Abstract
Structural studies of viral proteins most often use high-resolution techniques such as X-ray crystallography, nuclear magnetic resonance, single particle negative stain, or cryo-electron microscopy (EM) to reveal atomic interactions of soluble, homogeneous viral proteins or viral protein complexes. Once viral proteins or complexes are separated from their host’s cellular environment, their natural in-situ structure and details of how they interact with other cellular components may be lost. EM has been an invaluable tool in virology since its introduction in the late 1940’s and subsequent application to cells in the 1950’s. EM studies have expanded our knowledge of viral entry, viral replication, alteration of cellular components, and viral lysis. Most of these early studies were focused on conspicuous morphological cellular changes, because classic EM metal stains were designed to highlight classes of cellular structures rather than specific molecular structures. Much later, to identify viral proteins inducing specific structural configurations at the cellular level, immunostaining with a primary antibody followed by colloidal gold secondary antibody was employed to mark the location of specific viral proteins. This technique can suffer from artifacts in cellular ultrastructure due to compromises required to provide access to the immuno-reagents. Immunolocalization methods also require the generation of highly specific antibodies, which may not be available for every viral protein. Here we discuss new methods to visualize viral proteins and structures at high resolutions in-situ using correlated light and electron microscopy (CLEM). We discuss the use of genetically encoded protein fusions that oxidize diaminobenzidine (DAB) into an osmiophilic polymer that can be visualized by EM. Detailed protocols for applying the genetically encoded photo-oxidizing protein MiniSOG to a viral protein, photo-oxidation of the fusion protein to yield DAB polymer staining, and preparation of photo-oxidized samples for TEM and serial block-face scanning EM (SBEM) for large-scale volume EM data acquisition are also presented. As an example, we discuss the recent multi-scale analysis of Adenoviral protein E4-ORF3 that reveals a new type of multi-functional polymer that disrupts multiple cellular proteins. This new capability to visualize unambiguously specific viral protein structures at high resolutions in the native cellular environment is revealing new insights into how they usurp host proteins and functions to drive pathological viral replication.
Keywords: photo-oxidation, electron microscopy, Adenoviral proteins, E4-ORF3 polymer, MiniSOG, serial block face scanning electron microscopy
Introduction
1.1. Visualizing in-situ viral protein ultrastructure
Viruses are efficient nano-machines with a relatively small genome size in comparison to their host. Yet despite their small size and limited number of proteins, viruses usurp host cellular pathways with ease in order to promote their own survival. Strikingly, a survey of all human specific viral proteins (5289) from the UniProt database shows that 1487 proteins (28%) have less than 200 residues, 1986 proteins (38%) are between 200 to 500 residues, and 1314 proteins (25%) are between 500 to 1000 residues [1]. This suggests that viruses utilize multi-functional small proteins to disrupt normal cellular functions [2]. For example, the small DNA tumor virus Adenovirus (genome size < 40 kbp) is dependent on only eleven proteins expressed early (within 12 hours) in infection to transform quiescent human cells into replication factories that propagate the viral genome prior to lysis [3]. Adenovirus early proteins (E1 and E4 early viral proteins) usurp the interactions of critical growth regulatory hubs and have led to the discovery of many key tumor targets and mechanisms [2]. Furthermore, the majority of adenoviral early proteins are less than 20 kDa and have limited sequence homology to human proteins [4]. Thus, how the structures of these small adenoviral early proteins mediate dominant and disparate interactions with such apparently limited surface area and how these small proteins alter the cellular environment so that it is conducive to viral replication remains to be answered. Understanding viral protein structures and their localization in the context of an entire cell may shed light on the general principles that enable viruses to manipulate complex regulatory networks and uncover novel cellular pathways that are essential for normal cellular growth.
A protein’s three dimensional (3D) structure determines its interaction, such as the ordered self-association of individual proteins, the assembly of filamentous polymers or capsids that give a virus protection and shape, and protein interaction hubs that integrate multiple signals. Several biophysical tools have been applied to elucidate protein structures, with each tool yielding structures at different resolutions. At the extreme end of the range of spatial scales, nuclear magnetic resonance and X-ray crystallography can be applied to reveal protein structures at angstrom resolution, determining details of the intricate interactions between atoms and residues that facilitate both an understanding of protein-protein interactions and generate potential drug designs to target these interactions. At the other end of the spatial scale, light microscopy (LM), in particular fluorescence LM and the application of a genetically encoded GFP tag or other GFP variants [5], can reveal protein localization and organization within the context of an entire cell or organism at 200 nm lateral resolution or uncover the dynamics of protein movements through live-imaging. Although advancements in super resolution microscopy techniques such as SIM, STORM and PALM are pushing the boundary of light microscopy resolution to the order of 10–20 nm [6], they can only reveal structures labeled by fluorescent probes without the context of other unlabeled subcellular structures. Human viruses such as Parvovirus, Papillomavirus, HIV-1, Herpesvirus, and Variola virus, which range in size from 20 nm to 360 nm [7], fall into this gray zone of intermediate resolution, the mesoscale, that can only be investigated by electron microscopy (EM) [8]. Furthermore, since viral proteins may contain intrinsic disordered regions [2] or form insoluble oligomers, they are elusive to conventional high-resolution structural studies or biochemical characterization of their functions. However, EM does not have these constraints and has the power to reveal the ultrastructure and interactions of viral proteins in their native cellular environment that is not possible using any other biophysical methods.
The study of virus structure has been associated with EM since nearly the beginning when Ernst Ruska and Max Knoll constructed the first transmission electron microscope (TEM) in 1932 and Helmut Ruska acquired the first image of bacteria and viruses with TEM in 1939 [9]. Since then, almost all characterizations of viruses have involved the use of EM. In the early era of EM, isolated viruses were imaged with metal shadowing or negative staining, which mostly captured the organization of viral capsids, but no structural information was obtained for the viral proteins and cellular environment. After the development of thin sectioning methods for cutting resin-embedded tissues and cells with knives in the 1960’s [10], a burst of EM studies brought forth beautiful images of various stages of viral life cycles within a cell including viral entry, viral replication, viral protein-host protein interactions, and abnormal structural assemblies within a cell [11]. This could be called the golden age of virology, in which new viral protein structures and their effects on cellular environment were being discovered. Further, the finding that viruses could introduce oncogenes into human cells made virology a trending research topic. Despite these advancements, EM studies of viruses and viral proteins were mostly based on distinctive cellular morphologies since the classic EM stains (osmium tetroxide, uranyl acetate, lead citrate) indiscriminately stain lipids, RNA, and proteins. For example, extensive studies of Adenovirus by EM in the 1960’s focused on viral replication within the cell. Large and distinctive macromolecular structures such as viral capsids [12], paracrystalline protein lattices [13], and viral replication domains in the nucleus can be readily discriminated by EM [14]. A consistent theme among all the EM images is that Adenovirus infection dramatically transforms the cell nucleus and inducing the formation of numerous distinct structures and compartments that presumably facilitate viral replication. However, the specific viral and cellular protein structures that induce these distinct cellular morphologies proves to be a more difficult task.
One of the early tools developed to visualize a specific viral protein by EM is post-embedding colloidal gold immunostaining [15]. Cells are fixed, embedded in a resin and then cut into 70–100 nm thin sections. The thin sections are incubated with a primary antibody against a specific antigen, followed by a secondary antibody conjugated to colloidal gold, usually between 5–10 nm, to detect the primary antibody. If successful, gold particles decorate the antigen of interest, which can reveal a proteins subcellular localization and its organization into distinctive macromolecular structure. This technique was recently applied in 2008 by Franqueville and coworkers to elucidate which adenoviral proteins assemble paracrystalline lattices in the nucleus that were first observed in 1957 [13, 16]. Previous immuno-EM experiments had suggested several candidates: viral hexon, penton base, fiber, and basic core proteins, as well as cellular proteins such as CK2 and PKR [17]. However, mutant adenoviruses that do not express hexon viral proteins still form the paracrystalline lattice [18]. The controversy was resolved, using adenovirus mutants and immunogold labeling of monoclonal antibodies that recognize the fiber protein, which showed that the paracrystalline lattice comprises homopentamer of the penton base protein and a homotrimer of the fiber protein [16]. This report illustrates the caveats of immuno-gold labeling in identifying specific viral proteins. First, the requirement of an antibody to probe against a specific viral protein is not simple, and the specificity of the antibody needs to be rigorously tested to ensure only the antigens being probed are detected and with low background. Second, post-embedded immunostaining may not work for every antibody since the antibody can only stain antigens at the surface of the section and the antigens might be blocked or destroyed by the resin. However, this problem can be alleviated by immunostaining sections prepared with the Tokuyasu method [19–21]. The Tokuyasu method preserves the ultrastructure and antigenicity of the specimen by infiltrating the specimen in sucrose, freezing in liquid nitrogen, and sectioning into thin sections for immunostaining. Finally, while immuno-gold labeling identifies the location of a protein of interest, it does not reveal the underlying protein structure unless the immuno-gold shows a specific pattern and the epitope position of the antigen in a macromolecular structure is known. Thus, there is a need to develop a method to easily identify viral proteins within the cellular milieu to observe their structures and effects on other cellular components in EM.
1.2. DAB staining for visualization of ultrastructure in EM
Since contrast in the EM is most often based on differential scattering of electrons by heavy metal-based reagents that stain cellular components, a staining method that will specifically localize to a region of interest and provide EM contrast is very desirable, as it would enable identification of a specific protein or complex structure. One small organic molecule, DAB (3,3′-diaminobenzidine), is well suited for such specific labeling. In 1966, Graham and Karnovsky discovered that tissues injected with horseradish peroxidase, then incubated with DAB solution and hydrogen peroxide (H2O2) resulted in formation of insoluble osmiophilic polymer that appeared opaque in EM upon staining with osmium tetroxide [22]. This staining technique not only enables the localization of catalase, peroxidase, cytochrome oxidase to be determined, but also allows for the labeling of the organelles in which these enzymes reside (peroxisome, mitochondria) in diverse organisms such as mammalian cells, plants, or parasites. Unlike EM stains such as osmium tetroxide, uranyl acetate, or lead citrate, which stain broadly throughout the specimen, DAB-based staining provides an opportunity for localized staining to study a specific ultrastructure within a cell if one can manipulate the location of the oxidizing agent.
1.3. Localized DAB-based staining through enzymatic oxidation
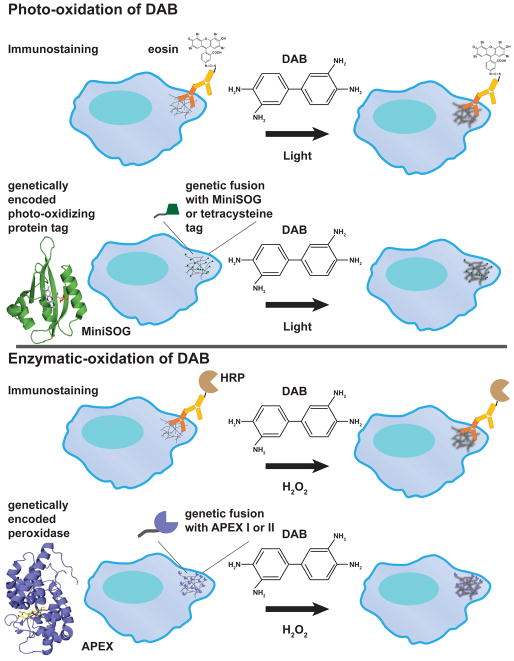
The application of using enzymes to oxidize DAB can be extended using secondary antibodies conjugated to peroxidase (Fig. 1) [23]. The antigen of interest is labeled using specific primary antibodies; however, instead of gold, the secondary antibody is conjugated to horseradish peroxidase (HRP), which can oxidize DAB into a polymer upon the addition of hydrogen peroxide. This technique was successfully applied as a diagnostic tool to detect viruses [11]. However this technique is strictly dependent on the availability of a primary antibody to label the target of interest and the accessibility of the antigen. Also, results need to be interpreted with caution due to the disruption of native cellular ultrastructure during the immunostaining process.
Fig. 1. Localized DAB polymer generation either through photo-oxidation or enzymatic oxidation to identify in-situ structures by EM.
Monomeric diaminobenzidine near an oxidation source will form an osmiophilic polymer that is electron opaque in EM after osmium tetroxide staining. DAB can be photo-oxidized with certain fluorophores or through enzymatic oxidation with the addition of hydrogen peroxide. Immunostaining by antibody or genetically encoded tags can be used to position the photo-oxidizer or enzyme near the protein of interest. As an example, the figure shows how a hypothetical fibrous structure in the cytoplasm of a cell can be detected by DAB-based staining through either photo-oxidation (antibody-conjugated with eosin or a MiniSOG or a tetracysteine tag) or enzymatic oxidation (antibody-conjugated with horseradish peroxidase (HRP) or an engineered peroxidase protein APEX tag). Structural homologs of MiniSOG (PDB: 4EEP) and APEX (PDB: 3V0H) are shown.
With the advent of molecular biology, attempts were made to create a genetically encoded HRP that could be fused with a target of interest. It was found that genetically encoded HRP required a KDEL peptide sequence in order for it to function in the cell [24]. This limits its usefulness as a genetic fusion tag since the KDEL sequence retains HRP protein fusions in the endoplasmic reticulum [25]. To function, HRP requires four disulfide bonds and two calcium binding sites that are inactivated in the reducing environment of the cytosol. To overcome these issues, Martell et al. re-engineered ascorbate peroxidase (APEX) from a class I cytosolic plant peroxidase to a monomer of 28 kDa that can be fused to either the N- or C- terminus of a protein and still retain peroxidase activity (Fig. 1) [26]. This enables the addition of APEX to a protein of interest, and following addition of H2O2 and DAB, induces localized polymerization of DAB around the tagged fusion protein. APEX was further improved by directed evolution to select for a mutant (APEX2) that has higher sensitivity to oxidize DAB even when expressed at a low level [27]. Since APEX and GFP (27 kDa) have similar molecular weights, it is relatively easy to confirm the fusion construct with the GFP tag to ensure proper localization and function of the viral protein before applying the APEX tag for labeling in EM. The use of the genetically encoded tag circumvents the availability of antibodies for colloidal gold immunostaining and avoids alteration to ultrastructure due to the immunostaining protocol. The APEX tag has been used successfully in labeling a large number of proteins, including histone H2B, vimentin, connexin43, actin, and tubulin by DAB polymer for EM. However, the use of this relatively large tag might be limited for labeling viral proteins that have less than 200 residues. Additionally the lack of autofluorescence in APEX does not allow the use of CLEM.
1.4. Localized DAB-based staining through photo-oxidation
In addition to enzymatic based oxidation of the DAB polymer, the next advancement in using the DAB polymer as a specific EM stain was the discovery of the photo-oxidation of DAB by Maranto [28]. By injecting the fluorescent marker Lucifer yellow into neurons, he discovered that irradiation with intense blue light in a DAB solution results in the formation of an opaque DAB polymer. He further showed that the staining of neurons by Lucifer yellow-induced DAB polymer is similar to HRP-oxidized DAB polymer, thus indicating that irradiation of a fluorescent dye is another method to oxidize DAB and catalyze DAB polymerization. Later, it was shown that Lucifer yellow is a photo-sensitizer that produces superoxide and hydrogen peroxide upon irradiation, the two main agents in DAB oxidation and polymerization [29]. The capability of using light to induce the oxidation of DAB implies that DAB polymerization could be fine-tuned to a small region if a fluorescence dye could be localized to a specific spot.
Based on this hypothesis, Deerinck et al. demonstrated that a brominated derivative of fluorescein, eosin-5-isothiocyanate (eosin), is able to photo-oxidize DAB into an electron opaque polymer upon irradiation at 515 nm [30]. To label microtubules with DAB polymer, the cells were immunostained with β-tubulin primary antibody followed by secondary antibody conjugated to eosin (Fig. 1). Upon light irradiation of cells bathed in DAB solution, eosin is able to photo-oxidize DAB into a polymer and reveals high contrast staining of microtubules in EM micrographs. Since eosin is a fluorescent dye, immunofluorescence images of the microtubules can be correlated with the EM ultrastructure labeled by DAB polymers, further confirming the identity of the labeled structure. The mechanism by which eosin oxidized DAB was suggested to involve the generation of singlet oxygen, since the singlet oxygen scavenger β-carotene can inhibit the photo-oxidation of DAB. Thus, eosin is a singlet oxygen generator, upon absorption of light, decays from the excited singlet state by intersystem crossing to a longer-lived triplet state [31]. The relaxation of the triplet state exchanges energy with ground state oxygen to generate singlet state oxygen, measured as a singlet oxygen quantum yield of 0.57 for eosin [31]. In comparison to the enzymatic based DAB oxidation, the photo-oxidation of DAB by eosin limits the diffusion of the DAB reaction product due to highly crosslinking of the sample and a smaller diffusion radius of reactive oxygen species from the fluorophore in comparison to HRP or APEX, thus yielding high spatial resolution of DAB stained structures [8]. The ease of inducing DAB oxidation by light, high singlet oxygen quantum yield, and high spatial resolution of DAB stained structures make this method attractive in labeling ultrastructure in EM. However, one major caveat is that immunostaining usually requires weak primary chemical fixation to retain antigenicity and detergent permeabilization to provide accessibility to intracellular antigens, which may alter or introduce artifacts to the ultrastructure. Thus, a non-invasive photo-oxidation technique is needed to label regions of interest in EM.
Like the development of APEX, a genetically encoded photo-oxidizable tag would open a new world in studying protein localization and organization by EM, especially if the tag were small enough not to perturb the native structure and function of small viral proteins. A photo-oxidizable genetically encoded tetracysteine tag (6 to 20 residues) was demonstrated by Gaietta et al. to enable both light imaging and photo-oxidation of DAB (Fig. 1) [32]. The tetracysteine tag has a high affinity toward two biarsenical compounds, FlAsH and ReAsH [33, 34]. Upon covalent linkage of four cysteines from the tag with the paired arsenic atoms of FlAsH or ReAsH, the fluorophore becomes fluorescent and has either a green emission (508 and 528 nm) for FlAsH or red emission (593 and 608 nm) for ReAsH. With two different fluorophore colors, Gaietta et al. was able to use pulse chase labeling of connexin43 with FlAsH and ReAsH to show gap junction dynamics in which new connexin43 were added at the periphery of the gap junction and old connexin43 remained in the middle of the gap junction. The tetracysteine tag sequence was later improved to increase both fluorescence and affinity toward FlAsH and ReAsH [35], and this tag was extensively used in studying viral entry, viral protein localization, and viral assembly by live-imaging and immunofluorescence [36]. Of the two fluorophores, only the ReAsH bound tetracysteine tag can photo-oxidize DAB under continuous light illumination. Tagged connexin43 was successfully photo-oxidized and showed dense packing of connexin43 within the gap junction in contrast to sparse labeling of connexin43 by immunogold. The tetracysteine tag was also applied to the coat protein of the flock house virus (FHV) to study its cellular location in viral assembly and to distinguish the viral particles from ribosomes due to their similar size [37]. In contrast to the GFP fusion tag, which perturbs FHV infection, the tetracysteine tag enables correlated light/EM to identify FHV capsids in the cytoplasm without altering their structure. Although a ReAsH bound tetracysteine tag is a small genetic encoded photo-oxidizing agent, cellular toxicity from ReAsH loading, stringent labeling requirements, and modest photo-oxidation sensitivity make this tag suboptimal.
The modest reactive oxygen quantum yield of the ReAsH bound tetracysteine tag provided the motivation to search for another genetically encoded tag that enables more efficient photo-oxidation of DAB without the need for an extraneous labeling reagent. This was achieved in 2011 by Shu and co-workers [38], in which they engineered the LOV2 domain of Arabidopsis thaliana phototropin 2 to produce singlet oxygen upon light irradiation at 488 nm. This domain is called MiniSOG (Mini Singlet Oxygen Generator) and is comprised of 106 residues that bind a flavin mononucleotide cofactor with a weak green emission fluorescence signal (Fig. 1). The singlet oxygen quantum yield of MiniSOG is 20 times higher than the ReAsH bound tetra-cysteine tag [38], although the higher photo-oxidation efficiency of MiniSOG is likely due to a combination of singlet oxygen production and a singlet oxygen independent pathway [39]. Regardless of which pathway MiniSOG utilizes to oxidize DAB, MiniSOG is a far more sensitive and robust photo-oxidizer than a ReAsH bound tetracysteine tag [40]. Additionally, since MiniSOG oxidizes DAB through light excitation and reactive oxygen species, it can tolerate strong primary chemical fixation with glutaraldehyde and does not require detergent permeabilization. Thus, it provides higher spatial resolution than enzymatic based DAB oxidation to define the ultrastructure of stained proteins.
1.5. Potential issues from DAB staining
The identification of a target of interest by DAB staining is based on localized oxidation of DAB monomer to form an osmiophilic polymer near the oxidizing agent. Its success is dependent on whether DAB polymerization is truly ‘local’ due to the target, and not due to background auto-polymerization of DAB. Mitochondrial proteins, even after fixation, can still oxidize DAB. The addition of potassium cyanide can alleviate the latter phenomenon [30]. Another source of non-specific DAB oxidation is the use of excitation wavelengths below 400 nm in photo-oxidation, which alone can induce polymerization of DAB and should be avoided [30, 41]. Another source that affects the resolution of DAB staining is the diffusion distance of the reactive oxygen species diffuse away from the target. The farther the reactive oxygen species diffuse away from the source, the localized DAB staining becomes less precise. The most common solution to this issue is to conduct oxidation of DAB at low temperature (4°C), thus limiting the diffusion of both reactive oxygen species and the reaction products from the oxidizing agent. Additionally, crosslinking with glutaraldehyde can reduce the diffusion of the reaction products [30]. Also, it has been shown that reactive oxygen species generated from a point source (a fluorophore, MiniSOG) can yield higher resolution DAB staining and less ‘fuzzy’ staining than an enzyme-based (peroxidase, APEX) oxidizing agent, as shown in the staining of microtubules either by eosin-mediated photo-oxidation or a peroxidase-mediated oxidation [8]. Finally, the DAB oxidation time has an inverse relationship to the precision of localized DAB staining: the longer the oxidation time, the more diffuse the DAB polymer will appear and yield lower resolution. However, a short DAB oxidation time may not yield sufficient signals to be detected in EM. Unfortunately this is an empirical variable that is case dependent and needs to be optimized by the investigator for the best resolution that suits the goal of the experiment.
1.6. Visualization of adenoviral E4-ORF3 non-repeating polymer using an internal MiniSOG fusion
Adenovirus is a lytic dsDNA tumor virus with a life cycle of 48 hours. The genome has a size of 36 kbp and expresses 39 viral proteins throughout its life cycle. Expression of these proteins can be separated into early expressed proteins that promote cellular and viral genome replication and inhibit the cellular anti-viral response, and late expressed proteins that are important for viral capsid assembly [42]. One of the early cellular anti-viral response genes is p53 [43, 44], a tumor suppressor whose expression is up-regulated upon Adenovirus infection and induces cell cycle arrest or apoptosis [45, 46]. To counter this effect, Adenovirus expresses two early viral proteins, E1B-55K and E4-ORF3, which inactivate p53. E1B-55K functions analogously to the cellular protein MDM2, an E3 ubiquitin ligase. It recruits Cullin-5, Elongins B and C, Rbx1, and E4-ORF6 resulting in the ubiquitination and degradation of p53 [47]. Our group has shown that E4-ORF3 inactivates p53 by preventing p53 from accessing its target genes by inducing heterochromatin assembly at p53 target genes [45]. E4-ORF3 is a small 13 kDa protein that assembles a nuclear polymer that weaves through the nucleus and inactivates multiple disparate tumor suppressors, such as PML [48], MRE11/RAD50/NBS DNA repair complex (MRN) [49], TRIM24 [50], and p53 [45]. Using dominant negative oligomerization mutants, we solved the crystal structure of an E4-ORF3 dimer subunit at 2.1 Å. In contrast to the archetypal adenovirus oncoprotein, E1A, E4-ORF3 has a discrete ordered structure and is not a homologue of known cellular polymers or oncogenes. Structural and functional assays for polymer assembly support a model in which E4-ORF3 dimers assemble with each other in different permutations via reciprocal or non-reciprocal swapping of their β4 strands. This non-repeating assembly mechanism creates a matrix with multiplicative and emergent protein interaction surfaces [4]. E4-ORF3 polymerization is required for binding and disrupting PML bodies, the MRN complex, TRIM24, and silencing p53 target genes. We showed that the higher order assembly of E4-ORF3 dimers is necessary to create avidity driven interactions with PML and an emergent MRN binding interface.
Our model predicted that E4-ORF3 polymerizes in multiple ways to form a disordered protein superstructure. The striking E4-ORF3 polymer structures observed in immunofluorescence suggested it may be visible in EM micrographs (Fig. 2A). However, using conventional EM stains, the E4-ORF3 polymer is not immediately apparent (Fig. 2B). One possible reason for the apparent invisibility of E4-ORF3 in EM could be that the tortuous polymer that weaves through the nucleus as seen by immunofluorescence may appear as punctate structures in thin-section EM micrographs. It is also possible that the E4-ORF3 polymer has a loose weave and limited contrast with respect to other nuclear protein structures. Immunogold labeling is of limited value in elucidating the ultrastructure of polymers. Therefore, to reveal the ultrastructure of E4-ORF3 polymers in infected cells, we engineered viruses that express a MiniSOG-E4-ORF3 fusion.
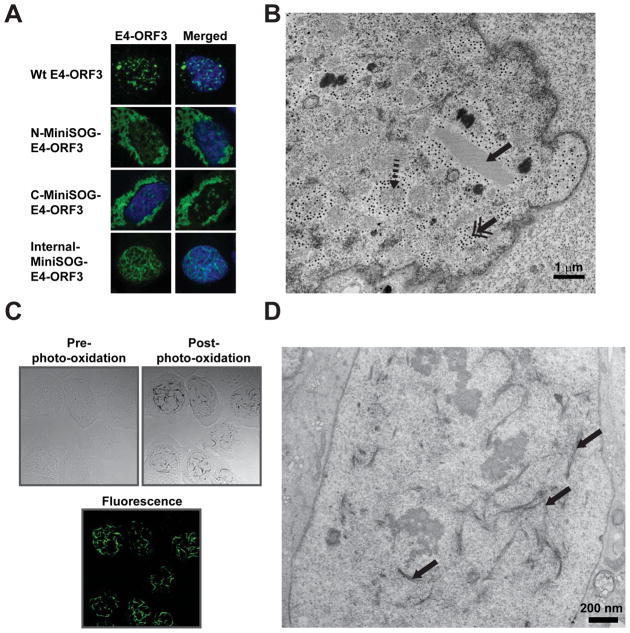
Fig. 2. Photo-oxidation of MiniSOG-E4-ORF3 to reveal in-situ ultrastructure in EM.
(A) U2OS cells were transfected with different MiniSOG-E4-ORF3 constructs fused at different positions and immunostained for E4-ORF3 (green). Wt-E4-ORF3 was transfected as a control. Only internal insertion of MiniSOG to E4-ORF3 displayed the correct nuclear polymer similar to Wt-E4-ORF3. DNA is stained with Hoechst. (B) U2OS cells infected with Wt Adenovirus expressing E4-ORF3 were fixed at 36 hours post infection, stained with osmium tetroxide and uranyl acetate, embedded in plastic resin, sectioned into 70–80 nm thin sections and imaged with TEM. A sample image shows many distinctive cellular and viral structures in an Adenovirus infected cell: the paracrystalline lattice (solid arrow), round, grainy viral replication domains (dashed arrow), and electron-dense particles (double arrowhead). Strikingly, no polymer structure is observed that resembles the E4-ORF3 immunofluorescence image. (C) U2OS cells infected with Adenovirus expressing MiniSOG-E4-ORF3 were photo-oxidized for 6 minutes in DAB buffer. MiniSOG-E4-ORF3 was not visible in the bright field image before photo-oxidation (left panel), but highly visible after photo-oxidation (right panel). Lower panel shows the fluorescence image of MiniSOG-E4-ORF3. (D) Thin sectioned TEM image of a photo-oxidized U2OS cell infected with Adenovirus expressing MiniSOG-E4-ORF3. The E4-ORF3 polymer (arrow) is highly visible in comparison with a conventionally stained infected U2OS cell in (B) due to DAB polymer precipitation along the E4-ORF3 polymer.
Material and Methods
The MiniSOG-E4-ORF3 fusion was integrated into the endogenous locus in the Adenoviral genome and the ultrastructure of E4-ORF3 polymer was investigated by both TEM, EM tomography, and serial block face scanning EM (SBEM) [4]. In this section, we describe the generation of the MiniSOG-E4-ORF3 fusion, the photo-oxidation station setup, and protocols for MiniSOG photo-oxidation and sample preparation for TEM and SBEM.
2.1. Generation of genetically encoded adenoviral MiniSOG-E4-ORF3 protein
One advantage of the MiniSOG protein as an EM reactive oxygen generator is its relatively small size (106 versus 238 residues for GFP) such that a genetic fusion with the targeted protein of interest would have minimal perturbation to the native protein structure. Shu et al. and other subsequent publications have shown that both N-terminal and C-terminal fusions of MiniSOG can preserve the structure and function of MiniSOG and the structure of the fused protein [38]. For proteins that assemble into higher order structures (i.e. actin filaments, microtubules, viral polymers), it is possible that the addition of an extraneous protein domain at the N- or C- terminus may affect higher order assembly. We encountered this problem when we tried to tag E4-ORF3 (13 kDa monomer, 116 residues) with MiniSOG at either the N- or the C- terminus (Fig. 2A) [4]. Both MiniSOG N- and C- terminal fusions resulted in polymer aggregates in the cytoplasm instead of the normal polymer formation in the nucleoplasm. To overcome this we used a flexible loop region between the α2 helix and the β2 strand identified in the E4-ORF3 crystal structure. MiniSOG was inserted in the middle of the E4-ORF3 protein after glycine residue 46 without the loss of E4-ORF3 polymer assembly and MiniSOG function (Figs. 2A and 2C). In post-photo-oxidized bright field imaging, the E4-ORF3 polymer is clearly labeled by polymerized DAB identical to the fluorescence signal (Fig. 2C). Thus MiniSOG can be genetically fused with the protein of interest at the N-, C-terminus, or within the protein to generate a DAB photo-oxidizing protein.
2.2. Photo-oxidation station
Here we describe the photo-oxidation station setup we employ as an example. The station includes a Leica/SPE II inverted epifluorescence/confocal microscope, a custom designed cold stage that fits a 35mm glass bottom MatTek dish (MatTek Corp.), an oxygen tank that delivers a stream of oxygen above the specimen during photo-oxidation, and an optional camera to record real-time imaging of the photo-oxidation process.
Leica SPE II confocal microscope system (Fig. 3A)
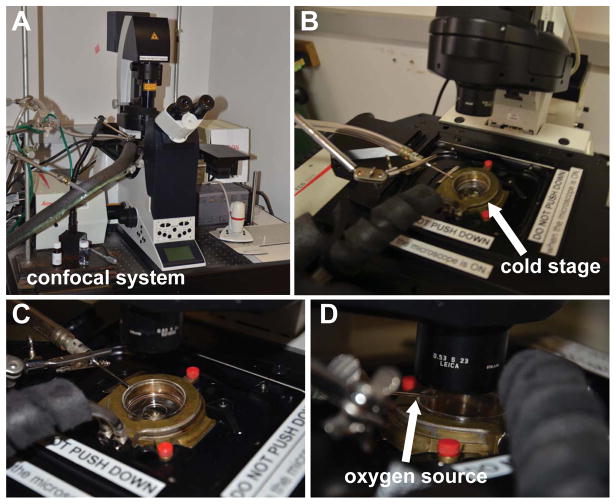
Fig. 3. Microscopy setup of a photo-oxidation station.
(A) A Leica/SP II confocal microscope for photo-oxidation. (B–C) A brass cold stage connected to a water cooler to maintain sample at 4°C. (D) A tube delivers a stream of oxygen above the sample during photo-oxidation to maintain an oxygenated environment to facilitate photo-oxidation of DAB.
Photo-oxidation requires an intense light source and the microscope is customized so that a 150W Xenon lamp house can be mounted directly to prevent power loss. The microscope has a multi-immersion - water, glycerol, oil - 20x lens and either a 63x oil immersion lens (NA 1.30) or a 63x water immersion lens (NA 1.15) for imaging. MiniSOG is excited using a 150W Xenon lamp with a Leica FITC/GFP filter set (11513878) or Semrock Lucifer Yellow filter set (Lucifer Yellow-B-L01). All excitation filters used in the system block UV from the lamp house to prevent auto-polymerization of DAB.
Cold stage (Figs. 3B and 3C)
To keep cells cold (~4°C) in order to improve membrane preservation, reduce reaction product diffusion, and improve oxygen saturation in the reacting solution, the sample dish is placed on a brass stage which is connected to a circulated water cooler that maintains the temperature of the stage at 4°C. If a cold stage setup is not available, an alternative method is to replace the cold chilled DAB solution every few minutes to maintain cells at 4°C.
Oxygen source (Fig. 3D)
A tube is connected to an oxygen tank that allows a stream of oxygen to flow above the dish but should not generate large ripples to the DAB solution in the dish. Photo-oxidation of DAB without an additional oxygen source will catalyze DAB polymerization at lower efficiency (longer photo-oxidation time). Alternatively, one can prepare an oxygenated DAB solution by bubbling oxygen into cold (4°C) DAB and replace the DAB solution frequently during the photo-oxidation process.
2.3. Photo-oxidation of MiniSOG-fused protein and conventional TEM
Except for step 1 of this protocol that is specific to Adenovirus infection of U2OS cells, the protocol can be generalized to sample preparation of other cultured cells. All solutions should be kept cold unless otherwise noted. Most of the solutions are highly toxic, and care should be taken in their handling and disposal.
Grow U2OS cells on a 35 mm glass bottom MatTek dish to 90% confluency in DMEM media with 10% FBS. Infect cells with Adenovirus carrying MiniSOG-E4-ORF3 at its endogenous genome position at MOI of 30 in DMEM media with 2% FBS.
Wash cultured cells 3 times in HBSS buffer (Life Technologies), and fix in glutaraldehyde (Electron Microscopy Sciences) (2% in 0.1 M sodium cacodylate, pH 7.4, containing 2 mM CaCl2) for 60 minutes (5 minutes at room temperature and 55 minutes on ice).
Wash cells 5 x 2 minutes in cold 0.1 M cacodylate (Ted Pella, Inc.) buffer.
Block free aldehydes and inhibit background DAB reactivity by incubating cells with blocking buffer (20 mM glycine, 10 mM KCN, 20 mM aminotriazole in 0.1 M cacodylate buffer) on ice for 30 minutes.
During the blocking step, make the DAB solution by dissolving 5.4 mg of free base DAB (Sigma-Aldrich) in 1 mL of 0.1 M HCl, then dilute the solution with 9 mL of 0.1 M cacodylate or blocking buffer (final DAB concentration 2.5 mM). Filter the DAB solution through a 0.22 μm syringe filter and keep on ice.
Place cells on the cold microscope stage at 4°C. Image the area of interest with fluorescent light and DIC. Once the region of interest is located, replace the solution with chilled DAB solution and turn on the oxygen tank to allow a stream of oxygen to gently blow across the top of the solution, taking care not to splash the DAB solution onto the microscope.
Illuminate the cells continuously with a 63x objective lens using a 488 nm filter at maximum intensity for 2–6 minutes depending on the level of protein expression and fluorescence intensity. The reaction is monitored by transmitted light and a light brown reaction product will gradually appear during photo-oxidation. The timing of the reaction needs to be determined empirically by the investigator, and the reaction is stopped by halting the illumination. One can also use a lower power objective; however, the length of the reaction will increase due to the reduced photo-intensity. DIC images of the cells can be taken after photo-oxidation to compare with the pre-photo-oxidize state. Multiple areas on each dish can be sequentially photo-oxidized.
After photo-oxidation, wash cells immediately on ice for 5 x 2 minutes in 0.1 M cacodylate buffer.
Post fix cells in 2% osmium tetroxide in 0.1 M sodium cacodylate on ice for 30 minutes. Optional to enhance membrane contrast: Right before use, prepare a solution containing 3% potassium ferrocyanide in 0.2M cacodylate buffer with 2 mM calcium chloride and combine with an equal volume of 4% aqueous osmium tetroxide (Electron Microscopy Sciences), and incubate cells in this solution for 30 minutes on ice.
Wash cells 5 x 1 minute in cold ddH2O.
Stain cells with 2% uranyl acetate (aqueous) for 1 hour on ice or incubate overnight at 4°C. After staining, wash cells 5 x 1 minute with ddH2O.
Prepare Durcupan ACM resin (Sigma-Aldrich) formulated by weight as follows: 11.4 g part A, 10 g part B, 0.3 g part C and 0.05–0.1 g part D, which yields a hard resin when polymerized. Mix resin thoroughly before use.
Dehydrate cells by preparing ice-cold solutions of fresh 20%, 50%, 70%, 90%, 100%, 100% ethanol (anhydrous), and a 3 minutes incubation for each ethanol concentration on ice. Then place cells in anhydrous 100% ethanol at room temp for 3 minutes.
After ethanol dehydration, infiltrate samples with a mixture of 1:1 Durcupan:ethanol for 30 minutes. Remove the resin and replace with fresh 100% Durcupan on cells and incubate for 30–60 minutes. Repeat this step 3 more times, taking care to remove the excess resin between changes.
Add fresh 100% Durcupan resin to cells (fill dish approximately 1/3 full) and place in a vacuum oven for 48 hours at 60°C to polymerize the resin.
2.4. Preparation of cultured cells for serial block face scanning electron microscopy
Samples for serial block face SEM require much more intense heavy metal staining than the protocol given above. This is to improve specimen contrast and conductivity. The following protocol is used immediately following the photo-oxidation process detailed above and employs the osmium-thiocarbohydrazide-osmium technique as well as en bloc uranyl acetate and lead aspartate staining.
Following step 7 above, and right before use, prepare a solution containing 3% potassium ferrocyanide in 0.2 M cacodylate buffer with 2 mM calcium chloride combined with an equal volume of 4% aqueous osmium tetroxide (EMS) and incubate cells in this solution for 30 minutes on ice.
While the initial osmium incubation (step 1 above) is occurring, prepare the following thiocarbohydrazide (TCH) solution. This reagent needs to be fresh and available right at the end of step 5. Add 0.1 g of thiocarbohydrazide (Ted Pella Inc.) to 10 ml ddH2O and place in a 60°C oven for 20 minutes (agitate by swirling gently every 10 minutes to facilitate dissolving). Filter this solution through a 0.22 μm Millipore syringe filter right before use and allow to cool to room temperature.
At the end of the first osmium tetroxide incubation described in step 1 (before adding the TCH), wash cells with ddH2O at room temperature 5 x 2 minutes.
Incubate cells with the filtered TCH solution for 10 minutes at room temperature.
Rinsed cells again 5 x 2 minutes in ddH2O at room temperature and thereafter place in 2% osmium tetroxide (NOT osmium-ferrocyanide) in ddH2O for 10 minutes on ice.
Following this second exposure to osmium, wash cells 5 x 2 minutes at room temperature in ddH2O, then place in 1% uranyl acetate (aqueous) and incubate overnight at 4°C.
The next day, perform en bloc Walton’s lead aspartate staining. First, prepare an aspartic acid stock solution by dissolving 0.998 g of L-aspartic acid (Sigma-Aldrich) in 250 ml of ddH2O. (Note: the aspartic acid will dissolve more quickly if the pH is raised to 3.8. This stock solution is stable for 1–2 months if refrigerated). To make the Walton’s lead aspartate stain, dissolve 0.066 g of lead nitrate in 10 ml of aspartic acid stock and adjust pH to 5.5 with 1N KOH. Place the lead aspartate solution in a 60°C oven for 30 minutes (no precipitate should form). Wash the cells 5 x 2 minutes in ddH2O at room temperature, then incubate cells with the lead aspartate solution in the 60°C oven for 5 minutes.
Wash cells 5 x 2 minutes at room temperature ddH2O and dehydrate using ice-cold solutions of freshly prepared 20%, 50%, 70%, 90%, 100% ethanol (anhydrous), 3 minutes incubation for each ethanol concentration, then place in anhydrous 100% ethanol at room temp for 3 minutes.
Durcupan ACM resin (Sigma-Aldrich) is formulated by weight as follows: 11.4 g part A, 10 g part B, 0.3 g part C and 0.05–0.1 g part D, which yields a hard resin when polymerized. Mix the resin thoroughly and incubate samples in 50%/50% Durcupan:ethanol for 30 minutes, follow by 2 x 2 hour 100% Durcupan incubations. Take care to remove excess resin between changes.
Place a small amount of resin over the cells and place a square piece of aclar (large enough to cover the hole) over the top opening to create a flat surface and place in a 60°C vacuum oven for 48 hours.
2.5. Image acquisition and Volume Reconstruction
Conventional TEM
Photo-oxidized areas of cultured cells are identified by transmitted light, the areas of interest are removed using a jeweler’s saw, and the glass coverslip is removed and mounted on acrylic blocks with cyanoacrylate adhesive. The block is trimmed with a razor blade, and ultrathin sections (70–80 nm) are cut using a conventional ultramicrotome and electron micrographs are recorded using a JEOL 1200 TEM (JEOL, Inc.) operating at 80 keV.
SBEM
Regions of interest are identified by transmitted light and removed as above, but the sample is mounted on an aluminum pin support (Gatan Inc, Pleasanton, CA) with cyanoacrylate adhesive and trimmed to a ~1mm2 block face. A 3View system (Gatan) mounted in a Quanta FEG variable pressure scanning electron microscope (FEI Company, Eindhoven, the Netherlands) or Merlin Compact (Zeiss) is employed. Block face imaging is performed at 2.5 keV at 20–50 pascals chamber pressure and material is removed from the block face at 60 nm increments using an oscillating diamond knife. Each image is scanned with a resolution of 8K by 6K pixels or larger.
2.6. Segmentation of 3View EM volume
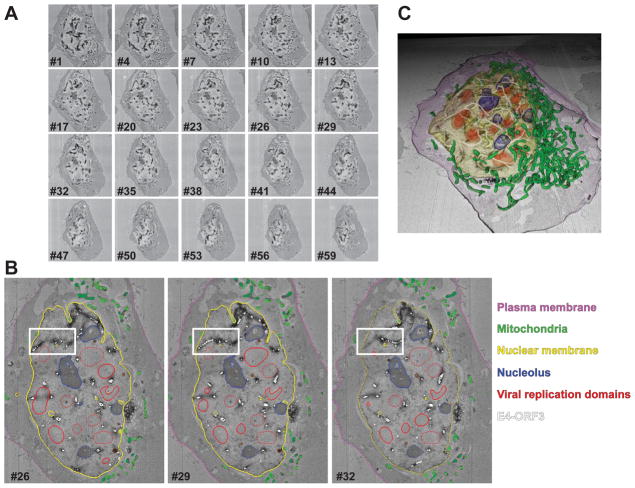
The 3D EM volume acquired from serial block face SEM (Fig. 4A) contains a wealth of information on how viral proteins alter the nuclear environment. Photo-oxidation of the MiniSOG-E4-ORF3 polymer reveals the irregular structure of the polymer that transverses throughout the nucleus. To semi-quantitate the diameter and distribution of the E4-ORF3 polymer, we used IMOD software (University of Colorado, Boulder) to view and segment the EM volume obtained from SBEM (Fig. 4B) [51]. The high contrast from the photo-oxidation of MiniSOG-E4-ORF3 allows facile tracing of the E4-ORF3 polymer. To simplify the tracing of the polymer structure and obtain an estimate of the E4-ORF3 diameter and length, a series of connected centered circles were drawn along the length of the polymer [52]. E4-ORF3 assemblies with diameters less than 60 nm were not segmented as they could not be traced with sufficient accuracy. Contrast from osmium tetroxide and uranium acetate staining and distinct morphology allowed identification of the nuclear membrane, nucleolus (round electron dense structures), mitochondria (elongated closed tube like structure), and viral replication centers (irregular oval structures with distinct contrast from nucleoplasm), which were segmented by tracing closed contours on every image plane with a mouse, and the plasma membrane was manually traced on every tenth slice and interpolation was used to segment the plasma membrane on every image plane. Segmented labels from IMOD were rendered in Cinema 4D and exported as movies to visualize E4-ORF3 polymer within the nuclear environment context (Fig. 4C) [4]. The segmentation reveals that E4-ORF3 polymer does not resemble other polymers like actin, microtubules, or amyloids. The E4-ORF3 diameter ranges from 60–710 nm and it forms an inter-connected network throughout the nucleus that assembles around the nucleoli and the viral replication centers.
Fig. 4. 3D EM ultrastructure of MiniSOG-E4-ORF3 polymer.
(A) U2OS cells infected with Adenovirus expressing MiniSOG-E4-ORF3 (dark structures) were fixed at 36 hours post infection, photo-oxidized and prepared for serial block face SEM imaging (SBEM). An array of individual images is shown. Each image plane was recorded at 60 nm intervals, and the entire cell was recorded using 150 image planes. (B) Segmentation of E4-ORF3 and other cellular structures in a cell from the SBEM volume from (A). Cell membrane (purple), nuclear membrane (yellow), E4-ORF3 (white), viral replication domains (red), nucleolus (blue), and mitochondria (green) were manually segmented in IMOD. Three sample slices are shown here. White box marks the tracing of E4-ORF3 polymer. (C) Rendering of the segmented structures from (B) showing E4-ORF3 polymer form a network that circumnavigates around nucleoli and the viral replication domains. All rendered surfaces are labeled in the same color as (B).
In addition EM tomography studies showed that E4-ORF3 polymer assemblies are a weave of thin linear and branched oligomer threads that are not stacked with one another throughout their lengths but associate at multiple points in no fixed geometric arrangement. Thus, E4-ORF3 assembles in multiple ways to form a disordered protein superstructure and 3D polymer network that physically partitions the nucleus. The interactions of such a polyvalent matrix with its binding partners could exceed that of avidity and affinity driven interactions.
Discussion
EM is a powerful technique to capture a snapshot of cellular ultrastructure at a specific time, although the standard practice is to image a single section between 50–70 nm thickness by TEM. The development of large 3D EM volume acquisition techniques such as SBEM [53] or focused ion beam-SEM (FIB-SEM) [54] has extended the window of observation beyond just a 70 nm thin section. Instead, the ultrastructure of an entire cell can be imaged, ranging from 19 μm3 for human pancreatic carcinoid cells [55] to 6,300,000 μm3 of mouse retina [56]. Both techniques use scanning EM to scan the surface of a sample block to form an image with secondary and back-scattered electrons, follow by sectioning off a slice of the sample block and re-scanning the new surface, and this scanning/sectioning process is reiterated through the sample block to obtain the entire 3D EM volume of the block. The two techniques differ in their sectioning method that yields different lateral and axial resolution to the final EM volume [57]: SBEM cuts the sample block with a diamond knife and the best reported resolution is 4.7 nm/pixel in XY and 25 nm/slice in Z; FIB-SEM mills the sample block with a gallium ion beam and the best reported resolution is 3 nm/pixel in XY and 3 nm/slice in Z. The ability to capture high resolution ultrastructure in a large volume by these new techniques will bring a holistic view of a virus infected cell by seeing the 3D morphological changes of an entire cell, not just local changes observed in a thin section. As we have discussed, there are several DAB-based staining methods that enable identification of specific proteins or structure within their natural cellular environment using EM. The development of genetically encoded tags such as tetracysteine, MiniSOG, and APEX are powerful tools for revealing the ultrastructure and interactions of viral and cellular protein interactions at high resolutions and entire cell volumes in the physiological relevant context of viral infection. This is clearly demonstrated in an Adenovirus-infected cell in which the virus reorganizes the nucleus into a new landscape with multiple new and distinct features that are as beautiful as they are enigmatic (Fig. 2B). The EM volumes imaged from SBEM or FIB-SEM will allow researchers to ask questions that were previously prohibitive and technically infeasible. It is foreseeable that one could replace each viral protein in a virus with these new genetically encoded tags and study the function and localization of each viral protein and uncover new pathways and biology. The newly invented genome editing CRISPR/Cas9 system [58] will enable endogenous knock-in of tags into the viral genome as shown already in Vaccinia virus [59], Epstein-Barr virus [60], Adenovirus [61], and Herpes simplex virus [62]. Virology and EM may be entering another golden age of discovery.
Conclusions
Within the past 20 years, great progress has been made in EM probe development [26, 27, 35, 38], instrumentation [53, 54, 63], image acquisition [57, 64], image reconstruction [65, 66], and segmentation [67, 68]. In contrast to conventional EM stains in which most objects are not stained specifically, now we have a diverse set of powerful tools to identify a specific protein or structure in EM by localized oxidation of DAB into osmiophilic polymer that selectively contrasts the protein of interest, either through photo-oxidation or enzymatic-oxidation. The availability of small genetically encoded tags such as tetracysteine and MiniSOG now enable the investigation of in-situ viral protein organization with minimal perturbation to their native structures that would not be possible with large genetically encoded fusion tags (GFP or APEX). Conversely, large viral proteins or cellular proteins could be labeled by APEX and reveal viral-host protein interactions and show how viral proteins alter cellular proteins to usurp cellular pathways and achieve viral replication. The combination of DAB labeling with a genetically encoded tag and recent advancement in EM volume image acquisition by SBEM or FIB-SEM permit investigation of viral protein localization and function in the context of the entire cellular environment. This can lead to a better understanding not only of how viral proteins alter cellular environment, but also the interactions with other viral proteins that ensure proper viral propagation. An example of this multi-scale approach to studying virus protein structure and function is E4-ORF3, which reveals a new type of multifunctional hub and protein polymer that redefines the possibilities and potential for such structures.
Highlights.
DAB is a small molecule that assembles into osmiophilic polymer upon oxidation
Localized formation of DAB polymer enables identification of proteins inside cells in EM
DAB can be oxidized by either photo-oxidation or enzymatic catalysis
Immunostaining or genetically encoded tags can generate localized DAB polymer
Protocols for photo-oxidation and sample preparation for serial block face SEM
Acknowledgments
This work was supported by the National Institutes of Health R01CA137094, and P30CA014195 from the National Cancer Institute, The Leona M. and Harry B. Helmsley Charitable Trust grant #2012-PG-MED002. C.C.O. is supported by the William Scandling Trust and the Price Family Foundation. NIH Grant GM103412 was awarded to Mark Ellisman, and NIH/NIGMS R01GM086197-06 was awarded to Mark Ellisman and Roger Tsien.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mulder NJ, Kersey P, Pruess M, Apweiler R. Molecular biotechnology. 2008;38:165–177. doi: 10.1007/s12033-007-9003-x. [DOI] [PubMed] [Google Scholar]
- 2.Ou HD, May AP, O’Shea CC. Wiley Interdiscip Rev Syst Biol Med. 2011;3:48–73. doi: 10.1002/wsbm.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berk AJ. Oncogene. 2005;24:7673–7685. doi: 10.1038/sj.onc.1209040. [DOI] [PubMed] [Google Scholar]
- 4.Ou HD, Kwiatkowski W, Deerinck TJ, Noske A, Blain KY, Land HS, Soria C, Powers CJ, May AP, Shu X, Tsien RY, Fitzpatrick JA, Long JA, Ellisman MH, Choe S, O’Shea CC. Cell. 2012;151:304–319. doi: 10.1016/j.cell.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shaner NC, Steinbach PA, Tsien RY. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 6.Huang B, Bates M, Zhuang X. Annu Rev Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hulo C, de Castro E, Masson P, Bougueleret L, Bairoch A, Xenarios I, Le Mercier P. Nucleic Acids Res. 2011;39:D576–582. doi: 10.1093/nar/gkq901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sosinsky GE, Giepmans BN, Deerinck TJ, Gaietta GM, Ellisman MH. Methods in cell biology. 2007;79:575–591. doi: 10.1016/S0091-679X(06)79023-9. [DOI] [PubMed] [Google Scholar]
- 9.Knott G, Genoud C. J Cell Sci. 2013;126:4545–4552. doi: 10.1242/jcs.124123. [DOI] [PubMed] [Google Scholar]
- 10.Harris JR. Archives of Biochemistry and Biophysics. 2015 [Google Scholar]
- 11.Curry A, Appleton H, Dowsett B. Micron. 2006;37:91–106. doi: 10.1016/j.micron.2005.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horne RW. Ann N Y Acad Sci. 1962;101:475–484. doi: 10.1111/j.1749-6632.1962.tb18888.x. [DOI] [PubMed] [Google Scholar]
- 13.Morgan C, Godman GC, Rose HM, Howe C, Huang JS. The Journal of biophysical and biochemical cytology. 1957;3:505–508. doi: 10.1083/jcb.3.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez-Palomo A, Le Buis J, Bernhard W. J Virol. 1967;1:817–829. doi: 10.1128/jvi.1.4.817-829.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faulk WP, Taylor GM. Immunochemistry. 1971;8:1081–1083. doi: 10.1016/0019-2791(71)90496-4. [DOI] [PubMed] [Google Scholar]
- 16.Franqueville L, Henning P, Magnusson M, Vigne E, Schoehn G, Blair-Zajdel ME, Habib N, Lindholm L, Blair GE, Hong SS, Boulanger P. PLoS ONE. 2008;3:e2894. doi: 10.1371/journal.pone.0002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Souquere-Besse S, Pichard E, Filhol O, Legrand V, Rosa-Calatrava M, Hovanessian AG, Cochet C, Puvion-Dutilleul F. Microscopy research and technique. 2002;56:465–478. doi: 10.1002/jemt.10060. [DOI] [PubMed] [Google Scholar]
- 18.Wills EJ, Russell WC, Williams JF. J Gen Virol. 1973;20:407–412. doi: 10.1099/0022-1317-20-3-407. [DOI] [PubMed] [Google Scholar]
- 19.Tokuyasu KT. J Cell Biol. 1973;57:551–565. doi: 10.1083/jcb.57.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bos E, Hussaarts L, van Weering JR, Ellisman MH, de Wit H, Koster AJ. J Struct Biol. 2014;186:273–282. doi: 10.1016/j.jsb.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 21.Oorschot VM, Sztal TE, Bryson-Richardson RJ, Ramm G. Methods in cell biology. 2014;124:241–258. doi: 10.1016/B978-0-12-801075-4.00011-2. [DOI] [PubMed] [Google Scholar]
- 22.Graham RC, Jr, Karnovsky MJ. The journal of histochemistry and cytochemistry: official journal of the Histochemistry Society. 1966;14:291–302. doi: 10.1177/14.4.291. [DOI] [PubMed] [Google Scholar]
- 23.Nakane PK, Pierce GB., Jr J Cell Biol. 1967;33:307–318. doi: 10.1083/jcb.33.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hopkins C, Gibson A, Stinchcombe J, Futter C. Methods Enzymol. 2000;327:35–45. doi: 10.1016/s0076-6879(00)27265-0. [DOI] [PubMed] [Google Scholar]
- 25.Munro S, Pelham HR. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 26.Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, Ellisman MH, Ting AY. Nat Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK, Ting AY. Nat Methods. 2015;12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maranto AR. Science. 1982;217:953–955. doi: 10.1126/science.7112109. [DOI] [PubMed] [Google Scholar]
- 29.Martin JP, Logsdon N. Photochemistry and photobiology. 1987;46:45–53. doi: 10.1111/j.1751-1097.1987.tb04734.x. [DOI] [PubMed] [Google Scholar]
- 30.Deerinck TJ, Martone ME, Lev-Ram V, Green DP, Tsien RY, Spector DL, Huang S, Ellisman MH. J Cell Biol. 1994;126:901–910. doi: 10.1083/jcb.126.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gandin E, Lion Y, Van de Vorst A. Photochemistry and photobiology. 1983;37:271–278. [Google Scholar]
- 32.Gaietta G, Deerinck TJ, Adams SR, Bouwer J, Tour O, Laird DW, Sosinsky GE, Tsien RY, Ellisman MH. Science. 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- 33.Griffin BA, Adams SR, Tsien RY. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 34.Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Llopis J, Tsien RY. J Am Chem Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 35.Martin BR, Giepmans BN, Adams SR, Tsien RY. Nat Biotechnol. 2005;23:1308–1314. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- 36.Whitt MA, Mire CE. Methods. 2011;55:127–136. doi: 10.1016/j.ymeth.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Lanman J, Crum J, Deerinck TJ, Gaietta GM, Schneemann A, Sosinsky GE, Ellisman MH, Johnson JE. J Struct Biol. 2008;161:439–446. doi: 10.1016/j.jsb.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shu X, Lev-Ram V, Deerinck TJ, Qi Y, Ramko EB, Davidson MW, Jin Y, Ellisman MH, Tsien RY. PLoS Biol. 2011;9:e1001041. doi: 10.1371/journal.pbio.1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruiz-Gonzalez R, Cortajarena AL, Mejias SH, Agut M, Nonell S, Flors C. J Am Chem Soc. 2013;135:9564–9567. doi: 10.1021/ja4020524. [DOI] [PubMed] [Google Scholar]
- 40.Boassa D, Berlanga ML, Yang MA, Terada M, Hu J, Bushong EA, Hwang M, Masliah E, George JM, Ellisman MH. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2013;33:2605–2615. doi: 10.1523/JNEUROSCI.2898-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirai K. The journal of histochemistry and cytochemistry: official journal of the Histochemistry Society. 1971;19:434–442. doi: 10.1177/19.7.434. [DOI] [PubMed] [Google Scholar]
- 42.Berk AJ. In: Fields Virology. Knipe DM, Howley PM, editors. Lippincott Williams & Wilkins and Wolters Kluwer; Philadelphia: 2007. pp. 2355–2394. [Google Scholar]
- 43.Lane DP, Crawford LV. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 44.Linzer DI, Levine AJ. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 45.Soria C, Estermann FE, Espantman KC, O’Shea CC. Nature. 2010;466:1076–1081. doi: 10.1038/nature09307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Shea C, Johnson L, Bagus B, Choi S, Nicholas C, Shen A, Boyle L, Pandey K, Soria C, Kunich J, Shen Y, Habets G, Ginzinger D, McCormick F. Cancer Cell. 2004;6:611–623. doi: 10.1016/j.ccr.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 47.Harada JN, Shevchenko A, Pallas DC, Berk AJ. J Virol. 2002;76:9194–9206. doi: 10.1128/JVI.76.18.9194-9206.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doucas V, Ishov AM, Romo A, Juguilon H, Weitzman MD, Evans RM, Maul GG. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 49.Stracker TH, Carson CT, Weitzman MD. Nature. 2002;418:348–352. doi: 10.1038/nature00863. [DOI] [PubMed] [Google Scholar]
- 50.Yondola MA, Hearing P. J Virol. 2007;81:4264–4271. doi: 10.1128/JVI.02629-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kremer JR, Mastronarde DN, McIntosh JR. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 52.Noske AB, Costin AJ, Morgan GP, Marsh BJ. J Struct Biol. 2008;161:298–313. doi: 10.1016/j.jsb.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Denk W, Horstmann H. PLoS Biol. 2004;2:e329. doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heymann JA, Hayles M, Gestmann I, Giannuzzi LA, Lich B, Subramaniam S. J Struct Biol. 2006;155:63–73. doi: 10.1016/j.jsb.2006.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Villinger C, Gregorius H, Kranz C, Hohn K, Munzberg C, von Wichert G, Mizaikoff B, Wanner G, Walther P. Histochem Cell Biol. 2012;138:549–556. doi: 10.1007/s00418-012-1020-6. [DOI] [PubMed] [Google Scholar]
- 56.Briggman KL, Helmstaedter M, Denk W. Nature. 2011;471:183–188. doi: 10.1038/nature09818. [DOI] [PubMed] [Google Scholar]
- 57.Peddie CJ, Collinson LM. Micron. 2014;61:9–19. doi: 10.1016/j.micron.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 58.Doudna JA, Charpentier E. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 59.Yuan M, Zhang W, Wang J, Yaghchi CA, Ahmed J, Chard L, Lemoine NR, Wang Y. J Virol. 2015 doi: 10.1128/JVI.00339-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuen KS, Chan CP, Wong NH, Ho CH, Ho TH, Lei T, Deng W, Tsao SW, Chen H, Kok KH, Jin DY. J Gen Virol. 2015;96:626–636. doi: 10.1099/jgv.0.000012. [DOI] [PubMed] [Google Scholar]
- 61.Holkers M, Maggio I, Henriques SF, Janssen JM, Cathomen T, Goncalves MA. Nat Methods. 2014;11:1051–1057. doi: 10.1038/nmeth.3075. [DOI] [PubMed] [Google Scholar]
- 62.Suenaga T, Kohyama M, Hirayasu K, Arase H. Microbiol Immunol. 2014;58:513–522. doi: 10.1111/1348-0421.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bushong EA, Johnson DD, Kim KY, Terada M, Hatori M, Peltier ST, Panda S, Merkle A, Ellisman MH. Microscopy and microanalysis: the official journal of Microscopy Society of America. Microbeam Analysis Society, Microscopical Society of Canada; 2014. pp. 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gan L, Jensen GJ. Quarterly reviews of biophysics. 2012;45:27–56. doi: 10.1017/S0033583511000102. [DOI] [PubMed] [Google Scholar]
- 65.Phan S, Lawrence A, Molina T, Lanman J, Berlanga M, Terada M, Kulungowski A, Obayashi J, Ellisman M. J Struct Biol. 2012;180:154–164. doi: 10.1016/j.jsb.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 66.Miranda K, Girard-Dias W, Attias M, de Souza W, Ramos I. Molecular reproduction and development. 2015 doi: 10.1002/mrd.22455. [DOI] [PubMed] [Google Scholar]
- 67.Volkmann N. Methods Enzymol. 2010;483:31–46. doi: 10.1016/S0076-6879(10)83002-2. [DOI] [PubMed] [Google Scholar]
- 68.Perez AJ, Seyedhosseini M, Deerinck TJ, Bushong EA, Panda S, Tasdizen T, Ellisman MH. Frontiers in neuroanatomy. 2014;8:126. doi: 10.3389/fnana.2014.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]