Abstract
The loss of chromosome 9 open reading frame 72 (C9ORF72) expression, associated with C9ORF72 repeat expansions, has not been examined systematically. Three C9ORF72 transcript variants have been described thus far; the GGGGCC repeat is located between two non-coding exons (exon 1a and exon 1b) in the promoter region of transcript variant 2 (NM_018325.4) or in the first intron of variant 1 (NM_145005.6) and variant 3 (NM_001256054.2). We studied C9ORF72 expression in expansion carriers (n = 56) for whom cerebellum and/or frontal cortex was available. Using quantitative real-time PCR and digital molecular barcoding techniques, we assessed total C9ORF72 transcripts, variant 1, variant 2, variant 3, and intron containing transcripts [upstream of the expansion (intron 1a) and downstream of the expansion (intron 1b)]; the latter were correlated with levels of poly(GP) and poly(GA) proteins aberrantly translated from the expansion as measured by immunoassay (n = 50). We detected a decrease in expansion carriers as compared to controls for total C9ORF72 transcripts, variant 1, and variant 2: the strongest association was observed for variant 2 (quantitative real-time PCR cerebellum: median 43 %, p = 1.26e-06, and frontal cortex: median 58 %, p = 1.11e-05; digital molecular barcoding cerebellum: median 31 %, p = 5.23e-10, and frontal cortex: median 53 %, p = 5.07e-10). Importantly, we revealed that variant 1 levels greater than the 25th percentile conferred a survival advantage [digital molecular barcoding cerebellum: hazard ratio (HR) 0.31, p = 0.003, and frontal cortex: HR 0.23, p = 0.0001]. When focusing on intron containing transcripts, analysis of the frontal cortex revealed an increase of potentially truncated transcripts in expansion carriers as compared to controls [digital molecular barcoding frontal cortex (intron 1a): median 272 %, p = 0.003], with the highest levels in patients pathologically diagnosed with frontotemporal lobar degeneration. In the cerebellum, our analysis suggested that transcripts were less likely to be truncated and, excitingly, we discovered that intron containing transcripts were associated with poly(GP) levels [digital molecular barcoding cerebellum (intron 1a): r = 0.33, p = 0.02, and (intron 1b): r = 0.49, p = 0.0004] and poly(GA) levels [digital molecular barcoding cerebellum (intron 1a): r = 0.34, p = 0.02, and (intron 1b): r = 0.38, p = 0.007]. In summary, we report decreased expression of specific C9ORF72 transcripts and provide support for the presence of truncated transcripts as well as pre-mRNAs that may serve as templates for RAN translation. We further show that higher C9ORF72 levels may have beneficial effects, which warrants caution in the development of new therapeutic approaches.
Keywords: C9ORF72, Frontotemporal dementia, Frontotemporal lobar degeneration, Motor neuron disease, Amyotrophic lateral sclerosis, Disease modifier
Introduction
The most frequent genetic cause of frontotemporal lobar degeneration (FTLD) and motor neuron disease (MND) is a hexanucleotide repeat expansion in chromosome 9 open reading frame 72 (C9ORF72) [8, 22], which is pathogenic due to the formation of RNA foci, repeat-associated non-ATG (RAN) translation resulting in dipeptide-repeat proteins (c9RAN proteins), and/or a loss of C9ORF72 expression [2, 8, 21]. It has been shown that C9ORF72 proteins localize to the nuclear membrane in healthy neurons and interact with components of the nuclear pore complex (i.e., Importin β1 and Ran-GTPase), implicating C9ORF72 proteins in nucleocytoplasmic import [29]. The presence of a repeat expansion in C9ORF72 is also thought to compromise nucleocytoplasmic transport [11, 14, 30].
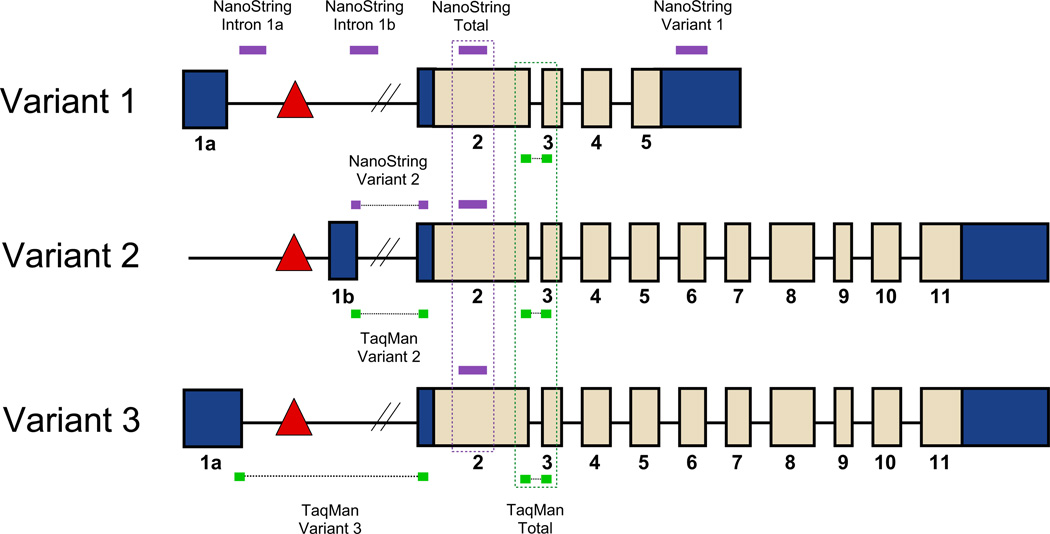
Although models have validated toxic gain-of-function mechanisms [5], they have been unable to confirm or exclude reduced C9ORF72 expression as a pathogenic mechanism [6, 15, 16, 23]. At least three transcripts are generated from the C9ORF72 gene: the GGGGCC repeat is located between two non-coding exons (exon 1a and exon 1b) either in the promoter region of transcript variant 2 (NM_018325.4), or in the first intron of variant 1 (NM_145005.6) and variant 3 (NM_001256054.2, Fig. 1). Whereas variant 1 is predicted to result in a 222 amino acid long protein (exon 2–5, isoform b), variant 2 and variant 3 appear to result in 481 amino acid long proteins (exon 2–11, isoform a).
Fig. 1.
Localization of C9ORF72 probes. The three known C9ORF72 transcripts are visualized; the repeat expansion in C9ORF72 is represented by a red triangle and located in the promoter region of variant 2 (NM_018325.4) or in the first intron of variant 1 (NM_145005.6) and variant 3 (NM_001256054.2). Whereas variant 1 is predicted to result in a 222 amino acid long protein (exon 2–5, isoform b), variant 2 and variant 3 appear to result in 481 amino acid long proteins (exon 2–11, isoform a). For our TaqMan assays, probes were used targeting total C9ORF72 transcripts, variant 2, and variant 3 (green). Our digital molecular barcoding experiment contained probes for total C9ORF72 transcripts, variant 1, and variant 2 as well as probes that target the region between exon 1a and the expansion (transcripts containing intron 1a) and between exon 1b and exon 2 (transcripts containing intron 1b, purple)
In C9ORF72 expansion carriers, decreased levels of C9ORF72 mRNA have been reported [8], which was confirmed by other studies. In the fontal cortex, for instance, reduced expression levels were detected for total C9ORF72 transcripts [3, 6, 12, 28], variant 1 [12], and variant 2 [8, 10, 28]. A reduction was also described in the cerebellum for total C9ORF72 transcripts [3] and variant 2 [28]. Interestingly, other experiments suggested that a repeat expansion in C9ORF72 can lead to truncated expansion-containing abortive transcripts [13], and that the expansion may cause a shift from transcription of exon 1b (variant 2) towards transcription of exon 1a (variant 1 and variant 3) [23]. Nevertheless, studies focusing on C9ORF72 expression have also revealed inconsistencies [3, 8, 12, 13, 17, 21, 22], potentially due to variability in sample size, nomenclature, methodology, tissue type, and RNA integrity.
Since large-scale studies have yet to be reported that evaluate C9ORF72 expression systematically, we examined all known C9ORF72 transcripts in a unique pathological cohort of C9ORF72 expansion carriers. To further investigate intron 1 containing transcripts, we also assessed two intronic regions, one upstream of the repeat expansion (intron 1a), and one downstream of the repeat expansion (intron 1b). Our study uncovers decreased expression of specific C9ORF72 transcripts, reveals novel associations with survival after onset, and provides evidence of truncated transcripts as well as pre-mRNAs that act as templates for RAN translation.
Materials and methods
Subjects
From the Mayo Clinic Florida Brain Bank, we selected all patients with C9ORF72 repeat expansions, and a subset of patients with FTLD and/or MND without repeat expansions and control subjects without neurological diseases, for whom frozen cerebellum and/or frontal cortex were available, and for whom sufficient high quality RNA could be obtained for C9ORF72 expression studies [RNA integrity number (RIN) above seven, Online Resource Materials and Methods]. To measure C9ORF72 expression levels, we used two approaches: TaqMan gene expression assays (Life Technologies) and digital molecular barcoding (NanoString Technologies). For our C9ORF72 expression studies using TaqMan assays, we were able to include a cohort of 56 C9ORF72 expansion carriers [23 FTLD, 16 FTLD/MND, 14 MND, and 3 others (i.e., Alzheimer’s disease and depressive pseudodementia)] [4], 31 patients without those expansions (9 FTLD, 13 FTLD/MND, and 9 MND), and 20 control subjects without any neurological disease (Table 1). The same cohort of expansion carriers was included in our expression studies using NanoString technologies (Online Resource Table 1). We evaluated associations with poly(GP) and poly(GA) levels for a total of 50 expansion carriers from this cohort (22 FTLD, 14 FTLD/MND, and 14 MND, Online Resource Table 2). Written informed consent was obtained before study entry from all subjects or their legal next of kin if they were unable to give written consent, and biological samples were obtained with ethical committee approval.
Table 1.
Subject characteristics in C9ORF72 expression cohort (TaqMan; n = 107)
| Variable | C9Plus cohort (n = 56) | C9Minus cohort (n = 31) | Control cohort (n = 20) |
|---|---|---|---|
| Subject characteristics | |||
| Gender (male) | 32 (57 %) | 14 (45 %) | 7 (35 %) |
| Age at onset (years) | 62.0 (53.1–66.8) | – | – |
| Age at death (years) | 66.5 (60.0–72.8) | 72.0 (65.5–83.0) | 87.5 (81.8–93.0) |
| Survival after onset (years) | 5.0 (2.8–8.1) | – | – |
| RIN cerebellum (value) | 9.4 (8.9–9.6) | 9.4 (9.2–9.5) | 9.3 (8.6–9.5) |
| RIN frontal cortex (value) | 9.1 (8.6–9.7) | 9.5 (9.1–9.7) | 8.9 (8.6–9.2) |
| Repeat length cerebellum (kb) | 12.2 (10.5–13.6) | – | – |
| Repeat length frontal cortex (kb) | 25.6 (21.5–39.8) | – | – |
| Diagnosis | |||
| FTLD | 23 (41 %) | 9 (29 %) | – |
| FTLD/MND | 16 (29 %) | 13 (42 %) | – |
| MND | 14 (25 %) | 9 (29 %) | – |
| Other | 3 (5 %) | – | – |
| Available cerebellum | 54 (96 %) | 29 (94 %) | 19 (95 %) |
| Available frontal cortex | 52 (93 %) | 29 (94 %) | 20 (100 %) |
Data are sample median (IQR) or number (%). Information was obtained for patients with (C9Plus) and without (C9Minus) expansions in C9ORF72, as well as for control subjects (Control); information was unavailable regarding age at onset (n = 5, C9Plus), age at death (n = 1, C9Plus), survival after onset (n = 5, C9Plus), and repeat length in frontal cortex (n = 2, C9Plus). We used “–” when information was not available for an entire group. This C9ORF72 expression study was performed in both the cerebellum and frontal cortex. Cerebellum was acquired for 54 expansion carriers (23 FTLD, 15 FTLD/MND, 14 MND, and 2 other diseases), 29 patients without expansions (8 FTLD, 13 FTLD/MND, and 8 MND), and 19 control subjects. We obtained frontal cortex for 52 expansion carriers (22 FTLD, 14 FTLD/MND, 13 MND, and 3 other diseases), 29 patients without expansions (7 FTLD, 13 FTLD/MND, and 9 MND), and 20 control subjects
Examination of C9ORF72 transcripts
Total RNA was extracted from frozen cerebellum and frontal cortex using the RNeasy Plus mini kit (Qiagen), which includes a step to remove genomic DNA (gDNA) with gDNA Eliminator columns. RNA quality and quantity were determined with an Agilent 2100 Bioanalyzer using the RNA Nano Chip (Agilent Technologies). We performed a reverse transcription reaction with 300 ng of RNA as template, using an equal ratio of Random Hexamers and Oligo dT primers and the SuperScript III Kit (Invitrogen). Quantitative real-time PCR was then done on an ABI7900 PCR system (Applied Biosystems) using TaqMan gene expression assays. We assessed neuronal markers synaptophysin (SYP, Hs00300531_m1) and microtubule-associated protein 2 (MAP2, Hs00258900_m1), and endogenous control genes ribosomal protein, large, P0 (RPLP0, Hs00420895_gh) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, Hs02758991_g1). Additionally, we examined total C9ORF72 transcripts (Hs00376619_m1), C9ORF72 variant 2 transcripts (custom) [8], and C9ORF72 variant 3 transcripts (Hs00948764_m1). Values of C9ORF72 variant 1 transcripts (Hs00331877_m1) were too low when using a TaqMan gene expression assay to allow inclusion of this variant, as reported previously [3]. Using R Statistical Software (R Foundation for Statistical Computing, version 3.2.0), we calculated the median of replicates and took the geometric mean of neuronal markers and endogenous control genes, and subsequently, determined expression levels relative to control subjects utilizing the ΔΔ Ct method.
Digital molecular barcoding was done in house by the manufacturer using 100 ng of RNA as template. We examined neuronal markers SYP and MAP2, and endogenous control genes RPLP0 and GAPDH. The following targets were included in our analysis: total C9ORF72 transcripts, C9ORF72 variant 1 transcripts, C9ORF72 variant 2 transcripts, C9ORF72 intron 1a containing transcripts, and C9ORF72 intron 1b containing transcripts (Fig. 1, Online Resource Table 3). C9ORF72 variant 3 values were around background level when using digital molecular barcoding (consistent with another report [9]), and consequently, variant 3 was not included in our digital molecular barcoding analysis. The NanoStringNorm R package and nSolver Analysis Software (NanoString Technologies, version 2.5) were used to account for technical assay variation using the geometric mean method. Maximum background subtraction was then performed, and data were normalized to the geometric mean of neuronal markers and endogenous control genes.
Poly(GP) and poly(GA) immunoassays
Brain homogenates were prepared as previously described [1]. In brief, approximately 50 mg of cerebellum or frontal cortex was lysed in cold radio immunoprecipitation assay (RIPA) buffer and sonicated on ice. Lysates were cleared by centrifugation at 100,000×g for 30 min at 4 °C. The supernatant was collected and the protein concentration was determined by BCA assay. To prevent carry-over, the resulting pellet was resuspended in RIPA buffer, re-sonicated, and re-centrifuged. The RIPA-insoluble pellet was then extracted using 7 M urea buffer, sonicated and centrifuged at 100,000×g for 30 min at 22 °C. The protein concentration of the urea-soluble supernatant was determined by Bradford assay. Homogenates were diluted in Trisbuffered saline (TBS) and equal amounts of protein for all samples from the same neuroanatomical region and fraction were tested in duplicate wells. Poly(GP) levels in each fraction were measured using a previously described sandwich immunoassay that utilizes an affinity purified rabbit polyclonal poly(GP) antibody (Rb9259) and Meso Scale Discovery (MSD) electrochemiluminescence detection technology [25]. We used serial dilutions of recombinant (GP)8 to prepare the standard curve. Response values, corresponding to the intensity of emitted light upon electrochemical stimulation of the assay plate using the MSD QUICKPLEX SQ120, were acquired and background corrected prior to interpolation of poly(GP) levels using the standard curve. Poly(GA) levels in cerebellar lysates were similarly measured with an MSD-based poly(GA) sandwich immunoassay that utilizes a purified rabbit polyclonal poly(GA) antibody (Rb4333) provided by the Cleveland laboratory and a mouse monoclonal poly(GA) antibody (clone 5F2) provided by the Edbauer laboratory [18]. Serial dilutions of recombinant (GA)50 were used for the standard curve. Total levels in a given tissue sample were calculated from levels in soluble and insoluble fractions and used for examining associations of intron 1 containing transcripts with poly(GP) or poly(GA) levels.
Statistical analysis
We summarized data with median and interquartile range (IQR). To compare expression levels of transcripts between groups (e.g. C9ORF72 expansion carriers, patients without those expansions, and control subjects without neurological diseases), we used a Kruskal–Wallis rank sum test, and when this test was significant, we used a Wilcoxon rank sum test for each pairwise comparison (p < 0.017 considered significant after Bonferroni correction). Associations between transcripts measured by gene expression assays and digital molecular barcoding techniques (i.e., total C9ORF72 transcripts and variant 2) were assessed using a Spearman’s test of correlation; Spearman’s correlation coefficient r and 95 % confidence intervals (CIs) were estimated (p < 0.025 considered significant after Bonferroni correction). Because intron 1a containing transcripts and intron 1b containing transcript were only detectable in a subset of samples, we also compared the number of subjects with and without those transcripts for each pairwise comparison with a Fisher’s exact test, and we estimated odds ratios (ORs) and 95 % CIs (p < 0.017 considered significant after Bonferroni correction).
To assess the presence of associations within our overall cohort of expansion carriers, and within each disease subgroup (i.e., FTLD, FTLD/MND, and MND), associations of expression levels with variables were evaluated using a Spearman’s test of correlation, a Wilcoxon rank sum test, or a Kruskal–Wallis rank sum test, as appropriate for the nature of the given variable. In addition, we investigated associations between expression levels and survival after onset in our overall cohort of patients with C9ORF72 repeat expansions, and in each disease subgroup separately, using Cox proportional hazards regression models. Hazard ratios (HRs) and 95 % CIs were estimated, and deaths of any cause were utilized as our survival endpoint. All models in the overall cohort were adjusted for age at onset and disease subgroup. When investigating separate disease subgroups, models were adjusted for age at onset. In our Cox regression analysis, we considered the RNA expression level as a dichotomous categorical variable using three different cutoff points (median, 25th percentile, and 75th percentile) in the given subject group. To adjust for multiple testing and to control the family-wise error rate at 5 %, we utilized a Bonferroni correction separately for each outcome and for each disease subgroup; p values <0.0063 were considered significant when evaluating associations of each outcome with variables studied, including disease subgroup, gender, age at onset, age at death, repeat length, and survival after onset (using three different cutoff points).
Given that intron 1a containing transcripts and/or intron 1b containing transcripts may serve as templates for RAN translation, we also determined whether those transcripts were associated with poly(GP) or poly(GA) levels using a Spearman’s test of correlation. For each transcript, two tests were performed in the cerebellum [i.e., poly(GP) and poly(GA)] and one test in the frontal cortex [i.e., poly(GP)], and thus, p values below 0.025 and 0.050 were considered significant after Bonferroni correction, respectively. R Statistical Software was used for all statistical analyses.
Results
Reduction of specific C9ORF72 transcripts
C9ORF72 expression levels differed significantly between C9ORF72 expansion carriers, patients without expansions, and control subjects for total C9ORF72 transcripts (p ≤ 5.65e-07) as well as for variant 1 (p ≤ 3.86e-05) and for variant 2 (p ≤ 1.86e-10, Tables 2, 3). For simplicity, we focused on comparisons with C9ORF72 expansion carriers (Tables 2, 3); all results are shown in Online Resource Tables 4–5.
Table 2.
Expression of C9ORF72 transcripts using TaqMan assays
| TaqMan |
C9Plus versus C9Minus cohort |
C9Plus versus Control cohort |
|||||
|---|---|---|---|---|---|---|---|
| Group | p valuea | C9Plus | C9Minus | p value | C9Plus | Control | p value |
| Cerebellum | |||||||
| Total | 5.65e-07 | 0.67 (0.53–0.82) | 0.98 (0.81–1.21) | 9.14e-06 | 0.67 (0.53–0.82) | 1.00 (0.86–1.18) | 2.61e-05 |
| Variant 2 | 1.86e-10 | 0.43 (0.33–0.55) | 0.94 (0.71–1.08) | 5.20e-09 | 0.43 (0.33–0.55) | 1.00 (0.78–1.20) | 1.26e-06 |
| Variant 3 | 0.12 | 0.92 (0.63–1.25) | 1.10 (0.93–1.41) | – | 0.92 (0.63–1.25) | 1.00 (0.76–1.30) | – |
| Frontal cortex | |||||||
| Total | 1.97e-07 | 0.78 (0.64–1.05) | 1.30 (1.13–1.61) | 2.03e-07 | 0.78 (0.64–1.05) | 1.00 (0.85–1.22) | 0.02 |
| Variant 2 | 8.53e-13 | 0.58 (0.48–0.67) | 1.31 (1.14–1.66) | 9.63e-12 | 0.58 (0.48–0.67) | 1.00 (0.73–1.28) | 1.11e-05 |
| Variant 3 | 0.02 | 0.99 (0.74–1.41) | 1.39 (0.97–2.01) | – | 0.99 (0.74–1.41) | 1.00 (0.80–1.54) | – |
Data are sample median (IQR) or p value and obtained using TaqMan assays; normalized to the geometric mean of synaptophysin (SYP) and microtubule-associated protein 2 (MAP2). For simplicity, only comparisons with C9ORF72 expansion carriers are shown, an extended version of this table is displayed in Online Resource Table 4. Information was obtained for patients with (C9Plus) and without (C9Minus) expansions in C9ORF72, as well as for control subjects (Control). For each brain region, three tests were performed (i.e., total C9ORF72 transcripts, variant 2 transcripts, and variant 3 transcripts), and thus, p values below 0.017 (bold) were considered significant after Bonferroni correction
A Kruskal–Wallis rank sum test was performed to determine whether expression levels differed between groups (p < 0.017 considered significant after Bonferroni correction); when significant differences were detected a Wilcoxon rank sum test was used for pairwise comparisons (p < 0.017 considered significant after Bonferroni correction); when no significant differences were detected a Wilcoxon rank sum test was not performed for pairwise comparisons (indicated with “–”)
Table 3.
Expression of C9ORF72 transcripts using NanoString technologies
| NanoString |
C9Plus versus C9Minus cohort |
C9Plus versus Control cohort |
|||||
|---|---|---|---|---|---|---|---|
| Group | p valuea | C9Plus | C9Minus | p value | C9Plus | Control | p value |
| Cerebellum | |||||||
| Total | 1.18e-07 | 0.58 (0.42–0.76) | 0.80 (0.67–0.93) | 0.003 | 0.58 (0.42–0.76) | 1.00 (0.88–1.19) | 1.59e-07 |
| Variant 1 | 9.45e-06 | 0.47 (0.31–0.63) | 0.61 (0.55–0.75) | 0.01 | 0.47 (0.31–0.63) | 1.00 (0.72–1.49) | 9.04e-06 |
| Variant 2 | 2.19e-12 | 0.31 (0.19–0.47) | 0.70 (0.55–0.90) | 7.27e-07 | 0.31 (0.19–0.47) | 1.00 (0.87–1.13) | 5.23e-10 |
| Intron 1a | 0.20 | 1.08 (0.50–1.86) | 0.76 (0.26–1.40) | – | 1.08 (0.50–1.86) | 1.00 (0.44–1.88) | – |
| Intron 1b | 0.91 | 1.00 (0.66–1.59) | 0.98 (0.55–1.50) | – | 1.00 (0.66–1.59) | 1.00 (0.66–1.57) | – |
| Frontal cortex | |||||||
| Total | 3.44e-11 | 0.72 (0.62–0.83) | 1.13 (1.04–1.34) | 1.42e-09 | 0.72 (0.62–0.83) | 1.00 (0.90–1.18) | 2.35e-06 |
| Variant 1 | 3.86e-05 | 0.69 (0.49–0.90) | 1.10 (0.88–1.38) | 0.0001 | 0.69 (0.49–0.90) | 1.00 (0.78–1.39) | 0.001 |
| Variant 2 | 1.04e-12 | 0.53 (0.34–0.63) | 1.21 (0.94–1.43) | 1.84e-08 | 0.53 (0.34–0.63) | 1.00 (0.90–1.31) | 5.07e-10 |
| Intron 1a | 0.0002 | 2.72 (0.98–4.81) | 0.00 (0.00–1.08) | 0.0005 | 2.72 (0.98–4.81) | 1.00 (0.00–2.34) | 0.003 |
| Intron 1b | 0.11 | 1.57 (0.96–2.36) | 1.24 (0.44–2.10) | – | 1.57 (0.96–2.36) | 1.00 (0.37–1.74) | – |
Data are sample median (IQR) or p value and obtained using NanoString technologies; normalized to the geometric mean of synaptophysin (SYP) and microtubule-associated protein 2 (MAP2). For simplicity, only comparisons with C9ORF72 expansion carriers are shown, an extended version of this table is displayed in Online Resource Table 5. Information was obtained for patients with (C9Plus) and without (C9Minus) expansions in C9ORF72, as well as for control subjects (Control). For each brain region, five tests were performed (i.e., total C9ORF72 transcripts, variant 1 transcripts, variant 2 transcripts, intron 1a containing transcripts, and intron 1b containing transcripts), and thus, p values below 0.010 (bold) were considered significant after Bonferroni correction
A Kruskal–Wallis rank sum test was performed to determine whether expression levels differed between groups (p < 0.010 considered significant after Bonferroni correction); when significant differences were detected a Wilcoxon rank sum test was used for pairwise comparisons (p < 0.017 considered significant after Bonferroni correction); when no significant differences were detected a Wilcoxon a rank sum test was not performed for pairwise comparisons (indicated with “–”)
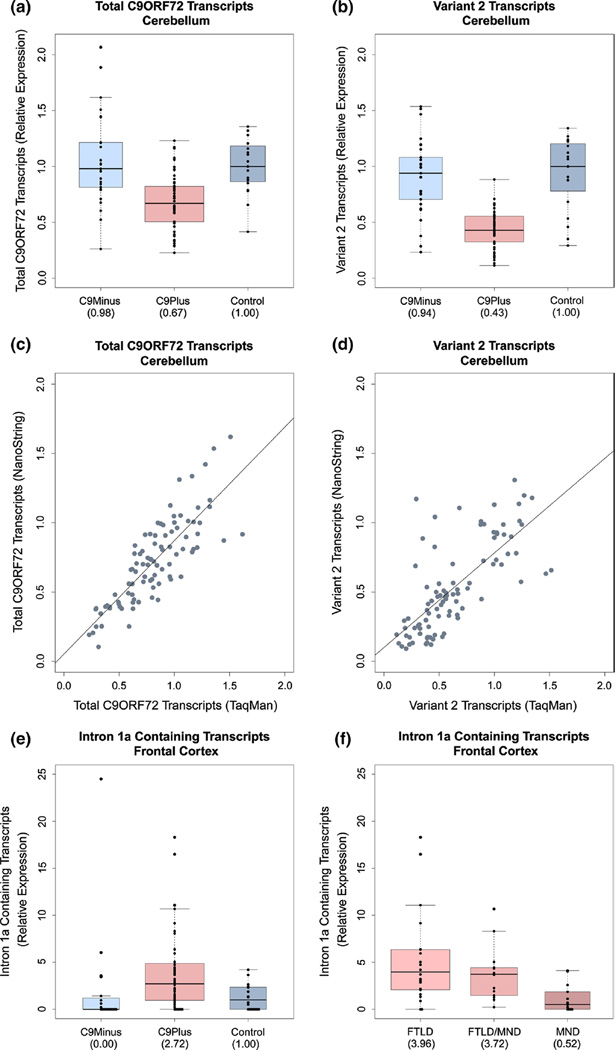
When using TaqMan assays, total C9ORF72 transcript levels in expansion carriers were 67 % (IQR 53–82 %) in the cerebellum, which was significantly lower than those in patients without expansions (98 %, IQR 81–121 %, p = 9.14e-06) or those in control subjects (100 %, IQR 86–118 %, p = 2.61e-05, Table 2, Fig. 2). The cerebellar expression of variant 2 was 43 % (IQR 33–55 %) in expansion carriers as compared to 94 % (IQR 71–108 %, p = 5.20e-09) in patients without expansions, and 100 % (IQR 78–120 %, p = 1.26e-06, Table 2, Fig. 2) in control subjects. No significant difference was detected for variant 3 (p = 0.12). In the frontal cortex, total C9ORF72 transcript levels in expansion carriers were 78 % (IQR 64–105 %), significantly lower than in patients without expansions (130 %, IQR 113–161 %, p = 2.03e-07) or in control subjects (100 %, IQR 85–122 %, p = 0.02, Table 2). Expression levels of variant 2 were 58 % (IQR 48–67 %) in expansion carriers as compared to 131 % (IQR 114–166 %, p = 9.63e-12) in patients without expansions, and 100 % (IQR 73–128 %, p = 1.11e-05, Table 2) in control subjects. After adjustment for multiple testing, no significant difference was observed for variant 3 (p = 0.02, Table 2).
Fig. 2.
Expression of specific C9ORF72 transcripts. C9Plus patients with C9ORF72 repeat expansions, C9Minus patients without C9ORF72 repeat expansions, and Control control subjects without neurological diseases. For each box plot, the median is represented by a solid line (values are specified between brackets below panels), and each box spans the IQR (25th percentile to 75th percentile). In the cerebellum, a reduction in total C9ORF72 transcripts is observed in patients with C9ORF72 repeat expansions as compared to patients without these expansions or to control subjects (a using TaqMan assays, Table 2). Additionally, a decrease in variant 2 is detected in the cerebellum of C9ORF72 expansion carriers as compared to patients without expansions or to control subjects (b using TaqMan assays, Table 2). In the cerebellum, there is an association between expression levels measured using TaqMan assays and NanoString technologies, both for total C9ORF72 transcripts (c Online Resource Table 6) and for variant 2 (d Online Resource Table 6). In the frontal cortex, levels of intron 1a containing transcripts are higher in expansion carriers than in patients without expansions or in control subjects (e using NanoString technologies, Table 3), and within our cohort of expansion carriers, the highest levels appear to be present in patients with FTLD or FTLD/MND (f using NanoString technologies, Table 4)
NanoString technologies were utilized to examine expression levels of total C9ORF72 transcripts, variant 1, and variant 2. In the cerebellum, total C9ORF72 transcripts were significantly lower in expansion carriers (58 %, IQR 42–76 %) than in patients without expansions (80 %, IQR 67–93 %, p = 0.003) or in control subjects (100 %, IQR 88–119 %, p = 1.59e-07, Table 3). Cerebellar levels of variant 1 were also reduced in expansion carriers (47 %, IQR 31–63 %) as compared to patients without expansions (61 %, IQR 55–75 %, p = 0.01) and to control subjects (100 %, IQR 72–149 %, p = 9.04e-06, Table 3). In C9ORF72 expansion carriers, the lowest levels were detected for variant 2: 31 % (IQR 19–47 %) in expansion carriers, 70 % (IQR 55–90 %, p = 7.27e-07) in patients without expansions, and 100 % (IQR 87–113 %, p = 5.23e-10, Table 3) in control subjects. Reduced levels of total C9ORF72 transcripts were also observed in the frontal cortex of expansion carriers (72 %, IQR 62–83 %) versus patients without expansions (113 %, IQR 104–134 %, p = 1.42e-09) or versus control subjects (100 %, IQR 90–118 %, p = 2.35e-06, Table 3). For variant 1, expression levels were 69 % (IQR 49–90 %) in expansion carriers as compared to 110 % (IQR 88–138 %, p = 0.0001) in patients without expansions, and 100 % (IQR 78–139 %, p = 0.001, Table 3) in control subjects. Again, the most prominent decrease was observed for variant 2, which demonstrated expression levels of 53 % (IQR 34–63 %) in expansions carriers, 121 % (IQR 94–143 %, p = 1.84e-08) in patients without expansions, and 100 % (IQR 90–131 %, p = 5.07e-10, Table 3) in control subjects.
Subsequently, we assessed associations between expression levels obtained using TaqMan assays and NanoString technologies for transcripts measured by both methods (i.e., total C9ORF72 transcripts and variant 2). We detected significant associations for total C9ORF72 transcripts in the cerebellum (r = 0.84, 95 % CI 0.75–0.90, p < 2.20e-16) and frontal cortex (r = 0.72, 95 % CI 0.58–0.82, p = 1.58e-15, Fig. 2, Online Resource Table 6). Similar associations were detected for variant 2 in the cerebellum (r = 0.75, 95 % CI 0.61–0.84, p < 2.20e-16) and frontal cortex (r = 0.80, 95 % CI 0.71–0.86, p < 2.20e-16, Fig. 2, Online Resource Table 6).
Elevation of C9ORF72 transcripts containing intron 1a
In the frontal cortex, we also discovered a significant difference in transcripts containing intron 1a (upstream of expansion, p = 0.0002, Table 3, Figs. 1, 2) when using NanoString technologies. For those transcripts, the relative expression was 272 % (IQR 98–481 %) in expansion carriers as compared to 0 % (IQR 0–108 %, p = 0.0005) in patients without expansions, and 100 % (IQR 0–234 %, p = 0.003) in control subjects. No such difference was detected in the cerebellum (p = 0.20). Intriguingly, for intron 1b containing transcripts (downstream of expansion) no significant differences were noted in the frontal cortex or in the cerebellum (p ≥ 0.11, Table 3). Since intron containing transcripts were undetectable in a subset of individuals, we also performed an alternative analysis using a Fisher’s exact test, which aligned with aforementioned results: in the frontal cortex, intron 1a containing transcripts were undetectable in 12 % of expansion carriers as compared to 55 % of patients without expansions (OR 0.11, 95 % CI 0.03–0.42, p = 0.0003), and 40 % of control subjects (OR 0.20, 95 % CI 0.05–0.80, p = 0.02, Online Resource Table 7). We did not observe a significant difference in the cerebellum (p ≥ 0.06), nor did we note significant differences for intron 1b containing transcripts in the frontal cortex or in the cerebellum (p = 1, Online Resource Table 7).
Associations of specific C9ORF72 transcripts with clinical features
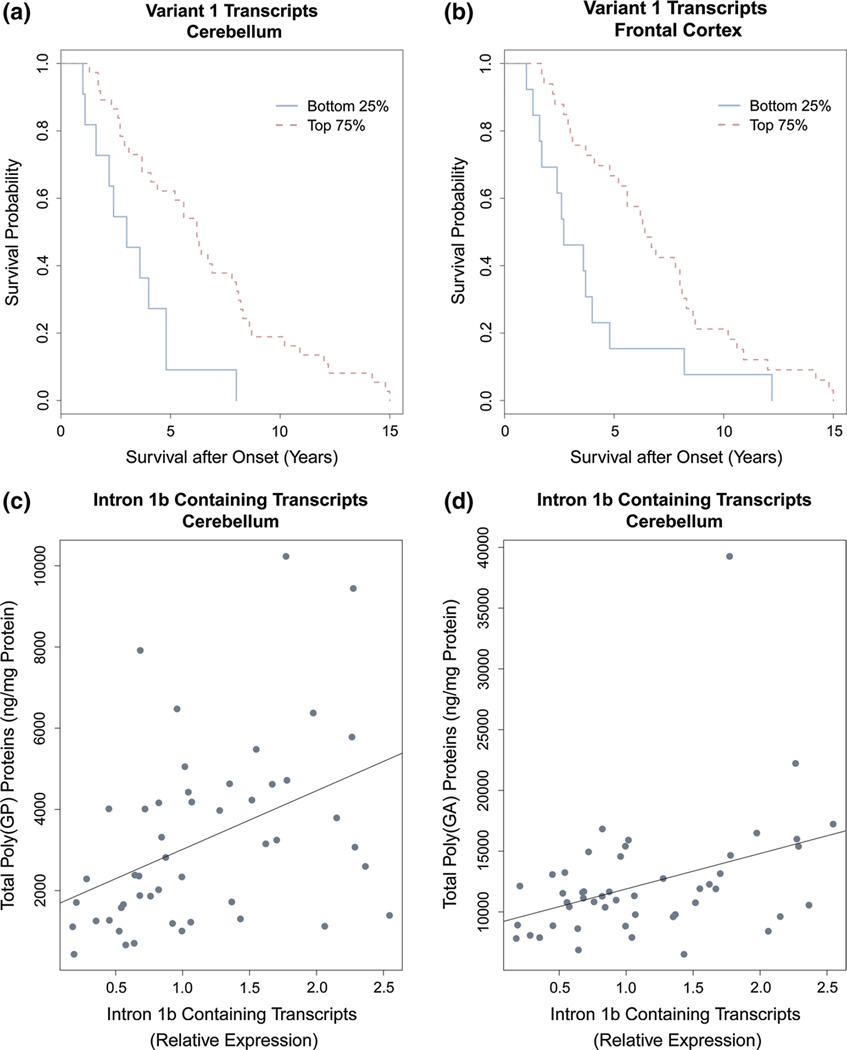
Next, we explored C9ORF72 expression within our group of expansion carriers. In both neuroanatomical regions, we noticed that expression levels of total C9ORF72 transcripts, variant 1, variant 2, variant 3, and intron 1b containing transcripts did not differ significantly between FTLD, FTLD/MND and MND patients (Online Resource Tables 8–9), nor did we detect associations with gender, age at onset, age at death or repeat length (Online Resource Tables 10–13). Importantly, we did detect a significant difference between disease subgroups for transcripts containing intron 1a in the frontal cortex (p = 0.002, Table 4, Fig. 2): for those transcripts, higher levels were observed in patients with FTLD (396 %, IQR 210–625 %) or FTLD/MND (373 %, IQR 158–441 %) as compared to patients with MND (52 %, IQR 0–186 %, p = 0.001 and p = 0.006, respectively). Interestingly, there was no significant difference between disease subgroups for transcripts containing intron 1a in the cerebellum. We did detect a significant association with survival after onset for variant 1 in both neuroanatomical regions: expression levels greater than the 25th percentile conferred a survival advantage (models adjusted for age at onset and disease subgroup, Table 5, Online Resource Tables 14–15), both in the cerebellum (HR 0.31, 95 % CI 0.14–0.67, p = 0.003) and in the frontal cortex (HR 0.23, 95 % CI 0.11–0.49, p = 0.0001). In our expansion carriers, median survival was reduced in patients with lower variant 1 levels (cerebellum: 3.0 years, IQR 1.9–4.4 years, frontal cortex: 2.7 years, IQR 1.7–4.0 years, Fig. 3) as compared to patients with higher variant 1 levels (cerebellum: 6.2 years, IQR 3.1–8.3 years, frontal cortex: 6.4 years, IQR 3.7–8.6 years, Fig. 3). We did not detect significant associations between other C9ORF72 transcripts and survival after onset in our expansion carriers (Online Resource Tables 14–15). Of note, for simplicity, we focused on our overall cohort of expansion carriers; we have, however, also provided tables that contain analyses for disease subgroups (e.g. Online Resource Tables 12–13).
Table 4.
Associations of C9ORF72 transcripts with disease subgroups using NanoString technologies
| NanoString |
FTLD versus FTLD/MND cohort |
FTLD versus MND cohort |
FTLD/MND versus MND cohort |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Group | p valuea | FTLD | FTLD/MND | p value | FTLD | MND | p value | FTLD/MND | MND | p value |
| Cerebellum | ||||||||||
| Intron 1a | 0.45 | 1.29 (0.66–2.09) | 0.93 (0.53–1.35) | – | 1.29 (0.66–2.09) | 0.96 (0.25–1.53) | – | 0.93 (0.53–1.35) | 0.96 (0.25–1.53) | – |
| Intron 1b | 0.33 | 1.06 (0.82–1.63) | 0.87 (0.68–1.33) | – | 1.06 (0.82–1.63) | 0.82 (0.32–1.42) | – | 0.87 (0.68–1.33) | 0.82 (0.32–1.42) | – |
| Frontal cortex | ||||||||||
| Intron 1a | 0.002 | 3.96 (2.10–6.25) | 3.73 (1.58–4.41) | 0.49 | 3.96 (2.10–6.25) | 0.52 (0.00–1.86) | 0.001 | 3.73 (1.58–4.41) | 0.52 (0.00–1.86) | 0.006 |
| Intron 1b | 0.61 | 1.83 (0.97–2.51) | 1.53 (1.04–2.24) | – | 1.83 (0.97–2.51) | 1.27 (0.55–2.30) | – | 1.53 (1.04–2.24) | 1.27 (0.55–2.30) | – |
Data are sample median (IQR) or p value and obtained using NanoString technologies; normalized to the geometric mean of synaptophysin (SYP) and microtubule-associated protein 2 (MAP2). In total, within our cohort of C9ORF72 expansion carriers, we examined eight different associations [i.e., disease subgroup, gender, age at onset, age at death, repeat length, and survival after onset (using three different cutoff points)] for each outcome, and thus, p values below 0.0063 (bold) were considered significant after Bonferroni correction; in this table, only one of those eight associations is displayed (disease subgroup)
A Kruskal–Wallis rank sum test was performed to determine whether expression levels differed between groups (p < 0.0063 considered significant after Bonferroni correction); when significant differences were detected a Wilcoxon rank sum test was used for pairwise comparisons (p < 0.017 considered significant after Bonferroni correction); when no significant differences were detected a Wilcoxon a rank sum test was not performed for pairwise comparisons (indicated with “–”)
Table 5.
Associations of C9ORF72 transcripts with survival after onset using NanoString technologies
| NanoString |
Overall cohort |
FTLD cohort |
FTLD/MND cohort |
MND cohort |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Group | Survival after onset | Hazard ratio (95 % CI) |
p value | Hazard ratio (95 % CI) |
p value | Hazard ratio (95 % CI) |
p value | Hazard ratio (95 % CI) |
p value |
| Cerebellum | |||||||||
| Variant 1 | >Median | 0.49 (0.26 to 0.93) | 0.03 | 0.43 (0.16 to 1.14) | 0.09 | 0.93 (0.29 to 2.97) | 0.90 | 0.20 (0.04 to 1.01) | 0.05 |
| >25th Percentile | 0.31 (0.14 to 0.67) | 0.003 | 0.04 (0.004 to 0.40) | 0.006 | 0.77 (0.18 to 3.28) | 0.72 | 0.23 (0.06 to 0.88) | 0.03 | |
| >75th Percentile | 0.65 (0.31 to 1.40) | 0.28 | 1.14 (0.43 to 3.04) | 0.79 | 2.09 (0.49 to 8.86) | 0.32 | – | – | |
| Frontal cortex | |||||||||
| Variant 1 | >Median | 0.82 (0.44 to 1.51) | 0.52 | 1.39 (0.52 to 3.71) | 0.52 | 0.50 (0.16 to 1.64) | 0.26 | 0.45 (0.12 to 1.76) | 0.25 |
| >25th Percentile | 0.23 (0.11 to 0.49) | 0.0001 | 0.37 (0.12 to 1.14) | 0.08 | 0.04 (0.004 to 0.42) | 0.007 | 0.17 (0.04 to 0.79) | 0.02 | |
| >75th Percentile | 0.90 (0.45 to 1.81) | 0.77 | 1.07 (0.38 to 3.03) | 0.89 | 0.97 (0.18 to 5.13) | 0.97 | 0.27 (0.03 to 2.15) | 0.21 | |
Data are hazard ratio (HR) with 95 % confidence interval (CI) or p value and obtained using NanoString technologies; normalized to the geometric mean of synaptophysin (SYP) and microtubule-associated protein 2 (MAP2). In total, within our cohort of C9ORF72 expansion carriers, we examined eight different associations [i.e., disease subgroup, gender, age at onset, age at death, repeat length, and survival after onset (using three different cutoff points)] for each outcome, and thus, p values below 0.0063 (bold) were considered significant after Bonferroni correction; in this table, only three of those eight associations are displayed (survival after onset with three cutoff points). A Cox proportional hazards regression model was used, using a dichotomous categorical variable with three different cutoff points (median, 25th percentile, and 75th percentile) in the given subject group. We used “–” to indicate that the number of samples in a given group was too low to allow p value calculation
Fig. 3.
Associations with clinical and pathological characteristics. Associations with clinical and pathological variables are shown for our overall cohort of C9ORF72 expansion carriers. Survival following disease onset, according to 25th percentile variant 1 levels, is displayed for the cerebellum (a using NanoString technologies, Table 5) and for the frontal cortex (b using NanoString technologies, Table 5). Additionally, associations between intron 1b containing transcripts and poly(GP) levels (c using NanoString technologies, Table 6) or poly(GA) levels (d using NanoString technologies, Table 6) are shown for the cerebellum
Associations of C9ORF72 transcripts containing intron 1 with poly(GP) and poly (GA) levels
To further investigate our findings related to intron 1 containing transcripts, we evaluated associations between those transcripts and poly(GP) c9RAN proteins. Although we did not detect a significant association in the frontal cortex (p ≥ 0.20), we noticed a significant association in the cerebellum for intron 1a containing transcripts (r = 0.33, 95 % CI 0.04–0.59, p = 0.02) and, more prominently, for intron 1b containing transcripts (r = 0.49, 95 % CI 0.23–0.70, p = 0.0004, Table 6, Fig. 3). Based on these significant findings in the cerebellum, we subsequently assessed associations between intron 1 containing transcripts and cerebellar poly(GA) levels: we also detected significant associations for both intron 1a containing transcripts (r = 0.34, 95 % CI 0.03–0.60, p = 0.02) and intron 1b containing transcripts (r = 0.38, 95 % CI 0.08–0.62, p = 0.007, Table 6, Fig. 3).
Table 6.
Associations of C9ORF72 transcripts with poly(GP) and poly(GA) levels using NanoString technologies
| NanoString |
Overall cohort |
FTLD cohort |
FTLD/MND cohort |
MND cohort |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Group | Association | Spearman’s r (95 % CI) |
p value | Spearman’s r (95 % CI) |
p value | Spearman’s r (95 % CI) |
p value | Spearman’s r (95 % CI) |
p value |
| Cerebellum | |||||||||
| Intron 1a | Poly(GP) | 0.33 (0.04 to 0.59) | 0.02 | 0.24 (−0.21 to 0.59) | 0.29 | 0.52 (−0.11 to 0.83) | 0.07 | 0.25 (−0.35 to 0.64) | 0.38 |
| Poly(GA) | 0.34 (0.03 to 0.60) | 0.02 | 0.57 (0.11 to 0.87) | 0.006 | 0.36 (−0.27 to 0.79) | 0.23 | 0.03 (−0.62 to 0.67) | 0.92 | |
| Intron 1b | Poly(GP) | 0.49 (0.23 to 0.70) | 0.0004 | 0.48 (0.05 to 0.78) | 0.03 | 0.80 (0.35 to 0.96) | 0.001 | 0.35 (−0.23 to 0.73) | 0.22 |
| Poly(GA) | 0.38 (0.08 to 0.62) | 0.007 | 0.61 (0.21 to 0.86) | 0.003 | 0.49 (−0.18 to 0.90) | 0.08 | 0.01 (−0.61 to 0.58) | 0.96 | |
| Frontal cortex | |||||||||
| Intron 1a | Poly(GP) | 0.19 (−0.12 to 0.47) | 0.20 | 0.19 (−0.29 to 0.59) | 0.40 | 0.59 (0.08 to 0.89) | 0.03 | 0.49 (−0.10 to 0.84) | 0.09 |
| Intron 1b | Poly(GP) | 0.11 (−0.19 to 0.40) | 0.45 | −0.21 (−0.59 to 0.22) | 0.36 | 0.47 (−0.16 to 0.85) | 0.09 | 0.20 (−0.48 to 0.74) | 0.51 |
Data are Spearman’s correlation coefficient r [95 % confidence interval (CI)] or p value and obtained using NanoString technologies; normalized to the geometric mean of synaptophysin (SYP) and microtubule-associated protein 2 (MAP2). For each transcript, two tests were performed in the cerebellum [i.e., poly(GP) and poly(GP)] and one test in the frontal cortex [i.e., poly(GP)], and thus, p values below 0.025 and 0.050 (bold) were considered significant after Bonferroni correction, respectively. A Spearman’s test of correlation was used (p < 0.025 and p < 0.050 considered significant after Bonferroni correction for the cerebellum and frontal cortex, respectively)
Discussion
To evaluate the contribution of C9ORF72 transcripts to disease pathogenesis, we set out to determine the expression of specific C9ORF72 transcripts in patients with repeat expansions in C9ORF72. Our study revealed a decreased expression of total C9ORF72 transcripts, variant 1, and variant 2, but not of variant 3, in expansion carriers as compared to non-expansion carriers and controls. Moreover, we discovered that higher levels of variant 1 were associated with prolonged survival after onset in expansion carriers. Interestingly, we also detected an increase in transcripts containing intron 1a, specifically in the frontal cortex of expansion carriers with an FTLD phenotype, while in the cerebellum intron 1 containing transcripts were significantly associated with poly(GP) and poly(GA) levels.
Previous studies focusing on C9ORF72 expression in expansion carriers revealed inconsistencies between laboratories and samples [3, 8, 12, 17, 21, 22], which could be attributed to relatively small sample sizes, investigated transcripts, differences in methods, variability between tissue or cell types, and the integrity of extracted RNA. For these reasons, we examined one of the world’s largest cohorts of pathologically confirmed expansion carriers, evaluated all known C9ORF72 transcripts, used TaqMan and NanoString techniques, assessed two brain regions, and only included samples with relatively high RIN values (Table 1).
We detected a significant decrease in expansion carriers as compared to patients without repeat expansions and to control subjects without neurological diseases for total C9ORF72 transcripts, variant 1, and variant 2. The strongest association was observed for variant 2, whereas no significant difference was observed for variant 3. Excitingly, we also discovered an association between variant 1 and survival after onset. Only a few factors are currently known to be associated with survival after onset in patients with C9ORF72 repeat expansions, including repeat length and genetic variants [e.g. rs5848 in granulin (GRN)] [26, 27]. The novel association we identified with variant 1 suggests that higher C9ORF72 levels may have beneficial effects, which could have major implications for the development of new therapeutic approaches targeting C9ORF72 (e.g. antisense oligonucleotides [ASOs]). Based on our findings, researchers should focus on ASOs that reduce GGGGCC-containing RNA foci and/or c9RAN proteins without altering C9ORF72 expression [16]. Additionally, future studies could investigate variant 1 in patients for whom RNA has been extracted from blood to determine whether its transcript levels could also be used as a prognostic indicator.
We further detected elevated levels of intron 1 containing transcripts (i.e., intron 1a), in agreement with recent reports [9, 13, 17, 21, 31]. Importantly, we identified significant differences between disease subgroups, with the most prominent elevation in the frontal cortex of FTLD patients, which is the primary affected region in those patients. One could speculate that a similar increase in intronic transcripts might be present in the spinal cord of MND patients, but this requires additional study.
Given the fact that we noted an increased expression of transcripts containing sequences upstream of the repeat (intron 1a), but not of transcripts downstream of the repeat (intron 1b), our data suggest that at least a subset of intron containing transcripts in C9ORF72 expansion carriers is truncated. It seems unlikely, however, that truncation occurs in all intron containing transcripts, especially since groups have been able to detect c9RAN proteins using antibodies flanking the repeat [31]. c9RAN protein pathology, in particular inclusions of poly(GP) and poly(GA), is abundant in neocortical regions, hippocampus, thalamus, and cerebellum [2, 18– 21, 24, 31]. While previous studies utilized immunohistochemical methods to investigate associations between c9RAN protein pathology and other features of disease [7, 18, 19, 24], we have now employed immunoassays to quantify poly(GP) and poly(GA) burden. Interestingly, our experiments showed a significant association of intron 1 containing transcripts (both intron 1a and intron 1b) with poly(GP) and poly(GA) levels in the cerebellum of C9ORF72 expansion carriers. As mentioned above, one could hypothesize that the increase of intron 1a (but not intron 1b) containing transcripts in the frontal cortex is a reflection of truncated transcripts that contain only part of the expansion, and therefore, those transcripts may not serve as templates for RAN translation. This would explain the absence of significant associations between intron 1 containing transcripts and poly(GP) levels in the frontal cortex. In contrast, the cerebellum likely contains less truncated transcripts as no relative increase in intron 1a containing transcripts was seen in expansion carriers. Moreover, the significant associations of poly(GP) and poly(GA) levels with both intron 1a containing transcripts and intron 1b containing transcripts in this region (most profoundly for intron 1b) suggest that transcripts containing the entire first intron may serve as templates for RAN translation in the cerebellum.
In summary, our present study reveals associations with specific C9ORF72 transcripts and provides support for the presence of truncated transcripts and pre-mRNAs that may serve as templates for RAN translation. Although these promising findings do require further investigation, they shed new light on disease pathogenesis and may point towards novel therapeutic approaches for C9ORF72-related diseases as well as the larger FTLD and MND disease continuum.
Supplementary Material
Acknowledgments
The research leading to these results has received funding by NIH grants R01 NS080882, P50 AG016574, P01 NS084974, R21 NS089979, the ALS Association, and the ALS Therapy Alliance. Dr. Van Blitterswijk is supported by the Milton Safenowitz Post Doctoral Fellowship for ALS research from the ALS Association, and the Clinical Research in ALS and Related Disorders for Therapeutic Development (CReATe) Clinical Research Fellowship (U54 NS092091).
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00401-015-1480-6) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Conflicts of interest MDJ and RR hold a patent on methods to screen for the hexanucleotide repeat expansion in the C9ORF72 gene. All other authors declare that they have no conflicts of interest.
References
- 1.Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, Degroot S, Tapper AR, Sellier C, Charlet-Berguerand N, Karydas A, Seeley WW, Boxer AL, Petrucelli L, Miller BL, Gao FB. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126(3):385–399. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, 3rd, Rademakers R, Boylan KB, Dickson DW, Petrucelli L. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77(4):639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belzil VV, Bauer PO, Prudencio M, Gendron TF, Stetler CT, Yan IK, Pregent L, Daughrity L, Baker MC, Rademakers R, Boylan K, Patel TC, Dickson DW, Petrucelli L. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013;126(6):895–905. doi: 10.1007/s00401-013-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieniek KF, van Blitterswijk M, Baker MC, Petrucelli L, Rademakers R, Dickson DW. Expanded C9ORF72 hexanucleotide repeat in depressive pseudodementia. JAMA Neurol. 2014;71(6):775–781. doi: 10.1001/jamaneurol.2013.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chew J, Gendron TF, Prudencio M, Sasaguri H, Zhang YJ, Castanedes-Casey M, Lee CW, Jansen-West K, Kurti A, Murray ME, Bieniek KF, Bauer PO, Whitelaw EC, Rousseau L, Stankowski JN, Stetler C, Daughrity LM, Perkerson EA, Desaro P, Johnston A, Overstreet K, Edbauer D, Rademakers R, Boylan KB, Dickson DW, Fryer JD, Petrucelli L. Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science. 2015;348(6239):1151–1154. doi: 10.1126/science.aaa9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciura S, Lattante S, Le Ber I, Latouche M, Tostivint H, Brice A, Kabashi E. Loss of function of C9orf72 causes motor deficits in a zebrafish model of amyotrophic lateral sclerosis. Ann Neurol. 2013;74(2):180–187. doi: 10.1002/ana.23946. [DOI] [PubMed] [Google Scholar]
- 7.Davidson YS, Barker H, Robinson AC, Thompson JC, Harris J, Troakes C, Smith B, Al-Saraj S, Shaw C, Rollinson S, Masuda-Suzukake M, Hasegawa M, Pickering-Brown S, Snowden JS, Mann DM. Brain distribution of dipeptide repeat proteins in frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun. 2014;2:70. doi: 10.1186/2051-5960-2-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, Maragakis N, Tienari PJ, Petrucelli L, Traynor BJ, Wang J, Rigo F, Bennett CF, Blackshaw S, Sattler R, Rothstein JD. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80(2):415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fratta P, Poulter M, Lashley T, Rohrer JD, Polke JM, Beck J, Ryan N, Hensman D, Mizielinska S, Waite AJ, Lai MC, Gendron TF, Petrucelli L, Fisher EM, Revesz T, Warren JD, Collinge J, Isaacs AM, Mead S. Homozygosity for the C9orf72 GGGGCC repeat expansion in frontotemporal dementia. Acta Neuropathol. 2013;126(3):401–409. doi: 10.1007/s00401-013-1147-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC, Petrucelli L, Kim HJ, Gao FB, Taylor JP. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;25(7567):129–133. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gijselinck I, Van Langenhove T, van der Zee J, Sleegers K, Philtjens S, Kleinberger G, Janssens J, Bettens K, Van Cauwenberghe C, Pereson S, Engelborghs S, Sieben A, De Jonghe P, Vandenberghe R, Santens P, De Bleecker J, Maes G, Baumer V, Dillen L, Joris G, Cuijt I, Corsmit E, Elinck E, Van Dongen J, Vermeulen S, Van den Broeck M, Vaerenberg C, Mattheijssens M, Peeters K, Robberecht W, Cras P, Martin JJ, De Deyn PP, Cruts M, Van Broeckhoven C. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012;11(1):54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- 13.Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, Rothstein JD, Wang J. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507(7491):195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW, 3rd, Sun S, Herdy JR, Bieri G, Kramer NJ, Gage FH, Van Den Bosch L, Robberecht W, Gitler AD. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci. 2015;18(9):1226–1229. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koppers M, Blokhuis AM, Westeneng HJ, Terpstra ML, Zundel CA, Vieira de Sa R, Schellevis RD, Waite AJ, Blake DJ, Veldink JH, van den Berg LH, Pasterkamp RJ. C9orf72 ablation in mice does not cause motor neuron degeneration or motor deficits. Ann Neurol. 2015;78(3):426–438. doi: 10.1002/ana.24453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li HR, Jiang J, Watt AT, Chun S, Katz M, Qiu J, Sun Y, Ling SC, Zhu Q, Polymenidou M, Drenner K, Artates JW, McAlonis-Downes M, Markmiller S, Hutt KR, Pizzo DP, Cady J, Harms MB, Baloh RH, Vandenberg SR, Yeo GW, Fu XD, Bennett CF, Cleveland DW, Ravits J. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci USA. 2013;110(47):E4530–E4539. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu EY, Russ J, Wu K, Neal D, Suh E, McNally AG, Irwin DJ, Van Deerlin VM, Lee EB. C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD. Acta Neuropathol. 2014;128(4):525–541. doi: 10.1007/s00401-014-1286-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, Weng SM, Haass C, Kretzschmar HA, Edbauer D, Neumann M. Dipeptide repeat protein pathology in C9ORF72 mutation cases: clinico-pathological correlations. Acta Neuropathol. 2013;126(6):859–879. doi: 10.1007/s00401-013-1181-y. [DOI] [PubMed] [Google Scholar]
- 19.Mann DM, Rollinson S, Robinson A, Bennion Callister J, Thompson JC, Snowden JS, Gendron T, Petrucelli L, Masuda-Suzukake M, Hasegawa M, Davidson Y, Pickering-Brown S. Dipeptide repeat proteins are present in the p62 positive inclusions in patients with frontotemporal lobar degeneration and motor neurone disease associated with expansions in C9ORF72. Acta Neuropathol Commun. 2013;1(1):68. doi: 10.1186/2051-5960-1-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mori K, Arzberger T, Grasser FA, Gijselinck I, May S, Rentzsch K, Weng SM, Schludi MH, van der Zee J, Cruts M, Van Broeckhoven C, Kremmer E, Kretzschmar HA, Haass C, Edbauer D. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 2013;126(6):881–893. doi: 10.1007/s00401-013-1189-3. [DOI] [PubMed] [Google Scholar]
- 21.Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, Haass C, Edbauer D. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339(6125):1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 22.Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Holtta-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chio A, Restagno G, Borghero G, Sabatelli M, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sareen D, O’Rourke JG, Meera P, Muhammad AK, Grant S, Simpkinson M, Bell S, Carmona S, Ornelas L, Sahabian A, Gendron T, Petrucelli L, Baughn M, Ravits J, Harms MB, Rigo F, Bennett CF, Otis TS, Svendsen CN, Baloh RH. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med. 2013;5(208):208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schludi MH, May S, Grasser FA, Rentzsch K, Kremmer E, Kupper C, Klopstock T, Arzberger T, Edbauer D. Distribution of dipeptide repeat proteins in cellular models and C9orf72 mutation cases suggests link to transcriptional silencing. Acta Neuropathol. 2015;130(4):537–555. doi: 10.1007/s00401-015-1450-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su Z, Zhang Y, Gendron TF, Bauer PO, Chew J, Yang WY, Fostvedt E, Jansen-West K, Belzil VV, Desaro P, Johnston A, Overstreet K, Oh SY, Todd PK, Berry JD, Cudkowicz ME, Boeve BF, Dickson D, Floeter MK, Traynor BJ, Morelli C, Ratti A, Silani V, Rademakers R, Brown RH, Rothstein JD, Boylan KB, Petrucelli L, Disney MD. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron. 2014;83(5):1043–1050. doi: 10.1016/j.neuron.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Blitterswijk M, DeJesus-Hernandez M, Niemantsverdriet E, Murray ME, Heckman MG, Diehl NN, Brown PH, Baker MC, Finch NA, Bauer PO, Serrano G, Beach TG, Josephs KA, Knopman DS, Petersen RC, Boeve BF, Graff-Radford NR, Boylan KB, Petrucelli L, Dickson DW, Rademakers R. Association between repeat sizes and clinical and pathological characteristics in carriers of C9ORF72 repeat expansions (Xpansize-72): a cross-sectional cohort study. Lancet Neurol. 2013;12(10):978–988. doi: 10.1016/S1474-4422(13)70210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Blitterswijk M, Mullen B, Wojtas A, Heckman MG, Diehl NN, Baker MC, DeJesus-Hernandez M, Brown PH, Murray ME, Hsiung GY, Stewart H, Karydas AM, Finger E, Kertesz A, Bigio EH, Weintraub S, Mesulam M, Hatanpaa KJ, White CL, 3rd, Neumann M, Strong MJ, Beach TG, Wszolek ZK, Lippa C, Caselli R, Petrucelli L, Josephs KA, Parisi JE, Knopman DS, Petersen RC, Mackenzie IR, Seeley WW, Grinberg LT, Miller BL, Boylan KB, Graff-Radford NR, Boeve BF, Dickson DW, Rademakers R. Genetic modifiers in carriers of repeat expansions in the C9ORF72 gene. Molecular neurodegeneration. 2014;9:38. doi: 10.1186/1750-1326-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waite AJ, Baumer D, East S, Neal J, Morris HR, Ansorge O, Blake DJ. Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiol Aging. 2014;35(7):1779, e1775–e1779, e1713. doi: 10.1016/j.neurobiolaging.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao S, MacNair L, McGoldrick P, McKeever PM, McLean JR, Zhang M, Keith J, Zinman L, Rogaeva E, Robertson J. Isoform Specific Antibodies Reveal Distinct Subcellular Localizations of C9orf72 in Amyotrophic Lateral Sclerosis. Ann Neurol. 2015 doi: 10.1002/ana.24469. [DOI] [PubMed] [Google Scholar]
- 30.Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, Gupta S, Thomas MA, Hong I, Chiu SL, Huganir RL, Ostrow LW, Matunis MJ, Wang J, Sattler R, Lloyd TE, Rothstein JD. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525(7567):56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, Ostrow LW, Rothstein JD, Troncoso JC, Ranum LP. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA. 2013;110(51):E4968–E4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.