Abstract
The expression of a subset of genes including mesoderm specific transcript (Mest), secreted frizzled-related protein 5 (Sfrp5) and bone morphogenetic protein 3 (Bmp3) in adipose tissue biopsies of C57BL/6J mice before exposure to an obesogenic diet were shown to be predictive for the development of obesity in mice after feeding a high fat diet for 8 weeks. This observation led to the supposition that adipose tissue expression of this subset of genes within inbred strains of mice could be associated with their susceptibility in the development of adiposity when fed a low fat diet. The analyses of male mice from 5 inbred strains showed average bodyweights ranging from 25.82–36.58 grams at 16 weeks of age. Bodyweight was highest for AKR/J and adiposity correlated highly with bodyweight for all strains. Analyses of epididymal fat gene expression showed Mest, Sfrp5 and Bmp3 to be highly concomitant with adiposity across all strains of mice. Naked 1 (Nkd1), a gene previously shown to be associated with variations of adiposity in mice fed a high fat diet, but not predictive for the development of adiposity, showed no correlation with adiposity. In addition, the expression of Mest and Sfrp5 were tightly associated across the 5 mouse strains with the highest and lowest expression occurring in DBA/2J and C57BL/6J (B6) respectively suggesting a common mechanism for their regulation. Surprisingly, when independent cohorts for these 2 strains were fed high fat diet for 8 weeks, DBA/2J showed no further increase in Sfrp5 expression whereas expression levels for B6 mice were induced almost 20-fold. Analyses of (B6 × DBA2/J) F1 mice fed a low fat diet for 8 weeks showed intermediate levels of adiposity and gene expression for Sfrp5 and Mest suggesting a strong genetic basis for these differences.
Keywords: Mest, Sfrp5, Bmp3, adiposity, obesity, epigenetics, genetics, inbred strains, diet-induced obesity
1. Introduction
Experiments using genetically homogeneous inbred mouse strains have demonstrated the emergence of large variations in obesity and diabetes phenotypes when fed a high fat diet. [1, 2]. In addition, phenotypic variability in traits such as adiposity and fat mass expansion within inbred populations of mice are highly stable over time suggesting that an epigenetic etiology, possibly involving developmental changes caused by pre- or postnatal programming events, is involved in phenotypic variation of diet-induced obesity. [2]. Previous studies using global gene expression analyses were able to identify genes that are expressed in adipose tissue that were positively associated with the development of adiposity in a population of inbred C57BL/6J (B6) male mice that included imprinted genes, as well as genes involved in Wnt and Tgf-β signaling pathways [2]. A subset of these genes, including the maternally imprinted mesoderm specific transcript (Mest), secreted-related frizzled protein 5 (Sfrp5) and bone morphogenetic protein 3 (Bmp3) were also shown to be highly predictive for future susceptibility to the development of adiposity in mice when expression was measured in fat biopsies prior to feeding mice an obesogenic diet [2].
There is large body of compelling evidence for a role of Mest in mediating fat accumulation in adipocytes and adipose tissue [2–6]; however, the mechanisms that regulate Mest and its catalytic function remain elusive. MEST is localized in the endoplasmic reticulum/Golgi apparatus of the adipocyte, a cellular component essential for lipid storage and metabolism where it may act to facilitate fat uptake in adipocytes and storage of fat in lipid droplets [6]. Although the association and role for Sfrp5 in adipose tissue remains controversial, it has been demonstrated that addition of exogenous recombinant Sfrp1 or Sfrp2, molecules with strong sequence homology with Sfrp5 [7], to 3T3-L1 preadipocytes was shown to promote spontaneous differentiation into adipocytes [8]. Studies demonstrating increased adipose tissue Sfrp5 expression associated with enhanced adiposity in B6 mice after a high fat diet are consistent for a role of these soluble inhibitors of Wnt signaling in the development of adiposity in mice [2]. Sfrp5 may act to stimulate adipocyte hypertrophy via inhibition of oxidative metabolism [9]; and, in addition has been suggested to act as an anti-inflammatory adipokine that regulates metabolic dysfunction and inflammation [10, 11].
Bone morphogenetic proteins (Bmp’s), members of the Tgf-β superfamily, were originally identified as peptides that induce bone and cartilage formation but have since been shown to be involved in a wide variety of morphogenetic processes during development [12–16]. The addition of recombinant Bmp2 and Bmp7, or ectopic expression of Bmp2 and Bmp4, is able to convert pluripotent mouse fibroblast cell line C3H10T1/2 into osteoblasts, chondroblasts or adipocytes [17–21]. Bmp3, unlike other Bmp’s, antagonizes Bmp2 signaling and osteogenesis via activation of the Tgf-β/activin pathway [22, 23]. Since evidence strongly indicates a reciprocal relationship in pathways leading to osteogenesis and adipogenesis, the antagonistic effect of Bmp3 on osteogenesis is consistent with a morphogenetic role for Bmp3 in adipogenesis [23–25].
Previous studies show Mest, Sfrp5 and Bmp3 in adipose tissue biopsies of C57BL/6J mice prior to feeding mice an obesogenic diet is highly predictive for future development of adiposity in an obesogenic environment; however, very little is known in regards to how non-dietary fat-induced ‘genetic’ differences in the expression of these genes correspond to the development of adiposity among unique inbred mouse strains. In this study we evaluated body composition and adipose tissue expression of Mest, Sfrp5 and Bmp3 in 5 inbred mouse strains to determine the relationship of these molecular correlates with the development of adiposity.
2. Materials and Methods
2.1 Animals and phenotyping
All in vivo experiments were carried out with male A/J, C57BL/6J, AKR/J, DBA/2J, 129SVImJ and (B6 × DBA/2J)F1 mice purchased from the Jackson Laboratory (Bar Harbor, ME). Mice were maintained in a temperature-controlled room (23°C) with a 12-h light/12-hr dark cycle. The mice were reared under conventional conditions and fed PicoLab Rodent Diet 20 (Lab Diet; 13% kcal fat) until 16 weeks of age. A subset of mice was fed high fat diet (D12331; 58 kcal% fat) for 8 weeks (8–16 weeks of age). Body composition (body fat, lean mass and free fluid) was analyzed by Minispec NMR (Bruker) which uses the contrasting hydrogen density and/or hydrogen spin properties from adipose tissue and muscle for estimating body composition. A quality control check of NMR parameters using a standard provided by the manufacturer was performed at the beginning of each day of testing. All animal experiments were approved by the Pennington Biomedical Research Center and Maine Medical Center Research Institute Institutional Animal Care and Use Committees and in accordance with National Institutes of Health guidelines for care and use of laboratory animals.
2.2 Gene expression analyses
Epididymal fat of mice was isolated from euthanized mice and quickly frozen in liquid nitrogen. RNA was extracted from tissue homogenized in TriReagent (Molecular Research Center, Inc.) and then purified using RNeasy Mini Kit and RNase-free DNAse (Qiagen). Isolated RNA was protected from RNAse contamination with SUPERase-In (Life Technologies). RNA quantity and quality was determined using Nanodrop 1000 spectrophotometer. Quantitative reverse transcription-PCR was performed using total RNA with specific primers and probes designed using Primer Express software v3.0.1 (Life Technologies) as previously described [6]. Gene expression data were normalized to cyclophilin b (Ppib).
2.3 Statistics
Statistical calculations were performed using GraphPad Prism software V.6 and Microsoft Excel 2010. Statistical differences between 2 groups were calculated using two-tailed unpaired parametric t-test with confidence level at 95%. Significance among multiple groups was calculated with ordinary one-way ANOVA followed by Tukey’s multiple comparisons test with an alpha of 0.5. Pearson correlation coefficients (Two-tailed; assuming Gaussian distribution) were calculated with a confidence interval of 95%. Data is presented as the mean ± SEM. P<0.05 was considered to be significant.
3. Results
3.1 Body weight and composition of inbred strains fed low fat (LFD) chow diet
Bodyweights (BWTs) of male mice from 5 unique strains (n=9–10 per strain) fed a LFD was measured at 8 and 16 weeks of age and body composition measured at 16 weeks of age. Data shown in Table 1 demonstrates a broad range of BWTs with differences ranging from ~7g (8 weeks) and ~10g (16 weeks) between strains with AKR/J being significantly larger than all other strains at both 8 and 16 weeks of age. BWT variation across all strains at 16 weeks corresponded strongly with lean mass (R=0.972; P<10−30) as measured via NMR suggesting that strain-specific differences in longitudinal (lean mass) growth contributes highly to the diversity in BWT across inbred strains fed a LFD. The contribution of fat mass to overall bodyweight is lower but is a significant component of overall BWT in mice fed a LFD (R=0.852; P<10−14) and shows a higher degree of variation between strains (~1.8-fold) compared to lean mass (~1.3-fold). Fat mass was highest in AKR/J and DBA/2J mice with both strains having >15% fat whereas B6 mice had less than 10% fat (Table 1). In addition, body composition profiles of DBA/2J and B6 mice (Table 1) show no difference in lean mass (P=0.62) whereas fat mass is significantly higher in DBA/2J mice (P<0.0005) indicating that adiposity is a principal component for differences in BWT between these two inbred strains. It is also important to note that at 8 weeks of age, DBA/2J had the lowest BWT of all strains (Table 1) but showed a ~32% increase in BWT during the subsequent 8 weeks suggesting that significant proportion of weight gain in this strain is likely due to increased fat mass. Epididymal (EPI) fat pad weights across all mouse strains were consistent with NMR-based measurements of fat mass (R=0.963; P<10−27) providing confidence for the body composition measurements in Table 1.
Table 1.
Body weights and body composition of 5 inbred strains of mice fed standard chow diet for 8 weeks. Data in columns sharing the same superscript indicate no significant difference between strains.
| 8 weeks | 16 weeks | |||||
|---|---|---|---|---|---|---|
| Strain | n | BWT (g) | BWT (g) | Lean Mass (g) | Fat Mass (g) | % Fat |
| A/J | 9 | 23.22 ± 0.33a | 25.90 ± 0.40a | 19.53 ± 0.26a | 2.64 ± 0.15a | 10.17 ± 0.46ab |
| C57BL/6J (B6) | 10 | 21.66 ± 0.53a | 25.82 ± 0.52a | 19.98 ± 0.42a | 2.26 ± 0.09a | 8.72 ± 0.25a |
| AKR/J | 10 | 28.66 ± 0.58b | 36.58 ± 1.16b | 25.76 ± 0.62b | 5.83 ± 0.64b | 15.66 ± 1.31c |
| DBA/2J | 10 | 21.50 ± 0.73a | 28.39 ± 0.86a | 20.34 ± 0.59a | 4.40 ± 0.34c | 15.42 ± 0.87c |
| 129SV/ImJ | 10 | 22.14 ± 0.84a | 27.42 ± 0.96a | 20.46 ± 0.71a | 3.28 ± 0.15ac | 12.01 ± 0.55b |
3.2 Strain-dependent variation in gene expression
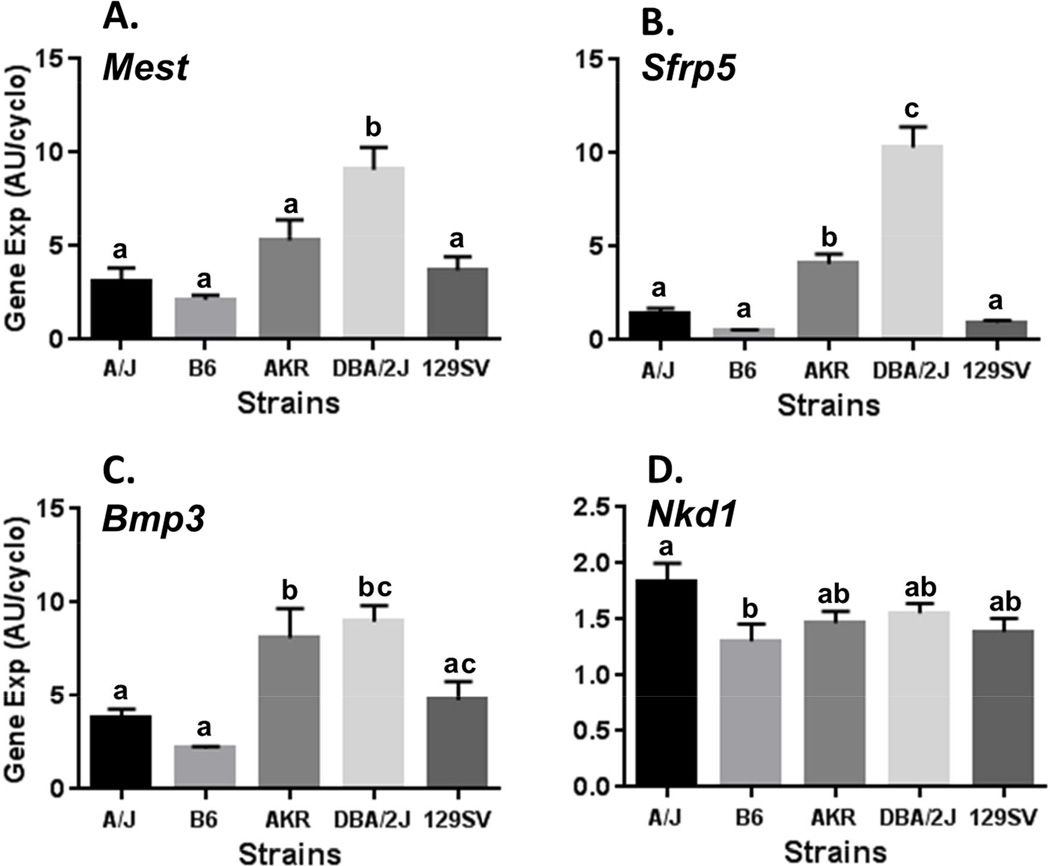
Gene expression for Mest, Sfrp5, Bmp3 and Nkd1 was measured in RNA isolated from EPI fat from each of the 5 strains as shown in Figure 1. The selection of EPI fat for these analyses was based on previous studies that analyzed the same subset of genes in EPI fat biopsies in 7 week old mice fed a LFD to determine whether pre-HFD levels of expression could predict susceptibility for the development of adiposity in C57BL/6J mice fed an obesogenic diet [2]. Mest expression was significantly higher in DBA/2J compared to all of the other mouse strains with almost a 4-fold increase compared to C57BL/6J (Fig. 1A). Importantly, EPI fat Mest expression in the LFD-fed mice in this study were consistent with levels detected in past experiments using cohorts of LFD-fed B6 mice as controls to determine effects of dietary fat on the induction of Mest [6]. Surprisingly, the strain-dependent pattern for EPI fat Mest expression was very similar to that of both Sfrp5 (Fig. 1B) and Bmp3 (Fig. 1C). EPI fat Sfrp5 expression in DBA/2J was more than 20-fold higher than B6 mice and more that 2-fold higher than all of the other mouse strains. EPI fat Nkd1 expression showed only a modest difference between A/J and B6 (P=0.04) but was essentially consistent among all strains. These data are consistent with previous studies showing the lack of association of Nkd1 expression in EPI fat biopsies with susceptibility for the development of adiposity following an obesogenic diet [2].
Figure 1.
Analyses of Mest (A), Sfrp5 (B), Bmp3 (C) and Nkd1 (D) mRNA expression in epididymal fat (EPI) of 5 inbred mouse strains fed a low fat diet (LFD) until 16 weeks of age. Gene expression measured by TaqMan QRT-PCR is represented as arbitrary units (AU) normalized to cyclophilin b. Each strain represents the mean ± SEM of at least 9 mice. Datasets annotated with the same letter indicate no significant differences between groups. Mest expression (A) is significantly higher in DBA/2J compared with B6 (P<0.0001), A/J (P=0.0002), 129SV (P=0.0007) and AKR (P=0.029). Sfrp5 expression (B) is significantly higher in DBA/2J compared with all strains (P<0.0001); and, AKR is higher than B6 (P=0.0007), 129SV (P=0.004) and A/J (P=0.02). Bmp3 expression (C) is significantly higher in DBA/2J compared with B6 (P=0.0003), A/J (P=0.006), 129SV (P=0.03); and higher in AKR compared with B6 (P=0.002) and A/J (P=0.03). Nkd1 expression (D) is significantly higher in A/J compared to B6 (P=0.04).
3.3 Molecular correlates for strain-dependent variation of adiposity
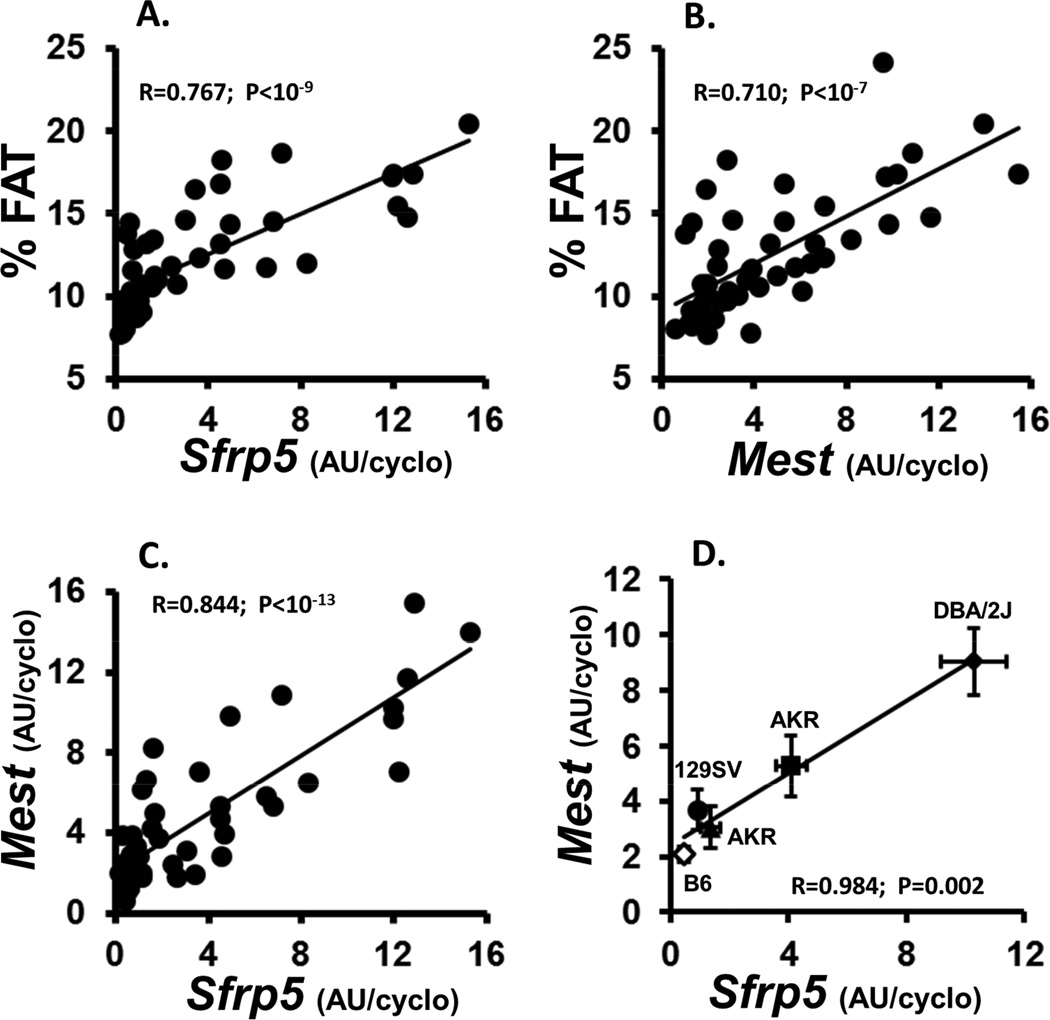
Analyses of the correlation of EPI fat gene expression and phenotypic parameters across the entire cohort of mice from the 5 strains showed very strong positive associations between adiposity (% fat) vs Sfrp5 (Fig. 2A) and Mest (Fig. 2B) with P values <10−9 and 10−7 respectively. Bmp3 was also significantly associated with adiposity (data not shown; R=0.644; P<10−6). Nkd1 showed no significant correlations with adiposity or any other phenotypic measurements. The strong correlation between Mest and Sfrp5 across the entire cohort of mice (Fig. 1C; R=0.844; P<10−13) or when plotted as the mean for each gene with respect to strain (Fig. 1D; R=0.984; P=0.002) suggesting the likelihood that a common mechanism is at least partially responsible for the regulation of these 2 genes. A correlation between Bmp3 vs Mest (P<10−5) and Sfrp5 (P<10−4) was also evident but was significantly reduced compared to that observed between Mest and Sfrp5.
Figure 2.
Data represents the correlation between indices of adiposity (% FAT) with Sfrp5 (A) and Mest (B) gene expression; and, the correlation between Mest and Sfrp5 gene expression in all individuals from the 5 mouse strains (C) or by the mean ± SEM of each of the strains (D). Each strain represents the mean ± SEM of at least 9 mice. Body composition was measured via NMR. Gene expression measured by TaqMan QRT-PCR is represented as arbitrary units (AU) normalized to cyclophilin b.
3.4 Genetic diversity in the regulation of Sfrp5 and Mest between B6 and DBA/2J mice
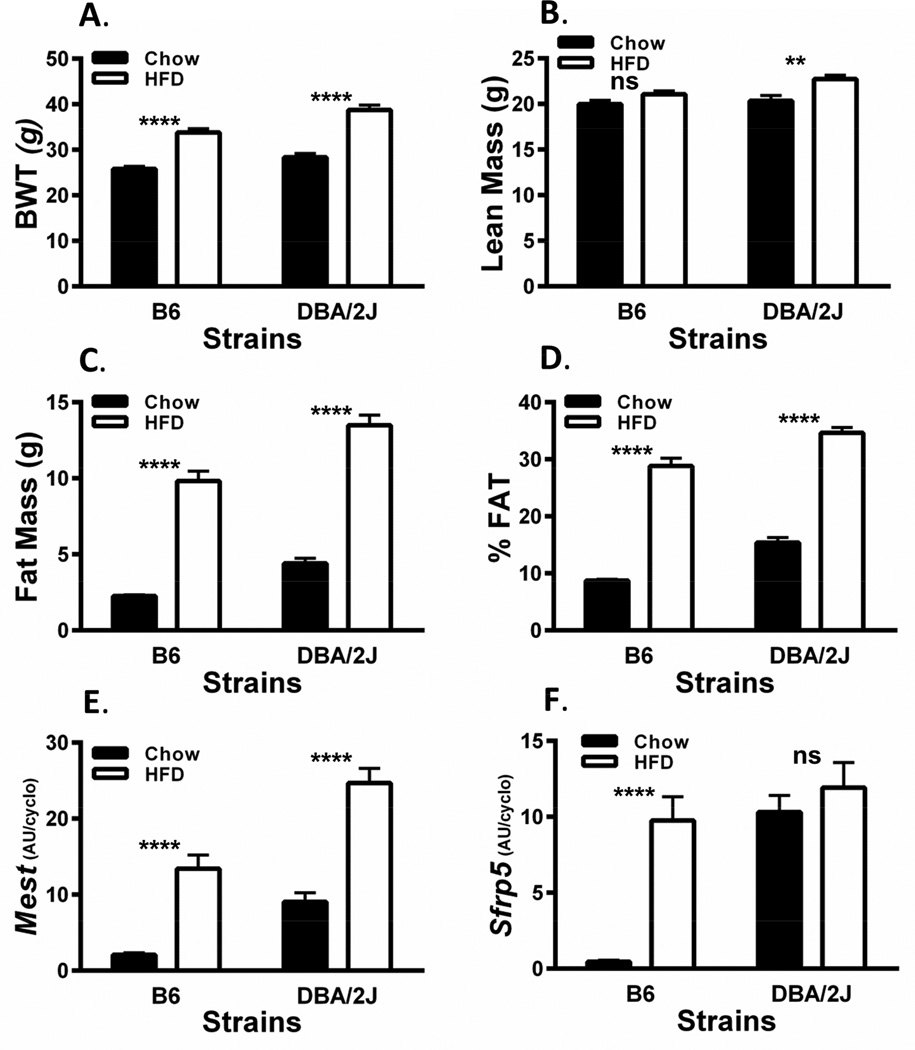
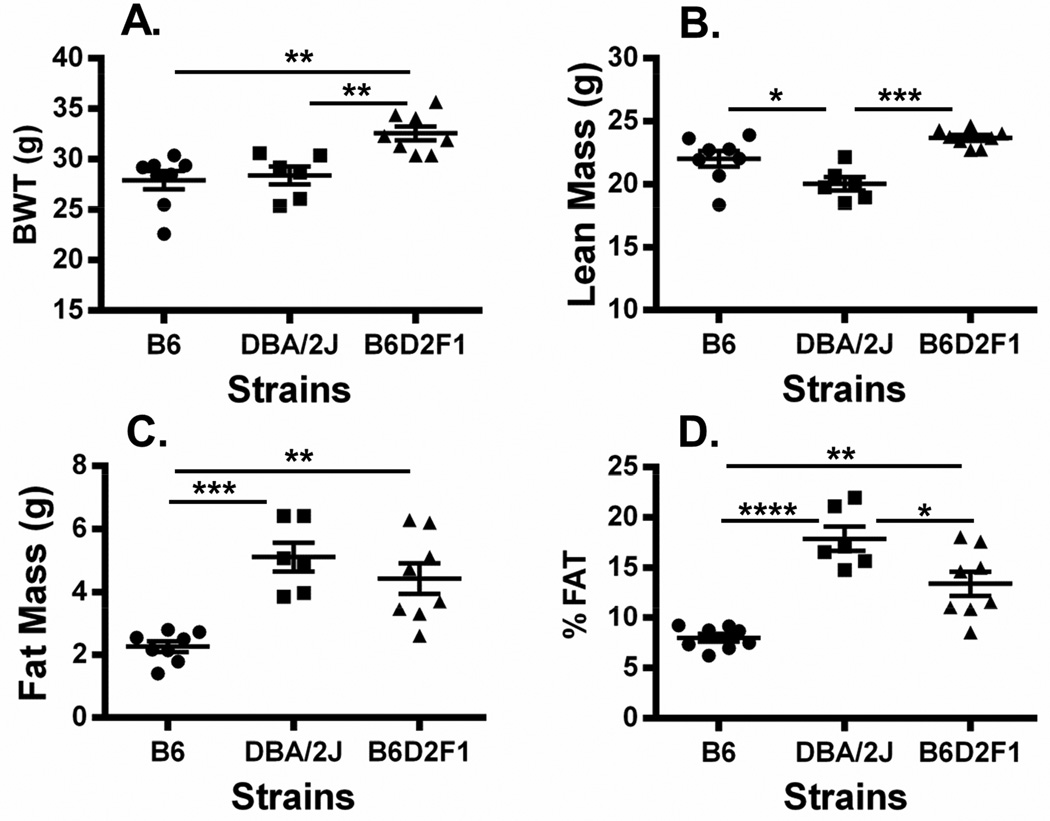
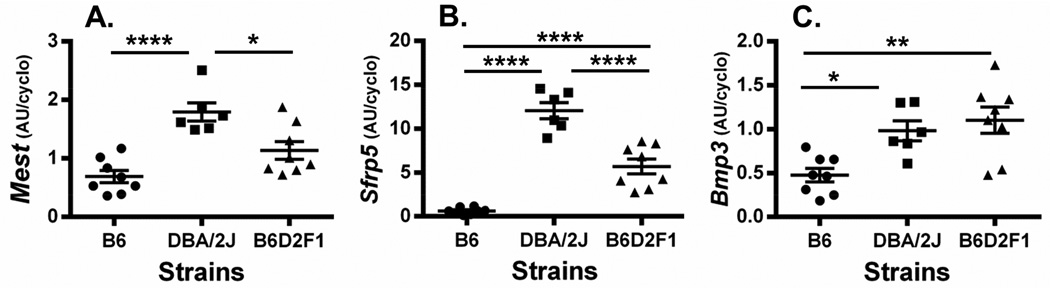
Since past studies have shown that feeding mice an obesogenic diet for as short of a period as 1 week can induce the expression of adipose tissue Sfrp5 and Mest in mice [2] we performed a study to determine whether the strikingly high expression of Sfrp5 in EPI fat of chow-fed DBA/2J mice can be further induced by HFD. Two additional cohorts of DBA/2J (n=9) and B6 (n=9) male mice were placed onto a HFD at 8 weeks of age for 8 weeks (until 16 weeks of age). It was anticipated that EPI fat expression of Sfrp5 and Mest would be robustly induced in B6 during this time [2, 6]. Physiological parameters of HFD-fed B6 and DBA/2J mice, as expected, showed highly significant increases in BWT (Fig. 3A) and adiposity (Fig. 3C and 3D) compared to the LFD-fed cohorts; however, lean mass was either not changed or only modestly increased by the HFD in B6 and DBA/2J mice respectively (Fig. 3B). EPI fat expression of Mest in HFD-fed B6 and DBA/2J mice shown in Figure 3E was more highly induced in B6 mice (>6-fold) compared with DBA/2J mice (~2.5-fold) when compared to LFD-fed mice but was still significantly higher in the HFD-fed DBA/2J mice compared to B6 (P=0.0006). In contrast, EPI fat Sfrp5 expression was induced over 20-fold in HFD-fed B6 mice but remained unchanged in HFD-fed DBA/2J mice compared to their LFD-fed cohort. Interestingly, EPI fat Sfrp5 mRNA in the HFD-fed B6 mice was not significantly different from either LFD or HFD-fed DBA/2J mice. To further test the genetic predisposition for the differences in EPI fat Sfrp5 expression in B6 vs DBA/2J mice, an independent cohort of B6 and DBA/2J was compared with an F1 hybrid; (B6 × DBA/2J)F1 (B6D2F1), derived from the two parental strains of mice. The 3 cohorts of mice were fed a LFD until 16 weeks of age and physiological parameters were measured. Data shows significantly higher BWT (Fig. 4A) in the B6D2F1 compared to both parental strains and significantly (P<0.001) and modestly (P=0.058) increased lean mass (Fig. 4B) compared to DBA/2J and B6 respectively suggesting slightly increased longitudinal growth. Adiposity measured by fat mass (Fig. 4C) and %FAT (Fig. 4D) in the B6D2F1 mice showed an intermediate phenotype compared to the B6 and DBA/2J parental strains which was consistent with the expression of EPI fat Mest (Fig. 5A) and Sfrp5 (Fig. 5B) among the 3 strains of mice. EPI fat Bmp3 expression (Fig. 5C) did not show the same expression profile and was similarly expressed in both DBA/2J and B6D2F1 mice. In addition, the association of EPI fat Sfrp5 with Mest (R2=0.78; P<10−7) across the 3 strains in this study was several orders of magnitude more significant than with Bmp3 (R2=0.45; P<0.001). These results suggest that intermediate levels of EPI fat Sfrp5 in B6D2F1 mice may be genetically regulated via a codominant or incompletely dominant manner which could lead to coordinated effects on the regulation of adipose tissue Mest expression and susceptibility for the development of adiposity.
Figure 3.
Analyses of bodyweight (BWT; A), lean mass (B), fat mass (C), adiposity (%FAT; D); and, epididymal (EPI) fat Mest (E) and Sfrp5 (F) mRNA expression in B6 and DBA/2J inbred mice fed a low fat diet (LFD) until 8 weeks of age and then fed either the LFD or a high fat diet (HFD; 58% kcal fat) for an additional 8 weeks until 16 weeks of age. Body composition was measured via NMR. Gene expression measured by TaqMan QRT-PCR is represented as arbitrary units (AU) normalized to cyclophilin b. Each group represents the mean ± SEM of at least 9 mice. Datasets annotated with ns, ** and **** indicate no significant differences, P<0.01 and P<0.0001 between groups respectively.
Figure 4.
Data shows a scatterplot of bodyweight (BWT; A), lean mass (B), fat mass (C) and adiposity (% FAT; B) in of B6, DBA/2J and (B6 × DBA/2J)F1 hybrid (B6D2F1) mice fed a low fat diet (LFD) until 16 weeks of age. Body composition was measured via NMR. Each group represents the mean ± SEM of 6–8 mice. Datasets annotated with 1, 2, 3 or 4 asterisks indicate significant differences of P<0.05, P<0.01, P<0.001 and P<0.0001 respectively.
Figure 5.
Data shows a scatterplot of Mest (A), Sfrp5 (B) and Bmp3 (C) mRNA expression in epididymal fat (EPI) of B6 (n=8), DBA/2J (n=6) and (B6 × DBA/2J)F1 (n=8) hybrid (B6D2F1) mice fed a low fat diet (LFD) until 16 weeks of age. Gene expression measured by TaqMan QRT-PCR is represented as arbitrary units (AU) normalized to cyclophilin b. Each group represents the mean ± SEM of 6–8 mice. Datasets annotated with 1, 2, and 4 asterisks indicate significant differences of P<0.05, P<0.01, and P<0.0001 respectively.
4. Discussion/Conclusion
Our previous investigations were aimed at identifying epigenetic determinants of obesity utilizing the high degree of variability in adiposity phenotypes after B6 mice have been exposed to an obesogenic environment. The four genes whose expression was measured in this present study; Mest, Sfrp5, Bmp3 and Nkd1 all showed a highly variable expression with expression ranging almost 80-fold (Mest) in mice fed a HFD for 4 weeks [2]. In addition, the expression of all of these genes was significantly elevated in adipose tissue in HFD-fed mice compared to basal expression levels in mice maintained on a LFD. The effects of dietary fat on Mest was particularly striking with some mice exhibiting substantial increases of expression after feeding mice a HFD for only 2 days whereas other individuals showed no increase [6]. Of the four genes showing highly variable HFD-mediated expression three of these; Mest, Sfrp5 and Bmp3 was also predictive of future development of obesity based on their expression in EPI fat biopsies of B6 mice prior to feeding a HFD [2]. Because of the predictive relationship between Mest, Sfrp5 and Bmp3 in mice fed LFD with susceptibility for development of adiposity, we became interested in gaining further functional insight for these genes by determining how their ‘non-fat-induced’ expression in adipose tissue is associated with strain-specific variations in adiposity related phenotypes.
Data from 5 strains of mice fed a LFD for 16 weeks in Table 1 and Figures 1 and 2 showed remarkable consistency between the expression of EPI fat Mest, Sfrp5 and Bmp3 with indices of adiposity, as well as strong associations between EPI fat Mest and Sfrp5 across the strains (Fig. 2D). A similar close association between adipose tissue Mest and Sfrp5 was also observed for B6 mice after feeding a HFD for 4 weeks [2]. Although the consistent association of Mest with Sfrp5 is not well understood in the context of fat mass gain in adult mice, it’s possible that Sfrp5 inhibition of oxidative metabolism [9] could lead to physiological conditions that promote adipocyte hypertrophy and fat mass expansion. Interestingly however, other studies have shown that Mest is most highly expressed in early post-natal development when fat mass is rapidly expanding whereas Sfrp5 expression in adipose tissue is expressed at very low levels during this time [6, 26].
One of the most intriguing outcomes of this present study is the high expression of Sfrp5 in EPI fat in DBA/2J mice which ranged almost 20-fold higher than B6 (Fig. 1B and 3F). Interestingly, a second cohort of DBA/2J and B6 mice fed a HFD from 8 to 16 weeks of age showed no further increase in DBA/2J EPI fat Sfrp5 expression compared with DBA/2J fed a LFD, whereas B6 mice showed a ~20-fold increase in EPI fat Sfrp5 with levels equivalent to DBA/2J mice fed either LFD or HFD (Fig. 3F). Additional cohorts of B6, DBA/2J and a F1 hybrid generated from a cross between B6 and DBA/2J (B6D2F1) fed LFD until 16 weeks of age showed intermediate phenotypes for adiposity (Fig. 4C and 4D), Sfrp5 and Mest (Fig. 5) in the B6D2F1 compared to the parental strains. The analyses of EPI fat Bmp3 in B6, DBA/2J and B6D2F1 mice (Fig. 5) showed a reduced association of Bmp3 with Sfrp5 compared with that of Mest. This observation was somewhat surprising since Sfrp5 has been shown to modulate BMP signaling in the regulation of organogenesis [27] and retinal development [28] in zebrafish. However, although it is well recognized that significant cross-talk occurs between the Wnt and BMP/Smad signaling pathways [29, 30] and BMP and Wnt signaling are important mediators of adipogenesis [31], very little is known in regards to specific Sfrp5-mediated effects on BMP signaling in mammals. In a genetics context, the intermediate expression of EPI fat Sfrp5 mRNA in the F1 progeny (B6D2F1) derived from B6 and DBA/2J suggests a codominant or incompletely dominant genetic contribution to this quantitative phenotype. Differentiating between codominance vs incomplete dominance with quantitative traits such as EPI fat Sfrp5 mRNA expression is difficult since the complexity of the genetics regulating Sfrp5 is not well defined. Since both B6 and DBA/2J do express Sfrp5, albeit at different levels when fed a HFD, it is more likely that the intermediate phenotype in the B6D2F1 is caused by codominant contribution of the two alleles [32]. Future quantitative analyses of EPI fat Sfrp5 mRNA in backcross, intercross, and/or recombinant inbred lines (BXD) derived from the B6 and DBA/2J parental strains will provide significant insight into the genetic complexity in the regulation of adipose Sfrp5. A query of MGI (The Jackson Laboratory) revealed only a single nucleotide polymorphism (SNP) within the 3’UTR of Sfrp5 (Chr 19) between B6 and DBA/2J. However, this same SNP also occurs in A/J mice which had levels of EPI fat Sfrp5 level almost as low as B6 (Fig. 1B) reducing the potential for this polymorphism in having a significant impact on the regulation of Sfrp5. In conclusion, our studies show that adipose tissue expression of Mest, Bmp3 and Sfrp5 show positive strong association with the development of adiposity across multiple inbred strains of mice in the absence of a HFD. These results are consistent with previous studies of these same genes in the context of variations in adiposity phenotypes within a population of HFD-fed inbred B6 mice. In addition, we have identified a mouse model, DBA/2J, with constitutively high expression of EPI fat Sfrp5 that is not further up-regulated via dietary fat. This model will be particularly useful for studies of the regulation of Sfrp5 and its role in adipose tissue function, energetics, inflammation, and glucose homeostasis.
Highlights.
Adipose Mest, Sfrp5 and Bmp3 are concomitant with strain-differences in adiposity.
Strong correlation between Mest and Sfrp5 suggests a common regulatory mechanism.
DBA/2J mice express high adipose tissue Sfrp5 compared to other mouse strains.
DBA/2J, unlike B6 mice, shows no fat-inducible adipose Sfrp5 expression.
Characteristics of DBA/2J provide an excellent model to study SFRP5 function.
Acknowledgments
We would like to thank Tamra Mendoza and Justin Manuel (Pennington Biomedical Research Center) for technical support during the early stages of the study. Support was also provided by the Genomics Core Facility (partly supported by NIH P20-RR021945 and P30-DK072476) at the Pennington Biomedical Research Center and the Molecular Phenotyping Core Facility (supported by NIGM 8P30-GM106391) at the Maine Medical Research Institute. These studies were partially supported by NIH R21DK074951 (R.A.K.) and NIH R01DK090361 (R.A.K.).
Abbreviations
- Mest
mesoderm specific transcript
- Sfrp5
secreted related protein 5
- Bmp3
bone morphogenetic protein 3
- Nkd1
Naked 1
- HFD
high fat diet; Surwit D12331
- LFD
low fat diet; PicoLab 20
- EPI
epididymal
- SNP
single nucleotide polymorphism
- Tgf-β
transforming growth factor beta
- NMR
nuclear magnetic resonance
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare that there are no conflicts of interest.
References
- 1.Burcelin R, et al. Heterogeneous metabolic adaptation of C57BL/6J mice to high-fat diet. Am J Physiol Endocrinol Metab. 2002;282(4):E834–E842. doi: 10.1152/ajpendo.00332.2001. [DOI] [PubMed] [Google Scholar]
- 2.Koza RA, et al. Changes in gene expression foreshadow diet-induced obesity in genetically identical mice. PLoS Genet. 2006;2(5):e81. doi: 10.1371/journal.pgen.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahashi M, Kamei Y, Ezaki O. Mest/Peg1 imprinted gene enlarges adipocytes and is a marker of adipocyte size. Am J Physiol Endocrinol Metab. 2004 doi: 10.1152/ajpendo.00244.2004. [DOI] [PubMed] [Google Scholar]
- 4.Moraes RC, et al. Study of the alteration of gene expression in adipose tissue of diet-induced obese mice by microarray and reverse transcription-polymerase chain reaction analyses. Endocrinology. 2003;144(11):4773–4782. doi: 10.1210/en.2003-0456. [DOI] [PubMed] [Google Scholar]
- 5.Soukas A, et al. Leptin-specific patterns of gene expression in white adipose tissue. Genes Dev. 2000;14(8):963–980. [PMC free article] [PubMed] [Google Scholar]
- 6.Nikonova L, et al. Mesoderm-specific transcript is associated with fat mass expansion in response to a positive energy balance. FASEB J. 2008;22(11):3925–3937. doi: 10.1096/fj.08-108266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116(Pt 13):2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 8.Bennett CN, et al. Regulation of Wnt signaling during adipogenesis. J Biol Chem. 2002;277(34):30998–31004. doi: 10.1074/jbc.M204527200. [DOI] [PubMed] [Google Scholar]
- 9.Mori H, et al. Secreted frizzled-related protein 5 suppresses adipocyte mitochondrial metabolism through WNT inhibition. J Clin Invest. 2012;122(7):2405–2416. doi: 10.1172/JCI63604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schulte DM, et al. Pro-inflammatory wnt5a and anti-inflammatory sFRP5 are differentially regulated by nutritional factors in obese human subjects. PLoS One. 2012;7(2):e32437. doi: 10.1371/journal.pone.0032437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouchi N, et al. Sfrp5 is an anti-inflammatory adipokine that modulates metabolic dysfunction in obesity. Science. 2010;329(5990):454–457. doi: 10.1126/science.1188280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hogan BL. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 1996;10(13):1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 13.Wang EA, et al. Purification and characterization of other distinct bone-inducing factors. Proc Natl Acad Sci U S A. 1988;85(24):9484–9488. doi: 10.1073/pnas.85.24.9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang EA, et al. Recombinant human bone morphogenetic protein induces bone formation. Proc Natl Acad Sci U S A. 1990;87(6):2220–2224. doi: 10.1073/pnas.87.6.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winnier G, et al. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9(17):2105–2116. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 16.Wozney JM, et al. Novel regulators of bone formation: molecular clones and activities. Science. 1988;242(4885):1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- 17.Ahrens M, et al. Expression of human bone morphogenetic proteins-2 or -4 in murine mesenchymal progenitor C3H10T1/2 cells induces differentiation into distinct mesenchymal cell lineages. DNA Cell Biol. 1993;12(10):871–880. doi: 10.1089/dna.1993.12.871. [DOI] [PubMed] [Google Scholar]
- 18.Asahina I, Sampath TK, Hauschka PV. Human osteogenic protein-1 induces chondroblastic, osteoblastic, and/or adipocytic differentiation of clonal murine target cells. Exp Cell Res. 1996;222(1):38–47. doi: 10.1006/excr.1996.0005. [DOI] [PubMed] [Google Scholar]
- 19.Skillington J, Choy L, Derynck R. Bone morphogenetic protein and retinoic acid signaling cooperate to induce osteoblast differentiation of preadipocytes. J Cell Biol. 2002;159(1):135–146. doi: 10.1083/jcb.200204060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang QQ, Otto TC, Lane MD. Commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc Natl Acad Sci U S A. 2004;101(26):9607–9611. doi: 10.1073/pnas.0403100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang EA, et al. Bone morphogenetic protein-2 causes commitment and differentiation in C3H10T1/2 and 3T3 cells. Growth Factors. 1993;9(1):57–71. doi: 10.3109/08977199308991582. [DOI] [PubMed] [Google Scholar]
- 22.Bahamonde ME, Lyons KM. BMP3: to be or not to be a BMP. J Bone Joint Surg Am. 2001;83-A(Pt 1) Suppl 1:S56–S62. [PubMed] [Google Scholar]
- 23.Daluiski A, et al. Bone morphogenetic protein-3 is a negative regulator of bone density. Nat Genet. 2001;27(1):84–88. doi: 10.1038/83810. [DOI] [PubMed] [Google Scholar]
- 24.Beresford JN, et al. Evidence for an inverse relationship between the differentiation of adipocytic and osteogenic cells in rat marrow stromal cell cultures. J Cell Sci. 1992;102(Pt 2):341–351. doi: 10.1242/jcs.102.2.341. (Pt 2) [DOI] [PubMed] [Google Scholar]
- 25.Cheng SL, et al. MSX2 promotes osteogenesis and suppresses adipogenic differentiation of multipotent mesenchymal progenitors. J Biol Chem. 2003;278(46):45969–45977. doi: 10.1074/jbc.M306972200. [DOI] [PubMed] [Google Scholar]
- 26.Kozak LP, et al. The early nutritional environment of mice determines the capacity for adipose tissue expansion by modulating genes of caveolae structure. PLoS One. 2010;5(6):e11015. doi: 10.1371/journal.pone.0011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stuckenholz C, et al. Sfrp5 modulates both Wnt and BMP signaling and regulates gastrointestinal organogenesis [corrected] in the zebrafish, Danio rerio. PLoS One. 2013;8(4):e62470. doi: 10.1371/journal.pone.0062470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holly VL, et al. Sfrp1a and Sfrp5 function as positive regulators of Wnt and BMP signaling during early retinal development. Dev Biol. 2014;388(2):192–204. doi: 10.1016/j.ydbio.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Guo X, Wang XF. Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 2009;19(1):71–88. doi: 10.1038/cr.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Itasaki N, Hoppler S. Crosstalk between Wnt and bone morphogenic protein signaling: a turbulent relationship. Dev Dyn. 2010;239(1):16–33. doi: 10.1002/dvdy.22009. [DOI] [PubMed] [Google Scholar]
- 31.Tang QQ, Lane MD. Adipogenesis: from stem cell to adipocyte. Annu Rev Biochem. 2012;81:715–736. doi: 10.1146/annurev-biochem-052110-115718. [DOI] [PubMed] [Google Scholar]
- 32.Silver LM. Mouse Genetics: Concepts and Applications. New York: Oxford University Press; 1995. pp. 253–257. [Google Scholar]