Abstract
We describe the presentation, treatment, clinical outcome, and targeted genome analysis of a metastatic salivary acinic cell carcinoma (AciCC). A 71-year-old male presented with a 3 cm right tail of a parotid lesion, first detected as a nodule by the patient seven months earlier. He had a right total parotidectomy with cranial nerve VII resection, right facial nerve resection and grafting, resection of the right conchal cartilage, and right modified radical neck dissection. The primary tumor revealed AciCC with two distinct areas: a well-differentiated component with glandular architecture and a dedifferentiated component with infiltrative growth pattern associated with prominent stromal response, necrosis, perineural invasion, and cellular pleomorphism. Tumor staging was pT4 N0 MX. Immunohistochemistry staining showed pankeratin (+), CD56 (−), and a Ki67 proliferation index of 15%. Upon microscopic inspection, 49 local lymph nodes resected during parotidectomy were negative for cancer cells. Targeted sequencing of the primary tumor revealed deletions of CDKN2A and CDKN2B, a nonsense mutation in ARID2, and single missense mutations of unknown significance in nine other genes. Despite postoperative localized radiation treatment, follow-up whole body PET/CT scan showed lung, soft tissue, bone, and liver metastases. The patient expired 9 months after resection of the primary tumor.
1. Introduction
The incidence of salivary gland cancers is approximately 0.6% of the incidence rate of all cancers in the United States [1, 2] and acinic cell carcinomas (AciCCs) account for approximately 2.4% of salivary gland cancers [3]. Most salivary gland AciCCs (86.3%) arise in the parotid gland and, to a much lesser extent, in the submandibular gland, other major and minor salivary glands, the parapharyngeal space, and the sublingual gland [4, 5]. The median age of AciCC presentation is 52 years with 59% incidence in women and 41% incidence in men [5]. Histologically, most tumors tend to be of low grade; however, on occasion, poorly differentiated and high grade tumors occur. The mainstay of treatment entails surgery followed by postoperative radiation [6]. Chemotherapy is generally not considered an effective treatment. The five-year disease-specific survival rate of AciCC is 91% with worse prognosis in cases where there is high grade histology, an age at presentation of greater than 30 years, and evidence for metastatic disease [5]. Local recurrences and distant metastases occur frequently, with most metastases occurring in the lungs and bone and, to a lesser extent, in the central nervous system, mediastinum, and liver [7]. In the single previous mutational spectrum report of a patient with AciCC whole exome analysis revealed deletions in CDKN2A, MTAP, and PPP1R13B along with somatic nonsynonymous point mutations in 14 other genes [8].
We reviewed the medical records and targeted genomic profile of a male who presented with AciCC of the parotid gland. Histopathology and immunohistochemistry were performed at Kaiser Permanente, CA. The patient underwent right total parotidectomy with cranial nerve VII resection, right facial nerve resection and grafting, resection of the right conchal cartilage, and right modified radical neck dissection. Because AciCC of the parotid gland is typically recognized as a low grade and indolent cancer, we sought to further study the genetic profiling of this specific case due to its aggressive clinical nature. Targeted genomic profiling of the parotid tumor using clinical next generation sequencing (NGS) to a minimum coverage depth of 500x was carried out on a FoundationOne platform in a Clinical Laboratory Improvement Amendments (CLIA) certified lab (Foundation Medicine, Cambridge, MA) (Table S1 in Supplementary Material available online at http://dx.doi.org/10.1155/2015/893694) [9]. FoundationOne uses targeted next generation sequencing to interrogate the coding regions of 315 cancer related genes and the introns of 28 genes that are reportedly involved in structural rearrangements (Tables S2 and S3).
2. Case Report
2.1. Case History
We describe a 71-year-old male who presented with a 2 cm indurated nontender subcutaneous nodule over the right angle of mandible with no redness, warmth, or drainage. According to the patient, the nodule appeared one month earlier and decreased in mass upon eating. His previous medical history was essential hypertension, primary open angle glaucoma, and chronic kidney disease stage 3. One month after presentation, needle aspiration of mass at right tail of the parotid revealed a salivary gland neoplasm. MRI revealed a 2.4 cm anterior-posterior × 2.4 cm transverse × 2.6 cm cranial caudal septated cystic mass in the right parotid lobe. A second fine needle aspiration performed four months after presentation revealed another 1 cm approximate diameter growing mass within the parotid. Five months after presentation, just prior to scheduled surgery, pathology analyses of computed tomography-guided biopsy results were consistent with high grade AciCC. The patient's presurgical cancer workup included complete blood count, coagulation panel, chemistry profile-20, and chest radiographs that were all negative for metastatic cancer.
The patient underwent a right total parotidectomy with cranial nerve VII resection, right facial nerve resection and grafting, resection of the right conchal cartilage, and right modified radical neck dissection. Intraoperatively, the tumor was noted to encase the right facial nerve and extend to ulcerated overlying skin. The face was rehabilitated with a right AlloDerm facial sling and placement of right upper eyelid gold weight. The facial defect was reconstructed with a cervicofacial advancement flap. Pathology analysis revealed a 3.0 × 3.0 × 3.0 cm AciCC with two distinct areas: a well-differentiated component with glandular architecture and a dedifferentiated component with infiltrative growth pattern associated with prominent stromal response, necrosis, perineural invasion, and cellular pleomorphism (Figure 1). At the time of resection, the staging was pT4, N0, MX. Immunohistochemistry staining showed pankeratin (+), CD56 (−), and a proliferation index of ~15% based on Ki67. The surgical margins were clear with the closest showing tumor 0.2 cm from anterior inked margin.
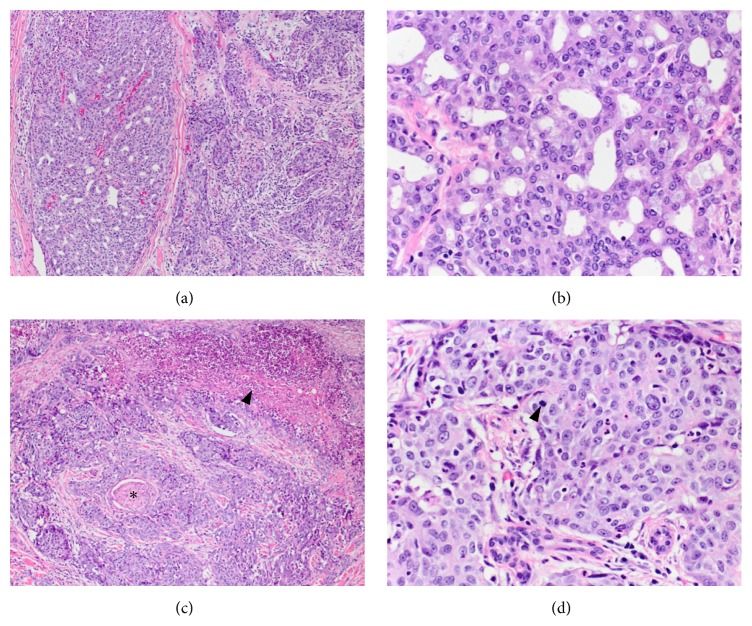
Figure 1.
Acinic cell carcinoma of the parotid gland. (a) The tumor exhibits two distinct areas on histologic examination. The two areas are juxtaposed to each other and there is no evidence of transition. On the left, the well-differentiated area shows glandular architecture and the area on the right side of the image shows an infiltrative growth pattern associated with prominent stromal response. H&E, 10x. (b) Higher magnification of the well-differentiated area demonstrates polygonal cells without significant cytologic atypia, variably basophilic granular cytoplasm, minimal pleomorphism, and inconspicuous mitotic activity. H&E, 40x. (c) Dedifferentiated regions of the tumor show focal necrosis (arrowhead) and multiple foci of perineural invasion (star) H&E, 10x. (d) Cytologically, the dedifferentiated areas display more pronounced pleomorphism and increased mitotic activity (arrowhead) H&E, 40x.
Postoperatively, the patient received 6000 cGy in 30 fractions using intensity modulated radiation therapy (IMRT) to the parotid bed and right neck. Three months after completing IMRT, a whole body PET/CT scan revealed multiple fluorodeoxyglucose- (FDG-) avid pulmonary nodules, a 2.3 cm mass in the right infrahilar region with a maximum standardized uptake value (SUV) of 5.74, a 2 × 2.6 cm subcarinal lymph node (SUV 6.08), multiple FDG-avid liver lesions, a soft tissue lesion in the lateral left upper quadrant 3 × 2.7 cm, and osseous metastases to the tenth thoracic vertebrae (T10) (maximum SUV 8.4) and proximal right femur (Figure 2). The patient developed dermal metastases along the neck incision site. A CT-guided biopsy of a pulmonary nodule confirmed metastatic AciCC with immunohistochemistry negative for thyroid transcription factor-1 (TTF-1), cytokeratin 7 (CK7), and cytokeratin 20 (CK20).
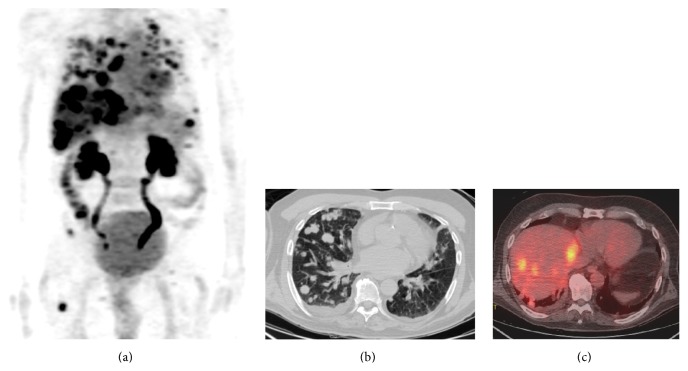
Figure 2.
Widespread metastatic acinic cell carcinoma by positron-emission tomography/computed tomography (PET/CT). (a) Whole body PET images demonstrate FDG-avid lesion in the lungs, liver, and bones. (b) Multiple metastatic pulmonary lesions were seen on noncontrast chest CT. (c) Fused PET/CT images demonstrate multiple FDG-avid hepatic metastases.
The patient underwent palliative radiotherapy to a painful left calcaneal bone metastasis and right femur metastasis. He was placed on zoledronic acid for the osseous metastasis. He was seen by medical oncology to discuss systemic treatment options, but the patient declined the treatment. He was ultimately placed on hospice due to progressive metastatic disease and worsening functional status.
2.2. Genomic Profiling
The NGS analysis reported deletions of two tumor suppressor genes on chromosome 9p21 that encode three tumor suppressor proteins and a nonsense mutation in another tumor suppressor gene on chromosome 12q12 (Table 1). One tumor suppressor gene is CDKN2B (cyclin-dependent kinase inhibitor 2B), which encodes p15INK4B [10–13] (Figure 3). The second tumor suppressor gene is CDKN2A, which encodes two proteins, p16INK4A and p14ARF. p16INK4A and p15INK4B are CDK inhibitors that separately form complexes with cyclin-dependent kinase 4 (CDK4) and cyclin-dependent kinase 6 (CDK6) to inhibit their interaction with cyclin D. These CDK inhibitors induce cell cycle arrest at G1 checkpoints, preventing phosphorylation of retinoblastoma protein (Rb), thus halting the cell cycle. p14ARF is created from a unique splicing event and uses alternative reading frame in exons 2 and 3 of CDKN2A. p14ARF stabilizes the tumor suppressor p53 by inhibiting its negative regulator MDM2. Deletion of CDKN2A leads to loss of p14ARF, which causes abrogation of p53-dependent cell cycle arrest, apoptosis, and other tumor suppressor functions [14, 15].
Table 1.
Somatic genomic alterations.
| Gene | Chr | Coding change | Base change | Pathway |
|---|---|---|---|---|
| CDKN2A | 9 | Loss | Loss | Cyclin-dependent kinase inhibitor, Rb tumor suppressor |
| CDKN2B | 9 | Loss | Loss | Cyclin-dependent kinase inhibitor, Rb and p53 tumor suppressors |
| ARID2 | 12 | E267∗ | 799G>T | Chromatin remodeling, tumor suppressor |
| AXL | 19 | T112M | 335C>T | Receptor tyrosine kinase signaling |
| ESR1 | 6 | G90R | 268G>C | Transcription factor |
| FBXW7 | 4 | E192A | 575A>C | Phosphorylation-dependent ubiquitination |
| GNA13 | 17 | T365S | 1093A>T | G-protein signaling |
| KEAP1 | 19 | C624Y | 1871G>C | Sensor of oxidative stress |
| KLHL6 | 3 | S497T | 1490G>C | Receptor signaling |
| LRP1B | 2 | N1724Y | 5170A>T | Receptor-mediated endocytosis signaling |
| PLCG2 | 16 | F528C | 1583T>G | Transmembrane signaling enzyme |
| PRSS8 | 16 | A209T | 625G>A | Proteolytic enzymes |
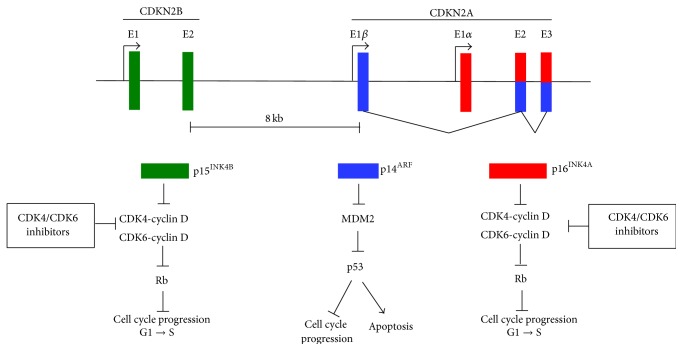
Figure 3.
The CDKN2A-CDKN2B locus regulates the cell cycle. Genomic organization of the 9p21 locus containing CDKN2B encoding p15INK4B and CDKN2A encoding p14ARF and p16INK4A. p14ARF is coded by exons E1β, E2, and E3 and p16INK4A is coded by exons E1α, E2, and E3. E2 and E3 are shared by p14ARF and p16INK4A, but they use alternate reading frames to produce unique protein sequences. p15INK4B and p16INK4A induce cell cycle arrest by complexing to CDK4 and CDK6 and preventing their complex formation with cyclin D. Rb remains active (dephosphorylated) and prevents E2F-mediated transcription of target genes necessary for cell cycle progression from G1 to S phase. p14ARF inhibits cell cycle progression and activates apoptosis by inhibiting MDM2, which leads to activation of p53.
Another tumor suppressor gene mutated in the patient's AciCC was AT-Rich Interactive Domain 2 (ARID2), which encodes AT-Rich Interactive Domain 2 protein (ARID2 protein), also known as BRG1-associated factor 200 (BAF200). ARID2 protein is a subunit of the polybromo- and BRG1-associated factor chromatin remodeling complex (PBAF) and appears to be necessary for complex stability [16]. In general, PBAF is responsible for nuclear receptor ligand-dependent transcriptional activation and is part of the SWI/SNF family of chromosome remodeling proteins [17]. A lack of ARID2 protein alters cellular transcription response to α-interferon [16] and inhibits osteoblast differentiation [18]. ARID2 protein has four domains: AT-rich interaction domain (ARID), RFX-like DNA binding motif (RFX), proline- and glutamine-rich region (GLN), and two C-terminal C2H2 zinc finger motifs (ZN) [19] (Figure 4). The patient harbored a loss-of-function (LoF) nonsense E267∗ mutation resulting in a truncated protein deficient in the RFX, GLN, and ZN.
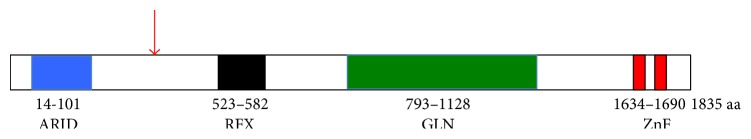
Figure 4.
The protein domain arrangement in ARID2 protein. ARID2 has four discrete domains: the ARID domain (residues 14–101), the RFX domain (residues 523–582), the GLN domain (residues 793–1128), and a domain that contains two ZnFs (1634–1690). The red arrow indicates the approximate location of the E267∗ nonsense mutation identified in the AciCC of the patient.
Nine missense mutations of unknown significance in AciCC were identified in AXL, ESR1, FBXW7, GNA13, KEAP1, KLHL6, LRP1B, PLCG2, and PRSS8 (Table 1). Other mutations in these nine genes have been reported in many cancer types underscoring the need for functional genomics to ascertain the significance of these particular changes in AciCC [20]. Interestingly, 9 of the 12 documented genomic insults are in genes involved in signaling pathways or regulation of the cell cycle (Table 1). Of the 10 point mutations observed in the sample, 8 were transversions.
3. Discussion
The case presented herein is of a highly aggressive AciCC in which both local recurrence and distant metastatic disease occurred within six months of curative intent surgery and adjuvant radiotherapy. Histologically, the presence of dedifferentiation was a poor prognostic feature. Furthermore, we detected two components juxtaposed to one another; one is well-differentiated with glandular architecture and the second shows infiltrative growth pattern associated with stromal response. The second area has dedifferentiated regions with focal necrosis, multiple foci of perineural invasion, pronounced pleomorphism, and increased mitotic activity. Interestingly, the juxtaposition of the two areas is a feature found in a subset of AciCC (n = 25) that failed to respond to extensive resection and adjuvant therapy [21]. In this subset, local recurrence or distant metastasis occurred in most patients with 72.7% dead with disease with a mean of 2.9 years.
In addition, targeted next generation sequencing revealed loss of tumor suppressor genes, CDKN2A, CDKN2B, and ARID2, as well as several mutations in genes involved in cell signaling, ubiquitination, cell cycle regulation, and endocytosis. The presence of the multitude of genetic alterations underscores the high grade, poorly differentiated nature of the tumor and, conversely, may help to explain its highly aggressive nature. Of course, because of the targeted nature of this analysis, it is quite possible that there are mutations in other critical genes not analyzed.
A single other report has been published where extensive genetic analysis of a salivary gland AciCC primary tumor was conducted [8]. In that report, whole exome sequencing analysis was performed. A comparison with our targeted sequencing study may shed light on features shared in the two cases. It was reported that a 58-year-old woman presented with a right parotid mass consistent with Warthin's tumor. An initial CT scan revealed a right parotid mass. A year later, another CT scan showed a mass centered in the right parotid effacing the jugular vein and abutting the mandible and skull base with extension along the facial nerve to the geniculate ganglion. Pathologic examination revealed a low grade AciCC with extensive perineural and lymphovascular spread and the lymph nodes negative for malignancy. In some respects, this contrasts with our patient who had a high grade AciCC and developed multiple metastases. However, similar to the previous case, the lymph nodes of our patient were negative. The previous case study reported that 35 genes were identified with somatic copy number aberrations and 14 with somatic single nucleotide variations. Somatic deletion of CDKN2A was the only genomic aberration detected in both patients, although we must consider that in our study only a fraction of the exome was analyzed.
In both salivary gland AciCC tumors, the CDK4/6/Rb and p53 pathways were targeted. It is worth noting that deletion of CDKN2A is a frequent event in heat and neck tumors. In one report, 22% of head and neck tumors (n = 279) have a deletion in CDKN2A [22]. Of the 27 active clinical trials underway for CDK4/6 inhibitors, four are specific for solid tumors with CDK4/6 pathway activation [23]. The FDA recently approved the CDK4/6 inhibitor palbociclib for treatment of metastatic breast cancer in combination with the aromatase inhibitor, letrozole [24]. In cancers that have genomic driver insults in CDKN2A or CDKN2B, inhibition of CDK4/6 is likely to be a therapeutic option worth investigating.
The other clinical target relevant to this study is MDM2 [25]. MDM2 binds to the p53 transactivation domain and mediates polyubiquitination of p53, leading to p53 proteolysis by the 26S proteasome [26]. Seven drugs that prevent complex formation between MDM2 and p53 are in phase I clinical trials. Because the drugs will also likely activate p53 in normal cells, it is expected that some side effects will occur. Indeed, early results from clinical trials show that two MDM2 targeting drugs cause thrombocytopenia and one drug causes apoptosis of megakaryocyte progenitor cells [25, 27, 28]. Other approaches to identify MDM2 inhibitors without severe side effects are being explored [29].
This case study is the first report of the ARID2 nonsense mutation (E267∗) in AciCC. Two other cases with this specific genomic insult (AA E267∗; nuc c799G>T) have been reported in hepatocellular carcinoma [30, 31]. A recent study revealed that 18.2% of patients with HCV-associated hepatocellular carcinoma have inactivating mutations in ARID2 [32]. ARID2 mutations have also been reported in pancreatic cancer, colorectal cancer, melanoma, and non-small-cell lung cancer [32–42]. Furthermore, in 60 hepatocellular samples evaluated by whole exome or genome sequencing, 35% harbored alterations in chromatin remodeling genes, including ARID1, another member of the ARID superfamily [32]. A study identified ARID2 loss-of-function mutations in 5% of non-small-cell lung cancers, making it the 6th most frequently mutated genes in this cancer type after TP53, KRAS, EGFR, CDKN2A, and STK11 [35].
In our AciCC case, we also observed missense mutations in AXL, ESR1, FBXW7, KEAP1, KLHL6, GNA13, PLCG2, and LRP1B. Mutations in these genes have been reported in non-head and neck cancers [30, 31]. AXL has been implicated in epithelial-mesenchymal transition (EMT) and has been hypothesized to play a key role not only in development of metastasis, but also in resistance to chemotherapy and targeted therapies, including lapatinib resistance in HER2-positive breast cancer [43] and in erlotinib resistance in EGFR-mutant non-small-cell lung cancer [44]. Among other salivary gland neoplasms, genomic alterations in FBXW7, KLHL6, and LRP1B have been identified in adenoid cystic carcinoma [33, 45]. Alterations of the oxidative stress gene KEAP1 was reported in human papilloma virus negative head-and-neck squamous cell carcinomas (HNSCCs) (5%) [46].
FBXW7 codes for F-box and WD40 repeat domain containing 7 protein (Fbxw7), a substrate recognition component of the SCF (Skp1-Cul1-F-box protein) type ubiquitin ligase complex. Fbxw7 plays a critical role in cell division, growth and differentiation by binding to proto-oncoproteins (c-Myc, Notch1, Notch4, c-Jun, cyclin E, KLF5, and mTOR) [47]. The binding domain within Fbxw7 is a conserved phosphorylated domain known as the Cdc4 phosphodegron and leads to ubiquitin-dependent proteolysis of the above proto-oncoproteins. Since most of its substrates are growth-promoting, Fbxw7 is thought of as a tumor suppressor. Overall, approximately 6% of multiple samples from 15 different human tumor types surveyed harbor FBXW7 mutations [48]. Most of these mutations result in amino-acid substitutions at key positions in the WD40 repeats with consequential disruption of substrate binding. Hot spot mutations were observed in the third WD40 repeat (nucleotides 1,393, 1,394 and 1,436). The remaining mutations were either nonsense mutations or mutations of unknown significance. The tumor in our case study harbored one of these mutations of unknown significance, E192A. A fibrolamellar HCC patient harboring the same mutation was treated with vorinostat and sirolimus as part of a phase I clinical trial for mTOR inhibitors. The time to treatment failure was 6.8+ months [49]. Thirty-one percent of T cell acute lymphocytic leukemia (T-ALL) patients have FBXW7 mutations with one reported case harboring the E192A mutation [50].
In this report, targeted next generation sequencing was performed on the excised primary parotid tumor. Recently, researchers used whole genome sequencing data to develop and validate “mutant-allele tumor heterogeneity” (MATH) as a measure of intratumor heterogeneity in HNSCC suggesting that it should be considered as a biomarker for survival in these cancers [51]. Given that high intratumor heterogeneity leads to worse clinical outcomes, one limitation of this study is that repeated sampling of the tumor with whole exome sequencing was not performed.
Recent cancer sequencing studies have unleashed an ever-expanding catalog of somatic aberrations, involving novel genes and pathways in many cases [46, 52–56]. This extensive mutational heterogeneity can inform genome forward treatment algorithms. This case study underscores the need to sequence more AciCC samples multiple times for the genomic, transcriptome, epigenomic, and proteomic annotation of molecular alterations. Such information will deepen our understanding of the pathophysiology, risk stratification, prognosis of AciCC, and guide precision genome forward treatment.
Supplementary Material
Table S1 of Supplementary Material presents the sequencing pipeline performance specifications and quality metrics. Table S2 lists the 315 genes (exons only) assayed by Foundation One. Table S3 lists the introns of 28 genes commonly involved in rearrangements assayed by Foundation One. These genes were confirmed to be somatically compromised in human solid tumors as evidenced by their validation for therapy (approved or clinical trials), and/or reported as unambiguous drivers of oncogenesis.
Acknowledgments
The authors would like to thank the patient and his family for their vision and altruism in making the medical records available. Their hope is that this case report will contribute to the available medical repository on this rare disease to the benefit of other patients. The authors acknowledge Lukas Wartman for advice on targeted gene sequencing. Wayne A. Warner was supported by Washington University School of Medicine, GSAS/CGFP Fund 94028C. Jamil Momand was supported by Grants NIGMS R01GM105898 and 5R25GM061331-12.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Speight P. M., Barrett A. W. Salivary gland tumours. Oral Diseases. 2002;8(5):229–240. doi: 10.1034/j.1601-0825.2002.02870.x. [DOI] [PubMed] [Google Scholar]
- 2.Surveillance. Seer∗Stat Database: Populations—Total U.S. (1969–2013) <Single Ages to 85+, Katrina/Rita Adjustment>—Linked to County Attributes—Total U.S., 1969–2013 Counties. National Cancer Institute, DCCPS, Surveillance Research Program, Surveillance Systems Branch; 2014. http://www.seer.cancer.gov. [Google Scholar]
- 3.Spiro R. H., Huvos A. G., Strong E. W. Acinic cell carcinoma of salivary origin. A clinicopathologic study of 67 cases. Cancer. 1978;41(3):924–935. doi: 10.1002/1097-0142(197803)41:3<924::aid-cncr2820410321>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 4.Dorn M. T., Wetherington R. W., Williams M. F. Pathologic quiz case 1. Acinic cell carcinoma of the deep lobe of the parotid gland involving the right parapharyngeal space. Archives of Otolaryngology—Head and Neck Surgery. 1999;125(6):694, 696–697. doi: 10.1001/archotol.125.6.694. [DOI] [PubMed] [Google Scholar]
- 5.Hoffman H. T., Karnell L. H., Robinson R. A., Pinkston J. A., Menck H. R. National cancer data base report on cancer of the head and neck: acinic cell carcinoma. Head and Neck. 1999;21(4):297–309. doi: 10.1002/(sici)1097-0347(199907)21:4lt;297::aid-hed2>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 6.North C. A., Lee D.-J., Piantadosi S., Zahurak M., Johns M. E. Carcinoma of the major salivary glands treated by surgery or surgery plus postoperative radiotherapy. International Journal of Radiation Oncology, Biology, Physics. 1990;18(6):1319–1326. doi: 10.1016/0360-3016(90)90304-3. [DOI] [PubMed] [Google Scholar]
- 7.DeVita V. T., Lawrence T. S., Rosenberg S. A. Devita, Hellman, and Rosenberg's Cancer: Principles & Practice of Oncology. Philadelphia, Pa, USA: Wolters Kluwer; 2015. [Google Scholar]
- 8.Nichols A. C., Chan-Seng-Yue M., Yoo J., et al. A case report and genetic characterization of a massive acinic cell carcinoma of the parotid with delayed distant metastases. Case Reports in Oncological Medicine. 2013;2013:7. doi: 10.1155/2013/270362.270362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frampton G. M., Fichtenholtz A., Otto G. A., et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nature Biotechnology. 2013;31(11):1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quelle D. E., Zindy F., Ashmun R. A., Sherr C. J. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83(6):993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 11.Sharpless N. E. INK4a/ARF: a multifunctional tumor suppressor locus. Mutation Research. 2005;576(1-2):22–38. doi: 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 12.Ozenne P., Eymin B., Brambilla E., Gazzeri S. The ARF tumor suppressor: structure, functions and status in cancer. International Journal of Cancer. 2010;127(10):2239–2247. doi: 10.1002/ijc.25511. [DOI] [PubMed] [Google Scholar]
- 13.Sherr C. J. Ink4-Arf locus in cancer and aging. Wiley Interdisciplinary Reviews: Developmental Biology. 2012;1(5):731–741. doi: 10.1002/wdev.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levine A. J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88(3):323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 15.Momand J., Wu H.-H., Dasgupta G. MDM2—master regulator of the p53 tumor suppressor protein. Gene. 2000;242(1-2):15–29. doi: 10.1016/s0378-1119(99)00487-4. [DOI] [PubMed] [Google Scholar]
- 16.Yan Z., Cui K., Murray D. M., et al. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes & Development. 2005;19(14):1662–1667. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemon B., Inouye C., King D. S., Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414(6866):924–928. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- 18.Xu F., Flowers S., Moran E. Essential role of ARID2 protein-containing SWI/SNF complex in tissue-specific gene expression. Journal of Biological Chemistry. 2012;287(7):5033–5041. doi: 10.1074/jbc.m111.279968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao H., Wang J., Han Y., et al. ARID2: a new tumor suppressor gene in hepatocellular carcinoma. Oncotarget. 2011;2(11):886–891. doi: 10.18632/oncotarget.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forbes S. A., Beare D., Gunasekaran P., et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Research. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson L. D., Aslam M. N., Stall J. N., Udager A. M., Chiosea S., McHugh J. B. Clinicopathologic and immunophenotypic characterization of 25 cases of acinic cell carcinoma with high-grade transformation. Head and Neck Pathology. 2015 doi: 10.1007/s12105-015-0645-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Clinicaltrials.Gov, US National Institutes of Health, http://www.clinicaltrials.gov/ct2/home.
- 24.Finn R. S., Crown J. P., Lang I., et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. The Lancet Oncology. 2015;16(1):25–35. doi: 10.1016/s1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Y., Aguilar A., Bernard D., Wang S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction (MDM2 inhibitors) in clinical trials for cancer treatment. Journal of Medicinal Chemistry. 2015;58(3):1038–1052. doi: 10.1021/jm501092z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendoza M., Mandani G., Momand J. The MDM2 gene family. Biomolecular Concepts. 2014;5(1):9–19. doi: 10.1515/bmc-2013-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iancu-Rubin C., Mosoyan G., Glenn K., Gordon R. E., Nichols G. L., Hoffman R. Activation of p53 by the MDM2 inhibitor RG7112 impairs thrombopoiesis. Experimental Hematology. 2014;42(2):137.e5–145.e5. doi: 10.1016/j.exphem.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 28.Ray-Coquard I., Blay J.-Y., Italiano A., et al. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. The Lancet Oncology. 2012;13(11):1133–1140. doi: 10.1016/s1470-2045(12)70474-6. [DOI] [PubMed] [Google Scholar]
- 29.Warner W. A., Sanchez R., Dawoodian A., Li E., Momand J. Identification of FDA-approved drugs that computationally bind to MDM2 . Chemical Biology and Drug Design. 2012;80(4):631–637. doi: 10.1111/j.1747-0285.2012.01428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forbes S. A., Tang G., Bindal N., et al. COSMIC (the catalogue of somatic mutations in cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Research. 2009;38(1):D652–D657. doi: 10.1093/nar/gkp995.gkp995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forbes S. A., Bhamra G., Bamford S., et al. Current Protocols in Human Genetics. chapter 10. John Wiley & Sons; 2008. UNIT 10.11 The catalogue of somatic mutations in cancer (COSMIC) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li M., Zhao H., Zhang X., et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nature Genetics. 2011;43(9):828–829. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho A. S., Kannan K., Roy D. M., et al. The mutational landscape of adenoid cystic carcinoma. Nature Genetics. 2013;45(7):791–798. doi: 10.1038/ng.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hodis E., Watson I. R., Kryukov G. V., et al. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manceau G., Letouzé E., Guichard C., et al. Recurrent inactivating mutations of ARID2 in non-small cell lung carcinoma. International Journal of Cancer. 2013;132(9):2217–2221. doi: 10.1002/ijc.27900. [DOI] [PubMed] [Google Scholar]
- 36.Nikolaev S. I., Rimoldi D., Iseli C., et al. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nature Genetics. 2012;44(2):133–139. doi: 10.1038/ng.1026. [DOI] [PubMed] [Google Scholar]
- 37.Stark M. S., Woods S. L., Gartside M. G., et al. Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nature Genetics. 2012;44(2):165–169. doi: 10.1038/ng.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei X., Walia V., Lin J. C., et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nature Genetics. 2011;43(5):442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong R., Liu L., Tian Y., et al. Genetic variant in SWI/SNF complexes influences hepatocellular carcinoma risk: a new clue for the contribution of chromatin remodeling in carcinogenesis. Scientific Reports. 2014;4, article 4147 doi: 10.1038/srep04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biankin A. V., Waddell N., Kassahn K. S., et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491(7424):399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cajuso T., Hänninen U. A., Kondelin J., et al. Exome sequencing reveals frequent inactivating mutations in ARID1A, ARID1B, ARID2 and ARID4A in microsatellite unstable colorectal cancer. International Journal of Cancer. 2014;135(3):611–623. doi: 10.1002/ijc.28705. [DOI] [PubMed] [Google Scholar]
- 42.Zhu B., Tian J., Zhong R., et al. Genetic variants in the SWI/SNF complex and smoking collaborate to modify the risk of pancreatic cancer in a Chinese population. Molecular Carcinogenesis. 2015;54(9):761–768. doi: 10.1002/mc.22140. [DOI] [PubMed] [Google Scholar]
- 43.Liu L., Greger J., Shi H., et al. Novel mechanism of lapatinib resistance in HER2-positive breast tumor cells: activation of AXL. Cancer Research. 2009;69(17):6871–6878. doi: 10.1158/0008-5472.can-08-4490. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Z., Lee J. C., Lin L., et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nature Genetics. 2012;44(8):852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stephens P. J., Davies H. R., Mitani Y., et al. Whole exome sequencing of adenoid cystic carcinoma. The Journal of Clinical Investigation. 2013;123(7):2965–2968. doi: 10.1172/jci67201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakayama K. I., Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nature Reviews Cancer. 2006;6(5):369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 48.Akhoondi S., Sun D., von der Lehr N., et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Research. 2007;67(19):9006–9012. doi: 10.1158/0008-5472.can-07-1320. [DOI] [PubMed] [Google Scholar]
- 49.Jardim D. L., Wheler J. J., Hess K., et al. FBXW7 mutations in patients with advanced cancers: clinical and molecular characteristics and outcomes with mTOR inhibitors. PLoS ONE. 2014;9(2) doi: 10.1371/journal.pone.0089388.e89388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Vlierberghe P., Ambesi-Impiombato A., De Keersmaecker K., et al. Prognostic relevance of integrated genetic profiling in adult T-cell acute lymphoblastic leukemia. Blood. 2013;122(1):74–82. doi: 10.1182/blood-2013-03-491092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mroz E. A., Tward A. M., Hammon R. J., Ren Y., Rocco J. W., Beck A. H. Intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the cancer genome atlas. PLOS Medicine. 2015;12(2) doi: 10.1371/journal.pmed.1001786.e1001786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L., Xiao Y., Ping Y., et al. Integrating multi-omics for uncovering the architecture of cross-talking pathways in breast cancer. PLoS ONE. 2014;9(8) doi: 10.1371/journal.pone.0104282.e104282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parfenov M., Pedamallu C. S., Gehlenborg N., et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(43):15544–15549. doi: 10.1073/pnas.1416074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leiserson M. D. M., Vandin F., Wu H.-T., et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nature Genetics. 2015;47(2):106–114. doi: 10.1038/ng.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davis C. F., Ricketts C. J., Wang M., et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014;26(3):319–330. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.The Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159(3):676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 of Supplementary Material presents the sequencing pipeline performance specifications and quality metrics. Table S2 lists the 315 genes (exons only) assayed by Foundation One. Table S3 lists the introns of 28 genes commonly involved in rearrangements assayed by Foundation One. These genes were confirmed to be somatically compromised in human solid tumors as evidenced by their validation for therapy (approved or clinical trials), and/or reported as unambiguous drivers of oncogenesis.