Abstract
Genetic factors explain approximately half of the variance in smoking behaviors, but the molecular mechanism by which genetic variation influences behavior is poorly understood. SNPs in the putative promoter region of CHRNB3, the gene that encodes the β3 subunit of the nicotinic acetylcholine receptor (nAChR), have been repeatedly associated with tobacco behaviors. In this work we sought to identify putative function of three SNPs in the promoter region of CHRNB3 on in vitro gene expression. Additionally, we used β3 null mutant mice as a model of reduced gene expression to assess the effects on nicotine behaviors. The effect of rs13277254, rs6474413, and rs4950 on reporter gene expression was examined using a luciferase reporter assay. A major and minor parent haplotype served as the background on which alleles at the three SNPs were flipped onto different backgrounds (e.g. minor allele on major haplotype background). Constructs were tested in three human cell lines: BE(2)-C, SH-SY5Y and HEK 293T. In all cell types the major haplotype led to greater reporter gene expression compared to the minor haplotype, and results indicate that this effect is driven by rs6474413. Moreover, mice lacking the β3 subunit showed reduced voluntary nicotine consumption compared that of wildtype animals. These data provide evidence that the protective genetic variant at rs6474413 identified in human genetic studies reduces gene expression and that decreased β3 gene expression in mice reduces nicotine intake. This work contributes to our understanding of the molecular mechanisms that contribute to the human genetic associations of nicotine behaviors.
Keywords: Nicotine, nicotinic acetylcholine receptors, consumption, luciferase, gene expression
1. INTRODUCTION
Genetic factors explain between 40–70% of the variance for smoking behaviors, as has been documented in numerous twin and family studies (Madden and Heath, 2002; Maes et al, 2004; True et al, 1999). The genes that encode neuronal nicotinic acetylcholine receptor (nAChR) subunits (CHRN genes) are among the most highly replicated for their strong association with smoking behaviors. The CHRNA6/CHRNB3 gene cluster (the genes that code for the α6 and β3 subunit, respectively) on chromosome 8 has emerged multiple times from Genome Wide Association Studies (GWAS) and candidate gene approaches as being associated with nicotine behaviors (Ehringer et al, 2010; Hoft et al, 2009; Rice et al, 2012; Saccone et al, 2007; Thorgeirsson et al, 2010; Zeiger et al, 2008). A number of SNPs in this region have been associated with nicotine phenotypes, but most represent a single genetic signal due to high linkage disequilibrium. Having the minor allele at these SNPs is protective against nicotine dependence (Rice et al, 2012).
nAChRs are ligand-gated channels that allow the influx of cations (Arias et al, 2006; McGehee, 1999). Formed of five subunits, neuronal acetylcholine receptors come in two varieties. Homomeric receptors are comprised of the same subunit (i.e. α7 receptors) while heteromeric receptors are comprised of a combination of α and β subunits (i.e. α4β2 receptors). One important feature of these receptors relates to their carefully regulated expression both temporally throughout development and spatially in certain regions of the brain (Millar and Gotti, 2009).
Although genetic associations between CHRN genes and nicotine behaviors have been widely replicated in the literature, the function of associated variants is not well characterized. One of the most well studied genetic variants is in the CHRNA5 gene and leads to an amino acid change in the protein and confers differential function of the receptor (Bierut et al, 2008; Morel et al, 2014; Tammimaki et al, 2012). The region coding for this gene (the CHRNA5/CRHNA3/CHRNB4 cluster of genes) has been repeatedly associated with nicotine dependence (Bierut et al, 2008). However, multiple studies have demonstrated that this variant (rs16969968) is not the only important association signal in the CHRNA5/CHRNA3/CHRNB4 region, and that other possibly noncoding functional variants contribute to risk for nicotine phenotypes, including age of onset of regular use of tobacco (Saccone et al, 2010; Stephens et al, 2013).
Studies of noncoding variants in the promoter region of CHRNB2 (Hoft et al, 2011), CHRNA4 (Hutchison et al, 2007), CHRNB3 (Ehringer et al, 2010), and intergenic regions in the CHRNA5/CHRNA3/CHRNB4 gene cluster (Flora et al, 2013; Gallego et al, 2013; Wu et al, 2009) have provided support for a regulatory role of noncoding variants. Although luciferase-based assays support a role for SNPs in all of these regions in mediating gene transcription, the biological mechanism underlying many of these results remain undetermined. To date the interaction of three SNPs across these genes have yielded results suggesting that disruption of enhancer regions may lead to observed changes in expression (Flora et al, 2013; Hutchison et al, 2007; Wu et al, 2009), but for most variants the mechanism influencing expression are unknown. Furthermore, how SNPs influence expression are further complicated by cell-type specific effects (Flora et al, 2013), and additional work is necessary to delineate these effects.
The β3 subunit of the nAChR is not widely expressed in the brain, but it is expressed in key brain regions important in drug behaviors including the ventral tegmental area, substantia nigra (Grady et al, 2007), and medial habenula (Grady et al, 2009). It is important to note that the α5 (mentioned above as a top genetic association) and β3 subunits are the only two subunits that do not participate in ligand binding but their inclusion in functional receptors are believed to confer subtle differences in structural conformations leading to differences in the pharmacological properties of the receptor (Gotti et al, 2009; Wu and Lukas, 2011). Moreover, receptors containing the β3 subunit have been implicated in dopamine release from dopamine terminals in the mouse striatum (Grady et al, 2007). Given the regional pattern of expression in the brain, its involvement in dopamine responses, and the human genetic associations observed, the β3 subunit warrants further investigation.
There were two goals of the current study. First, we used in vitro gene expression assays to examine the function of three SNPs in the promoter region of CHRNB3. This expands on previous work implicating this region in gene expression regulation (Ehringer et al, 2010), and may be a mechanism by which these genetic variants modulate nicotine behaviors. Second, we examined nicotine behaviors in the β3 null mutant mice as a model of reduced gene expression.
2. MATERIALS AND METHODS
2.1 In Vitro Studies
2.1.1 Luciferase assays
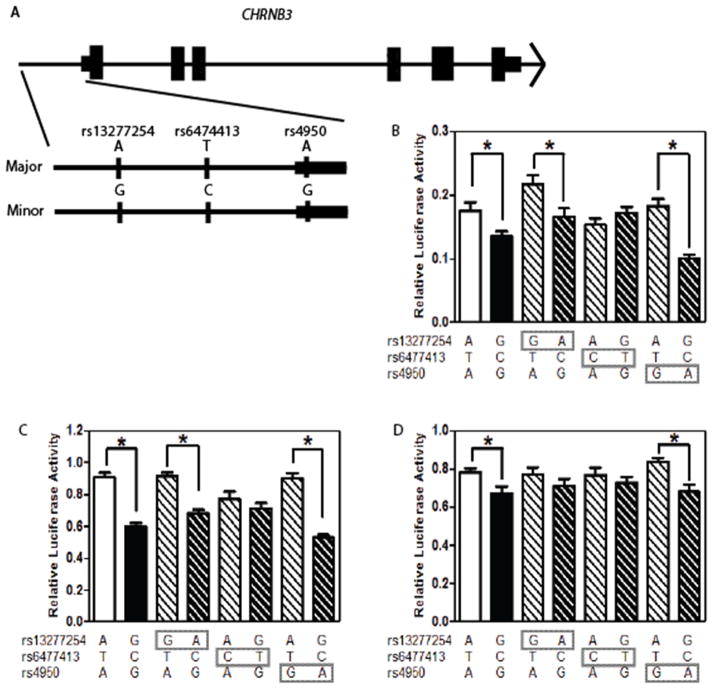
The development of the promoter region of CHRNB3 constructs has previously been described (Ehringer et al, 2010). Briefly, a 2,987 bp fragment of the human promoter region of CHRNB3 was amplified and introduced into a pGL3-basic vector (Promega, Madison, WI). The major and minor alleles at known SNPs in the region were identified and two haplotypes were developed: one containing the major allele SNPs and the other containing the minor allele SNPs (referred to as parent haplotypes). For investigation in this study three genetic variants were selected based on two criteria. First, SNPs associated with nicotine behaviors were prioritized. Second, we focused on SNPs predicted to change the binding of a transcription factor (in the Transcription Element Search System (TESS) database; http://www.cbil.upenn.edu/tess) that has been implicated in drug behavior and/or is expressed in the brain. For example, the POU3F2 transcription factor binds when the T allele is present at rs4950, but not the C allele, and is expressed in the cortical neurons (Dominguez et al, 2013). Six new constructs (Figure 1) were developed where the major/minor alleles at the three SNPs of interest (rs13277254, rs6474413, and rs4950) were flipped on the major and minor parent haplotypes using site directed mutagenesis (i.e., the major allele of rs13277254 was introduced on the minor parent haplotype; Agilent Technologies, Santa Clara, CA). In total, eight constructs were tested. The two parent haplotypes and the six newly created constructs.
Fig. 1.
SNPs upstream of CHRNB3 influence gene expression. (A) represents a graphical depiction of SNPs tested and corresponding location to the CHRNB3 gene. Briefly, our construct contained a 2,987bp fragment cloned from the region upstream of the CHRNB3 including the 5′UTR. The expanded region labeled major and minor depicts the 2,987bp fragment and location of 3 SNPs examined and allele present on the major and minor parent constructs. The effect of the genotype at rs13277254, rs6474413, and rs4950 on reporter gene expression was examined in three cell lines (B) BE(2)-C, (C) SH-SY5Y, and (D) HEK293T cells. The genotype at each SNP is depicted below the graph. The open white and closed black bars represent the major and minor parent constructs, respectively. Data (mean ± SEM) relative gene expression. * p <0.05
Constructs were tested in two human neuroblastoma cells lines, BE(2)-C and SH-SY5Y, in addition to human embryonic kidney cells, HEK293T cells (ATCC, Manassas, VA). Briefly, cells were grown in 24-well cell culture plates for 24 h (3.5 × 105 cells per well for HEK293T and 1 × 105 cells per well for BE(2)-C and SH-SY5Y). Cells were transfected using X-tremeGENE DNA transfection reagent (Roche, Indianapolis, IN) with 400 ng of test plasmid and 1 ng of the control vector pRL-CMV (Renilla Luciferase; Promega, Madison, WI). Control transfections included the empty pGL3-basic vector (Promega, Madison, WI) and the pRL-CMV vector. 48h after transfection, cells were harvested and the luciferase assay was performed in triplicate with the Dual-Luciferase Assay Reporter System (Promega, Madison, WI). Luminescence was quantified on a PerkinElmer Victor 3V plate reader (Perkin Elmer, Wellesley, MA). For each well, gene expression was quantified as the firefly luciferase luminescence values divided by renilla luciferase luminescence values to control for differences in transfection efficiency. Replicate reactions were averaged. Moreover, expression levels were normalized to the control construct (pGL3-basic). Each construct was tested in at least two experiments and two different maxi preps were used. The primary dependent variable, relative gene expression (luciferase/renilla luminescence), was analyzed with a 1-way ANOVA followed by appropriate post-hoc t-tests for specific comparisons of interest.
2.1.2 Electrophoretic mobility shift assay (EMSA)
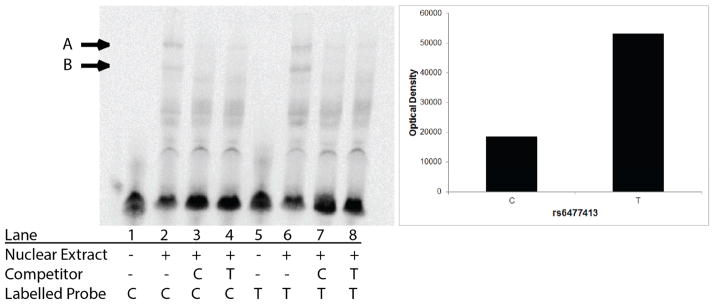
EMSA studies were performed to examine if the allele present at rs6477413 influenced the binding of nuclear extract proteins derived from both the BE(2)-C and SH-SY5Y cells. Double-stranded DNA oligonucleotides (36 bp) spanning the region around rs6474413 were synthesized by Integrated DNA Technologies [rs6474413 T-allele: 5′-TAAAGGTGAAACTTCCTGTACACTTGGATTGGCGGT-3′; rs6474423 C-allele: 5′-TAAAGGTGAAACTTCCTGCACACTTGGATTGGCGGT-3′]. Labeled oligonucleotides were synthesized with biotin on the 3′ end. Nuclear proteins were extracted from BE(2)-C or SHSY-5Y cells using the NE-PER kit (Thermo Scientific, Waltham, MA). The amount of protein was quantitated with the Pierce® BCA Protein Assay Kit (Thermo Scientific, Waltham, MA). EMSAs were performed using the LightShift Chemiluminescent EMSA kit (Thermo Scientific, Waltham, MA) under the manufacturers recommended protocol. Briefly, 0.0625 pMol labeled probe was incubated in the presence of cell nuclear extracts (10–15μg). To prevent non-specific binding poly[dI-dC] (1μg) and 20 mM EDTA (for BE(2)-C EMSAs only) were added to a 20μl total reaction volume. Specificity was established with the addition of 100-fold molar excess of the unlabeled oligonucleotide. The mixture was migrated on a gel using electrophoresis, blotted and detected with the Pierce® Chemiluminescent Detection Kit (Thermo Scientific, Waltham, MA). Images were acquired using a GelLogic 2200Pro CCD-camera and the density of the gel shift bands was quantified using Carestream Molecular Imaging Software v5.0.7.24 (Carestream Molecular Imaging, Woodbridge, CT).
2.1.3 Bioinformatics Analysis
To more extensively evaluate whether the SNP rs6474413 was located in a transcription factor binding site we queried four transcription factor databases Promo (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3), TFSEARCH (http://www.cbrc.jp/research/db/TFSEARCH.html), TESS (http://www.cbil.upenn.edu/tess) and Mapper (http://snpper.chip.org/mapper).
2.2 Behavioral Studies
2.2.1 Animals
Mice deficient of the β3 subunit were previously produced using homologous recombination technology by an interruption of exon 5 of the Chrnb3 gene (Cui et al, 2003). The mutation was produced in a 129Svj embryonic stem cell that was injected into a C57BL/J (C57) blastocyst (Cui et al, 2003). Mice carrying the null mutation were backcrossed independently to both 129S6/SvEvTac (129) and C57 mice for at least 10 generations to create two separate lines of mice harboring the mutation (herein described as β3-129 and β3-C57 mice). Wildtype, heterozygous, and homozygous null mutant animals tested in these experiments were produced from heterozygous breeder pairs mated at the Institute for Behavioral Genetics. Offspring of these breeders were housed 2–4 per cage in standard mouse cages with ad libitum water and rodent chow until testing. At the start of behavioral testing, animals were between 50 and 120 days old. Lighting in the animal colony was maintained on a 12-hour light/dark cycle with lights on at 0700 hours. All testing was approved by the University of Colorado’s Institutional Animal Care and Use Committee and follow the National Institutes of Health guide for the care and use of laboratory animals (NIH Publication No. 8023, revised 1978).
2.2.2 Locomotion and body temperature
Locomotor activity and body temperature were examined using a test battery developed at the Institute for Behavioral Genetics for measuring the behavioral effects of nicotine (Marks et al, 1985; Marks et al, 1989; Mexal et al, 2012). Briefly, locomotor activity was examined in a symmetrical Y-maze, consisting of three red Plexiglas arms (26 L X 6.1 W X 10.2 H; cm). Each arm had 2 photobeam detectors that measured locomotor activity. Body temperature was measured with a Bailey Instruments rectal probe lubricated with peanut oil.
A within-subjects design was used to measure the effect of nicotine on locomotion and body temperature. Briefly, each animal was tested following both an i.p. injection of saline and nicotine (0.25, 0.5, or 1 mg/kg; doses presented as freebase nicotine) (Marks et al, 1989). The nicotine and saline injections were counterbalanced and the two testing sessions occurred a week apart. Immediately following the injection, each mouse was placed in a holding cage. Three minutes after the injection the animal was placed into the Y-maze and locomotor activity was measured for 3 minutes. Following activity testing, the mice were returned to the holding cages before being tested for rectal body temperature 13 minutes after the nicotine injection. Nicotine doses and testing times were based on published methods (Marks et al, 1985; Marks et al, 1989; Mexal et al, 2012). Two primary dependent variables were evaluated: nicotine-induced locomotor activity (beam crosses) and change in body temperature (°C). Both variables were calculated as a within subject change score by subtracting the nicotine response from the saline response.
2.2.3 Two-bottle choice nicotine consumption
Nicotine intake was measured in a standard 2-bottle free choice paradigm (Butt et al, 2005; Wilking et al, 2010). A subset of animals tested for locomotor activity and body temperature were used for these experiments. Mice were allowed a one-week break prior to starting the nicotine drinking study. Animals were singly housed in standard mouse cages. Mice were provided access to two 25 ml graduate cylinders fitted with standard drinking spouts filled with water for the first four days to acclimate them to the test environment. On the first testing day one tube of water was replaced with a tube containing 25 μg/ml nicotine. Nicotine drinking solutions were made of free-base nicotine (Sigma-Aldrich, St. Louis, MO) diluted in tap water (Matta et al, 2007). Consistent with prior published reports (Butt et al, 2005; Wilking et al, 2010), no effort was made to mask the flavor of the nicotine or to adjust the pH of the solutions. The volume of fluid in the tubes was recorded at approximately the same time each day and the side of the cage on which they were presented was switched every other day. Every four days the concentration of nicotine presented increased (25, 50, 100 and 200 μg/ml). Since prior evidence suggests that the behavior of Chrnb3 mice can be influenced by stressful handling of mice (Booker et al, 2007), the animals were only weighed the first and last day of nicotine exposure and the average of these two weights was used to adjust consumption by body weight throughout the experiment. A measure of evaporation/leakage was obtained from two empty cages handled like the experimental cages. Three primary dependent variables were obtained: nicotine consumption (mg/kg), nicotine preference (ml of nicotine/total ml fluid) and total fluid consumed (ml). These dependent variables were derived from the average of all four days of each nicotine concentration (Kamens et al, 2006).
2.2.4 Two-bottle choice tastant consumption
One week after completion of the nicotine consumption study, tastant consumption was measured with a procedure identical to that described above for nicotine. Mice were offered access to saccharin (0.033 and 0.066%) and quinine (0.015 and 0.03 mM) in a two-bottle choice procedure (Kamens et al, 2010; Kamens et al, 2005). Briefly, each tastant was presented for 4 days with the lower concentration offered first. As with the nicotine consumption study, fluid was read every day and the side where the tastant was presented was switched every two days. Dependent variables included average 24 hour tastant consumption (mg/kg), tastant preference (ml of tastant/total ml fluid consumption), and total fluid consumption (ml) across all four days of each tastant concentration.
2.2.5 Statistics
All statistical analyses were carried out in SPSS (IBM, Armonk, NY). Dependent variables were analyzed using an analysis of variance (ANOVA) or a repeated measures ANOVA with sex, genotype, nicotine dose, or nicotine concentration as possible independent variables. α < 0.05 was considered significant.
3 RESULTS
3.1 Functional Study
3.1.1 Luciferase assays
Luciferase-based gene expression studies revealed that rs6474413, upstream of CHRNB3, influences gene expression (Figure 1). To determine the impact of specific genetic variants upstream of CHRNB3, eight constructs were examined in pairwise combinations based on the SNP being assessed. These constructs were tested in three cell lines. In BE(2)-C cells we observed a significant main effect of construct (F7,272 = 9.7, p < 0.001). There was a significant difference between the major (ATA) and minor (GCG) parent haplotypes (T66 = 2.6, p < 0.05), replicating our prior work (Ehringer et al, 2010) that the major haplotype leads to increased gene expression compared to the minor haplotype. The significant difference in expression observed between the parent constructs was retained when the SNPs at rs13277254 (T66 = 2.5, p < 0.05) or rs4950 (T66 = 7.0, p < 0.001) were reversed. In contrast, exchanging the SNPs for rs6474413 eliminated expression differences between constructs. These data provide evidence that rs6474413 is involved in modulating gene expression since altering the genotype at rs13277254 and rs4950 had no effect on expression compared to the parent haplotypes. In contrast, when the genotype of rs6474413 was altered it eliminated the expression difference observed between the parent major and minor haplotypes.
Similar results were obtained with HEK293T and SH-SY5Y cell lines. In the SH-SY5Y line there was a significant main effect of construct overall (F7,156 = 22.8, p < 0.001). Similar to the results in the BE(2)-C cell line there was a significant difference between the major and minor parent haplotypes (T38 = 7.0, p < 0.001), with the major (ATA) haplotype having more gene expression than the minor (GCG) haplotype. Expression differences between constructs were retained when we examined the rs13277254 genotype (T38 = 7.4, p < 0.001) and the rs4950 genotype (T34 = 8.9, p < 0.001), but not rs6474413, providing evidence that this allele influences gene expression.
In the HEK293T cell type we again saw a significant main effect when all constructs were examined (F7,168 = 3.1, p < 0.01). Similar to our previous work we showed that the major (ATA) haplotype was associated with higher levels of gene expression than the minor (GCG) haplotype (T40 = 2.7, p < 0.05). When the three SNPs in this region were examined, the significant main effect of construct was only retained with rs4950 (T40 = 3.7, p < 0.05). Thus in the HEK293T cell line both rs13277254 and rs6474413 were implicated in influencing gene expression.
3.1.2 Electrophoretic mobility shift assay (EMSA) study
Since the T allele at rs6474413 was shown to express luciferase significantly more than the C allele, we hypothesized that the allele present at this SNP would alter the binding affinity of a transcription factor. To explore this hypothesis, double-stranded, biotin-labeled DNA oligonucleotides containing the alternate SNP alleles of rs6474413 were synthesized and the binding of these oligonucleotides to nuclear extracts of BE(2)-C and SH-SY5Y cells were examined. Figure 2 shows a representative gel demonstrating the retardation of probe migration by nuclear proteins (lane 2, C allele, lane 6, T allele). There were two bands observed that bound to the probes, and could be out-competed by the inclusion of excess unlabeled probe (lanes 3–4 and 5–6, bands designated by “→”). Quantitation of the band densities indicated that the T allele had greater binding compared to the C allele when the lower band was examined (labeled band B in Figure 2). These results were replicated 5 times in extracts derived from BE(2)-C cells and 4 times in nuclear extracts from SH-SY5Y cells (data not shown). The binding to the top band (labeled A) was not consistent across experiments with the C allele binding more in some experiments and the T allele binding more in others.
Fig. 2.
There is increased nuclear protein binding when the T allele is present at rs6474413 compared to the C allele. Arrows indicate bands that show specific binding. Band A did not show consistent binding difference between the C and T allele across experiments. Band B, for which the quantitation is shown on the right, consistently displayed higher binding with the T allele compared to the C allele. This representative image of data was replicated 5 times in nuclear extracts derived from BE(2)-C cells and replicated 4 times in extracts obtained from SH-SY5Y cells.
3.1.3 Bioinformatics analysis
To examine transcription factors that are predicted to bind to the region around rs6474413 we used four transcription factor predictor programs. We evaluated the same 52 base pair sequence with the rs6474413 allele at position 27 using each program. There were many transcription factors predicted to bind at the rs6474413 locus when both the C and T alleles were present. When TFSearch was used Elk-1 was predicted to bind when the T allele was present, but not the C allele. With TESS both TCF-2α and GABP were predicted to bind when the T allele was present, but not the C allele. Finally when Mapper was used ELK4 and SPI-B were predicted to bind when T was present at rs6474413, but not the C allele. Although the prediction programs indicated that rs6474413 altered the binding of transcription factors, no single transcription factor was identified in more than one program, thus none received priority for additional consideration. In all cases, the T allele, which leads to higher gene expression, is predicted to bind a transcription factor while the lower-expressing C allele is not.
3.2 Behavioral Studies
3.2.1 Locomotor activity
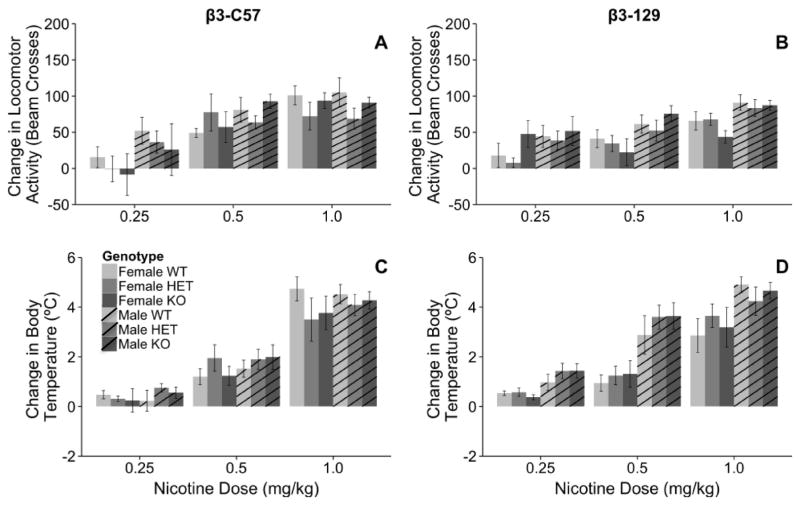
The β3 subunit of the nAChR did not influence nicotine-induced locomotor activity (Fig 3A and B). The genotypic effect of the β3 subunit was independently examined on both the 129 and C57 genetic backgrounds. When the mutation was on the C57 background, there was a significant main effect of sex (F1, 108=4.01, p < 0.05) and dose (F2, 108=21.0, p < 0.001), but no significant effects or interactions with genotype were observed. As expected, nicotine resulted in a dose-dependent reduction of locomotor activity as indicated by a larger change in locomotor activity (Fig 3A, all p < 0.05). Moreover, male mice were more sensitive to nicotine-induced locomotor depression compared to female mice (67.3 ± 6.0, 51.9 ± 8.0, respectively).
Fig. 3.
The Chrnb3 gene does not modulate nicotine-induced changes in locomotor activity or body temperature in either the β3-C57 or β3-129 mice. Data (mean ± SEM) represent nicotine-induced locomotor activity of (A) β3-C57 and (B) β3-129 Chrnb3 mice and nicotine-induced hypothermia of (C) β3-C57 and (D) β3-129 Chrnb3 mice. Bars represent nicotine response minus saline response, thus larger bars indicate greater locomotor sedation and hypothermia. N = 3–9/group of β3-C57 mice and N = 7–12/group of β3-129 mice.
Similar results were observed in the β3-129 animals, where the β3 genotype did not influence nicotine-induced locomotor activity (Fig 3B). A significant main effect of sex (F1, 165=18.5, p < 0.001) and dose (F2, 165=13.5, p < 0.001) were observed in the β3-129 animals. Male mice showed greater nicotine-induced locomotor depression than female animals (64.5 ± 4.7, 52.2 ± 4.5, respectively). Furthermore, locomotor depression occurred in a dose dependent manner (Figure 3B). There were no significant main effects or interactions with genotype.
3.2.2 Body temperature
The β3 subunit of the nAChR did not influence nicotine-induced hypothermia (Fig 3C and D). When mice on the C57 genetic background were examined there was a significant main effect of sex on body temperature (F2, 108=91.1, p < 0.001), such that males showed a greater hypothermic response than females (2.3 ± 0.2, 2.0 ± 0.3, respectively). No other significant main effects or interactions were observed.
When the β3 null mutation was examined on the 129 genetic background a significant sex X nicotine dose interaction (F2, 165=3.5, p < 0.05; Figure 3D) was observed. In female β3-129 mice, the 1 mg/kg dose of nicotine resulted in a greater hypothermic response compared to both the 0.25 and 0.5 mg/kg nicotine doses (0.25 mg/kg: 0.5 ± 0.1, 0.5 mg/kg: 1.2 ± 0.2, 0.5 mg/kg: 3.3 ± 0.4, p’s < 0.001). In male mice this response was also dose dependent, but all doses were significantly different from each other (0.25 mg/kg: 1.3 ± 0.2, 0.5 mg/kg: 3.9 ± 0.3, 0.5 mg/kg: 4.6 ± 0.3, p’s < 0.005). Similar to the β3-C57 mice, there were no significant effects of genotype on the hypothermic response.
3.2.3 Nicotine consumption
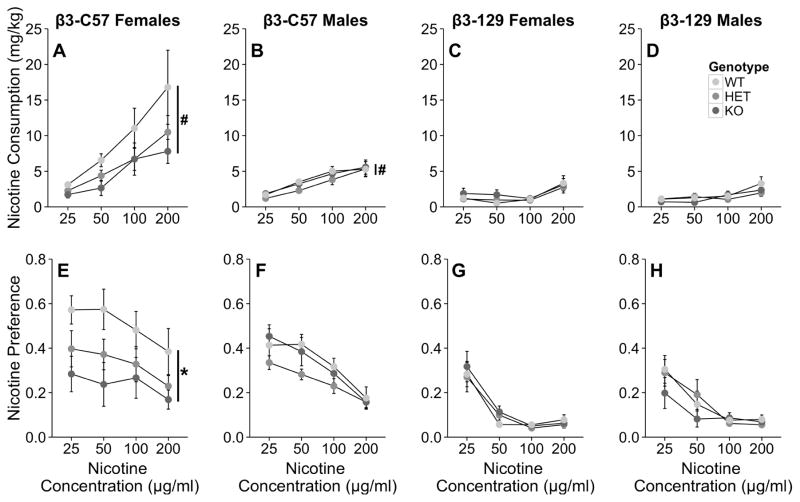
Deletion of the β3 subunit of the nAChR influenced nicotine consumption when the mutation was on the C57 background, but not on the 129 genetic background (Fig 4). In β3-C57 animals, there was a significant main effect of genotype on nicotine consumption (F2, 52=3.2, p < 0.05), such that heterozygous and knockout mice consumed significantly less nicotine than did wildtype mice (WT: 6.4 ± 0.9 mg/kg, HET 4.3 ± 0.4 mg/kg, KO 4.2 ± 0.4 mg/kg; all p < 0.05). Genotype did not interact with any other factors. Investigation of a significant concentration X sex interaction (F3, 156=8.0, p < 0.001) provided evidence that both male and female mice consume more nicotine with the availability of increasing nicotine concentrations, but female mice increase their consumption to a greater extent (female 25 μg/ml: 2.3 ± 0.3, 50 μg/ml: 4.4 ± 0.6, 100 μg/ml: 7.9 ± 1.3, 200 μg/ml: 11.3 ± 1.9, versus male 25 μg/ml: 1.6 ± 0.1, 50 μg/ml: 2.9 ± 0.2, 100 μg/ml: 4.4 ± 0.4, 200 μg/ml: 5.4 ± 0.6). Nicotine preference was analyzed within each sex, due to a significant sex X genotype interaction (F2, 52=3.4, p < 0.05). In female mice, there was a significant main effect of concentration (F3, 66=5.0, p < 0.05). Moreover, β3 knockout mice preferred less nicotine than did wildtype animals as evidenced by a significant main effect of genotype (F2, 22=4.0, p < 0.05). β3 heterozygous mice also tended (p = 0.08) to prefer less nicotine compared to wildtype animals. Overall, female mice preferred significantly more nicotine when the 25 and 50 μg/ml concentrations of nicotine were available compared to the 100 and 200 μg/ml concentrations (25 μg/ml: 0.41 ± 0.05, 50 μg/ml: 0.38 ± 0.06, 100 μg/ml: 0.35 ± 0.05, 200 μg/ml: 0.25 ± 0.04, versus, all p <0.05). In male mice, a significant main effect of concentration was observed (F3, 90=36.7, p < 0.001). Male mice preference for nicotine was similar when the 25 and 50 ug/ml nicotine concentrations were available, but after that preference decreased in a concentration-dependent manner (25 μg/ml: 0.40 ± 0.03, 50μg/ml: 0.35 ± 0.03, 100 μg/ml: 0.27 ± 0.02, 200 μg/ml: 0.16 ± 0.02, all p < 0.01). Females consumed significantly more fluid than did male mice (F1, 52=5.1, p < 0.05) and fluid consumption varied slightly across nicotine concentrations (F3, 156=5.0, p < 0.01).
Fig. 4.
The Chrnb3 gene influences nicotine consumption in β3-C57, but not β3-129 mice. Data (mean ± SEM) represent average 24 hour nicotine consumption of (A) β3-C57 female, (B) β3-C57 male, (C) β3-129 female, and (D) β3-129 male Chrnb3 mice and preference for nicotine of (E) β3-C57 female, (F) β3-C57 male, (G) β3-129 female, and (H) β3-129 male Chrnb3 mice. * p <0.05 depicting a significant main effect of strain nicotine preference. # denotes the significant main effect of strain (p <0.05) observed when both male and female data are combined as there were no main effect or interactions with sex on nicotine consumption. N = 7–13/group of β3-C57 mice and N = 6–8/group of β3-129 mice.
When the influence of the β3 gene on nicotine consumption was examined on the 129 background, genotype did not influence nicotine consumption (Fig 4C and 4D). When nicotine consumption (mg/kg), nicotine preference, and total fluid consumed were examined in all cases a significant main effect of nicotine concentration was observed (F3,108=15.9, p < 0.001, F3,108=50.0, p < 0.001, F3,108=28.2, p < 0.001, respectively). Mice consumed significantly more nicotine when the 200 ug/ml concentration of nicotine was available compared to all other concentrations (25 μg/ml: 1.2 ± 0.2, 50 μg/ml: 1.1 ± 0.2, 100 μg/ml: 1.2 ± 0.1, 200 μg/ml: 2.8 ± 0.3, all p < 0.005) and preference for the nicotine solution decreased in a dose dependent manner to 100 μg/ml, at which it leveled off (25 μg/ml: 0.28 ± 0.03, 50 μg/ml: 0.12 ± 0.02, 100 μg/ml: 0.06 ± 0.01, 200 μg/ml: 0.07 ± 0.01, all p < 0.05). Total fluid consumption was significantly lower when the first concentration of nicotine (25 μg/ml) was presented compared to all other nicotine concentrations (25 μg/ml: 4.2 ± 0.3, 50 μg/ml: 4.7 ± 0.3, 100 μg/ml: 4.8 ± 0.3, 200 μg/ml: −4.7 ± 0.2, all p < 001).
3.2.4 Tastant consumption
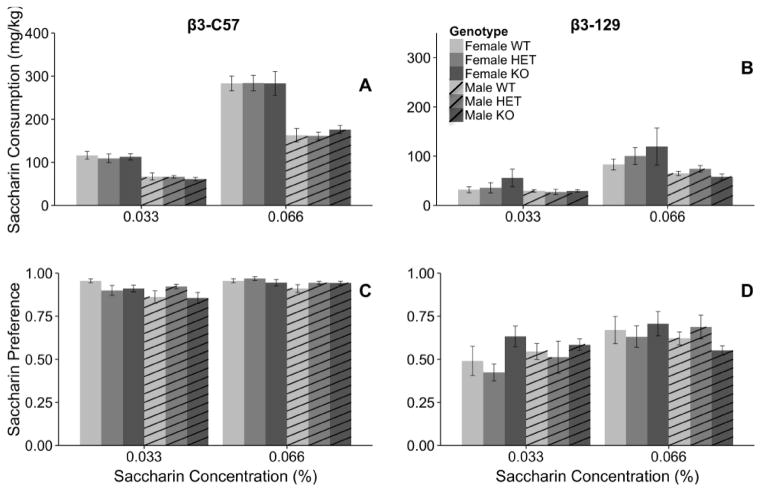
To determine if genotypic differences in nicotine consumption could be due to taste sensitivity we examined the consumption of a sweet (saccharin) and bitter (quinine) tastant. The β3 subunit of the nAChR did not modulate saccharin consumption (Fig 5). Male and female β3-C57 mice were analyzed for saccharin consumption independently because of a significant sex X concentration interaction (F1,52=53.4, p < 0.001). Female β3-C57 mice consumed significantly more saccharin when 0.066% saccharin was available compared to 0.033% saccharin (F1, 22=349.7, p < 0.001; 112.7 ± 5.2 vs. 283.4 ± 12.9, respectively). This pattern was similar in male mice with saccharin consumption being greater when the higher concentration of saccharin was available (F1, 30=534.5, p < 0.001; 0.033% saccharin: 64.9 ± 2.8, 0.066% saccharin 166.8 ± 6.4, respectively). When saccharin preference was examined there was a significant main effect of sex (F1, 52=7.3, p < 0.01) and concentration (F1, 12.7=3.6, p < 0.01). Female mice preferred significantly more saccharin than did male mice (0.94 ± 0.01, 0.91 ± 0.01, respectively). Additionally, mice across both sexes preferred significantly more saccharin when the higher concentration was available compared to the lower concentration (0.033%: 0.90 ± 0.01, 0.066%: 0.94 ± 0.01). Evaluation of total fluid consumption in β3-C57 mice revealed a significant main effect of concentration (F1, 52 =120.5, p < 0.001) and sex (F1, 52=25.9, p < 0.001). β3-C57 mice consumed significantly more total fluid when the 0.066% concentration of saccharin was available compared to the 0.033% solution (7.3 ± 0.8 vs. 8.9 ± 0.2, respectively). Furthermore, female mice consumed more fluid compared to male mice (8.3 ± 0.2, 6.4 ± 0.1, respectively), but neither of these variables interacted with genotype.
Fig. 5.
The Chrnb3 gene does not modulate saccharin consumption in either the β3-C57 or β3-129 mice. Data (mean ± SEM) represent average 24 hour saccharin consumption of (A) β3-C57 and (B) β3-129 Chrnb3 mice and saccharin preference of (C) β3-C57 and (D) β3-129 Chrnb3 mice. N = 7–13/group of β3-C57 mice and N = 6–8/group of β3-129 mice.
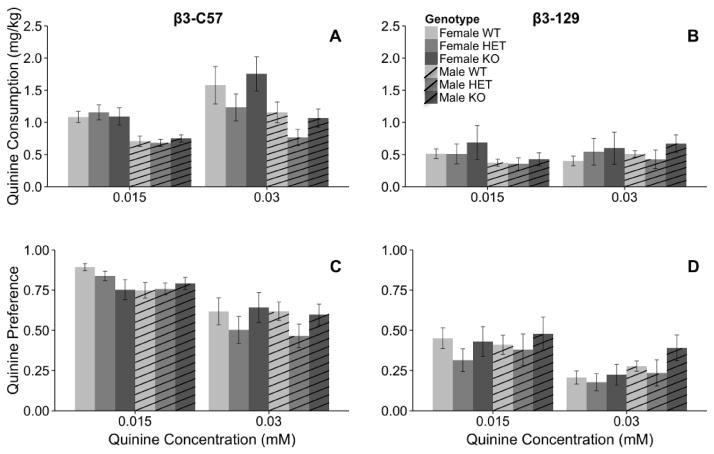
Presence or absence of the β3 subunit of the nAChR was not associated with consumption of the bitter tastant quinine when examined in β3-C57 animals (Fig 6). Female mice consumed significantly more quinine than did male mice as evidenced by a significant main effect of sex (F1, 52=20.0, p < 0.001; 1.3 ± 0.09, 0.8 ± 0.05, respectively). Although a significant concentration X genotype interaction (F2, 52=3.9, p < 0.05) was detected, when consumption was examined at each quinine concentration independently no significant main effects of genotype were observed. Similar statistical results were also observed when quinine preference was examined. A significant concentration X genotype interaction was detected (F2, 52=4.0, p < 0.05), but follow-up analyses within each concentration provided no statistical evidence of a difference among the genotypes. Total fluid consumption was influenced by sex (F2, 52=5.7, p < 0.05) and concentration (F1, 52=5.1, p < 0.05). Female mice drank significantly more fluid compared to male mice (0.015 mM: 5.0 ± 0.2, 0.03 mM: 4.4 ± 0.1). Furthermore mice consumed more fluid when the lower concentration of quinine was available (0.015 mM: 4.8 ± 0.1, 0.03 mM: 4.6 ± 0.1).
Fig. 6.
The Chrnb3 gene does not modulate quinine consumption in either the β3-C57 or β3-129 mice. Data (mean ± SEM) represent quinine consumption of (A) β3-C57 and (B) β3-129 Chrnb3 mice and quinine preference of (C) β3-C57 and (D) β3-129 Chrnb3 mice. N = 7–13/group of β3-C57 mice and N = 6–8/group of β3-129 mice.
There was no effect of the β3 gene on either saccharin or quinine consumption in β3-129 animals (Fig 5). There was a significant main effect of sex on saccharin consumption (F1,36=4.6, p < 0.05) and this interacted with concentration (F1,36=6.3, p < 0.05), thus males and females were examined independently. Female mice consumed significantly more saccharin when the 0.066% concentration of saccharin was presented compared to when the 0.033% concentration was presented (F1,19=56.3, p < 0.001; 41.1 ± 7.1 vs. 101.0 ± 14.0, respectively). Male mice also consumed more saccharin when the high concentration of saccharin was presented compared to the low concentration (F1,17=107.1, p < 0.001; 28.7 ± 2.2, 65.8 ± 3.3, respectively). When saccharin preference was examined there was significant main effect of concentration (F1,36=14.9, p < 0.001), but no other significant effects or interactions. Mice preferred more saccharin when the 0.066% concentration was available compared to the 0.033% concentration (0.53 ± 0.03, 0.65 ± 0.03, respectively). There were no significant effects on total fluid consumption.
When consumption of the bitter tastant quinine was examined in the β3-129 animals, there was no significant effect of genotype on consumption (Fig 6). There was a significant sex X concentration interaction observed for quinine consumption (F1,36=10.6, p < 0.005), thus we examined each sex independently. In female β3-129 animals, there were no significant main effects or interactions observed. In male mice, there was a significant main effect of quinine concentration (F1,17=13.9, p < 0.005), such that mice drank significant more quinine when the 0.03 mM quinine was available compared to 0.015 mM quinine (0.39 ± 0.05, 0.54 ± 0.07), respectively. When quinine preference was examined a significant main effect of concentration (F1,36=55.4, p < 0.001) indicated that preference for quinine decreased when the 0.03 mM quinine was available compared to 0.015 mM quinine (0.41 ± 0.03, 0.25 ± 0.03, respectively). There were no significant main effects or interactions when total fluid consumption was examined.
4. DISCUSSION
This study sought to examine the influence of three CHRNB3 SNPs associated with nicotine traits on gene expression. Further, we hoped to characterize the importance of β3 gene expression on nicotine behaviors. We found that rs6474413, a SNP upstream of CHRNB3, influenced the expression of a reporter gene using in vitro assays. These findings provide a possible mechanism by which genetic variants upstream of CHRNB3, that are associated with nicotine phenotypes, may influence nicotine behaviors. Additionally, we were able to use β3 null mutant animals as a proxy for how differences in the expression of this gene may influence nicotine behaviors in vivo. Mice lacking one or more copies of the gene coding for the β3 subunit consumed significantly less nicotine than wildtype animals when the null mutation was present on the C57 background, but no significant effects were observed when the mutation was on 129 genetic background. Together these data provide evidence of the importance of the β3 subunit in nicotine behaviors and provide a possible mechanism through which this occurs. Moreover, these results highlight the importance that genetic background can play in the overall complexity of drug behaviors.
Nicotine binds to nAChRs to exert its effects. Human genetic studies have provided support that CHRNB3 is associated with nicotine behaviors (Bierut et al, 2007; Chen et al, 2012; Culverhouse et al, 2014; Ehringer et al, 2010; Rice et al, 2012; Saccone et al, 2007; Thorgeirsson et al, 2010; Zeiger et al, 2008). Critical for the interpretation of the current study, the minor allele, C, at rs6474413 was shown to be protective against nicotine dependence (Rice et al, 2012).
This study provides insight into how genetic variation upstream of CHRNB3 may influence nicotine behaviors. We tested three SNPs located upstream of CHRNB3. The SNPs were selected because of prior association with nicotine traits (Bierut et al, 2007; Saccone et al, 2007; Zeiger et al, 2008) as well as bioinformatics analysis indicating they were predicted to change the binding of a transcription factor. Importantly, these three variants are all in high linkage disequilibrium (LD; r2 = 0.957 https://www.broadinstitute.org/mpg/snap) with each other, making it difficult to isolate a putative functional variant using human genetic studies. To determine the impact of any individual SNP tested one must compare the results to those of the major/minor parent haplotypes. When the allele at rs6474413 was changed, the gene expression differences observed between the parent haplotypes was eliminated in all three cell types tested. These data suggest that rs6474413 may be a causal variant.
In contrast to rs6474413, neither rs13277254 nor rs4950 consistently influenced gene expression in this assay. In HEK293T cells, the reversal of the allele at rs13277254 eliminated the gene expression differences observed in the parent constructs, but this finding was not observed in BE(2)-C or SH-SY5Y cells. There are likely different combinations of transcription factors driving gene expression between these cell lines, a finding that is consistent with studies done on genetic variation in the CHRNA5/CHRNA3/CHRNB4 gene region (Flora et al, 2013).
The genetic variant rs6474413 appears of be a good candidate for regulation of gene expression, and potentially nicotine’s behavioral effects, but the protein(s) that interact with this genomic region to cause these effects is/are yet to be determined. Given that this SNP is located upstream of CHRNB3 and influences gene expression, we hypothesized that the allele present at this genetic location would influence the binding of a transcription factor. While bioinformatics analysis suggested that the allele present at rs6474413 is predicted to change the binding of transcription factors (ELK-1, TCF-2α, GABP, ELK4 and SPI-B), no consistent transcription factor emerged across the different prediction programs. Proteomics approaches may prove useful for identification of the protein(s) that interact at this location.
Although no specific transcription factor was identified in these experiments, it is interesting to note that across prediction software programs having the C (minor) allele present at rs6474413, which confers protection against nicotine dependence (Rice et al, 2012), always disrupted the binding of a transcription factor that bound when the T allele was present. In our luciferase assay the C allele was associated with reduced gene expression. Additionally, in the EMSA assay the T allele appears to bind more strongly to nuclear proteins. Taken together, these data suggest that an activating transcription factor most likely binds the site including rs6474413.
An important caveat to the functional experiments is that they represent an in vitro model and examined the expression of a reporter gene, not the β3 subunit. Further work is needed to determine if rs6474413 alters gene expression in vivo. Allele-specific gene expression has been used to examine the relative importance of SNPs on gene expression in vivo for other CHRN genes (Smith et al, 2011; Wang et al, 2013). Future studies investigating rs6474413 and CHRNB3 mRNA expression in human brain tissues are merited. Moreover, we know changes in mRNA levels do not always represent a change in protein level (Huang and Winzer-Serhan, 2006; Marks et al, 1992; Pauly et al, 1996), thus future work also needs to address protein levels.
Given that rs6474413 was the only variant to consistently alter reporter gene expression patterns, we chose to focus on this SNP. Although rs6474413 appears to modulate gene expression, the data provided here suggest it is not the only genetic variant in this region to influence gene expression. If rs6474413 were the only SNP to modify expression in this region one would expect to see a complete reversal of the expression pattern compared to the parent haplotype constructs. Instead, an incomplete reversal was observed in all three cell types, suggesting there are other genetic factors in this region that also modulate gene expression. Examination of the human genetic association signals indicates that there are SNPs in two LD blocks in CHRNB3 that contribute to nicotine phenotypes. The first linkage block is represented by the SNPs examined in this study and has been associated with nicotine dependence, number of cigarettes smoked per day, and initial sensitivity to nicotine (Ehringer et al, 2010; Saccone et al, 2009; Thorgeirsson et al, 2010). There are at least 17 SNPs in high LD (https://www.broadinstitute.org/mpg/snap), and it is possible that other SNPs in this block not examined may also be important. Within the second LD block of SNPs associated with nicotine traits in CHRNB3, many SNPs result in synonymous changes in the coding region (Saccone et al, 2007; Zeiger et al, 2008). We cannot rule out the possibility that these other genetic variants may influence the expression differences we observe herein.
In this study, we used β3 null mutant mice as a model of reduced gene expression to test the effect of expression of this subunit on nicotine behavioral responses. Mice lacking the β3 subunit on the C57 background showed reduced nicotine consumption compared to that of their wildtype littermates, but did not differ in consumption of the non-caloric tastants saccharin or quinine. These data provide support that β3-containing nAChRs are important for nicotine behaviors in vivo. Nicotinic receptors containing this subunit have been implicated in dopamine release from striatal nerve terminals (Grady et al, 2007) providing a potential biological mechanism that may be driving this behavior. No differences in behavioral response to an acute nicotine injection (locomotor and hypothermia) were observed.
To our knowledge, this is the first evidence in animal models that the β3 subunit is involved in nicotine consumption. The effect on nicotine consumption was only observed when the mutation was on the C57 background. β3-129 mice consumed very little nicotine at the concentrations tested (Fig 4C and 4D). It is known that there are strain differences in nicotine consumption (Robinson et al, 1996), and testing these mice with lower concentrations of nicotine may be warranted. A study with lower nicotine concentrations may differentiate if the lack of effect is due to a floor effect or because the β3 subunit does not influence nicotine intake when harbored on a 129 genetic background. If the latter was true, this would provide evidence of an epistatic interaction that should be explored further. An alternative hypothesis is that natural genetic variation in the Chrnb3 promoter between B6 and 129 may lead to changes in β3 expression similar to what we observed with the luciferase assays. Further, this variation may contribute to the baseline differences in nicotine consumption observed between these strains. Unfortunately, the 129 strain (129S6/SvEvTac) to which the β3 null mutation has been crossed has not been sequenced. Examination of known mouse polymorphisms in the region upstream (within 10 kb) of Chrnb3 between the 129S6 strain and B6 (Mouse Phenome Database; http://phenome.jax.org/) does not provide support for this hypothesis at this time.
The reduction in nicotine consumption was accompanied by a significant reduction in nicotine preference in both heterozygous and knockout animals when the full sample was analyzed. Preference data were analyzed further within each sex due to a significant sex X genotype interaction. When analyzed by sex this effect on preference appeared to be driven by the females. Within female mice β3 knockout animals displayed reduced nicotine preference compared to wildtype animals. Heterozygous animals also tended (p = 0.08) to show a reduced preference (Fig 4E). In male mice there was no significant effect of strain on nicotine preference (Fig 4F). The lack of significant results when the sample is split by sex of the animal is likely due to reduced statistical power. The sample size within each sex was only 7–13 animals per group. Our findings are consistent with prior work demonstrating that female C57 female mice consume and prefer significantly more nicotine than male mice (Klein et al, 2004). Moreover, female mice metabolism nicotine faster than male mice (Hatchell and Collins, 1980; Siu et al, 2006) and this difference in metabolism could be driving the observed sex-dependent effects.
Consistent with nicotine consumption paradigms in mice (Klein et al, 2004; Robinson et al, 1996), we did not show an overt preference for nicotine. In most cases the mice avoided nicotine as defined by having a less than 50% preference for the nicotine bottle (Fig 4A–D). Future research should examine whether the β3 subunit influences nicotine reward as measured by conditioned place preference.
As previously mentioned the gene that codes for the β3 subunit is located adjacent to the gene coding the α6 subunit on human chromosome 8. Not only do these genes physically map next to each other, these subunits co-assemble to form functional receptors in reward relevant brain regions (Cui et al, 2003; Gotti et al, 2005). Work with genetically modified animals and pharmacologic tools have supported a role for α6* nicotinic acetylcholine receptors in nicotine conditioned place preference (Jackson et al, 2009; Sanjakdar et al, 2015) and self-administration (Pons et al, 2008). While β3 heterozygous and homozygous null mutant mice show reduced Chrnb3 mRNA compared to wildtype animals in striatum and the ventral tegmental area, α6 mRNA levels were unchanged in these brain regions (Cui et al, 2003). Despite no apparent change in α6 mRNA in β3 null mutant mice, these animals have reduced α6-containing nAChR receptors and reduced α6 receptor-mediated striatal dopamine release (Salminen et al, 2004). However, β3 heterozygous animals have no changes in α6 receptors (Gotti et al, 2005) or α6 receptor function in the striatum (Salminen et al, 2004). Given that we observe differences in consumption in β3 heterozygous animals these data provide further support that these findings are a result of the null mutation in β3 and not due to changes in the α6 subunit. Furthermore, it is possible that the effect of β3 deletion on nicotine intake is the result of disruption β3-containing nAChR such as the α3β4β3 nAChR expressed in the habenular-interpeduncular nucleus pathway (Grady et al., 2009).
The behavioral results in β3 null mutant mice are interesting to consider in the context of the functional luciferase results. Taken together these data lead to the hypothesis that subjects having the rs6474413 C (minor) allele may have reduced β3 gene expression, leading to reduced nicotine intake and/or dependence. In fact, this is what is observed in the literature. Examination of associations with rs6474413, or a proxy SNP in high LD, reveal that the minor C allele is protective against nicotine dependence (Chen et al, 2012; Culverhouse et al, 2014; Rice et al, 2012). Moreover, in a large meta-analysis that examined associations with the number of cigarettes smoked per day, the major T allele conferred risk (Thorgeirsson et al, 2010). Thus, altering expression of the β3 subunit may be how rs6474413 influences nicotine behaviors.
The conclusions made by comparing the functional gene expression results and the behavior observed in β3 null mutant animals is not without limitations. The functional work examined the influence of the human gene sequence and found only decreases on gene expression, not total lack of expression. Behavioral experiments in β3 null mutant mice examine the importance of the mouse β3 gene and how complete elimination of the gene influences behavior. Heterozygous mice having only one functional β3 gene may provide a better model of the reduced level of gene expression we might expect to see in human populations. Our minor construct had a 14–34% reduction in reporter gene expression compared to the major construct (Figure 1). Interestingly, compared to wildtype mice β3 heterozygous animals have an approximate 17–34% reduction in Chrnb3 mRNA (as measured by in situ hybridization) in the ventral tegmental area, substantia nigra, and medial habenula (Cui et al, 2003). In the current study, β3 heterozygous mice showed decrease nicotine consumption compared to wildtype animals. While speculative, these data provide suggestive evidence that this reduction in β3 expression may be relevant to nicotine behaviors in vivo.
Human genetic association studies have repeatedly implicated genetic variation in CHRNB3 in nicotine behaviors. This study provides data on one potential biological mechanism by which these genetic variants may influence biology and thus behavior.
HIGHLIGHTS.
rs6474413 upstream of CHRNB3 influences reporter gene expression
Chrnb3 influences nicotine consumption and preference
Chrnb3 does not influence acute locomotor activity or body temperature
Acknowledgments
These studies were performed with support from AA019447 (HMK), AA017889 (MAE), DA015663 (Michael Marks).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arias HR, Bhumireddy P, Bouzat C. Molecular mechanisms and binding site locations for noncompetitive antagonists of nicotinic acetylcholine receptors. Int J Biochem Cell Biol. 2006;38(8):1254–1276. doi: 10.1016/j.biocel.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Human molecular genetics. 2007;16(1):24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, Xuei X, et al. Variants in nicotinic receptors and risk for nicotine dependence. The American journal of psychiatry. 2008;165(9):1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booker TK, Butt CM, Wehner JM, Heinemann SF, Collins AC. Decreased anxiety-like behavior in beta3 nicotinic receptor subunit knockout mice. Pharmacol Biochem Behav. 2007;87(1):146–157. doi: 10.1016/j.pbb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Butt CM, King NM, Hutton SR, Collins AC, Stitzel JA. Modulation of nicotine but not ethanol preference by the mouse Chrna4 A529T polymorphism. Behav Neurosci. 2005;119(1):26–37. doi: 10.1037/0735-7044.119.1.26. [DOI] [PubMed] [Google Scholar]
- Chen LS, Baker TB, Grucza R, Wang JC, Johnson EO, Breslau N, et al. Dissection of the phenotypic and genotypic associations with nicotinic dependence. Nicotine & tobacco research : official journal of the Society for Research on Nicotine and Tobacco. 2012;14(4):425–433. doi: 10.1093/ntr/ntr231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C, Booker TK, Allen RS, Grady SR, Whiteaker P, Marks MJ, et al. The beta3 nicotinic receptor subunit: a component of alpha-conotoxin MII-binding nicotinic acetylcholine receptors that modulate dopamine release and related behaviors. J Neurosci. 2003;23(35):11045–11053. doi: 10.1523/JNEUROSCI.23-35-11045.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culverhouse RC, Johnson EO, Breslau N, Hatsukami DK, Sadler B, Brooks AI, et al. Multiple distinct CHRNB3-CHRNA6 variants are genetic risk factors for nicotine dependence in African Americans and European Americans. Addiction. 2014 doi: 10.1111/add.12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez MH, Ayoub AE, Rakic P. POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cerebral cortex. 2013;23(11):2632–2643. doi: 10.1093/cercor/bhs252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehringer MA, McQueen MB, Hoft NR, Saccone NL, Stitzel JA, Wang JC, et al. Association of CHRN genes with “dizziness” to tobacco. Am J Med Genet B Neuropsychiatr Genet. 2010;153B(2):600–609. doi: 10.1002/ajmg.b.31027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flora AV, Zambrano CA, Gallego X, Miyamoto JH, Johnson KA, Cowan KA, et al. Functional characterization of SNPs in CHRNA3/B4 intergenic region associated with drug behaviors. Brain Res. 2013;1529:1–15. doi: 10.1016/j.brainres.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego X, Cox RJ, Laughlin JR, Stitzel JA, Ehringer MA. Alternative CHRNB4 3′-UTRs mediate the allelic effects of SNP rs1948 on gene expression. PloS one. 2013;8(5):e63699. doi: 10.1371/journal.pone.0063699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotti C, Clementi F, Fornari A, Gaimarri A, Guiducci S, Manfredi I, et al. Structural and functional diversity of native brain neuronal nicotinic receptors. Biochemical pharmacology. 2009;78(7):703–711. doi: 10.1016/j.bcp.2009.05.024. [DOI] [PubMed] [Google Scholar]
- Gotti C, Moretti M, Clementi F, Riganti L, McIntosh JM, Collins AC, et al. Expression of nigrostriatal alpha 6-containing nicotinic acetylcholine receptors is selectively reduced, but not eliminated, by beta 3 subunit gene deletion. Molecular pharmacology. 2005;67(6):2007–2015. doi: 10.1124/mol.105.011940. [DOI] [PubMed] [Google Scholar]
- Grady SR, Moretti M, Zoli M, Marks MJ, Zanardi A, Pucci L, et al. Rodent habenulo-interpeduncular pathway expresses a large variety of uncommon nAChR subtypes, but only the alpha3beta4* and alpha3beta3beta4* subtypes mediate acetylcholine release. J Neurosci. 2009;29(7):2272–2282. doi: 10.1523/JNEUROSCI.5121-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady SR, Salminen O, Laverty DC, Whiteaker P, McIntosh JM, Collins AC, et al. The subtypes of nicotinic acetylcholine receptors on dopaminergic terminals of mouse striatum. Biochem Pharmacol. 2007;74(8):1235–1246. doi: 10.1016/j.bcp.2007.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatchell PC, Collins AC. The influence of genotype and sex on behavioral sensitivity to nicotine in mice. Psychopharmacology (Berl) 1980;71(1):45–49. doi: 10.1007/BF00433251. [DOI] [PubMed] [Google Scholar]
- Hoft NR, Corley RP, McQueen MB, Schlaepfer IR, Huizinga D, Ehringer MA. Genetic association of the CHRNA6 and CHRNB3 genes with tobacco dependence in a nationally representative sample. Neuropsychopharmacology. 2009;34(3):698–706. doi: 10.1038/npp.2008.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoft NR, Stitzel JA, Hutchison KE, Ehringer MA. CHRNB2 promoter region: association with subjective effects to nicotine and gene expression differences. Genes Brain Behav. 2011;10(2):176–185. doi: 10.1111/j.1601-183X.2010.00650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LZ, Winzer-Serhan UH. Chronic neonatal nicotine upregulates heteromeric nicotinic acetylcholine receptor binding without change in subunit mRNA expression. Brain Res. 2006;1113(1):94–109. doi: 10.1016/j.brainres.2006.06.084. [DOI] [PubMed] [Google Scholar]
- Hutchison KE, Allen DL, Filbey FM, Jepson C, Lerman C, Benowitz NL, et al. CHRNA4 and tobacco dependence: from gene regulation to treatment outcome. Arch Gen Psychiatry. 2007;64(9):1078–1086. doi: 10.1001/archpsyc.64.9.1078. [DOI] [PubMed] [Google Scholar]
- Jackson KJ, McIntosh JM, Brunzell DH, Sanjakdar SS, Damaj MI. The role of alpha6-containing nicotinic acetylcholine receptors in nicotine reward and withdrawal. J Pharmacol Exp Ther. 2009;331(2):547–554. doi: 10.1124/jpet.109.155457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamens HM, Andersen J, Picciotto MR. Modulation of ethanol consumption by genetic and pharmacological manipulation of nicotinic acetylcholine receptors in mice. Psychopharmacology (Berl) 2010 doi: 10.1007/s00213-009-1759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamens HM, Burkhart-Kasch S, McKinnon CS, Li N, Reed C, Phillips TJ. Sensitivity to psychostimulants in mice bred for high and low stimulation to methamphetamine. Genes Brain Behav. 2005;4(2):110–125. doi: 10.1111/j.1601-183X.2004.00101.x. [DOI] [PubMed] [Google Scholar]
- Kamens HM, Burkhart-Kasch S, McKinnon CS, Li N, Reed C, Phillips TJ. Ethanol-related traits in mice selectively bred for differential sensitivity to methamphetamine-induced activation. Behav Neurosci. 2006;120(6):1356–1366. doi: 10.1037/0735-7044.120.6.1356. [DOI] [PubMed] [Google Scholar]
- Klein LC, Stine MM, Vandenbergh DJ, Whetzel CA, Kamens HM. Sex differences in voluntary oral nicotine consumption by adolescent mice: a dose-response experiment. Pharmacol Biochem Behav. 2004;78(1):13–25. doi: 10.1016/j.pbb.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Madden PA, Heath AC. Shared genetic vulnerability in alcohol and cigarette use and dependence. Alcohol Clin Exp Res. 2002;26(12):1919–1921. doi: 10.1097/01.ALC.0000040960.15151.30. [DOI] [PubMed] [Google Scholar]
- Maes HH, Sullivan PF, Bulik CM, Neale MC, Prescott CA, Eaves LJ, et al. A twin study of genetic and environmental influences on tobacco initiation, regular tobacco use and nicotine dependence. Psychol Med. 2004;34(7):1251–1261. doi: 10.1017/s0033291704002405. [DOI] [PubMed] [Google Scholar]
- Marks MJ, Pauly JR, Gross SD, Deneris ES, Hermans-Borgmeyer I, Heinemann SF, et al. Nicotine binding and nicotinic receptor subunit RNA after chronic nicotine treatment. J Neurosci. 1992;12(7):2765–2784. doi: 10.1523/JNEUROSCI.12-07-02765.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks MJ, Romm E, Bealer SM, Collins AC. A test battery for measuring nicotine effects in mice. Pharmacol Biochem Behav. 1985;23(2):325–330. doi: 10.1016/0091-3057(85)90577-5. [DOI] [PubMed] [Google Scholar]
- Marks MJ, Stitzel JA, Collins AC. Genetic influences on nicotine responses. Pharmacol Biochem Behav. 1989;33(3):667–678. doi: 10.1016/0091-3057(89)90406-1. [DOI] [PubMed] [Google Scholar]
- Matta SG, Balfour DJ, Benowitz NL, Boyd RT, Buccafusco JJ, Caggiula AR, et al. Guidelines on nicotine dose selection for in vivo research. Psychopharmacology (Berl) 2007;190(3):269–319. doi: 10.1007/s00213-006-0441-0. [DOI] [PubMed] [Google Scholar]
- McGehee DS. Molecular diversity of neuronal nicotinic acetylcholine receptors. Ann N Y Acad Sci. 1999;868:565–577. doi: 10.1111/j.1749-6632.1999.tb11330.x. [DOI] [PubMed] [Google Scholar]
- Mexal S, Horton WJ, Crouch EL, Maier SI, Wilkinson AL, Marsolek M, et al. Diurnal variation in nicotine sensitivity in mice: role of genetic background and melatonin. Neuropharmacology. 2012;63(6):966–973. doi: 10.1016/j.neuropharm.2012.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar NS, Gotti C. Diversity of vertebrate nicotinic acetylcholine receptors. Neuropharmacology. 2009;56(1):237–246. doi: 10.1016/j.neuropharm.2008.07.041. [DOI] [PubMed] [Google Scholar]
- Morel C, Fattore L, Pons S, Hay YA, Marti F, Lambolez B, et al. Nicotine consumption is regulated by a human polymorphism in dopamine neurons. Molecular psychiatry. 2014;19(8):930–936. doi: 10.1038/mp.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly JR, Marks MJ, Robinson SF, van de Kamp JL, Collins AC. Chronic nicotine and mecamylamine treatment increase brain nicotinic receptor binding without changing alpha 4 or beta 2 mRNA levels. J Pharmacol Exp Ther. 1996;278(1):361–369. [PubMed] [Google Scholar]
- Pons S, Fattore L, Cossu G, Tolu S, Porcu E, McIntosh JM, et al. Crucial role of alpha4 and alpha6 nicotinic acetylcholine receptor subunits from ventral tegmental area in systemic nicotine self-administration. J Neurosci. 2008;28(47):12318–12327. doi: 10.1523/JNEUROSCI.3918-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JP, Hartz SM, Agrawal A, Almasy L, Bennett S, Breslau N, et al. CHRNB3 is more strongly associated with Fagerstrom test for cigarette dependence-based nicotine dependence than cigarettes per day: phenotype definition changes genome-wide association studies results. Addiction. 2012;107(11):2019–2028. doi: 10.1111/j.1360-0443.2012.03922.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SF, Marks MJ, Collins AC. Inbred mouse strains vary in oral self-selection of nicotine. Psychopharmacology (Berl) 1996;124(4):332–339. doi: 10.1007/BF02247438. [DOI] [PubMed] [Google Scholar]
- Saccone NL, Culverhouse RC, Schwantes-An TH, Cannon DS, Chen X, Cichon S, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS genetics. 2010;6(8) doi: 10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone NL, Saccone SF, Hinrichs AL, Stitzel JA, Duan W, Pergadia ML, et al. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(4):453–466. doi: 10.1002/ajmg.b.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Human molecular genetics. 2007;16(1):36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen O, Murphy KL, McIntosh JM, Drago J, Marks MJ, Collins AC, et al. Subunit composition and pharmacology of two classes of striatal presynaptic nicotinic acetylcholine receptors mediating dopamine release in mice. Molecular pharmacology. 2004;65(6):1526–1535. doi: 10.1124/mol.65.6.1526. [DOI] [PubMed] [Google Scholar]
- Sanjakdar SS, Maldoon PP, Marks MJ, Brunzell DH, Maskos U, McIntosh JM, et al. Differential roles of alpha6beta2* and alpha4beta2* neuronal nicotinic receptors in nicotine- and cocaine-conditioned reward in mice. Neuropsychopharmacology. 2015;40(2):350–360. doi: 10.1038/npp.2014.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu EC, Wildenauer DB, Tyndale RF. Nicotine self-administration in mice is associated with rates of nicotine inactivation by CYP2A5. Psychopharmacology (Berl) 2006;184(3–4):401–408. doi: 10.1007/s00213-006-0306-6. [DOI] [PubMed] [Google Scholar]
- Smith RM, Alachkar H, Papp AC, Wang D, Mash DC, Wang JC, et al. Nicotinic alpha5 receptor subunit mRNA expression is associated with distant 5′ upstream polymorphisms. European journal of human genetics : EJHG. 2011;19(1):76–83. doi: 10.1038/ejhg.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens SH, Hartz SM, Hoft NR, Saccone NL, Corley RC, Hewitt JK, et al. Distinct Loci in the CHRNA5/CHRNA3/CHRNB4 Gene Cluster Are Associated With Onset of Regular Smoking. Genet Epidemiol. 2013;37(8):846–859. doi: 10.1002/gepi.21760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tammimaki A, Herder P, Li P, Esch C, Laughlin JR, Akk G, et al. Impact of human D398N single nucleotide polymorphism on intracellular calcium response mediated by alpha3beta4alpha5 nicotinic acetylcholine receptors. Neuropharmacology. 2012;63(6):1002–1011. doi: 10.1016/j.neuropharm.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nature genetics. 2010;42(5):448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True WR, Xian H, Scherrer JF, Madden PA, Bucholz KK, Heath AC, et al. Common genetic vulnerability for nicotine and alcohol dependence in men. Arch Gen Psychiatry. 1999;56(7):655–661. doi: 10.1001/archpsyc.56.7.655. [DOI] [PubMed] [Google Scholar]
- Wang JC, Spiegel N, Bertelsen S, Le N, McKenna N, Budde JP, et al. Cis-regulatory variants affect CHRNA5 mRNA expression in populations of African and European ancestry. PloS one. 2013;8(11):e80204. doi: 10.1371/journal.pone.0080204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilking JA, Hesterberg KG, Crouch EL, Homanics GE, Stitzel JA. Chrna4 A529 knock-in mice exhibit altered nicotine sensitivity. Pharmacogenetics and genomics. 2010;20(2):121–130. doi: 10.1097/FPC.0b013e3283369347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Hu Z, Yu D, Huang L, Jin G, Liang J, et al. Genetic variants on chromosome 15q25 associated with lung cancer risk in Chinese populations. Cancer research. 2009;69(12):5065–5072. doi: 10.1158/0008-5472.CAN-09-0081. [DOI] [PubMed] [Google Scholar]
- Wu J, Lukas RJ. Naturally-expressed nicotinic acetylcholine receptor subtypes. Biochemical pharmacology. 2011;82(8):800–807. doi: 10.1016/j.bcp.2011.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiger JS, Haberstick BC, Schlaepfer I, Collins AC, Corley RP, Crowley TJ, et al. The neuronal nicotinic receptor subunit genes (CHRNA6 and CHRNB3) are associated with subjective responses to tobacco. Human molecular genetics. 2008;17(5):724–734. doi: 10.1093/hmg/ddm344. [DOI] [PubMed] [Google Scholar]