Summary
Broadly neutralizing antibodies (bnAbs) directed to the V2 apex of the HIV envelope (Env) trimer isolated from individual HIV-infected donors potently neutralize diverse HIV strains, but strategies for designing immunogens to elicit bnAbs have not been identified. Here, we compared four prototypes (PG9, CH01, PGT145 and CAP256.VRC26.09) of V2 apex bnAbs and showed that all recognized a core epitope of basic V2 residues and the glycan-N160. Two prototype bnAbs were derived from VH-germlines that were 99% identical and used a common germline D-gene encoded YYD-motif to interact with the V2-epitope. We identified viruses that were neutralized by iGL from three prototype bnAbs and soluble Env derived from one of the isolates was shown to form a well-ordered Env trimer that mimics that on the surface of virions and could serve to initiate a V2-apex bnAb response. These studies illustrate a strategy to transition from panels of bnAbs to vaccine candidates.
Graphical abstract
INTRODUCTION
Much HIV vaccine research has recently begun to focus on how to induce broadly neutralizing antibodies (bnAbs), as they can neutralize multiple HIV strains and provide broad protection in animal models. There is general agreement that the study of bnAbs arising in natural infection is crucial in this endeavor (Burton and Mascola, 2015; Klein et al., 2013b). In particular, understanding how bnAbs from different individuals recognize the same target on the virus promises to yield valuable information for immunogen design and immunization strategies.
The surface HIV-1 envelope (Env) spike, a heterodimeric trimer (gp120-gp41)3, is the sole target of bnAbs (Julien et al., 2013a; Lyumkis et al., 2013; Pancera et al., 2014). Although, the Env spike employs a variety of mechanisms for immune evasion, bnAbs effective against a wide spectrum of viruses develop over time in natural infection (reviewed in Burton and Mascola, 2015). Thus far, several major target specificities of these bnAbs have been reported and mapped to various sites on Env, which include; the CD4 binding site (CD4bs), the second variable loop (V2) and the N160 glycan (V2 apex), the third variable loop (V3) and glycan N332 (N332-V3 or high-mannose patch) of gp120, the membrane proximal external region (MPER) of gp41 and finally the gp120/41 interface region (reviewed in Ward & Wilson, 2015). These antibodies are highly potent and broadly neutralizing against a diverse panel of HIV-1. Moreover, passive transfer experiments in animal models have shown bnAbs to be protective and therapeutic (reviewed in (Burton and Mascola, 2015; Klein et al., 2013b).
BnAbs typically take years to appear in natural HIV infection in contrast to strain-specific anti-Env neutralizing Abs, which emerge in the first few months of infection. Of the bnAbs, those directed to the V2 apex emerge relatively early (Doria-Rose et al., 2014; Moore et al., 2011; Wibmer et al., 2013). Further, these bnAbs are elicited relatively frequently compared to other bnAb specificities as shown in a number of studies involving large cohorts of HIV infected donors (Georgiev et al., 2013; Gray et al., 2011; Walker et al., 2010). In total, these findings suggest that the V2 apex region could serve as an important target for HIV vaccine development.
The V1V2 region is highly variable in terms of sequence, length, and the extent of glycosylation (Zolla-Pazner and Cardozo, 2010). Due to sequence variation, antibody responses to V1V2 tend to be strain specific and furthermore the loops appear to sterically obstruct antibody access to the CD4bs on the Env spike (Julien et al., 2013a; Pinter et al., 2004). Structurally, the V1V2 region forms a four anti-parallel β-stranded sheet or a five β-stranded barrel (McLellan et al., 2011; Pancera et al., 2013; Pancera et al., 2014). V1V2 stabilizes the Env spike forming the trimer apex (Julien et al., 2013a; Lyumkis et al., 2013; Pancera et al., 2014). Further, the V1V2 region harbors the epitopes that are recognized by V2 apex directed bnAbs (Bonsignori et al., 2011; Doria-Rose et al., 2014; Walker et al., 2011; Walker et al., 2009). Indeed, among the major vulnerable target sites for bnAbs on HIV Env, only Abs to the V2 apex display cross-neutralizing activity with viruses from other groups of HIV-1 and Simian Immunodeficiency Viruses (SIV), which infect gorillas and chimpanzees, suggesting a cross-group and cross-species conservation of this epitope (Barbian et al., 2015; Braibant et al., 2013).
To date, four prototypes of V2 apex bnAbs (PG9, CH01, PGT145, and CAP256.09) have been isolated from individual HIV-1 infected donors (Bonsignori et al., 2011; Doria-Rose et al., 2014; Walker et al., 2011; Walker et al., 2009). All recognize the N-linked glycan at residue 160 (N160) to varying degrees and a protein surface of the V2 domain of gp120. These antibodies are either trimer-specific or trimer-preferring, frequently binding with nM affinity to Env trimers but failing to show any significant binding to monomeric gp120 (Doria-Rose et al., 2014; Sanders et al., 2013; Sok et al., 2014; Walker et al., 2011; Walker et al., 2009). Since these bnAbs appear early in infection they frequently possess relatively low levels of somatic mutation compared to other bnAbs (Doria-Rose et al., 2014; Klein et al., 2013a; Walker et al., 2011; Walker et al., 2009; West et al., 2012). The antibodies are highly potent and broadly cross-reactive and do not generally display autoreactivity. An important characteristic of this group of antibodies is their extraordinarily long CDRH3 loop (≥24 amino acids), which helps them to penetrate the glycan shield on the Env spike trimer and access the protein surface below (Julien et al., 2013b; McLellan et al., 2011; Pejchal et al., 2010). The CDRH3 is anionic in nature and possesses sulfated tyrosines, except the CH01 Ab prototype, that appear to contribute to antibody binding (Pejchal et al., 2010).
In this study, we compared the four prototype V2 apex bnAbs in terms of the epitopes recognized and characteristics of the Abs using a variety of approaches including mutagenesis of the viral Env and antibody engineering. We highlighted common features of Env recognized by all the bnAb prototypes and showed that two of the prototypes possessed some common features that were present in inferred germline versions of the antibodies. We identified immunogens that bound to inferred germline Abs and propose potential immunization strategies that will facilitate HIV vaccine development.
RESULTS
The N-linked glycan at N160 and lysine-rich strand C residues in the V2 domain form a core epitope for all V2 apex prototype bnAbs
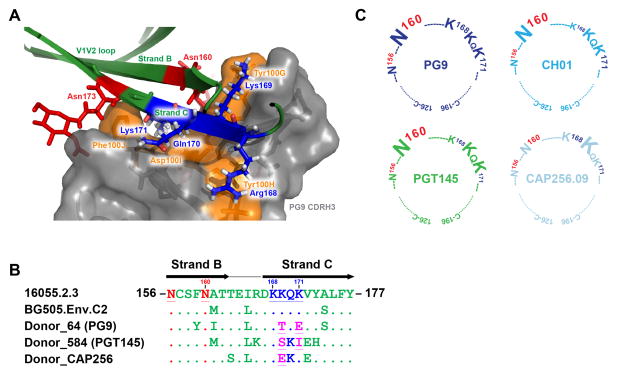
The trimeric nature of the V2 apex bnAb epitopes has hampered their full structural characterization and determination of the core epitope on the Env spike. The co-crystal structures of PG9 and PG16 antibodies with ZM109F.PB4 and CAP45.00.G3 V1V2 scaffolds shows that PG9 and PG16 CDRH3s primarily interact with N-linked glycans at N160 and N156 or N173 and positively charged amino acid residues in strand C (KKQK: HXB2 numbering 168–171) of the V2 domain (Figure 1A)(McLellan et al., 2011; Pancera et al., 2013). Studies have shown that neutralization mediated by V2 apex bnAbs is highly dependent on the overall charge in strand C of the V2 domain (Doores and Burton, 2010; Doria-Rose et al., 2012). Additionally, a recent longitudinal study of donor CAP256 (from whom CAP256-VRC26.09 Ab was isolated, which we refer here as CAP256.09) yielded a neutralization escape virus with mutation at the K169 residue of strand C (Figure 1B) (Doria-Rose et al., 2014). In this study, we isolated viruses from two IAVI protocol G HIV infected donors, 64 and 584, from whom PG9 and PGT145 bnAbs were isolated respectively (Walker et al., 2011; Walker et al., 2009). We found that these viruses were resistant to neutralization by PG9 and PGT145 bnAbs (data not shown), indicative of viral escape. The sequence analysis of these escape variants revealed mutations at the K169 and K171 strand C residues (Figure 1B), again suggestive of antibody selection pressure in this region (Moore et al., 2011; Moore et al., 2013).
Figure 1. Epitope recognition by V2 apex bnAbs (See also Figure S1 and Table S1).
A. PG9 CDRH3 interaction with glycans and strand B and C of the V1V2 domain presented on a scaffold shown as a ribbon representation (modified from (McLellan et al., 2011)). PG9 CDRH3-interacting amino acid residues are shown in orange with side chains as sticks. The V2 glycans N160 and N173 are depicted in red, and the four key residues from the lysine-rich strand C are shown in blue with side chains as sticks.
B. Amino acid sequence alignment of strands B and C of isolate 16055.2.3, BG505.Env.C2, IAVI donor 64 PG9 antibody escape virus, IAVI donor 584 PGT145 antibody escape virus and donor CAP256 CAP256.09 antibody escape virus (HXB2 numbering: 156–177). The sequences were aligned with clustalX in the BioEdit program. The N-linked glycan sites N156 and N160 are shown in red, the four amino acids in the lysine rich region on strand C are shown in blue and the escape mutations at position 169 and 171 in the donor escape viruses are highlighted in pink.
C. Graphic representation of the epitopes for four (PG9, CH01, PGT145 and CAP256.09) V2 apex bnAb prototypes. Each cartoon represents the cysteine-linked V1V2 loops (HXB2: 126–196), highlighting the core epitope for all the V2 apex bnAb prototypes. The epitope includes the glycans N156, N160 and four strand C amino acid residues. The size of each amino acid letter is proportional to the dependence on that residue for neutralization.
Based on these observations, we generated a series of variants of the 16055.2.3 Env by substitution of residues in the strand C region and elimination of the glycans at N156 and N160. The Env was chosen as it derives from a subtype C virus that is sensitive to neutralization by all four V2 apex bnAb prototypes. The type of substitutions made were chosen on the basis of amino acid variability around the strand C residues in different HIV isolates and included the escape mutations at residues K169T and K171E found in the PG9 donor escape virus (Figure S1, Table S1, Figure 1A). We tested two Abs from each V2 apex bnAb prototype against the Env pseudovirus variants in the standard TZM-bl cell neutralization assay (Seaman et al., 2010). The results revealed that all of the V2 apex antibody prototypes preferred positively charged residues at positions 168, 169 and 171 (HXB2 numbering). Apart from the CAP256.09 Ab prototype, which seemed to be more dependent on K169, a combination of two substitutions was required to completely abrogate neutralizing activity, suggesting avid antibody binding to the epitope (Table S1). Further, the N160 glycan was obligatory for all V2 apex bnAbs except the CAP256.09 prototype, which showed a partial dependence on N160, in agreement with earlier findings (Doores and Burton, 2010; Doria-Rose et al., 2014; Walker et al., 2011; Walker et al., 2009). To rule out any virus envelope-specific bias, we tested the most critical mutations on a subtype A HIV envelope, BG505.Env.C2 and found a similar trend (Table S1). Overall, we observed that all of the V2 apex bnAb prototypes were sensitive to mutations at glycan N160 and four strand C residues with subtle differences in terms of their epitope requirement, K169 being the common requisite for all prototypes (Figure 1C).
The glycan at N160 is crucial for all V2 apex bnAbs but there are differences in fine specificity for glycan recognition
In order to investigate the nature of glycans recognized by the V2 apex bnAbs, we employed three approaches. First, we generated knock-outs of the glycans at N156 and N160 in the BG505.Env.C2 SOSIP.664 soluble Env trimer to complement those previously generated in the 16055.2.3 virus and shown in figure 1. We tested two Abs from each V2 apex bnAb prototype against the Env trimer variants for ELISA binding. Consistent with the 16055.2.3 virus neutralization results, binding of the V2 apex bnAbs was highly dependent on the N160 glycan (Figure 2A, B, Table S2). The exception was the CAP256.09 Ab prototype, whose binding showed partial dependence on N160. Second, we produced the 16055.2.3 virus and soluble SOSIP.664 Env trimer in the presence of the glycosidase inhibitors kifunensine (ER α - mannosidase I inhibitor) and swainsonine (Golgi α - mannosidase II inhibitor). Kifunensine and swainsonine are expected to modify glycan processing of sugars on the Env trimer to yield mostly high mannose (Man9GlcNAc2) and hybrid-type glycans with a Man5GlcNAc2 core, respectively (Doores and Burton, 2010). The results revealed that neutralization by all the V2 apex bnAbs was generally sensitive to kifunensine treatment in an isolate and context dependent manner as observed previously (Doores and Burton, 2010; Doria-Rose et al., 2014; Walker et al., 2011). Swainsonine treatment had minimal effects (Figure 2A, B, Table S2). These results indicated that V2 apex bnAbs were possibly unable to tolerate extra high-mannose sugars (Man9GlcNAc2) at glycan N160. Third, we assessed the reactivity of the bnAbs to different types of glycans on glycan microarrays (Table S3). Antibodies from PG9 and CAP256.09 Ab prototypes, showed reactivity to glycans while PGT145 and CH01 Ab prototypes did not display any binding. PG9 and CAP256.09 Ab prototypes showed reactivity to a variety of diverse glycans with terminally linked α-2–3 or α-2–6 sialic acid residues (Figure 2C, Table S3). Notably, PG9 Ab bound preferentially to glycans with a α-2–3 linked sialic acid while PG16 preferred a α-2–6 linkage, consistent with previous findings (Pancera et al., 2013). Overall, the results reveal that glycan at position N160 is important for all V2 apex bnAbs and two of the antibody prototypes PG9 and CAP256.09 can bind diverse glycan types with sialic acid as the terminal sugar.
Figure 2. Glycan specificity of V2 apex bnAbs (See also Tables S2-S3).
A. V2 apex bnAb neutralization of Env variants displaying differing glycan composition in the context of the 16055.2.3 isolate. Two Abs from each V2 apex bnAb prototype were tested for neutralization against 16055.2.3 wild type (WT), N156K, N160K glycan variants, and viruses produced in the presence of glycosidase inhibitors kifunensine (Kif) and swainsonine (Swain). B. The four V2 apex bnAb prototypes were tested in ELISA binding to Gallanthus Nivalis Lectin (GNL)-purified BG505.Env.C2.SOSIP.664 soluble trimer glycovariants. The gp120-gp41 glycan dependent Ab PGT151, the CD4bs Ab VRC01 and a Dengue Ab (DEN3) were used as controls.
C. Reactivity of V2 apex bnAbs with glycans on a glycan microarray. The V2 apex bnAbs were tested for reactivity on a glycan array. The data are shown as Relative Fluorescence Units (RFU) and the error bars represent the average percentage error for all data points reported. The symbol for each monosaccharide is indicated. The glycan dependent antibody PGT151 was used as control.
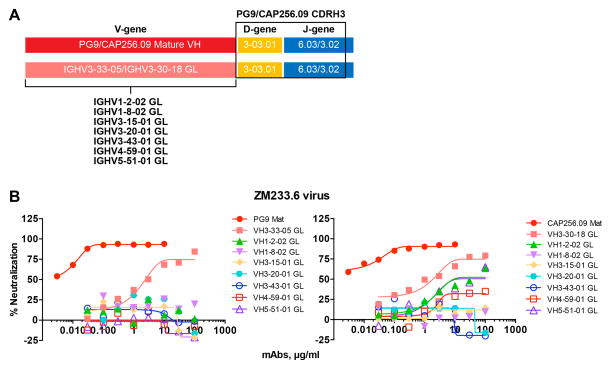
PG9/PG16 and CAP256.09 are derived from similar VH germline-gene families and contain a conserved YYD motif in CDRH3 that interacts with the V2 domain
In order to identify common features used by different V2 apex bnAbs, we compared the closest inferred variable heavy (VH) and variable light (VL) chain germline sequences (predicted by the ImMunoGeneTics (IMGT) human immunoglobulin G (IgG) germline database) (Brochet et al., 2008), (http://www.imgt.org) from each prototype. We found that PG9/PG16 and CAP256.09 antibodies were derived from different VH-germline gene families but these families shared 99% sequence identity at the amino acid level as depicted in Figure 3A (Figure S2A). In addition, the D-gene usage for these two Ab prototypes was identical (Figure 3A, Table S4). The CH01 and PGT145 Ab prototypes were derived from different VH gene families with many differences in amino acid sequence (Figure 3A, Table S4). The VL germline sequences for all the Ab prototypes were derived from diverse gene families (Table S4). The extent of somatic mutation for different Ab prototypes ranged from 14 to 17% in VH and from 9 to 17% in VL chains at nucleotide level (Table S4). Comparison of CDRH3 sequences showed conservation of a tyrosine-tyrosine-aspartic acid (YYD) motif in PG9, PG16 and CAP256.09 Abs (Figure 3B). The YYD motif in these antibodies originated from the germline D-gene and remained unchanged in the mature antibody (Brochet et al., 2008; Doria-Rose et al., 2014; Walker et al., 2009). We created single amino acid substitutions in PG9 and PG16 antibodies in the YYD motif and tested for the ability of the mutant antibodies to neutralize four viruses with varying degrees of neutralization sensitivity. The results showed partial or complete loss of antibody neutralizing activity (Figure 3C). Substitution of the D112.8 (IMGT numbering) residue showed the most profound effect on antibody activity. A similar result was observed on the ELISA binding capacity of these antibody variants to recombinant gp120 proteins when this could be assessed (Figure 3D). Overall, these results suggest that PG9, PG16 and CAP256.09 Abs were derived from similar VH genes and possess a motif in their CDRH3 encoded by a common germline-D region. PG9 Ab structural studies have shown this motif (McLellan et al., 2011; Pancera et al., 2013) to recognize the basic strand C residues in the V2 domain, however it remains to be seen whether CAP256.09 Ab structurally uses a similar or distinct mode for epitope recognition.
Figure 3. Variable (V), Diversity (D) and Joining (J) gene families encoding V2 apex bnAbs and conservation of the YYD motif in PG9 and CAP256.09 prototype Abs (See also Figure S2 and Table S4).
A. Representation based on the amino acid sequence alignment of the inferred germline-encoding heavy chain variable regions (VH). The V, D and J gene usage for various V2 apex bnAb prototypes (denoted on the right) is listed in the boxes. The V-gene sequence alignment shows that PG9 and CAP256.09 Ab prototypes, derived from different gene families share 99% sequence similarity at amino acid level (shown in red) and use same germline D-gene. Different gene families encode the J-genes for each of the Ab prototypes. Abs possess (N)-nucleotide insertions of variable lengths at V-D and D-J junctions in the CDRH3.
B. Sequence alignment of CDRH3 sequences of V2 apex bnAbs shows conservation of the YYD motif (underlined bold) in PG9, PG16 and CAP256.09 antibody prototypes. IMGT numbering is shown.
C. The effects of substitutions in the YYD motif amino acid residues in PG9 and PG16 Abs on neutralization of viruses. The IC50 neutralization titers of mature (WT) PG9 and PG16 Abs and variant antibodies against a 4-virus panel are represented as bars.
D. Binding of WT PG9 and PG16 Abs and their variants to recombinant GNL-purified gp120 proteins by ELISA. Antibodies to the high mannose patch (PGT128), CD4bs (b6, b12 and PGV04) and Dengue (DEN3) were used as control mAbs.
Inferred germline (iGL) versions of V2 apex bnAbs can neutralize a number of viruses
Several studies indicate the importance of a long CDRH3 for binding of the V2 apex bnAbs to Env and for neutralization. Indeed, the PG9 and PG16 antibody CDRH3s are thought to form a subdomain (“hammerhead”) that permits these antibodies to penetrate through the glycan shield to reach the protein surface (McLellan et al., 2011; Pancera et al., 2010; Pejchal et al., 2010). Further, the ability to exchange the CDRH3s of PG9 and PG16 with exchange of neutralization specificity has previously been shown (Pancera et al., 2010; Pejchal et al., 2010). As one approach to investigating the role of CDRH3 and other antibody sequence elements in V2 apex bnAbs in general, we generated a series of VH and/or VL chain reverted inferred germline (iGL) antibodies according to the schematic shown in Figure 4A in which the mature CDRH3 and CDRL3 were retained. Using the mature CDRH3s in the reverted Abs remains one of the limitations of this study. These Abs, referred to as “iGL Abs”, were generated by replacing the VH or VL gene segments in the corresponding mature (Mat) antibodies with their closest iGL sequence (IMGT). The antibodies were tested against a panel of 18 tier 2 viruses from different HIV clades (Table S5). We found that PG9, CH01 and CAP256.09 VH-VL iGL Ab prototypes exhibited neutralization of a number of viruses while PGT145 did not. There was a rough inverse correlation between the number of somatic mutations in the VH and VL regions and neutralization IC50 but with considerable variation so that potent neutralization was seen on occasion with no or a limited number of mutations, especially for the CH01 and CAP256.09 Abs (Figure 4B, 4C) (Table S5). Since the iGL Abs have mature CDRH3s, Figure 4C illustrates that the breadth of PGT145 neutralization was particularly dependent on somatic mutations in the VH and VL chains. The inferred GL Abs showed similar glycan dependence in virus neutralization as compared to their mature versions (Table S6) and did not display autoreactivity (Figure S4). To gain more insight about the role of somatic mutations in the PGT145 Ab prototype, we compared the PGT143 and PGT145 mature VH sequences with their germline sequence and identified a substantial number of somatic mutations in the CDRH1, CDRH2 and FR3 regions that could be important for antibody neutralization (Figure S3A).
Figure 4. Neutralization breadth and potency of mature, chimeric and inferred germline reverted V2 apex bnAbs (See also Figures S3-S4 and Table S5-S6).
A. A schematic representation of chimeric and inferred germline (iGL) reverted antibodies. The mature antibody, referred to as ‘Mat’ shows constant region (grey), the variable heavy (VH) chain (red) and variable light (VL) chain (green). The third Complementarity Determining Regions of VH (CDRH3) and VL (CDRL3) chains are shown at the end of each variable region. The VH and VL genes are further divided into framework regions (FR1-3) and other CDRs (CDR1-2) shown with ImMunoGeneTics (IMGT) numbering. To generate the VH or VL regions in iGL Abs were replaced in with their closest inferred germline sequence predicted by in the IMGT human IgG germline database (Brochet et al., 2008). The CDRH3 and CDRL3 from the Mat antibodies were retained in the reverted iGL Abs.
B. Association of IC50 neutralization titers with number of somatic mutations in VH and VL for the four prototype Abs, PG9, CH01, CAP256.09 and PGT145.
C. Neutralization breadth of Mat, chimeric and iGL reverted V2 apex bnAbs. The number of viruses in an 18-virus panel neutralized with an IC50 <10 μg/ml is shown for Mat and chimeric Abs. For the less potent iGL Abs, neutralization is scored as positive with IC50 <100 μg/ml.
To evaluate the role of these mutations, we generated PGT145 antibody variants by independently replacing the three regions described in the mature VH with the corresponding germline sequence, paired with mature VL, and tested the resulting antibodies for neutralization. The results revealed a strong dependence of PGT145 antibody activity on the CDRH2 region and a similar effect was confirmed for PGT143 (Figure S3B). Overall, these results suggest that the PGT145 Abs represent a discrete group of V2 apex bnAbs.
Neutralization of isolates by iGL versions of PG9 and CAP256.09 is not due to the CDRH3 alone
The iGL reverted versions of PG9 and CH01 Abs neutralized only a fraction of the viruses that were neutralized by their respective mature Abs, which highlights the significance of somatic mutations in the variable HC and LC regions for Ab function. However, the CAP256.09 iGL reverted Ab, albeit less potently, neutralized all the viruses that were sensitive to its mature version in the panel used, implying that the neutralization activity could be almost entirely mediated by CDRH3. To investigate the role of CDRH3 and the VH germline gene segment in V2 apex bnAb neutralization, we employed antibody engineering to generate a panel of PG9 and CAP256.09 iGL Ab variants with diverse VH-GL genes and a common CDRH3 from either PG9 or CAP256.09 that are termed “inferred germline-VH Ab variants” (Figure 5A). The choice of VH GL genes was based on the gene families used by HIV bnAbs targeting different sites on Env (Bonsignori et al., 2011; Gorny et al., 2005; Walker et al., 2011; Walker et al., 2009; West et al., 2012). We paired these VH iGL variants with the respective Ab (PG9 or CAP256.09) light chain iGLs and tested them against ZM233M.PB6 as a prototype virus since it was effectively neutralized by both iGL Abs. We observed a complete loss of neutralization for PG9 VH-GL Ab variants while some of the CAP256.09 VH-GL Ab variants neutralized the virus with very low potency (Figure 5B). The results suggest that it is a combination of VH germline and CDRH3 are required for the corresponding germline antibody activity.
Figure 5. Neutralization of ZM233M.PB6 by PG9 and CAP256.09 heavy chain inferred germline antibody variants (See also Table S7).
A. Schematic representation of PG9 and CAP256.09 inferred germline-VH Ab variants. The PG9 or CAP256.09 VH-germline gene was replaced by germline-VH encoding sequences from diverse HIV-1 specific Abs. The selected germline genes are used by envelope specific bnAbs that include IGHV1-2-02 (CD4bs), IGHV1-8-02, IGHV3-15-01, IGHV3-20-01, IGHV3-43-01 (V2 apex), IGHV4-59-01 (N332-V3), IGHV5-51-01 (V3).
B. Neutralization of ZM233M.PB6 virus by PG9 and CAP256.09 iGL variant antibodies in which the VH germline has been substituted as in (A) above.
To further validate this finding, we transplanted the CDRH3 regions from mature PG9 and CAP256.09 antibodies on to the framework of different V2 apex mature antibodies (2909, PGT145, PG9 or CAP256.09) (Doria-Rose et al., 2014; Gorny et al., 2005; Walker et al., 2011; Walker et al., 2009). The basis for using these antibodies as scaffolds for PG9 and CAP256.09 CDRH3 was to partially retain the configuration of antibody structure that would naturally align with the quaternary V2 epitope. Furthermore, these antibodies use different somatic mutation patterns in their VH and VL chains that, in concert with the respective CDRH3, define their mode of neutralization, and thus can provide a means to assess the precise role of CDRH3. For example, the antibody 2909 can only neutralize viruses that lack a glycan at the N160 position (Gorny et al., 2005). Hence neutralization of any virus that has a glycan at N160 by antibody 2909 in which the native CDRH3 has been replaced with that from PG9 or CAP256.09 can be attributed to the transplanted CDRH3. A panel of transplanted antibodies with PG9 and CAP256.09 CDRH3s showed no neutralizing activity against a panel of viruses with a single exception (Table S7). Together, these results suggest that the VH germline genes in PG9 and CAP256.09 iGL Abs play a vital role in neutralization of sensitive viruses and their ability to neutralize these viruses is not a function of CDRH3 alone.
Common patterns of somatic mutation in PG9, PG16 and CAP256.09 contribute to broad neutralization
The fact that two of the V2 apex bnAb prototypes, PG9, PG16 and CAP256.09, are derived from almost identical VH germline precursor gene sequences at the amino acid level implies the selection of VH structural features that favor interaction with a V2 antigenic structure. These features may be present in the germline configurations of the antibodies and/or emerge during affinity maturation. To investigate, we compared the somatic mutation patterns in the VH genes of PG9, PG16 and CAP256.09 by aligning the mature HC sequences with their respective iGL sequences. The alignment revealed 8 common positions in VH at which all 3 bnAbs had mutations away from germ line. For three of these positions, namely Y67, K72 and Y103 in the FR3 region, common mutations, to yield H67, W72 and F103 were noted in all three antibodies (Figure 6A). To confirm the role of somatic mutation in neutralization, we incorporated the mutations observed in PG9 at the 8 positions described into the PG9 heavy chain iGL, “PG9 VH iGL-modified Ab”, and found up to a 46-fold improvement in IC50 neutralization for 4 viruses that PG9 antibody neutralized as an iGL Ab (Figure 6C, Figure S5). In addition, the VH iGL-modified PG9 Ab gained 50% (IC50) neutralizing activity against two new viruses (CAP45.00.G3 and CRF02_AG_250) suggesting that the common sites of somatic mutation in VH in PG9, PG16 and CAP256.09 Abs were important for their function. Since PG9 LC somatic mutations were essential for antibody neutralizing activity (Figure 4B, C), we hypothesized that combining the somatic mutations in VH with VL mutations might have a complementary effect, especially residues E37, S38 and D56 that are critical for glycan recognition (McLellan et al., 2011; Pancera et al., 2013). Hence, we generated a PG9 iGL variant “PG9 VH-VL iGL-modified Ab” in which 8 mutations were introduced into both the HC and the LC of iGL PG9 (Figure 6B). Since CAP256.09 was encoded by a different LC germline gene, it was not investigated. Glycan reactivity was partially restored in the PG9 VH-VL iGL-modified Ab variant, as determined by glycan microarray (Figure S5), and the variant neutralized 89% (16/18) of virus isolates, albeit less potently than the mature PG9 antibody, in spite of having only about half the level of SHM of the mature antibody (Figure 6C, Figure S5). The somatic mutations in VH and VL chains of PG9 are depicted as surface rendering in Figure 6D. The location of somatic mutations in the VH and VL chains suggests that the gain of antibody function can be attributed to either a stabilizing effect or to direct interaction with the V2 epitope, possibly with glycans not fully described in the scaffold structures. Overall, the findings suggest that V2 apex bnAbs, particularly, PG9 and CAP256.09 Ab prototypes, use similar, antigen-driven developmental pathways to accumulate somatic mutations.
Figure 6. Comparison of mutations in PG9 and CAP256.09 contributing to broad neutralization (See also Figure S5).
A–B. Sequence alignment of mature heavy (A) and light (B) chain sequences of PG9, PG16 and CAP256.09 antibodies with their corresponding inferred germline (iGL) encoding sequences. Each VH and VL gene sequence is divided into FR1-3 and CDR1-2 with junctions underlined. A. Common positions at which somatic mutations occur in the mature HC of PG9, PG16 and CAP256.09 Abs are highlighted in cyan, common mutations shown in red. B. Common somatic mutations at 8 positions in mature PG9 and PG16 VL are highlighted in cyan. E37 in PG9, which is D in PG16, essential for glycan recognition (McLellan et al., 2011; Pancera et al., 2013) is included. C. The VH or VH-VL iGL-modified PG9 Ab variants were tested against the 18-virus panel and the median IC50 neutralization titers are shown for each.
D. Ribbon representation of PG9 antibody in complex with a ZM109F V1V2 loop (green) scaffold (modified from (McLellan et al., 2011)). The glycans at N160 and N173 positions (red; represented as sticks) and strand C residues (blue) in V2 domain are labeled. The PG9 variable heavy (cyan) and light (aqua) chains illustrate common positions (shown as surface rendering) at which somatic mutations occur in PG9, PG16 and CAP256.09 HC (denoted by residues in black) and PG9 and PG16 LC (denoted by residues in red).
The CRF02_AG_250 virus is neutralized by 3 iGL prototype bnAbs and a well-ordered trimer Env from this isolate can be constructed that is a potential immunogen
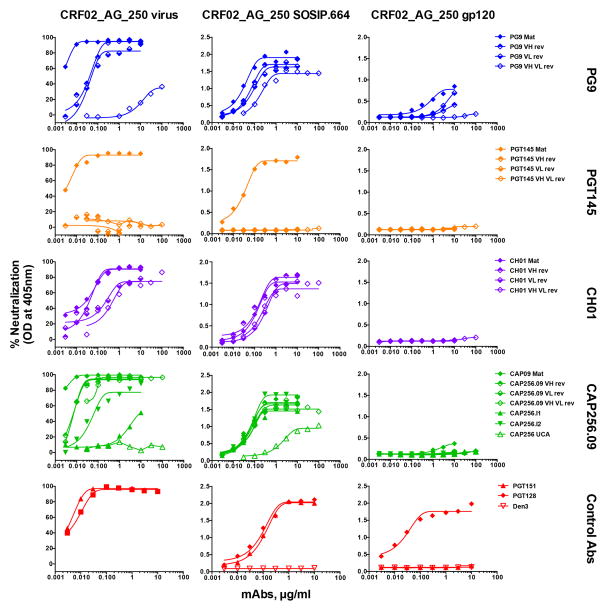
In this study, we identified 5 HIV isolates across different HIV clades that are neutralized by iGL Abs from two or more V2 apex bnAb prototypes (Table S5). Of the 5 HIV isolates, one isolate, ZM233M.PB6, has been previously shown to be sensitive to PG9 and CH01 iGL Ab prototypes (Bonsignori et al., 2011; Pancera et al., 2010). Three of the isolates, CRF02_AG_250 (subtype AG), ZM233M.PB6 (subtype C) and C1080.c03 (subtype AE), were neutralized by 3 of the 4 iGL Ab prototypes (PG9, CH01 and CAP256.09) and the neutralization curves are presented for CRF02_AG_250 virus (Fig 7A (left)). These results suggest that the CRF02_AG_250, ZM233M.PB6 and C1080.c03 isolates presented the V2 epitope differently as compared to other isolates that are equivalently sensitive to neutralization by the mature V2 apex bnAbs but were not neutralized by the corresponding iGL Abs (Table S5). The unusual sensitivity of these isolates to neutralization by iGL Abs from different V2 apex Ab prototypes suggests that the corresponding Env trimers may be useful in priming appropriate B-cell responses. Accordingly, we decided to investigate a well-ordered recombinant Env trimer from isolate CRF02_AG_250.
Figure 7. The CRF02_AG_250 virus is neutralized by 3 iGL prototype bnAbs and forms well-ordered recombinant Env trimers. (See also Figure S6-S7).
A. V2 apex bnAb neutralization of CRF02_AG_250 virus and binding to the corresponding SOSIP.664 and gp120 proteins. (A. Left column) the mature (Mat), VH or VL reverted chimeric and VH-VL iGL antibodies from each Ab prototype, an inferred unmutated common ancestor (CAP256 UCA) and two intermediate (CAP256.I1 and CAP256.I2) Abs derived from the CAP256 donor, were tested for neutralization against the CRF02_AG_250 virus. The neutralization curves show the activity of Abs, Mat (filled diamond), VH or VL reverted chimeras (half-filled diamond), VH-VL iGL (empty diamond). (A. Middle column) Abs were assessed for binding to the PGT145 antibody affinity purified soluble trimeric Env form (SOSIP.664) of CRF02_AG_250 by ELISA. (A. Right column) Abs were further tested for binding to the GNL-affinity purified monomeric gp120 protein corresponding to CRF02_AG_250 envelope, by ELISA. Antibodies, PGT128, PGT151 and DEN3 were used as controls.
We constructed a SOSIP.664 Env trimer from the CRF02_AG_250 sequence and purified this protein with a trimer-specific Ab PGT145 affinity column to enrich for a well-ordered Env trimer population. PGT145 antibody is a strictly trimer-specific antibody and enriches for highly homogeneous and consistently folded well-ordered native-like envelope trimers (Sanders et al., 2013). ELISA binding titrations revealed that the iGL Abs exhibited a strong reactivity with the CRF02_AG_250 SOSIP.664 trimer (Figure 7A (middle)). In addition, the CRF02_AG_250 SOSIP.664 trimer displayed strong binding to the CAP256 derived less mutated Ab intermediates, CAP256.I1 and CAP256.I2, and an inferred unmutated common ancestor Ab (CAP256 UCA) that was identified by 454 next-generation sequencing (NGS) from the CAP256 donor and is believed to have initiated the CAP256 Ab lineage (Doria-Rose et al., 2014). Remarkably, CRF02_AG_250 envelope showed notable similarities to CAP256 superinfecting donor virus (CAP256.SU), including amino acid sequence similarity in the strand B and C of V2 loop and the neutralization sensitivity pattern to various members of the CAP256 family of Abs (Figure S6, S7). We also assessed the binding of iGL Abs to monomeric gp120 from the CRF02_AG_250 isolate; with the exception of some weak binding by mature PG9 (and its chimeric variants), the mature and reverted iGL Abs from all 4 antibody prototypes failed to bind the gp120 protein (Figure 7A (right)). Finally, to evaluate the antigenic profile of the CRF02_AG_250 SOSIP.664 trimer, we tested its binding to a range of neutralizing and non-neutralizing Abs (Figure S7). The CRF02_AG_250 trimer showed little or no reactivity with non-neutralizing Abs but robust binding to neutralizing Abs consistent with a well-ordered trimer configuration (Sanders et al., 2013). The binding pattern of Abs to the SOSIP.664 trimer was highly correlated with virus neutralization (Figure S7) and overall the data suggests that the trimer is a promising immunogen to prime V2 apex bnAb responses.
DISCUSSION
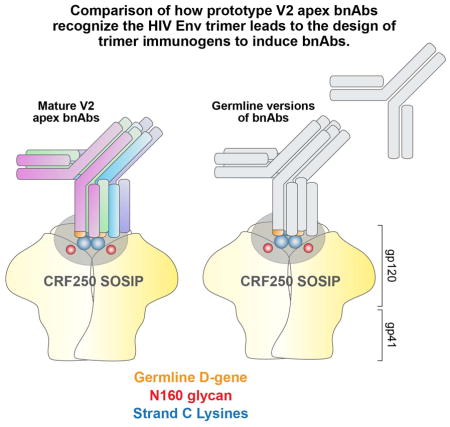
The interaction of bnAbs with Env trimer offers valuable information on which to base rational HIV vaccine design strategies. Study of multiple mAbs isolated from several infected donors provides an opportunity to gain a better understanding of the requirements for recognition of a given broadly neutralizing region and should lead to more reliable immunogen design (Jardine et al., 2013; Zhou et al., 2013). In essence, a better appreciation is gained of the features in the immunogen and the bnAb that are most critical for neutralization are obtained allowing for greater precision in immuogen design. The first of the new generation of more potent HIV bnAbs targeted an epitope associated with the V2 region of the Env trimer (Walker et al., 2009) and since then it has become clear that this region is a major target for bnAbs in natural HIV infection. It is estimated that roughly 21–42% of bnAbs in several cohorts are reactive with the region (Georgiev et al., 2013; Gray et al., 2011; Walker et al., 2010) and several bnAbs have been isolated (Bonsignori et al., 2011; Doria-Rose et al., 2014; Sok et al., 2014; Walker et al., 2011; Walker et al., 2009). In this study, we have compared bnAbs to the region, determined common features of Abs and Ab recognition and designed immunogens that may favor triggering of B cell responses to the region.
Structural studies have shown that the V2 bnAbs target a quaternary epitope at the apex of the Env trimer and we refer to them here as “V2 apex bnAbs”. We show that all 4 prototypes of V2 apex bnAbs bind to a core protein epitope involving the lysine-rich beta-strand C in the V2 domain, with Lys169 being the common requirement, and an N-linked glycan at position 160. Interestingly, within a relatively conserved region of V2 domain (HXB2: 155–183 (http://www.hiv.lanl.gov)), the Lys169 and Lys171 residues in strand C, which form the core epitope for V2 apex bnAbs, are the most variable residues in viral isolates across diverse HIV-1 subtypes (Figure S1). Thus the V2 apex bnAbs have acquired the ability to recognize, within certain boundaries, variation in the core epitope. Presumably, a successful immunization regime therefore would need to induce this ability, possibly by use of multiple immunogens with differing core protein epitopes.
Access to the core V2 protein epitope is reached by penetrating between glycans, notably glycans at N160 and N156. Structural and functional studies have previously shown that PG9 and PG16 antibodies make extensive contacts with Man5GlcNAc2 at N160 and a hybrid-type glycan at N156 or N173 positions (Doores and Burton, 2010; McLellan et al., 2011; Pancera et al., 2013; Walker et al., 2009). The PG9 and PG16 clonal variants have been shown to subtly differ in terms of their preference for the terminal glycosidic linkages (Pancera et al., 2013). The two prototypes of V2 apex bnAbs, PG9 and CAP256.09, appear to recognize a large spectrum of glycans with terminally linked α-2–3 or α-2–6 sialic acid residues. These findings suggest that these antibodies have evolved to recognize diverse sugars to counter the glycan diversity, which the virus uses for immune evasion.
The structural feature of the V2 apex bnAbs that allows penetration of the glycan shield to reach the core protein epitope is an extraordinarily long CDRH3 loop (McLellan et al., 2011; Pejchal et al., 2010). Therefore it might have been anticipated that the inferred germline antibodies (iGL) from three V2 apex bnAb prototypes (PG9, CAP256.09 and CH01) that maintain the CDRH3 from the mature antibody are able to neutralize some viruses, consistent with the importance of the CDRH3 for antibody activity. In contrast, despite having a long CDRH3, PGT145 was shown to be also dependent on CDRH2 and the VL chain. Overall, it appears that the PGT145 prototype Ab is distinct from the other 3 Ab prototypes. Further, the 3 V2 apex bnAb prototypes show higher levels of somatic mutation in their HCs as compared to LCs, whereas PGT145 shows similar levels in both chains (Table S4). In general, it seems the induction of PGT145-like Abs, which includes some of the most potent V2 apex bnAbs (Sok et al., 2014; Walker et al., 2011), may require immunization strategies distinct from those for the other 3 prototypes.
Within the group of 3 prototype bnAbs described above, PG9 and CAP256.09 show many notable similarities. The two prototypes are derived from distinct VH germ line gene families but with almost identical amino acid sequences. Inferred germ line versions (iGL) of these Abs do neutralize a number of isolates. Formally, the occurrence of these VH genes could have been random or due to selection by a V2 apex structure because of the strong neutralization dependence on CDRH3, which is not encoded by the VH gene. Antibody engineering experiments strongly supported the selection theory since replacement of the VH germline gene in iGL versions of PG9 and CAP256.09 with other VH genes was not associated with neutralizing activity. Presumably a B-cell receptor (BCR) shape associated with this V gene favorably interacts with a part of the V2 apex epitope. Epitope-specific VH germ line gene preference has been previously observed for other HIV envelope target sites (Jardine et al., 2013; Klein et al., 2013a).
As well as having a 99% amino acid identical germ line VH gene, PG9 and CAP256.09 also both retain a conserved tripeptide motif (YYD) in their CDRH3, which originates from the same germline D-gene and remains unmodified in the mature antibodies. We observed this motif to be essential for antibody neutralization and it has been shown previously to be critical for interaction with the Lys-rich strand C residues (McLellan et al., 2011; Pancera et al., 2013). Therefore overall, it appears that PG9 and CAP256.09- like responses could be initiated when the appropriate V gene in combination with a long CDRH3 incorporating the appropriate D gene is triggered. Following triggering, there is a suggestion from study of PG9 and CAP256.09 of a common maturation pathway as observed in a number of somatic mutations at common positions in both VH genes. A rationally designed immunogen employed following triggering might seek to shepherd maturation along this pathway.
Considering the initiation of a V2 apex bnAb response, we showed here that CRF02_AG_250, a subtype AG isolate, is recognized by 3 V2 apex iGL Ab prototypes. Additionally, in this study, we identified a number of envelopes that displayed an antigenic profile similar to that from the CRF02_AG_250 isolate (Table S3), including one isolate that was shown to be neutralized by PG9 and CH01 iGL Ab (Bonsignori et al., 2011; Pancera et al., 2010). The CRF02_AG_250 isolate showed a few noteworthy similarities with the CAP256.SU envelope that presumably triggered the CAP256 V2 apex Ab lineage. The Env trimer molecule (SOSIP.664) from the CRF02_AG_250 isolate could be constructed and purified with the Env trimer-specific Ab, PGT145, with a protein yield that is comparable to BG505.Env.C2 SOSIP.664 trimer (Sanders et al., 2013). A soluble Env trimer derived from the CRF02_AG_250 isolate showed a favorable antigenic profile in that it displayed strong reactivity with bnAbs but did not bind to non-neutralizing antibodies directed to various antigenic sites on envelope. These findings suggest that CRF02_AG_250 and related trimer phenotypes might be used as a germline targeting immunogen to trigger V2 apex bnAbs.
Overall, the results suggest a potential strategy for the elicitation of bnAbs, particularly of the PG9, CAP256.09 type. First, the immune system should be primed with a motif- targeting molecule that can engage B-cells with the YYD motif in the CDRH3. Interestingly, the YYD motif can originate independently from four major germline D- genes (3–3, 3–9, 3–16 and 3–22) (Brochet et al., 2008) and hence should constitute a significant precursor B-cell pool. Ideally, one would couple selection of the YYD motif with that for a long HCDR3. This might be accomplished using a conformationally constrained molecule such as a trimeric V2 scaffold or trimeric V2 apex structure presented on a virus like particle (VLP) that is capable of enriching YYD motif possessing B-cells. Alternatively, the CRF02_AG_250-like trimers might be directly used to prime the YYD motif-bearing long CDRH3 precursor B cell. The next step would involve boosting with trimer variants that can specifically enrich for somatic mutations in VH and VL chain variable regions. Such boosting may involve cocktail or sequential immunization with Env variants (Bonsignori et al., 2011; Pancera et al., 2010; Wang et al., 2015). The combination of designed immunogens and immunization strategies based on the type of data presented here together with empirical evaluation of immunogens in animal systems, particularly those expressing human antibody repertoires such as humanized or knock-in mice (Jardine et al., 2015; Lee et al., 2014), is most likely to be successful in inducing HIV bnAbs.
EXPERIMENTAL PROCEDURES
Antibodies and viruses
Antibodies belonging to four V2 apex bnAb prototypes, PG9 (PG9, PG16), CH01 (CH01-CH04), PGT145 (PGT141, PGT145) CAP256.09 (CAP256.01-CAP256.12 and CAP256I1–12, CAP256 UCA) were included in this study. The antibodies have been previously isolated from four individual HIV-1 infected donors (Bonsignori et al., 2011; Doria-Rose et al., 2014; Walker et al., 2011; Walker et al., 2009). In addition, we used a panel of antibodies and viruses for different experiments and the details of those are provided in the Supplemental Experimental Procedures.
Site directed mutagenesis
Amino acid point mutations in HIV envelope and antibody encoding plasmids were made by using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturers instructions. All the mutations were verified by DNA sequence analysis (Eton Bioscience, San Diego, CA).
Germline antibody design and antibody engineering
The variable heavy and light chain inferred germlines (iGL) of four V2 apex bnAbs prototypes (PG9, CH01, CAP256.09 and PGT145) were designed according to the method described previously (Jardine et al., 2013; Pancera et al., 2010) (For more details please refer to Figure 4 and Supplemental Experimental Procedures).
Recombinant envelope protein design
For this study, two envelope sequences (CRF02_AG_250, BG505.Env.C2) were used to generate soluble gp140 trimeric forms of Env spike trimer (SOSIP.664 gp140). The env gene sequence of CRF02_AG_250 was retrieved from Genbank (Accession number: EU513189), a subtype AG virus isolated previously from the peripheral blood mononuclear cells (PBMCs) of an HIV-1 infected donor from Cameroon (isolated by Ellenberger et al., at CDC, Atlanta, GA) (Seaman et al., 2010). To generate the SOSIP.664 gp140 constructs, we introduced the changes in the Env sequences as mentioned elsewhere (Sanders et al., 2013). (See Supplemental Experimental Procedures for more information).
Expression and purification of recombinant envelope proteins
For production of trimers with varied glycan composition, the α-mannosidase I and II inhibitors, kifunensine (Tocris) and swainsonine (Cayman) respectively, were added to the HEK293F cell cultures at a final concentration of 25 μM at the time of transfection (Doores and Burton, 2010). CRF02_AG_250 SOSIP.664 gp140 trimer was purified from the supernatants using PGT145 bnAb antibody affinity column as described elsewhere (Sanders et al., 2013). The BG505.Env.C2 SOSIP.664 gp140 trimer, its glycan variants and gp120 proteins were purified using a Galanthus Nivalis Lectin (GNL) (Vector Labs) column. The affinity-purified proteins were size exclusion chromatography (SEC) purified with Superdex 200 10–300 GL column (GE Healthcare) in PBS.
Antibody binding assay (ELISA)
Enzyme Linked ImmunoSorbent Assay (ELISA) was performed as described previously (Sanders et al., 2013). The 50% of maximal binding (EC50) was represented as the antibody concentrations required to reach half of maximum binding.
Virus production and Neutralization assay
To produce pseudoviruses, plasmids encoding Env were cotransfected with an Env-deficient backbone plasmid (pSG3ΔEnv) in a 1:2 ratio with the transfection reagent Fugene 6 (Promega). Kifunensine and swainsonine treated pseudoviruses were produced by adding these inhibitors to 293T cells at a final concentration of 25 μM on the day of transfection. Pseudoviruses were harvested 48–72 h posttransfection for use in neutralization assays. Neutralization was measured by using single-round-of-infection HIV-1 Env-pseudoviruses and TZM-bl target cells, as described previously (Seaman et al., 2010).
Wong Glycan Microarray
The Abs were assessed for glycan reactivity with amine functional glycans printed onto NHS-activated glass slides as described previously (Falkowska et al., 2014)(more details in Supplemental Experimental Procedures).
Autoreactivity assay
Antibodies were assayed at 100 μg/mL for autoreactivity to HEp-2 cells (Aesku Diagnostics) by immunofluorescence according to the manufacturer’s instructions.
Statistical analysis
Statistical analyses were performed using Graph Pad Prism 6 for Windows, Graph Pad Software, San Diego, California, USA.
Supplementary Material
Acknowledgments
We thank, John Mascola, Michel Nussenzweig, James Robinson, Miroslaw K. Gorny and Susan Zolla-Pazner for providing antibodies directly or through the IAVI Neutralizing Antibody Consortium (Scripps, La Jolla) or NIH AIDS Research and Reference Reagent Program (ARRRP). This work was supported by the International AIDS Vaccine Initiative (IAVI) Neutralizing Antibody Consortium SFP2120/2121 (D.R.B.); Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery Grant UM1AI100663 (to D.R.B.). This study was made possible by the generous support of the Bill and Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery and the American people through USAID. We thank Christina Corbaci for her help in the preparation of figures.
Footnotes
ACCESSION NUMBERS
The Genbank accession numbers for the nucleotide sequences of Donor_64 and Donor_584 escape viruses are KT252544 and KT252545 respectively.
AUTHOR CONTRIBUITIONS
R.A. and D.R.B. designed the experiments. R.A., J.E.V., C.H.L., B.B., L.E.M. performed the experiments. C.Y.W., C.H.W. and P.P. contributed critical reagents. R.A. and D.R.B. analyzed the data and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barbian HJ, Decker JM, Bibollet-Ruche F, Galimidi RP, West AP, Jr, Learn GH, Parrish NF, Iyer SS, Li Y, Pace CS, et al. Neutralization properties of simian immunodeficiency viruses infecting chimpanzees and gorillas. mBio. 2015;6 doi: 10.1128/mBio.00296-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. Journal of virology. 2011;85:9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braibant M, Gong EY, Plantier JC, Moreau T, Alessandri E, Simon F, Barin F. Cross-group neutralization of HIV-1 and evidence for conservation of the PG9/PG16 epitopes within divergent groups. Aids. 2013;27:1239–1244. doi: 10.1097/QAD.0b013e32835ecb42. [DOI] [PubMed] [Google Scholar]
- Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic acids research. 2008;36:W503–508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nature immunology. 2015;16:571–576. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores KJ, Burton DR. Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. Journal of virology. 2010;84:10510–10521. doi: 10.1128/JVI.00552-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Georgiev I, O’Dell S, Chuang GY, Staupe RP, McLellan JS, Gorman J, Pancera M, Bonsignori M, Haynes BF, et al. A short segment of the HIV-1 gp120 V1/V2 region is a major determinant of resistance to V1/V2 neutralizing antibodies. Journal of virology. 2012;86:8319–8323. doi: 10.1128/JVI.00696-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowska E, Le KM, Ramos A, Doores KJ, Lee JH, Blattner C, Ramirez A, Derking R, van Gils MJ, Liang CH, et al. Broadly neutralizing HIV antibodies define a glycan-dependent epitope on the prefusion conformation of gp41 on cleaved envelope trimers. Immunity. 2014;40:657–668. doi: 10.1016/j.immuni.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev IS, Doria-Rose NA, Zhou T, Kwon YD, Staupe RP, Moquin S, Chuang GY, Louder MK, Schmidt SD, Altae-Tran HR, et al. Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science. 2013;340:751–756. doi: 10.1126/science.1233989. [DOI] [PubMed] [Google Scholar]
- Gorny MK, Stamatatos L, Volsky B, Revesz K, Williams C, Wang XH, Cohen S, Staudinger R, Zolla-Pazner S. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. Journal of virology. 2005;79:5232–5237. doi: 10.1128/JVI.79.8.5232-5237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, et al. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. Journal of virology. 2011;85:4828–4840. doi: 10.1128/JVI.00198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, Kalyuzhniy O, Skog PD, Thinnes TC, Bhullar D, Briney B, et al. HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science. 2015;349:156–161. doi: 10.1126/science.aac5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013a;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ, et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proceedings of the National Academy of Sciences of the United States of America. 2013b;110:4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013a;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Antibodies in HIV-1 vaccine development and therapy. Science. 2013b;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EC, Liang Q, Ali H, Bayliss L, Beasley A, Bloomfield-Gerdes T, Bonoli L, Brown R, Campbell J, Carpenter A, et al. Complete humanization of the mouse immunoglobulin loci enables efficient therapeutic antibody discovery. Nature biotechnology. 2014;32:356–363. doi: 10.1038/nbt.2825. [DOI] [PubMed] [Google Scholar]
- Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Gray ES, Sheward D, Madiga M, Ranchobe N, Lai Z, Honnen WJ, Nonyane M, Tumba N, Hermanus T, et al. Potent and broad neutralization of HIV-1 subtype C by plasma antibodies targeting a quaternary epitope including residues in the V2 loop. Journal of virology. 2011;85:3128–3141. doi: 10.1128/JVI.02658-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore PL, Sheward D, Nonyane M, Ranchobe N, Hermanus T, Gray ES, Abdool Karim SS, Williamson C, Morris L. Multiple pathways of escape from HIV broadly cross-neutralizing V2-dependent antibodies. Journal of virology. 2013;87:4882–4894. doi: 10.1128/JVI.03424-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, McLellan JS, Wu X, Zhu J, Changela A, Schmidt SD, Yang Y, Zhou T, Phogat S, Mascola JR, Kwong PD. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. Journal of virology. 2010;84:8098–8110. doi: 10.1128/JVI.00966-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, McLellan JS, Bailer RT, Dai K, Loesgen S, Louder MK, Staupe RP, Yang Y, et al. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nature structural & molecular biology. 2013;20:804–813. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R, Walker LM, Stanfield RL, Phogat SK, Koff WC, Poignard P, Burton DR, Wilson IA. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11483–11488. doi: 10.1073/pnas.1004600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter A, Honnen WJ, He Y, Gorny MK, Zolla-Pazner S, Kayman SC. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. Journal of virology. 2004;78:5205–5215. doi: 10.1128/JVI.78.10.5205-5215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS pathogens. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, et al. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. Journal of virology. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, van Gils MJ, Pauthner M, Julien JP, Saye-Francisco KL, Hsueh J, Briney B, Lee JH, Le KM, Lee PS, et al. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:17624–17629. doi: 10.1073/pnas.1415789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS pathogens. 2010;6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Mata-Fink J, Kriegsman B, Hanson M, Irvine DJ, Eisen HN, Burton DR, Wittrup KD, Kardar M, Chakraborty AK. Manipulating the Selection Forces during Affinity Maturation to Generate Cross-Reactive HIV Antibodies. Cell. 2015;160:785–797. doi: 10.1016/j.cell.2015.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Jr, Diskin R, Nussenzweig MC, Bjorkman PJ. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2083–2090. doi: 10.1073/pnas.1208984109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer CK, Bhiman JN, Gray ES, Tumba N, Abdool Karim SS, Williamson C, Morris L, Moore PL. Viral escape from HIV-1 neutralizing antibodies drives increased plasma neutralization breadth through sequential recognition of multiple epitopes and immunotypes. PLoS pathogens. 2013;9:e1003738. doi: 10.1371/journal.ppat.1003738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Zhu J, Wu X, Moquin S, Zhang B, Acharya P, Georgiev IS, Altae-Tran HR, Chuang GY, Joyce MG, et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39:245–258. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S, Cardozo T. Structure-function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design. Nature reviews Immunology. 2010;10:527–535. doi: 10.1038/nri2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.