Abstract
As imaging mass spectrometry (IMS) has grown in popularity in recent years, the applications of this technique have become increasingly diverse. Currently there is a need for sophisticated data processing strategies that maximize the information gained from large IMS data sets. Traditional two-dimensional heat maps of single ions generated in IMS experiments lack analytical detail, yet manual analysis of multiple peaks across hundreds of pixels within an entire image is time-consuming, tedious and subjective. Here, various chemometric methods were used to analyze data sets obtained by matrix-assisted laser desorption/ionization (MALDI) IMS of multicellular spheroids. HT-29 colon carcinoma multicellular spheroids are an excellent in vitro model system that mimic the three dimensional morphology of tumors in vivo. These data are especially challenging to process because, while different microenvironments exist, the cells are clonal which can result in strong similarities in the mass spectral profiles within the image. In this proof-of-concept study, a combination of principal component analysis (PCA), clustering methods, and linear discriminant analysis was used to identify unique spectral features present in spatially heterogeneous locations within the image. Overall, the application of these exploratory data analysis tools allowed for the isolation and detection of proteomic changes within IMS data sets in an easy, rapid, and unsupervised manner. Furthermore, a simplified, non-mathematical theoretical introduction to the techniques is provided in addition to full command routines within the MATLAB programming environment, allowing others to easily utilize and adapt this approach.
Keywords: MALDI imaging, chemometrics, 3D cell culture, MATLAB, Principal Component Analysis
Introduction
Matrix-assisted laser desorption/ionization imaging mass spectrometry (MALDI IMS) is becoming an increasingly popular method for a variety of imaging applications ranging from organ sections of animals to disease and disease-free clinical samples1-4. MALDI IMS is a label-free technique with the ability to detect a variety of molecules including proteins, peptides, lipids, and small molecules over a wide mass range making it the most versatile of the MS imaging techniques5. This method of imaging provides extremely rich data sets yielding ion heat maps containing both spatial and spectral information of the analytes found within the sample. Recent years have seen a remarkable increase in the application of IMS of all types to systems beyond traditional tissue imaging. A growing area of research is the application of IMS to three dimensional cell culture (3DCC) systems3, 6-8. These models occupy a distinct niche between conventional two dimensional cell culture systems and animal models9-11. Advantages to 3DCC include rapid analysis time, relatively high throughput and experimental flexibility12, as compared to animal models.
The 3DCC system examined in this study is the HT-29 colon carcinoma spheroid tumor model. When plated under specific growth conditions, these cells grow into spheres that can reach up to 1 mm in diameter. At this size, these models develop nutrient, oxygen and transport gradients that generate biologically distinct microenvironments within the spheroids3, 13. Importantly, these microenvironments closely resemble different populations of cells found in large tumors in vivo. The outer proliferative region of the spheroid mimics regions of tumors situated closest to blood vessels or other nutrient sources. An intermediate quiescent region is populated by cells that have restricted access to oxygen and nutrients and are alive but are no longer actively dividing. The cell culture models also contain an inner necrotic core of dead and dying cells which resembles the population of cells in a tumor furthest from blood vessels with little or no access to oxygen and fresh nutrients. 3DCC models allow researchers a unique opportunity to study the relationships between these heterogeneous cellular populations, and how changes in one region affect the others as well as how these structures are affected by drug or small molecule treatments5, 6. The combination of MALDI IMS and 3DCC takes advantage of the experimental flexibility of 3DCC systems and the wide mass range of MALDI mass spectrometry to yield highly complex and informative data sets.
Most 3DCC models lack visible tissue structures that separate biologically distinct regions of interest, further complicating their analysis. In particular, images acquired from this cell culture system represent a unique processing challenge because of the clonal nature of cell lines14, 15. HT-29 spheroid cells arose from a single progenitor cell or group of progenitor cells, therefore all cells have the same genetic background16. Additionally, the expression of many highly abundant housekeeping proteins is consistent within each of the microdomains in the spheroid. In sum, each of the heterogeneous spheroid microenvironments exhibit unique protein expression patterns with strong mass spectral overlap3.
While single-ion heat maps can provide useful information, the application of various data processing approaches is required in order to take full advantage of all the benefits offered by IMS17, 18. The use of multivariate statistical analyses on IMS data sets can help highlight otherwise undetected differences among distinct regions and can be especially useful for elucidating changes over time in cell populations as a result of drug treatment or other cell culture perturbation6, 8, 11, 19, 20. These methods are especially advantageous when considering the complexity of IMS images (i.e. hundreds of mass spectra containing hundreds of peaks in spatially relevant regions). As an example, multivariate analysis techniques have been applied to secondary ion mass spectrometry (SIMS) images of rat heart tissue and heterogeneous cellular cultures8, 20-23. Barnes and colleagues used SIMS imaging to identify cell types in a heterogeneous mixture of two different cell types grown together. The authors relied on multivariate analysis of the acquired images to differentiate between the two cell types in absence of distinct visual markers. This study demonstrated the usefulness of chemometrics and IMS in distinguishing phenotypic changes in a heterogeneous cell population lacking visible morphological features. Fornai et al. used metal-assisted SIMS imaging to construct a three-dimensional ion image of serially sectioned rat heart tissue. While discreet structures such as the coronary arteries, ventricular valves and ventricles are easily visible using the hematoxylin and eosin (H&E) staining technique, the authors were also able to detect distinct molecular signatures from each region of the heart tissue using SIMS imaging and principal component analysis (PCA). When these two imaging modalities were combined, the information gained from the SIMS imaging experiment added an entirely new dimension not seen with conventional H&E or other immuno-based staining techniques.
MALDI IMS presents several unique experimental difficulties that can be corrected using multivariate analyses including matrix interference, apparent mass discrepancies due to sample height differences and non-uniform ionization efficiency24. There is no consensus on a universally recognized approach for multivariate analysis of IMS data because each data set presents different challenges. However, each data analysis workflow seeks to accomplish three main goals: the removal of experimental artifacts from the imaging process, reduction of the complex data sets into more computationally manageable formats (without significantly compromising the experimental information contained within) and the successful application of multivariate analyses to capitalize on what these complex data sets have to offer25, 26. In this scheme, each processing step insures that any subsequent multivariate analyses would examine trends present in the data arising from biological variation instead of experimental artifacts. As an example, pixels containing only noise-related or high-intensity matrix-related spectral features are removed because they could bias subsequent multivariate analyses against detecting more subtle changes in protein expression.
Fonville, et al. described the effects of different normalization strategies on subsequent multivariate analysis of MALDI IMS performed on rat brain tissue in the small molecule mass range27. They followed the same basic blueprint for the processing of IMS data: isolation and removal of noise-related or matrix-related pixels, transformation of the remaining biologically relevant pixels into a multivariate space and further multivariate analysis of the data to reveal spectral trends identifying different regions of tissue based on patterns in the spectra. In one particular case, after identifying and removing pixels containing only noise or matrix-related peaks from the data, the authors applied logarithmic scaling to the remaining pixels. This logarithmic transformation reduced the dynamic range of the measurements, allowing the low intensity features in the mass spectra to become more pronounced relative to large peaks. As a result, small but relevant changes in the measured intensities of the pixels could be appropriately included into the final analysis28.
In a study by McCombie et al., the authors focused on the combination of PCA, clustering methods, and linear discriminant analysis (LDA) to reveal biologically important regions of mouse brain sections analyzed by MALDI IMS29. This particular approach made use of the iterative application of multiple chemometric techniques to highlight biologically relevant spectral features in the data in an unbiased manner. PCA was first used to reduce the dimensionality of the data set and identify noise and matrix related pixels, which were then removed. Next, the authors applied unsupervised clustering methods, including k-means and hierarchical clustering analysis (HCA), to find and aggregate groups of pixels that exhibited similarities in protein expression (demonstrated by strong spectral overlap). Finally, LDA was used to generate discriminant spectra that identified specific m/z ratios differentially expressed within distinct regions of the brain tissue.
Here we integrate several chemometric methods into a single workflow for the processing of MALDI IMS data generated from HT-29 spheroids. Adapting from the workflows described above, PCA was first used to pinpoint and isolate only the biologically relevant pixels from the entire MALDI IMS image. Second, after some spectral processing steps, a second iteration of PCA was used to isolate relevant features within the mass spectra of the image. Next, clustering methods including k-means and HCA were used to identify and group the relevant pixels into multiple distinct cellular populations based on similarity in the recorded mass spectra. Finally, a combined PCA-LDA approach was used to highlight specific m/z values differentially expressed within the unique cellular populations of this clonal system.
Results and Discussion
Workflow Overview
The overall research goal was to utilize various multivariate data analysis methods for the identification of spectral features associated with distinct cellular subpopulations within multicellular spheroids, a system that yields mass spectra with significant spectral overlap. The data shown here is taken from a single HT-29 spheroid slice to show proof-of-concept. An overview of the approach is presented first, followed by a detailed analysis. If needed, the reader is directed to the Supporting Information for a theoretical introduction on each of the chemometric techniques.
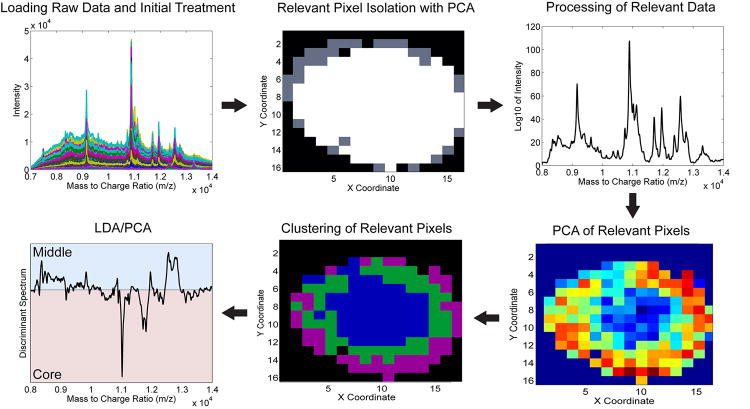
Figure 1 illustrates the statistical workflow for the analysis of a MALDI image of an HT-29 spheroid section. First, raw data files for each pixel within the image were converted into the mzXML format using CompassXport 5.0, imported into the MATLAB programming environment and concatenated into a single data matrix. After a few initial data processing steps, PCA was then performed on this mass spectra data matrix. Using a two-dimensional score plot, the spatial distribution pattern of the principal component (PC) scores was then used to isolate physiologically-relevant spectra originating from the HT-29 cells from pixels corresponding to the surrounding gelatin support. The mass spectra of the relevant pixels were then further processed to better highlight potentially discriminating spectral features of low abundance, remove baseline shifts, reduce noise, correct for differences in apparent peak m/z, and normalize for differences in absolute inter-pixel intensity. A second iteration of PCA was then performed on the processed data of the relevant pixels to highlight spatially-localized mass spectral features within the data set, further remove noise, and reduce the dimensionality of the data prior to subsequent chemometric techniques. Next, cluster analysis was performed on the scores of the relevant PCs to identify and group pixels associated with distinct cellular subpopulations that exhibited spectral similarity within the spheroid section. Finally, PCA-LDA was used to identify specific m/z values associated with proteins differentially expressed within the distinct regions of interest.
Figure 1.
Summary of data workflow. MALDI IMS data is first imported into the MATLAB programming environment. PCA is then used to isolate relevant pixels within the image. The mass spectra of the isolated pixels are then further processed before a second round of PCA that highlights general spectral features associated within the sample. Clustering analysis is then performed using the scores of the relevant pcs from this second iteration of PCA to reveal the spatial distribution pattern of distinct cellular populations. Finally, the results of both the PCA and clustering of relevant pixels are used as inputs into an LDA routine that identifies specific m/z values selectively associated within each of the distinct cellular microenvironments in the spheroid.
Isolation and Initial Processing of Relevant Pixels
The mass spectra were initially recorded encompassing an m/z range of 4 to 20 kDa. In the first step of the statistical workflow, the recorded spectra were trimmed to a relevant mass range spanning 8-14 kDa, Gaussian filtered to remove noise, and subsequently downsampled. m/z values within this smaller 8-14 kDa window encompassed the peaks that were clearly distinguishable from noise. Trimming the low and high ends of the mass spectra improves computational processing time and ensures that multivariate analyses will be focused on changes in protein expression instead of noise. Gaussian-filtering is a common default processing step available in many commercial software packages and was used to remove noise and make peaks more symmetrical30, 31. While downsampling reduces spectral resolution, it significantly increases processing efficiency and prevents errors associated with the limited memory capacity of the computer by reducing the number of points per spectrum. Here, spectra were downsampled to an m/z resolution of 10 m/z. Because distinct protein species should be more than 10 m/z apart within this mass range, misidentification errors should be minimized by this downsampling rate. Trimming and downsampling allowed the large IMS data set to be more computationally manageable for further chemometric processing and these steps are commonly used by other researchers25, 32, 33.
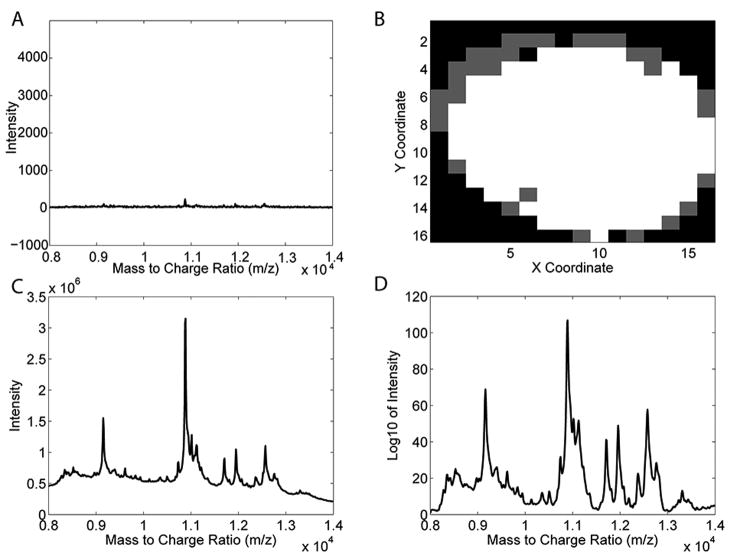
Once trimmed, it is necessary to select pixels that arise directly from the sample and discard those that contain mostly noise and matrix related peaks. The original MALDI IMS image contained 208 pixels, the majority of which were associated with the spheroid cells while others were associated with the gelatin support and/or experimental artifacts. These latter irrelevant pixels generally exhibited featureless mass spectra for pixels associated with gelatin, mass spectra containing matrix artifacts, or mass spectra that are excessively noisy for accurate analyses as shown in Figure 2A. For these reasons, removing the irrelevant pixels made it easier for the subsequent clustering algorithms to identify distinct regions of interest within the spheroid section (see below).
Figure 2.
Isolation of relevant pixels and spectral processing. A) Example mass spectrum of a pixel discarded after an initial round of pca on the unprocessed data. B) Isolation of mass spectra from pixels associated with spheroid cells (white) using PCA. Spectra arising from pixels outside of the sample (grey) were discarded. C) The total ion spectrum of all relevant pixels identified in B (white pixels). D) The total ion spectrum after log transformation and baseline removal.
A first iteration of PCA was performed on the trimmed and downsampled data set to isolate only the relevant pixels from the entire MALDI image. PCA is an exploratory analysis tool that deconstructs a multivariate data set into a set of PCs (also termed eigenvectors or loadings) and scores. For MALDI IMS data sets, the loadings highlight independent sources of spectral variation within the data set while the scores describe the importance of each of those spectral features to each imaged pixel. Pixels with similar mass spectral profiles should have similar PCA score values and aggregate together within a two-dimensional PCA score plot34. In this case, pixels that were associated with the relevant spheroid cells grouped separately from those pixels associated with the gelatin support and experimental artifacts (see Supporting Information user guide for a visualization of this score plot). Based on the clustering pattern of a two-dimensional score plot, the relevant pixels associated with the spheroid cells were identified and retained for further processing while pixels associated with artifacts were discarded. Figure 2B shows the relevant pixels identified using this PCA approach. Specifically, 50 pixels from outside the cell mass or those pixels associated with technical/instrumental artifacts (grey) were successfully separated from 158 relevant pixels associated with the spheroid cells (white). While PCA could have also been used to discard noise by removing the PCs that describe only noise, all PCs were retained such that the entire image could be processed subsequently.
Once relevant pixels were isolated, other standard data processing steps for removing MALDI experimental artifacts were applied25. A top-hat filter function was used to remove baseline changes associated with MALDI matrix within the spectra. Like Gaussian filtering, top-hat filtering is a routine step in the analysis of protein mass spectra and is also available as a default processing option within commercial mass spectrometry software packages30. Because a time-of-flight (TOF) instrument was used to image the spheroid slice, differences in sample height can cause shifts in the apparent mass of peaks26. The m/z alignment of mass spectra is an essential pre-processing step necessary for TOF data to ensure that regions of interest are identified based on physiological spectral trends associated with protein expression patterns29. If spectra are not aligned, subsequent clustering algorithms may falsely identify regions of interest based on topographical artifacts associated with the imaging experiment including sample roughness or sample height differences caused by the spheroid sectioning process. Here, two peaks present in the mass spectra were used for alignment of the HT-29 spheroid section data; the peaks that appeared at approximately 9160 m/z and 12,550 m/z were chosen for the aligning process as the majority of the spectral features in all mass spectra across all pixels were encompassed within this range. In each spectrum, the apparent masses of these two peaks were aligned to the median m/z values recorded for those two peaks across all pixels using an interpolation-based algorithm35. The summed spectra across all relevant pixels after baseline subtraction and spectral alignment is shown in Figure 2C.
The intensities of peaks within an individual mass spectrum can span several orders of magnitude. This large variation makes it difficult for subsequent chemometric methods such as PCA to identify low intensity peaks that offer high discrimination power between different regions of interest within a MALDI image27. This issue is typically subverted using variance scaling prior to PCA, a process that allows PCA to assess all m/z values as equally important36. Unfortunately, this approach also dramatically increases the influence of m/z values that describe only noise and can thus degrade the quality of the results obtained. As a compromise, Fonville et al. used an empirical approach whereby all spectra underwent a logarithmic transformation prior to PCA; their approach successfully allowed PCA to detect small intensity discriminating peaks without significantly increasing the influence of noise27. Here, the same logarithmic-transformation procedure was used to identify as many discriminating peaks as possible within this data set that showed an unusually high degree of spectral similarity (as shown in Figure 1)3. Figure 2D shows a summed spectrum after baseline removal, spectral alignment, and logarithmic scaling. Compared to the summed spectrum shown in Figure 2C, the summed spectrum in Figure 2D shows the ability of the logarithmic scaling to successfully enhance peaks of lower relative abundance without significantly increasing noise. The final step of spectral processing was normalizing the mass spectra to the total area under all peaks. This final step removed any remaining variation in the data due to the MALDI measurement process (i.e., uneven ionization of analytes) instead of relevant biological changes in the sample.
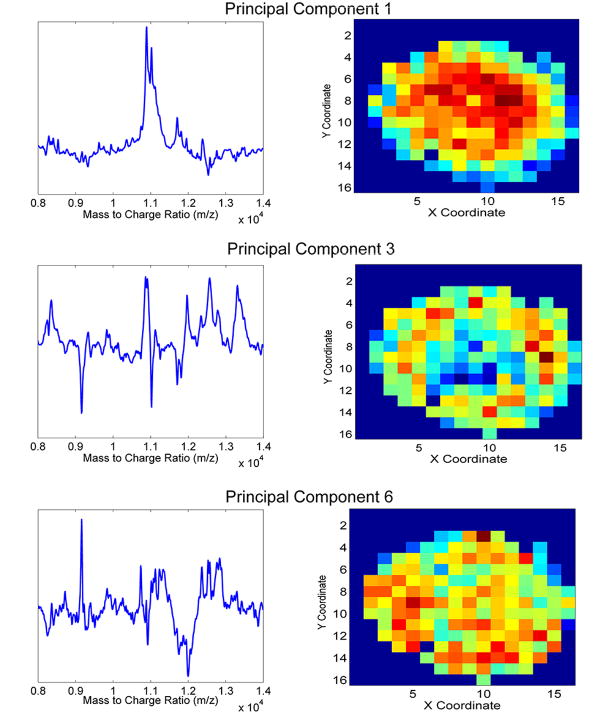
A second iteration of PCA was performed to further remove spectral noise, reduce data dimensionality prior to LDA, and identify spatially-localized spectral trends present in the MALDI-IMS data set. After PCA was performed for a second time, the PCs describing relevant spectral features were retained while PCs that described only noise were discarded. Figure 3 shows data obtained from this second iteration of PCA performed on the processed data of the relevant pixels. As examples, the loadings from PCs one, three and six are shown on the left side of the figure while the corresponding score heat map for each PC is shown on the right side. Examining PC 1, the loading plot shows several features of the spectra between 10,000 and 12,000 m/z in the top half of the plot, meaning these features are strongly associated with red/yellow-colored pixels towards the center of the spheroid slice. Examples of other spatially-localized features associated with other PCs are also shown in Figure 3. Despite the fact that a specific pixel may have a high score value for one particular PC, spectral characteristics identified by other PCs may also contribute significantly to its recorded mass spectrum. As such, these “pseudo-spectra” must be interpreted cautiously because, while they include m/z information of high discriminatory power, one particular PC does not solely describe all relevant information within a specific pixel37. Thus, because PC loadings can sometimes be difficult to interpret38, 39, further processing is needed to identify all of the spectrally-relevant features that discriminate between distinct regions of interest within the spheroid section.
Figure 3.
PCA of isolated spectra. Principal component loadings (left) and spatially-localized pixel score plots (right) for principal components 1 (top), 3 (middle), and 6 (bottom).
Cluster analysis
The purpose of the previous steps in the overall workflow was to identify potentially discriminating spectral features within the relevant pixels of the MALDI image. Cluster analysis was next used to identify groups of pixels associated with distinct cellular populations within the heterogeneous spheroid structure. Cluster analysis was performed on the scores of the relevant retained PCs of the processed data. While strong spectral overlap exists because of the clonal nature of the cells within the spheroid section, the distinct cellular phenotypes also exhibit some unique features in their mass spectral profiles3. As the nutrient gradient across the spheroid grows more pronounced concomitant with an increase in spheroid size, a gradual phenotypic transition becomes apparent, yielding changes in the mass spectral profiles among the different microenvironments. Because PC scores describe the relative importance of certain spectral features to each pixel within the MALDI image, the score values can be used by cluster analysis to segregate pixels into distinct cellular populations40. In addition, by removing the noise-containing PCs, the ability of clustering methods to assign pixels to specific groups is improved29.
Two different methods of cluster analysis were tested here: HCA and k-means clustering. In HCA, inter-point distances between all data elements are calculated and the two closest points are aggregated and treated as a cluster. Inter-element distances are again calculated and data elements are again joined (either separate data points are joined together, data points are connected to existing clusters, or existing clusters are linked together). The process is repeated until all data elements are linked together in an interconnected hierarchy. A distance cutoff can then be used to split the data elements into several distinct clusters41. In k-means clustering, k number of points are first randomly chosen to be cluster centers and all points are then assigned to the group with the nearest cluster center; the resulting cluster center is recalculated and the process is repeated iteratively until convergence42. Generally, cluster analysis typically requires that the number of distinct groups be known before the analysis begins. While three general microenvironments are known to exist in the HT-29 spheroids, to eliminate subjective bias, the gap statistic was used here to estimate the number of distinct groups for both HCA and k-means clustering as this empirical approach does not require any a priori knowledge of the dataset24,43.
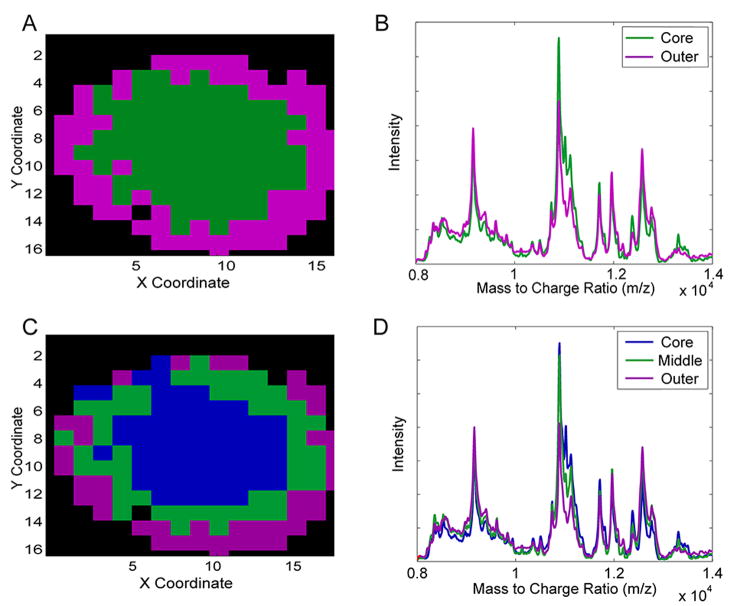
The results of the cluster analysis on the relevant PC scores are shown in Figure 4. Using the gap-statistic, HCA identified two distinct groups of pixels (Figure 4A and B) and k-means identified three distinct groups of pixels (Figure 4C and D). The cluster maps represent distinct groupings of pixels identified by the clustering algorithms and the traces represent the average of the processed mass spectra for all the pixels within the particular group. While both approaches identified distinct populations within the expected concentric pattern, in this case only k-means clustering was able to successfully identify three separate groups associated with the three well-known microenvironments of 3DCC systems. The algorithm was able to correctly identify all three regions expected in the spheroid. Furthermore, this clustering was done in an unsupervised fashion and yielded separate regions falling within the expected spatial distribution of the spheroid structure: the outer proliferative cells were highlighted as a ring around the outside of the spheroid, the intermediate quiescent cells were a distinct group just inside the first ring of cells and finally, the necrotic core was shown as a single population occupying the center of the structure.
Figure 4.
Cluster analysis. A) Regions of interest identified by HCA. B) The average processed spectra of each region in A. The purple and green traces correspond to the average processed spectra of all purple and green pixels, respectively. C) Regions of interest identified by k-means clustering. D) The average processed spectra of each region in C. The purple, green, and blue traces correspond to the average processed spectra of all purple, green, and blue pixels, respectively.
The combination of the cluster map in Figure 4C and the summed processed spectra from each region in 4D give an approximation of the similarities and differences in protein expression among the distinct biological environments. While the PC loadings shown in Figure 3 offer some insight into these relationships, those pseudo-spectra are not directly interpretable. Figure 4 offers directly interpretable mass traces with corresponding cluster maps that are directly translatable to biological changes within the sample. Figure 4D also highlights the difficulty inherent with the analyses of these systems and the success of this workflow. In general, the same analytes (within this measured m/z range) were detected in each of the different microenvironments; this behavior is somewhat expected since all cells are clonal. However, much of what discriminates these microenvironments into separate populations are the relative ratios of these analytes. These subtle differences present a difficult analytical challenge that requires both finesse and sophistication for proper identification.
PCA-LDA
The results shown in Figure 4D can be used to help identify changes in protein expression patterns within the distinct cellular populations of 3DCC systems. However, a combination of PCA and LDA was used as a final step to clearly visualize m/z values selectively associated within distinct microenvironments within the spheroid.
The field of multivariate discriminant analysis is traditionally used to classify future data observations into different categories based on the similarity to known, well-characterized data. One goal of LDA is to easily visualize the differences amongst groups of data that have previously been clustered into separate groups. To achieve this objective, a series of linear boundaries are calculated and plotted in the same coordinate system, segregating a multivariate data space into separate regions associated with different groups of data. These boundaries represent a way to discriminate between different clusters of data observations because future observations are classified into a particular data group depending on which of the separate regions of a multivariate space they are localized (see Supporting Information).
Specifically, LDA mathematically calculates a boundary that maximizes intergroup separation using a linear combination of independent variables:
| (Eq 1) |
where X1, X2, and X3 are the independent variables and A, B, and C are scalar coefficients used to determine direction44. In a combined PCA-LDA analysis, the independent variables used to calculate the linear boundaries between groups are the retained PCs. In terms of MALDI IMS data, LDA would normally require that the number of points within a mass spectrum to be less than the number of pixels imaged29. By performing PCA prior to LDA, the number of independent variables decreases from many m/z values to a few relevant PCs, enabling the calculation of a unique solution for the linear boundaries by LDA45. Moreover, noise can be removed from the spectra in the PCA step performed prior to LDA by discarding the non-relevant principal components, thereby improving the linear boundary calculations29.
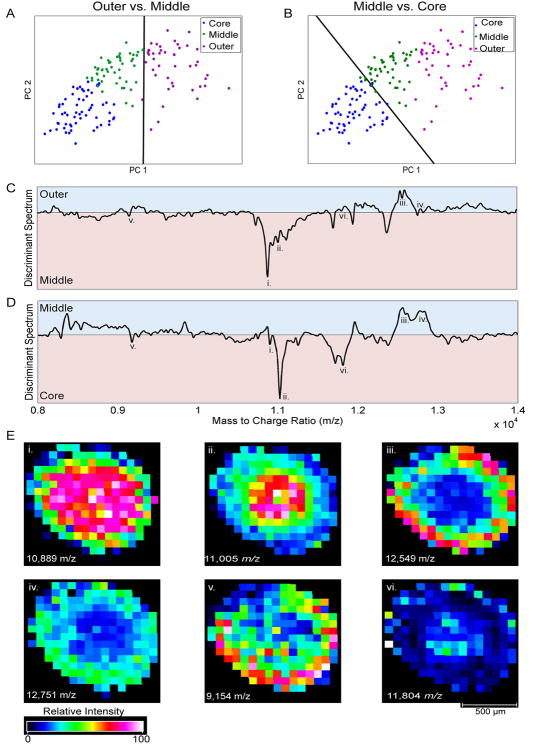
Figure 5 demonstrates the combined PCA-LDA analysis. Figures 5A and 5B show PCA score plots of the pixels clustered by k-means and the calculated linear boundaries. In both cases, the boundaries successfully discriminate amongst the different spheroid microenvironments (outer versus middle in 5A and middle versus core in 5B). While a simple line is shown on these two-dimensional score plots for clarity, it is important to note that the linear boundary actually exists as a bisecting hyperplane within the multivariate hyperspace that includes all of the retained PCs (see Supporting Information).
Figure 5.
PCA-LDA. A) and B) show score plots of the clustered pixels and the calculated linear boundaries. A) The calculated linear boundary separates the pixels associated with the outer and middle regions of the spheroid. B) The calculated linear boundary separates the pixels associated with the middle and core regions of the spheroid. C) The calculated discriminant spectrum differentiating between the proliferative outer region (shaded blue) and the quiescent middle region (shaded pink). D) The calculated discriminant spectrum differentiating between the quiescent middle region (shaded blue) and the necrotic core region (shaded pink). The lower case roman numerals highlight several m/z values discussed in the text. E) Single ion heat maps of m/z values identified using the discriminant spectra. The m/z value and its corresponding roman numeral notation is inset within each two-dimensional heat map.
Equation 1 shows that each of the calculated PCA-LDA discriminating boundaries is simply a linear combination of PCs. Much the same way that a recorded mass spectrum can be calculated using the linear combination of PC loadings and scores, a discriminant spectrum can be calculated by the linear combination of PC loadings and the scalar coefficients from the linear boundary that separates two groups of data using Equation 1. A separate discriminant spectrum can be calculated for each linear boundary. Here, two boundaries were calculated which generated two discriminant spectra (outer versus middle and middle versus core). Because the linear boundary drawn maximally separates two groups of data with different spectral features, the resulting discriminant spectrum highlights spectral features that maximally differentiates the two groups of data29.
These calculated discriminant spectra are shown in Figures 5C and 5D. The spectral features that are more strongly associated with one group of pixels (specific microenvironment) versus another are on either side of the mean value within a discriminant spectrum. Because mean-centering was used prior to PCA, the mean value within a discriminant spectrum will be zero, greatly improving interpretation because the sign (positive or negative) of the calculated discriminant spectrum at a specified m/z can be used to determine the selective association of an analyte with a particular group29. m/z ratios that exhibit positive intensities in the discriminant spectra are more strongly associated with one group of pixels, while m/z ratios that exhibit negative intensities are more strongly associated with the other group of pixels.
The calculated discriminant spectra highlight several species that are selectively distributed in the different microenvironments within the spheroid. Perhaps the easiest to notice first is a large negative peak at approximately 10,890 m/z (labeled i) in the discriminant spectrum shown in Figure 5C which indicates that this analyte is more dominant in the middle region of the spheroid as compared to the outer region. This same m/z value also appears as a small negative peak in the discriminant spectrum in Figure 5D, indicating that this species is slightly more abundant in the core region as compared to the middle region. Taken together, both discriminant spectra suggest that there exists a species at approximately 10,890 m/z whose abundance increases dramatically from the outer to the middle regions of the spheroid, but only a small amount when moving from the middle region into the core. This molecule could indicate a protein being expressed in relation to the cells in the inner regions of the spheroid undergoing some sort of metabolic stress. Figure 5E shows a two-dimensional heat map from flexImaging for the species at 10,889 m/z. Indeed, the abundance of this species increases towards the center of the spheroid, with the largest increase being between the outer and the middle regions. This behavior was also shown in the average traces shown in Figure 4D. While this difference appears subtle, it is not easily noticeable when examining only the standard ion maps generated by the imaging software and indeed small changes in low abundance proteins could portend more drastic phenotypic changes in the cell populations. The ability to discern these small changes is very important for understanding the biochemical differences in these populations of cells.
The discriminant spectra also offer clues as to the biological status of the different groups of cells. For example, at an m/z of 11,005 (ii), there is a moderately-sized peak in the negative region of the outer versus middle discriminant spectrum and a large negative peak in the middle versus core discriminant spectrum. This indicates that the protein at m/z 11,005 is increasing in abundance towards the necrotic center of the spheroid. Both the two-dimensional heat map shown in Figure 5E and the average traces shown in Figure 4D for 11,005 m/z confirm that the discriminant spectra were correct. Because it is known that the cells in the necrotic core are deprived of nutrients and oxygen, this protein species may be an indicator of dead or dying cells. Conversely, there is a peak at approximately 12,550 m/z (iii) that is positive in both discriminant spectra indicating that this species is more abundant in the middle region as compared to the core region, yet also more abundant in the outer region than the middle region; in other words, the abundance of the species at 12,550 m/z is increasing in abundance outward from the center of the spheroid. Because the cells in the intermediate and outer regions of the spheroid are exposed to oxygen and fresh nutrients, they are generally healthier, with the population in the very outer region still actively dividing. Therefore the protein species at 12,550 m/z could be associated with healthy and/or actively dividing cells. The two-dimensional heat map of this ion shown in Figure 5E and the average traces shown in Figure 4D again confirm the results obtained by the discriminant spectra.
The average traces in Figure 4D show that there is a species at approximately 12,750 m/z whose abundance is similar in the outer and middle regions, but is also reduced in the necrotic core. There is no peak in the discriminant spectrum of Figure 5C at this m/z (iv), indicating that there is no significant difference in the abundance of this species between the outer and middle regions. However, the discriminant spectrum between the middle and core regions shown in Figure 5D yield a positive peak, indicating that this species is less abundant in the core. Taken together, the discriminant spectra confirm the behavior shown in Figure 4D. The two-dimensional heat map at this m/z shown in Figure 5E also confirms a moderate abundance of this species in both the outer and middle regions. Interestingly, both discriminant spectra lack a significant peak in the region of 9,155 m/z (v) even though this analyte is prominent throughout the entire image, suggesting that this highly-abundant species offers little in the way of determining differences in cellular populations. Indeed, while this species is of high abundance, the average traces in Figure 4D show its average intensity is relatively consistent in all three regions of the spheroid. The two-dimensional heat map in Figure 5E also shows that there are areas of high, moderate, and low relative abundance of the 9,155 m/z peak in each of the three regions of the spheroid.
As a final example, the discriminant spectrum in Figure 5D shows a negative peak at approximately 11,805 m/z (vi), indicating that this species is more abundant in the necrotic core as compared to the quiescent middle region. This fact is confirmed by the two-dimensional heat map shown in Figure 5E. While this species originally appeared insignificant in the original spectra (Figure 2) and even as a small shoulder of the more-intense 11,700 m/z peak within the processed data (Figure 4), the discriminant spectra were successfully able to identify this analyte as one whose abundance yields high contrast within different microenvironments of the spheroid. It is likely that a peak of such low relative abundance in the naïve data set could have been overlooked during traditional analysis of this MALDI IMS data set. Moreover, while each of the single ion heat maps shown in Figure 5E can be observed in the native data analysis program accompanying the mass spectrometer, discriminant spectra shown in Figures 5C and 5D allow the user to easily simultaneously visualize how multiple ions relate to one another in each microenvironment of the sample.
There are several caveats in interpreting discriminant spectra. First, while very powerful, discriminant spectra still represent calculated best estimates of spectral features that distinguish between separate groups of pixels. Second, if groups of points cluster in such a way that a linear boundary does not ideally distinguish between two groups, the discriminant spectrum calculated using this boundary could contain erroneous information44, 45. Third, the quality of the results obtained by the PCA-LDA approach is highly dependent on the number of relevant PCs retained. If too few principal components are retained, potentially discriminating features would be left out of the calculated discriminant spectra. Conversely, the quality of the discriminant spectra can also deteriorate if too many noise principal components are retained. If these less important PCs are factored into the PCA-LDA analysis, the noise-containing PCs will show significant overlap in the cluster analysis that would cause the boundary calculated by the algorithm to falsely represent randomly distributed noise as a distinguishing feature of the dataset. Therefore, it is essential to appropriately ascertain the proper number of relevant PCs to retain in the multivariate model prior to the LDA step.
Conclusions
The advent of mathematical programming environments such as MATLAB allows complex data analysis methodologies to become streamlined and automated. Indeed, excluding the loading of data into the MATLAB programming environment, the execution of the entire analysis workflow presented here takes less than one hour for the experienced user as compared to days-worth of individual peak analysis within individual pixels. In addition, this semi-automated approach can remove potential bias associated with manual evaluation of data sets.
Chemometric methods including those shown here can dramatically improve the quality of the conclusions obtained from MALDI IMS data sets and their use should certainly continue. In this proof-of-concept demonstration, the combination of PCA, clustering methods, and LDA allowed for the easy visualization of differences in protein expression levels across multiple cellular subpopulations within a single MALDI image of a 3DCC section. While it is known that there are distinct regions within the 3DCC models, the cluster maps and discriminant spectra easily identified the spatial relationship among multiple m/z values within the different cellular microenvironments simultaneously. This is important because in most cases a single protein species does not adequately describe all biological variation within a system. Unfortunately, chemometric techniques can appear theoretically obtuse, are sometimes described in the literature with advanced statistical sophistication, and can seem daunting to execute for the inexperienced, yet interested user. To confront these challenges and engender more widespread use among other researchers, we have provided a simplified, non-mathematically intensive introduction to the techniques as well as the MATLAB command lines necessary to perform them (Supporting Information).
The authors acknowledge that there are many options for each of the steps presented here and again no single workflow is optimal for all MALDI IMS data sets. Because of the flexibility of the MATLAB programming environment, any IMS data set that can be converted into the .mzXML file format can, with some small adjustments, be processed using the scripts provided in the Supporting Information. As noted throughout the paper, many of these steps have previously been used on other IMS samples27, 29. The workflow presented in this paper was determined to be optimal for the processing of 3DCC IMS data, a system which presents several unique analytical challenges specific to multicellular spheroid systems based on clonal populations of cells which exhibit a high degree of spectral overlap. With workflows such as the one described here in place, detailed studies of the proteomic effects of various genetic manipulations, drug treatments or co-culture experiments can be undertaken in the future.
Experimental
Cell Culture
HT-29 cells obtained from American Type Culture Collection (ATCC, Manassas, VA) were cultured in 5% CO2 at 37 °C in McCoy's 5A media (Life Technologies, Grand Island, NY) supplemented with 10% fetal bovine serum (Hyclone Laboratories, Logan, UT) and 2 mM L-Glutamine (Invitrogen, San Diego, CA) as described previously3. Cell lines were used within 3 months after receipt or resuscitation of frozen aliquots thawed from liquid nitrogen. The provider assured the authentication of these cell lines by cytogenetic analysis. To induce three-dimensional growth, approximately 6000 cells were seeded in each well of the inner 60 wells in 96-well culture plates (Thermo, Rockford IL) and incubated with complete media changes 10 and 14 days after seeding.
Sectioning and Sample Preparation for MALDI Imaging
Spheroids were harvested according to the gelatin-assisted sectioning method as described previously3. Briefly, spheroids were removed from media and washed in 1x PBS (Gibco). A thin layer of gelatin was applied to the bottom of several wells of a 24-well cell culture plate (Thermo) and allowed to cool. After cooling, the washed spheroids were gently placed on the gelatin in the wells and then quickly covered in a layer of warm, liquid gelatin. The cell culture tray was immediately placed at -80°C until sectioning.
Prior to sectioning, the gelatin blocks were removed from the wells and freeze mounted to sectioning mounts using deionized water and cooled in the cryomicrotome chamber (Leica Biosystems, Nussloch, Germany) set at -30°C. Once samples equilibrated, they were sliced in 14 μm-thick sections. Intact slices were thaw-mounted to a room temperature ITO-coated glass slide (Delta Technologies, Loveland, CO) which was immediately transferred to a desiccator where the slides were stored until MALDI matrix application.
The matrix used was sinapic acid (Sigma, St. Louis, MO) mixed in 50:50 vol/vol HPLC grade water (Burdick and Jackson, Phoenix, AZ) w/0.1% trifluoroacetic acid (Sigma) and HPLC grade acetonitrile (Burdick and Jackson). Matrix was applied by hand using a 1.0 μl GC syringe (Hamilton, Reno, NV) with the aid of a dissecting microscope. Slides were washed twice for 30 seconds in cold acetone (EMD Millipore, Billerica, MA) before matrix application. Washing serves to fix protein and remove lipids and other small organic contaminants46. Slides were dried in a vacuum desiccator after each wash. Matrix coverage was inspected with a microscope after each application and was repeated as needed. Slides were dried in a vacuum desiccator between each application and stored in the dark in a desiccator until imaging.
MALDI Imaging
MALDI spectra were acquired with a Bruker Autoflex III Smartbeam MALDI-TOF mass spectrometer (Bruker Daltonics, Billerica MA) in linear positive ion mode in the 4-20 kDa mass range. The laser spot was set to “Ultra” and laser attenuator was manually adjusted prior to each experiment for optimal fluence. Spectra were acquired by flexImaging 2.1 (Bruker) with a raster increment of 70 μm.
Data Analysis
The flexImaging (Bruker) and MATLAB (version R2013a, MathWorks, Natick, MA) software packages were used for all data analyses. flexImaging was used to generate and visualize semi-quantitative, two-dimensional ion density maps. Raw data files were converted into the mzXML format using CompassXport 5.0 (Bruker). MATLAB was then used for all other statistical and chemometric data processing. Specifically, in-house MATLAB command lines were written to open/read the mzXML data, process and transform data (trimming, down-sampling, filtering, aligning, normalization, etc.), perform PCA, and perform PCA-LDA. The MATLAB command lines used for clustering analysis were adapted from Martinez, Martinez, and Solka47. A MATLAB function freely available from MATLAB Central's File Exchange was adapted to isolate data points visually presented within the PCA score plot (Supporting Information)48. Specifically, this routine was used to select and isolate relevant pixels from within the entire MALDI image. MATLAB command lines and a user guide necessary to perform all of the chemometric techniques used here are included in Supporting Information.
Singular value decomposition was used to mathematically perform PCA49. Data sets were mean-centered prior to both iterations of PCA, but variance scaling was not used to ensure that noise present within the mass spectra would be of minimal influence in the PCA calculation. For the second iteration of PCA, the Scree graph was used to determine the proper number of relevant principal components to retain38, 50. An interpolation-based algorithm was used to align all mass spectra35. As a general overview, in this process mass spectra were first moved to align with a specific m/z of a peak of low m/z and then stretched or shrunk to align around a second, higher m/z peak. For HCA and k-means clustering, Euclidean distances were used to calculate inter-point distances. In addition, complete (furthest-neighbor) linkages were used by HCA to calculate inter-cluster distances.
Supplementary Material
Acknowledgments
Funding for R.B.K. was provided by Roanoke College. Salary support for A.B.H was provided by the Walther Cancer Foundation and this research was supported by National Institutes of Health (1R01GM110406-01) for A.B.H.
References
- 1.Reyzer ML, Caldwell RL, Dugger TC, Forbes JT, Ritter CA, Guix M, Arteaga CL, Caprioli RM. Cancer research. 2004;64:9093–9100. doi: 10.1158/0008-5472.CAN-04-2231. [DOI] [PubMed] [Google Scholar]
- 2.Walch A, Rauser S, Deininger SO, Hofler H. Histochemistry and cell biology. 2008;130:421–434. doi: 10.1007/s00418-008-0469-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H, Hummon AB. Analytical chemistry. 2011;83:8794–8801. doi: 10.1021/ac202356g. [DOI] [PubMed] [Google Scholar]
- 4.Khatib-Shahidi S, Andersson M, Herman JL, Gillespie TA, Caprioli RM. Analytical chemistry. 2006;78:6448–6456. doi: 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- 5.Weaver EM, Hummon AB. Advanced drug delivery reviews. 2013;65:1039–1055. doi: 10.1016/j.addr.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, Weaver EM, Hummon AB. Anal. Chem. 2013;85:6295–6302. doi: 10.1021/ac400519c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmerman TA, Rubakhin SS, Sweedler JV. Journal of the American Society for Mass Spectrometry. 2011;22:828–836. doi: 10.1007/s13361-011-0111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnes CA, Brison J, Robinson M, Graham DJ, Castner DG, Ratner BD. Analytical chemistry. 2012;84:893–900. doi: 10.1021/ac201179t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Debnath J, Brugge JS. Nature Reviews Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- 10.Weiswald LB, Richon S, Validire P, Briffod M, Lai-Kuen R, Cordelieres FP, Bertrand F, Dargere D, Massonnet G, Marangoni E, Gayet B, Pocard M, Bieche I, Poupon MF, Bellet D, Dangles-Marie V. British journal of cancer. 2009;101:473–482. doi: 10.1038/sj.bjc.6605173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirschhaeuser F, Menne H, Dittfeld C, West J, Mueller-Klieser W, Kunz-Schughart LA. Journal of Biotechnology. 2010;148:3–15. doi: 10.1016/j.jbiotec.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Friedrich J, Seidel C, Ebner R, Kunz-Schughart LA. Nature Protocols. 2009;4:309–324. doi: 10.1038/nprot.2008.226. [DOI] [PubMed] [Google Scholar]
- 13.Keithley RB, Weaver EM, Rosado AM, Metzinger MP, Hummon AB, Dovichi NJ. Analytical chemistry. 2013;85:8910–8918. doi: 10.1021/ac402262e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leibovitz A, Stinson JC, McCombs WB, 3rd, McCoy CE, Mazur KC, Mabry ND. Cancer research. 1976;36:4562–4569. [PubMed] [Google Scholar]
- 15.Sottoriva A, Kang H, Ma Z, Graham TA, Salomon MP, Zhao J, Marjoram P, Siegmund K, Press MF, Shibata D, Curtis C. Nature genetics. 2015;47:209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fogh J. Human tumor cells in vitro. Plenum Press; New York: 1975. [Google Scholar]
- 17.Trede D, Schiffler S, Becker M, Wirtz S, Steinhorst K, Strehlow J, Aichler M, Kobarg JH, Oetjen J, Dyatlov A, Heldmann S, Walch A, Thiele H, Maass P, Alexandrov T. Analytical chemistry. 2012;84:6079–6087. doi: 10.1021/ac300673y. [DOI] [PubMed] [Google Scholar]
- 18.Andersson M, Groseclose MR, Deutch AY, Caprioli RM. Nature methods. 2008;5:101–108. doi: 10.1038/nmeth1145. [DOI] [PubMed] [Google Scholar]
- 19.Lanni EJ, Rubakhin SS, Sweedler JV. J. Proteomics. 2012;75:5036–51. doi: 10.1016/j.jprot.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jerigova M, Biro C, Kirchnerova J, Chorvatova A, Chorvat D, Jr, Lorenc D, Velic D. Molecular imaging and biology : MIB : the official publication of the Academy of Molecular Imaging. 2011;13:1067–1076. doi: 10.1007/s11307-010-0460-4. [DOI] [PubMed] [Google Scholar]
- 21.Grey AC, Gelasco AK, Section J, Moreno-Rodriguez RA, Krug EL, Schey KL. Anat Rec (Hoboken) 2010;293:821–828. doi: 10.1002/ar.21103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fornai L, Angelini A, Klinkert I, Giskes F, Kiss A, Eijkel G, Amstalden-van Hove EA, Klerk LA, Fedrigo M, Pieraccini G, Moneti G, Valente M, Thiene G, Heeren RM. Analytical and bioanalytical chemistry. 2012;404:2927–2938. doi: 10.1007/s00216-012-6451-3. [DOI] [PubMed] [Google Scholar]
- 23.Brulet M, Seyer A, Edelman A, Brunelle A, Fritsch J, Ollero M, Laprevote O. Journal of lipid research. 2010;51:3034–3045. doi: 10.1194/jlr.M008870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonnell LA, Heeren RM. Mass spectrometry reviews. 2007;26:606–643. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 25.Jones EA, Deininger SO, Hogendoorn PC, Deelder AM, McDonnell LA. J. Proteomics. 2012;75:4962–89. doi: 10.1016/j.jprot.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 26.Norris JL, Cornett DS, Mobley JA, Andersson M, Seeley EH, Chaurand P, Caprioli RM. International journal of mass spectrometry. 2007;260:212–221. doi: 10.1016/j.ijms.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fonville JM, Carter C, Cloarec O, Nicholson JK, Lindon JC, Bunch J, Holmes E. Analytical chemistry. 2012;84:1310–1319. doi: 10.1021/ac201767g. [DOI] [PubMed] [Google Scholar]
- 28.Wagner MS, Graham DJ, Ratner BD, Castner DG. Surf Sci. 2004;570:78–97. [Google Scholar]
- 29.McCombie G, Staab D, Stoeckli M, Knochenmuss R. Analytical chemistry. 2005;77:6118–6124. doi: 10.1021/ac051081q. [DOI] [PubMed] [Google Scholar]
- 30.Bauer C, Cramer R, Schuchhardt J. Methods Mol Biol. 2011;696:341–352. doi: 10.1007/978-1-60761-987-1_22. [DOI] [PubMed] [Google Scholar]
- 31.Waldron KC, Dovichi NJ. Analytical chemistry. 1992;64:1396–1399. [Google Scholar]
- 32.Trede D, Kobarg JH, Oetjen J, Thiele H, Maass P, Alexandrov T. Journal of integrative bioinformatics. 2012;9:189. doi: 10.2390/biecoll-jib-2012-189. [DOI] [PubMed] [Google Scholar]
- 33.Alexandrov T, Meding S, Trede D, Kobarg JH, Balluff B, Walch A, Thiele H, Maass P. Journal of proteomics. 2011;75:237–245. doi: 10.1016/j.jprot.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Gemperline P. Practical guide to chemometrics. 2nd. CRC/Taylor & Francis; Boca Raton: 2006. [Google Scholar]
- 35.Li XF, Ren HJ, Le XC, Qi M, Ireland ID, Dovichi NJ. J Chromatogr A. 2000;869:375–384. doi: 10.1016/s0021-9673(99)00893-6. [DOI] [PubMed] [Google Scholar]
- 36.Kramer R. Chemometric techniques for quantitative analysis. Marcel Dekker; New York: 1998. [Google Scholar]
- 37.Vickerman JC, Briggs JCVD, Briggs D. TOF-SIMS: Materials Analysis by Mass Spectrometry. IM Publications; 2013. [Google Scholar]
- 38.Jolliffe IT. Principal component analysis. 2nd. Springer; New York: 2002. [Google Scholar]
- 39.Jackson JE. A user's guide to principal components. Wiley-Interscience; Hoboken, N.J: 2003. [Google Scholar]
- 40.Deininger SO, Ebert MP, Futterer A, Gerhard M, Rocken C. Journal of proteome research. 2008;7:5230–5236. doi: 10.1021/pr8005777. [DOI] [PubMed] [Google Scholar]
- 41.Beebe KR, Pell RJ, Seasholtz MB. Chemometrics : a practical guide. Wiley; New York: 1998. [Google Scholar]
- 42.Hanrahan G. Key concepts in environmental chemistry. Academic Press; Waltham, MA: 2012. [Google Scholar]
- 43.Hastie T, Tibshirani R, Friedman JH. The elements of statistical learning : data mining, inference, and prediction. 2nd. Springer; New York, NY: 2009. [Google Scholar]
- 44.Dixon SJ, Brereton RG. Chemometrics and Intelligent Laboratory Systems. 2009;95:1–17. [Google Scholar]
- 45.Tominaga Y. Chemometrics and Intelligent Laboratory Systems. 1999;49:105–115. [Google Scholar]
- 46.Kaletas BK, van der Wiel IM, Stauber J, Guzel C, Kros JM, Luider TM, Heeren RM. Proteomics. 2009;9:2622–2633. doi: 10.1002/pmic.200800364. [DOI] [PubMed] [Google Scholar]
- 47.Martinez WL, Martinez AR, Solka JL. Exploratory data analysis with MATLAB. 2nd. CRC Press; Boca Raton, Fla: 2011. [Google Scholar]
- 48.Matlab Central File Exchange. http://www.mathworks.com/matlabcentral/fileexchange/
- 49.Hendler RW, Shrager RI. Journal of biochemical and biophysical methods. 1994;28:1–33. doi: 10.1016/0165-022x(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 50.Cattell RB. Multivar Behav Res. 1966;1:245–276. doi: 10.1207/s15327906mbr0102_10. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.