 15, coat tissue culture-treated plates with 0.33% fibronectin at 4°C overnight. 24- or 48- well plates are recommended.
-
23
Prepare d16 medium (see Reagent Setup).
-
24
Replating: Collect and save the d15 medium from the culture plate (do a few wells at a time to avoid drying of the wells). Briefly digest with warm (0.05%) trypsin for 1 minute. Aspirate trypsin and add the collected day 15 medium back to the wells. Mechanically detach the cells with 1000μL barrier tips and transfer to a microcentrifuge tube (one to two wells at a time) or a conical tube (multiple wells at a time). Pipette up and down with medium force for a few times, and let settle for a few minutes. Aspirate gently to remove the “cloudy” supernatant. Resuspend the clumps in d16 medium. Re-plate into a new 0.33 % fibronectin-coated plate at 1:3 – 1:5 ratio (depending on the cell line, initial confluence of the culture, and whether the endpoint is d25 or d50). Return to 5% O2/5% CO2 tissue culture incubator.
Critical step: This step is critical for multiple reasons. First, in some lines, lung progenitor induction is not complete until ~ d25. In these cases, re-plating is essential for post-d15 lung specification. Second, this step is critical for removing non-endodermal cells in the culture. Typically these cells are either neurectodermal cells, or FOXA2−P63+NKX2.1−SOX2−EpCAM+PAX6−PAX8− cells of which the identity is unclear2. Trypsin preferentially digests these into single cells or small cell clusters that stay in the supernatant and are removed by aspiration. Third, dissociation into single cells compromises viability compared to clump passaging. Viability can be improved by plating at confluent or over-confluent density, suggesting that the lung progenitors require close cell contact for survival. Furthermore, single cell replating promotes the outgrowth of Tuj1+ neuronal cells.
Pause point: the clumps harvested at this point can be frozen in DMEM+10% DMSO+50% FBS, and thawed and plated onto 0.33% fibronectin coated tissue culture plates at later time points.
-
25
Culture for 10 additional days, changing medium every 2nd day (daily medium change might be necessary for confluent cultures). In general, it takes 4–6 days for the clumps to completely attach. In the first 4 days after re-plating, it might be necessary to manually remove the medium with pipet tips (instead of using vacuum suction) while changing medium to avoid detaching the clumps. Collect the remaining suspension clumps (should be few), re-plate in a new 0.33 % fibronectin-coated plate or redistribute back to the original plate.
-
26
On day 25, set wells aside for RNA extraction and IF staining. If non-endodermal monolayers grow back at this stage, repeat the “re-plating” technique (Step 25) and proceed to Step 27.
Troubleshooting
Lung/airway epithelial maturation
-
27
Prepare d26 medium (see Reagent setup).
-
28
Aspirate medium, and replace with d26 medium.
-
29
Continue culturing for additional 25 days to up to ~1 year, change medium every 2nd day.
-
30
Terminate the culture for RNA extraction and IF staining.
TIMING
Steps 1–5, 3 days (preparation of Matrigel plates and hPSCs cells: 30min – 1hr active time)
Step 6, time of hPSCs cells on Matrigel: 12 – 24 h
Steps 7–9, Primitive Streak Induction: 18 – 24 h
Steps 10–13, DE Induction: 3 – 4 d
Steps 14–17, Anterior Foregut Patterning of Definitive Endoderm: 48 h
Steps 18–26, Lung progenitor induction and expansion: 18 – 20 d
Steps 27–30, Lung/airway epithelial maturation: 20 – 25 d. The optimal timing of addition of maturation components is around day 25 – 35 of this protocol. This is the time after maximum lung progenitor specification is achieved. Addition of maturation medium at day 35 did not significantly affect the efficiency of lineage maturation at day 48. Cells at this differentiation stage can be maintained up to a year in the presence of maturation components.
TROUBLESHOOTING
Troubleshooting advice can be found in Table 2.
Table 2.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 14 | Low DE yield | Cultures were examined too early in differentiation | Check later time points with 6- hour intervals |
|
| |||
| Maintenance in pluripotent state was not optimized for specific hPSC line. | Check pluripotency by staining for pluripotency markers | ||
| Take an early passage and improve the maintenance (See Reagent Setup – Human Pluripotent Stem Cell Culture) | |||
|
| |||
| A suboptimal lot of Activin A, N2 or B27 | Systematic lot testing highly recommended | ||
|
| |||
| Insufficient feeding at day 2 or 3 of differentiation | Replace a larger fraction of the medium | ||
|
| |||
| Suboptimal CXCR4+c- KIT+EpCAM+ expression, but good (>80%) expression of FOXA2 | Repeated forceful pipetting with 1000μL pipette tips while cells are still in trypsin appears to destroy CXCR4 epitope (Supplementary Figure 1) | ||
|
| |||
| Mycoplasma contamination | Verify Mycoplasma in hPSCs every 6 months | ||
|
| |||
| Abnormal karyotype | Verify karyotype of hPSCs every 6 months | ||
|
| |||
| Insufficient depletion of MEFs prior to DE induction | See Step 3, start differentiation from a high-density hPSC culture (nearly confluent). | ||
|
| |||
| 21 | Poor DE expansion at day 15: the culture appears as small clumps of FOXA2+ cells sitting in between flat FOXA2−P63+ cells | Occurs in some hiPSCs | Use 100ng/ml of NOGGIN instead of 2 μM Dorsomorphin dihydrochloride (Step 14) |
|
| |||
| 26 | Good DE yield but low lung induction efficiency between d15 and 25 (Figure 3a,b,e,f) | Duration of Activin A treatment was too long and addition of AFE induction medium occurred too late. | Start AFE patterning within 6–9 hr after the DE yield maximizes. |
|
| |||
| Decay of ATRA, which is light-sensitive and easily oxidized | Use fresh ATRA solution – Measure the optical density of the ATRA solution periodically using a spectrophotometer at OD350nm to verify if there is a decay of the solution. | ||
|
| |||
| Overgrowth of contaminating neuronal or other non-endodermal lineages in post d25 | Differentiation started from an endoderm with low purity (i.e., <85%) | Improve DE yield (see above) | |
|
| |||
| Poor attachment after d15 re-plating may favor non-endodermal lineage overgrowth | Use freshly prepared fibronectin plate Increase fibronectin concentration to 0.5–1% if necessary Avoid over trypsinization at Step 24 |
||
|
| |||
| Insufficient feeding | Increase the frequency of medium replacement | ||
|
| |||
| Replating of single cells rather than large clumps at day15 of differentiation | Pay attention to Critical Step 24 | ||
ANTICIPATED RESULTS
Variability among hPSC differentiation potential is well documented. Cell line-dependent modifications include timing of DE dissociation (Figure 3), concentration of DSM/NOGGIN, and of RA (Ref 2, Supplementary Figure 1)2. If all the steps are performed according to the procedures described above, with “Critical Steps” taken into account, the purity of FOXA2+ SOX2+NKX2.1+PAX8− lung progenitors at ~ day 15 of differentiation (Step 21) is expected to be ~80% or more for RUES2 (Figure 4a), and ~ 30–60% or higher for most hiPSCs2 (Figure 3d,h). For multiple hiPSC lines however, FOXA2+SOX2+NKX2.1+PAX8− lung progenitor induction may not be complete at this stage (Step 21) and reaches ~70–90% by ~day 25 (Step 26) (Figure 3e,i). For lines such as RUES2, the induction is near 100% by ~day 25 (Figure 4b).
Figure 3. Variability in hPSC differentiation potential associated with DE dissociation timing.
Representative 10× tile scan images (2×2) of the expression of FOXA2 (goat)/NKX2.1 (rabbit) (Table 1) in two hiPSC lines (SVhiPS1 and SVhiPS2) cultured according to the protocol shown in panel (a) at the top of the figure. (b,c) d15 (b) and d25 (c) differentiation of SVhiPS1 cells from d5 DE. (d,e) d15 (d) and d25 (e) differentiation of SVhiPS1 cells from d4 DE, showing improved generation of NKX2.1+ cells. (f,g) d15 (f) and d25 (g) differentiation of SVhiPS2 cells from d5.5 DE. (h,i) d15 (h) and d25 (i) differentiation of SVhiPS1 cells from d4.5 DE, again showing improved generation of NKX2.1+ cells. Scale bar insets = 40 μm
Figure 4. Expression of FOXA2 NKX2.1 SOX2in RUES2 cells at day 15 and 25 of differentiation.
(a) Day 15 of differentiation examined by IF for FOXA2 (goat), NKX2.1 (mouse) and SOX2 (rabbit) (Table 1) in RUES2 cells cultured according to the protocol shown at the bottom of the panels. Left panels, 10× tile scan images (2×2). Right panels, enlarged view of the select fields from the left panels. (b) Idem as panel (a) for cultures at d25.
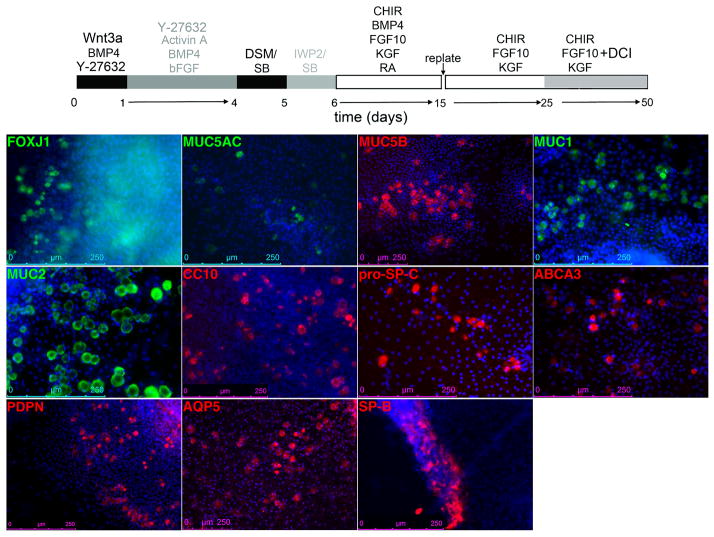
At d25, FOXA2+NKX2.1+P63+ cells, a phenotype of putative basal cell precursors, are typically seen at the periphery of the clumps (Supplementary Figure 2). Specification of other lung/airway epithelial lineages as determined by expression of markers or differentiated epithelial cells by IF begins at d25 with the appearance of sporadic Mucin5AC+ cells (ref 2, Figure 4d)2. Most markers of differentiated cells appear at ~ day 352. The abundance of each of these lineages is ~ 1–4% at day 48 (Step 30) (Figure 5 and ref 2, Figure 5b-c)2. At this stage, the majority of the cells are FOXA2+NKX2.1+ (Figure 6a-c) with a majority expressing SP-B (Figure 6d-f), and capable of SP-B uptake and release (ref 2, Supplementary Figure 11)2. Cells expand ~35–50 fold over the course of 50 days of differentiation (Original paper, Figure 5d)2. The cell types generated by this protocol most closely recapitulate the characteristics of cultured fetal human lung cells (ref 2, Supplementary Figure 10)2.
Figure 5. Differentiation of hPSCS-derived lung and airway progenitors at ~d50.
Representative example of the expression of markers Foxj1, Mucin1, Mucin2, Mucin5AC, Mucin5B, CC-10, pro-SP-C, ABCA3, PDPN, AQP5, mature SP-B (Table 1) of mature lung and airway epithelial cells after culturing SVhiPS1 cells according to the protocol shown at the top of the panel.
Figure 6. Expression of NKX2.1, FOXA2and SP-B at ~d50 in RUES2, SVhiPS1 and SVhiPS2 cells.
(a–c) 10× tile scan images (3×3) of NKX2.1 (rabbit) and FOXA2 (goat) (Table 1) expression in RUES2 (a), sviPS1 (b) and sviPS2 (c) differentiated according to the protocol shown at the top of Figure 6. (d–f) 20× tile scan images (6×6) of SP-B (Table 1) expression RUES2 (a), sviPS1 (b) and sviPS2 (c) differentiated according to the protocol shown at the top of Figure 6. Scale bar insets = 40 μm
Cells at this differentiation stage can be maintained up to a year. Interestingly, after day 50 and for up to 3 months of further culture, a specific regional organization arises, with areas of proliferation surrounding areas where cells detach (Figure 7b,c). Cells in these areas express the ATII marker, SP-B or co-express SP-B and the ATI marker, podoplanin (PDPN) (Table 1), suggestive of bipotential alveolar progenitors29, or of ATII cells transitioning to an ATI fate (Figure 7d,e). On the other hand, apoptosis occurs in areas with cells expressing airway markers, such as CC10 (Table 1) (Figure 7f). Thus, the dynamics of the culture likely explain the predominance of cells expressing the ATII marker, SP-B, a population that includes a majority of SP-B+PDPN− cells, and minority of SP-B+PDPN+ cells, a phenotype that was recently proposed to represent bipotential development alveolar progenitors29.
Supplementary Material
Acknowledgments
This works was supported by grant 1R01HL120046 to HWS.
Footnotes
Author contributions statement
S.X.L.H. developed the lung and airway differentiation protocol and co-wrote the manuscript; M.D.G. developed the AFE generation protocol; A.T.C. M.M., Y.-W.C., contributed to the development of the protocol; S.L.D. provided cells used in differentiation assays; H.-W.S. developed the concept, contributed to protocol development and co-wrote the manuscript with S.X.L.H.
Competing financial interests
The authors have filed patent applications PCT/US11/33751 and IRCU13340.
References
- 1.Green MD, et al. Generation of anterior foregut endoderm from human embryonic and induced pluripotent stem cells. Nature biotechnology. 2011;29:267–272. doi: 10.1038/nbt.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang SX, et al. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nature biotechnology. 2014;32:84–91. doi: 10.1038/nbt.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. S0092-8674(08)00216-X [pii] [DOI] [PubMed] [Google Scholar]
- 4.Morrisey EE, Hogan BL. Preparing for the first breath: genetic and cellular mechanisms in lung development. Developmental cell. 2010;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green MD, Huang SX, Snoeck HW. Stem cells of the respiratory system: from identification to differentiation into functional epithelium. BioEssays : news and reviews in molecular, cellular and developmental biology. 2013;35:261–270. doi: 10.1002/bies.201200090. [DOI] [PubMed] [Google Scholar]
- 6.Whitsett JA, Wert SE, Weaver TE. Alveolar surfactant homeostasis and the pathogenesis of pulmonary disease. Annual review of medicine. 2010;61:105–119. doi: 10.1146/annurev.med.60.041807.123500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitsett JA, Wert SE, Trapnell BC. Genetic disorders influencing lung formation and function at birth. Human molecular genetics. 2004;13(Spec No 2):R207–215. doi: 10.1093/hmg/ddh252. [DOI] [PubMed] [Google Scholar]
- 8.Xu X, et al. Evidence for type II cells as cells of origin of K-Ras-induced distal lung adenocarcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:4910–4915. doi: 10.1073/pnas.1112499109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ware LB, Matthay MA. The acute respiratory distress syndrome. The New England journal of medicine. 2000;342:1334–1349. doi: 10.1056/NEJM200005043421806. [DOI] [PubMed] [Google Scholar]
- 10.Nkadi PO, Merritt TA, Pillers DA. An overview of pulmonary surfactant in the neonate: genetics, metabolism, and the role of surfactant in health and disease. Molecular genetics and metabolism. 2009;97:95–101. doi: 10.1016/j.ymgme.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winkler ME, et al. Serum-free differentiation of murine embryonic stem cells into alveolar type II epithelial cells. Cloning Stem Cells. 2008;10:49–64. doi: 10.1089/clo.2007.0075. [DOI] [PubMed] [Google Scholar]
- 12.Wang D, Haviland DL, Burns AR, Zsigmond E, Wetsel RA. A pure population of lung alveolar epithelial type II cells derived from human embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4449–4454. doi: 10.1073/pnas.0700052104. 0700052104 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Longmire TA, et al. Efficient derivation of purified lung and thyroid progenitors from embryonic stem cells. Cell stem cell. 2012;10:398–411. doi: 10.1016/j.stem.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghaedi M, et al. Human iPS cell-derived alveolar epithelium repopulates lung extracellular matrix. The Journal of clinical investigation. 2013;123:4950–4962. doi: 10.1172/JCI68793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mou H, et al. Generation of Multipotent Lung and Airway Progenitors from Mouse ESCs and Patient-Specific Cystic Fibrosis iPSCs. Cell stem cell. 2012;10:385–397. doi: 10.1016/j.stem.2012.01.018. S1934-5909(12)00056-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Firth AL, et al. Generation of multiciliated cells in functional airway epithelia from human induced pluripotent stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E1723–1730. doi: 10.1073/pnas.1403470111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gotoh S, et al. Generation of alveolar epithelial spheroids via isolated progenitor cells from human pluripotent stem cells. Stem cell reports. 2014;3:394–403. doi: 10.1016/j.stemcr.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong AP, et al. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTRTR protein. Nature biotechnology. 2012;30:876–882. doi: 10.1038/nbt.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domyan ET, et al. Signaling through BMP receptors promotes respiratory identity in the foregut via repression of Sox2. Development. 2011;138:971–981. doi: 10.1242/dev.053694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Gordon J, Manley NR, Litingtung Y, Chiang C. Bmp4 is required for tracheal formation: a novel mouse model for tracheal agenesis. Developmental biology. 2008;322:145–155. doi: 10.1016/j.ydbio.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubo A, et al. Development of definitive endoderm from embryonic stem cells in culture. Development. 2004;131:1651–1662. doi: 10.1242/dev.01044. [DOI] [PubMed] [Google Scholar]
- 22.Nostro MC, et al. Stage-specific signaling through TGFbeta family members and WNT regulates patterning and pancreatic specification of human pluripotent stem cells. Development. 2011;138:861–871. doi: 10.1242/dev.055236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goss AM, et al. Wnt2/2b and beta-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Developmental cell. 2009;17:290–298. doi: 10.1016/j.devcel.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bellusci S, Henderson R, Winnier G, Oikawa T, Hogan BL. Evidence from normal expression and targeted misexpression that bone morphogenetic protein (Bmp-4) plays a role in mouse embryonic lung morphogenesis. Development. 1996;122:1693–1702. doi: 10.1242/dev.122.6.1693. [DOI] [PubMed] [Google Scholar]
- 25.Chen F, et al. A retinoic acid-dependent network in the foregut controls formation of the mouse lung primordium. The Journal of clinical investigation. 2010;120:2040–2048. doi: 10.1172/JCI40253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzales LW, Guttentag SH, Wade KC, Postle AD, Ballard PL. Differentiation of human pulmonary type II cells in vitro by glucocorticoid plus cAMP. American journal of physiology. Lung cellular and molecular physiology. 2002;283:L940–951. doi: 10.1152/ajplung.00127.2002. [DOI] [PubMed] [Google Scholar]
- 27.Gouon-Evans V, et al. BMP-4 is required for hepatic specification of mouse embryonic stem cell-derived definitive endoderm. Nature biotechnology. 2006;24:1402–1411. doi: 10.1038/nbt1258. [DOI] [PubMed] [Google Scholar]
- 28.Rock JR, et al. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Desai TJ, Brownfield DG, Krasnow MA. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 2014;507:190–194. doi: 10.1038/nature12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osafune K, et al. Marked differences in differentiation propensity among human embryonic stem cell lines. Nature biotechnology. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 31.Boulting GL, et al. A functionally characterized test set of human induced pluripotent stem cells. Nature biotechnology. 2011;29:279–286. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bock C, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gafni O, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- 34.Takashima Y, et al. Resetting Transcription Factor Control Circuitry toward Ground-State Pluripotency in Human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.