Abstract
Hepatocellular carcinoma (HCC) occurs predominantly in patients with liver cirrhosis. Here, we show an innovative RNA-based targeted approach to enhance endogenous albumin production whilst reducing liver tumour burden. We designed short-activating RNAs (saRNA) to enhance expression of C/EBPα (CCAAT/enhancer-binding protein-α), a transcriptional regulator and activator of albumin gene expression. Increased levels of both C/EBPα and albumin mRNA in addition to a 3-fold increase in albumin secretion and 50% decrease in cell proliferation was observed in C/EBPα-saRNA transfected HepG2 cells.
Intravenous injection of C/EBPα-saRNA in a cirrhotic rat model with multifocal liver tumours increased circulating serum albumin by over 30% showing evidence of improved liver function. Tumour burden decreased by 80% (p = 0.003) with a 40% reduction in a marker of pre-neoplastic transformation.
Since C/EBPα has known anti-proliferative activities via retinoblastoma, p21 and cyclins; we used mRNA expression liver cancer specific microarray in C/EBPα-saRNA transfected HepG2 cells to confirm down-regulation of genes strongly enriched for negative regulation of apoptosis, angiogenesis and metastasis. Up-regulated genes were enriched for tumour suppressors and positive regulators of cell differentiation. A quantitative PCR and Western-blot analysis of C/EBPα-saRNA transfected cells suggested that in addition to the known anti-proliferative targets of C/EBPα, we also observed suppression of IL6R, c-Myc and reduced STAT3 phosphorylation.
Conclusion
We demonstrate for the first time that a novel injectable saRNA-oligonucleotide that enhances C/EBPα expression successfully reduces tumour burden and simultaneously improves liver function in a clinically relevant liver cirrhosis/HCC model.
Keywords: Liver cirrhosis, Hepatocellular Carcinoma, Short-activating RNA, PAMPAM dendrimer nanoparticles, C/EBPα transcription factor
INTRODUCTION
Human hepatocellular carcinoma (HCC) is currently the third most common cause of cancer related mortality worldwide.1 The majority of patients with HCC develop malignant tumours from a background of liver cirrhosis. Currently most patients are diagnosed at an advanced disease stage and therefore the 5 year survival for the majority of HCC patients remain dismal.2 Surgical resection, loco-regional ablation and liver transplantation are currently the only therapeutic options which have the potential to cure HCC. However, based on the evaluation of individual liver function and tumour burden only about 5–15% of patients are eligible for surgical intervention.3
Most eukaryotic cells use RNA-complementarity as a mechanism for regulating gene expression. One example is the classic RNA interference (RNAi) pathway which uses double stranded short interfering RNAs to knockdown gene expression via the RNA-induced silencing complex (RISC).4 It is now established that short duplex RNA oligonucleotides also have the ability to target the promoter regions of genes and mediate transcriptional activation of these genes and they have been referred to as RNA activation (RNAa), antigene RNAs (agRNAs), short-activating RNA (saRNA).5–8 SaRNA induced activation of genes appears to be conserved in other mammalian species including mouse, rat, and non-human primates and is fast becoming a popular method for studying the effects of endogenous up-regulation of genes.5 SaRNAs have recently been designed to activate expression of genes such as p21 as potential therapy for the treatment of HCC or prostate cancer thus demonstrating a promising novel approach for adjuvant therapy.9,10
With the same iterative approach that we previously used to design saRNAs specific for Kruppel-like factor 4 (Klf4), c-Myc and MafA7,11, we generated saRNA molecules to up-regulate transcript levels of the CCAAT/enhancer-binding protein alpha (C/EBPα) gene.
C/EBPα is a leucine zipper protein that is conserved across humans and rats. This transcription factor is enriched in hepatocytes, myelomonocytes, adipocytes, as well as mammary epithelial cells including other cell types.12 In the adult liver, C/EBPα is defined as functioning in terminally differentiated hepatocytes whilst rapidly proliferating hepatoma cells express only a fraction of C/EBPα.13 C/EBPα is known to up-regulate p21, a strong inhibitor of cell proliferation through the up-regulation of retinoblastoma and inhibition of Cdk2 and Cdk414,15. Since the binding sites for the family of C/EBP transcription factors are present in the promoter regions of numerous genes that are involved in the maintenance of normal hepatocyte function and response to injury (including albumin, interleukin 6 response, energy homeostasis, ornithine cycle regulation and serum amyloid A expression)16–20; we determined the therapeutic benefit of up-regulating expression of C/EBPα in cirrhotic rats with compromised liver function and HCC by using saRNA as a safe and potentially clinically translatable method of targeted gene up-regulation.
For targeted in vivo delivery, we complexed C/EBPα-saRNA into the structurally flexible triethanolamine (TEA)-core poly (amidoamine) (PAMAM) dendrimer.21 The in vivo efficacy of these nanoparticles have previously been evaluated where biodistribution studies show that the dendrimers preferentially accumulate in peripheral blood mononuclear cells and liver with no discernible toxicity.21 Here we demonstrate the therapeutic effect of intravenously injecting C/EBPα-saRNA-dendrimers in a clinically relevant rat liver tumour model.44
After three doses through tail vein injection at 48hour intervals, the treated cirrhotic rats showed significantly increased serum albumin levels within one week. More important was the unexpected observation that liver tumour burden significantly decreased in the C/EBPα-saRNA-dendrimer treated groups. This study demonstrates, for the first time, that gene targeting by small RNA molecules can be used by systemic intravenous administration to simultaneously ameliorate liver function and reduce tumour burden in cirrhotic rats with HCC.
EXPERIMENTAL PROCEDURES
Full methods for designing short-activating RNA, animal experiments, nuclease activity, assessment of tumour burden, immuno-staining, qRT-PCR, gene microarray profiling, ChIP-seq analysis, gene ontology enrichment analysis and gene methylation analysis are available in the Supporting Information.
Design of short activating RNA oligonucleotides
The gene sequence of albumin and C/EBPα was selected for designing short activating RNA molecules for its specific activation using the parameters previously described.7
Transfection of saRNA oligonucleotides into HepG2 and rat-liver epithelial cell lines
HepG2 is a liver cell line derived from a human hepatoblastoma that is free of known hepatotropic viral agents and expresses genes involved in a wide variety of liver-specific metabolic functions.22 HepG2 cells were cultured in Roswell Park Memorial Institute medium (RPMI) supplemented with 100 units/ml penicillin, 0.1mg/ml streptomycin, 2mmol/L glutamine (Sigma) and 10% fetal bovine serum (Labtech International). For C/EBPα-saRNA transfection, cells were grown to 60% confluency in 24 well plates prior to transfection of 5, 10 and 20 nmoles of saRNA using Nanofectamine (PAA, UK) following the manufacturer's protocol. This process was repeated three times at 16 hours intervals before cells were harvested for isolation of total RNA for mRNA analysis.
Albumin ELISA
Rat liver epithelial cells and HepG2 cells were cultured in phenol-red free RPMI media in the presence of charcoal stripped FCS. Following three sets of saRNA transfections at 8 hours, 16 hours and 24 hours, the culture media was collected for total murine albumin ELISA (Assay Max, Albumin ELISA, Assay Pro USA) following the manufacturer’s instructions.
WST-1 assay
Cell proliferation was quantified at 16, 24 and 96 hours following C/EBPα-saRNA transfection by mitochondrial dehydrogenase expression analysis, using WST-1 reagent following the manufacturer’s guideline (Roche, UK). Briefly, the WST-1 reagent was used at 1:100 dilution to plates and incubated for one hour. The enzymatic reaction was measured at 450 nm using Bio-Tek ELISA reader.
Isolation of total RNA from cell lines
Total RNA extraction from cell lines was performed using the RNAqueous-Micro kit (Ambion, UK) following the manufacturer's instructions. Briefly, the cells were gently centrifuged followed by 3 pulses of sonication at Output 3 in Lysis buffer (Ambion, UK). The cell lysates were then processed through an RNA binding column, followed by multiple washes and elution. The total RNA isolated was quantified by a Nanodrop 2000 spectrophotometer. 500ng of total extracted RNA was processed for elimination of genomic DNA followed by reverse transcription using the QuantiTect® Reverse Transcription kit from Qiagen.
Animal experiments
We used a clinically relevant rat liver tumour model previously described.23 For in vivo therapy C/EBPα-saRNA was reconstituted with 100µl of RNase/Dnase free H2O; 50 µl of 20nM saRNA oligonucleotide and 50 µl of (TEA) core PAMAM dendrimer, previously described.24,25 10 cirrhotic animals were treated with 3 × doses via tail vein injections in the 1st week. Control animals (n=10) were injected with equal volume of PBS or scramble-saRNA. All animals received humane care according to the criteria outlined in the "Guide for the Care and Use of Laboratory Animals" prepared by the National Academy of Sciences and published by the National Institutes of Health (NIH publication 86-23 revised 1985).
RESULTS
Expression level of C/EBPα and Albumin in HepG2 Cells transfected withC/EBPα-saRNA
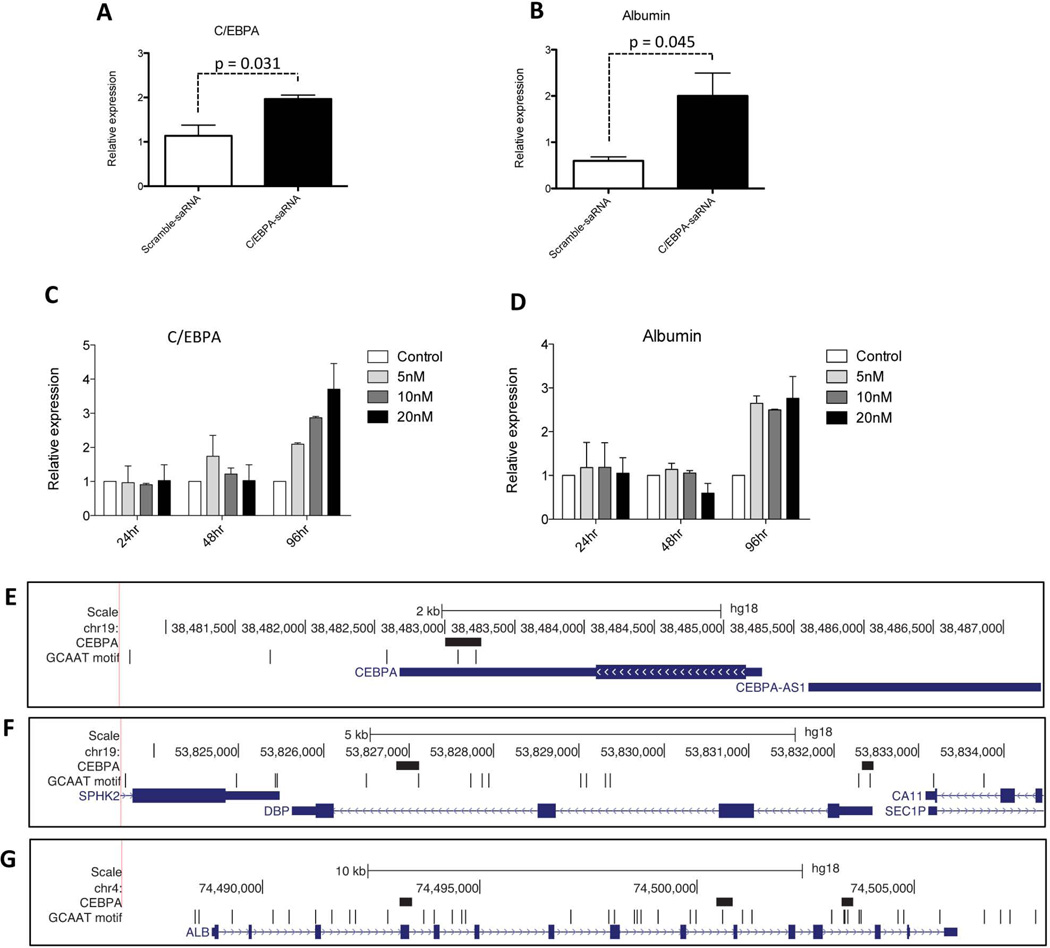
We assessed the effect of transfecting C/EBPα-saRNA on C/EBPα and albumin transcript levels. Both C/EBPα (Fig. 1A) and albumin transcripts (Fig. 1B) increased over two fold. Increasing the amounts of C/EBPα-saRNA (5, 10, and 20 nM (nanomoles) dose dependently enhanced C/EBPα transcript levels (Fig. 1C). The maximum expression of albumin was achieved with 5 nM of C/EBPα-saRNA, with no further dose dependent increase at higher saRNA levels (Fig. 1D). Analysis of the promoter regions of C/EBPα (Fig. 1E), the binding box of albumin (DBP) (Fig. 1F) and albumin (Fig. 1G) showed the presence of the core C/EBPα binding motifs (GCAAT) thus supporting targeting of both transcripts by C/EBPα-saRNA induced up-regulation of C/EBPα. An EpiTect Methyl PCR assay also demonstrated reduced methylation at the CpG-island of both C/EBPA and DBP promoters following transfection of C/EBPα-saRNA (Figs. 2A and 2B).
Fig. 1. Transfection of C/EBPα-saRNA in HepG2 cells regulates expression of C/EBPα and albumin.
(A) HepG2 cells were transfected with 20 nM C/EBPα-saRNA and harvested for total RNA extraction and reverse transcription for quantitative analysis of C/EBPα and (B) albumin gene expression. (C) A dose escalation of C/EBPα-saRNA demonstrates its effect over 96 hours (hr) on C/EBPα gene expression and (D) albumin gene expression. Data represents mean ± SD. The panels show the genomic region containing (E) C/EBPα, (F) DBP and (G) Albumin (ALB) 2000 nucleotides upstream and downstream of each gene where all have one or more C/EBPα binding sites. The figure panels show the chromosomal coordinates ("Scale" and chromosome identifier), C/EBPα binding sites ("C/EBPA"; black boxes), occurrence of the C/EBPα binding motif ("GCAAT motif"; black vertical lines), and RefSeq genes (blue boxes and lines) within the genomic regions.
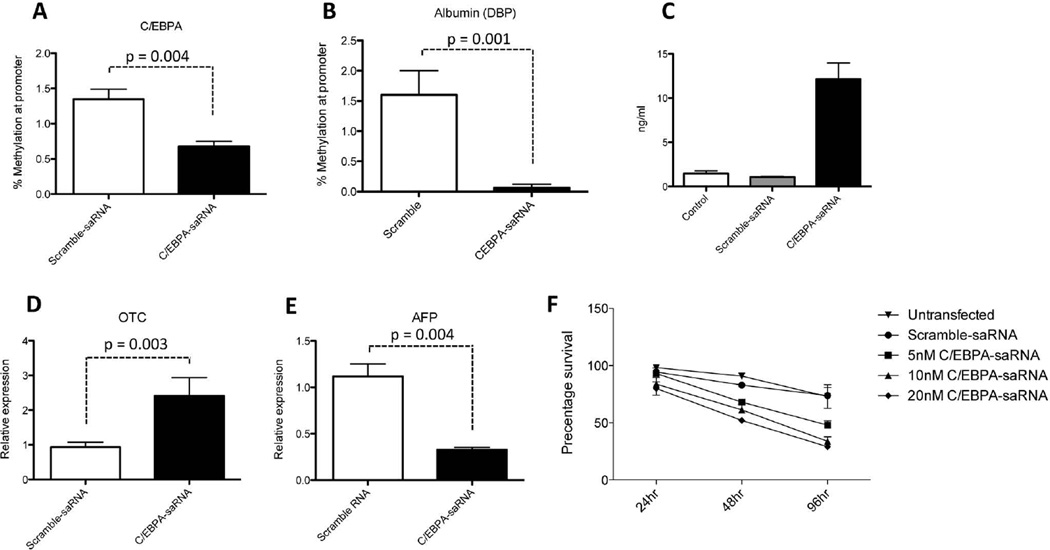
Fig. 2. Transfection of C/EBPα-saRNA in HepG2 cells regulates hepatocyte function and affects cell proliferation.
(A) Methylation assay of the CpG islands at the promoter regions of C/EBPA and (B) DBP demonstrated reduction in methylation when compared to control. Data represents mean ± SD. (C) An enzyme linked immunosorbent assay (ELISA) specific for human albumin detected a significant increase of albumin secretion following transfection of 20 nM CEPBA-saRNA. Data represents mean ± SD. (D) Expression of the gene encoding ornithine cycle enzyme Ornithine transcarbamylase (OTC) increased in C/EBPα-saRNA transfected cells suggesting an improved ability of urea production. (E) Decreased expression of the gene encoding alphafetoprotein (AFP) suggested improved regulation of cell differentiation. Data represents mean ± SD. (F) A WST-1 cell proliferation assay on HepG2 cells over 96 hr transfected with increasing amounts of C/EBPα-saRNA. Results show a dose dependent reduction in cell proliferation. Data represents mean ± SD.
To determine the biological relevance of increased albumin mRNA transcripts in C/EBPα-saRNA transfected HepG2 cells, a human albumin specific enzyme-linked-immunosorbent assay (ELISA) was performed. Secreted albumin peptide was detected in the culture media of the transfected cells (Fig. 2C).
To establish if enhanced albumin secretion in HepG2 cells by C/EBPα-saRNA also affected other hepatocytes specific functions and maintenance of hepatocyte differentiation, we measured expression levels of the ornithine cycle enzyme ornithine transcarbamylase (OTC) and alpha-fetoprotein (AFP). C/EBPα-saRNA caused an increase in OTC levels (Fig. 2D) suggesting an improved ability of urea production. The expression level of AFP decreased (Fig. 2E) indicative of the negative regulation typically observed with normal hepatocytes.26 In addition to the observed gene changes described, we also observed that C/EBPα-saRNA caused a marked down-regulation of HepG2 cell proliferation (Fig. 2F). This observation confirms the known anti-proliferative effects of C/EBPα.14,27
Intravenous injection of C/EBPα-saRNA in male Wistar rats bearing liver cirrhosis/HCC promoted increased circulating levels of albumin, amelioration of liver function and a reduced tumour burden
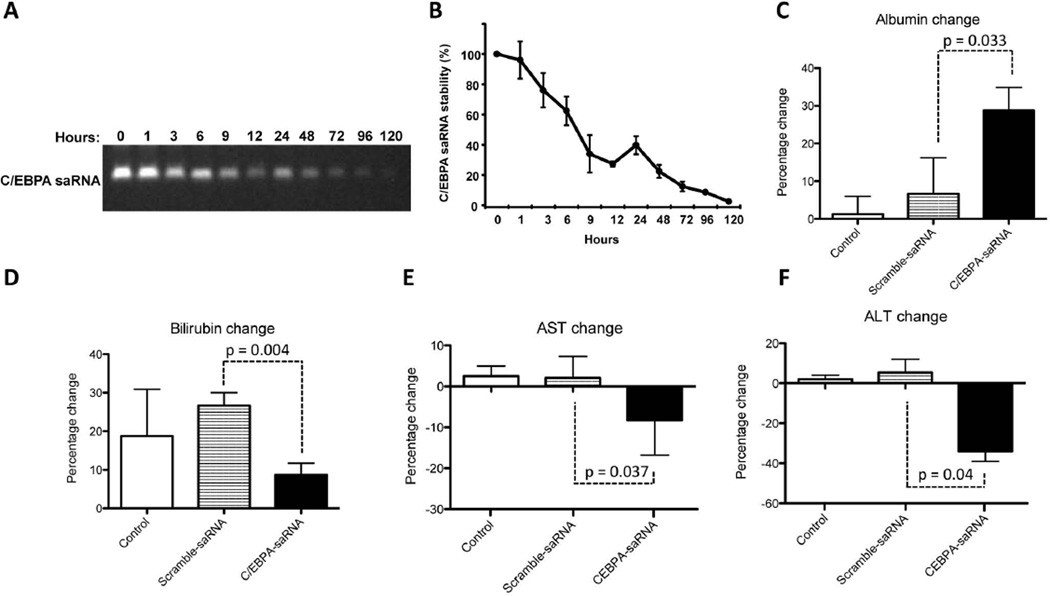
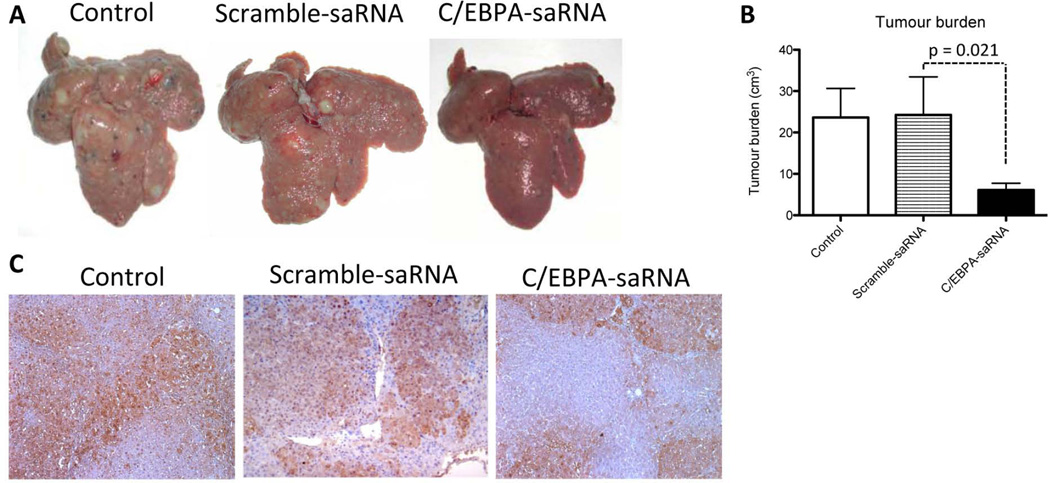
The stability of C/EBPα-saRNA was initially tested in circulating serum by performing a nuclease activity assay using blood samples from C/EBPα-saRNA treated rats. We observed a significant reduction in the stability of C/EBPα-saRNA duplex by 48 hours (Figs. 3A and 3B). We thus injected cirrhotic rats over a period of one week with repeat doses of C/EBPα-saRNA-dendrimer. Measurement of circulating albumin showed a significant increase of over 30% after three doses of C/EBPα-saRNA-dendrimer injection when compared to PBS control or scramble-saRNA-dendrimer control groups (Fig. 3C). Further blood analysis demonstrated that the worsening of bilirubin levels was significantly less in the C/EBPα-saRNA-dendrimer treated group by at least 17% when compared to both control groups (Fig 3D). There was also a significant drop in levels of the liver enzymes aspartate aminotransferase (AST) and alanine aminotransferase (ALT) by at least 10% and 30% respectively in the C/EBPα-saRNA-dendrimer treated group when compare to both control groups (Fig 3E and 3F). Histological examination of the liver showed a significant reduction in tumour nodules from C/EBPα-saRNA-dendrimer injected rats when compared to both control groups (Fig 4A and 4B). These results were consistent with immunohistology studies of tissue sections from C/EBPα-saRNA treated rat liver stained for placenta-form of glutathione S-transferase (GST-p). Independent conclusions by two pathologists suggested that there was evidence of reduced carcinogenesis by treatment of C/EBPα-saRNA-dendrimer when compared to the PBS control or scramble-saRNA-dendrimer control groups. Furthermore there were no differences in liver fibrosis between the PBS control; scramble-saRNA-dendrimer or C/EBPα-saRNA-dendrimer treated groups (Fig. 4C). The average density of positive staining for GST-p from control groups was 70 (± 5.0 %), and that from C/EBPα-saRNA-dendrimer injected rats was 32 (± 6.5%). Since overexpression of GST-p is observed during rat liver pre-neoplastic state and neoplastic transformation28,29, this data suggests that C/EBPα-saRNA-dendrimer treatment may reduce this process.
Fig. 3. Intravenous injection of C/EBPα-saRNA-dendrimer in male Wistar rats with liver cirrhosis and HCC shows improved liver function.
(A) C/EBPα-saRNA-dendrimer was tested for nuclease sensitivity in rat serum for the indicated times. RNA stability was visualised on a 2% denaturing agarose gel and (B) quantified by densitometry analysis. (Data represents mean ± SD, n=3). (C) C/EBPα-saRNA injected rats showed a significant change in circulating levels of albumin when compared to PBS control (Control) or scramble-saRNA-dendrimer control groups. (D) Changes in bilirubin levels suggested that C/EBPα-saRNA-dendrimer injected rats had at least a 10% improvement when compared to both control groups. Changes in (E) aspartate aminotransferase (F) (AST) and alanine aminotransferase (ALT) demonstrated at least a 10% and 30% improvement in values when compared to the control groups. Data represents mean ± SD.
Fig. 4. Intravenous injection of C/EBPα-saRNA-dendrimer in male Wistar rats with liver cirrhosis and HCC shows reduced tumour burden.
(A) Liver tumour nodules were visibly reduced in C/EBPα-saRNA injected rats when compared to both PBS control and Scramble-saRNA control groups. (B) Tumour burden was assessed by the volume of all tumour nodules with a diameter in excess of 3 mm. C/EBPα-saRNA injected rats had significantly reduced tumour burden after two weeks of treatment when compared to both control groups. Data represents mean ± SD. (C) 2 µm liver sections from PBS control, scramble-saRNA-dendrimer control and C/EBPα-saRNA-dendrimer injected rats were immunostained for expression of placenta-form glutathione-S-transferase (GST-p). PBS control rats showed 70% (± 5.0 %) of positive staining for the pre-neoplastic marker, scramble-saRNA-dendrimer injected rats showed 64% (± 10.0 %) of positive staining whilst C/EBPα-saRNA-dendrimer injected rats only showed 32% (± 6.5 %) of positive staining.
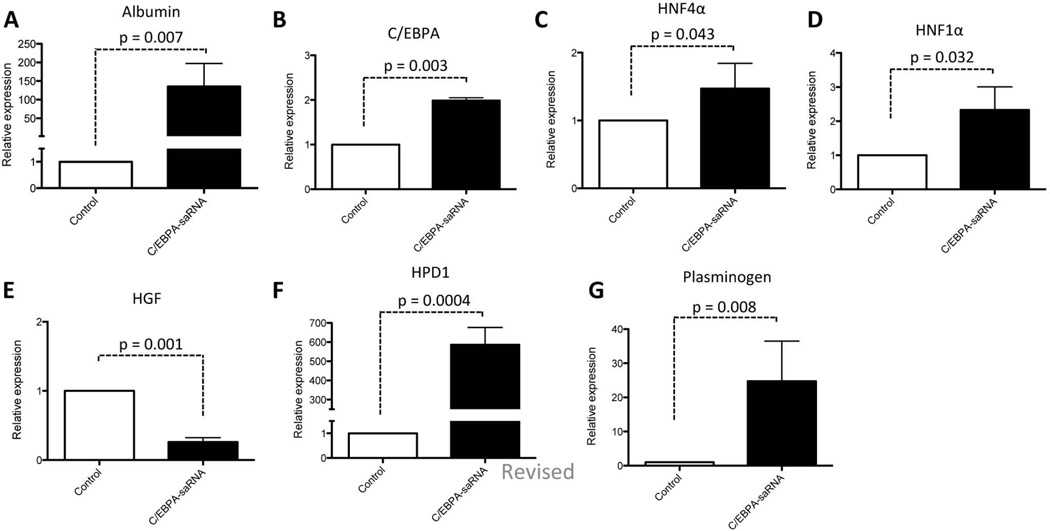
Total RNA extracted from liver biopsies of 7 animals from each group were screened for transcript levels of albumin (Fig. 5A), C/EBPα (Fig. 5B), hepatocyte nuclear factor 4-alpha (HNF4α) (Fig. 5C) and hepatocyte nuclear factor 1-alpha (HNF1α) (Fig. 5D). A significant increase in mRNA level was observed for all the factors, consistent with the role of HNF4α in hepatocyte differentiation together with C/EBPα and HNF1α in promoting expression of albumin. Taken together, lower mRNA levels of hepatocyte growth factor (HGF) (Fig. 5E) and increased levels of 4-hydroxyphenylpyruvic acid dioxygenase (HPD1) (Fig. 5F) and plasminogen (Fig. 5G) are suggestive of improved liver function in these cirrhotic rats treated with C/EBPα-saRNA-dendrimer.30
Fig. 5. Intravenous injection of C/EBPα-saRNA-dendrimer in male Wistar rats with liver cirrhosis and HCC positively regulates expression of factors for liver function.
(A) Total RNA extracts from 7 control rats vs 7 C/EBPα-saRNA injected rats were analysed for albumin gene expression, (B) C/EBPα gene expression, (C) HFN4α gene expression and (D) HNF1α gene expression showed an increase in these factors. (E) Decreased mRNA levels encoding HGF and (F) increased levels of hydroxyphenylpyruvic acid dioxygenase (HPD1) and (G) plasminogen indicated positive regulation of cell proliferation and improved liver function. Data represents mean ± SD.
Pathway gene microarray analysis suggests that C/EBPα-saRNA contributes to up-regulation of tumour suppressor genes and down-regulation of genes involved in liver cancer
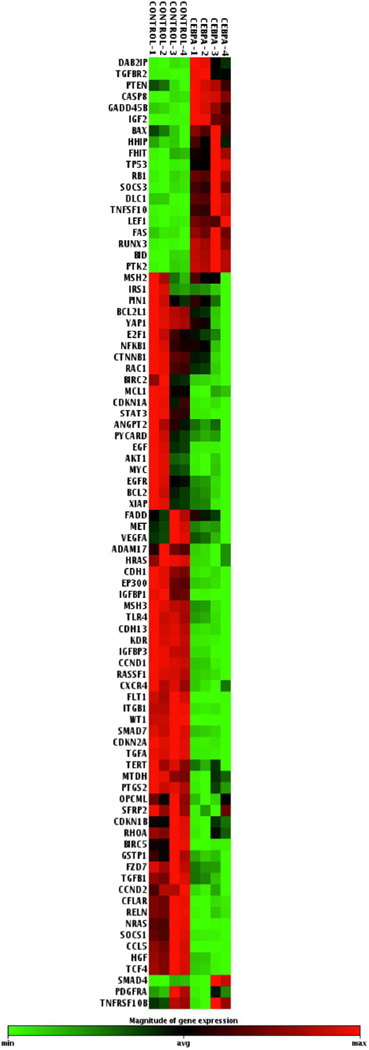
To investigate other liver specific factors that might be affected in response to C/EBPα-saRNA; we analysed the gene expression profile of a panel of 84 liver cancer specific genes (Qiagen/SABiosciences Human Liver Cancer RT2 profiler™) in C/EBPα-saRNA transfected HepG2 cells (Fig 6). Of particular interest was the observed up-regulation of 20 genes (Table 1), 18 of which are known tumour suppressor genes in HCC (Table 3) including RB. The most significantly up-regulated (over 3 fold) included the death agonist gene BH3-interacting domain (BID), and tumour protein 53 gene (TP53), encoding p53. BID interacts with BCl2-associated X protein (BAX) which in turn is up-regulated by wild type p53 to regulate cell cycle arrest and apoptosis in response to DNA damage.31,32
Fig. 6. Microarray analysis of 84 liver cancer pathway genes.
Negative regulation of genes (in green) or positive regulation of genes (in red) following transfection of 20 nM C/EBPα-saRNA in HepG2 cells.
Control (CONTROL-1 to 4) and C/EBPA-saRNA transfected (C/EBPA-1 to 4) are shown as 4 repeats.
Table 1. Gene expression (up-regulated by C/EBPA-saRNA).
Clustered analysis of genes positively regulated following transfection of C/EBPα-saRNA in HepG2 cells.
| Gene Symbol | Fold Up-Regulation | T-TEST |
|---|---|---|
| C/EBPAsaRNA/Control Group | p value | |
| BAX | 1.12 | 0.0027 |
| BID | 13.58 | 0.0001 |
| CASP8 | 6.69 | 0.0000 |
| DAB2IP | 2.59 | 0.0042 |
| DLC1 | 4.84 | 0.0001 |
| FAS | 1.64 | 0.0004 |
| FHIT | 2.84 | 0.0021 |
| GADD45B | 3.35 | 0.0001 |
| HHIP | 1.59 | 0.0054 |
| IGF2 | 9.75 | 0.0001 |
| LEF1 | 17.86 | 0.0001 |
| PTEN | 1.28 | 0.0013 |
| PTK2 | 2.87 | 0.0001 |
| RB1 | 1.96 | 0.0001 |
| RUNX3 | 6.01 | 0.0002 |
| SMAD4 | 1.72 | 0.0019 |
| SOCS3 | 6.52 | 0.0003 |
| TGFBR2 | 3.71 | 0.0025 |
| TNFSF10 | 3.53 | 0.0003 |
| P53 | 3.91 | 0.0018 |
Table 3. Up-regulation of tumour supressor genes by C/EBPA-saRNA.
Analysis of tumour suppressor genes up-regulated following transfection of C/EBPα-saRNA in HepG2 cells.
| Gene Symbol | Gene Function | Fold Up-Regulation | T-TEST |
|---|---|---|---|
| C/EBPAsaRNA/Control Group | p value | ||
| BAX | Apoptosis | 1.12 | 0.0027 |
| BID | 13.58 | 0.0001 | |
| CASP8 | Apoptosis, angiogenesis | 6.69 | 0.0000 |
| DLC1 | Apoptosis, Ras/Raf/MEK/ERK, small GTPase-mediated signalling | 4.84 | 0.0001 |
| FAS | 1.64 | 0.0004 | |
| FHIT | 2.84 | 0.0021 | |
| GADD45B | Apoptosis, cell cycle | 3.35 | 0.0001 |
| RUNX3 | 6.01 | 0.0002 | |
| SOCS3 | Apoptosis, adhesion & proteolysis | 6.52 | 0.0003 |
| TNFSF10 | 3.53 | 0.0003 | |
| PTEN | Cell cycle, PI3K/AKT, adhesion & proteolysis, angiogenesis | 1.28 | 0.0013 |
| RB1 | Cell cycle, classical WNT, Ras/Raf/MEK/ERK, small GTPase-mediated signalling | 1.96 | 0.0001 |
| IGF2 | Cell cycle, IGF/IGFR signalling | 9.75 | 0.0001 |
| TP53 | DNA damage, Ras/Raf/MEK/ERK, small GTPase-mediated signalling | 3.91 | 0.0018 |
| DAB2IP | Small GTPase-mediated signalling | 2.59 | 0.0042 |
| HHIP | Hedgehog signalling | 1.59 | 0.0054 |
| SMAD4 | TGFβ signalling, epithelial to mesenchymal transition | 1.72 | 0.0019 |
| TGFBR2 | TGFβ signalling, angiogenesis | 3.71 | 0.0025 |
Growth arrest and DNA-damage-inducible, 45 beta (GADD45B), also up-regulated, is a member of the growth arrest DNA damage inducible gene family associated with cell growth control where together with p53 induces hepatoprotection in HepG2 cells.33 Deleted in Liver Cancer 1 (DLC1) gene is a reported tumour suppressor for human liver cancer inhibiting cell growth and proliferation, as well as inducing apoptosis.34 Our data suggests that DLC1 is up-regulated in C/EBPα-saRNA transfected HepG2 cells (Table 3).
Runt-related transcription factor-3 (RUNX3) is a member of the runt domain family of transcription factor and has been frequently been observed in HCC where its expression is significantly lower than in surrounding normal tissue.35 Since ectopic expression of RUNX3 reverses epithelial-mesenchymal transition (EMT) in HCC cells36, we also observed, in the C/EBPα-saRNA transfected HepG2 cells, an up-regulation of RUNX3 (Table 3) and down-regulation of 4 genes involved in EMT. These included CTNB1 (encoding β-catenin), Hepatocyte growth factor (HGF), Small body size mothers against decapentaplegic homolog 7 (SMAD7), and Transforming factor beta 1 (TGFB1) (Table 4).
Table 4. Analysis of genes down-regulated by C/EBPA-saRNA.
Analysis of genes down-regulated following transfection of C/EBPα-saRNA in HepG2 cells.
| Gene Symbol | Gene Function | Fold Down-Regulation | T-TEST |
|---|---|---|---|
| C/EBPAsaRNA/Control Group | p value | ||
| CCND1 | Classical Wnt, cell cycle, DNA damage | −6.45 | 0.0001 |
| CDKN2A | Classical Wnt, Ras/Raf/MEK/ERK & Small GTPase-mediated signalling, cell cycle | −6.75 | 0.0001 |
| CTNNB1 | Classical Wnt, epithelial to mesenchymal transition (EMT), angiogenesis | −1.59 | 0.0081 |
| FZD7 | Classical Wnt | −2.00 | 0.0004 |
| MTDH | −1.73 | 0.0014 | |
| PIN1 | −1.15 | 0.0012 | |
| TCF4 | −9.96 | 0.0008 | |
| SMAD7 | TGFβ signalling, EMT, adhesion & proteolysis | −3.46 | 0.0002 |
| TGFB1 | TGFβ signalling, EGFR signalling, EMT, immune & inflammatory response | −1.80 | 0.0002 |
| AKT1 | PI3K/AKT signalling, adhesion & proteolysis | −1.78 | 0.0220 |
| IRS1 | PI3K/AKT signalling | −1.61 | 0.0010 |
| IGFBP1 | IGF/IGFR signalling | −7.36 | 0.0001 |
| IGFBP3 | −17.56 | 0.0001 | |
| IRS1 | −1.61 | 0.0010 | |
| YAP1 | Hippo signalling | −1.58 | 0.0058 |
| CDKN1A | Ras/Raf/MEK/ERK & small GTPase-mediated signalling, cell cycle, CDKN1A | −2.53 | 0.0016 |
| HRAS | Ras/Raf/MEK/ERK & small GTPase-mediated signalling | −3.69 | 0.0001 |
| NRAS | −29.47 | 0.0001 | |
| RAC1 | Ras/Raf/MEK/ERK & small GTPase-mediated signalling, immune & inflammatory response, adhesion & proteolysis | −1.46 | 0.0069 |
| RHOA | −1.46 | 0.0019 | |
| RASSF1 | Ras/Raf/MEK/ERK & small GTPase-mediated signalling, cell cycle | −4.59 | 0.0001 |
| ADAM17 | EGFR signalling, adhension & proteolysis | −2.19 | 0.0007 |
| CDH13 | EGFR & small GTPase-mediated signalling, adhesion & proteolysis, angiogenesis | −5.52 | 0.0001 |
| EGF | EGFR signalling, angiogenesis | −8.63 | 0.0074 |
| EGFR | EGFR signalling, adhesion & proteolysis | −2.58 | 0.0064 |
| TGFA | EGFR signalling | −9.27 | 0.0001 |
| HGF | MET/HGF signalling, EMT | −11.07 | 0.0001 |
| MET | MET/HGF signalling | −1.30 | 0.0247 |
| RELN | Small GTPase-mediated signalling, adhesion & proteolysis | −2.09 | 0.0005 |
| CDKN1B | Cell cycle | −1.74 | 0.0145 |
| MYC | −1.76 | 0.0132 | |
| E2F1 | Cell cycle, apoptosis | −1.40 | 0.0174 |
| EP300 | Cell cycle, apoptosis, adhesion & proteolysis | −3.28 | 0.0001 |
| BCL2 | Apoptosis | −2.77 | 0.0177 |
| BCL2L1 | −1.77 | 0.0014 | |
| BIRC2 | −2.83 | 0.0054 | |
| BIRC5 | −18.53 | 0.0014 | |
| FADD | −1.15 | 0.0001 | |
| GSTP1 | −1.97 | 0.0019 | |
| MCL1 | −2.47 | 0.0031 | |
| TERT | −1.65 | 0.0005 | |
| TNFRSF10B | −1.07 | 0.0063 | |
| WT1 | −6.674 | 0.0010 | |
| CFLAR | Apoptosis, adhesion & proteolysis | −7.12 | 0.0003 |
| MSH2 | Apoptosis, DNA damage | −1.03 | 0.0057 |
| PYCARD | Apoptosis, adhesion & proteolysis | −1.49 | 0.0178 |
| CCL5 | Immune & inflammatory response | −35.1 | 0.0001 |
| CXCR4 | −2.73 | 0.0004 | |
| NFKB1 | −1.48 | 0.0365 | |
| PTGS2 | −2.76 | 0.0001 | |
| TLR4 | −4.81 | 0.0007 | |
| CDH1 | Adhesion & proteolysis | −3.04 | 0.0002 |
| EGFR | −2.58 | 0.0064 | |
| EP300 | −3.28 | 0.0001 | |
| HGF | −11.07 | 0.0001 | |
| ITGB1 | −2.48 | 0.0001 | |
| OPCML | −1.51 | 0.0074 | |
| SOCS1 | −7.04 | 0.0001 | |
| TERT | −1.65 | 0.0005 | |
| ANGPT2 | Angiogenesis | −1.61 | 0.0097 |
| FLT1 | −36.26 | 0.0001 | |
| KDR | −13.5 | 0.0001 | |
| PDGFRA | −1.54 | 0.0019 | |
| VEGFA | −1.30 | 0.0429 | |
Suppression of cytokine signalling 3 (SOCS3) was also detected. SOCS3 is a member of the STAT-induced STAT inhibitor (SSI) which functions as negative regulators of cytokine signalling. Decreased expression of SOCS3 is correlated with increased phosphorylation of STAT3 in HCC.37 SOCS3 furthermore has been implicated in negatively regulating cyclin D1 (CCND1), and anti-apoptotic genes including XIAP, survivin (BIRC5), and myeloid leukaemia cell differentiation protein (MCL1).38 Here, we observed a significant increase in expression of SOCS3 (Table 3) and a significant decrease in STAT3, CCND1, XIAP, BIRC5 and MCL1 expression (Table 4). Similar to the in vivo observations of reduction in GST-p (Fig. 2D), the array data also confirmed down-regulation in expression of GSTP1 (Table 4).
Overall, the down-regulated genes were strongly enriched for functions related to negative regulation of apoptosis and cell death (gene ontology (GO) terms GO:0043066 and GO:0060548; p-values 2×10−9 and 2×10−9, respectively), whereas the up-regulated genes were enriched for functions related to positive regulation of cell differentiation (GO:0045597; p = 5×10−3).
Transfection of C/EBPα-saRNA in HepG2 suppresses STAT3, IL6R and cMyc in HepG2 cells
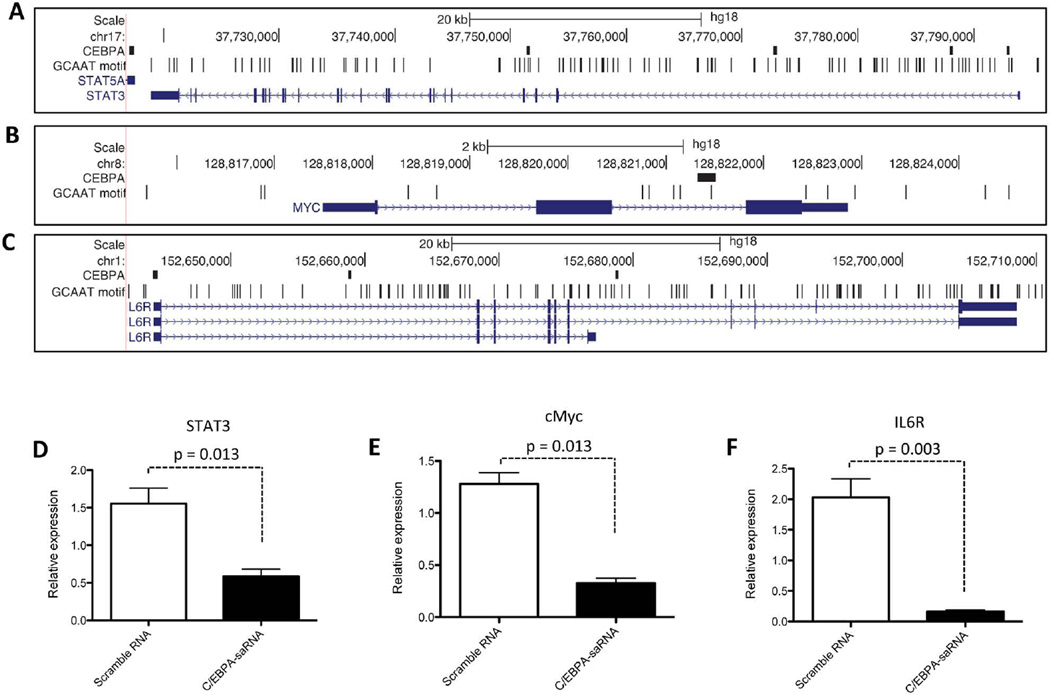
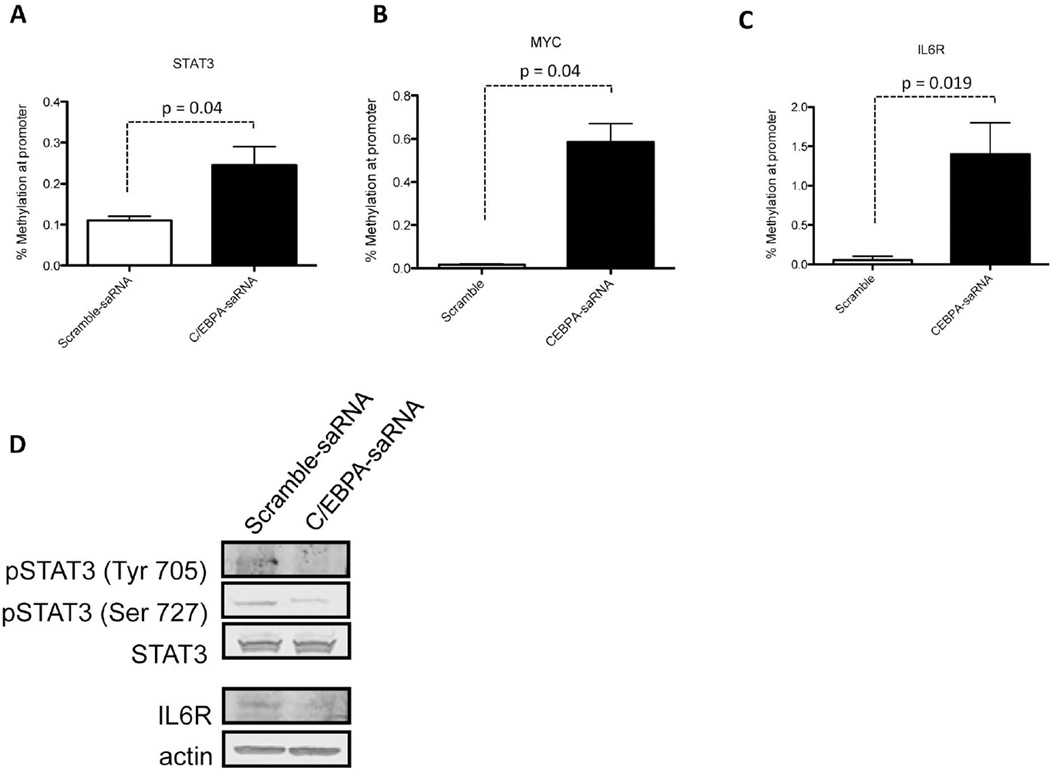
Previously published reports demonstrate that IL6R promote hepatic oncogenesis by directly activating STAT3 and in turn up-regulating expression of c-Myc.39 Since a ChIP-Seq analysis of these 3 genes show the presence of C/EBPα binding sites within their promoter regions (Figs. 7A, 7B, and 7C), we assessed whether transfection of C/EBPα-saRNA in HepG2 cells would affect expression levels of these three factors. We observed a significant reduction in mRNA levels of STAT3 (Fig. 7D), cMyc (Fig. 7E) and IL6R (Fig. 7F) when compared to untransfected cells. This trend in gene reduction was also observed for MYC and STAT in our previously described gene expression array (Table 2, in bold). When the methylation status of the CpG islands at the promoter regions of STAT3 (Fig. 8A), MYC (Fig. 8B) and IL6R (Fig. 8C) were assessed using EpiTect Methyl II PCR assay (Qiagen), an increase in methylation state at the promoters of all three genes was detected. A Western blot also confirmed a reduction in the phosphorylation status of STAT3 and in the protein level of IL6R (Fig. 8D). Collectively, we show that in vivo delivery of C/EBPα might have a positive effect in assisting liver function and decreasing aberrant cell proliferation in a cirrhotic/HCC setting.
Fig. 7. Transfection of C/EBPα-saRNA in HepG2 cells results in negative regulation of cell proliferating factors.
STAT3, c-Myc (MYC) and IL6R have one or more C/EBPα binding sites. The panels show the genomic region 2000 nucleotides upstream and downstream of (A) STAT3 (B) MYC and (C) IL6R. Figure panels shown are as described in (Figure 1 E–G). C/EBPα-saRNA transfected HepG2 cells show negative regulation in mRNA expression levels of genes encoding (D) STAT3, (E) c-Myc and (F) Interleukin 6 receptor (IL6R). Data represents mean ± SD.
Table 2. Gene expression (down-regulated by C/EBPA-saRNA).
Clustered analysis of genes negatively regulated following transfection of C/EBPα-saRNA in HepG2 cells.
| Gene Symbol | Fold Down-Regulation | T-TEST |
|---|---|---|
| C/EBPAsaRNA/Control Group | p value | |
| ADAM17 | −2.19 | 0.0007 |
| AKT1 | −1.78 | 0.0220 |
| ANGPT2 | −1.61 | 0.0097 |
| BCL2 | −2.77 | 0.0177 |
| BCL2L1 | −1.77 | 0.0014 |
| BIRC2 | −2.83 | 0.0054 |
| BIRC5 | −18.53 | 0.0014 |
| CCL5 | −35.1 | 0.0001 |
| CCND1 | −6.45 | 0.0001 |
| CCND2 | −2.40 | 0.0001 |
| CDH1 | −3.04 | 0.0002 |
| CDH13 | −5.52 | 0.0001 |
| CDKN1A | −2.53 | 0.0016 |
| CDKN1B | −1.74 | 0.0145 |
| CDKN2A | −6.75 | 0.0001 |
| CFLAR | −7.12 | 0.0003 |
| CTNNB1 | −1.59 | 0.0081 |
| CXCR4 | −2.73 | 0.0004 |
| E2F1 | −1.40 | 0.0174 |
| EGF | −8.63 | 0.0074 |
| EGFR | −2.58 | 0.0064 |
| EP300 | −3.28 | 0.0001 |
| FADD | −1.15 | 0.0001 |
| FLT1 | −36.26 | 0.0001 |
| FZD7 | −2.00 | 0.0004 |
| GSTP1 | −1.97 | 0.0019 |
| HGF | −11.07 | 0.0001 |
| HRAS | −3.69 | 0.0001 |
| IGFBP1 | −7.36 | 0.0001 |
| IGFBP3 | −17.56 | 0.0001 |
| IRS1 | −1.61 | 0.0010 |
| ITGB1 | −2.48 | 0.0001 |
| KDR | −13.5 | 0.0001 |
| MCL1 | −2.47 | 0.0031 |
| MET | −1.30 | 0.0247 |
| MSH2 | −1.03 | 0.0057 |
| MSH3 | −3.63 | 0.0002 |
| MTDH | −1.73 | 0.0014 |
| MYC | −1.76 | 0.0132 |
| NFKB1 | −1.48 | 0.0365 |
| NRAS | −29.47 | 0.0001 |
| OPCML | −1.51 | 0.0074 |
| PDGFRA | −1.54 | 0.0019 |
| PIN1 | −1.15 | 0.0012 |
| PTGS2 | −2.76 | 0.0001 |
| PYCARD | −1.49 | 0.0178 |
| RAC1 | −1.46 | 0.0069 |
| RASSF1 | −4.59 | 0.0001 |
| RELN | −2.09 | 0.0005 |
| RHOA | −1.46 | 0.0019 |
| SFRP2 | −1.73 | 0.0092 |
| SMAD7 | −3.46 | 0.0002 |
| SOCS1 | −7.04 | 0.0001 |
| STAT3 | −13.06 | 0.0006 |
| TCF4 | −9.96 | 0.0008 |
| TERT | −1.65 | 0.0005 |
| TGFA | −9.27 | 0.0001 |
| TGFB1 | −1.80 | 0.0002 |
| TLR4 | −4.81 | 0.0007 |
| TNFRSF10B | −1.07 | 0.0063 |
| VEGFA | −1.30 | 0.0429 |
| WT1 | −6.674 | 0.0010 |
| XIAP | −2.93 | 0.0228 |
| YAP1 | −1.58 | 0.0058 |
Fig. 8. Transfection of C/EBPα-saRNA in HepG2 cells targets STAT3, c-Myc and IL6R signalling.
A methylation assay of the CpG islands at the promoter regions of (A) STAT3, (B) MYC and (C) IL6R demonstrated hypermethylation when compared to control. Data represents mean ± SD. (D) A Western blot analysis showed decreased phosphorylation of STAT3 at residues 705 and 727 and down-regulation of IL6R in cells transfected with C/EBPα-saRNA.
DISCUSSION
Hepatocellular carcinoma (HCC) develops in most patients from a background of liver cirrhosis and accounts for 90% of all liver cancers. Although much progress has been made in targeting therapy to HCC, few of these treatments have had much impact in patient outcome.
The initial aim of this investigation was to study the therapeutic potential of using saRNAs to help ameliorate liver function in a clinically relevant rat model of liver cirrhosis with hepatocellular carcinoma. By enhancing expression of the gene encoding C/EPBα, a liver enriched transcription factor that enhances albumin and confers anti-mitotic activity; we primarily sought to increase circulating albumin in these rats. Using our previously published concept of designing short activating RNA oligonucleotide to increase the expression of a target gene7,11, C/EPBα-saRNA was generated. This was initially tested in the hepatocellular carcinoma line (HepG2) where introduction of the saRNA oligonucleotide led to increased transcript levels of C/EPBα and albumin. Both genes furthermore contained the recognition motif of C/EPBα, CGAAT within their promoter regions. It was therefore unsurprising to detect a loss in methylation status at their CpG islands following transfection of C/EPBα-saRNA.
The biological significance of increasing albumin transcript levels in C/EPBα-saRNA transfected cells corresponded well with the increased secretion of albumin. Interestingly, we found that the maximum albumin gene expression was achieved at 5 nM of C/EPBα-saRNA with no further increase at higher saRNA levels. In addition to the albumin gene, we also found increased gene expression in other important biological markers such as ornithine cycle enzyme ornithine transcarbamylase (OTC) and alpha-feto-protein (AFP).40
To test the potential therapeutic value of the C/EPBα-saRNA, we subsequently performed an in vivo study using an HCC rat model. For targeted delivery of C/EPBα-saRNA we linked the duplex RNA molecule to cationic PAMAM dendrimers. These nanoparticle have previously been evaluated where biodistribution studies demonstrate that they preferentially accumulate in peripheral blood mononuclear cells and the liver with no discernible toxicity.25 Intravenous injection of C/EPBα-saRNA-dendrimers over a course of one week showed a significant improvement by 30% in the circulating levels of albumin where compared to PBS control or scramble-saRNA-dendrimer control groups. Changes in bilirubin levels showed a 10% improvement in the C/EBPα-saRNA group when compared to the control groups. Additionally a 10% improvement in AST levels and 30% improvement in ALT levels were observed in the C/EBPα-saRNA treated group when compared to the control groups. More significant was the reduction in tumour burden and the inhibition of pre-neoplastic lesions as detected by a 40% reduction in GST-p staining in the liver sections from the C/EBPα-saRNA treated group. From a clinical perspective, this represents a very attractive therapeutic avenue since the expression level of C/EBPα in matched tumour tissues and non-tumour tissues of HCC patients is down-regulated in the majority of tumour specimens. Moreover, patients with tumour samples showing higher levels of C/EBPα have a longer survival rate than those patients with tumour samples in which the expression of the C/EBPα is lower.41 Our data supports this evidence suggesting that up-regulation of C/EBPα provides a strong anti-proliferative role in hepatocytes.14,42
To better understand the global molecular effect of C/EPBα-saRNA more specific to liver cancer, we performed a liver cancer pathway gene expression profile analysis. Such analysis of whole tumours is frequently confounded by the presence of cell types other than those with a transformed phenotype.43 Therefore we profiled the gene expression changes brought about by C/EPBα-saRNA in HepG2 cells.
The expression pattern of the liver cancer genes varied greatly between untransfected and C/EPBα-saRNA transfected HepG2 cells. After normalisation and cluster analysis, several important genes were significantly altered in expression. From the list of 20 genes that were up-regulated, 18 were known tumour suppressor genes. Of note was the up-regulation of RB, TP53, BID and BAX to regulate cell cycle and apoptosis. The down-regulation of key genes were also noted, in particular ADAM17, a metalloproteinase reported as being a pathological feature of HCC.44 ADAM17 is known to cause the shedding of receptor ligands such as epidermal growth factor (EGF) and tumour necrosis factor alpha (TNFα)45,46 thus preventing regulation of key signaling events for normal cell signaling.
Upon further analysis of the tumour suppressor genes, we noticed a pathway-defined trend where key effector genes of the tumour suppressors were down-regulated. Examples of this included repression of RHOA following up-regulation of the tumour suppressor DLC1; or up-regulation of RUNX3 to reverse expression of the oncogenes involved in epithelial-mesenchymal transition (EMT). Here we observed down-regulation of CTNB1 (β-catenin), HGF, SMAD7 and TGFB1. We also observed increased expression of the tumour suppressor SOC3, a known regulator of apoptosis and cell adhesion. Concomitantly, we also observed down-regulation of the associated SOC3 oncogenes including STAT3, cyclin-D1 (CCND1), XIAP, BIRC5 and MCL1. STAT3 activation together with IL6R is known to enhance hepatic oncogenesis as part of a feedback loop47, and moreover perturbation in any of the components from this network is sufficient to suppress HCC.39 Here, we demonstrated by gene expression analysis and detection of hypermethylation within the gene promoters, that both STAT3 and IL6R were down-regulated following C/EBPα-saRNA transfection. In addition to the well characterised anti-mitotic activity of C/EPBα involving retinoblastoma, p21 and the cyclin dependent proteins, our data here suggests that C/EPBα may regulate other liver specific oncogenic pathways including c-Myc (MYC).48 Our observed reduction in the EMT factors; the positive regulation of apoptosis and down-regulation of IL6R, STAT3 and MYC, and the presence of numerous C/EBPα binding motifs within the promoter regions of these three genes provide a novel landscape to further study the role of C/EPBα in improving the function of hepatocytes in a cirrhotic/HCC setting.
In summary, we initially designed saRNAs targeting the liver enriched transcription factor C/EBPα with the aim of addressing hypoalbuminemia. This was successfully done in vitro and in vivo. In the course of this work we also confirmed the well known anti-proliferative effects of C/EPBα in a clinically relevant cirrhotic/HCC model. In addition to regulating known targets of C/EPBα that controls cell proliferation we demonstrated using a liver cancer specific gene array analysis that C/EPBα potentially targets numerous other oncogenes and tumour suppressor genes which must be further investigated. C/EPBα-saRNAs therefore may have a profound effect at the transcriptional level for liver cancer. Currently, most therapeutic disciplines such as surgery, chemotherapy, radiotherapy and biologics are associated with variable decrease of liver dysfunction.49,50 The data presented here offers a new approach to targeting liver cancer cells.
Supplementary Material
ACKNOWLEDGEMENT
We are sincerely grateful to Dr Albert Deisseroth and Professor Farzin Farzaneh for their valuable input to the construction of this manuscript.
Dr. Rossi’s NIH grant should have been acknowledged in both papers: NIH R01 HL074704.
List of abbreviations
- HCC
Hepatocellular carcinoma
- C/EBPα
CCAAT/enhancer binding protein alpha
- saRNA
short activating RNA
- OTC
Ornithine transcarbamylase
- AFP
Alphafetoprotein
- AST
Aspartate aminotransferase
- ALT
Alanine aminotransferase
REFERENCES
- 1.Waly Raphael S, Yangde Z, YuXiang C. Hepatocellular Carcinoma: Focus on Different Aspects of Management. ISRN Oncology. 2012;2012:1–12. doi: 10.5402/2012/421673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imai N, et al. Persistent elevated C-reactive protein after treatment is an independent marker of a poor prognosis in patients with hepatocellular carcinoma. Clin Transl Oncol. 2012:1–7. doi: 10.1007/s12094-012-0976-y. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB, Marrero JA, Rudolph L, Reddy KR. Diagnosis and treatment of hepatocellular carcinoma. Gastroenterology. 2008;134:1752–1763. doi: 10.1053/j.gastro.2008.02.090. [DOI] [PubMed] [Google Scholar]
- 4.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L-C, et al. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl. Acad. Sci. U.S.A. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janowski BA, et al. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat Chem Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 7.Voutila J, et al. Gene Expression Profile Changes After Short-activating RNA-mediated Induction of Endogenous Pluripotency Factors in Human Mesenchymal Stem Cells. Mol Ther Nucleic Acids. 2012;1:e35. doi: 10.1038/mtna.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu JJ, et al. Promoter-associated small double-stranded RNA interacts with heterogeneous nuclear ribonucleoprotein A2/B1 to induce transcriptional activation. Biochem. J. 2012;447:407–416. doi: 10.1042/BJ20120256. [DOI] [PubMed] [Google Scholar]
- 9.Kosaka M, Kang MR, Yang G, Li L-C. Targeted p21WAF1/CIP1 Activation by RNAa Inhibits Hepatocellular Carcinoma Cells. Nucleic Acid Ther. 2012 doi: 10.1089/nat.2012.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Place RF, et al. Formulation of Small Activating RNA Into Lipidoid Nanoparticles Inhibits Xenograft Prostate Tumor Growth by Inducing p21 Expression. Mol Ther Nucleic Acids. 2012;1:e15. doi: 10.1038/mtna.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.V, et al. A Short-activating RNA Oligonucleotide Targeting the Islet β-cell Transcriptional Factor MafA in CD34(+) Cells. Mol Ther Nucleic Acids. 2013;2:e97. doi: 10.1038/mtna.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lekstrom-Himes J, Xanthopoulos KG. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J Biol Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 13.Umek RM, Friedman AD, McKnight SL. CCAAT-enhancer binding protein: a component of a differentiation switch. Science. 1991;251:288–292. doi: 10.1126/science.1987644. [DOI] [PubMed] [Google Scholar]
- 14.Timchenko NA, Wilde M, Nakanishi M, Smith JR, Darlington GJ. CCAAT/enhancer-binding protein alpha (C/EBP alpha) inhibits cell proliferation through the p21 (WAF-1/CIP-1/SDI-1) protein. Genes & Development. 1996;10:804–815. doi: 10.1101/gad.10.7.804. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, et al. C/EBPalpha arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Molecular Cell. 2001;8:817–828. doi: 10.1016/s1097-2765(01)00366-5. [DOI] [PubMed] [Google Scholar]
- 16.Nerlov CC, Ziff EBE. Three levels of functional interaction determine the activity of CCAAT/enhancer binding protein-alpha on the serum albumin promoter. Genes & Development. 1994;8:350–362. doi: 10.1101/gad.8.3.350. [DOI] [PubMed] [Google Scholar]
- 17.Poli VV, Mancini FPF, Cortese RR. IL-6DBP, a nuclear protein involved in interleukin-6 signal transduction, defines a new family of leucine zipper proteins related to C/EBP. Cell. 1990;63:643–653. doi: 10.1016/0092-8674(90)90459-r. [DOI] [PubMed] [Google Scholar]
- 18.Wang ND, et al. Impaired energy homeostasis in C/EBP alpha knockout mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 19.Kimura T, et al. Hypoglycemia-associated hyperammonemia caused by impaired expression of ornithine cycle enzyme genes in C/EBPalpha knockout mice. J Biol Chem. 1998;273:27505–27510. doi: 10.1074/jbc.273.42.27505. [DOI] [PubMed] [Google Scholar]
- 20.Ray A, Ray BK. Serum amyloid A gene expression under acute-phase conditions involves participation of inducible C/EBP-beta and C/EBP-delta and their activation by phosphorylation. Molecular and Cellular Biology. 1994;14:4324–4332. doi: 10.1128/mcb.14.6.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou J, et al. Systemic administration of combinatorial dsiRNAs via nanoparticles efficiently suppresses HIV-1 infection in humanized mice. Mol Ther. 2011;19:2228–2238. doi: 10.1038/mt.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Javitt NB. Hep G2 cells as a resource for metabolic studies: lipoprotein, cholesterol, and bile acids. The FASEB Journal. 1990;4:161–168. doi: 10.1096/fasebj.4.2.2153592. [DOI] [PubMed] [Google Scholar]
- 23.Huang K-W, et al. Dual Therapeutic Effects of Interferon-α Gene Therapy in a Rat Hepatocellular Carcinoma Model With Liver Cirrhosis. Mol Ther. 2008;16:1681–1687. doi: 10.1038/mt.2008.160. [DOI] [PubMed] [Google Scholar]
- 24.Zhou J, et al. PAMAM dendrimers for efficient siRNA delivery and potent gene silencing. Chemical Communications. 2006;0:2362–2364. doi: 10.1039/b601381c. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, et al. Efficient delivery of sticky siRNA and potent gene silencing in a prostate cancer model using a generation 5 triethanolamine-core PAMAM dendrimer. Mol. Pharm. 2012;9:470–481. doi: 10.1021/mp2006104. [DOI] [PubMed] [Google Scholar]
- 26.Semenkova LN, Dudich EI, Dudich IV. Induction of apoptosis in human hepatoma cells by alpha-fetoprotein. Tumour Biol. 1997;18:261–273. doi: 10.1159/000218039. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, et al. C/EBPalpha arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Molecular Cell. 2001;8:817–828. doi: 10.1016/s1097-2765(01)00366-5. [DOI] [PubMed] [Google Scholar]
- 28.Tatematsu MM, et al. Stable phenotypic expression of glutathione S-transferase placental type and unstable phenotypic expression of gamma-glutamyltransferase in rat liver preneoplastic and neoplastic lesions. Carcinogenesis. 1988;9:215–220. doi: 10.1093/carcin/9.2.215. [DOI] [PubMed] [Google Scholar]
- 29.Harrison DJ, May L, Hayes JD, Neal GE. Glutathione S-transferase localization in aflatoxin B1-treated rat livers. Carcinogenesis. 1990;11:927–931. doi: 10.1093/carcin/11.6.927. [DOI] [PubMed] [Google Scholar]
- 30.Currier AR, et al. Plasminogen directs the pleiotropic effects of uPA in liver injury and repair. Am. J. Physiol. Gastrointest. Liver Physiol. 2003;284:G508–G515. doi: 10.1152/ajpgi.00336.2002. [DOI] [PubMed] [Google Scholar]
- 31.Song G, Chen G, Yun JP, Lai P. Association of p53 with Bid Induces Cell Death in Response to Etoposide Treatment in Hepatocellular Carcinoma. CCDT. 2009;9:871–880. doi: 10.2174/156800909789760302. [DOI] [PubMed] [Google Scholar]
- 32.Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 33.Seewoo V, et al. The Different Induction Mechanisms of Growth Arrest DNA Damage Inducible Gene 45 β in Human Hepatoma Cell Lines. Chemotherapy. 2012;58:165–174. doi: 10.1159/000338386. [DOI] [PubMed] [Google Scholar]
- 34.Kim TY, et al. DLC-1, a GTPase-activating protein for Rho, is associated with cell proliferation, morphology, and migration in human hepatocellular carcinoma. Biochemical and Biophysical Research Communications. 2007;355:72–77. doi: 10.1016/j.bbrc.2007.01.121. [DOI] [PubMed] [Google Scholar]
- 35.Shiraha H, Nishina S-I, Yamamoto K. Loss of runt-related transcription factor 3 causes development and progression of hepatocellular carcinoma. J. Cell. Biochem. 2011;112:745–749. doi: 10.1002/jcb.22973. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka S, et al. Runt-related transcription factor 3 reverses epithelial-mesenchymal transition in hepatocellular carcinoma. Int J Cancer. 2012;131:2537–2546. doi: 10.1002/ijc.27575. [DOI] [PubMed] [Google Scholar]
- 37.Wu W-Y, et al. Prognostic significance of phosphorylated signal transducer and activator of transcription 3 and suppressor of cytokine signaling 3 expression in hepatocellular carcinoma. Exp Ther Med. 2011;2:647–653. doi: 10.3892/etm.2011.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei R-C, et al. Augmenting the Antitumor Effect of TRAIL by SOCS3 with Double-Regulated Replicating Oncolytic Adenovirus in Hepatocellular Carcinoma. Human Gene Therapy. 2011;22:1109–1119. doi: 10.1089/hum.2010.219. [DOI] [PubMed] [Google Scholar]
- 39.Hatziapostolou M, et al. An HNF4α-miRNA Inflammatory Feedback Circuit Regulates Hepatocellular Oncogenesis. Cell. 2011;147:1233–1247. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan EH, Ma FJ, Gopinadhan S, Sakban RB, Wang N-D. C/EBPα knock-in hepatocytes exhibit increased albumin secretion and urea production. Cell Tissue Res. 2007;330:427–435. doi: 10.1007/s00441-007-0505-4. [DOI] [PubMed] [Google Scholar]
- 41.Tomizawa M, Watanabe K, Saisho H, Nakagawara A, Tagawa M. Down-regulated expression of the CCAAT/enhancer binding protein alpha and beta genes in human hepatocellular carcinoma: a possible prognostic marker. Anticancer Res. 2003;23:351–354. [PubMed] [Google Scholar]
- 42.Mischoulon D, Rana B, Bucher NL, Farmer SR. Growth-dependent inhibition of CCAAT enhancer-binding protein (C/EBP alpha) gene expression during hepatocyte proliferation in the regenerating liver and in culture. Molecular and Cellular Biology. 1992;12:2553–2560. doi: 10.1128/mcb.12.6.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meng F, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 44.Ding X, Yang L-Y, Huang G-W, Wang W, Lu W-Q. ADAM17 mRNA expression and pathological features of hepatocellular carcinoma. WJG. 2004;10:2735–2739. doi: 10.3748/wjg.v10.i18.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kutay H, et al. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J. Cell. Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mochizuki S, Okada Y. ADAMs in cancer cell proliferation and progression. Cancer Sci. 2007;98:621–628. doi: 10.1111/j.1349-7006.2007.00434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He G, et al. Hepatocyte IKKbeta/NF-kappaB inhibits tumor promotion and progression by preventing oxidative stress-driven STAT3 activation. Cancer Cell. 2010;17:286–297. doi: 10.1016/j.ccr.2009.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shachaf CM, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431:1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 49.Wörns MA, et al. The impact of patient and tumour baseline characteristics on the overall survival of patients with advanced hepatocellular carcinoma treated with sorafenib. Biochemical and Biophysical Research Communications. 2012 doi: 10.1016/j.dld.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 50.Carr BI. Some New Approaches to the Management of Hepatocellular Carcinoma. Seminars in Oncology. 2012;39:369–373. doi: 10.1053/j.seminoncol.2012.05.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.